Abstract
Background
As a histone methyltransferase, suppressor of variegation 3–9 homolog 1 (SUV39H1) plays an important role in the occurrence and development of cancer. To explore the mechanism and biological function of SUV39H1 in hepatitis B virus-associated hepatocellular carcinoma (HBV-HCC) can gain an insight into the pathogenesis of HBV-HCC.
Methods
The effect of HBV infection on SUV39H1 in hepatoma cells was detected. CCK-8, colony growth assay and wound healing assay were used to assess the proliferation and migration of HBV-positive hepatoma cells. RNA sequencing (RNA-seq) was applied to find differential genes and enriched pathways. The serum SUV39H1 level in HBV-HCC patients was detected and its correlation with clinical indicators was analyzed.
Results
SUV39H1 was increased by HBV infection and promoted the proliferation and migration of hepatoma cells. SUV39H1 could upregulate the expression of mitochondrial oxidative phosphorylation (OXPHOS) pathway-related genes. OXPHOS pathway inhibitors could reduce the capacity of proliferation and migration of hepatoma cells after overexpressing SUV39H1. Serum SUV39H1 levels were higher in chronic hepatitis B (CHB) patients than in healthy controls and higher in HBV-HCC patients than in CHB patients. In the diagnosis of HCC, the predictive value of SUV39H1 combined with alpha-fetoprotein (AFP) was better than that of AFP alone.
Conclusion
SUV39H1 is regulated by HBV infection and promotes the proliferation and migration of hepatoma cells by targeting OXPHOS pathway. It indicates that SUV39H1 may be a new biomarker of the diagnosis of HCC.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-023-11633-4.
Keywords: SUV39H1, Hepatocellular carcinoma, Hepatitis B virus, Oxidative phosphorylation, Biomarker
Introduction
Primary liver cancer is one of the most common cancers worldwide and the third leading cause of cancer-related death [1]. Hepatocellular carcinoma (HCC) accounts for about 90% of the total cases of primary liver cancer [2]. Known major risk factors for HCC include hepatitis B virus (HBV) infection, hepatitis C virus (HCV) infection, excessive alcohol consumption, aflatoxin, obesity, diabetes mellitus, and non-alcoholic fatty liver disease, among which HBV infection plays an essential role [3, 4]. Therefore, it is particularly imperative to explore the pathogenesis of HBV-associated HCC (HBV-HCC).
Histone methylation is one of the most complex histones post-translational modifications. It can be methylated in lysine and arginine residues, playing an important role in the progression of HCC [5]. Suppressor of variegation 3–9 homolog 1 (SUV39H1) is a histone lysine methyltransferase responsible for the trimethylation of histone 3 lysine 9 (H3K9) and regulates heterochromatin formation to inhibit transcription [6, 7]. It has been reported that the role of SUV39H1 in cancer is a double-edged sword. SUV39H1 acts as a tumor suppressor in cervical cancer, parathyroid cancer and leukemia, but plays a cancer-promoting role in colorectal cancer and melanoma [8–12]. Recently, studies have shown that SUV39H1 is upregulated in HCC patient samples and is associated with HCC recurrence rate [13, 14]. It has been reported that AIFM2 can enhance mitochondrial biogenesis through activation of SIRT1/PGC-1α signaling to promote hepatocellular carcinoma metastasis [15]. Moreover, SIRT1 can interact directly with SUV39H1, and contribute to elevated levels of SUV39H1 activity [16]. However, the role and mechanism of SUV39H1 in the progression of liver cancer remains largely elusive.
Tumor initiation and progression require metabolic reprogramming [17]. However, previous studies mainly focused on the changes in tumor metabolism, such as glycolysis, while oxidative phosphorylation (OXPHOS) was rarely reported. OXPHOS is a fundamental mitochondrial metabolic process that produces adenosine triphosphate (ATP) by transporting electrons to a series of transmembrane protein complexes, called electron transport chains (ETC) in the inner mitochondrial membrane [18]. Recent studies have shown that mitochondrial energy pathways are reprogrammed and tumor cells metabolic signals are upregulated to promote and meet the metabolic needs of cancer growth and proliferation in highly aggressive tumors [19]. Therefore, OXPHOS is also being considered as an emerging target for cancer therapy. We found that SUV39H1 overexpression led to increased transcript levels of most genes in the oxidative phosphorylation pathway in HCC by transcriptome sequencing. In addition, increased intracellular ATP production was detected after SUV39H1 overexpression in HCC cells, implying altered cellular metabolism. Since the relationship between SUV39H1 and oxidative phosphorylation pathway has not been reported, we investigated the effect of SUV39H1 on the oxidative phosphorylation pathway of HCC to provide a new pathogenic mechanism for HCC.
In this study, we examined SUV39H1 expression in HBV infected hepatoma cells and determined the effect of SUV39H1 on proliferation and migration of hepatoma cells. Further, we explored the underlying molecular pathway mediated by SUV39H1 in HCC progression. In addition, we investigated serum SUV39H1 levels in healthy controls, CHB patients and HBV-HCC patients. And we analyzed the correlation of various liver function indicators with SUV39H1. Moreover, we compared the diagnostic potential of AFP and SUV39H1 in patients with HBV-HCC. In conclusion, our work demonstrated the function and molecular mechanism of SUV39H1 and gained an insight into the diagnostic potential of SUV39H1in HBV-HCC patients.
Materials and methods
Microarray data
The gene expression profiling dataset GSE121248 was obtained from the gene expression omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/), which based on Platforms GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array). Totally 70 tumor tissues from chronic hepatitis B (CHB) induced HCC and 37 adjacent normal tissues were obtained.
Survival analysis
Kaplan-Meier analysis was performed for disease free survival (DFS) in the Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia2.cancer-pku.cn/#analysis). And the low SUV39H1 group was based on 20% cutoff-low expression value of SUV39H1 in HCC patients, whereas the high SUV39H1 group based on 40% cutoff-high expression value.
Plasmids and small interference RNA (siRNA)
The HBV plasmid was a pGEM-4Z vector carrying 1.3 copies of the HBV genome. And the SUV39H1 overexpression plasmid was that SUV39H1 (NM_001282166.1) subcloned into pcDNA3.1 vector. They were obtained from MiaoLing Plasmid Platform (Wuhan, China). SUV39H1 siRNA (si-SUV39H1) and the negative control siRNA (si-NC) were designed and synthesized by Genepharm (Shanghai, China). The siRNA sequences were listed in Table S1.
Cell culture and transfection
HepG2 cells were derived from human hepatoblastoma and HepG2.215 cells were HepG2 cells that stabilized the whole gene sequence of HBV, which could stably produce infectious HBV particles. They were both purchased from iCell Bioscience Inc (Shanghai, China). Hep3B cells were human hepatocellular carcinoma cells, which contained an integrated hepatitis B virus genome and could synthesize hepatitis B surface antigen (HBsAg) [20]. All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, USA) containing 10% fetal bovine serum (FBS) (Gibco, USA) and 1% penicillin-streptomycin (Gibco, USA) and maintained in a 5% CO2 atmosphere at 37℃. Plasmids and a final concentration of 100 nM siRNA was transfected into the cells with Lipofectamine 2000 (Invitrogen, USA).
Lentiviruses containing LV-NC (negative control) and LV-oe-SUV39H1 were constructed using pLV[Exp]-EGFP:T2A:Puro-EF1A vector by VectorBuilder Inc (Guangzhou, China). HepG2 cells were exposed to recombinant lentivirus with 8 µg/mL polybrene. The successfully infected cells were selected with 2 µg/mL puromycin. Chaetocin was an inhibitor of the SUV39 family to target H3K9. The cells were treated with Chaetocin (Selleckchem, USA) 50 nM for 24 h. Rotenone and Oligomycin were inhibitors of OXPHOS pathway and cells were treated with Rotenone (MCE, USA) 0.5 µM for 24 h or Oligomycin (MCE, USA) 1 µM for 24 h.
Quantitative real-time PCR (qPCR)
Total RNA samples were extracted from treated cells using TRIzol (Invitrogen, USA). For each sample, 500 ng of total RNA was reverse transcribed into cDNA with Evo M-MLV RT Premix for qPCR (Accurate Biology, China), and amplified with SYBR Green premix Pro Taq HS qPCR Kit (Accurate Biology, China) using LightCycler 96 instrument (Roche, Switzerland). The PCR primers used were listed in Table S1 [21]. The relative expression was determined using the 2−ΔΔCT method and transcript levels were normalised to the levels of GAPDH mRNA expression.
Western blot (WB)
Total protein samples were separated on a 10% SDS-PAGE gel and transferred to PVDF membrane (Millipore, USA), the membranes were blocked with 5% skim milk. The blots were cut and then incubated with the primary antibodies of anti-SUV39H1 (Cell Signaling Technology, USA, #8729) and anti-GAPDH (Cell Signaling Technology, USA, #5174) respectively. Horseradish peroxidase (HRP)-conjugated anti-rabbit IgG antibody was used as a secondary antibody, and SuperSignal West Femto Maximum Sensitivity Substrate (Thermo, USA) was used for chemiluminescent detection. All blots including all replicates with clear membrane edges were provided in the Supplementary Information file.
Cell counting Kit-8 (CCK-8)
Hep3B cells were seeded into 96-well plates with 5,000 cells each well. The CCK-8 reagent (Bioscience, China) was added and incubated for 1–2 h. The OD values were measured at 450 nm using a microplate spectrophotometer (Thermo, USA) at 0 h, 24 h, 48 h, 72 h, 96 h, respectively.
Colony growth assay
Hep3B cells were seeded into 6-well plates with 1000 or 2500 cells each well and cultured in an incubator with medium changes every 3 days. When visible white granules of cells are observed, the colonies were fixed, stained, photographed and counted.
Wound healing assay
Hep3B cells were seeded into 6-well plates and transfected with the plasmids and siRNAs of SUV39H1 for 48 h. The cell monolayer was scraped by sterile 10 µL plastic pipette tips, and then cells were washed twice. 1% FBS culture medium was added at different time periods (0 h, 24 h, or 48 h). Images of wound closure were recorded by an inverted microscope and evaluated using ImageJ software (National Institutes of Health, USA).
RNA sequencing (RNA-seq)
Total RNA of sample was isolated by TRIzol. RNA-seq was performed on the Illumina NovaSeq 6000. Index of the reference genome was built using Hisat2 (v2.0.5) and paired-end reads were aligned to the reference genome using Hisat2 (v2.0.5). The R package “DESeq2” and “edgeR” were used to analyze the data, using a |log2FC| >1 and a P value < 0.05 as the cut-off values, respectively.
Enrichment analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Genome Ontology (GO) analysis was applied to systematically analyze enrichment genes [22–24]. The R packages named clusterProfiler, org.Hs.eg.db, GOplot, stringr and tinyarray were used to implement GO enrichment analysis and KEGG annotations. Using analysis tool (http://www.broadinstitute.org/gsea/index.jsp), Gene Set Enrichment Analysis (GSEA) revealed the overall representation of differential genes in a pathway. A P value < 0.05 was considered significantly different.
Hub genes analysis
Protein and protein interaction (PPI) networks were constructed into Seach Tool for the Retrieval of Interacting Genes (STRING) (https://cn.string-db.org). Then the PPI network was visualized in Cytoscape software. MCC algorithm of the Cyto-Hubba software was used to screen out hub genes.
Measurement of ATP
The cellular ATP levels were detected by the ATP Assay Kit (Beyotime, China). The cells transfected were lysed by cell lysis buffer and the supernatant was collected after centrifugation at 4℃, 12,000 g. The concentration of the protein was detected by BCA Assay Kit (Beyotime, China) and the assay solution was added and Luminance was measured by multifunctional microplate reader (Tecan Italia Srl, Italy), immediately.
Serum samples
This study was approved by the Ethics Committee of the Eighth Affiliated Hospital of Sun Yat-sen University and performed according to recommendations of the International Committee of Medical Journal Editors. Written informed consent was obtained from all patients involved. The serum samples of 96 participants were enrolled at the Eighth Affiliated Hospital of Sun Yat-sen University between July 2022 and December 2022, including 35 healthy controls, 34 CHB patients and 27 HBV-HCC patients. Healthy controls were normal liver function, without viral hepatitis infection and other liver-related or malignant disease. The CHB group consisted of patients with chronic HBV infection who have not developed HCC. HBV-HCC group was a histologically confirmed HCC patient diagnosed with CHB (Table S2).
ELISA
The serum SUV39H1 of participants was measured by the SUV39H1 ELISA kit (LMAI Bio, China). According to the manufacturer’s instructions, the absorption value was read at 450 nm. The SUV39H1 concentration was calculated with reference to standard curves.
Statistical analysis
SPSS software was used to carry out Spearman’s correlation coefficient. Student’s t test was used for two groups’ analyses and one-way ANOVA was for comparing more than two groups to evaluate the statistical significance. In addition, the chi-square test was used to analyze gender differences among three groups. And a receiver operating characteristic (ROC) curve was used to evaluate diagnostic values of SUV39H1 and alpha-fetoprotein (AFP). A P value < 0.05 was identified as a significant difference. Analyses were carried out using the GraphPad Prism 8.0.2 software (La Jolla, CA, USA). All cell experiments were repeated three times.
Results
SUV39H1 expression is upregulated in HBV-HCC
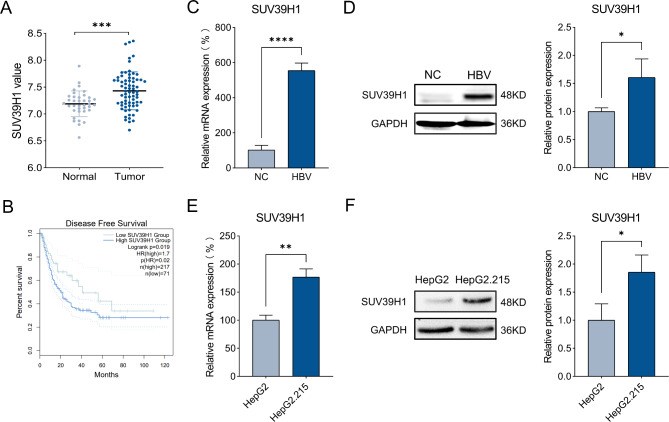
To determine whether HBV infection has a regulatory effect on SUV39H1, the GSE121248 dataset was selected for analysis. The expression of SUV39H1 was significantly increased in HBV-HCC tissues compared with adjacent normal tissues (Fig. 1A). Kaplan-Meier analysis showed that high SUV39H1 expression was correlated with a worse prognosis in HCC patients (Fig. 1B). HBV whole genome plasmid transfection in HepG2 cells significantly increased SUV39H1 mRNA and protein levels compared with the control group (NC) (Fig. 1C-D). Meanwhile, comparing the levels of SUV39H1 in HepG2 cells and HepG2.215 cells also proved that HBV could promote the expression of SUV39H1 (Fig. 1E-F).
Fig. 1.
HBV induced SUV39H1 expression in HCC tissues and cells. (A) Differential expression of SUV39H1 in HBV-HCC tissues (Tumor) and adjacent normal tissues (Normal). (B) The prognostic value of mRNA expression of SUV39H1 in HCC was analyzed by using Kaplan-Meier analysis. (C, D) Expression of SUV39H1 mRNA and protein after transfection with HBV plasmid and HBV control plasmid (NC) in HepG2 cells. (E, F) Relative level of SUV39H1 mRNA and protein in HepG2 cells and HepG2.215 cells. Data presented as mean ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 VS Normal, NC or HepG2 cells group
SUV39H1 enhances the proliferation and migration of hepatoma cells
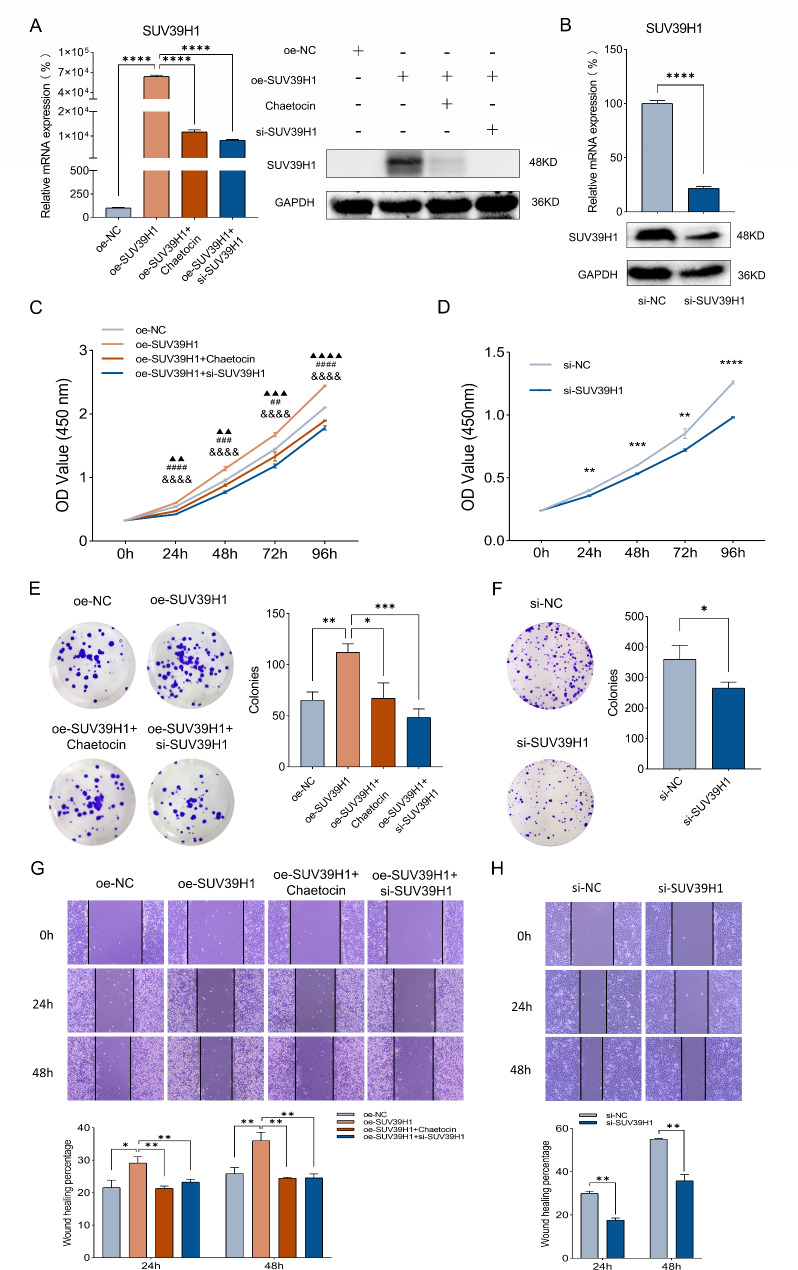
The function of SUV39H1 in HCC progression was next explored. We established SUV39H1 overexpression and knockdown models in Hep3B cells (Fig. 2A-B). CCK-8 assay and colony growth assay showed that overexpression of SUV39H1 significantly promoted the proliferation of Hep3B cells, while the inhibition of SUV39H1 by Chaetocin or si-SUV39H1 for cells transfected oe-SUV39H1 plasmid reduced the promotion effect (Fig. 2C, E). In contrast, SUV39H1 knockdown markedly inhibited the proliferation of Hep3B cells (Fig. 2D, F). Moreover, wound healing assay revealed that SUV39H1 overexpression significantly promoted the migration of Hep3B cells, while SUV39H1 knockdown inhibited the cell migration (Fig. 2G-H). These results suggest that SUV39H1 is critical for the proliferation and migration of HCC cells.
Fig. 2.
SUV39H1 regulates proliferation and migration of Hep3B cells. (A) Expression of SUV39H1 mRNA and protein after transfection with oe-NC, oe-SUV39H1, oe-SUV39H1 + Chaetocin or oe-SUV39H1 + si-SUV39H1 were examined by qPCR and WB. (B) The relative expression of SUV39H1 mRNA and protein after transfection with si-NC or si-SUV39H1. (C–F) CCK-8 assay and colony growth assay revealed the changes in proliferation ability after above transfection. (G, H) Wound healing assay was performed to examine the migration ability. ▲P value representing oe-SUV39H1 vs. oe-NC, #oe-SUV39H1 vs. oe-SUV39H1 + Chaetocin, &oe-SUV39H1 vs. oe-SUV39H1 + si-SUV39H1. Data presented as mean ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ▲▲P < 0.01, ▲▲▲P < 0.001, ▲▲▲▲P < 0.0001, ##P < 0.01, ###P < 0.001, ####P < 0.0001, &&&&P < 0.0001
OXPHOS is essential pathway regulated by SUV39H1
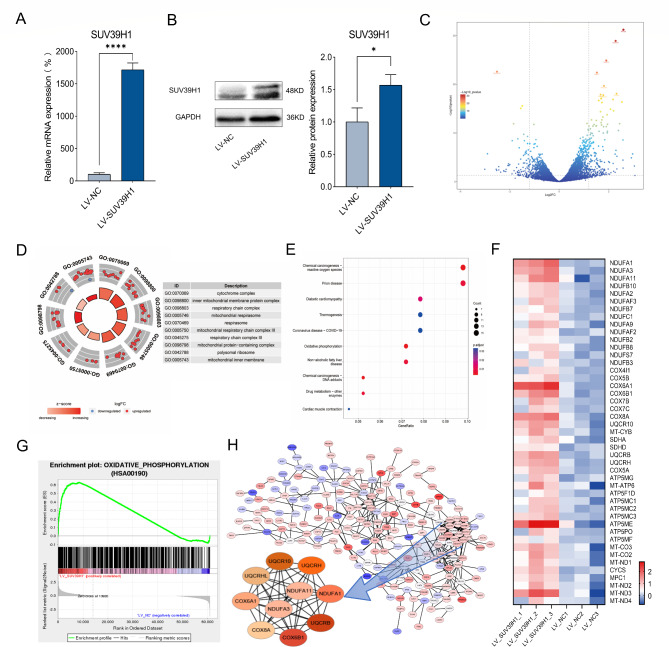
To investigate the molecular mechanism of SUV39H1 promoting proliferation and migration in HCC cells, HepG2 cell stably overexpressing SUV39H1 were established (Fig. 3A-B). The RNA-seq was applied to screen differentially expressed genes (DEGs) in HepG2-LV-SUV39H1 cells versus HepG2-LV-NC cells, including 444 upregulated genes and 218 downregulated genes (Fig. 3C). GO enrichment analysis showed that its function was mainly concentrated in respiratory chain (Fig. 3D). KEGG enrichment analysis also revealed significance in OXPHOS, which including respiratory chain (Fig. 3E). These data showed an important role for OXPHOS in HepG2-LV-SUV39H1 cells. The analysis of OXPHOS pathway genes indicated that 45 genes were upregulated (Fig. 3F). GSEA also confirmed a trend of elevated OXPHOS pathway genes (Fig. 3G). Further, we made PPI network maps of differential genes using Cytoscape software. The top 10 hub genes of DEGs were COX6A1, COX6B1, UQCRB, UQCR10, UQCRH, NDUFA1, NDUFA3, NDUFA11, UQCRHL and COX8A, all associated with genes involved in the mitochondrial respiratory chain (Fig. 3H). These results suggest that SUV39H1 may regulate OXPHOS pathway genes in the mitochondrial respiratory chain.
Fig. 3.
DEGs and enrichment pathway was performed in HepG2 cells stably overexpressing SUV39H1. (A, B) Construction of HepG2-LV-SUV39H1, qPCR and WB were used to detect the mRNA and protein of SUV39H1. (C) Volcano map showed the differentially expressed genes. Enrichment pathways were analyzed by GO (D) and KEGG (E) analysis. (F) Heatmap revealed that DEGs enriched in the OXPHOS pathway. (G) GSEA found a significant difference in OXPHOS pathway. (H) Hub genes of PPI network were screened by Cytoscape software
SUV39H1 promotes metabolic reprogramming in HCC
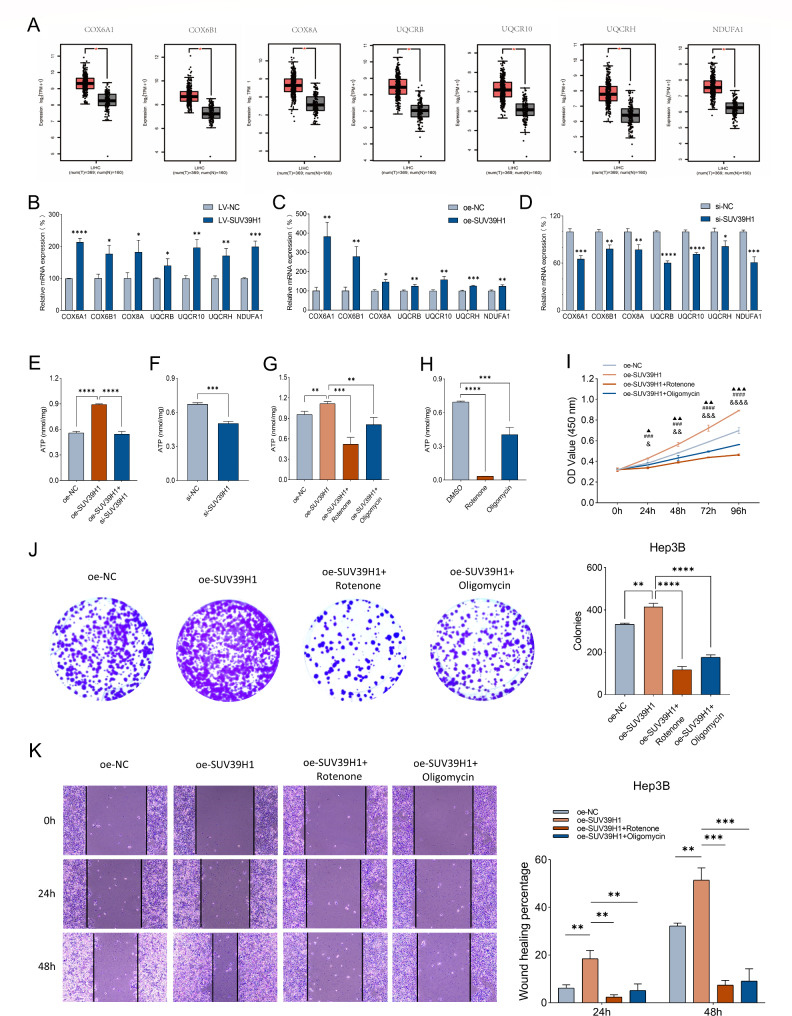
By searching the expression characteristics of hub genes, we found that COX6A1, COX6B1, COX8A, UQCRB, UQCR10, UQCRH and NDUFA1 were highly expressed in HCC tissues with statistical significance in the TCGA database (Fig. 4A). The mRNA expression of these seven genes in HepG2-LV-SUV39H1 group was proved to be higher than control one (Fig. 4B). Overexpression of SUV39H1 also caused their increase, while silence of SUV39H1 led to their decrease in HepG2 cells (Fig. 4C-D). To investigate the changes of mitochondrial respiratory function, intracellular ATP levels were detected. Compared with control group, ATP levels increased of overexpressing SUV39H1, while co-transfection with si-SUV39H1 restored the increased ATP level in HepG2 cells (Fig. 4E), suggesting that SUV39H1 promotes ATP production in vitro. And ATP level reduced by silencing SUV39H1 in HepG2 cells (Fig. 4F). Both Rotenone and Oligomycin, which were mitochondrial respiratory chain inhibitors, reduced the ATP concentration after overexpression of SUV39H1 in Hep3B cells, and the reduction effect of Rotenone was more obvious (Fig. 4G). In addition, we also examined ATP concentrations in Hep3B cells using Rotenone or Oligomycin alone and found that inhibitors of OXPHOS significantly inhibited ATP production compared to DMSO controls (Fig. 4H).
Fig. 4.
SUV39H1 regulates metabolic reprogramming. (A) Difference box plot of hub genes: COX6A1, COX6B1, COX8A, UQCRB, UQCR10, UQCRH, and NDUFA1. Red box means tumors and black box means normal controls. Relative mRNA expression of COX6A1, COX6B1, COX8A, UQCRB, UQCR10, UQCRH, and NDUFA1 in HepG2 cells: (B) LV-NC and LV-SUV39H1, (C) oe-NC and oe-SUV39H1, (D) si-NC and si-SUV39H1. Intracellular ATP detection of HepG2 cells transfected with oe-NC, oe-SUV39H1 or oe-SUV39H1 + si-SUV39H1 for 48 h (E), and transfected si-NC or si-SUV39H1(F). Intracellular ATP detection of Hep3B cells transfected with oe-NC, oe-SUV39H1, oe-SUV39H1 + Rotenone or oe-SUV39H1 + Oligomycin (G), and transfected with DMSO, Rotenone or Oligomycin (H). Hep3B cells transfected with oe-NC, oe-SUV39H1, oe-SUV39H1 + Rotenone or oe-SUV39H1 + Oligomycin: (I) CCK-8 to detect the proliferation ability; (J) Colony growth assay to detect clonal formation; (K) Wound healing assay to detect the migration ability. ▲P value representing oe-SUV39H1 vs. oe-NC, #oe-SUV39H1 vs. oe-SUV39H1 + Rotenone, &oe-SUV39H1 vs. oe-SUV39H1 + Oligomycin. Data presented as mean ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ▲P < 0.05, ▲▲P < 0.01, ▲▲▲P < 0.001, ###P < 0.001, ####P < 0.0001, &P < 0.05, &&P < 0.01, &&&P < 0.001, &&&&P < 0.0001
However, whether SUV39H1 has an effect on hepatoma cells through the OXPHOS pathway remains to be explored. The Hep3B cells transfected with SUV39H1 overexpressed plasmid were treated with Rotenone and Oligomycin, respectively, and CCK-8 and colony growth assays showed that their proliferative capacity was lower than those in the untreated overexpression group (Fig. 4I-J). And the wound healing assay also showed that the ability to migrate was lower than that in the untreated overexpression group (Fig. 4K). It suggested that SUV39H1 might promote metabolic reprogramming by regulating the OXPHOS pathway, thereby improving the proliferation and migration of hepatoma cells.
Diagnostic value of serum SUV39H1 for HBV-HCC
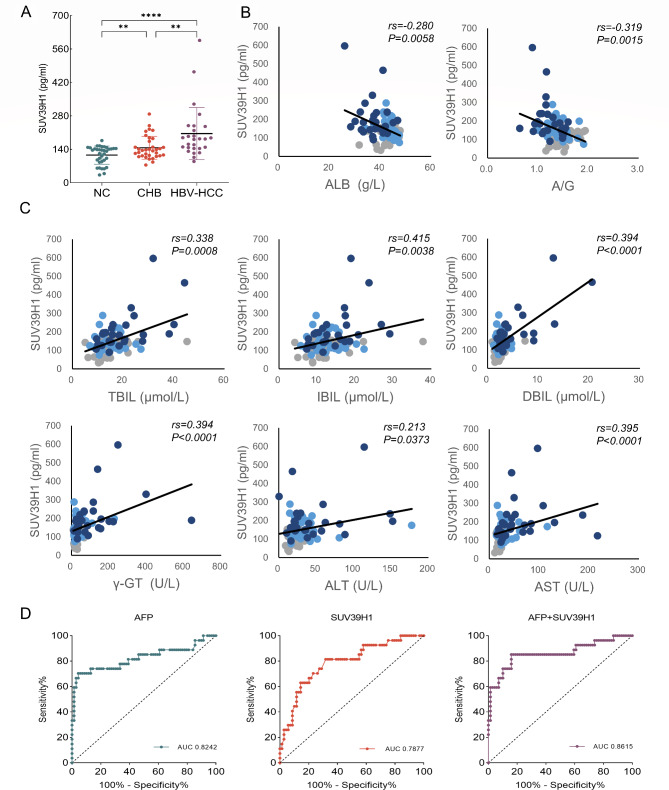
To further determine the association between SUV39H1 and HBV-HCC, we examined serum SUV39H1 levels in healthy controls, CHB patients, and HBV-HCC patients. The results showed that the level of SUV39H1 in CHB patients was higher than that in healthy controls, while SUV39H1 expression of HBV-HCC patients was higher than that of CHB patients and healthy controls (Fig. 5A). As shown in Table 1, there were significant decreases in TP, A/G and ALB levels of HBV-HCC group compared with CHB group, while A/G and ALB levels of HBV-HCC group obviously declined contrasted to healthy group. DBIL and γ-GT levels were significantly elevated in HBV-HCC group in contrast to healthy group and CHB group. Comparing with heathy group, ALT and AST levels were increased in CHB group and rose markedly in HBV-HCC group. But only AST displayed a significant increase between CHB group and HBV-HCC group.
Fig. 5.
Serum SUV39H1 expression in CHB and HBV-HCC patients and its relationship with clinical characteristics. (A) The level of serum SUV39H1 in healthy group (NC), CHB group and HBV-HCC group was determined by ELISA. (B, C) Correlation analysis of serum SUV39H1 expression level with biochemical indexes, n = 96. Gray indicates NC group, light blue indicates CHB group and dark blue indicates HBV-HCC group. (D) ROC curve showed the distinguishing capability of AFP, SUV39H1 or AFP + SUV39H1 for HBV-HCC patients. **P < 0.01, ****P < 0.0001
Table 1.
Comparison of characterization between the Healthy group, the CHB group and the HBV-HCC group
| Indexes | Healthy group (n = 35) |
CHB group(n = 34) | HBV-HCC group (n = 27) |
Healthy vs. CHB P value |
Healthy vs. HBV-HCC P value |
CHB vs. HBV-HCC P value |
|---|---|---|---|---|---|---|
| TP | 71.91 ± 0.90 | 73.92 ± 0.56 | 68.46 ± 1.31 | 0.1790 | 0.1017 | 0.0016 ** |
| A/G | 1.48 ± 0.04 | 1.43 ± 0.03 | 1.22 ± 0.05 | 0.7007 | 0.0000 **** | 0.0013 ** |
| ALB | 42.69 ± 0.57 | 43.35 ± 0.44 | 37.23 ± 0.96 | 0.7412 | 0.0000 **** | 0.0000 **** |
| TBIL | 15.36 ± 1.17 | 15.13 ± 0.79 | 20.07 ± 1.83 | 0.9977 | 0.1022 | 0.0524 |
| DBIL | 2.57 ± 0.19 | 2.61 ± 0.15 | 5.29 ± 0.84 | 0.9973 | 0.0111 * | 0.0119 * |
| IBIL | 12.79 ± 0.98 | 12.51 ± 0.66 | 14.78 ± 1.10 | 1.0000 | 0.4123 | 0.2779 |
| γ-GT | 28.78 ± 2.34 | 34.92 ± 2.84 | 121.87 ± 26.21 | 0.7847 | 0.0045 ** | 0.0095 ** |
| ALT | 20.23 ± 1.25 | 34.02 ± 4.99 | 45.61 ± 7.63 | 0.0323 * | 0.0084 ** | 0.5070 |
| AST | 20.99 ± 0.76 | 31.07 ± 3.24 | 60.33 ± 9,74 | 0.0133 * | 0.0013 ** | 0.0226 * |
Values are mean ± SD
TP, total protein; A/G, albumin to globulin; ALB, albumin; TBIL, total bilirubin; DBIL, direct bilirubin; IBIL, indirect bilirubin; γ-GT, γ-glutamyl transpeptidase; ALT, alanine aminotransferase; AST, aspartate transaminase. *P < 0.05, **P < 0.01, ****P < 0.0001
The serum SUV39H1 expression was negatively correlated with ALB, A/G and positively correlated with TBIL, DBIL, IBIL, γ-GT, ALT and AST, indicating that serum SUV39H1 levels had inherent predictive ability for liver injury (Fig. 5B-C). In addition, the SUV39H1 level was positively correlated with serum AFP (Fig. S1).
To validate the diagnostic potential of SUV39H1 and compare it with the commonly used biomarker AFP, we performed ROC curve analysis. As shown in Fig. 5D, the area under the curve (AUC) differentiating HBV-HCC patients from healthy controls and CHB patients was 0.8242 (95%CI: 0.7101–0.9383) for AFP and 0.7877 (95%CI: 0.6848–0.8906) for SUV39H1, indicating that both SUV39H1 and AFP have high diagnostic potential. When SUV39H1 was combined with AFP, the AUC was 0.8615 (95%CI: 0.7644–0.9586), indicating the predictive ability of the two combined diagnoses SUV39H1 and AFP was higher than either one alone. Therefore, SUV39H1 may be an assistant diagnostic marker for AFP.
Discussion
HBV infection is a major risk factor for HCC, and although large-scale vaccination has greatly reduced HBV infection in children, HBV infection still causes more than 40% of liver cancer [25, 26]. Many mechanisms are involved in this process, including viral DNA integration, viral genome or host gene mutation, epigenetic modification and changes in tumor signaling pathways. Under the synergistic action of these mechanisms, the evolution of HBV infection from inflammation to tumorigenesis is accelerated [27]. Among them, targeted epigenetic modification is considered a promising therapeutic approach. HBV infection can cause epigenetic remodeling through DNA methylation and histone modification, leading to malignant transformation of HCC [28].
SUV39H1 mainly catalyzes H3K9me3 which is generally considered as an epigenetic marker of gene inhibition [6, 7]. Abnormal expression of SUV39H1 has been found in many diseases. For instance, SUV39H1 is highly expressed in renal tubular cells of patients [29]. And the significantly increased expression of SUV39H1 in patients with idiopathic scoliosis can promote the proliferation chondrocytes and Alpinetin can improve colitis by decreasing the expression of SUV39H1 [30, 31]. Previous studies have suggested that SUV39H1 plays a role in tumor inhibition. However, SUV39H1 has also been reported as a tumor promoter in recent years [32]. The inhibition of SUV39H1 in renal clear cell carcinoma can induce iron accumulation and lipid peroxidation, leading to the ferroptosis of cancer cells [33]. It is reported that SUV39H1 enhances the migration ability of HCC cells, and SUV39H1 knockdown impairs the growth and spheroid formation of HCC cells [13]. Besides, SUV39H1 interacts with hepatitis B virus X protein (HBx) to enhance each other’s activity, leading to HBx-mediated hepatocarcinogenesis [14]. However, the carcinogenic mechanism of SUV39H1 in HBV-HCC has not been fully clarified. In this study, we also confirmed that HBV infection could increase the expression of SUV39H1 promoting the proliferation and migration of HBV positive hepatoma cells. We reported that SUV39H1 regulated the pathway genes of the mitochondrial respiratory chain by analyzing the data of LV-oe-SUV39H1 RNA-seq and experimental validation for the first time.
Warburg effect suggests that tumor cells still rely on glycolysis, for energy supply under aerobic conditions, instead of using efficient OXPHOS [34]. However, with the deepening of cancer research, the key role of mitochondrial OXPHOS in tumor progression is being confirmed by more studies [35]. Inhibition of mitochondrial biogenesis in resistant melanoma can induce mitochondrial dysfunction and inhibit tumor bioenergetics, thereby erating intrinsically resistant cells [36]. When the OXPHOS of cancer stem cells (CSCs) was upregulated, the malignant biological behavior of liver cancer stem cells was enhanced [37]. Knockdown of MALAT1 can induce a variety of abnormalities in mitochondrial function, including the reduction of OXPHOS and ATP production, and play a role in promoting HCC [38]. CR6-interacting factor 1 is a mitochondrial protein required for the assembly of OXPHOS complexes and plays an important role in HCC progression [39]. FH535 and Y3 are inhibitors of mitochondrial OXPHOS that disrupt transmembrane potential and electron transport chains, reduce ATP production, and lead to apoptotic hepatoma cells death [40, 41]. However, the role of SUV39H1 in the OXPHOS has not been reported. Through data analysis, we found that the differential genes regulated by SUV39H1 were mainly enriched in OXPHOS pathway.
Since the early diagnosis rate of hepatocellular carcinoma is only 40-50%, and the median survival of advanced hepatocellular carcinoma is only 15 months, the development of early diagnostic markers is the key to improving the prognosis of hepatocellular carcinoma [42]. AFP is currently the most commonly used serum biomarker and the only clinically validated biomarker for HCC, while the sensitivity of AFP for early screening is only 39-64% [43, 44]. Hepatitis B core-related antigen correlates with serum HBV DNA and intrahepatic cccDNA, predicting the occurrence or recurrence of HBV-HCC [45]. Mac-2 binding protein glycosylation isomer (M2BPGi) is a Mac-2 binding protein (M2BP) that is more effective than AFP in forecasting the development of HCC in patients with CHB [46]. Exosomes are intracellularly formed vesicles, encapsulating cell-derived nucleic acid fragments secreted into body fluids such as microRNAs. MicroRNA-21 was significantly higher in the serum of HCC patients than in CHB patients and healthy controls [47]. Serum exosomal heterogeneous nuclear ribonucleoprotein H1 (hnRNPH1) mRNA is a biomarker for HCC, and its diagnostic and predictive power is further increased when coupled with AFP [48]. In addition, blood-free DNA and circulating tumor DNA can also be used as biomarkers to predict the early recurrence of HBV-HCC after HCC resection [49]. Therefore, there is an urgent need for a new highly sensitive serum biomarker for early diagnostic of HBV-HCC. Our data showed that serum SUV39H1 levels in HBV-HCC patients were significantly higher than those in CHB patients and healthy controls. Serum SUV39H1 levels were found to be correlated with liver function indicators and AFP, and the predictive ability of SUV39H1 combined with AFP in the diagnosis of HBV-HCC patients was higher than that of AFP alone. Therefore, serum SUV39H1 may be a diagnostic biomarker for HBV-HCC.
There is no denying the fact that our research also has several limitations. One concern about our findings was the lack of in vivo experiments. Meanwhile, we did not further explore that how SUV39H1 regulates the oxidative phosphorylation pathway. Therefore, future research should be undertaken to explore the regulatory mechanism of SUV39H1 on oxidative phosphorylation pathway through Chromatin Immunoprecipitation and Mass Spectrometry and to make our results more credible by trials in animals.
Our study demonstrated for the first time that elevated SUV39H1 expression could regulate oxidative phosphorylation pathway and the serum level of SUV39H1 indicated the diagnosis of HBV-HCC. These findings provide new sights for the treatment of HBV-HCC. At present, the treatment of HCC is mainly surgery, but there are many newly diagnosed patients with unresectable advanced HCC, resulting in poor survival rate and prognosis. After further exploration, we may be able to use mitochondrial oxidative phosphorylation inhibitors in combination with chemotherapeutic agents to treat advanced HCC patients with high SUV39H1 levels.
Conclusion
In summary, we confirmed that SUV39H1 is essential in HBV-HCC progression and identified a mechanism by which SUV39H1 targets the OXPHOS pathway. Our data suggest that serum SUV39H1 level may be a biomarker for the diagnosis of HBV-HCC. We are further exploring the role of SUV39H1 in HBV-HCC through animal experiments and investigating other potential targets for SUV39H1 in future studies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge that Hep3B cell line was obtained from Dr. Shi Liang of the Eighth Affiliated Hospital of Sun Yat-sen University.
Abbreviations
- SUV39H1
Suppressor of variegation 3–9 homolog 1
- HBV-HCC
Hepatitis B virus-associated hepatocellular carcinoma
- RNA-seq
RNA sequencing
- OXPHOS
Oxidative phosphorylation
- CHB
Chronic hepatitis B
- AFP
Alpha-fetoprotein
- HCC
Hepatocellular carcinoma
- HBV
Hepatitis B virus
- HCV
Hepatitis C virus
- H3K9
Histone 3 lysine 9
- ATP
Adenosine triphosphate
- ETC
Electron transport chains
- DFS
Disease free survival
- GEPIA
Gene Expression Profiling Interactive Analysis
- siRNA
small interference RNA
- HBsAg
hepatitis B surface antigen
- DMEM
Dulbecco’s modified Eagle’s medium
- FBS
Fetal bovine serum
- qPCR
Quantitative real-time PCR
- WB
Western blot
- HRP
Horseradish peroxidase
- CCK-8
Cell Counting Kit-8
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PPI
Protein and protein interaction
- STRING
Seach Tool for the Retrieval of Interacting Genes
- ROC
Receiver operating characteristic
- HBsAb
Hepatitis B surface antibody
- HBeAg
Hepatitis B e antigen
- HBeAb
Hepatitis B e antibody
- HBcAb
Hepatitis B core antibody
- DEGs
Differentially expressed genes
- AUC
Area under the curve
- TP
Total protein
- A/G
Albumin to globulin
- ALB
Albumin
- TBIL
Total bilirubin
- DBIL
Direct bilirubin
- IBIL
Indirect bilirubin
- γ-GT
γ-glutamyl transpeptidase
- ALT
Alanine aminotransferase
- AST
Aspartate transaminase
- HBx
Hepatitis B virus X protein
- CSCs
Cancer stem cells
- M2BPGi
Mac-2 binding protein glycosylation isomer
- M2BP
Mac-2 binding protein
- hnRNPH1
heterogeneous nuclear ribonucleoprotein H1
Author contributions
The experiments were conceptualised and designed by Aixia Zhai, Chao Wu and Changlong Bi. Yanping Zhang performed the experiments and manuscript writing. Wanwen Lao and Kaming Yang contributed to data analysis and manuscript writing. Xinyi Kong, Yuetong Li, Xin Yu, Xumeng Wang, Yang Liu, Yilin Deng and Shuping Nie collected samples and clinical data. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China under Grant (82072267, 82272271, 82172215 and 81871562), GuangDong Basic and Applied Basic Research Foundation (2021A1515011396, 2021A1515012109 and 2019A1515110073), and Shenzhen Futian District Public Health Research Project under Grant (FTWS2020017, FTWS2020008).
Data Availability
The datasets generated or analyzed in this study are available in open access databases. In this study we used the following databases for analysis, data acquisition and visualization: GEO (https://www.ncbi.nlm.nih.gov/geo/), GEPIA (http://gepia2.cancer-pku.cn/#analysis). For the GEO database, we used the dataset which was coded as GSE121248 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE121248). And our data of RNA-sequencing was coded as GSE237514 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23751). All data are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
The study has been performed in accordance with the Declaration of Helsinki and was approved by the Research Ethics Board of the Eighth Affiliated Hospital of Sun Yat-sen University (Approval number: ZB-KYIRB-AF/SC-06/01.0), and written informed consent was obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yanping Zhang, Wanwen Lao and Kaming Yang contributed equally to this work.
Contributor Information
Changlong Bi, Email: BCL163@163.com, Email: bichlong@mail.sysu.edu.cn.
Chao Wu, Email: wuchao99261@163.com.
Aixia Zhai, Email: aixiazhai@126.com, Email: zhaiaix@mail.sysu.edu.cn.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Foerster F, Gairing SJ, Ilyas SI, Galle PR. Emerging immunotherapy for HCC: a guide for hepatologists. Hepatology. 2022;75(6):1604–26. doi: 10.1002/hep.32447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kulik L, El-Serag HB. Epidemiology and management of Hepatocellular Carcinoma. Gastroenterology. 2019;156(2):477–491e471. doi: 10.1053/j.gastro.2018.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–14. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 5.Fernández-Barrena MG, Arechederra M, Colyn L, Berasain C, Avila MA. Epigenetics in hepatocellular carcinoma development and therapy: the tip of the iceberg. JHEP Rep. 2020;2(6):100167. doi: 10.1016/j.jhepr.2020.100167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–9. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 7.Liu B, Wang Z, Zhang L, Ghosh S, Zheng H, Zhou Z. Depleting the methyltransferase Suv39h1 improves DNA repair and extends lifespan in a progeria mouse model. Nat Commun. 2013;4:1868. doi: 10.1038/ncomms2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodrigues C, Pattabiraman C, Vijaykumar A, Arora R, Narayana SM, Kumar RV, Notani D, Varga-Weisz P, Krishna S. A SUV39H1-low chromatin state characterises and promotes migratory properties of Cervical cancer cells. Exp Cell Res. 2019;378(2):206–16. doi: 10.1016/j.yexcr.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Yang YJ, Han JW, Youn HD, Cho EJ. The Tumor suppressor, parafibromin, mediates histone H3 K9 methylation for cyclin D1 repression. Nucleic Acids Res. 2010;38(2):382–90. doi: 10.1093/nar/gkp991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu Y, Chen Y, Guo H, Li M, Wang B, Shi D, Cheng X, Guan J, Wang X, Xue C, et al. SUV39H1 regulates the progression of MLL-AF9-induced acute Myeloid Leukemia. Oncogene. 2020;39(50):7239–52. doi: 10.1038/s41388-020-01495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu C, Klement JD, Yang D, Albers T, Lebedyeva IO, Waller JL, Liu K. SUV39H1 regulates human colon carcinoma apoptosis and cell cycle to promote Tumor growth. Cancer Lett. 2020;476:87–96. doi: 10.1016/j.canlet.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim G, Kim JY, Lim SC, Lee KY, Kim O, Choi HS. SUV39H1/DNMT3A-dependent methylation of the RB1 promoter stimulates PIN1 expression and Melanoma development. Faseb j. 2018;32(10):5647–60. doi: 10.1096/fj.201700645RRRRR. [DOI] [PubMed] [Google Scholar]
- 13.Chiba T, Saito T, Yuki K, Zen Y, Koide S, Kanogawa N, Motoyama T, Ogasawara S, Suzuki E, Ooka Y, et al. Histone lysine methyltransferase SUV39H1 is a potent target for epigenetic therapy of hepatocellular carcinoma. Int J Cancer. 2015;136(2):289–98. doi: 10.1002/ijc.28985. [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi Y, Tsuge M, Tsushima K, Suehiro Y, Fujino H, Ono A, Yamauchi M, Makokha GN, Nakahara T, Murakami E, et al. Signal activation of Hepatitis B Virus-Related Hepatocarcinogenesis by Up-regulation of SUV39h1. J Infect Dis. 2020;222(12):2061–70. doi: 10.1093/infdis/jiaa317. [DOI] [PubMed] [Google Scholar]
- 15.Guo S, Li F, Liang Y, Zheng Y, Mo Y, Zhao D, Jiang Z, Cui M, Qi L, Chen J, et al. AIFM2 promotes hepatocellular carcinoma Metastasis by enhancing mitochondrial biogenesis through activation of SIRT1/PGC-1α signaling. Oncogenesis. 2023;12(1):46. doi: 10.1038/s41389-023-00491-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450(7168):440–4. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 17.Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer. 2021;21(10):669–80. doi: 10.1038/s41568-021-00378-6. [DOI] [PubMed] [Google Scholar]
- 18.Schöpf B, Weissensteiner H, Schäfer G, Fazzini F, Charoentong P, Naschberger A, Rupp B, Fendt L, Bukur V, Giese I, et al. OXPHOS remodeling in high-grade Prostate cancer involves mtDNA mutations and increased succinate oxidation. Nat Commun. 2020;11(1):1487. doi: 10.1038/s41467-020-15237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roth KG, Mambetsariev I, Kulkarni P, Salgia R. The Mitochondrion as an emerging therapeutic target in Cancer. Trends Mol Med. 2020;26(1):119–34. doi: 10.1016/j.molmed.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and Hepatitis B surface antigen. Science. 1980;209(4455):497–9. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Chen Y, Cui L, Yang K, Wang X, Lei L, Zhang Y, Kong X, Lao W, Li Z, et al. CXCL8, CXCL9, CXCL10, and CXCL11 as biomarkers of liver injury caused by chronic Hepatitis B. Front Microbiol. 2022;13:1052917. doi: 10.3389/fmicb.2022.1052917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–51. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51(D1):D587–d592. doi: 10.1093/nar/gkac963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui F, Shen L, Li L, Wang H, Wang F, Bi S, Liu J, Zhang G, Wang F, Zheng H, et al. Prevention of Chronic Hepatitis B after 3 decades of escalating Vaccination Policy, China. Emerg Infect Dis. 2017;23(5):765–72. doi: 10.3201/eid2305.161477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z, Jiang Y, Yuan H, Fang Q, Cai N, Suo C, Jin L, Zhang T, Chen X. The trends in incidence of primary Liver cancer caused by specific etiologies: results from the global burden of Disease Study 2016 and implications for Liver cancer prevention. J Hepatol. 2019;70(4):674–83. doi: 10.1016/j.jhep.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Jiang Y, Han Q, Zhao H, Zhang J. The mechanisms of HBV-Induced Hepatocellular Carcinoma. J Hepatocell Carcinoma. 2021;8:435–50. doi: 10.2147/JHC.S307962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian Y, Ou JH. Genetic and epigenetic alterations in Hepatitis B virus-associated hepatocellular carcinoma. Virol Sin. 2015;30(2):85–91. doi: 10.1007/s12250-015-3582-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Yan W, Peng X, Jiang Y, He L, Peng Y, Chen X, Ye M, Zhuo H. Functional role of SUV39H1 in human renal tubular epithelial cells under high-glucose ambiance. Inflammation. 2018;41(1):1–10. doi: 10.1007/s10753-017-0657-7. [DOI] [PubMed] [Google Scholar]
- 30.Li J, Yang G, Liu S, Wang L, Liang Z, Zhang H. Suv39h1 promotes facet joint chondrocyte proliferation by targeting miR-15a/Bcl2 in idiopathic scoliosis patients. Clin Epigenetics. 2019;11(1):107. doi: 10.1186/s13148-019-0706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao Y, Lv Q, Qiao S, Yang L, Tao Y, Yan W, Wang P, Cao N, Dai Y, Wei Z. Alpinetin improves intestinal barrier homeostasis via regulating AhR/suv39h1/TSC2/mTORC1/autophagy pathway. Toxicol Appl Pharmacol. 2019;384:114772. doi: 10.1016/j.taap.2019.114772. [DOI] [PubMed] [Google Scholar]
- 32.Saha N, Muntean AG. Insight into the multi-faceted role of the SUV family of H3K9 methyltransferases in carcinogenesis and cancer progression. Biochim Biophys Acta Rev Cancer. 2021;1875(1):188498. doi: 10.1016/j.bbcan.2020.188498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Yin X, He W, Xue W, Zhang J, Huang Y. SUV39H1 deficiency suppresses clear cell renal cell carcinoma growth by inducing ferroptosis. Acta Pharm Sin B. 2021;11(2):406–19. doi: 10.1016/j.apsb.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–37. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 35.Burke PJ. Mitochondria, Bioenergetics and apoptosis in Cancer. Trends Cancer. 2017;3(12):857–70. doi: 10.1016/j.trecan.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang G, Frederick DT, Wu L, Wei Z, Krepler C, Srinivasan S, Chae YC, Xu X, Choi H, Dimwamwa E, et al. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J Clin Invest. 2016;126(5):1834–56. doi: 10.1172/JCI82661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jagust P, de Luxán-Delgado B, Parejo-Alonso B, Sancho P. Metabolism-based therapeutic strategies targeting Cancer Stem cells. Front Pharmacol. 2019;10:203. doi: 10.3389/fphar.2019.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y, Zhou L, Li H, Sun T, Wen X, Li X, Meng Y, Li Y, Liu M, Liu S, et al. Nuclear-encoded lncRNA MALAT1 epigenetically controls metabolic reprogramming in HCC cells through the Mitophagy Pathway. Mol Ther Nucleic Acids. 2021;23:264–76. doi: 10.1016/j.omtn.2020.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang H, Li J, Qu K, Wan Y, Liu S, Zheng W, Zhang Z, Liu C. CRIF1 overexpression facilitates Tumor growth and Metastasis through inducing ROS/NFκB pathway in hepatocellular carcinoma. Cell Death Dis. 2020;11(5):332. doi: 10.1038/s41419-020-2528-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turcios L, Vilchez V, Acosta LF, Poyil P, Butterfield DA, Mitov M, Marti F, Gedaly R. Sorafenib and FH535 in combination act synergistically on hepatocellular carcinoma by targeting cell bioenergetics and mitochondrial function. Dig Liver Dis. 2017;49(6):697–704. doi: 10.1016/j.dld.2017.01.146. [DOI] [PubMed] [Google Scholar]
- 41.Turcios L, Marti F, Watt DS, Kril LM, Khurana A, Chapelin F, Liu C, Zwischenberger JB, Evers BM, Gedaly R. Mitochondrial uncoupling and the disruption of the metabolic network in hepatocellular carcinoma. Oncotarget. 2020;11(31):3013–24. doi: 10.18632/oncotarget.27680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsuchiya N, Sawada Y, Endo I, Saito K, Uemura Y, Nakatsura T. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2015;21(37):10573–83. doi: 10.3748/wjg.v21.i37.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trevisani F, D’Intino PE, Morselli-Labate AM, Mazzella G, Accogli E, Caraceni P, Domenicali M, De Notariis S, Roda E, Bernardi M. Serum alpha-fetoprotein for diagnosis of hepatocellular carcinoma in patients with chronic Liver Disease: influence of HBsAg and anti-HCV status. J Hepatol. 2001;34(4):570–5. doi: 10.1016/S0168-8278(00)00053-2. [DOI] [PubMed] [Google Scholar]
- 44.Lok AS, Sterling RK, Everhart JE, Wright EC, Hoefs JC, Di Bisceglie AM, Morgan TR, Kim HY, Lee WM, Bonkovsky HL, et al. Des-gamma-carboxy prothrombin and alpha-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology. 2010;138(2):493–502. doi: 10.1053/j.gastro.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baudi I, Inoue T, Tanaka Y. Novel biomarkers of Hepatitis B and Hepatocellular Carcinoma: clinical significance of HBcrAg and M2BPGi. Int J Mol Sci. 2020;21(3):949. doi: 10.3390/ijms21030949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jun T, Hsu YC, Ogawa S, Huang YT, Yeh ML, Tseng CH, Huang CF, Tai CM, Dai CY, Huang JF, et al. Mac-2 binding protein glycosylation isomer as a Hepatocellular Carcinoma Marker in patients with chronic Hepatitis B or C Infection. Hepatol Commun. 2019;3(4):493–503. doi: 10.1002/hep4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res Int. 2014;2014:864894. doi: 10.1155/2014/864894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu H, Dong X, Chen Y, Wang X. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin Chem Lab Med. 2018;56(3):479–84. doi: 10.1515/cclm-2017-0327. [DOI] [PubMed] [Google Scholar]
- 49.Wang D, Hu X, Long G, Xiao L, Wang ZM, Zhou LD. The clinical value of total plasma cell-free DNA in Hepatitis B virus-related hepatocellular carcinoma. Ann Transl Med. 2019;7(22):650. doi: 10.21037/atm.2019.10.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated or analyzed in this study are available in open access databases. In this study we used the following databases for analysis, data acquisition and visualization: GEO (https://www.ncbi.nlm.nih.gov/geo/), GEPIA (http://gepia2.cancer-pku.cn/#analysis). For the GEO database, we used the dataset which was coded as GSE121248 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE121248). And our data of RNA-sequencing was coded as GSE237514 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23751). All data are available from the corresponding author upon reasonable request.