Abstract
In an era of increasing need for precision medicine, machine learning has shown promise in making accurate acute myocardial infarction outcome predictions. The accurate assessment of high-risk patients is a crucial component of clinical practice. Type 2 diabetes mellitus (T2DM) complicates ST-segment elevation myocardial infarction (STEMI), and currently, there is no practical method for predicting or monitoring patient prognosis. The objective of the study was to compare the ability of machine learning models to predict in-hospital mortality among STEMI patients with T2DM. We compared six machine learning models, including random forest (RF), CatBoost classifier (CatBoost), naive Bayes (NB), extreme gradient boosting (XGBoost), gradient boosting classifier (GBC), and logistic regression (LR), with the Global Registry of Acute Coronary Events (GRACE) risk score. From January 2016 to January 2020, we enrolled patients aged > 18 years with STEMI and T2DM at the Affiliated Hospital of Zunyi Medical University. Overall, 438 patients were enrolled in the study [median age, 62 years; male, 312 (71%); death, 42 (9.5%]). All patients underwent emergency percutaneous coronary intervention (PCI), and 306 patients with STEMI who underwent PCI were enrolled as the training cohort. Six machine learning algorithms were used to establish the best-fit risk model. An additional 132 patients were recruited as a test cohort to validate the model. The ability of the GRACE score and six algorithm models to predict in-hospital mortality was evaluated. Seven models, including the GRACE risk model, showed an area under the curve (AUC) between 0.73 and 0.91. Among all models, with an accuracy of 0.93, AUC of 0.92, precision of 0.79, and F1 value of 0.57, the CatBoost model demonstrated the best predictive performance. A machine learning algorithm, such as the CatBoost model, may prove clinically beneficial and assist clinicians in tailoring precise management of STEMI patients and predicting in-hospital mortality complicated by T2DM.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12872-023-03626-9.
Keywords: ST-segment elevation myocardial infarction, Diabetes mellitus, Machine learning, Mortality
Introduction
ST-segment elevation myocardial infarction (STEMI) is a severe type of acute myocardial infarction (AMI) with a poor prognosis and an association with high morbidity and mortality [1–3]. There are multiple risk factors for STEMI, including tobacco use, dyslipidemia, hypertension, diabetes mellitus (DM), and familial history of coronary artery disease (CAD) [4–6]. Particularly, STEMI patients with type 2 diabetes mellitus (T2DM) face an increased risk of cardiovascular complications, with a myocardial infarction rate 2–4 times higher than that in non-diabetic patients [7]. Although recent studies have shown that proactive management of T2DM significantly reduces cardiovascular complications and mortality, the overall prognosis for STEMI patients with T2DM remains poor [8–11].
Traditional models, such as the GRACE score based on clinical data and the TIMI score based on coronary angiography information, have been used to predict patient outcomes [12, 13]. However, the performance of these scores in prognosis prediction has certain limitations, and there is a risk of delayed scoring. Machine Learning (ML), in contrast, demonstrates superior predictive power in identifying interaction patterns between variables, especially in predicting in-hospital mortality and short-term outcomes for acute myocardial infarction patients, compared to conventional statistical methods [14, 15].
Currently, there is no ML model capable of detecting in-hospital mortality for STEMI patients with T2DM. Therefore, this study aims to develop an accurate and effective ML model to predict outcomes for STEMI patients with T2DM, offering better treatment options and reducing perioperative complications in these patients.
Methods
Study population
Between January 2016 and June 2020, patients from the affiliated hospital of Zunyi Medical University were recruited [5]. All patients met the diagnostic criteria for STEMI and underwent primary PCI according to the current guidelines [16]. Throughout this study, all procedures involving human participants were in accordance with the Declaration of Helsinki. The present study was approved by the Ethical Evaluation Committee of Zunyi Medical Hospital (ZMU〔2022〕1-177).
Definitions and data collection
Data about demographics, clinical outcomes, and procedural characteristics were collected using a standardized form. Baseline data on patient characteristics were collected from both medical records and standardized in-person interviews during the index admission for AMI. In this study, T2DM was defined as a chart diagnosis of diabetes or the use of glucose-lowering medication at AMI presentation [17]. Delay was defined as the upper limit of time from onset to the hospital first medical contact > 12 h. The cohort studies were reported in accordance with Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD). The electronic health records were used to collect and analyze the complete case data. In our study, the assessment of predictive factors was performed without the knowledge of the participants’ outcomes. An independent observer recorded in-hospital deaths.
ML algorithm methods
We developed six ML algorithms to model our data: random forest (RF), CatBoost classifier (CatBoost), Naive Bayes (NB), Logistic Regression (LR), gradient boosting classifier (GBC), and extreme gradient boosting (XGBoost). Using a randomization process, we split our dataset into two groups: a training set (70%) to develop the ML models and a validation set (30%) to examine model performance. Using 10-fold cross-validation to ensure the robustness of validation set results.
Missing data
Complete case data were collected from the electronic health records (EHRs) and analyzed. To simplify the review and ensure accuracy, variables with more than 20% of observations missing were also removed.
Statistical analysis
Continuous variables are presented as median (IQR), and categorical variables are presented as n (%). During training, the ML-based models were tuned to avoid overfitting, and the models were internally validated using all data via 10-fold cross-validation. The following indicators were used to define model performance: area under the curve (AUC), recall, precision, and F1 value. In the ROC analysis of the entire dataset, a 95% confidence interval was used to assess statistical significance and compare models. All statistical analyses were conducted using Python (version 3.7) and R (version 4.0.2).
Results
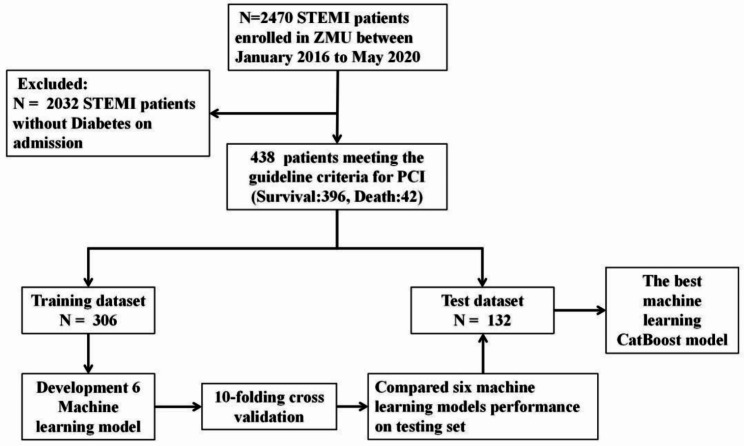
A total of 438 patients with STEMI registered in the database between January 2016 and January 2020 were included (Fig. 1). The median patient age was 62 (52–71) years, 71% were male, and 42(9.5%)patients died in the hospital. All patients underwent emergency PCI. A comparison of the demographic data and baseline characteristics between patients is shown in Table 1. Six ML models (LR, RF, CatBoost, XGBoost, GBC, and NB) were developed to predict in-hospital mortality rates based on all available features. A comparison of the predictive performances of the six ML algorithm models in the validation set is presented in Table 2. CatBoost (AUC = 0.92 [95%CI:0.909–0.922]),XGBoost(AUC = 0.88[95%CI:0.875–0.891]),RF(AUC = 0.89[95%CI:0.883–0.904]),GBC(AUC = 0.91[95%CI:0.903–0.916]),NB(AUC = 0.87[95%CI:0.859–0.878]),and LR(AUC = 0.84[95%CI:0.833–0.855]) provided similar accuracy values in our study. The GRACE risk assessment tool (AUC = 0.83[95%CI:0.789–0.862]) also demonstrated good discriminatory ability. We investigated the possibility of ensembling and combining different models to further optimize the models, including bagging, boosting, and stacker methods, and found that the performance of the ML models did not improve significantly.
Fig. 1.
Flow diagram outlining the study process
Table 1.
Demographics and clinical characteristics of patients with and without mortality in the cohort
| Variables | Total (n = 438) | Death (n = 42) | Survival (n = 396) | P | statistic |
|---|---|---|---|---|---|
| Demographic characteristics | |||||
| Sex, n (%) | 0.021 | 5.293 | |||
| female | 126 (29) | 19 (45) | 107 (27) | ||
| male | 312 (71) | 23 (55) | 289 (73) | ||
| Age, Median (Q1,Q3) | 62 (52, 71) | 71 (62.2, 76.8) | 60 (52, 70) | < 0.001 | 11,548 |
| Weekend on admission, n (%) | 159 (36) | 12 (29) | 147 (37) | 0.354 | 0.859 |
| Vascular risk factors | |||||
| Hypertension, n (%) | 273 (62) | 24 (57) | 249 (63) | 0.574 | 0.316 |
| Smoke, n (%) | 249 (57) | 18 (43) | 231 (58) | 0.078 | 3.103 |
| Stroke, n (%) | 28 (6) | 4 (10) | 24 (6) | 0.33 | Fisher |
| CKD, n (%) | 58 (13) | 12 (29) | 46 (12) | 0.004 | 8.083 |
| Delay, n (%) | 126 (29) | 11 (26) | 115 (29) | 0.835 | 0.044 |
| Electrocardiographic diagnosis | |||||
| Inferior wall STEMI, n (%) | 191 (44) | 12 (29) | 179 (45) | 0.057 | 3.621 |
| Anterior wall STEMI, n (%) | 230 (53) | 26 (62) | 204 (52) | 0.263 | 1.253 |
| Other STEMI, n (%) | 12 (3) | 1 (2) | 11 (3) | 1 | Fisher |
| Right ventricular STEMI, n (%) | 7 (2) | 1 (2) | 6 (2) | 0.509 | Fisher |
| Clinical data | |||||
| Heart rate, Median (Q1,Q3) | 81.5 (73, 95) | 90.5 (75, 111) | 81 (73, 93) | 0.018 | 10166.5 |
| SBP, Median (Q1,Q3) | 130 (113, 142) | 128 (105.2, 139.8) | 130 (114, 142) | 0.24 | 7399 |
| DBP, Mean ± SD | 81 ± 16.3 | 75.9 ± 19.2 | 81.5 ± 15.9 | 0.073 | -1.831 |
| Shock index, Median (Q1,Q3) | 0.6 (0.6, 0.7) | 0.7 (0.6, 0.8) | 0.6 (0.5, 0.7) | 0.037 | 9940 |
| Killip, n (%) | < 0.001 | Fisher | |||
| 1 | 349 (80) | 16 (38) | 333 (84) | ||
| 2 | 47 (11) | 5 (12) | 42 (11) | ||
| 3 | 19 (4) | 6 (14) | 13 (3) | ||
| 4 | 23 (5) | 15 (36) | 8 (2) | ||
| OHCA, n (%) | 22 (5) | 17 (40) | 5 (1) | < 0.001 | Fisher |
| Laboratory examinations on admission | |||||
| WBC, Median (Q1,Q3) | 10.4 (8.5, 13.4) | 12.3 (9.5, 18.7) | 10.4 (8.3, 13.1) | 0.001 | 10,789 |
| Neutrophil count, Median (Q1,Q3) | 8.2 (6, 10.9) | 10.1 (8.1, 15.3) | 8 (5.9, 10.6) | < 0.001 | 11360.5 |
| NLR, Median (Q1,Q3) | 5.9 (3.3, 9.5) | 8.6 (5.8, 13.1) | 5.8 (3.1, 9.3) | < 0.001 | 11,351 |
| PLR, Median (Q1,Q3) | 143.6 (95.1, 204.3) | 181.2 (95.5, 276) | 142.1 (95.6, 195.6) | 0.013 | 10,205 |
| MLR, Median (Q1,Q3) | 0.5 (0.3, 0.7) | 0.6 (0.4, 0.9) | 0.5 (0.3, 0.7) | 0.022 | 10058.5 |
| SIRI, Median (Q1,Q3) | 3.8 (2.2, 6.7) | 5.6 (3.6, 9.9) | 3.6 (2.2, 6.4) | < 0.001 | 11100.5 |
| SII, Median (Q1,Q3) | 1136.5 (635.1, 1968.1) | 1823.1 (1243.6, 3138.4) | 1078.1 (629, 1879.5) | < 0.001 | 11,205 |
| HB, Mean ± SD | 137.4 ± 21.4 | 124.7 ± 22.8 | 138.8 ± 20.8 | < 0.001 | -3.827 |
| RBC, Median (Q1,Q3) | 4.6 (4.1, 5) | 4.1 (3.7, 4.8) | 4.6 (4.1, 5) | 0.002 | 5856.5 |
| PLT, Median (Q1,Q3) | 199 (162, 247) | 200.5 (163.5, 274.5) | 199 (162.5, 245) | 0.593 | 8711.5 |
| ALT, Median (Q1,Q3) | 31 (21, 48) | 37 (20, 69) | 30.5 (21, 47.2) | 0.09 | 9328 |
| AST, Median (Q1,Q3) | 63 (34.5, 134) | 128 (47, 241) | 59.5 (34, 121.2) | 0.01 | 10048.5 |
| GGT, Median (Q1,Q3) | 42 (26, 70.2) | 35 (25, 74) | 42 (27, 70) | 0.846 | 7867 |
| BUN, Median (Q1,Q3) | 5.7 (4.7, 7.6) | 8.9 (5.5, 11.6) | 5.6 (4.6, 7.3) | < 0.001 | 11,523 |
| Creatinine, Median (Q1,Q3) | 80 (63, 101) | 112 (84, 162) | 78 (62, 98) | < 0.001 | 11892.5 |
| UA, Median (Q1,Q3) | 361 (287, 435) | 449 (368, 545) | 355 (280, 419.2) | < 0.001 | 11,609 |
| CyC, Median (Q1,Q3) | 1 (0.8, 1.3) | 1.6 (1.1, 1.9) | 1 (0.8, 1.2) | < 0.001 | 11852.5 |
| CK, Median (Q1,Q3) | 454 (175, 1153.2) | 574.5 (256.5, 1379.2) | 423 (174.8, 1105.5) | 0.187 | 9346 |
| CKMB, Median (Q1,Q3) | 44 (22, 101) | 66 (29.8, 107.8) | 43 (21, 101) | 0.095 | 9618 |
| LDH, Median (Q1,Q3) | 366.5 (262.2, 581.2) | 530 (391, 829.2) | 344 (258.8, 561.5) | < 0.001 | 11,641 |
| HBDH, Median (Q1,Q3) | 257 (172, 447.8) | 393 (241.5, 631.8) | 249.5 (166, 424) | < 0.001 | 11,338 |
| CTnT, Median (Q1,Q3) | 809.8 (175.2, 2413.2) | 2294 (648.6, 5431) | 746.4 (145.7, 2195.5) | < 0.001 | 11192.5 |
| myoglobin, Median (Q1,Q3) | 233.6 (62.5, 728.4) | 705.9 (141.1, 1912) | 216.4 (55.8, 659.7) | < 0.001 | 10,777 |
| BNP, Median (Q1,Q3) | 840.9 (236.8, 2616.8) | 5483.5 (1445, 11624.5) | 753.4 (194.3, 2269.5) | < 0.001 | 11536.5 |
| Diseased vessel during procedure | |||||
| LM, n(%) | 16 (4) | 1 (2) | 15 (4) | 1 | Fisher |
| LAD, n(%) | 317 (88) | 22 (79) | 295 (89) | 0.119 | Fisher |
| LCX, n(%) | 212 (75) | 18 (78) | 194 (75) | 0.916 | Fisher |
| RCA, n(%) | 262 (85) | 18 (78) | 244 (86) | 0.354 | Fisher |
| Risk assessment | |||||
| GRACE, Median (Q1,Q3) | 118 (99, 139) | 174.5 (134.2, 198.8) | 115 (98, 133.2) | < 0.001 | 13771.5 |
Abbreviations: Shock index, ratio of HR to SBP; SIRI, systemic inflammatory response index; SII, systemic inflammatory reaction index; PLR, platelet-to-lymphocyte ratio; NLR, neutrophil-to-lymphocyte ratio; MLR, monocyte-to-lymphocyte ratio; OHCA, out-of-hospital cardiac arrest; GRACE, Global Registry of Acute Coronary Events score; α-HBDH, α-hydroxybutyrate dehydrogenase; BNP, B-type natriuretic peptide; CTnT, cardiac troponin T; CyC, Cystatin C; RCA, right coronary artery; LCX, left circumflex artery; LAD, left anterior descending artery; LM, left main artery; RBC, red blood cell; PLT platelet; AST, Aspartate aminotransferase; ALT, alanine aminotransferase; GGT, gamma-glutamyltransferase; BUN, blood urea nitrogen; UA, uric acid
Table 2.
Comparison of validation set results of the machine learning models
| Model | Accuracy [95%CI] |
AUC [95%CI] |
Recall [95%CI] |
Precision [95%CI] |
F1 [95%CI] |
|---|---|---|---|---|---|
| CatBoost Classifier |
0.93 [0.925–0.932] |
0.92 [0.909–0.922] |
0.52 [0.485–0.548] |
0.79 [0.755–0.815] |
0.57 [0.544–0.599] |
| Gradient Boosting Classifier |
0.91 [0.908–0.916] |
0.91 [0.903–0.916] |
0.46 [0.431–0.486] |
0.67 [0.639–0.694] |
0.52 [0.493–0.543] |
| Random Forest Classifier |
0.92 [0.918–0.926] |
0.89 [0.883–0.904] |
0.54[0.511–0.572] |
0.74 [0.707–0.766] |
0.58 [0.551–0.604] |
| Extreme Gradient Boosting |
0.91 [0.909–0.916] |
0.88 [0.875–0.891] |
0.52 [0.483–0.55] |
0.56 [0.531–0.592] |
0.51 [0.478–0.536] |
| Naive Bayes |
0.9 [0.895–0.902] |
0.87 [0.859–0.878] |
0.73 [0.704–0.746] |
0.58 [0.563–0.596] |
0.62 [0.609–0.634] |
| Logistic Regression |
0.9 [0.891-0.9] |
0.84 [0.833–0.855] |
0.4 [0.373–0.427] |
0.55 [0.516–0.584] |
0.39 [0.36–0.414] |
| GRACE |
0.83 [0.789–0.862] |
- | - | - | |
| Bagging-Catboost |
0.93 [0.925–0.932] |
0.92 [0.91–0.923] |
0.52 [0.485–0.548] |
0.79 [0.755–0.815] |
0.57 [0.52–0.622] |
| Boosting-Catboost |
0.92 [0.914–0.923] |
0.9 [0.894–0.912] |
0.49 [0.461–0.523] |
0.68 [0.645–0.705] |
0.54 [0.513–0.57] |
| Blend model-ALL model |
0.92 [0.918–0.926] |
0.92 [0.91–0.924] |
0.52 [0.485–0.548] |
0.74 [0.705–0.765] |
0.57 [0.537–0.593] |
| Stacker model-ALL model |
0.93 [0.922–0.929] |
0.92 [0.918–0.93] |
0.48 [0.455–0.512] |
0.79 [0.755–0.815] |
0.55 [0.526–0.577] |
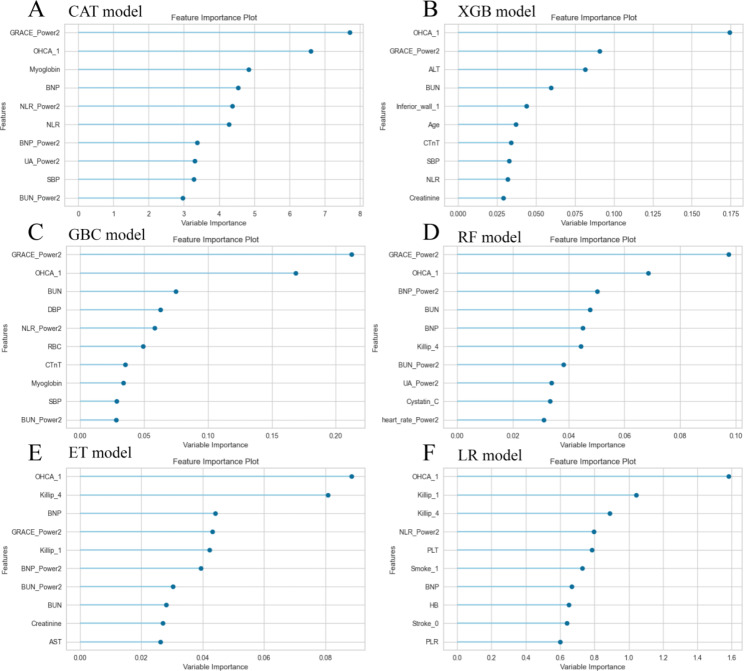
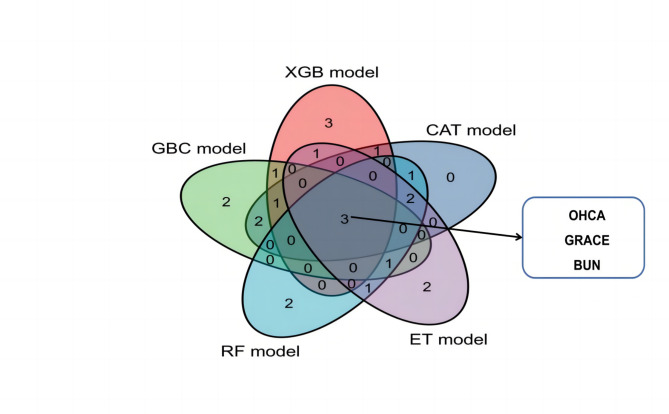
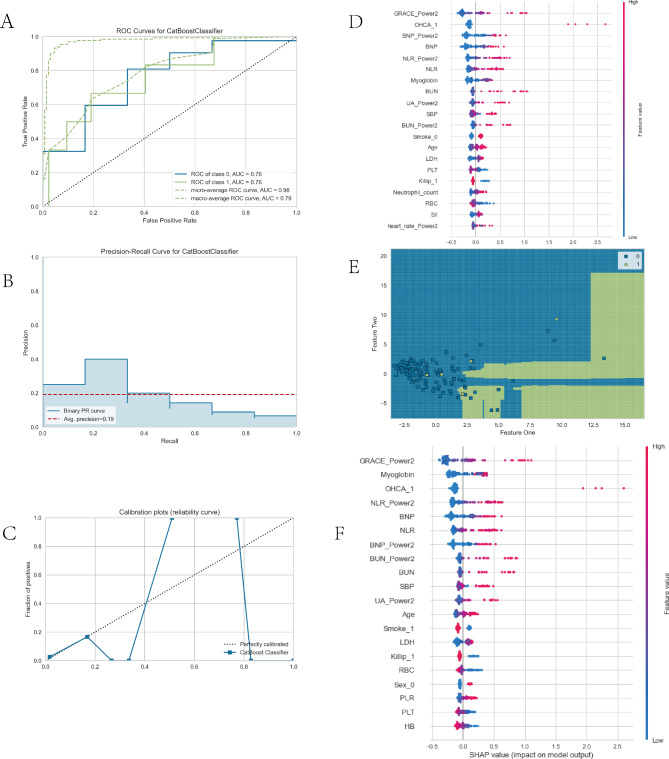
Figure 2 illustrates the relative importance of the variables in each ML algorithm. Excluding the logistics model, the top 10 most important intersection variables of the other five excellent ML models are shown in Fig. 3. We can see a general trend in the evidence: despite the slight difference in the importance of variables in these ML algorithms, factors including out-of-hospital cardiac arrest (OHCA), GRACE, and blood urea nitrogen (BUN) ranked in the top five (Fig. 3). The neutrophil-to-lymphocyte ratio (NLR), B-type natriuretic peptide (BNP), and cystatin C were ranked in the top ten. Conversely, the cardiac troponin T and creatinine variables contributed little to the prediction. The significance of the high-ranking variables in the CatBoost model decreased in the following order: GRACE, OHCA, myoglobin, BNP, and NLR. There are challenges involved in saving patients who were hypoperfused when they were admitted due to STEMI. The CatBoost model demonstrated the highest performance of the predictive models, with an AUC of 0.92, precision of 0.79, and accuracy of 0.93. Therefore, we selected the CatBoost model as the final predictive model for application to the validation set (Fig. 4).
Fig. 2.
The relative importance of variants in machine learning algorithms
Fig. 3.
Cross-verification in the top 10 variables of the machine learning models
Fig. 4.
Visualization of CatBoost model performance in the validation set. (A) AUCROC curve. (B) Precision-recall curve. (C) Calibration curve. (D) Classification report. (E) Decision boundary. (F) Feature SHAP value
Discussion
This study aimed to develop an accurate and user-friendly prediction model for STEMI patients with T2DM to enhance therapeutic decision-making and reduce periprocedural complications. Our approach leveraged machine learning (ML) algorithms, known for their exceptional performance over traditional regression methods in large-scale outcome prediction [14, 15, 18, 19]. We focused on integrating ML techniques with preoperative clinical data for this patient group.
We addressed the challenge of potential data lag in the risk model, which incorporates preoperative, intraoperative, and postoperative clinical data, as this can delay the prediction of adverse events. While logistic regression is often chosen in traditional binary outcome prediction models for its strong predictive power and model interpretability, in our study, CatBoost showed superior performance. Among the five ML models, the GRACE variable and OHCA were the strongest predictors of almost all the analysis methods. Coronary artery disease, particularly acute coronary syndrome, is the leading cause of OHCA in Asians. The survival rate for Asians with OHCA is 3.0% [20]. The treatment of post-resuscitation is dependent on emergency coronary angiography and PCI. Therefore, this study again validated the effectiveness of the GRACE score in a cohort of patients with T2DM and STEMI while finding that ML models had better model discrimination and accuracy. Moreover, it is worth mentioning that, in the case of medical sample imbalance, some studies propose that the model integration technology can significantly improve model performance and capability [14, 21].
ML in clinical medicine faces challenges like data quality and processing complexity, with ML model features often being subtle and difficult for clinicians to interpret. Despite these challenges, ML has achieved significant breakthroughs in biomedicine. For example, ML algorithms have been utilized to analyze complex, high-dimensional data in 12-lead ECGs, predicting artery occlusion in myocardial infarction cases [22]. Liang et al.‘s study effectively used ML to predict heart failure risk during hospitalization for patients with acute anterior wall ST-segment elevation myocardial infarction, employing parameters such as VF, CAP, age, LVEF, and NT-pro-BNP peak levels. This approach enabled the identification of high-risk patients, guiding personalized and proactive management strategies [23]. Further, research by Tofighi et al. demonstrated the efficacy of ML in identifying high-risk STEMI patients for adverse events during follow-up, aiding in crafting individualized treatment plans to improve outcomes and lessen disease burden [24]. Avvisato’s study illustrated ML’s powerful capacity to dissect individual variations in the whole brain functional connectome, aiding in the diagnosis of neurological diseases and predicting clinical outcomes [25]. Those predictive models can be seamlessly integrated into the emergency visit process to aid physicians in determining the prognosis of patients’ conditions. By prioritizing higher-risk patients for more immediate and comprehensive care and monitoring or scheduling follow-ups for lower-risk patients, this approach not only optimizes emergency physicians’ workflow but also ensures that patients receive care tailored to their specific risk profiles. Therefore, understanding the synthetic significance of clinical data samples and the models and methods to accurately diagnose diseases or predict the occurrence of adverse events will remain a persistent focus in medical integration.
Limitations
This study also has some limitations. Firstly, being a single-center study, the generalizability of our results might be limited. Additionally, its retrospective design raises the possibility of selection bias, which could affect the interpretation of results and the applicability of prediction models. Secondly, like other similar studies, ours relied on clinical data elements such as medical history, physical examination, and lab findings as input features. This reliance potentially restricts the model’s applicability in real-world settings and results in some models lacking interpretability. Thirdly, akin to most studies [26], ours used a single dataset for training and testing the model, but the sample size was insufficient for precise testing and training, mainly due to a lack of adequate positive samples. Even though synthetic sampling techniques were employed, the improvement was not significant, indicating a need for studies with larger sample sizes in this field. Fourthly, the absence of external validation in our study could lead to overfitting. Fifth, While the GRACE score’s inclusion might enhance the model’s predictive accuracy, it can also complicate the interpretation, as the score itself is a composite measure. This layer of abstraction might obscure the individual impact of the clinical variables constituting the GRACE score. Relying heavily on an existing score can affect the robustness of the model, potentially making it more sensitive to any biases or limitations inherent in the GRACE score. Lastly, the indicators included in the analysis were not comprehensive, missing crucial variables such as a timeline of medical visits, medication usage, etc., all of which could influence the outcomes. Therefore, future research should employ larger sample sizes and more scientific methodologies.
Conclusion
In this study, various ML methods were used to establish in-hospital death prediction models for AMI complicated by diabetes. Compared with the traditional model and GRACE score, the predictive discrimination of the ML model based on the CatBoost algorithm was more accurate. Although this ML algorithm presently has a good prediction ability, it still requires further scientific and reasonable external verification.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: sFig1.ROC curve analysis of the GRACE score and six machine learning models in the overall dataset
Acknowledgements
We express our gratitude to all patients who participated in this study.
Author Contributions
Panke Chen, Li Zhao, and Bine Wang contributed equally to this work and served as co-first authors. Conceptualization: Zhixun Bai and Bei Shi; Methodology: Panke Chen and Zhixun Bai; Formal analysis and investigation: Bine Wang, Li Zhao, Panke Chen and Shuai Ma; Writing-original draft preparation: Panke Chen, Yanping Wang, Yunyue Zhu, Xin Zeng and Zhixun Bai; Writing-review and editing: Zhixun Bai and Bei Shi; Funding acquisition: Zhixun Bai; Supervision: Zhixun Bai and Bei Shi. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 82260106). The Special Projects of Innovation and Exploration in Zunyi Medical University (2020-024), the Project of Guizhou Provincial Health Commission (gzwkj2021-138), the Zunyi Medical University College Student Innovation and Entrepreneurship Project (ZYDC2022119/S202210661301), and the Technological Project of Zunyi Science and Technology Bureau (HZ-2021- No.218).
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and informed consent
The present study was approved by the Ethical Evaluation Committee of Zunyi Medical Hospital (ZMU〔2022〕1-177). This study is a retrospective data analysis. In accordance with the nature of this research, it has received approval from the relevant ethics committee and has been confirmed to comply with the ethical principles of the Declaration of Helsinki. Given the retrospective design and the use of pre-existing anonymized datasets, the ethics committee has granted a waiver for the requirement of informed consent. All patient data have been rigorously anonymized to ensure that individual identities cannot be traced, thus safeguarding participant privacy and data confidentiality.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Panke Chen, Bine Wang and Li Zhao contributed equally to this work.
Contributor Information
Zhixun Bai, Email: zhixunbai@zmu.edu.cn.
Bei Shi, Email: shibei2147@163.com.
References
- 1.Ralapanawa U, Sivakanesan R. Epidemiology and the Magnitude of Coronary Artery Disease and Acute Coronary Syndrome: a narrative review. J Epidemiol Glob Health. 2021;11(2):169–77. doi: 10.2991/jegh.k.201217.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasprzak D, Rzezniczak J, Ganowicz T, Luczak T, Slomczynski M, Hiczkiewicz J, et al. A review of Acute Coronary Syndrome and its potential impact on cognitive function. Glob Heart. 2021;16(1):53. doi: 10.5334/gh.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman AR, Shah A, Lee KK, Anand A, Francis O, Adamson P, et al. Long-term outcomes in patients with type 2 Myocardial Infarction and myocardial Injury. Circulation. 2018;137(12):1236–45. doi: 10.1161/CIRCULATIONAHA.117.031806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchis-Gomar F, Perez-Quilis C, Leischik R, Lucia A. Epidemiology of coronary Heart Disease and acute coronary syndrome. Ann Transl Med. 2016;4(13):256. doi: 10.21037/atm.2016.06.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai Z, Ma Y, Shi Z, Li T, Hu S, Shi B. Nomogram for the prediction of Intrahospital Mortality Risk of patients with ST-Segment Elevation Myocardial Infarction complicated with hyperuricemia: a Multicenter Retrospective Study. THER CLIN RISK MANAG. 2021;17:863–75. doi: 10.2147/TCRM.S320533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lacey B, Herrington WG, Preiss D, Lewington S, Armitage J. The role of emerging risk factors in Cardiovascular outcomes. CURR ATHEROSCLER REP. 2017;19(6):28. doi: 10.1007/s11883-017-0661-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rawshani A, Rawshani A, Franzen S, Eliasson B, Svensson AM, Miftaraj M, et al. Mortality and Cardiovascular Disease in Type 1 and type 2 Diabetes. N Engl J Med. 2017;376(15):1407–18. doi: 10.1056/NEJMoa1608664. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhury M, Nevitt S, Eleftheriadou A, Kanagala P, Esa H, Cuthbertson DJ et al. Cardiac autonomic neuropathy and risk of Cardiovascular Disease and mortality in type 1 and type 2 Diabetes: a meta-analysis. BMJ Open Diabetes Res Care 2021, 9(2). [DOI] [PMC free article] [PubMed]
- 9.Tancredi M, Rosengren A, Svensson AM, Kosiborod M, Pivodic A, Gudbjornsdottir S, et al. Excess mortality among persons with type 2 Diabetes. N Engl J Med. 2015;373(18):1720–32. doi: 10.1056/NEJMoa1504347. [DOI] [PubMed] [Google Scholar]
- 10.Norhammar A, Mellbin L, Cosentino F, Diabetes Prevalence, prognosis and management of a potent cardiovascular risk factor. EUR J PREV CARDIOL. 2017;24(3suppl):52–60. doi: 10.1177/2047487317709554. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann R, James SK, Jernberg T, Lindahl B, Erlinge D, Witt N, et al. Oxygen therapy in suspected Acute Myocardial Infarction. N Engl J Med. 2017;377(13):1240–9. doi: 10.1056/NEJMoa1706222. [DOI] [PubMed] [Google Scholar]
- 12.Yanqiao L, Shen L, Yutong M, Linghong S, Ben H. Comparison of GRACE and TIMI risk scores in the prediction of in-hospital and long-term outcomes among east Asian non-ST-elevation Myocardial Infarction patients. BMC Cardiovasc Disord. 2022;22(1):4. doi: 10.1186/s12872-021-02311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torralba F, Navarro A, la Hoz JC, Ortiz C, Botero A, Alarcon F, et al. HEART, TIMI, and GRACE scores for prediction of 30-Day major adverse Cardiovascular events in the era of high-sensitivity troponin. ARQ BRAS CARDIOL. 2020;114(5):795–802. doi: 10.36660/abc.20190206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai Z, Lu J, Li T, Ma Y, Liu Z, Zhao R et al. Clinical Feature-Based Machine Learning Model for 1-Year Mortality Risk Prediction of ST-Segment Elevation Myocardial Infarction in Patients with Hyperuricemia: A Retrospective Study. Comput Math Methods Med 2021, 2021: 7252280. [DOI] [PMC free article] [PubMed]
- 15.Bai Z, Hu S, Wang Y, Deng W, Gu N, Zhao R, et al. Development of a machine learning model to predict the risk of late cardiogenic shock in patients with ST-segment elevation Myocardial Infarction. Ann Transl Med. 2021;9(14):1162. doi: 10.21037/atm-21-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibanez B, James S, Agewall S, Antunes MJ, Bucciarelli-Ducci C, Bueno H, et al. 2017 ESC guidelines for the management of acute Myocardial Infarction in patients presenting with ST-segment elevation: the Task Force for the management of acute Myocardial Infarction in patients presenting with ST-segment elevation of the European Society of Cardiology (ESC) EUR HEART J. 2018;39(2):119–77. doi: 10.1093/eurheartj/ehx393. [DOI] [PubMed] [Google Scholar]
- 17.Ding Q, Spatz ES, Lipska KJ, Lin H, Spertus JA, Dreyer RP, et al. Newly diagnosed Diabetes and outcomes after acute Myocardial Infarction in young adults. Heart. 2021;107(8):657–66. doi: 10.1136/heartjnl-2020-317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hulsen T, Jamuar SS, Moody AR, Karnes JH, Varga O, Hedensted S, et al. From Big Data to Precision Medicine. Front Med (Lausanne) 2019;6:34. doi: 10.3389/fmed.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ngiam KY, Khor IW. Big data and machine learning algorithms for health-care delivery. LANCET ONCOL. 2019;20(5):e262–73. doi: 10.1016/S1470-2045(19)30149-4. [DOI] [PubMed] [Google Scholar]
- 20.Hassager C, Nagao K, Hildick-Smith D. Out-of-hospital Cardiac Arrest: in-hospital intervention strategies. Lancet. 2018;391(10124):989–98. doi: 10.1016/S0140-6736(18)30315-5. [DOI] [PubMed] [Google Scholar]
- 21.Idakwo G, Thangapandian S, Luttrell J, Li Y, Wang N, Zhou Z, et al. Structure-activity relationship-based chemical classification of highly imbalanced Tox21 datasets. J Cheminform. 2020;12(1):66. doi: 10.1186/s13321-020-00468-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Zaiti SS, Martin-Gill C, Zegre-Hemsey JK, Bouzid Z, Faramand Z, Alrawashdeh MO, et al. Machine learning for ECG diagnosis and risk stratification of occlusion Myocardial Infarction. NAT MED. 2023;29(7):1804–13. doi: 10.1038/s41591-023-02396-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang J, Zhang Z. Predictors of in-hospital Heart Failure in patients with acute anterior wall ST-segment elevation Myocardial Infarction. INT J CARDIOL. 2023;375:104–9. doi: 10.1016/j.ijcard.2023.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Tofighi S, Poorhosseini H, Jenab Y, Alidoosti M, Sadeghian M, Mehrani M, et al. Comparison of machine-learning models for the prediction of 1-year adverse outcomes of patients undergoing primary percutaneous coronary intervention for acute ST-elevation Myocardial Infarction. CLIN CARDIOL; 2023. [DOI] [PMC free article] [PubMed]
- 25.Avvisato R, Forzano I, Varzideh F, Mone P, Santulli G. A machine learning model identifies a functional connectome signature that predicts blood pressure levels: imaging insights from a large population of 35 882 patients. CARDIOVASC RES. 2023;119(7):1458–60. doi: 10.1093/cvr/cvad065. [DOI] [PubMed] [Google Scholar]
- 26.Li AX, Yan K, Chandramouli LL, Hu C, Jin R. Machine learning-based prediction of infarct size in patients with ST-segment elevation Myocardial Infarction: a multi-center study. INT J CARDIOL. 2023;375:131–41. doi: 10.1016/j.ijcard.2022.12.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: sFig1.ROC curve analysis of the GRACE score and six machine learning models in the overall dataset
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.