Abstract
Having a previous history of sexually transmitted diseases (STDs) such as gonorrhea and chlamydia increases the chance of developing prostate cancer, the second most frequent malignant cancer among men. However, the molecular functions that cause the development of prostate cancer in persons with gonorrhea and chlamydia are yet unknown. In this study, we studied RNA-seq gene expression profiles using computational biology methods to find out potential biomarkers that could help us in understanding the patho-biological mechanisms of gonorrhea, chlamydia, and prostate cancer. Using statistical methods on the Gene Expression Omnibus (GEO) data sets, it was found that a total of 22 distinct differentially expressed genes were shared among these 3 diseases of which 14 were up-regulated (PGRMC1, TSC22D1, SH3BGRL, NNT, CTSC, FRMD3, CCR2, FAM210B, VCL, PTGS1, SLFN11, SLC40A1, PROS1, and DSE) and the remaining 8 genes were down-regulated (PRNP, HINT3, MARCKSL1, TMED10, SH3KBP1, ENSA, DERL1, and KMT2B). Investigation on these 22 unique dysregulated genes using Gene Ontology, BioCarta, KEGG, and Reactome revealed multiple altered molecular pathways, including regulation of amyloid precursor protein catabolic process, ferroptosis, effects on gene expression of Homo sapiens PPAR pathway, and innate immune system R-HSA-168249. Four significant hub proteins namely VCL, SH3KBP1, PRNP, and PGRMC1 were revealed by protein-protein interaction network analysis. By analyzing gene-transcription factors and gene-miRNAs interactions, significant transcription factors (POU2F2, POU2F1, GATA6, and HIVEP1) and posttranscriptional regulator microRNAs (hsa-miR-7-5p) were also identified. Three potential therapeutic compounds namely INCB3284, CCX915, and MLN-1202 were found to interact with up-regulated protein C-C chemokine receptor type 2 (CCR2) in protein-drug interaction analysis. The proposed biomarkers and therapeutic potential molecules could be investigated for potential pharmacological targets and activity in the fight against in patients with gonorrhea, chlamydia, and prostate cancer.
Keywords: Prostate cancer, gonorrhea, chlamydia, STD, biomarker identification, drug candidate
Introduction
Gonorrhea is a widely known sexually transmitted disease (STD) all over the world which is caused by a bacterium named Neisseria gonorrhoeae that infects the mucosa of exposed anatomic areas, including urogenital tract, rectum, mouth, and conjunctiva. Without proper treatment, cervical gonorrhea can have serious effects on reproductive health.1,2 The symptoms in females include abnormal uterine bleeding, vaginal discharge, dyspareunia, dysuria, lower abdominal and/or rectal pain while in male dysuria, urethral discharge and/or itching, and testicular or rectal pain are common. The urethra and cervix, followed by the anal and pharyngeal regions, are the anatomical sites that are the most frequently impacted. 3 From roughly 13 cases per 100 people in 2008 to up to 29 cases per 100 people in 2017, the Public Health Agency of Canada (PHAC) reported that the prevalence of gonorrhea has more than doubled since 2013. 4 The reported rate in the United States seems to be much higher with 171.9 cases per 100 000 people in 2017. 5 World Health Organization (WHO) reported 82 million new infections of gonorrhea in 2022 alone. 6
The infection of a bacterium Chlamydia trachomatis results in STD named chlamydia. In females, urethritis, cervicitis, pelvic inflammatory disease, proctitis, and perihepatitis are some of the symptoms of this disease and if proper treatment is not received in a timely manner, it might result in infertility and ectopic pregnancy. In case of men, the bacterium can cause epididymitis, urethritis, proctitis, prostatitis, or reactive arthritis. 7 Besides, this pathogen is the most common preventable cause of blindness in the endemic regions such as Africa and the Middle East. 8 According to Centers for Disease Control and Prevention (CDC) in 2018, almost 4 million people are diagnosed with chlamydia in the United States. 9 According to WHO, 129 million new infections of chlamydia were reported in 2022 alone. 6 The PHAC, Canada, reported a national rate of 345.7 cases of chlamydia per 100 000 populations in 2017. 4 In men, the symptoms of chlamydia include pain in testicles, burning sensation during urination, yellow or green discharge from the urinating part, pain in lower abdomen while in case of women, painful feeling during sexual intercourse, vaginal discharge, inflammation of cervix, burning sensation when urinating and most importantly bleeding between menstrual cycles are observed. 10
Prostate cancer is the second most frequent malignant cancer among men. According to the American Cancer Society, about 288 300 new cases were reported and about 34 700 people died from prostate cancer in 2023. 11 It was stated that this cancer affects more than 35 000 men in the United Kingdom alone every year, and 10 000 men pass away as a result of the condition. Although the patient may be asymptomatic, the main complications with this disease are micturition, straining to start, frequency, and nocturia. 12 It is evident that access to diagnosis and treatment affects prostate cancer rates significantly. Males of African heritage have greater prostate cancer rates and lower prognosis. 13
Having a previous history of STDs like chlamydia or gonorrhea may increase the possibility of developing prostate cancer. A research funded by California Cancer Research Program, and Community Benefits Program of Kaiser Permanente found that 26.3% of males reported a history of any STD, with 14.7% of those men having experienced 2 or more STDs before being diagnosed with prostate cancer. The most prevalent STD among them was gonorrhea (17.1%), and the percentage of chlamydia was 3.6%. 14 Another study related to chlamydia shows that, in human prostate epithelial cells, the expression of inflammatory tumorigenic cytokines and chemokines is promoted by the infection of C. trachomatis, including the components of the Toll-like receptor and NF-kB pathways. According to this research, epithelial cells from human prostate cancer are prone to C. trachomatis infection and increase more pro-inflammatory markers when infected. 15 Although it is well-thought-out that history of previous STDs like chlamydia, and gonorrhea may impact the development of prostate cancer, the markers which are often dysregulated in those 3 diseases have not been well considered at molecular levels till now using RNA data sets and microarray data which are available on online databases like NCBI.
In our research, a system biology approach has been employed to reveal differentially expressed genes (DEGs) and related molecular pathways identical among patients with gonorrhea, chlamydia, and prostate cancer. The DEGs shared among the 3 diseases were studied in interaction networks using the following tactics: (1) study of gene enrichment to determine cellular components, molecular functions, and biological processes in which a gene is associated, (2) finding hub genes using the protein-protein interaction (PPI) network, (3) identification of transcription factors (TFs) and miRNAs that are associated with the common DEGs, and (4) promising drug candidates screening using protein-drug interaction networks. The scientific findings of this study will aid in revealing biomarkers for disease diagnosis and molecular targets for drug development which will be used to treat these diseases.
Methods
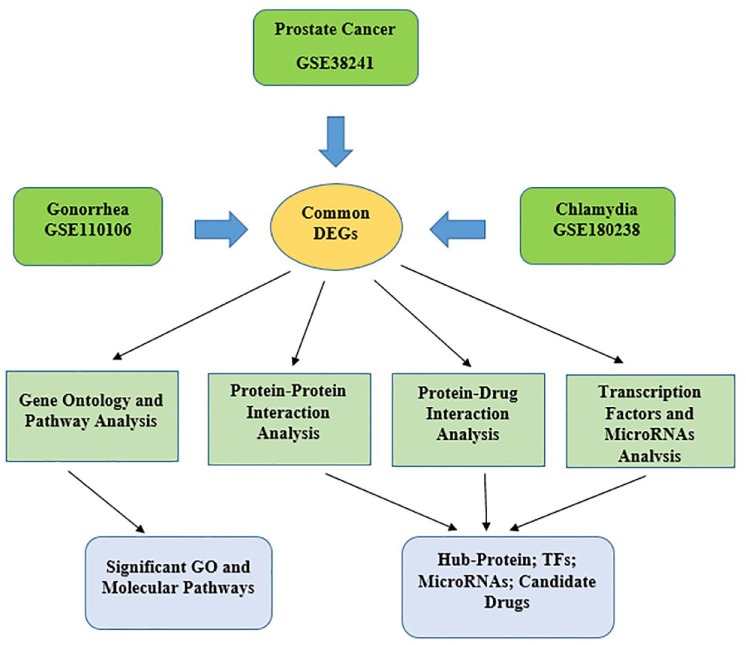
For biomarkers and drug target identification, data from NCBI-Gene Expression Omnibus (GEO) database were used and statistical approaches were applied to select shared dysregulated genes among the 3 disorders, namely gonorrhea, chlamydia, and prostate cancer. Overall procedures are illustrated in Figure 1.
Figure 1.
The entire methodology employed in this study. Differentially expressed genes for gonorrhea, chlamydia, and prostate cancer were identified, and then, statistical approaches were applied to select shared dysregulated genes among these 3 disorders. To identify important shared pathways and GO keywords, gene enrichment analysis was carried out. PPI analysis was performed to identify hub proteins, TFs, and miRNAs that regulate those DEGs. Finally, protein-drug interactions were used to identify potential therapeutic candidates.
DEGs indicates differentially expressed genes; miRNAs, microRNA; GO, gene ontology; PPI, protein-protein interaction.
Microarray data sets
This study used data from the NCBI-GEO database, GSE110106, GSE180238, and GSE38241 with a total sample size of 49. The 4 samples from GSE110106 come from 2 healthy individuals (GSM2977717 and GSM2977718) and 2 gonorrhea affected individuals with biopsy-confirmed N gonorrhoeae (GSM3093828 and GSM3093832). In case of chlamydia, there were 6 individuals with 3 patients and 3 healthy come from GSE180238 where C. trachomatis-infected human leukaemia (HL-60) samples were considered as patient and uninfected HL-60 samples were considered as control. For prostate cancer, 39 samples come from GSE38241 with 18 patients and 21 healthy individuals where normal prostate from organ donors considered as control and samples from lethal metastatic prostate cancer considered as patient. The control groups, which served as a reference in the data sets, didn’t show any inflammation.
Analysis of differential gene expression
Differentially expressed genes were recognized using GEO2R by comparing patients to their respective control groups in all 3 GEO data sets. Statistically significant DEGs were identified by considering log2 (Fold Change) ⩾0.5 for up-regulated genes and log2 (Fold Change) ⩽−0.5 for down-regulated genes with a P value less than .05 and the Benjamini and Hochberg (false-discovery rate) criterion for P value adjustment. The Venny tool was used to find DEGs that were common among gonorrhea, chlamydia, and prostate cancer (https://bioinfogp.cnb.csic.es/tools/venny/).
Gene ontology and DEG pathway enrichment
To learn more about the DEGs, gene enrichment analysis was carried out. Enrichr (https://maayanlab.cloud/Enrichr/) was used to obtain the cellular components, molecular activities, and biological processes to which a gene contributes using P < .05 and also common pathways among gonorrhea, chlamydia, and prostate cancer were determined through KEGG, Reactome, and BioCarta pathways (https://maayanlab.cloud/Enrichr/).
PPI network study
Protein-protein interaction networks were created using STRING protein interaction database, and Network Analyst web resource, with a confidence score of 700 (https://www.networkanalyst.ca/).
Investigation of DEG interaction with transcriptional and posttranscriptional regulators
Using TRANSFAC, and miRTarBase databases, prominent transcription factors and microRNAs that regulate DEGs of importance at both the transcriptional and posttranscriptional levels, respectively, were mined with a P value < .05 (https://maayanlab.cloud/Enrichr/). Top 10 miRNAs were selected, and their interactions with target genes were validated using RNAhybrid tool (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid) to calculate minimum free energy on hybridization with target mRNA.
Assessment of protein-drug interactions
Effective therapeutic candidates for gonorrhea, chlamydia, and prostate cancer were revealed by analyzing protein-drug interactions with Network Analyst (https://www.networkanalyst.ca/) tool with the help of Drug Bank database (version 5.0).
Results
Identification of DEGs shared among gonorrhea, chlamydia, and prostate cancer patients
Gene expressions of microarray data sets were studied for gonorrhea, chlamydia, and prostate cancer, and significant DEGs were identified using statistical methods. It was found that a total of 22 distinct genes were discovered to be shared among the gonorrhea DEGs, chlamydia DEGs, and prostate cancer DEGs of which 14 were up-regulated (PGRMC1, TSC22D1, SH3BGRL, NNT, CTSC, FRMD3, CCR2, FAM210B, VCL, PTGS1, SLFN11, SLC40A1, PROS1, and DSE). The remaining 8 genes were down-regulated (PRNP, HINT3, MARCKSL1, TMED10, SH3KBP1, ENSA, DERL1, and KMT2B) (Figures 2 and 3).
Figure 2.
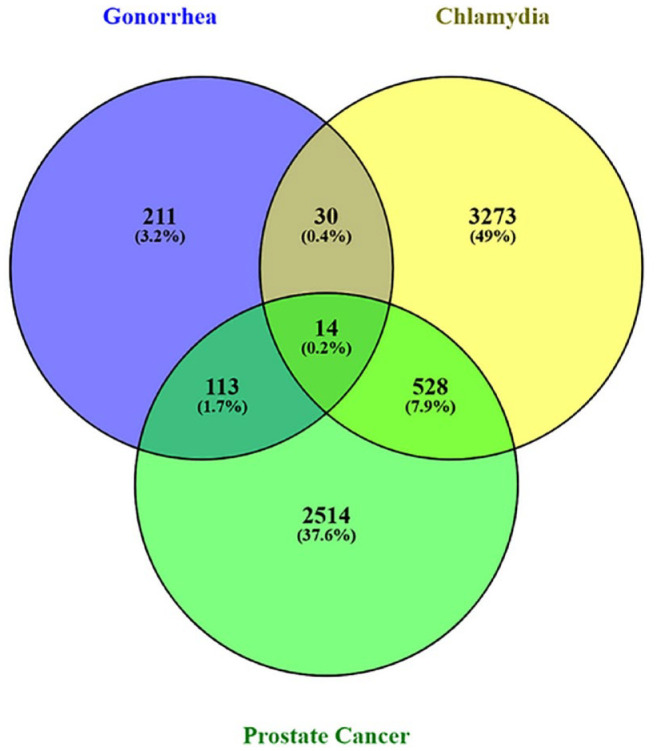
The Venny tool (www.bioinfogp.cnb.csic.es/tools/venny/) was used to identify genes that are commonly up-regulated in gonorrhea, chlamydia, and prostate cancer. In gonorrhea, chlamydia, and prostate cancer, a total of 14 genes were discovered to be frequently up-regulated.
Figure 3.
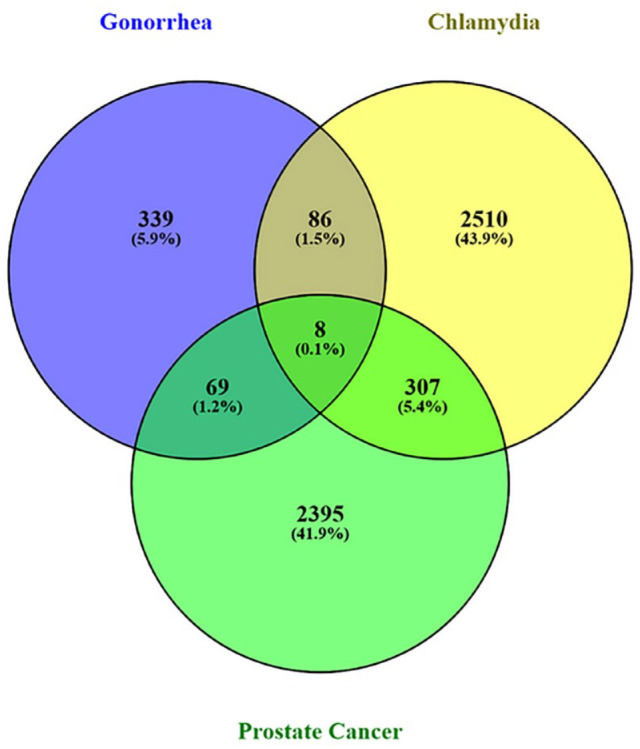
The Venny tool (www.bioinfogp.cnb.csic.es/tools/venny/) was used to identify genes that are commonly down-regulated in gonorrhea, chlamydia, and prostate cancer. A total of 8 genes were found to be frequently down-regulated in gonorrhea, chlamydia, and prostate cancer.
These shared 22 DEGs were chosen for gene set enrichment analysis to understand more about the biological processes, molecular functions, and cellular components that a gene associates. The enriched biological processes are regulation of amyloid precursor protein catabolic process, regulation of amyloid-beta formation, protein destabilization, positive regulation of lymphocyte activation, regulated exocytosis, ER to Golgi vesicle-mediated transport, and others.
The significant molecular functions of DEGs shared by gonorrhea, chlamydia, and prostate cancer were amyloid-beta binding, endopeptidase inhibitor activity, protease binding, signal recognition particle binding, and others, and the important cellular components were found as zymogen granule ER-Golgi intermediate compartment membrane, COPII-coated ER to Golgi transport vesicle coated vesicle, secretory granule lumen, and others as shown in Table 1. Common pathways are identified by KEGG. BioCarta and Reactome are summarized in Table 2.
Table 1.
Significant gene ontology (GO) terms related to common differentially expressed genes in gonorrhea, chlamydia, and prostate cancer.
| Group | GO term | P |
|---|---|---|
| Biological process | Regulation of amyloid precursor protein catabolic process | .000076 |
| Regulation of amyloid-beta formation | .000634 | |
| Protein destabilization | .000634 | |
| Positive regulation of lymphocyte activation | .000672 | |
| Regulated exocytosis | .000973 | |
| Endoplasmic reticulum to Golgi vesicle-mediated transport | .001053 | |
| Regulation of interleukin-2 production | .001263 | |
| COPII vesicle coating | .002166 | |
| Vesicle coating | .002166 | |
| Vesicle targeting, rough endoplasmic reticulum (ER) to cis-Golgi | .002166 | |
| Molecular function | Amyloid-beta binding | .003465 |
| Endopeptidase inhibitor activity | .007023 | |
| Protease binding | .007381 | |
| Aspartic-type endopeptidase inhibitor activity | .007676 | |
| Signal recognition particle binding | .007676 | |
| Cuprous ion binding | .008768 | |
| G protein-coupled glutamate receptor binding | .008768 | |
| Iron ion transmembrane transporter activity | .008768 | |
| Unmethylated CpG binding | .008768 | |
| Potassium channel inhibitor activity | .009858 | |
| Cellular component | Endoplasmic reticulum-Golgi intermediate compartment membrane | .001316 |
| COPII-coated endoplasmic reticulum (ER) to Golgi transport vesicle | .003380 | |
| Coated vesicle | .003813 | |
| Secretory granule lumen | .004816 | |
| Zymogen granule | .006582 | |
| Zymogen granule membrane | .006582 | |
| Endoplasmic reticulum membrane | .006986 | |
| Gamma-secretase complex | .007676 | |
| COPI-coated vesicle | .007676 | |
| Mitochondrial outer membrane | .008376 |
P < .05.
Table 2.
Genes that are variably expressed and are common among gonorrhea, chlamydia, and prostate cancer patients contribute to various molecular pathways (P < .05).
| Pathways | P | Genes involved in pathways |
|---|---|---|
| KEGG (Kyoto Encyclopedia of Genes and Genomes) | ||
| Ferroptosis | 9.23E-04 | PRNP and SLC40A1 |
| Nicotinate and nicotinamide metabolism | .037820 | NNT |
| BioCarta | ||
| Basic mechanism of action of PPARa, PPARb(d), and PPARg and effects on gene expression Homo sapiens h PPAR pathway | .005488 | PTGS1 |
| Mechanism of acetaminophen activity and toxicity Homo sapiens h acetaminophen pathway | .007676 | PTGS1 |
| CBL mediated ligand-induced downregulation of EGF receptors Homo sapiens h cbl pathway | .008768 | SH3KBP1 |
| Cell-to-cell adhesion signaling Homo sapiens h cell-to-cell pathway | .013124 | VCL |
| Extrinsic prothrombin activation pathway Homo sapiens h extrinsic pathway | .014210 | PROS1 |
| Aspirin blocks signaling pathway involved in platelet activation Homo sapiens h spaa pathway | .018544 | PTGS1 |
| Prion pathway Homo sapiens h prion pathway | .019624 | PRNP |
| Sprouty regulation of tyrosine kinase signals Homo sapiens h spry pathway | .021782 | SH3KBP1 |
| Eicosanoid metabolism Homo sapiens h eicosanoid pathway | .025009 | PTGS1 |
| Intrinsic prothrombin activation pathway Homo sapiens h intrinsic pathway | .025009 | PROS1 |
| Reactome | ||
| Innate immune system R-HSA-168249 | .004629 | PGRMC1, PROS1, CTSC, VCL, and CCR2 |
| Reelin signaling pathway R-HSA-8866376 | .005488 | SH3KBP1 |
| Platelet degranulation R-HSA-114608 | .008248 | PROS1 and VCL |
| Response to elevated platelet cytosolic Ca2 + R-HSA-76005 | .008895 | PROS1 and VCL |
| POU5F1 (OCT4), SOX2, NANOG repress genes related to differentiation R-HSA-2892245 | .009858 | TSC22D1 |
| Transport of gamma-carboxylated protein precursors from endoplasmic reticulum to Golgi apparatus R-HSA-159763 | .009858 | PROS1 |
| Removal of amino terminal propeptides from gamma-carboxylated proteins R-HSA-159782 | .010948 | PROS1 |
| MASTL facilitates mitotic progression R-HSA-2465910 | .010948 | ENSA |
| Gamma-carboxylation of protein precursors R-HSA-159740 | .010948 | PROS1 |
| Gamma-carboxylation, transport, and amino-terminal cleavage of proteins R-HSA-159854 | .012036 | PROS1 |
Hub protein identification from PPI network
The PPI network generated by STRING consists of 4 hub nodes from shared DEGs and 187 edges (Figure 4) that uncovered 4 hub proteins, namely VCL, SH3KBP1, PRNP, and PGRMC1 (Table 3). Hub proteins were selected based on the higher number of connectivity with shared DEGs in the PPI network (degree of connectivity >10).
Figure 4.

PPI network includes frequently DEGs plus additional STRING database genes. This network contains 187 nodes (5 hub nodes from shared DEGs) and 187 edges.
DEGs indicates differentially expressed genes; PPI, protein-protein interaction.
Table 3.
List of biomarker representatives (proteins and transcription factors) with their biological functions.
| Biomarker representatives | Full form | Role of biomarkers |
|---|---|---|
| Hub proteins | ||
| VCL | Vinculin | VCL, a cytoskeletal protein, associated with cell-cell and cell-matrix junctions involved in anchoring F-actin to the membrane. Diseases associated with VCL gene include congestive heart failure and cardiomyopathy. |
| SH3KBP1 | SH3 domain containing kinase-binding protein 1 | The encoded protein by SH3KBP1 gene promotes protein-protein interactions and has been linked to a variety of cellular processes such as apoptosis, cytoskeletal reorganization, cell adhesion, and the control of clathrin-dependent endocytosis. Diseases associated with SH3KBP1 gene include adrenal cortical adenocarcinoma and immunodeficiency (an X-linked recessive primary immunologic disorder). |
| PRNP | Prion protein | This gene encodes a membrane glycosylphosphatidylinositol-anchored glycoprotein that aggregates into rod-like structure. Diseases associated with PRNP gene include Creutzfeldt-Jakob disease and Huntington disease-like 1. |
| PGRMC1 | Progesterone receptor membrane component 1 | A putative membrane-associated progesterone steroid receptor encodes by this gene. Diseases associated with PGRMC1 gene include premature menopause and cataract. |
| Transcription factor; P < .05 | ||
| POU2F2 | POU class 2 homeobox 2 | A homeobox-containing transcription factor of the POU domain family is encoded by this gene. Diseases associated with POU2F2 gene include B-cell lymphoma and prion disease. |
| POU2F1 | POU class 2 homeobox 1 | Diseases associated with POU2F1 gene include herpes simplex and inflammatory bowel disease. |
| GATA6 | GATA-binding protein 6 | This gene belongs to a small family of zinc finger transcription factors that regulate cellular differentiation and organogenesis during vertebrate development. Diseases associated with GATA6 include atrioventricular septal defect and congenital anomalies. |
| HIVEP1 | HIVEP zinc finger 1 | Isoforms 2 and 3 of this gene may be involved in apoptosis. Diseases associated with HIVEP1 include attention-deficit hyperactivity disorder and Brugada syndrome. |
Revealing transcriptional and posttranscriptional regulators that interact with DEGs related to prostate cancer, gonorrhea, and chlamydia patient
Because the expression of genes is regulated at both the transcriptional and posttranscriptional levels, significant transcription factors and miRNAs that regulate expression of shared DEGs among gonorrhea, chlamydia, and prostate cancer were determined. We found significant TFs POU2F2 (targeting DEG PRNP, SH3BGRL, TMED10, TSC22D1, HINT3, PROS1, DSE, and CTSC), POU2F1 (targeting DEG PRNP, SH3BGRL, TMED10, PROS1, and DSE), GATA6 (targeting DEG TMED10, TSC22D1, ENSA, DERL1, and CCR2), and HIVEP1 (targeting DEG FRMD3 and TSC22D1) (Table 3). In addition, the top 10 statistically prominent miRNAs were found namely hsa-miR-1224-3p, hsa-miR-485-3p, hsa-miR-125a-5p, hsa-miR-7-5p, hsa-miR-5572, hsa-miR-4717-5p, hsa-miR-153-3p, hsa-miR-506-5p, hsa-miR-4518, and hsa-miR-1266-5p are presented in Table 4 of which all showed negative free energy change on binding with target mRNA indicating possibility of interaction and gene regulation.
Table 4.
Top 10 miRNAs that interact with DEGs generated from miRTarBase2017 database organized by P value.
| miRNA name | P | Target gene | Minimum free energy of hybridization (kcal/mol) |
|---|---|---|---|
| hsa-miR-1224-3p | .002429 | NNT | −37.3 |
| .002638 | KMT2B | −39.4 | |
| .002646 | ENSA | −35.2 | |
| hsa-miR-485-3p | .002775 | TMED10 | −25.4 |
| .002930 | SLC40A1 | −30.7 | |
| hsa-miR-125a-5p | .003236 | SLFN11 | −29.3 |
| .003557 | NNT | −30.8 | |
| .004152 | PTGS1 | −30.5 | |
| hsa-miR-7-5p | .004330 | MARCKSL1 | −22.6 |
| .002429 | TMED10 | −23.5 | |
| .002638 | SLFN11 | −22.6 | |
| ENSA | −21.6 | ||
| hsa-miR-5572 | .002646 | MARCKSL1 | −34.6 |
| .002775 | TMED10 | −30.8 | |
| hsa-miR-4717-5p | .002930 | SH3BGRL | −18.4 |
| .003236 | SLFN11 | −28.3 | |
| hsa-miR-153-3p | .003557 | FAM210B | −20.2 |
| .004152 | KMT2B | −25.3 | |
| hsa-miR-506-5p | .004330 | PRNP | −23.3 |
| SLFN11 | −24.3 | ||
| hsa-miR-4518 | .002429 | MARCKSL1 | −28.8 |
| .002638 | NNT | −28.7 | |
| hsa-miR-1266-5p | .002646 | MARCKSL1 | −30.4 |
| NNT | −29.4 |
Abbreviations: DEGs, differentially expressed genes; miRNAs, microRNA.
Micro RNA’s interactions with target genes were validated using RNAhybrid tool. A negative free energy change indicates spontaneous binding of miRNA with target genes.
Effective therapeutics identification by protein-drug interaction network
C-C chemokine receptor type 2 gene encodes a protein which acts as a functional receptor (C-C motif chemokine receptor 2) for CCL2 but also can bind with CC7 and CCL12.16-18 It has been reported that polymorphisms in CCR and its ligand monocyte chemoattractant protein-1 (MCP-1) are associated with various diseases.19-23 Several research reported CCR2 protein’s involvement with prostate cancer.24,25
As this CCR2 gene is up-regulated in gonorrhea, chlamydia, and prostate cancer, our objective was to reduce the expression of this gene when it binds with a potential therapeutic substances. By using Network Analyst tool, drug bank database search against DEGs found existing drugs namely INCB3284, CCX915, and MLN-1202 interactions with commonly up-regulated gene CCR2 (Table 5 and Figure 5). According to drug bank, INCB3284 is an antagonist for CCR2 and currently being investigated to be used inflammatory disorder treatment while CCX315 is in phase 1 of clinical trial for the treatment of multiple sclerosis and neurologic disorders. MLN-1202 is a humanized monoclonal antibody which has been investigated for the treatment of atherosclerosis.
Table 5.
Interactivity of the 3 well-known drug compounds of drug bank with the common up-regulated gene CCR2 obtained with the help of Network Analyst tool.
| ID | Label | Degree | Betweenness |
|---|---|---|---|
| 729230 | CCR2 | 3 | 3 |
| DB05130 | INCB3284 | 1 | 0 |
| DB05159 | CCX915 | 1 | 0 |
| DB05486 | MLN-1202/Plozalizumab | 1 | 0 |
Figure 5.

The protein-drug interaction network between hub-protein CCR2 and suggested therapeutics derived from the Network Analyst tool, where the area of each node indicates the degree of interaction. CCR2 indicates C-C chemokine receptor type 2.
Discussion
In this work, publicly accessible RNA-seq data of gonorrhea (GSE110106), chlamydia (GSE180238), and prostate cancer (GSE38241) patients were analyzed to determine potential common biomarkers and molecular targets in all 3 diseases. Primarily, those 3 data sets were examined statistically to categorize DEGs, and then, the DEGs of these data sets were taken into account for further investigation. Gene enrichment analysis found that the identified 22 DEGs were mostly responsible for regulation of amyloid precursor protein catabolic process, regulation of amyloid-beta formation, positive regulation of lymphocyte activation, amyloid-beta binding, endopeptidase inhibitor activity, protease binding, nicotinate and nicotinamide metabolism, glycosaminoglycan biosynthesis, regulation of lipolysis in adipocytes, mineral absorption, and so on (Tables 1 and 2). These DEGs were then investigated using the PPI network, which identified 4 hub proteins: VCL, SH3KBP1, PRNP, and PGRMC1 (Figure 4 and Table 3).
Based on information from gene cards database, 26 protein encoded by VCL gene acts as an interacting protein which is involved in anchoring F acting to the membrane. Defect in this gene could lead to dilated cardiomyopathy,27,28 a condition which leads to congestive heart failure and arrhythmias by ventricular dilation and impaired systolic action. It is also involved in obstructive hypertrophic cardiomyopathy. 29 SH3KBP1 gene encodes an adapter protein which facilitates PPI and is also engaged in several cellular processes including cytoskeleton rearrangement, apoptosis, and cellular adhesion, and disorder associated with this gene includes immunodeficiency. 30 Mutation in PRNP gene is associated with Creutzfeldt-Jakob disease.31,32 PGRMC1 gene encodes a membrane-associated progesterone steroid receptor which is predominantly expressed in liver and kidney. Disease associated with PGRMC1 gene includes pediatric cataract and breast cancer.33,34
As TFs and miRNAs control gene expression at the transcriptional and posttranscriptional levels, alteration in these biomolecules provides fundamental evidence for gene expression dysregulation. So in this work, we investigated common DEG-TF and DEG-miRNA interactions (Tables 3 and 4). Four transcription factors such as POU2F2, POU2F1, GATA6, and HIVEP1 are further examined for determining their association with disease. In accordance with the data from the gene cards database, 26 diseases associated with POU2F2 include B-cell lymphoma and classic Hodgkin lymphoma.35,36 Diseases associated with POU2F1 include herpes simplex and inflammatory bowel disease37,38 while diseases associated with GATA6 gene include congenital diaphragmatic hernia, inherited diabetes mellitus, and pancreatic agenesis.39-41 Mutation in this GATA6 gene also causes human cardiac outflow tract defects 42 and disorders associated with HIVEP1 gene includes immune deficiency diseases. 43
Among miRNAs (hsa-miR-1224-3p, hsa-miR-485-3p, hsa-miR-125a-5p, hsa-miR-7-5p, hsa-miR-5572, hsa-miR-4717-5p, hsa-miR-153-3p, hsa-miR-506-5p, hsa-miR-4518, and hsa-miR-1266-5p), hsa-miR-7-5p may have role in prostate cancer. 44 According to reports, miR-7 is a tumor suppressor that can stop prostate cancer cells from being stem cells and prevents carcinogenesis by inhibiting KLF4. 45 miR-7 also has been proposed as a potential biomarker in prognosis of prostate cancer. 46 Besides this miR-153 also plays a crucial role in cancer, 47 miR-153 has been proposed as a circulating biomarker for prostate cancer diagnosis. 48 In human prostate cancer, upregulation of miR-153 encourages cell proliferation through downregulation of the PTEN (phosphate and tensin homolog) tumor suppressor gene. 49 hsa-miR-1266-5p may be promising candidates for further research in prostate cancer treatment via the anti-apoptotic pathway. 50
For identification of drugs, we analyzed protein-drug interaction using the Network Analyst tool which revealed 3 possible therapeutic compounds, namely INCB3284, CCX915, and MLN-1202 (Figure 5). Several reports have been found that CCR2 protein has involvement with prostate cancer.24,25 According to chEMBL, INCB3284 is an antagonist for CCR2 and currently used to treat inflammatory disorders. It is a selective, potent, and orally bioavailable hCCR2 antagonist. 51 It blocks the stimulation of pro-inflammatory mediators. According to Adis-Insight, 52 CCX915 is a also CCR2 antagonist which is orally available and currently used to treat inflammatory-mediated autoimmune disorders. MLN-1202 is a humanized monoclonal antibody which is designed to treat diabetic nephropathy. It is a G-protein coupled receptor-blocking monoclonal antibody which also has been proposed as a therapeutic agent to treat cancers. 53 These 3 compounds are already in use to treat diseases and have good absorption, distribution, metabolism, and excretion values indicating promising candidates as drug against the gonorrhea, chlamydia, and prostate cancer.
Pathways and other network analysis could provide valuable understanding toward the development of diagnostic and therapeutic approaches. The found biomarkers and therapeutic potential molecules could be investigated for potential pharmacological targets and activity in the fight against patients with gonorrhea, chlamydia, and prostate cancer.
Conclusions
A large number of people are now suffering from different STDs such as gonorrhea and chlamydia all over the world, and previous history of STDs may influence the development of prostate cancer. In this study, we studied RNA-seq gene expression profiles using computational biology methods to find potential biomarkers that could help us in understanding the patho-biological mechanisms of gonorrhea, chlamydia, and prostate cancer. At the transcriptional and posttranscriptional levels, the identified biomolecules may serve as system biomarkers. Additional research may be necessary to confirm the effectiveness and safety of the compounds found in protein-drug interaction networks as potential treatments for patients with gonorrhea, chlamydia, and prostate cancer.
Acknowledgments
The authors acknowledge the Department of Biotechnology and Genetic Engineering, Noakhali Science and Technology University, Bangladesh, for providing support to conduct the research work.
Footnotes
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: MKI conceived and designed the experiments, made critical revisions, and approved the final version. AAN and MKI analyzed the data and wrote the first draft of the manuscript. MKI, AAN, TF, MMH, and MSKS reviewed the analysis and contributed to the preparation of the manuscript. All authors reviewed and approved the final manuscript.
References
- 1. Kirkcaldy RD, Weston E, Segurado AC, Hughes G. Epidemiology of gonorrhoea: a global perspective. Sex Health. 2019;16:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Piszczek J, St Jean R, Khaliq Y. Gonorrhea: treatment update for an increasingly resistant organism. Can Pharm J (Ottawa). 2015;148:82-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Public Health Agency of Canada. Executive summary—report on sexually transmitted infections in Canada: 2009. Government of Canada. Accessed January 16, 2023. https://www.canada.ca/en/public-health/services/publications/diseases-conditions/report-sexually-transmitted-infections-canada-2017.html [Google Scholar]
- 4. Report on sexually transmitted infections in Canada, 2017. Accessed March 6, 2023. https://www.canada.ca/en/public-health/services/infectious-diseases/sexual-health-sexually-transmitted-infections/canadian-guidelines/gonorrhea/risk-factors-clinical-manifestation.html
- 5. Braxton J, Davis D, Emerson B, et al. Sexually transmitted disease surveillance 2017. Published September 2018. https://www.cdc.gov/std/stats17/2017-STD-Surveillance-Report_CDC-clearance-9.10.18.pdf
- 6. Sexually transmitted infections (STIs). Accessed March 6, 2023. https://www.who.int/news-room/fact-sheets/detail/sexually-transmitted-infections-(stis)
- 7. Mohseni M, Sung S, Takov V. Chlamydia. StatPearls; 2019. [PubMed] [Google Scholar]
- 8. O’Connell CM, Ferone ME. Chlamydia trachomatis genital infections. Microb Cell. 2016;3:390-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Newman L, Rowley J, Vander Hoorn S, et al. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS ONE. 2015;10:e0143304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. The Healthline Editorial Team. Everything you need to know about chlamydia infection. Healthline. Accessed January 16, 2023. https://www.healthline.com/health/std/chlamydia
- 11. Key statistics for prostate cancer. Accessed March 3, 2023. https://www.cancer.org/cancer/prostate-cancer/about/key-statistics.html
- 12. Daniyal M, Siddiqui ZA, Akram M, Asif H, Sultana S, Khan A. Epidemiology, etiology, diagnosis and treatment of prostate cancer. Asian Pac J Cancer Prev. 2014;15:9575-9578. [DOI] [PubMed] [Google Scholar]
- 13. Rebbeck TR. Prostate cancer genetics: variation by race, ethnicity, and geography. Semin Radiat Oncol. 2017;27:3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng I, Witte JS, Jacobsen SJ, et al. Prostatitis, sexually transmitted diseases, and prostate cancer: the California Men’s Health Study. PLoS ONE. 2010;5:e8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sellami H, Said-Sadier N, Znazen A, Gdoura R, Ojcius DM, Hammami A. Chlamydia trachomatis infection increases the expression of inflammatory tumorigenic cytokines and chemokines as well as components of the Toll-like receptor and NF-κB pathways in human prostate epithelial cells. Mol Cell Probes. 2014;28:147-154. [DOI] [PubMed] [Google Scholar]
- 16. Charo IF, Myers SJ, Herman A, Franci C, Connolly AJ, Coughlin SR. Molecular cloning and functional expression of two monocyte chemoattractant protein 1 receptors reveals alternative splicing of the carboxyl-terminal tails. Proc Natl Acad Sci USA. 1994;91:2752-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamagami S, Tokuda Y, Ishii K, Tanaka H, Endo N. CDNA cloning and functional expression of a human monocyte chemoattractant protein 1 receptor. Biochem Biophys Res Commun. 1994;202:1156-1162. [DOI] [PubMed] [Google Scholar]
- 18. Tan JH, Ludeman JP, Wedderburn J, et al. Tyrosine sulfation of chemokine receptor CCR2 enhances interactions with both monomeric and dimeric forms of the chemokine monocyte chemoattractant protein-1 (MCP-1). J Biol Chem. 2013;288:10024-10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao J, Liu X, Wei L, et al. Genetic variants of MCP-1 and CCR2 genes and IgA nephropathy risk. Oncotarget. 2016;7:77950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Segerer S, Cui Y, Hudkins KL, et al. Expression of the chemokine monocyte chemoattractant protein-1 and its receptor chemokine receptor 2 in human crescentic glomerulonephritis. J Am Soc Nephrol. 2000;11:2231-2242. [DOI] [PubMed] [Google Scholar]
- 21. D e Lema GP, Maier H, Nieto E, et al. Chemokine expression precedes inflammatory cell infiltration and chemokine receptor and cytokine expression during the initiation of murine lupus nephritis. J Am Soc Nephrol. 2001;12:1369-1382. [DOI] [PubMed] [Google Scholar]
- 22. Kitagawa K, Wada T, Furuichi K, et al. Blockade of CCR2 ameliorates progressive fibrosis in kidney. Am J Pathol. 2004;165:237-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kashyap S, Warner GM, Hartono SP, et al. Blockade of CCR2 reduces macrophage influx and development of chronic renal damage in murine renovascular hypertension. Am J Physiol Renal Physiol. 2016;310:F372-F384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu Y, Cai Z, Xiao G, et al. CCR2 expression correlates with prostate cancer progression. J Cell Biochem. 2007;101:676-685. [DOI] [PubMed] [Google Scholar]
- 25. Zhang J, Patel L, Pienta KJ. Targeting chemokine (CC motif) ligand 2 (CCL2) as an example of translation of cancer molecular biology to the clinic. Prog Mol Biol Transl Sci. 2010;95:31-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. GeneCards—the human gene database. Accessed March 6, 2023. https://www.genecards.org/
- 27. Olson TM, Illenberger S, Kishimoto NY, Huttelmaier S, Keating MT, Jockusch BM. Metavinculin mutations alter actin interaction in dilated cardiomyopathy. Circulation. 2002;105:431-437. [DOI] [PubMed] [Google Scholar]
- 28. Zemljic-Harpf AE, Miller JC, Henderson SA, et al. Cardiac-myocyte-specific excision of the vinculin gene disrupts cellular junctions, causing sudden death or dilated cardiomyopathy. Mol Cell Biol. 2007;27:7522-7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vasile VC, Edwards WD, Ommen SR, Ackerman MJ. Obstructive hypertrophic cardiomyopathy is associated with reduced expression of vinculin in the intercalated disc. Biochem Biophys Res Commun. 2006;349:709-715. [DOI] [PubMed] [Google Scholar]
- 30. Keller B, Shoukier M, Schulz K, et al. Germline deletion of CIN85 in humans with X chromosome–linked antibody deficiency. J Exp Med. 2018;215:1327-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chapman J, Brown P, Rabey J, et al. Transmission of spongiform encephalopathy from a familial Creutzfeldt-Jakob disease patient of Jewish Libyan origin carrying the PRNP codon 200 mutation. Neurology. 1992;42:1249-1250. [DOI] [PubMed] [Google Scholar]
- 32. Goldfarb LG, Brown P, Haltia M, et al. Creutzfeldt-Jakob disease cosegregates with the codon 178Asn PRNP mutation in families of European origin. Ann Neurol. 1992;31:274-281. [DOI] [PubMed] [Google Scholar]
- 33. Jones JL, Corbett MA, Yeaman E, et al. A 127 kb truncating deletion of PGRMC1 is a novel cause of X-linked isolated paediatric cataract. Eur J Hum Genet. 2021;29:1206-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ahmed IS, Rohe HJ, Twist KE, Mattingly MN, Craven RJ. Progesterone receptor membrane component 1 (Pgrmc1): a heme-1 domain protein that promotes tumorigenesis and is inhibited by a small molecule. J Pharmacol Exp Ther. 2010;333:564-573. [DOI] [PubMed] [Google Scholar]
- 35. Hoefnagel JJ, Mulder MM, Dreef E, et al. Expression of B-cell transcription factors in primary cutaneous B-cell lymphoma. Mod Pathol. 2006;19:1270-1276. [DOI] [PubMed] [Google Scholar]
- 36. Browne P, Petrosyan K, Hernandez A, Chan JA. The B-cell transcription factors BSAP, Oct-2, and BOB.1 and the pan–B-cell markers CD20, CD22, and CD79a are useful in the differential diagnosis of classic Hodgkin lymphoma. Am J Clin Pathol. 2003;120:767-777 [DOI] [PubMed] [Google Scholar]
- 37. Cleary MA, Herr W. Mechanisms for flexibility in DNA sequence recognition and VP16-induced complex formation by the Oct-1 POU domain. Mol Cell Biol. 1995;15:2090-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Van Heel DA, Udalova IA, De Silva AP, et al. Inflammatory bowel disease is associated with a TNF polymorphism that affects an interaction between the OCT1 and NF-κB transcription factors. Hum Mol Genet. 2002;11:1281-1289. [DOI] [PubMed] [Google Scholar]
- 39. Yu L, Bennett JT, Wynn J, et al. Whole exome sequencing identifies de novo mutations in GATA6 associated with congenital diaphragmatic hernia. J Med Genet. 2014;51:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yorifuji T, Kawakita R, Hosokawa Y, Fujimaru R, Yamaguchi E, Tamagawa N. Dominantly inherited diabetes mellitus caused by GATA6 haploinsufficiency: variable intrafamilial presentation. J Med Genet. 2012;49:642-643. [DOI] [PubMed] [Google Scholar]
- 41. Allen HL, Flanagan SE, Shaw-Smith C, et al. GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nat Genet. 2012;44:20-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kodo K, Nishizawa T, Furutani M, et al. GATA6 mutations cause human cardiac outflow tract defects by disrupting semaphorin-plexin signaling. Proc Natl Acad Sci USA. 2009;106:13933-13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Muchardt C, Seeler J-S, Nirula A, Shurland D, Gaynor R. Regulation of human immunodeficiency virus enhancer function by PRDII-BF1 and c-rel gene products. J Virol. 1992;66:244-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morales-Martínez M, Vega MI. Role of microRNA-7 (MiR-7) in cancer physiopathology. Int J Mol Sci. 2022;23:9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chang Y-L, Zhou P-J, Wei L, et al. MicroRNA-7 inhibits the stemness of prostate cancer stem-like cells and tumorigenesis by repressing KLF4/PI3K/Akt/p21 pathway. Oncotarget. 2015;6:24017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Santos JI, Teixeira AL, Dias F, et al. Influence of peripheral whole-blood microRNA-7 and microRNA-221 high expression levels on the acquisition of castration-resistant prostate cancer: evidences from in vitro and in vivo studies. Tumour Biol. 2014;35:7105-7113. [DOI] [PubMed] [Google Scholar]
- 47. Yousefnia S. A comprehensive review on miR-153: mechanistic and controversial roles of miR-153 in tumorigenicity of cancer cells. Front Oncol. 2022;12:985897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bertoli G, Panio A, Cava C, et al. Secreted miR-153 Controls proliferation and invasion of higher Gleason score prostate cancer. Int J Mol Sci. 2022;23:6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu Z, He B, He J, Mao X. Upregulation of miR-153 promotes cell proliferation via downregulation of the PTEN tumor suppressor gene in human prostate cancer. Prostate. 2013;73:596-604. [DOI] [PubMed] [Google Scholar]
- 50. Ostadrahimi S, Valugerdi MA, Hassan M, et al. MiR-1266-5p and miR-185-5p promote cell apoptosis in human prostate cancer cell lines. Asian Pac J Cancer Prev. 2018;19:2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xue C-B, Feng H, Cao G, et al. Discovery of INCB3284, a potent, selective, and orally bioavailable hCCR2 antagonist. ACS Med Chem Lett. 2011;2:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Adis Insight. Accessed March 6, 2023. https://adisinsight.springer.com/drugs/800035649
- 53. Herr DR. Potential use of G protein-coupled receptor-blocking monoclonal antibodies as therapeutic agents for cancers. Int Rev Cell Mol Biol. 2012;297:45-81. [DOI] [PubMed] [Google Scholar]