ABSTRACT
Polyploid cells, which contain multiple copies of the typically diploid genome, are widespread in plants and animals. Polyploidization can be developmentally programmed or stress induced, and arises from either cell-cell fusion or a process known as endoreplication, in which cells replicate their DNA but either fail to complete cytokinesis or to progress through M phase entirely. Polyploidization offers cells several potential fitness benefits, including the ability to increase cell size and biomass production without disrupting cell and tissue structure, and allowing improved cell longevity through higher tolerance to genomic stress and apoptotic signals. Accordingly, recent studies have uncovered crucial roles for polyploidization in compensatory cell growth during tissue regeneration in the heart, liver, epidermis and intestine. Here, we review current knowledge of the molecular pathways that generate polyploidy and discuss how polyploidization is used in tissue repair and regeneration.
Keywords: Endomitosis, Endocycling, Polyploid, Wound, Healing, Hippo
Summary: In regenerating tissues, polyploidization is a commonly used strategy to compensate for cell loss. Such tissues are less dependent on resident stem cells, an advantage that may be exploited in regenerative therapy.
Introduction
Cells are considered polyploid if they possess three or more complete copies of the haploid genome. This condition can arise either through cell-cell fusion or via a process known as endoreplication. A polyploid genome may be transmitted through the germline, thus resulting in an organism that is wholly polyploid. This condition, which is termed ‘autopolyploidy’ (see Glossary, Box 1), is common in plants, both in the wild and in crop varieties, and is believed to result from chromosome mis-segregation during meiosis. Polyploid cells may also arise from diploid cells in somatic tissues, a condition defined as ‘endopolyploidy’, which is the topic of this Review.
Box 1. Glossary.
C ploidy values. C indicates chromatin amount or DNA content, as a multiple of the haploid genome (e.g. 4C is the chromatin amount of a diploid cell in G2 or a tetraploid cell in G1) (Brodsky et al., 1985; White, 1973a).
n (or N) ploidy values. The number of sets of chromosomes, as a multiple of the haploid genome: 1n is the chromosome content of a sperm or egg cell and 2n is the chromosomal content of a diploid cell (with two haploid sets of chromosomes – one maternal and one paternal).
Allopolyploid. An organism with more than two haploid sets of chromosomes that have been derived from two or more species by hybridization. Allopolyploidy usually arises from cell-cell fusion between gametes of two related species, where at least one gamete has a ploidy yn>1n (y=whole number) (White, 1937).
Aneuploid. A genome in which partial genomic content has been gained or lost (e.g. yn−x, yn+x), usually as a product of chromosome mis-segregation.
Autopolyploid. An organism with more than two haploid sets of chromosomes that have been derived from the same parent species. Autopolyploidy usually arises from cell-cell fusion between gametes, where at least one gamete has a ploidy yn>1n (White, 1937).
Endopolyploid. Somatic cells that are polyploid (yC>2C). Endopolyploidy usually occurs as a result of endoreplication, but also includes cell-cell fusion (White, 1973b).
Euploid. Genome with an exact multiple of the haploid genome (yn).
Polytene chromosomes. Chromosomes containing multiple parallel strands of DNA.
Endopolyploid cells are generated via endoreplication cell cycles in which cells successively replicate the genome without completing cytokinesis during mitosis (Fig. 1); such cells replicate their DNA during S phase and either revert back to a gap (G) phase, skipping mitosis completely (in the case of endocycling), or enter mitosis and fail to complete cytokinesis (in the case of endomitosis). Endoreplicating cells are known to reach ploidies greater than 200,000C (where C=total ‘chromatin’ value, as a multiple of the haploid genome; see Glossary, Box 1). This increase in genomic DNA content allows a higher transcriptional output, which can facilitate the growth of very large cells and/or enhance macromolecular secretion. Endoreplication is found in all eukaryotic kingdoms, i.e. in plants, fungi, protozoa and animals (Joubès and Chevalier, 2000; Rusch et al., 1966; Yin et al., 2010; Zielke et al., 2013). The sea hare Aplysia possesses giant neurons that can reach ploidies of 260,000C, and massive ploidy has also been noted in neurons, epidermal gland cells and digestive glands in other mollusks (Achatina, Helix and Lymnaea) (Anisimov, 2005; Lasek and Dower, 1971; Mandrioli et al., 2010). There are also several examples in which somatic growth depends upon increases in ploidy, such as in Caenorhabditis, Oikopleura and Drosophila (Edgar and Orr-Weaver, 2001; Ganot and Thompson, 2002; Sulston and Horvitz, 1977). Polyploidy is also widespread in plant tissues, including tomato fruit, peanuts, maize kernels and the leaf epidermis (Joubès and Chevalier, 2000). Owing to its widespread nature and ability to sustain increased cell growth, endoreplication has been estimated to contribute significantly to biological mass in nature, possibly even more than cell proliferation (Sugimoto-Shirasu and Roberts, 2003).
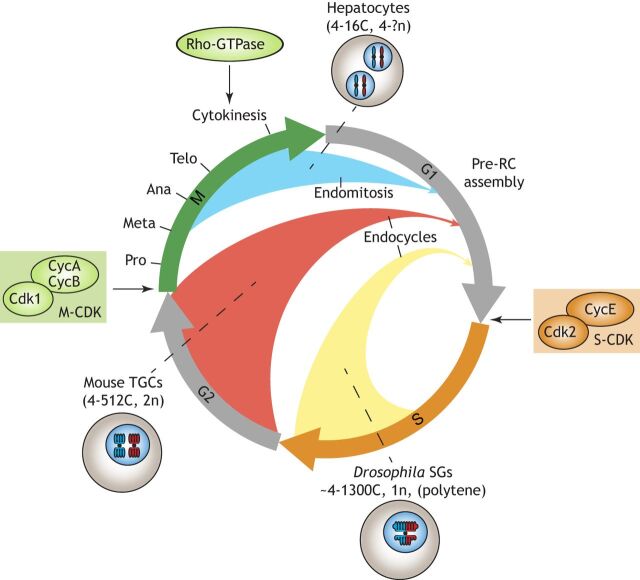
Fig. 1.
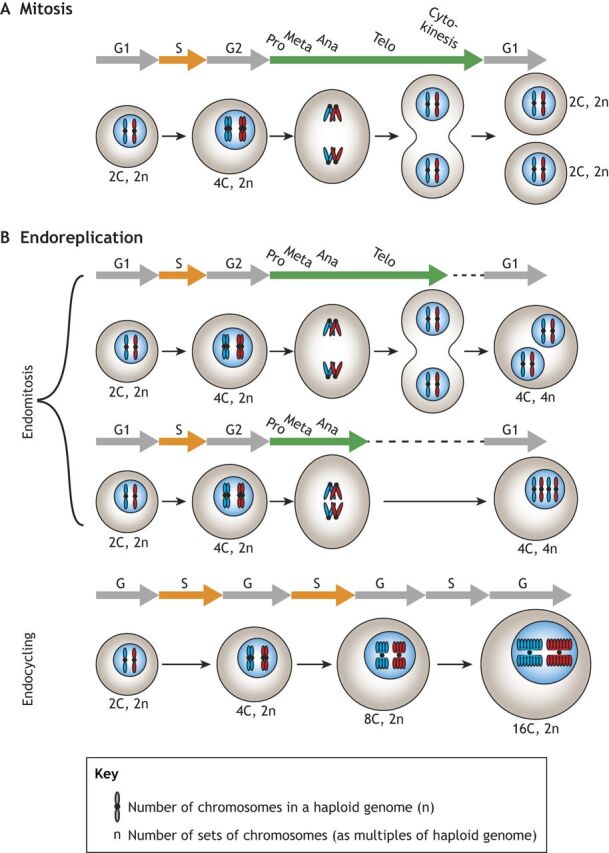
An overview of alternative cell cycles. (A,B) While mitosis (A) gives rise to diploid cells, a common path to polyploidy is endoreplication (B), which includes two subgroups: endomitosis and endocycling. Similar to mitotic cells, cells that undergo endomitosis enter the cell cycle, which consists of four canonical phases: G1, S, G2 and M. Endomitosis is characterized by incomplete cytokinesis, thus resulting in a polyploid binucleate cell or a polyploid mononucleate cell. By contrast, endocycling cells lack M phase altogether, resulting in a two-phase cell cycle consisting of alternating G and S phases. Endocycling cells often over- or under-amplify certain genomic regions, resulting in joined polytene chromosomes.
In mammals, polyploidy is represented to a variable extent in many tissues, and can be induced by developmental programming, as a response to tissue injury, or by failed mitosis (mitotic slippage). Transient polyploidization is also known to occur in tumorigenesis, giving rise to aneuploid cells (see Box 2). Because polyploid cells tend to possess a higher capacity for growth (Orr-Weaver, 2015), such cells are likely to support organ growth and tissue homeostasis in the absence of mitosis. This ability has been proposed to support regeneration of the mammalian liver upon chemotoxic stress and aging (Gentric et al., 2012; Gupta, 2000), and wound healing in Drosophila (Losick, 2016; Losick et al., 2013, 2016). The growth of polyploid cells is thought to be fueled by their increased gene copy number, supporting increased mRNA and protein synthesis (Zhurinsky et al., 2010). Other advantages associated with polyploidy include genomic buffering against mutations, resistance to apoptosis, increased lifespan and increased metabolism. Polyploid cells also tend to be more efficient in forming protective tissue envelopes and barriers (Orr-Weaver, 2015), possibly because tissues formed by larger cells require a lower density of cell-cell junctions. In addition, because mitosis involves a reorganization of the cytoskeleton and a loss of cell adhesion, compensatory polyploidization-mediated growth may be a preferred option to replace lost cell mass, or to relieve tissue tension, in the event of tissue injury or stress. As such, a better understanding of wound-induced polyploidization may potentially spawn novel therapeutic strategies for healing tissues with poor regenerative capacity.
Box 2. Polyploidy and cancer.
The majority of cancers display aneuploidy, with around 90% of solid tumors and 75% of hematopoietic cancers having abnormal chromosome numbers (Weaver and Cleveland, 2006). Aneuploidy in cancer cells likely originates from previously polyploid cells generated by either cell-cell fusion, which may be induced by viral infection (Duelli and Lazebnik, 2007; Duelli et al., 2007), or endoreplication, although some prefer the term ‘abortive cell cycle’ to distinguish it from developmentally programmed endoreplication (Davoli and de Lange, 2011; Storchova and Pellman, 2004). Tetraploidization has been observed to occur alongside upregulation of Mad2, which downregulates MKlp2, a kinesin required for cytokinesis (Lee et al., 2010). This may enhance the occurrence of abortive cell cycles. Developmentally programmed endoreplication usually occurs alongside irreversible differentiation and termination of proliferation. Normal exceptions include Drosophila rectal papillar cells and hepatocytes, which re-enter mitosis (Duncan et al., 2010; Fox et al., 2010). However, genetically unstable cells may be capable of continued proliferation, which may involve reduced stringency of postmitotic cell cycle checkpoints that prevent the proliferation of aneuploid/tetraploid cells (Andreassen et al., 1996; Lanni and Jacks, 1998; Minn et al., 1996). Because centrosomes are also duplicated in S phase, tetraploid cells obtain supernumerary centrosomes before the ensuing M phase. Cells with supernumerary centrosomes are quite prone to chromosome mis-segregation and aneuploidy (Storchova and Pellman, 2004). This hypothesis is supported by the observed enrichment of near-tetraploid aneuploid cells (∼4n) in tumors, and aneuploid tumor cells possessing supernumerary centrosomes (Kaneko and Knudson, 2000; Levine et al., 1991; Mitelman, 2005; Reid et al., 1996). Aneuploidy also contributes to rapid cancer cell evolution, because it can give rise to an increasingly heterogenous population of tumor cells that can undergo selection. Accordingly, aneuploidy in cancer is associated with poor prognosis (Carter et al., 2006; Coward and Harding, 2014; Oltmann et al., 2018; Sheffer et al., 2009; Walther et al., 2008) and increased tolerance to chemotherapy (Lee et al., 2011; Vargas-Rondon et al., 2018). We can draw a parallel with the heterogeneity and increased fitness created by mitosis of tetraploid hepatocytes, and speculate that aneuploidy in cancer cells turns this otherwise beneficial mechanism to its selfish advantage.
In this Review, we first summarize examples of polyploidy that are found in nature and then discuss the molecular mechanisms that can induce and regulate endoreplication in different organisms. Finally, we discuss recent discoveries highlighting how endocycling, endomitosis and/or cell-cell fusion can contribute to tissue homeostasis and regeneration.
Developmental endoreplication
Endoreplication has been reported in several groups of plants and animals, where it often contributes to increased cell and/or body size. In plants, endocycling is an essential aspect of normal development in many cell types (Breuer et al., 2010; Caro et al., 2008; De Veylder et al., 2011; Gutierrez, 2005, 2009; Harashima and Schnittger, 2010; Sabelli and Larkins, 2009). In Arabidopsis thaliana, most leaf cell types enter the endocycle and reach ploidies of up to 32C, such that ploidy and cell expansion, and consequently leaf growth, are correlated (Kondorosi et al., 2000; Melaragno et al., 1993; Sugimoto-Shirasu and Roberts, 2003). In metazoans, an example of consistent regulation of cell size and patterning by polyploid cells has been documented in the urochordate Oikopleura dioica, a pan-global species of marine zooplankton. This marine organism loses most mitotic features a few hours after hatching, as most cells transit to developmentally controlled endocycles, giving rise to ploidies ranging from 4C to 1300C, with cell sizes proportional to their ploidy (Campsteijn et al., 2012; Ganot and Thompson, 2002). As a result, Oikopleura grows 10-fold during its 1-week life cycle.
Endoreplication is also common in insects and has been extensively characterized in the fruit fly Drosophila melanogaster (Edgar and Orr-Weaver, 2001; Edgar et al., 2014; Fox and Duronio, 2013; Fox et al., 2010; Swanson et al., 2015; Zielke et al., 2013). Most differentiated larval tissues of Drosophila develop through endocycling, which has been studied in the salivary glands, fat body, gut, renal tubes, trachea and epidermis (Smith and Orr-Weaver, 1991). Mitotic proliferation is mostly retained in undifferentiated progenitor and stem cells, including imaginal discs and adult midgut progenitors of the larval gut. Several cell types in the adult fly are also polyploid, including neurons, sensory bristles, gut enterocytes and ovarian nurse and follicle cells, with ploidies ranging from 8C to 2000C (Audibert et al., 2005; Hammond and Laird, 1985; Unhavaithaya and Orr-Weaver, 2012). Endocycling is also required for increased growth in these cell types, and is therefore necessary to sustain normal cellular functions. For example, decreased ploidy of ovarian nurse cells causes female sterility, because endocycling is required for normal oocyte development (Lilly and Spradling, 1996; Maines et al., 2004).
Developmental endopolyploidy has also been reported in fish (Mandrioli et al., 2010), mice and humans, and likely occurs in most mammals. In fact, it has been estimated that mammalian tissues are generally composed of up to 20% polyploid cells (Biesterfeld et al., 1994), although some human tissues have been estimated to contain ∼50% and ∼70% polyploid cells (Gandarillas and Freije, 2014; Mollova et al., 2013). Vertebrate cells known to undergo developmental endoreplication include placental trophoblast giant cells (TGCs), endometrial stromal cells (Qi et al., 2015), cardiac myocytes (Mollova et al., 2013; Soonpaa et al., 1996), hepatocytes (Gupta, 2000), megakaryocytes (Ravid et al., 2002; Trakala et al., 2015), keratinocytes (Gandarillas, 2012; Gandarillas and Freije, 2014), epicardial cells and vascular smooth muscle cells (Cao et al., 2017; McCrann et al., 2008). In addition, mammary epithelial cells are bi-nucleate (Rios et al., 2016), but whether this polyploidy arises from cell-cell fusion or endoreplication is not clear. Thus, polyploidy may be of importance whether one is looking at cancer, aging, tissue homeostasis or wound healing.
Molecular control of endoreplication and polyploidization
Polyploidization and the switch from mitosis to endoreplication often take place as part of a developmental program that involves the differentiation of mitotic progenitor cells into more specialized differentiated cells. Although the cell cycle regulators controlling G1/S in endocycles and mitotic cycles are mostly the same, the mitotic regulators are typically suppressed in endocycling cells (reviewed by De Veylder et al., 2011; DePamphilis, 2016; Edgar and Orr-Weaver, 2001; Edgar et al., 2014; Fox and Duronio, 2013; Orr-Weaver, 2015; Zielke et al., 2013). At least two essential modifications of the mitotic cell cycle must be made in order to switch from a mitotic cell cycle to an endocycle. First, the key events of M phase, sister chromatid separation and cytokinesis, must be suppressed without blocking S phase. This is generally accomplished by downregulating the activity of mitotic cyclin-dependent kinase (M-CDK), which drives G2/M-phase progression, while the activity of the CDK responsible for S-phase entry (S-CDK) is maintained (Fig. 2). In animal cells, M-CDK activity is provided by CDK1 bound to one of its activating subunits, cyclin B or cyclin A, which also confer substrate specificity towards M-phase regulators. By contrast, S-CDK activity is normally provided by CDK2 that is bound and activated by cyclin E or cyclin A. In Drosophila salivary glands, Cyclin B, Cyclin A and CDK1 levels are reduced in endocycling cells, whereas they are elevated in mitotic cells (Zielke et al., 2011). In Oikopleura, the mitotic cyclins, cyclin A, cyclin B and cyclin B3, and a CDK1 paralog that is essential for oogenic M phase, CDK1d, are all transcriptionally downregulated in endocycling somatic tissue (Campsteijn et al., 2012; Øvrebø et al., 2015). M-CDK downregulation is also sufficient to induce endocycling in many cell types that are not pre-programmed to enter endocycling (Broek et al., 1991; Chen et al., 2016; Hassel et al., 2014; Hayashi, 1996; Hayles et al., 1994; Mihaylov et al., 2002; Sauer et al., 1995; Sigrist and Lehner, 1997; Weigmann et al., 1997), suggesting that M-CDK downregulation may be a widespread means by which to trigger endocycling. However, blocking the factors and pathways that are activated downstream of M-CDK may also be sufficient to induce endoreplication. For example, in the planarian S. mediterranea, endoreplication cycles can be induced by blocking chromosome condensation through RNAi-mediated knockdown of condensins (Lai et al., 2017).
Fig. 2.
Alternative cell cycles found in development and regeneration. The mitotic cell cycle is composed of four phases: G1, S, G2 and M. The G1/S and G2/M cell cycle transitions are controlled by S-CDK and M-CDK activities, respectively. Endopolyploidy arises from altered cell cycles in which different cell cycle phases are truncated or bypassed entirely (blue, red and yellow arrows). As examples, mouse hepatocytes skip only cytokinesis in a cell cycle variant known as endomitosis (blue arrow), mouse trophoblast giant cells (TGCs) enter G1 following G2 (red arrow) and Drosophila salivary gland cells (SGs) re-enter a G1-like phase before fully completing DNA replication in S phase (yellow arrow). Endocycles and endomitoses are frequently regulated through downregulation of M-CDK and cytokinesis, respectively.
The second requirement for mitosis to endoreplication switching is the re-assembly of the pre-replication complex (pre-RC) during successive G phases. During late M phase, Cdc6 is recruited to origin recognition complexes (ORCs), which are multi-protein complexes assembled at initiation sites for DNA replication (Bell and Stillman, 1992; Riera et al., 2017; Yuan et al., 2017). Cdc6-ORC facilitates the recruitment of two Cdt1-bound Mcm2-7 hexamers on replication origins, forming pre-RCs on licensed origins (Bell and Labib, 2016; Cocker et al., 1996). Origin firing, and the onset of DNA replication, is triggered by S-CDK activity, which prompts the recruitment of multiple additional factors and activates Mcm2-7 DNA helicase activity (Heller et al., 2011; Zegerman and Diffley, 2007). These events ultimately lead to the recruitment of DNA clamps known as proliferating cell nuclear antigen (PCNA) and DNA polymerase, marking the onset of S phase. Cdt1 recruitment and function is repressed by CDK activity (Chen and Bell, 2011; Sugimoto et al., 2004) and pre-RC assembly therefore requires a window of low CDK activity in G phase. Constitutive S-CDK activity can thus block endocycling (Follette et al., 1998; Remus and Diffley, 2009; Weiss et al., 1998). Although the mechanisms by which CDK activity suppresses pre-RCs have been established in budding yeast, how CDKs might do this in endocycling cells is less clear. Furthermore, and as we detail below, the mechanisms used to block M-CDK and retain S-CDK oscillations in endocycling cells vary widely between cell types and organisms, showing just how versatile the building blocks of the cell cycle are.
Drosophila endocycles
An informative example of the transition from mitosis to endocycling is observed in follicle cells of the Drosophila ovary. These cells, which form an epithelium that surrounds the developing Drosophila oocyte, proliferate by mitosis up until the 7th stage of oogenesis, at which point the oocyte and polyploid nurse cells that support the oocyte start expressing the Notch ligand Delta (Deng et al., 2001; Lopez-Schier and St Johnston, 2001). Expression of Delta in the germline activates Notch signaling in the surrounding follicle cells. This induces expression of Hindsight (Hnt), which encodes a transcription factor that represses expression of the essential CDK1 activator String/Cdc25 (Fig. 3). The loss of String/Cdc25 expression thus precludes M-phase entry and causes a prolonged arrest in G2 (Schaeffer et al., 2004; Shcherbata et al., 2004; Sun and Deng, 2007). In addition to repressing String/Cdc25 expression, Hnt represses expression of Cut, another transcription factor gene, and this allows accumulation of Fzr/Cdh1, an activating subunit of the anaphase promoting complex/cyclosome (APC/C) (Narbonne-Reveau et al., 2008; Sun and Deng, 2007). In contrast to the other activating subunit of APC/C, Fzy/Cdc20, Fzr/Cdh1 does not require activation by M-CDK activity and promotes degradation of the mitotic cyclins (A, B and B3), thus maintaining a window of low CDK activity during G1. Fzr/Cdh1 also targets Geminin, a protein that prevents DNA re-replication by binding Cdt1, for proteasomal degradation. The activity of Fzr/Cdh1 is thus sufficient to allow re-assembly of pre-RCs in G1, and guides G2-arrested cells back into a G1-like state ready for DNA replication (Sun and Deng, 2007). The follicle cells can therefore re-enter S phase once S-CDK activity reaches sufficient levels to fire pre-RCs. Similarly, Delta-Notch signaling promotes the mitosis-to-endocycle switch during the differentiation of enterocytes in the adult Drosophila midgut and of larval glial cells (Von Stetina et al., 2018; Xiang et al., 2017). However, Delta-Notch signaling has exactly the opposite effect in follicle cells in a distantly related insect, the flour beetle Tribolium (Bäumer et al., 2012), suggesting that upstream regulatory inputs into the mitotic-endocycle switch are not evolutionarily conserved. The mechanisms that promote downregulation of mitotic cyclins can also differ across tissues. For example, although mitotic cyclins are downregulated post-transcriptionally in Drosophila follicle cells, they are transcriptionally silenced in endocycling Drosophila salivary glands (Maqbool et al., 2010; Zielke et al., 2008).
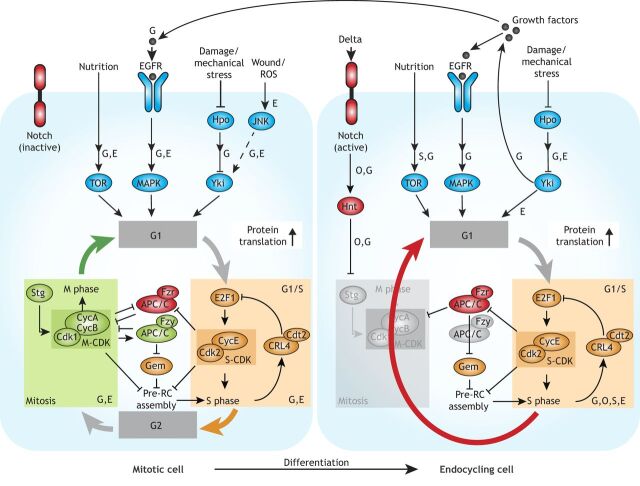
Fig. 3.
Mitotic-endocycle transitions in Drosophila. Cell growth in Drosophila is controlled by multiple pathways, including the PI3K/TOR, EGFR/MAPK, JAK/STAT, JNK and Hippo (Hpo)/Yki pathways; pathways shown to operate in ovarian follicle cells (O), adult midgut (G), salivary gland (S) and epidermis (E) are indicated. Cellular growth rates affect levels of E2F1 protein, which controls G1/S in a rate-limiting manner through transcriptional activation of Cyclin E (CycE), which binds to and activates CDK2, forming active S-CDK complexes. S phase triggers proteasomal degradation of E2F1 through activation of CRL4cdt2. The G2/M transition is regulated by CycA- and CycB-dependent CDK1 kinase activity (M-CDK), which requires activation by String (Stg; a Cdc25-type phosphatase). M-CDK kinase activity activates the APC/C subunit Fizzy (Fzy). The E3 ligase APC/CFzy targets Geminin (Gem), CycA and CycB for proteasomal degradation; the subsequent depletion of Geminin and M-CDK activity relieves inhibition of Cdt1 and creates a window of low CDK activity (not shown), respectively, which allows re-assembly of the pre-replication complex (pre-RC). In the adult midgut (G) and follicle cells of the ovary (O), mitosis-endocycle transitions are triggered through expression of the Notch ligand Delta in adjacent cells. In follicle cells, active Notch induces Hindsight (Hnt), which represses Stg expression and thereby blocks M-CDK activity. Upregulation of Fizzy-related (Fzr), another activating subunit of APC/C that does not require activation by M-CDK, ensures low M-CDK activity and mediates destruction of Geminin, which in turn allows pre-RC assembly while bypassing M phase. Increased endocycling can also be induced by mechanical stress via the Hippo pathway and JNK. The Hippo pathway stimulates increased ploidy non-cell-autonomously in enterocytes of the adult midgut (G) through expression and secretion of cytokines and growth factors, which activate the EGFR/Ras/MAPK and JAK/STAT pathways, increasing cell growth rates and decreasing the length of G phase. In the adult epidermis (E), Yorkie (Yki) is required for polyploidization cell-autonomously upon wound closure.
Transcriptional regulation is also important for continued progression through endocycling. Drosophila has a single activating member of the E2F family of transcription factors, E2F1, that binds DNA as a heterodimer with ‘dimerization partner’ (DP). Drosophila also has a repressor E2F, E2F2, that represses transcription of E2F1-DP targets as part of the Myb-MuvB complex. E2F1-DP regulates several M-phase genes and genes required for DNA replication, and dampened E2F1 activity is required for endocycles in fat bodies and salivary glands (Maqbool et al., 2010). Disturbing the balance of E2F1-DP/Myb-MuvB activity, either through gene silencing or overexpression, disrupts endocycles and reduces the ploidy of these tissues (Maqbool et al., 2010). This observation suggests that reduced E2F1-DP activity is required to dampen the expression of M-phase genes, whereas minimal activity is needed to maintain transcription of cyclin E and other S-phase regulators. Similar to cyclins, Drosophila E2F1 displays oscillating protein expression, such that its accumulation peaks by the end of G1 phase, followed by proteasomal degradation during S phase. This degradation of E2F1 is directly linked to DNA replication: chromatin-bound PCNA binds E2F1 through a ‘PCNA interacting protein (PIP) motif’, which mediates E2F1 proteolysis through the CRL4cdt2 ubiquitin ligase (Shibutani et al., 2008). Although mammalian E2F factors (E2F1-E2F3) are not known to be regulated by PIP-motif targeted degradation during S phase, human E2F1-E2F3 also display cyclic activity both through cyclic competition with the atypical E2Fs, E2F7/8, and via cyclic degradation by SCFSkp2 and APC/Ccdc20 in S/G2 and M phases, respectively (de Bruin et al., 2003; Maiti et al., 2005; Marti et al., 1999; Peart et al., 2010; Wong et al., 2011).
Mammalian endocycles
One of the best studied examples of endocycling in mammals is in murine trophoblast giant cells (TGCs). These cells form the outermost layer of the placental extraembryonic compartment and produce a number of pregnancy-specific cytokines and hormones. TGCs undergo a mitosis-endocycle switch during embryogenesis, and failure to endocycle compromises normal TGC development and affects fetal viability (Ouseph et al., 2012). They are believed to completely replicate their genome, in contrast to the polytene chromosomes of Drosophila salivary glands (Sher et al., 2013), although some genomic regions containing key placental genes are amplified (Hannibal and Baker, 2016). Diploid TGC progenitors and trophoblast stem cells (TSCs) can be cultured in vitro, where they can be experimentally induced to switch to an endocycle. Studies of such cultured cells have revealed that, similar to Drosophila endocycling cells, TGCs sustain downregulation of M-CDK activity as they switch to endocycling (Hochegger et al., 2007; Ullah et al., 2008).
The selective inhibition of CDK1 activity is sufficient for TSCs to differentiate into TGCs (Ogawa et al., 2016). TSCs remain undifferentiated through epigenetic modification by TET1 and TET2, which promote demethylation of chromatin (Chrysanthou et al., 2018; Tahiliani et al., 2009). As TSCs differentiate, TET1 and TET2 are downregulated, causing increased expression of the cyclin-dependent kinase inhibitor (CKI) p21Cip1 and enhanced cyclin B degradation (Chrysanthou et al., 2018). The induction of endocycling by withdrawal of the fibroblast growth factor FGF4 (Tanaka et al., 1998) also promotes the accumulation of p21Cip1 and another CKI, p57Kip2, which consequently inhibits CDK1 and suppresses mitosis (Ullah et al., 2008) (Fig. 4). In this context, upregulation of p57Kip2 and p21Cip1 appears to be controlled by checkpoint kinase-1 (Chk1), a component of the DNA damage checkpoint (Ullah et al., 2011). In mitotically proliferating TGCs, p57Kip2 and p21Cip1 are targeted for proteasomal degradation upon phosphorylation by Chk1. Upon FGF4 deprivation, however, Chk1 is silenced, allowing accumulation of p57Kip2 and p21Cip1. In this context, p57Kip2 translocates to the nucleus where it appears to be the main mediator of the endocycle switch, as evidenced by the fact that p21Cip1-deficient TGCs endocycle normally, whereas p57Kip2-deficient trophoblasts are hyper-proliferative and cause hyperplasia and placentomegaly (Ullah et al., 2008). In contrast to p57Kip2, p21Cip1 localizes to the cytoplasm, where it protects TGCs from DNA damage-induced apoptosis (de Renty et al., 2014).
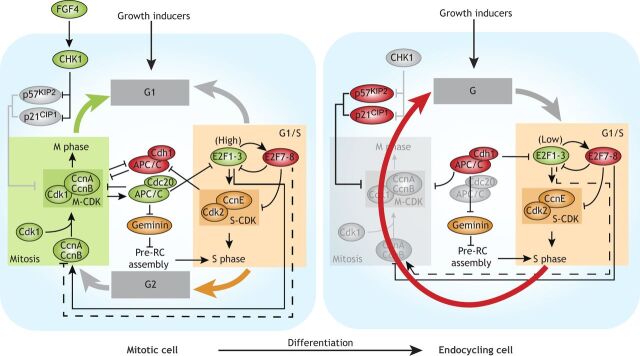
Fig. 4.
Mitotic-endocycle transitions in trophoblast giant cells. Cellular growth rates affect activation of E2F1-3, which control G1/S through transcriptional activation of S-CDK activity. E2F1-3 also activate expression of E2F7-8, which in turn repress expression of E2F1 and its targets, thus forming a negative-feedback loop. E2F1-3 are also required for the expression of cyclin A and cyclin B (CcnA and CcnB), which activate CDK1 (M-CDK) and are required for M-phase entry. M-CDK kinase activity activates the APC/C subunit Cdc20. The E3 ligase APC/CCdc20 targets geminin, CcnA and CcnB for proteasomal degradation. Depletion of geminin and M-CDK activity relieves inhibition of Cdt1 and creates a window of low CDK activity, respectively, which allows re-assembly of the pre-replication complex (pre-RC). In vitro, trophoblast stem cells are induced to enter endocycles upon FGF4 deprivation, which reduces CHK1 activity and relieves degradation of p57Kip2 and p21cip1. Accumulation of p57Kip2 and p21cip1 block M-CDK activity, which establish an M-phase bypass and onset of endocycles in TGCs. Further polyploidization is also affected by E2F7-8-dependent downregulation of E2F1-3.
Changes in G1/S transcriptional control also play an important role in the TGC endocycle switching. This control involves the E2F family of transcription factors, which in mammals includes the activator E2Fs (E2F1-E2F3), the repressor E2Fs (E2F4-E2F6), and two atypical repressor E2Fs (E2F7 and E2F8) (Chen et al., 2009; van den Heuvel and Dyson, 2008). Activity of the E2Fs is also modulated by their activating dimerization partners (Dp1, Dp2 and Dp4) and repressed by retinoblastoma-like pocket proteins (Rb, p107 and p130). Rb-deficient TGCs display excess mitotic proliferation, caused by elevated E2F3a activity (Chong et al., 2009; Wenzel et al., 2007), whereas deletion of all three activator E2Fs results in increased TGC ploidy (Chen et al., 2012). Conversely, deletion of the two atypical repressor E2Fs, E2F7 and E2F8, leads to reduced TGC ploidy, which favors mitotic proliferation. Interestingly, E2F8 is also indispensable for the polyploidization of endometrial stromal cells, which is associated with ERK- and STAT3-dependent E2F8 expression and suppressed CDK1 activity (Qi et al., 2015). These observations suggest that E2F1-E2F3 activity promotes mitosis and needs to be suppressed for correct switching to endocycling (Chen et al., 2012). This idea is supported by the fact that knockouts of E2F1 or E2F3 are sufficient to rescue endocycle defects caused by an E2F7/E2F8 double deletion (Chen et al., 2012; Ouseph et al., 2012). Considering this, it is somewhat puzzling that cyclin E1, a transcriptional target of E2F1-E2F3 (Parisi et al., 2003), is essential for endocycling in TGCs. This paradox might be explained by alternative regulation of cyclin E1 expression, for example by Myc, in parallel with E2F activity (Santoni-Rugiu et al., 2000). Overall, these observations imply that mammalian activator E2Fs restrict endocycling and underscore the notion that the balance between activator and repressor E2Fs contributes to the switch from mitotic cycling to endocycling.
The upstream regulation of endoreplication and ploidy
Endocycling rates can often be dictated by environmental factors, such as nutrition or stress. For example, sunlight can affect whether plant leaf cells undergo mitotic cycles or endocycles (Berckmans et al., 2011; Gendreau et al., 1998). However, superimposed upon such environmental regulation, developmentally programmed regulation often dictates the final ploidy achieved by endocycling (Audibert et al., 2005; Hammond and Laird, 1985; Unhavaithaya and Orr-Weaver, 2012). Thus, under optimal nutritional conditions, the larval salivary gland cells of Drosophila typically reach a ploidy of ∼1300C, while its fat body cells achieve ploidies of ∼256C and adult midgut enterocytes gain a maximum ploidy of 32C (Butterworth and Rasch, 1986; Hammond and Laird, 1985). In these cases, final ploidy is likely to be controlled by both a developmental time window and gap (G) phase durations. For example, the time window that supports endocycles starts when CDK1 activity is suppressed and lasts for as long as essential cell cycle regulators such as E2F1 and Cyclin E are expressed. In addition, the number of endocycles that occur within a particular developmental time window is likely determined by the length of each cycle, which is mostly dictated by G-phase length. Duration of G phase, in turn, is decided by the overall growth rate of the cell, which is decided by environmental factors such as nutrition, growth signals and stress. For example, it has been shown that in Drosophila salivary glands, increased growth rate is followed by an increased rate of protein synthesis, followed by increased accumulation of cell cycle regulators, including E2F1, which leads to faster transition to S phase and thus less time spent in G phase (Zielke et al., 2011). As the length of endocycle G phases appears to be controlled by the same gene products that control G1/S-phase progression in mitotic cells [namely CDK2/cyclin complexes, CDK inhibitors (CKIs) and E2F/Rb, etc.], it should come as no surprise that mitotic cycles and endocycles share the same upstream regulators of G1/S-phase progression. A number of such upstream regulators have been identified. The polyploidization of megakaryocytes, for example, is induced by thrombopoietin (Mcdonald, 1992), a glycoprotein hormone secreted from liver and kidney. Thrombopoietin regulates G1/S phase, by controlling cyclin E expression, in endomitotic megakaryocytes via the STAT pathway (Eliades et al., 2010; Kaushansky, 2016). Other pathways and factors affecting megakaryocyte polyploidization include PI3K/Akt, MAPK/ERK and Myc (Chanprasert et al., 2006). Earlier studies also demonstrated that nutrient availability and protein synthesis rates, which depend on Myc and TOR activity, are tightly linked to endocycle speed and final ploidies (Britton and Edgar, 1998; Britton et al., 2002; Demontis and Perrimon, 2009; Grewal et al., 2005; Pierce et al., 2004; Saucedo et al., 2003). In Drosophila larval salivary glands and adult midgut enterocytes, endocycle rates appear to be controlled, downstream of the TOR and EGFR pathways, respectively, by the post-transcriptional expression of the single Drosophila activator E2F: E2F1 (Xiang et al., 2017; Zielke et al., 2011). In TGCs, however, endocycles are promoted by elevated E2F8 expression (Qi et al., 2015), which is induced by progesterone through the EGFR/ERK/STAT3 signaling pathway. Paradoxically, the activator E2Fs are dispensable for TGC and hepatocyte endoreplication in triple knockout (E2F1-3) mice (Chen et al., 2012), where it remains unclear how S-CDK and DNA replication factor gene expression is sustained.
Although metabolism and growth may affect endocycle speed in many contexts, exit from the endocycle in some cases may be controlled separately by developmental induction of CKIs or transcriptional downregulation of S-CDK activity. This latter mechanism might be achieved through a switch from activating to repressive E2F activity, followed by a ‘lock down’ of G1/S-phase regulators by chromatin remodeling complexes (Buttitta and Edgar, 2007). In this way, upstream regulators may define a developmental time-window during which growth-dependent endocycling is allowed. For example, Drosophila ovarian follicle cells exit endocycles in response to developmentally controlled downregulation of Notch receptor activity, which coincides with decreasing levels of the Notch ligand Delta being expressed in oocytes. Loss of Notch activity then causes increased activity of Tramtrack, a transcriptional repressor downstream of the ecdysone receptor (EcR) signaling pathway, the activity of which is required for termination of endocycling (Sun et al., 2008).
Polyploidization via cell fusion
Sometimes polyploidy does not arise directly from cell cycle modifications, but instead occurs following the fusion of two neighboring cells to produce a cell with increased ploidy. A well-known example of cell-cell fusion is the fusion of two haploid germ cells, which gives rise to a diploid zygote. However, there are also examples of diploid somatic cells that fuse together to form polyploid multinuclear cells. These include vertebrate and Drosophila myoblasts and mammalian osteoclasts (Chen and Olson, 2004; Kim et al., 2015a; Xing et al., 2012).
Our best understanding of cell-cell fusion derives from studies of myoblasts in Drosophila, zebrafish and mice (Kim et al., 2015a). In this context, cell-cell fusion requires cell-cell adhesion followed by enhancement of cell membrane proximity and destabilization of lipid bilayers. In Drosophila, cell-cell adhesion and recognition are mediated by immunoglobulin domain-containing cell-adhesion molecules (CAMs): the formation of new muscle fibers is seeded by founder cells expressing the CAMs Dumbfounded/Kin-of-IrreC (Duf/Kirre) and Roughest (Rst), which are attracted by fusion-competent myoblasts (FCMs) expressing a CAM named Sticks and stones (Sns) (Bour et al., 2000; Ruiz-Gómez et al., 2000; Strunkelnberg et al., 2001). In zebrafish myoblasts, cell adhesion is established by the Duf/Kirre homolog Kirrel (Srinivas et al., 2007), but whether two different cell types are also required in vertebrates is not known. In Drosophila, membrane proximity is then enhanced by F-actin-enriched podosome-like structures formed in FCMs, which protrude towards the founder cell membrane. This protrusion is propelled by actin polymerization regulated by the Arp2/3 complex (Berger et al., 2008; Massarwa et al., 2007; Richardson et al., 2007). Membrane proximity is further enhanced by a mechanosensory response in founder cells, creating protrusion resistance through Myosin II-induced cortical tension. The accumulation of MyoII activity ultimately promotes the formation of a fusion pore, joining the two fusing cells. In mice, exposure of phosphatidylserines to the cell surface has been proposed to be involved in membrane destabilization during fusion pore formation (Jeong and Conboy, 2011; Kim et al., 2015b; van den Eijnde et al., 2001). Similar to several endocycling cell types in Drosophila, Delta-Notch signaling is involved in the differentiation of FCMs, whereas Ras controls the differentiation of founder cells (Artero et al., 2003).
Stress- and injury-induced polyploidy
Injury, as well as cellular stress, can cause loss of cells and hence loss of tissue integrity. In such cases, cell mass needs to be restored in order to maintain tissue function and homeostasis. In most regenerative tissues, lost cells are replaced by cell division of nearby progenitor cells or resident stem cells. However, recently reported examples show that some tissues also use endocycles and/or cell-cell fusion to compensate for losses of tissue mass. Together, these findings illuminate a new aspect of wound healing that has the potential to open up novel strategies for regenerative medicine. Below, we discuss how the control of ploidy contributes to tissue regeneration in the Drosophila gut and epidermis, and in the vertebrate heart and liver.
Polyploidy in the regenerating Drosophila gut
The Drosophila intestinal epithelium is composed predominantly of a monolayer of polyploid absorptive cells called enterocytes (ECs). These terminally differentiated cells arise from progenitor cells, known as enteroblasts (EBs) (Ohlstein and Spradling, 2007), that undergo up to four endocycles, giving rise to polyploid ECs with a final ploidy of 8-32C (Edgar et al., 2014). Upon EC loss and stress, which can be caused by cytotoxic exposure or enteric infection, tissue homeostasis is maintained by a pool of resident intestinal stem cells (ISCs) that divide to produce committed post-mitotic EBs. Specifically, damaged ECs stimulate the proliferation and differentiation of neighboring ISCs via the secretion of cytokines and EGFR ligands, which are regulated by signaling pathways such as the JNK and Hippo (Hpo) pathways (Huang et al., 2005; Shaw et al., 2010; Staley and Irvine, 2010; Zhou et al., 2017).
The Hpo pathway is an evolutionarily conserved pathway that senses structural integrity and changes in cell adhesion to regulate cell proliferation and survival (Dupont, 2016) (Fig. 3). The activity of the protein kinase Hpo is disrupted by tissue damage through a mechanism that involves signals derived from organization of the F-actin cytoskeleton, which modulates the activity of the Hpo pathway effector Yorkie (Yki) according to cell adhesion, cell density and actin filament tension (Rauskolb et al., 2014; Varelas et al., 2010; Zhao et al., 2012). In short, loss of Hpo activity allows nuclear localization of Yki and transcriptional activation of numerous genes involved in cell growth and other processes (Huang et al., 2005). In the fly gut, Yki also stimulates the transcription of secreted cytokines and growth factors, which stimulate Janus Kinase (JAK) and EGFR activity, respectively, promoting faster growth and endocycling in differentiating EBs (Furriols and Bray, 2001; Houtz et al., 2017; Ohlstein and Spradling, 2007; Ren et al., 2010; Shaw et al., 2010). Similarly, damage and ROS activate ISC proliferation through JNK activation in ECs, which stimulates the secretion of cytokines upon injury (Jiang et al., 2009; Santabarbara-Ruiz et al., 2015).
EBs endocycle in response to Notch signaling, although how M-CDK activity is suppressed by Notch remains obscure. In addition, although ECs from healthy guts are known to require TOR activity for scheduled endocycling (Xiang et al., 2017), EGFR signaling has been demonstrated to control damage-induced endocycling independently of TOR signaling, via post-transcriptional upregulation of E2F1 (Xiang et al., 2017). Thus, upon damage, EGFR signaling promotes compensatory polyploidization of ECs and, surprisingly, Insulin/PI3K/TOR signaling becomes dispensable. This example demonstrates a conditional ‘switching’ of pathways controlling endocycles, with one input pathway controlling endocycling during normal growth and another during stress-induced regeneration. E2F1 has also been demonstrated to be a rate-limiting G1/S-phase regulator in numerous Drosophila tissues, including the embryonic epidermis, larval salivary glands, fat body, gut, imaginal discs (epidermal progenitors) and the adult gut (Britton and Edgar, 1998; Duronio and O'Farrell, 1995; Follette et al., 1998; Maqbool et al., 2010; Xiang et al., 2017; Zielke et al., 2011). E2F1 therefore has a key role as a growth-sensing cell cycle regulator in Drosophila.
Wound healing and polyploidy in the Drosophila epidermis
There are many adult Drosophila tissues that lack stem cells and that therefore must accomplish wound healing via other mechanisms. The epidermis of adult Drosophila, located beneath the cuticle, is one such tissue. This tissue normally consists of a continuous layer of diploid ectodermal epithelial cells. However, if this epithelium is damaged by a puncture wound, it heals by complete wound closure in the absence of mitoses (Losick et al., 2016) (Fig. 5). As is the case for the regenerating Drosophila gut, this wound-healing process involves JNK and the Hpo pathway effector Yki. Interestingly, JNK has been reported to cross-regulate the Hpo pathway in wing discs (Enomoto et al., 2015).
Fig. 5.
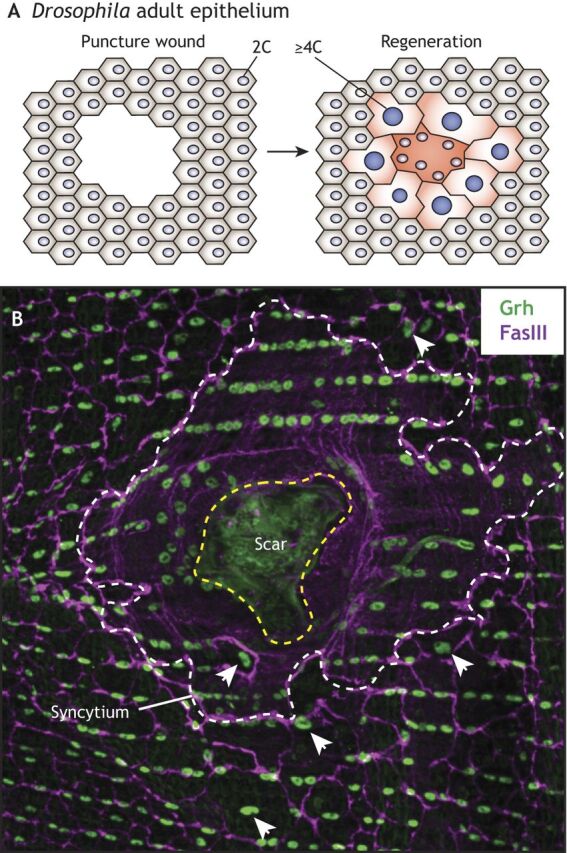
Wound-induced polyploidization in the adult Drosophila epithelium. (A) The adult Drosophila epithelium is composed of post-mitotic diploid cells. Upon epithelial puncture wounds, diploid epithelial cells are lost and the open wound is closed by surrounding cells that fuse together to form a syncytium. Cells surrounding the central syncytium undergo endocycles and promote compensatory growth. (B) Immunofluorescence image of regenerating Drosophila adult epithelium. Epidermal nuclei and cell-cell septate junctions, marked by Grainy-head (Grh, in green) and Fas3 (in magenta), respectively, are shown. The boundaries of the scar and syncytium are outlined in yellow and white dashed lines, respectively. Examples of large polyploid nuclei are indicated by arrowheads. Image courtesy of Vicki Losick (see Losick et al., 2013 for details).
Shortly after wounding, Yki and JNK activities increase in cells surrounding the wound site. Epithelial cells closest to the site of injury slowly migrate towards the center of the wound, and multiple cells fuse together to form a syncytium. This effect is dependent on Rac GTPase activity (Losick et al., 2013), which is also known to control cell fusion, myoblast migration and epithelial wound closure in the Drosophila embryo (Fernandes et al., 2005; Verboon and Parkhurst, 2015). As in the adult, wound closure in the Drosophila larval epithelium deploys cell-cell fusion around a wound site. In this context, cell-cell fusion is promoted by JNK activity, which is elevated around the wound site, while cell fusion is suppressed by JAK/STAT activity distal from the wound site (Lee et al., 2017). How JNK promotes cell-cell fusion is not well understood, but may involve JNK-dependent upregulation of integrins (Lee et al., 2017; Wang et al., 2015). In the adult fly epithelium, the downstream targets of JNK, Jra/Kay (Jun/Fos), modulate the activity of Yki by dampening its effect on polyploidization (Losick et al., 2016). However, JNK is also known to increase Yki activity in imaginal discs through inhibition of Warts, or through direct activation (Bunker et al., 2015; Sun and Irvine, 2013). It thus seems that JNK may affect Yki activity in several different tissue- or context-specific ways. In the adult epithelium, cells more distal to the wound site compensate for lost cells by endocycling and increasing cell growth, while leading-edge cells migrate toward the center of the wound. This endocycling response is dependent on Yki, the nuclear translocation and activation of which are likely induced by disruption of cell-cell contacts, and/or on stretching of the epithelium (Aragona et al., 2013; Gaspar and Tapon, 2014). Yki has previously been shown to upregulate Cyclin E in wing and eye imaginal discs, and both Yki and Cyclin E are required for compensatory proliferation in these contexts (Meserve and Duronio, 2015; Shu and Deng, 2017; Udan et al., 2003). Cyclin E may therefore be an essential target of Yki during compensatory endocycling and cell growth in Drosophila. Of note, Yki is also required for wound closure in the post-mitotic polyploid larval epithelium, which also occurs independently of mitotic proliferation. In this context, however, Yki stimulates wound closure through polymerization of actin filament cables and does not induce further polyploidization or cell fusion, again suggesting that the mode of contribution of Yki to wound healing is somewhat tissue specific (Tsai et al., 2017).
Polyploidization and heart regeneration
The contribution of polyploidization to wound healing has also been studied in the mammalian heart. The mammalian heart is known to contain polyploid cells: by adulthood, up to 70% of human and 85% of rodent cardiac myocytes are polyploid (Mollova et al., 2013; Soonpaa et al., 1996). Murine cardiomyocytes stop proliferation during postnatal development, when M-CDK activity is blocked through downregulation of its activator, Cdc25 (Kang et al., 1997; Soonpaa et al., 1996; Tane et al., 2014b). Both M- and S-CDK activity are also reduced by upregulation of CKIs (p21Cip1, p27Kip) (Poolman et al., 1998; Tane et al., 2014a). However, a wave of S- and M-CDK activity triggers one additional S phase at postnatal day 5, followed by an endomitotic event that results in binucleation of 80-90% of cardiomyocytes during the two first weeks after birth (Soonpaa et al., 1996). This process is dependent on the orphan cyclin, cyclin G1 (Liu et al., 2010). Like the murine heart, cells in the human heart become polyploid after birth, although in this case the cardiomyocytes remain mononucleate (Mollova et al., 2013).
Neonatal mammalian and adult zebrafish hearts display great regenerative capacity, through compensatory mitotic proliferation of pre-excising cardiomyocytes (Jopling et al., 2010; Kikuchi et al., 2010; Porrello et al., 2011). By contrast, postnatal mammalian hearts regenerate poorly and show limited cardiomyocyte proliferation. In this context, compensation for lost cells or cardiac stress, such as that caused by myocardial infarction, is therefore supported by further polyploidization and post-mitotic/hypertrophic growth (Ebert and Pfitzer, 1977; Senyo et al., 2013; Soonpaa and Field, 1997). Little is known about the signals that stimulate polyploidization in mammalian hearts, but a recent study in zebrafish has demonstrated that mechanical stretching of epicardial cells (the mesothelial cells that cover the heart) grown in an elastic growth chamber is sufficient to induce endocycling and endomitosis through tension (Cao et al., 2017). This artificial tension mimics the physical stretching suffered by epicardial cells, as they migrate towards a wound site, while retaining existing cell-cell adhesions, to seal the wound (Cao et al., 2017). This study also demonstrated that polyploid cells envelop the damaged heart more efficiently than diploid cells. Interestingly, in the zebrafish model, these polyploid cells are formed transiently upon heart injury, and apoptose once regeneration is complete, leaving behind diploid cells. Although polyploid cells facilitate regeneration of the zebrafish epicardium, it has recently been reported that ectopic polyploidization of the myocardium limits the regenerative capacity of zebrafish hearts (Gonzalez-Rosa et al., 2018). This observation suggests that polyploidization in general presents a physiological challenge to regeneration in heart tissues, thus mobilization of mitotic cell division, through cell reprogramming, may be promising as a therapeutic strategy (Srivastava and DeWitt, 2016; Tzahor and Poss, 2017).
Polyploidization during liver regeneration
The liver is also known for its remarkable regenerative capacity; this is not surprising, as it must endure chemotoxic stress from ingestion of toxins from various food sources. Liver polyploidization occurs postnatally and is responsive to both developmental and stress-linked inputs. As such, the postnatal liver becomes increasingly enriched in polyploid cells. Indeed, the human liver is composed of more than 20% polyploid cells at adulthood, whereas rodent livers are more than 70% polyploid (Wang et al., 2017), as a result of endoreplication. Developing hepatocytes undergo endomitosis, generating multi-nucleate cells, which can later divide to produce cells with a 4C or 8C DNA content (Guidotti et al., 2003; Margall-Ducos et al., 2007). Interestingly, blocking mitosis through conditional knockout of CDK1 in mouse liver does not impair regeneration upon 70% partial hepatectomy, indicating that regeneration can be maintained by hypertrophic growth of polyploid cells (Diril et al., 2012). Upon 30% partial hepatectomy, wild-type mouse livers regenerate primarily through compensatory growth by polyploid cells. Upon 70% hepatectomy, however, polyploid hepatocytes respond with hypertrophic growth, followed by cell division of binucleate hepatocytes to increase cell numbers (Miyaoka et al., 2012).
The molecular pathways and factors controlling liver polyploidization have been investigated. These studies have shown, for example, that oxidative stress, which is caused by pathological stress such as in non-alcoholic fatty liver disease, limits M-CDK activity and leads to the formation of highly polyploid (≥8C) mono-nucleated hepatocytes (Gentric et al., 2015). However, although endoreplication facilitates liver regeneration, this example also illustrates that excessive endoreplication can be associated with pathological conditions (Gentric and Desdouets, 2015). As with Drosophila endocycling cells, mouse hepatocytes need to downregulate mitotic regulators in order to bypass full mitosis, while maintaining expression of cyclin E1 to sustain DNA replication (Chen et al., 2012; Nevzorova et al., 2009). The literature indicates that E2F1-E2F3 promote expression of multiple mitotic genes in hepatocytes, including cyclin A and cyclin B, whereas E2F7 and E2F8 promote endoreplication by repressing expression of cyclin A1 and cyclin A2 (Chen et al., 2012). Thus, the double knockout of E2F7 and E2F8 causes increased cyclin A1 expression and a failure to endoreplicate, while endoreplication can be restored in E2F7- and E2F8-deficient hepatocytes when combined with the knockout of cyclin A1 and cyclin A2 or their transcriptional activator, E2F1 (Chen et al., 2012; Kim et al., 2016; Pandit et al., 2012). This is consistent with the reduced ploidy observed in E2F7/E2F8 double-knockout TGCs.
As with the Drosophila epidermis, the Hpo pathway appears to play a key role in liver polyploidization. The mammalian Yki homolog, yes-associated protein (YAP), is required for hepatocyte polyploidization (Zhang et al., 2017). YAP has been found to elevate the activity of Akt, which activates the acetyltransferase p300 by phosphorylation (Zhang et al., 2017) (Fig. 6). p300 then acetylates the F-box protein Skp2, which serves as a substrate recognition component of the Skp1, cullin 1, F-Box (SCF) E3 ligase complex. This acetylation displaces Skp2 from the nucleus to the cytoplasm, thus preventing the Skp2-dependent targeting of nuclear p27 for ubiquitylation and proteasomal degradation, and thereby promoting nuclear accumulation of p27. The net result of this cascade is that YAP elevates p27 levels to suppress M-CDK activity and block cell division (Zhang et al., 2017). How p27 allows enough CycE-CDK2 activity to promote S-phase entry, while at the same time preventing CycB/CDK1 activity and M-phase entry is not yet fully understood. One possible mechanism is that p27 limits the overall CDK activity to a level below the required M-phase threshold, while allowing a level sufficient for S-phase entry, consistent with a mechanism proposed earlier (Edgar et al., 2014; Stern and Nurse, 1996).
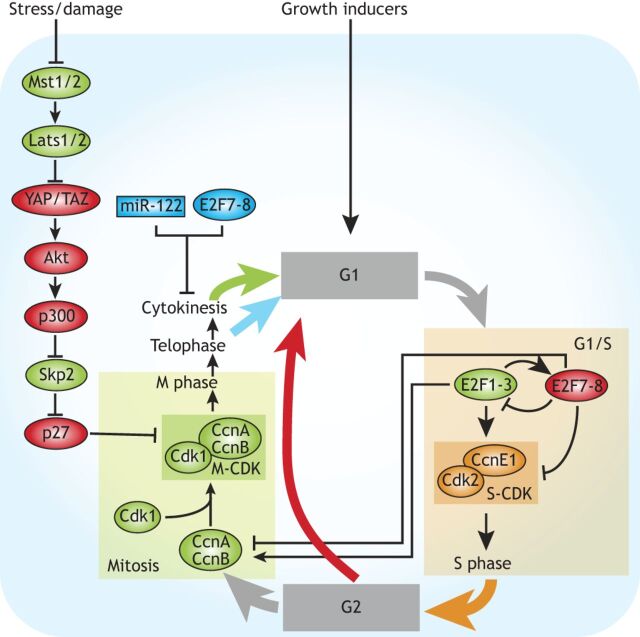
Fig. 6.
Polyploidization during liver regeneration. Hepatocyte growth rates affect the activation of E2F1-3, which controls G1/S through transcriptional activation of cyclin E1 (CcnE1)/Cdk2. E2F1-3 also activate expression of E2F7-8, which in turn repress expression of E2F1 and its targets, thus forming a negative-feedback loop. E2F1-3 are also required for expression of CcnA and CcnB, which activate Cdk1 (M-CDK) and are required for M-phase entry. In hepatocytes, E2F1-3 depletion promotes endoreplication (red arrow), whereas E2F7-8 depletion promotes mitosis (green arrow). Endoreplication is also induced through the Hippo pathway. The Hippo homologs Mst1/2 suppress activity of the Yorkie homologs YAP and TAZ through activation of Lats1/2. Upon Mst1/2 inactivation, YAP promotes Akt activity, which promotes activation of the acetyl transferase p300. The subsequent p300-dependent acetylation of Skp2, an F-box protein of the SCF ubiquitin ligase complex, sequesters Skp2 to the cytoplasm. This prevents proteasomal degradation of p27, an inhibitor of M-CDK activity. Hepatocytes also become polyploid via endomitosis (blue arrow) through downregulation of cytokinetic regulators such as Rho-GTPase. E2F7-8 and miR-122 are known suppressors of cytokinesis and thereby promote endomitosis.
In murine livers, E2F8 activity has also been shown to be required for hepatocyte binucleation, where it silences expression of cytokinetic regulators, such as Racgap, Ect2 and Mklp1 (Pandit et al., 2012). Recently, expression of the micro-RNA miR-122 was shown to be required for hepatocyte bi-nucleation, through silencing of a number of pro-cytokinetic effectors including Cux1 (Hsu et al., 2016), which also regulates expression of the E2F8 targets Racgap, Ect2 and Mklp1. Bi-nucleation can also be induced in cultured hepatocytes through stimulation by the cytokine TGFβ, which prevents midzone localization of RhoA-GTPase, a known regulator of cytokinesis (De Santis Puzzonia et al., 2016). Furthermore, the insulin-PI3K-Akt pathway has been shown to control bi-nucleation, possibly through TORC2-dependent regulation of Rho-GTPase activity (De Santis Puzzonia et al., 2016).
Once hepatocytes reach a ploidy of 8C, they enter senescence through upregulation of p16ink4A, p21 and p53 (Wang et al., 2014). However, senescence is reversible and polyploid hepatocytes may re-enter mitosis, giving rise to tetraploid and diploid cells, a dynamic process referred to as a ‘ploidy conveyor’ (Duncan et al., 2010). This suggests that senescent hepatocytes are programmed to allow re-establishment of M-CDK activity upon cell cycle entry. Ploidy reduction is prone to multipolar spindle formation during mitosis, and is therefore likely to give rise to aneuploid hepatocytes. However, hepatocytes manage to turn aneuploidy to the benefit of liver resilience, as aneuploidy creates a heterogeneous population of hepatocytes, some of which have increased fitness during chronic stress (Duncan et al., 2012a, 2010). Experiments in mice have revealed that livers with a heterogeneous population of hepatocytes can develop resistance against chronic injury through conditional cell selection, which gives rise to a less heterogeneous population of hepatocytes that share genotypes that endow specific stress resistances (Duncan et al., 2012a,b, 2010).
The multiple modes of polyploidization observed in the liver illustrate how plastic the cell cycle can be. Mitosis, endomitosis and endoreplication all require cell cycle entry followed by DNA replication through S-CDK activity. Blocking cytokinesis through miR-122 leads to endomitosis, whereas blocking M phase altogether, through M-CDK repression, gives rise to endoreplication. In contrast to TGCs and Drosophila polyploid cells, where the cell cycle type seems ‘hard wired’ through strict downregulation of M-CDK activity, hepatocytes appear to adopt a plastic mode of cell cycle regulation. M-CDK activity in hepatocytes is not completely squelched, as in Drosophila endocycling cells, but appears to be dampened and adjusted to various levels through CKIs and E2F7/8 activity. Moreover, a study in HeLa cells has shown that high concentrations of CDK1 inhibitors induce endocycles, whereas lower concentrations induce endomitosis (Chen et al., 2016), suggesting therefore that levels of M-CDK activity determine whether a cell performs mitosis, endomitosis or endocycling in an activity level-dependent manner. We therefore presume that promiscuous regulation of CDK in the liver allows sufficient M-CDK activity for sister chromatin separation and karyokinesis in endomitotic hepatocytes, whereas M-CDK activity is sufficiently restrained to block APC/Ccdc20 activation and anaphase in endoreplicating hepatocytes. Interestingly, cytokinesis and M-CDK activity appear to be regulated by some of the same upstream regulators (e.g. Akt, E2F8/7), and thus we speculate that the M-CDK/RhoGTP balance may control cell cycle decisions in the mammalian liver.
Conclusions and perspectives
In animals, the capacity for regeneration is often determined by the presence of mitotically capable stem cells, which can provide new cells as needed. This is typical in tissues with a high turnover rate, such as those exposed to harsh environments, like the intestine and skin. Cells in other tissues, however, are made to last and may be supported by few or even no stem cells. In extreme cases, cells that form a tissue during development must support tissue/organ function for the life of the organism, as is the case of human cardiomyocytes (Tzahor and Poss, 2017), the mammalian central nervous system and many organs in short-lived invertebrates. But what underlies these differences in regenerative modes and capacities? One possibility is that tissue function is the underlying factor. Cardiomyocyte function, for example, requires continuous contractile activity generated by actin-myosin sarcomeres. Mitotic proliferation requires disruption of sarcomeres, which would temporarily compromise cardiomyocyte function. Polyploidy in the heart may thus be beneficial because of acquired resistance to apoptosis, which ensures longevity, and because it allows growth while maintaining tissue function. In fact, polyploid cells, such as mammalian TGCs and Drosophila subperineurial glia, often form barrier tissues where growth of such barriers can be maintained continuously, without the loss of cell-cell junctions that occurs during mitosis (Unhavaithaya and Orr-Weaver, 2012; Von Stetina et al., 2018). However, polyploidization is not only an asset in wound healing: it can enhance cellular damage resistance. The protective cells that cover the skin, keratinocytes, undergo endomitosis and endocycling to a maximum ploidy of 12C (Gandarillas and Freije, 2014). Polyploidization of keratinocytes is induced by UV irradiation and is likely to increase tolerance to genotoxic stress (Gandarillas, 2012; Gandarillas et al., 2018), as reported in hepatocytes (Zhang et al., 2018). In fact, in plants, strains with increased ploidy are more resistant to UV radiation (Gegas et al., 2014). Similarly, the polyploid tissues of Drosophila larvae can survive high doses of irradiation that are sufficient to kill mitotic progenitor cells (Hassel et al., 2014; Zhang et al., 2014). As mentioned here, there seem to be several advantages to polyploidization in tissues where it is pre-programmed. It would thus be interesting to explore the extent to which these advantages can be conferred to ectopically induced polyploid cells.
We have learned a lot about how polyploidy is generated through endoreplication and cell-cell fusion, but there is still more to understand, for example about the damage sensors that induce regeneration through either mitosis or endoreplication. As discussed, several recent examples demonstrate the involvement of the Hpo pathway in the wound healing response. In mammals, the Hpo pathway is involved in contact inhibition of cell proliferation, where low cell density appears to lower the threshold for growth factor-sensitive proliferation (Gumbiner and Kim, 2014). This makes sense as a signal during the wound repair process, because cells adjacent to a wound have fewer cell contacts, and this condition would thus promote proliferation through the Hpo/Yap pathway. Tissue injury and cell death are also associated with ROS production, which stimulates JNK activity, and appear to have important roles in damage sensing (Mittal et al., 2014; Santabarbara-Ruiz et al., 2015). Further exploration of the primary damage sensors should be of great interest in fields involving regeneration.
As seen in mammalian hepatocytes, zebrafish cardiomyocytes and Drosophila epithelial cells, the generation of multinuclear cells during regeneration is a recurrent mechanism in wound healing. The advantage of multinuclear cells over mono-nuclear polyploid cells is not clear. Perhaps the added genome-to-nuclear surface ratio of multinuclear cells, or the dispersal of nuclei in a large cytoplasmic space, may be advantageous to cell growth and/or function. As reviewed here, recent studies of regeneration have highlighted the involvement of polyploid cells, but there are still many questions to be answered about how polyploidy is elaborated during wound healing, and what its advantages are. A potential future application of artificially induced polyploidy is to enhance the regeneration of postmitotic tissues that lack stem cells, for example during recovery from myocardial infarction. We are rapidly gaining the tools needed to trigger polyploidization, a simpler process than restoring the entire mitotic proliferation program, and it will be interesting to test these tools in various wound-healing and regeneration scenarios.
Acknowledgements
We thank Vicki Losick for the image presented in Fig. 5B, and Mahi Rahman and Liv Gansmo for providing helpful comments on the manuscript and figures.
Footnotes
Funding
The authors’ research is supported by Huntsman Cancer Foundation.
Contributor Information
Jan Inge Øvrebø, Huntsman Cancer Institute, Salt Lake City, UT 84112, USA.
Bruce A. Edgar, Huntsman Cancer Institute, Salt Lake City, UT 84112, USA.
References
- Andreassen, P. R., Martineau, S. N. and Margolis, R. L. (1996). Chemical induction of mitotic checkpoint override in mammalian cells results in aneuploidy following a transient tetraploid state. Mutat. Res. 372, 181-194. 10.1016/S0027-5107(96)00138-8 [DOI] [PubMed] [Google Scholar]
- Anisimov, A. P. (2005). Endopolyploidy as a morphogenetic factor of development. Cell Biol. Int. 29, 993-1004. 10.1016/j.cellbi.2005.10.013 [DOI] [PubMed] [Google Scholar]
- Aragona, M., Panciera, T., Manfrin, A., Giulitti, S., Michielin, F., Elvassore, N., Dupont, S. and Piccolo, S. (2013). A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047-1059. 10.1016/j.cell.2013.07.042 [DOI] [PubMed] [Google Scholar]
- Artero, R., Furlong, E. E., Beckett, K., Scott, M. P. and Baylies, M. (2003). Notch and Ras signaling pathway effector genes expressed in fusion competent and founder cells during Drosophila myogenesis. Development 130, 6257-6272. 10.1242/dev.00843 [DOI] [PubMed] [Google Scholar]
- Audibert, A., Simon, F. and Gho, M. (2005). Cell cycle diversity involves differential regulation of Cyclin E activity in the Drosophila bristle cell lineage. Development 132, 2287-2297. 10.1242/dev.01797 [DOI] [PubMed] [Google Scholar]
- Bäumer, D., Ströhlein, N. M. and Schoppmeier, M. (2012). Opposing effects of Notch-signaling in maintaining the proliferative state of follicle cells in the telotrophic ovary of the beetle Tribolium. Front. Zool. 9, 15. 10.1186/1742-9994-9-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, S. P. and Labib, K. (2016). Chromosome duplication in Saccharomyces cerevisiae. Genetics 203, 1027-1067. 10.1534/genetics.115.186452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, S. P. and Stillman, B. (1992). Atp-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357, 128-134. 10.1038/357128a0 [DOI] [PubMed] [Google Scholar]
- Berckmans, B., Lammens, T., Van Den Daele, H., Magyar, Z., Bogre, L. and De Veylder, L. (2011). Light-dependent regulation of DEL1 is determined by the antagonistic action of E2Fb and E2Fc. Plant Physiol. 157, 1440-1451. 10.1104/pp.111.183384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, S., Schafer, G., Kesper, D. A., Holz, A., Eriksson, T., Palmer, R. H., Beck, L., Klambt, C., Renkawitz-Pohl, R. and Onel, S.-F. (2008). WASP and SCAR have distinct roles in activating the Arp2/3 complex during myoblast fusion. J. Cell Sci. 121, 1303-1313. 10.1242/jcs.022269 [DOI] [PubMed] [Google Scholar]
- Biesterfeld, S., Gerres, K., Fischer-Wein, G. and Bocking, A. (1994). Polyploidy in non-neoplastic tissues. J. Clin. Pathol. 47, 38-42. 10.1136/jcp.47.1.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour, B. A., Chakravarti, M., West, J. M. and Abmayr, S. M. (2000). Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev. 14, 1498-1511. [PMC free article] [PubMed] [Google Scholar]
- Breuer, C., Ishida, T. and Sugimoto, K. (2010). Developmental control of endocycles and cell growth in plants. Curr. Opin. Plant Biol. 13, 654-660. 10.1016/j.pbi.2010.10.006 [DOI] [PubMed] [Google Scholar]
- Britton, J. S. and Edgar, B. A. (1998). Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development 125, 2149-2158. [DOI] [PubMed] [Google Scholar]
- Britton, J. S., Lockwood, W. K., Li, L., Cohen, S. M. and Edgar, B. A. (2002). Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2, 239-249. 10.1016/S1534-5807(02)00117-X [DOI] [PubMed] [Google Scholar]
- Brodsky, V. Y., Brodskiĭ, V. I. A., Brodskij, V. J., Brodskiĭ, V. I. A. and Uryuvaeva, I. V. (1985). Genome Multiplication in Growth and Development: Biology of Polyploid and Polytene Cells. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Broek, D., Bartlett, R., Crawford, K. and Nurse, P. (1991). Involvement of P34cdc2 in establishing the dependency of S phase on mitosis. Nature 349, 388-393. 10.1038/349388a0 [DOI] [PubMed] [Google Scholar]
- Bunker, B. D., Nellimoottil, T. T., Boileau, R. M., Classen, A. K. and Bilder, D. (2015). The transcriptional response to tumorigenic polarity loss in Drosophila. eLife 4, e03189. 10.7554/eLife.03189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterworth, F. M. and Rasch, E. M. (1986). Adipose-tissue of Drosophila-melanogaster 7. Distribution of nuclear-DNA amounts along the anterior posterior axis in the larval fat-body. J. Exp. Zool. 239, 77-85. 10.1002/jez.1402390110 [DOI] [PubMed] [Google Scholar]
- Buttitta, L. A. and Edgar, B. A. (2007). Mechanisms controlling cell cycle exit upon terminal differentiation. Curr. Opin. Cell Biol. 19, 697-704. 10.1016/j.ceb.2007.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campsteijn, C., Ovrebo, J. I., Karlsen, B. O. and Thompson, E. M. (2012). Expansion of cyclin D and CDK1 paralogs in Oikopleura dioica, a chordate employing diverse cell cycle variants. Mol. Biol. Evol. 29, 487-502. 10.1093/molbev/msr136 [DOI] [PubMed] [Google Scholar]
- Cao, J. L., Wang, J. H., Jackman, C. P., Cox, A. H., Trembley, M. A., Balowski, J. J., Cox, B. D., De Simone, A., Dickson, A. L., Di Talia, S., et al. (2017). Tension creates an endoreplication wavefront that leads regeneration of epicardial tissue. Dev. Cell 42, 600. 10.1016/j.devcel.2017.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro, E., Desvoyes, B., Ramirez-Parra, E., Sanchez, M. P. and Gutierrez, C. (2008). Endoreduplication control during plant development. SEB Exp. Biol. Ser. 59, 167-187. [PubMed] [Google Scholar]
- Carter, S. L., Eklund, A. C., Kohane, I. S., Harris, L. N. and Szallasi, Z. (2006). A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 38, 1043-1048. 10.1038/ng1861 [DOI] [PubMed] [Google Scholar]
- Chanprasert, S., Geddis, A. E., Barroga, C., Fox, N. E. and Kaushansky, K. (2006). Thrombopoietin (TPO) induces c-myc expression through a PI3K- and MAPK-dependent pathway that is not mediated by Akt, PKC zeta or mTOR in TPO-dependent cell lines and primary megakaryocytes. Cell. Signal. 18, 1212-1218. 10.1016/j.cellsig.2005.09.010 [DOI] [PubMed] [Google Scholar]
- Chen, S. Y. and Bell, S. P. (2011). CDK prevents Mcm2-7 helicase loading by inhibiting Cdt1 interaction with Orc6. Gene Dev 25, 363-372. 10.1101/gad.2011511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, E. H. and Olson, E. N. (2004). Towards a molecular pathway for myoblast fusion in Drosophila. Trends Cell Biol. 14, 452-460. 10.1016/j.tcb.2004.07.008 [DOI] [PubMed] [Google Scholar]
- Chen, H.-Z., Tsai, S.-Y. and Leone, G. (2009). Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer 9, 785-797. 10.1038/nrc2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.-Z., Ouseph, M. M., Li, J., Pécot, T., Chokshi, V., Kent, L., Bae, S., Byrne, M., Duran, C., Comstock, G., et al. (2012). Canonical and atypical E2Fs regulate the mammalian endocycle. Nat. Cell Biol. 14, 1192-1202. 10.1038/ncb2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Stout, J. R., Dharmaiah, S., Yde, S., Calvi, B. R. and Walczak, C. E. (2016). Transient endoreplication down-regulates the kinesin-14 HSET and contributes to genomic instability. Mol. Biol. Cell 27, 2911-2923. 10.1091/mbc.e16-03-0159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, J.-L., Tsai, S.-Y., Sharma, N., Opavsky, R., Price, R., Wu, L., Fernandez, S. A. and Leone, G. (2009). E2f3a and E2f3b contribute to the control of cell proliferation and mouse development. Mol. Cell. Biol. 29, 414-424. 10.1128/MCB.01161-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrysanthou, S., Senner, C. E., Woods, L., Fineberg, E., Okkenhaug, H., Burge, S., Perez-Garcia, V. and Hemberger, M. (2018). A critical role of TET1/2 proteins in cell-cycle progression of trophoblast stem cells. Stem Cell Rep. 10, 1355-1368. 10.1016/j.stemcr.2018.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocker, J. H., Piatti, S., Santocanale, C., Nasmyth, K. and Diffley, J. F. X. (1996). An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature 379, 180-182. 10.1038/379180a0 [DOI] [PubMed] [Google Scholar]
- Coward, J. and Harding, A. (2014). Size does matter: why polyploid tumor cells are critical drug targets in the war on cancer. Front. Oncol. 4, 123. 10.3389/fonc.2014.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli, T. and de Lange, T. (2011). The causes and consequences of polyploidy in normal development and cancer. Annu. Rev. Cell Dev. Biol. 27, 585-610. 10.1146/annurev-cellbio-092910-154234 [DOI] [PubMed] [Google Scholar]
- de Bruin, A., Maiti, B., Jakoi, L., Timmers, C., Buerki, R. and Leone, G. (2003). Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 278, 42041-42049. 10.1074/jbc.M308105200 [DOI] [PubMed] [Google Scholar]
- de Renty, C., DePamphilis, M. L. and Ullah, Z. (2014). Cytoplasmic localization of p21 protects trophoblast giant cells from DNA damage induced apoptosis. PLoS ONE 9, e97434. 10.1371/journal.pone.0097434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santis Puzzonia, M., Cozzolino, A. M., Grassi, G., Bisceglia, F., Strippoli, R., Guarguaglini, G., Citarella, F., Sacchetti, B., Tripodi, M., Marchetti, A., et al. (2016). TGFbeta induces binucleation/polyploidization in hepatocytes through a Src-dependent cytokinesis failure. PLos ONE 11, e0167158. 10.1371/journal.pone.0167158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Larkin, J. C. and Schnittger, A. (2011). Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 16, 624-634. 10.1016/j.tplants.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Demontis, F. and Perrimon, N. (2009). Integration of Insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development 136, 983-993. 10.1242/dev.027466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, W. M., Althauser, C. and Ruohola-Baker, H. (2001). Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development 128, 4737-4746. [DOI] [PubMed] [Google Scholar]
- DePamphilis, M. L. (2016). Genome duplication: the heartbeat of developing organisms. Curr. Top. Dev. Biol. 116, 201-229. 10.1016/bs.ctdb.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diril, M. K., Ratnacaram, C. K., Padmakumar, V. C., Du, T., Wasser, M., Coppola, V., Tessarollo, L. and Kaldis, P. (2012). Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc. Natl. Acad. Sci. USA 109, 3826-3831. 10.1073/pnas.1115201109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duelli, D. and Lazebnik, Y. (2007). Cell-to-cell fusion as a link between viruses and cancer. Nat. Rev. Cancer 7, 968-976. 10.1038/nrc2272 [DOI] [PubMed] [Google Scholar]
- Duelli, D. M., Padilla-Nash, H. M., Berman, D., Murphy, K. M., Ried, T. and Lazebnik, Y. (2007). A virus causes cancer by inducing massive chromosomal instability through cell fusion. Curr. Biol. 17, 431-437. 10.1016/j.cub.2007.01.049 [DOI] [PubMed] [Google Scholar]
- Duncan, A. W., Taylor, M. H., Hickey, R. D., Hanlon Newell, A. E., Lenzi, M. L., Olson, S. B., Finegold, M. J. and Grompe, M. (2010). The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467, 707-710. 10.1038/nature09414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, A. W., Hanlon Newell, A. E., Bi, W., Finegold, M. J., Olson, S. B., Beaudet, A. L. and Grompe, M. (2012a). Aneuploidy as a mechanism for stress-induced liver adaptation. J. Clin. Invest. 122, 3307-3315. 10.1172/JCI64026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, A. W., Hanlon Newell, A. E., Smith, L., Wilson, E. M., Olson, S. B., Thayer, M. J., Strom, S. C. and Grompe, M. (2012b). Frequent aneuploidy among normal human hepatocytes. Gastroenterology 142, 25-28. 10.1053/j.gastro.2011.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont, S. (2016). Role of YAP/TAZ in cell-matrix adhesion-mediated signalling and mechanotransduction. Exp. Cell Res. 343, 42-53. 10.1016/j.yexcr.2015.10.034 [DOI] [PubMed] [Google Scholar]
- Duronio, R. J. and O'Farrell, P. H. (1995). Developmental control of the G(1) to S transition in Drosophila: Cyclin E is a limiting downstream target of E2f. Gene Dev. 9, 1456-1468. 10.1101/gad.9.12.1456 [DOI] [PubMed] [Google Scholar]
- Ebert, L. and Pfitzer, P. (1977). Nuclear DNA of myocardial cells in the periphery of infarctions and scars. Virchows Arch. B Cell Pathol. 24, 209-217. [DOI] [PubMed] [Google Scholar]
- Edgar, B. A. and Orr-Weaver, T. L. (2001). Endoreplication cell cycles: more for less. Cell 105, 297-306. 10.1016/S0092-8674(01)00334-8 [DOI] [PubMed] [Google Scholar]
- Edgar, B. A., Zielke, N. and Gutierrez, C. (2014). Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell Biol. 15, 197-210. 10.1038/nrm3756 [DOI] [PubMed] [Google Scholar]
- Eliades, A., Papadantonakis, N. and Ravid, K. (2010). New roles for cyclin E in megakaryocytic polyploidization. J. Biol. Chem. 285, 18909-18917. 10.1074/jbc.M110.102145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto, M., Kizawa, D., Ohsawa, S. and Igaki, T. (2015). JNK signaling is converted from anti- to pro-tumor pathway by Ras-mediated switch of Warts activity. Dev. Biol. 403, 162-171. 10.1016/j.ydbio.2015.05.001 [DOI] [PubMed] [Google Scholar]
- Fernandes, J. J., Atreya, K. B., Desai, K. M., Hall, R. E., Patel, M. D., Desai, A. A., Benham, A. E., Mable, J. L. and Straessle, J. L. (2005). A dominant negative form of Rac1 affects myogenesis of adult thoracic muscles in Drosophila. Dev. Biol. 285, 11-27. 10.1016/j.ydbio.2005.05.040 [DOI] [PubMed] [Google Scholar]
- Follette, P. J., Duronio, R. J. and O'Farrell, P. H. (1998). Fluctuations in cyclin E levels are required for multiple rounds of endocycle S phase in Drosophila. Curr. Biol. 8, 235-238. 10.1016/S0960-9822(98)70089-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, D. T. and Duronio, R. J. (2013). Endoreplication and polyploidy: insights into development and disease. Development 140, 3-12. 10.1242/dev.080531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, D. T., Gall, J. G. and Spradling, A. C. (2010). Error-prone polyploid mitosis during normal Drosophila development. Genes Dev. 24, 2294-2302. 10.1101/gad.1952710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furriols, M. and Bray, S. (2001). A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr. Biol. 11, 60-64. 10.1016/S0960-9822(00)00044-0 [DOI] [PubMed] [Google Scholar]
- Gandarillas, A. (2012). The mysterious human epidermal cell cycle, or an oncogene-induced differentiation checkpoint. Cell Cycle 11, 4507-4516. 10.4161/cc.22529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandarillas, A. and Freije, A. (2014). Cycling up the epidermis: reconciling 100 years of debate. Exp. Dermatol. 23, 87-91. 10.1111/exd.12287 [DOI] [PubMed] [Google Scholar]
- Gandarillas, A., Molinuevo, R. and Sanz-Gomez, N. (2018). Mammalian endoreplication emerges to reveal a potential developmental timer. Cell Death Differ. 25, 471-476. 10.1038/s41418-017-0040-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot, P. and Thompson, E. M. (2002). Patterning through differential endoreduplication in epithelial organogenesis of the chordate, Oikopleura dioica. Dev. Biol. 252, 59-71. 10.1006/dbio.2002.0834 [DOI] [PubMed] [Google Scholar]
- Gaspar, P. and Tapon, N. (2014). Sensing the local environment: actin architecture and Hippo signalling. Curr. Opin. Cell Biol. 31, 74-83. 10.1016/j.ceb.2014.09.003 [DOI] [PubMed] [Google Scholar]
- Gegas, V. C., Wargent, J. J., Pesquet, E., Granqvist, E., Paul, N. D. and Doonan, J. H. (2014). Endopolyploidy as a potential alternative adaptive strategy for Arabidopsis leaf size variation in response to UV-B. J. Exp. Bot. 65, 2757-2766. 10.1093/jxb/ert473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau, E., Hofte, H., Grandjean, O., Brown, S. and Traas, J. (1998). Phytochrome controls the number of endoreduplication cycles in the Arabidopsis thaliana hypocotyl. Plant J. 13, 221-230. 10.1046/j.1365-313X.1998.00030.x [DOI] [PubMed] [Google Scholar]
- Gentric, G. and Desdouets, C. (2015). Liver polyploidy: Dr Jekyll or Mr Hide? Oncotarget 6, 8430-8431. 10.18632/oncotarget.3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentric, G., Celton-Morizur, S. and Desdouets, C. (2012). Polyploidy and liver proliferation. Clin. Res. Hepatol. Gastroenterol. 36, 29-34. 10.1016/j.clinre.2011.05.011 [DOI] [PubMed] [Google Scholar]
- Gentric, G., Maillet, V., Paradis, V., Couton, D., L'Hermitte, A., Panasyuk, G., Fromenty, B., Celton-Morizur, S. and Desdouets, C. (2015). Oxidative stress promotes pathologic polyploidization in nonalcoholic fatty liver disease. J. Clin. Invest. 125, 981-992. 10.1172/JCI73957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rosa, J. M., Sharpe, M., Field, D., Soonpaa, M. H., Field, L. J., Burns, C. E. and Burns, C. G. (2018). Myocardial polyploidization creates a barrier to heart regeneration in zebrafish. Dev. Cell 44, 433. 10.1016/j.devcel.2018.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal, S. S., Li, L., Orian, A., Eisenman, R. N. and Edgar, B. A. (2005). Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 7, 295-302. 10.1038/ncb1223 [DOI] [PubMed] [Google Scholar]
- Guidotti, J.-E., Brégerie, O., Robert, A., Debey, P., Brechot, C. and Desdouets, C. (2003). Liver cell polyploidization: a pivotal role for binuclear hepatocytes. J. Biol. Chem. 278, 19095-19101. 10.1074/jbc.M300982200 [DOI] [PubMed] [Google Scholar]
- Gumbiner, B. M. and Kim, N.-G. (2014). The Hippo-YAP signaling pathway and contact inhibition of growth. J. Cell Sci. 127, 709-717. 10.1242/jcs.140103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, N.-G. S. (2000). Hepatic polyploidy and liver growth control. Semin. Cancer Biol. 10, 161-171. 10.1006/scbi.2000.0317 [DOI] [PubMed] [Google Scholar]
- Gutierrez, C. (2005). Coupling cell proliferation and development in plants. Nat. Cell Biol. 7, 535-541. 10.1038/ncb0605-535 [DOI] [PubMed] [Google Scholar]
- Gutierrez, C. (2009). The Arabidopsis cell division cycle. Arabidopsis Book 7, e0120. 10.1199/tab.0120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, M. P. and Laird, C. D. (1985). Control of DNA replication and spatial distribution of defined DNA sequences in salivary gland cells of Drosophila melanogaster. Chromosoma 91, 279-286. 10.1007/BF00328223 [DOI] [PubMed] [Google Scholar]
- Hannibal, R. L. and Baker, J. C. (2016). Selective amplification of the genome surrounding key placental genes in trophoblast giant cells. Curr. Biol. 26, 230-236. 10.1016/j.cub.2015.11.060 [DOI] [PubMed] [Google Scholar]
- Harashima, H. and Schnittger, A. (2010). The integration of cell division, growth and differentiation. Curr. Opin. Plant Biol. 13, 66-74. 10.1016/j.pbi.2009.11.001 [DOI] [PubMed] [Google Scholar]
- Hassel, C., Zhang, B., Dixon, M. and Calvi, B. R. (2014). Induction of endocycles represses apoptosis independently of differentiation and predisposes cells to genome instability. Development 141, 112-123. 10.1242/dev.098871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, S. (1996). A Cdc2 dependent checkpoint maintains diploidy in Drosophila. Development 122, 1051-1058. [DOI] [PubMed] [Google Scholar]
- Hayles, J., Fisher, D., Woollard, A. and Nurse, P. (1994). Temporal order of S phase and mitosis in fission yeast is determined by the state of the P34(Cdc2) mitotic B-cyclin complex. Cell 78, 813-822. 10.1016/S0092-8674(94)90542-8 [DOI] [PubMed] [Google Scholar]
- Heller, R. C., Kang, S., Lam, W. M., Chen, S., Chan, C. S. and Bell, S. P. (2011). Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 146, 80-91. 10.1016/j.cell.2011.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochegger, H., Dejsuphong, D., Sonoda, E., Saberi, A., Rajendra, E., Kirk, J., Hunt, T. and Takeda, S. (2007). An essential role for Cdk1 in S phase control is revealed via chemical genetics in vertebrate cells. J. Cell Biol. 178, 257-268. 10.1083/jcb.200702034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtz, P., Bonfini, A., Liu, X., Revah, J., Guillou, A., Poidevin, M., Hens, K., Huang, H.-Y., Deplancke, B., Tsai, Y.-C., et al. (2017). Hippo, TGF-beta, and Src-MAPK pathways regulate transcription of the upd3 cytokine in Drosophila enterocytes upon bacterial infection. PLoS Genet. 13, e1007091. 10.1371/journal.pgen.1007091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, S.-H., Delgado, E. R., Otero, P. A., Teng, K.-Y., Kutay, H., Meehan, K. M., Moroney, J. B., Monga, J. K., Hand, N. J., Friedman, J. R., et al. (2016). MicroRNA-122 regulates polyploidization in the murine liver. Hepatology 64, 599-615. 10.1002/hep.28573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J., Wu, S., Barrera, J., Matthews, K. and Pan, D. (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421-434. 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Jeong, J. and Conboy, I. M. (2011). Phosphatidylserine directly and positively regulates fusion of myoblasts into myotubes. Biochem. Biophys. Res. Commun. 414, 9-13. 10.1016/j.bbrc.2011.08.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H. Q., Patel, P. H., Kohlmaier, A., Grenley, M. O., McEwen, D. G. and Edgar, B. A. (2009). Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343-1355. 10.1016/j.cell.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling, C., Sleep, E., Raya, M., Marti, M., Raya, A. and Izpisua Belmonte, J. C. (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606-609. 10.1038/nature08899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès, J. and Chevalier, C. (2000). Endoreduplication in higher plants. Plant Mol. Biol. 43, 735-745. 10.1023/A:1006446417196 [DOI] [PubMed] [Google Scholar]
- Kaneko, Y. and Knudson, A. G. (2000). Mechanism and relevance of ploidy in neuroblastoma. Gene Chromosome Canc 29, 89-95. [DOI] [PubMed] [Google Scholar]
- Kang, M. J., Kim, J. S., Chae, S. W., Koh, K. N. and Koh, G. Y. (1997). Cyclins and cyclin dependent kinases during cardiac development. Mol. Cells 7, 360-366. [PubMed] [Google Scholar]
- Kaushansky, K. (2016). Thrombopoietin and its receptor in normal and neoplastic hematopoiesis. Thromb. J. 14. 10.1186/s12959-016-0095-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, K., Holdway, J. E., Werdich, A. A., Anderson, R. M., Fang, Y., Egnaczyk, G. F., Evans, T., Macrae, C. A., Stainier, D. Y. R. and Poss, K. D. (2010). Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464, 601-605. 10.1038/nature08804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. H., Jin, P., Duan, R. and Chen, E. H. (2015a). Mechanisms of myoblast fusion during muscle development. Curr. Opin. Genet. Dev. 32, 162-170. 10.1016/j.gde.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. H., Ren, Y., Ng, W. P., Li, S., Son, S., Kee, Y.-S., Zhang, S., Zhang, G., Fletcher, D. A., Robinson, D. N., et al. (2015b). Mechanical tension drives cell membrane fusion. Dev. Cell 32, 561-573. 10.1016/j.devcel.2015.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.-H., Jeon, Y., Kim, H.-S., Lee, J.-K., Lim, H. J., Kang, D., Cho, H., Park, C.-K., Lee, H. and Lee, C.-W. (2016). Hepatocyte homeostasis for chromosome ploidization and liver function is regulated by Ssu72 protein phosphatase. Hepatology 63, 247-259. 10.1002/hep.28281 [DOI] [PubMed] [Google Scholar]
- Kondorosi, E., Roudier, F. and Gendreau, E. (2000). Plant cell-size control: growing by ploidy? Curr. Opin. Plant Biol. 3, 488-492. 10.1016/S1369-5266(00)00118-7 [DOI] [PubMed] [Google Scholar]
- Lai, A. G., Kosaka, N., Abnave, P., Sahu, S. and Aboobaker, A. A. (2017). The abrogation of condensin function provides independent evidence for defining the self-renewing population of pluripotent stem cells. Dev. Biol. 433, 218-226. 10.1016/j.ydbio.2017.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanni, J. S. and Jacks, T. (1998). Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol. Cell. Biol. 18, 1055-1064. 10.1128/MCB.18.2.1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasek, R. J. and Dower, W. J. (1971). Aplysia californica: analysis of nuclear DNA in individual nuclei of giant neurons. Science 172, 278-280. 10.1126/science.172.3980.278 [DOI] [PubMed] [Google Scholar]
- Lee, S. H., McCormick, F. and Saya, H. (2010). Mad2 inhibits the mitotic kinesin MKlp2. J. Cell Biol. 191, 1069-1077. 10.1083/jcb.201003095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, A. J. X., Endesfelder, D., Rowan, A. J., Walther, A., Birkbak, N. J., Futreal, P. A., Downward, J., Szallasi, Z., Tomlinson, I. P. M., Howell, M., et al. (2011). Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 71, 1858-1870. 10.1158/0008-5472.CAN-10-3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.-H., Lee, C.-W., Park, S.-H. and Choe, K.-M. (2017). Spatiotemporal regulation of cell fusion by JNK and JAK/STAT signaling during Drosophila wound healing. J. Cell Sci. 130, 1917-1928. 10.1242/jcs.187658 [DOI] [PubMed] [Google Scholar]
- Levine, D. S., Sanchez, C. A., Rabinovitch, P. S. and Reid, B. J. (1991). Formation of the tetraploid intermediate is associated with the development of cells with more than four centrioles in the elastase-simian virus 40 tumor antigen transgenic mouse model of pancreatic cancer. Proc. Natl. Acad. Sci. USA 88, 6427-6431. 10.1073/pnas.88.15.6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly, M. A. and Spradling, A. C. (1996). The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 10, 2514-2526. 10.1101/gad.10.19.2514 [DOI] [PubMed] [Google Scholar]
- Liu, Z., Yue, S., Chen, X., Kubin, T. and Braun, T. (2010). Regulation of cardiomyocyte polyploidy and multinucleation by CyclinG1. Circ. Res. 106, 1498-1506. 10.1161/CIRCRESAHA.109.211888 [DOI] [PubMed] [Google Scholar]
- Lopez-Schier, H. and St Johnston, D. (2001). Delta signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes Dev. 15, 1393-1405. 10.1101/gad.200901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick, V. P. (2016). Wound-induced polyploidy is required for tissue repair. Adv. Wound Care 5, 271-278. 10.1089/wound.2014.0545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick, V. P., Fox, D. T. and Spradling, A. C. (2013). Polyploidization and cell fusion contribute to wound healing in the adult Drosophila epithelium. Curr. Biol. 23, 2224-2232. 10.1016/j.cub.2013.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick, V. P., Jun, A. S. and Spradling, A. C. (2016). Wound-induced polyploidization: regulation by Hippo and JNK signaling and conservation in mammals. PLoS ONE 11, e0151251. 10.1371/journal.pone.0151251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines, J. Z., Stevens, L. M., Tong, X. L. and Stein, D. (2004). Drosophila dMyc is required for ovary cell growth and endoreplication. Development 131, 775-786. 10.1242/dev.00932 [DOI] [PubMed] [Google Scholar]
- Maiti, B., Li, J., de Bruin, A., Gordon, F., Timmers, C., Opavsky, R., Patil, K., Tuttle, J., Cleghorn, W. and Leone, G. (2005). Cloning and characterization of mouse E2F8, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 280, 18211-18220. 10.1074/jbc.M501410200 [DOI] [PubMed] [Google Scholar]
- Mandrioli, M., Mola, L., Cuoghi, B. and Sonetti, D. (2010). Endoreplication: a molecular trick during animal neuron evolution. Q. Rev. Biol. 85, 159-169. 10.1086/652341 [DOI] [PubMed] [Google Scholar]
- Maqbool, S. B., Mehrotra, S., Kolpakas, A., Durden, C., Zhang, B., Zhong, H. and Calvi, B. R. (2010). Dampened activity of E2F1-DP and Myb-MuvB transcription factors in Drosophila endocycling cells. J. Cell Sci. 123, 4095-4106. 10.1242/jcs.064519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margall-Ducos, G., Celton-Morizur, S., Couton, D., Bregerie, O. and Desdouets, C. (2007). Liver tetraploidization is controlled by a new process of incomplete cytokinesis. J. Cell Sci. 120, 3633-3639. 10.1242/jcs.016907 [DOI] [PubMed] [Google Scholar]
- Marti, A., Wirbelauer, C., Scheffner, M. and Krek, W. (1999). Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat. Cell Biol. 1, 14-19. 10.1038/8984 [DOI] [PubMed] [Google Scholar]
- Massarwa, R., Carmon, S., Shilo, B.-Z. and Schejter, E. D. (2007). WIP/WASp-based actin-polymerization machinery is essential for myoblast fusion in Drosophila. Dev. Cell 12, 557-569. 10.1016/j.devcel.2007.01.016 [DOI] [PubMed] [Google Scholar]
- McCrann, D. J., Nguyen, H. G., Jones, M. R. and Ravid, K. (2008). Vascular smooth muscle cell polyploidy: an adaptive or maladaptive response? J. Cell. Physiol. 215, 588-592. 10.1002/jcp.21363 [DOI] [PubMed] [Google Scholar]
- Mcdonald, T. P. (1992). Its biology, clinical aspects, and possibilities. Am. J. Pediat. Hematol. 14, 8-21. 10.1097/00043426-199221000-00002 [DOI] [PubMed] [Google Scholar]
- Melaragno, J. E., Mehrotra, B. and Coleman, A. W. (1993). Relationship between endopolyploidy and cell size in epidermal tissue of arabidopsis. Plant Cell 5, 1661-1668. 10.1105/tpc.5.11.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meserve, J. H. and Duronio, R. J. (2015). Scalloped and Yorkie are required for cell cycle re-entry of quiescent cells after tissue damage. Development 142, 2740-2751. 10.1242/dev.119339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylov, I. S., Kondo, T., Jones, L., Ryzhikov, S., Tanaka, J., Zheng, J., Higa, L. A., Minamino, N., Cooley, L. and Zhang, H. (2002). Control of DNA replication and chromosome ploidy by geminin and cyclin A. Mol. Cell. Biol. 22, 1868-1880. 10.1128/MCB.22.6.1868-1880.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn, A. J., Boise, L. H. and Thompson, C. B. (1996). Expression of Bcl-x(L) and loss of p53 can cooperate to overcome a cell cycle checkpoint induced by mitotic spindle damage. Genes Dev. 10, 2621-2631. 10.1101/gad.10.20.2621 [DOI] [PubMed] [Google Scholar]
- Mitelman, F. (2005). Database of chromosome aberrations in cancer. Chromosome Res. 13, 5. [Google Scholar]
- Mittal, M., Siddiqui, M. R., Tran, K., Reddy, S. P. and Malik, A. B. (2014). Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox. Signal. 20, 1126-1167. 10.1089/ars.2012.5149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka, Y., Ebato, K., Kato, H., Arakawa, S., Shimizu, S. and Miyajima, A. (2012). Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr. Biol. 22, 1166-1175. 10.1016/j.cub.2012.05.016 [DOI] [PubMed] [Google Scholar]
- Mollova, M., Bersell, K., Walsh, S., Savla, J., Das, L. T., Park, S.-Y., Silberstein, L. E., dos Remedios, C. G., Graham, D., Colan, S., et al. (2013). Cardiomyocyte proliferation contributes to heart growth in young humans. Proc. Natl. Acad. Sci. USA 110, 1446-1451. 10.1073/pnas.1214608110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbonne-Reveau, K., Senger, S., Pal, M., Herr, A., Richardson, H. E., Asano, M., Deak, P. and Lilly, M. A. (2008). APC/CFzr/Cdh1 promotes cell cycle progression during the Drosophila endocycle. Development 135, 1451-1461. 10.1242/dev.016295 [DOI] [PubMed] [Google Scholar]
- Nevzorova, Y. A., Tschaharganeh, D., Gassler, N., Geng, Y., Weiskirchen, R., Sicinski, P., Trautwein, C. and Liedtke, C. (2009). Aberrant cell cycle progression and endoreplication in regenerating livers of mice that lack a single E-type cyclin. Gastroenterology 137, 691-703, 703.e691-696. 10.1053/j.gastro.2009.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, H., Takyu, R., Morimoto, H., Toei, S., Sakon, H., Goto, S., Moriya, S. and Kono, T. (2016). Cell proliferation potency is independent of FGF4 signaling in trophoblast stem cells derived from androgenetic embryos. J. Reprod. Dev. 62, 51-58. 10.1262/jrd.2015-097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein, B. and Spradling, A. (2007). Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science 315, 988-992. 10.1126/science.1136606 [DOI] [PubMed] [Google Scholar]
- Oltmann, J., Heselmeyer-Haddad, K., Hernandez, L. S., Meyer, R., Torres, I., Hu, Y., Doberstein, N., Killian, J. K., Petersen, D., Zhu, Y. J., et al. (2018). Aneuploidy, TP53 mutation, and amplification of MYC correlate with increased intratumor heterogeneity and poor prognosis of breast cancer patients. Gene Chromosome Canc. 57, 165-175. 10.1002/gcc.22515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver, T. L. (2015). When bigger is better: the role of polyploidy in organogenesis. Trends Genet. 31, 307-315. 10.1016/j.tig.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouseph, M. M., Li, J., Chen, H.-Z., Pécot, T., Wenzel, P., Thompson, J. C., Comstock, G., Chokshi, V., Byrne, M., Forde, B., et al. (2012). Atypical E2F repressors and activators coordinate placental development. Dev. Cell 22, 849-862. 10.1016/j.devcel.2012.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Øvrebø, J. I., Campsteijn, C., Kourtesis, I., Hausen, H., Raasholm, M. and Thompson, E. M. (2015). Functional specialization of chordate CDK1 paralogs during oogenic meiosis. Cell Cycle 14, 880-893. 10.1080/15384101.2015.1006000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit, S. K., Westendorp, B., Nantasanti, S., van Liere, E., Tooten, P. C. J., Cornelissen, P. W. A., Toussaint, M. J. M., Lamers, W. H. and de Bruin, A. (2012). E2F8 is essential for polyploidization in mammalian cells. Nat. Cell Biol. 14, 1181-1191. 10.1038/ncb2585 [DOI] [PubMed] [Google Scholar]
- Parisi, T., Beck, A. R., Rougier, N., McNeil, T., Lucian, L., Werb, Z. and Amati, B. (2003). Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. EMBO J. 22, 4794-4803. 10.1093/emboj/cdg482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart, M. J., Poyurovsky, M. V., Kass, E. M., Urist, M., Verschuren, E., Summers, M. K., Jackson, P. K. and Prives, C. (2010). APC/C(Cdc20) targets E2F1 for degradation in prometaphase. Cell Cycle 9, 3956-3964. 10.4161/cc.9.19.13162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce, S. B., Yost, C., Britton, J. S., Loo, L. W., Flynn, E. M., Edgar, B. A. and Eisenman, R. N. (2004). dMyc is required for larval growth and endoreplication in Drosophila. Development 131, 2317-2327. 10.1242/dev.01108 [DOI] [PubMed] [Google Scholar]
- Poolman, R. A., Gilchrist, R. and Brooks, G. (1998). Cell cycle profiles and expressions of p21CIP1 AND P27KIP1 during myocyte development. Int. J. Cardiol. 67, 133-142. 10.1016/S0167-5273(98)00320-9 [DOI] [PubMed] [Google Scholar]
- Porrello, E. R., Mahmoud, A. I., Simpson, E., Hill, J. A., Richardson, J. A., Olson, E. N. and Sadek, H. A. (2011). Transient regenerative potential of the neonatal mouse heart. Science 331, 1078-1080. 10.1126/science.1200708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Q.-R., Zhao, X.-Y., Zuo, R.-J., Wang, T.-S., Gu, X.-W., Liu, J.-L. and Yang, Z.-M. (2015). Involvement of atypical transcription factor E2F8 in the polyploidization during mouse and human decidualization. Cell Cycle 14, 1842-1858. 10.1080/15384101.2015.1033593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb, C., Sun, S. G., Sun, G. P., Pan, Y. W. and Irvine, K. D. (2014). Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell 158, 143-156. 10.1016/j.cell.2014.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid, K., Lu, J., Zimmet, J. M. and Jones, M. R. (2002). Roads to polyploidy: the megakaryocyte example. J. Cell. Physiol. 190, 7-20. 10.1002/jcp.10035 [DOI] [PubMed] [Google Scholar]
- Reid, B. J., Barrett, M. T., Galipeau, P. C., Sanchez, C. A., Neshat, K., Cowan, D. S. and Levine, D. S. (1996). Barrett's esophagus: Ordering the events that lead to cancer. Eur. J. Cancer Prev. 5, 57-65. 10.1097/00008469-199612002-00009 [DOI] [PubMed] [Google Scholar]
- Remus, D. and Diffley, J. F. X. (2009). Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol. 21, 771-777. 10.1016/j.ceb.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Ren, F., Wang, B., Yue, T., Yun, E.-Y., Ip, Y. T. and Jiang, J. (2010). Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc. Natl. Acad. Sci. USA 107, 21064-21069. 10.1073/pnas.1012759107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, B. E., Beckett, K., Nowak, S. J. and Baylies, M. K. (2007). SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development 134, 4357-4367. 10.1242/dev.010678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera, A., Barbon, M., Noguchi, Y., Reuter, L. M., Schneider, S. and Speck, C. (2017). From structure to mechanism-understanding initiation of DNA replication. Gene Dev. 31, 1073-1088. 10.1101/gad.298232.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios, A. C., Fu, N. Y., Jamieson, P. R., Pal, B., Whitehead, L., Nicholas, K. R., Lindeman, G. J. and Visvader, J. E. (2016). Essential role for a novel population of binucleated mammary epithelial cells in lactation. Nat. Commun. 7, e11400.. 10.1038/ncomms11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gómez, M., Coutts, N., Price, A., Taylor, M. V. and Bate, M. (2000). Drosophila dumbfounded: a myoblast attractant essential for fusion. Cell 102, 189-198. 10.1016/S0092-8674(00)00024-6 [DOI] [PubMed] [Google Scholar]
- Rusch, H. P., Sachsenmaier, W., Behrens, K. and Gruter, V. (1966). Synchronization of mitosis by the fusion of the plasmodia of Physarum polycephalum. J. Cell Biol. 31, 204-209. 10.1083/jcb.31.1.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli, P. A. and Larkins, B. A. (2009). The contribution of cell cycle regulation to endosperm development. Sex. Plant Reprod. 22, 207-219. 10.1007/s00497-009-0105-4 [DOI] [PubMed] [Google Scholar]
- Santabarbara-Ruiz, P., Lopez-Santillan, M., Martinez-Rodriguez, I., Binagui-Casas, A., Perez, L., Milan, M., Corominas, M. and Serras, F. (2015). ROS-induced JNK and p38 signaling is required for unpaired cytokine activation during Drosophila regeneration. PLoS Genet. 11, e1005595. 10.1371/journal.pgen.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni-Rugiu, E., Falck, J., Mailand, N., Bartek, J. and Lukas, J. (2000). Involvement of Myc activity in a G(1)/S-promoting mechanism parallel to the pRb/E2F pathway. Mol. Cell. Biol. 20, 3497-3509. 10.1128/MCB.20.10.3497-3509.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo, L. J., Gao, X., Chiarelli, D. A., Li, L., Pan, D. and Edgar, B. A. (2003). Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5, 566-571. 10.1038/ncb996 [DOI] [PubMed] [Google Scholar]
- Sauer, K., Knoblich, J. A., Richardson, H. and Lehner, C. F. (1995). Distinct modes of cyclin E/cdc2c kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes Dev. 9, 1327-1339. 10.1101/gad.9.11.1327 [DOI] [PubMed] [Google Scholar]
- Schaeffer, V., Althauser, C., Shcherbata, H. R., Deng, W.-M. and Ruohola-Baker, H. (2004). Notch-dependent Fizzy-related/Hec1/Cdh1 expression is required for the mitotic-to-endocycle transition in Drosophila follicle cells. Curr. Biol. 14, 630-636. 10.1016/j.cub.2004.03.040 [DOI] [PubMed] [Google Scholar]
- Senyo, S. E., Steinhauser, M. L., Pizzimenti, C. L., Yang, V. K., Cai, L., Wang, M., Wu, T.-D., Guerquin-Kern, J.-L., Lechene, C. P. and Lee, R. T. (2013). Mammalian heart renewal by pre-existing cardiomyocytes. Nature 493, 433-436. 10.1038/nature11682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, R. L., Kohlmaier, A., Polesello, C., Veelken, C., Edgar, B. A. and Tapon, N. (2010). The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development 137, 4147-4158. 10.1242/dev.052506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbata, H. R., Althauser, C., Findley, S. D. and Ruohola-Baker, H. (2004). The mitotic-to-endocycle switch in Drosophila follicle cells is executed by Notch-dependent regulation of G1/S, G2/M and M/G1 cell-cycle transitions. Development 131, 3169-3181. 10.1242/dev.01172 [DOI] [PubMed] [Google Scholar]
- Sheffer, M., Bacolod, M. D., Zuk, O., Giardina, S. F., Pincas, H., Barany, F., Paty, P. B., Gerald, W. L., Notterman, D. A. and Domany, E. (2009). Association of survival and disease progression with chromosomal instability: A genomic exploration of colorectal cancer. Proc. Natl. Acad. Sci. USA 106, 7131-7136. 10.1073/pnas.0902232106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher, N., Von Stetina, J. R., Bell, G. W., Matsuura, S., Ravid, K. and Orr-Weaver, T. L. (2013). Fundamental differences in endoreplication in mammals and Drosophila revealed by analysis of endocycling and endomitotic cells. Proc. Natl. Acad. Sci. USA 110, 9368-9373. 10.1073/pnas.1304889110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani, S. T., de la Cruz, A. F. A., Tran, V., Turbyfill, W. J., III, Reis, T., Edgar, B. A. and Duronio, R. J. (2008). Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev. Cell 15, 890-900. 10.1016/j.devcel.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, Z. and Deng, W.-M. (2017). Differential Regulation of Cyclin E by Yorkie-Scalloped Signaling in Organ Development. G3 7, 1049-1060. 10.1534/g3.117.039065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist, S. J. and Lehner, C. F. (1997). Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell 90, 671-681. 10.1016/S0092-8674(00)80528-0 [DOI] [PubMed] [Google Scholar]
- Smith, A. V. and Orr-Weaver, T. L. (1991). The regulation of the cell cycle during Drosophila embryogenesis: the transition to polyteny. Development 112, 997-1008. [DOI] [PubMed] [Google Scholar]
- Soonpaa, M. H. and Field, L. J. (1997). Assessment of cardiomyocyte DNA synthesis in normal and injured adult mouse hearts. Am. J. Physiol. 272, H220-H226. 10.1152/ajpheart.1997.272.1.H220 [DOI] [PubMed] [Google Scholar]
- Soonpaa, M. H., Kim, K. K., Pajak, L., Franklin, M. and Field, L. J. (1996). Cardiomyocyte DNA synthesis and binucleation during murine development. Am. J. Physiol. 271, H2183-H2189. 10.1152/ajpheart.1996.271.5.H2183 [DOI] [PubMed] [Google Scholar]
- Srinivas, B. P., Woo, J., Leong, W. Y. and Roy, S. (2007). A conserved molecular pathway mediates myoblast fusion in insects and vertebrates. Nat. Genet. 39, 781-786. 10.1038/ng2055 [DOI] [PubMed] [Google Scholar]
- Srivastava, D. and DeWitt, N. (2016). In vivo cellular reprogramming: the next generation. Cell 166, 1386-1396. 10.1016/j.cell.2016.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley, B. K. and Irvine, K. D. (2010). Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr. Biol. 20, 1580-1587. 10.1016/j.cub.2010.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, B. and Nurse, P. (1996). A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet. 12, 345-350. 10.1016/S0168-9525(96)80016-3 [DOI] [PubMed] [Google Scholar]
- Storchova, Z. and Pellman, D. (2004). From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 5, 45-54. 10.1038/nrm1276 [DOI] [PubMed] [Google Scholar]
- Strunkelnberg, M., Bonengel, B., Moda, L. M., Hertenstein, A., de Couet, H. G., Ramos, R. G. and Fischbach, K. F. (2001). rst and its paralogue kirre act redundantly during embryonic muscle development in Drosophila. Development 128, 4229-4239. [DOI] [PubMed] [Google Scholar]
- Sugimoto, N., Tatsumi, Y., Tsurumi, T., Matsukage, A., Kiyono, T., Nishitani, H. and Fujita, M. (2004). Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J. Biol. Chem. 279, 19691-19697. 10.1074/jbc.M313175200 [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K. and Roberts, K. (2003). “Big it up”: endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6, 544-553. 10.1016/j.pbi.2003.09.009 [DOI] [PubMed] [Google Scholar]
- Sulston, J. E. and Horvitz, H. R. (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110-156. 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Sun, J. and Deng, W.-M. (2007). Hindsight mediates the role of notch in suppressing hedgehog signaling and cell proliferation. Dev. Cell 12, 431-442. 10.1016/j.devcel.2007.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, G. and Irvine, K. D. (2013). Ajuba family proteins link JNK to Hippo signaling. Sci. Signal. 6, ra81. 10.1126/scisignal.2004324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J., Smith, L., Armento, A. and Deng, W.-M. (2008). Regulation of the endocycle/gene amplification switch by Notch and ecdysone signaling. J. Cell Biol. 182, 885-896. 10.1083/jcb.200802084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, C. I., Meserve, J. H., McCarter, P. C., Thieme, A., Mathew, T., Elston, T. C. and Duronio, R. J. (2015). Expression of an S phase-stabilized version of the CDK inhibitor Dacapo can alter endoreplication. Development 142, 4288-4298. 10.1242/dev.115006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani, M., Koh, K. P., Shen, Y. H., Pastor, W. A., Bandukwala, H., Brudno, Y., Agarwal, S., Iyer, L. M., Liu, D. R., Aravind, L., et al. (2009). Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 324, 930-935. 10.1126/science.1170116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, S., Kunath, T., Hadjantonakis, A. K., Nagy, A. and Rossant, J. (1998). Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072-2075. 10.1126/science.282.5396.2072 [DOI] [PubMed] [Google Scholar]
- Tane, S., Ikenishi, A., Okayama, H., Iwamoto, N., Nakayama, K. I. and Takeuchi, T. (2014a). CDK inhibitors, p21(Cip1) and p27(Kip1), participate in cell cycle exit of mammalian cardiomyocytes. Biochem. Biophys. Res. Commun. 443, 1105-1109. 10.1016/j.bbrc.2013.12.109 [DOI] [PubMed] [Google Scholar]
- Tane, S., Kubota, M., Okayama, H., Ikenishi, A., Yoshitome, S., Iwamoto, N., Satoh, Y., Kusakabe, A., Ogawa, S., Kanai, A., et al. (2014b). Repression of cyclin D1 expression is necessary for the maintenance of cell cycle exit in adult mammalian cardiomyocytes. J. Biol. Chem. 289, 18033-18044. 10.1074/jbc.M113.541953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakala, M., Rodríguez-Acebes, S., Maroto, M., Symonds, C. E., Santamaría, D., Ortega, S., Barbacid, M., Méndez, J. and Malumbres, M. (2015). Functional reprogramming of polyploidization in megakaryocytes. Dev. Cell 32, 155-167. 10.1016/j.devcel.2014.12.015 [DOI] [PubMed] [Google Scholar]
- Tsai, C.-R., Anderson, A. E., Burra, S., Jo, J. and Galko, M. J. (2017). Yorkie regulates epidermal wound healing in Drosophila larvae independently of cell proliferation and apoptosis. Dev. Biol. 427, 61-71. 10.1016/j.ydbio.2017.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzahor, E. and Poss, K. D. (2017). Cardiac regeneration strategies: staying young at heart. Science 356, 1035-1039. 10.1126/science.aam5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan, R. S., Kango-Singh, M., Nolo, R., Tao, C. and Halder, G. (2003). Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5, 914-920. 10.1038/ncb1050 [DOI] [PubMed] [Google Scholar]
- Ullah, Z., Kohn, M. J., Yagi, R., Vassilev, L. T. and DePamphilis, M. L. (2008). Differentiation of trophoblast stem cells into giant cells is triggered by p57/Kip2 inhibition of CDK1 activity. Genes Dev. 22, 3024-3036. 10.1101/gad.1718108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah, Z., de Renty, C. and DePamphilis, M. L. (2011). Checkpoint kinase 1 prevents cell cycle exit linked to terminal cell differentiation. Mol. Cell. Biol. 31, 4129-4143. 10.1128/MCB.05723-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya, Y. and Orr-Weaver, T. L. (2012). Polyploidization of glia in neural development links tissue growth to blood-brain barrier integrity. Genes Dev. 26, 31-36. 10.1101/gad.177436.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Eijnde, S. M., van den Hoff, M. J., Reutelingsperger, C. P., van Heerde, W. L., Henfling, M. E., Vermeij-Keers, C., Schutte, B., Borgers, M. and Ramaekers, F. C. (2001). Transient expression of phosphatidylserine at cell-cell contact areas is required for myotube formation. J. Cell Sci. 114, 3631-3642. [DOI] [PubMed] [Google Scholar]
- van den Heuvel, S. and Dyson, N. J. (2008). Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 9, 713-724. 10.1038/nrm2469 [DOI] [PubMed] [Google Scholar]
- Varelas, X., Samavarchi-Tehrani, P., Narimatsu, M., Weiss, A., Cockburn, K., Larsen, B. G., Rossant, J. and Wrana, J. L. (2010). The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Dev. Cell 19, 831-844. 10.1016/j.devcel.2010.11.012 [DOI] [PubMed] [Google Scholar]
- Vargas-Rondon, N., Villegas, V. E. and Rondon-Lagos, M. (2018). The Role of Chromosomal Instability in Cancer and Therapeutic Responses. Cancers 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboon, J. M. and Parkhurst, S. M. (2015). Rho family GTPase functions in Drosophila epithelial wound repair. Small GTPases 6, 28-35. 10.4161/21541248.2014.982415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stetina, J. R., Frawley, L. E., Unhavaithaya, Y. and Orr-Weaver, T. L. (2018). Variant cell cycles regulated by Notch signaling control cell size and ensure a functional blood-brain barrier. Development 145, dev157115. 10.1242/dev.157115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, A., Houlston, R. and Tomlinson, I. (2008). Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut 57, 941-950. 10.1136/gut.2007.135004 [DOI] [PubMed] [Google Scholar]
- Wang, M.-J., Chen, F., Li, J.-X., Liu, C.-C., Zhang, H.-B., Xia, Y., Yu, B., You, P., Xiang, D., Lu, L., et al. (2014). Reversal of hepatocyte senescence after continuous in vivo cell proliferation. Hepatology 60, 349-361. 10.1002/hep.27094 [DOI] [PubMed] [Google Scholar]
- Wang, Y., Antunes, M., Anderson, A. E., Kadrmas, J. L., Jacinto, A. and Galko, M. J. (2015). Integrin adhesions suppress syncytium formation in the Drosophila larval epidermis. Curr. Biol. 25, 2215-2227. 10.1016/j.cub.2015.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M.-J., Chen, F., Lau, J. T. Y. and Hu, Y.-P. (2017). Hepatocyte polyploidization and its association with pathophysiological processes. Cell Death Dis. 8, e2805. 10.1038/cddis.2017.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, B. A. A. and Cleveland, D. W. (2006). Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 18, 658-667. 10.1016/j.ceb.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Weigmann, K., Cohen, S. M. and Lehner, C. F. (1997). Cell cycle progression, growth and patterning in imaginal discs despite inhibition of cell division after inactivation of Drosophila Cdc2 kinase. Development 124, 3555-3563. [DOI] [PubMed] [Google Scholar]
- Weiss, A., Herzig, A., Jacobs, H. and Lehner, C. F. (1998). Continuous Cyclin E expression inhibits progression through endoreduplication cycles in Drosophila. Curr. Biol. 8, 239-242. 10.1016/S0960-9822(98)70090-9 [DOI] [PubMed] [Google Scholar]
- Wenzel, P. L., Wu, L., de Bruin, A., Chong, J.-L., Chen, W.-Y., Dureska, G., Sites, E., Pan, T., Sharma, A., Huang, K., et al. (2007). Rb is critical in a mammalian tissue stem cell population. Genes Dev. 21, 85-97. 10.1101/gad.1485307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, M. J. D. (1937). The Chromosomes. New York: Chemical Pub. Co. [Google Scholar]
- White, M. J. D. (1973a). Animal Cytology and Evolution, 3rd edn. Cambridge, UK: University Press. [Google Scholar]
- White, M. J. D. (1973b). The Chromosomes, 6th edn. London, UK: Chapman and Hall. [Google Scholar]
- Wong, J. V., Dong, P., Nevins, J. R., Mathey-Prevot, B. and You, L. (2011). Network calisthenics: control of E2F dynamics in cell cycle entry. Cell Cycle 10, 3086-3094. 10.4161/cc.10.18.17350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, J., Bandura, J., Zhang, P., Jin, Y., Reuter, H. and Edgar, B. A. (2017). EGFR-dependent TOR-independent endocycles support Drosophila gut epithelial regeneration. Nat. Commun. 8, 15125. 10.1038/ncomms15125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, L., Xiu, Y. and Boyce, B. F. (2012). Osteoclast fusion and regulation by RANKL-dependent and independent factors. World J. Orthop. 3, 212-222. 10.5312/wjo.v3.i12.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, L. H., Gater, S. T. and Karrer, K. M. (2010). A developmentally regulated gene, ASI2, is required for endocycling in the macronuclear anlagen of tetrahymena. Eukaryot. Cell 9, 1343-1353. 10.1128/EC.00089-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Z. N., Riera, A., Bai, L., Sun, J. C., Nandi, S., Spanos, C., Chen, Z. A., Barbon, M., Rappsilber, J., Stillman, B., et al. (2017). Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1. Nat. Struct. Mol. Biol. 24, 316. 10.1038/nsmb.3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman, P. and Diffley, J. F. X. (2007). Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445, 281-285. 10.1038/nature05432 [DOI] [PubMed] [Google Scholar]
- Zhang, B., Mehrotra, S., Ng, W. L. and Calvi, B. R. (2014). Low levels of p53 protein and chromatin silencing of p53 target genes repress apoptosis in Drosophila endocycling cells. PLoS Genet. 10, e1004581. 10.1371/journal.pgen.1004581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Chen, Q., Liu, Q., Li, Y., Sun, X., Hong, L., Ji, S., Liu, C., Geng, J., Zhang, W., et al. (2017). Hippo signaling suppresses cell ploidy and tumorigenesis through Skp2. Cancer Cell 31, e669-e684.7. 10.1016/j.ccell.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Zhou, K., Luo, X., Li, L., Tu, H. C., Sehgal, A., Nguyen, L. H., Zhang, Y., Gopal, P., Tarlow, B. D., et al. (2018). The polyploid state plays a tumor-suppressive role in the liver. Dev. Cell 44, 447-459, e445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B., Li, L., Wang, L., Wang, C.-Y., Yu, J. and Guan, K.-L. (2012). Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 26, 54-68. 10.1101/gad.173435.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Edgar, B. A. and Boutros, M. (2017). ATF3 acts as a rheostat to control JNK signalling during intestinal regeneration. Nat. Commun. 8,14289. 10.1038/ncomms14289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurinsky, J., Leonhard, K., Watt, S., Marguerat, S., Bähler, J. and Nurse, P. (2010). A coordinated global control over cellular transcription. Curr. Biol. 20, 2010-2015. 10.1016/j.cub.2010.10.002 [DOI] [PubMed] [Google Scholar]
- Zielke, N., Querings, S., Rottig, C., Lehner, C. and Sprenger, F. (2008). The anaphase-promoting complex/cyclosome (APC/C) is required for rereplication control in endoreplication cycles. Genes Dev. 22, 1690-1703. 10.1101/gad.469108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke, N., Kim, K. J., Tran, V., Shibutani, S. T., Bravo, M.-J., Nagarajan, S., van Straaten, M., Woods, B., von Dassow, G., Rottig, C., et al. (2011). Control of Drosophila endocycles by E2F and CRL4(CDT2). Nature 480, 123-127. 10.1038/nature10579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke, N., Edgar, B. A. and DePamphilis, M. L. (2013). Endoreplication. Cold Spring Harb. Perspect. Biol. 5, a012948. 10.1101/cshperspect.a012948 [DOI] [PMC free article] [PubMed] [Google Scholar]