Abstract
We report here the development of two different sensing strategies based on the use of antigen-conjugated nucleic acid strands for the detection of a bispecific antibody against the tumor-related proteins Mucin1 and epidermal growth factor receptor. Both approaches work well in serum samples (nanomolar sensitivity), show high specificity against the two monospecific antibodies, and are rapid. The results presented here demonstrate the versatility of DNA-based platforms for the detection of bispecific antibodies and could represent a versatile alternative to other more reagent-intensive and time-consuming analytical approaches.
Keywords: bispecific antibody, antibody detection, synthetic nucleic acid, DNA nanotechnology
Therapeutic antibodies have emerged over the past two decades as innovative drugs for the treatment of various diseases.1−3 Since the first Food and Drug Administration (FDA) approval of rituximab, a monoclonal antibody targeting the CD20 protein, more than 100 antibodies have been approved as drugs for the treatment of various diseases, such as cancer, autoimmune diseases, and chronic inflammation, showing significant response and long-term benefit.4−7 Advances in antibody technology and biology have recently led to the development of novel antibody formats to create therapeutic drugs with better efficacy.1,8−11 In this regard, bispecific antibodies (BsAbs) which, unlike monoclonal antibodies (mAbs), can recognize and block two distinct epitopes on the cell surface, represent the most promising direction for future cancer immunotherapy development.12−14 BsAbs are engineered immunoglobulins produced in vitro by biochemical, biological, or genetic processes.15−19 The bifunctional binding ability of BsAbs enables higher tumor specificity than mAb, ultimately leading to better therapeutic efficacy.20−22
New methods for detecting bispecific antibodies in clinical fluids are increasingly needed to characterize their pharmacokinetics and toxicokinetics and to find the best conditions for their efficacy.23−26 Current methods for detecting bispecific antibodies are mostly laboratory-based approaches used for structural and binding characterization of BsAbs. For example, several assays based on the adaptation of an enzyme-linked immunosorbent assay (ELISA) have been described.27 These assays use a “bridging” format in which one recombinant antigen is immobilized on a solid phase and a biotinylated version of the second antigen is added to form a ternary complex with the target antibody.12,20,28−33 This approach allows for highly sensitive BsAb detection but requires multiple washing and reaction steps, ultimately increasing the cost and reaction time. Recently, two surface-based approaches were also proposed. In one, a surface plasmon resonance (SPR)-based method that provides real-time information on the kinetics and affinity of the BsAb/antigen interaction in a label-free format was described.28,31 In another example, a chip with a Y-shaped DNA nanostructure labeled with two antigens and two optical dyes was used to characterize the binding properties of BsAb.34 While both systems provide excellent sensitivity to the target BsAb and appear to be suitable for characterizing BsAb binding, the complexity of the instrumentation required makes them less suitable for point-of-care detection.
In recent years, we and other research groups have reported several DNA-based devices that use synthetic antigen-conjugated strands for the optical detection of a wide range of monoclonal target antibodies.35 Despite the many advantages provided by these platforms (i.e., sensitivity, versatility, specificity, etc.), their possible use for the detection of bispecific antibodies has not yet been reported. Motivated by the above considerations, we demonstrate here the use of antigen-conjugated synthetic nucleic acid strands to develop two general platforms for the rapid, sensitive, inexpensive, and quantitative detection of BsAbs.
Results and Discussion
In this work we selected as a model BsAb an antibody that is engineered to recognize with one arm (Single-chain variable fragment, scFv) the tumor-associated Mucin1 (MUC1) protein and, with the second arm (Fragment antigen-binding, Fab), the epidermal growth factor receptor (EGFR) (Figure 1A).14 To develop a DNA-based platform for the detection of this antibody, we first need to conjugate the relevant antigens to two synthetic nucleic acid strands. First, we conjugated the human EGFRvIII protein to the 5′ end of a synthetic 27-nt dibenzocyclooctyne (DBCO)-modified DNA strand. To do so, we used a 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide N-hydroxysuccinimide (EDC-NHS) ester coupling reaction (see also Supporting Information) followed by purification with ion exchange chromatography (Figure 1B). As antigen for the second binding site (targeting MUC1), we chose a 15 amino acid long exposed epitope recognized in tumor-associated MUC1 by Anti-MUC1 antibodies.36 Peptides are generally more difficult to conjugate to DNA strands and would lead to a more laborious purification of the conjugate. For this reason, we chose to conjugate the MUC1 peptide to a peptide nucleic acid (PNA) strand that, thanks to its pseudopeptide backbone, allows for easier conjugation while maintaining the same DNA sequencing ability. Specifically, the peptide residue at the N-terminus was conjugated to an 18-nt PNA strand by forming an amide bond by using a solid-phase synthesis method.
Figure 1.
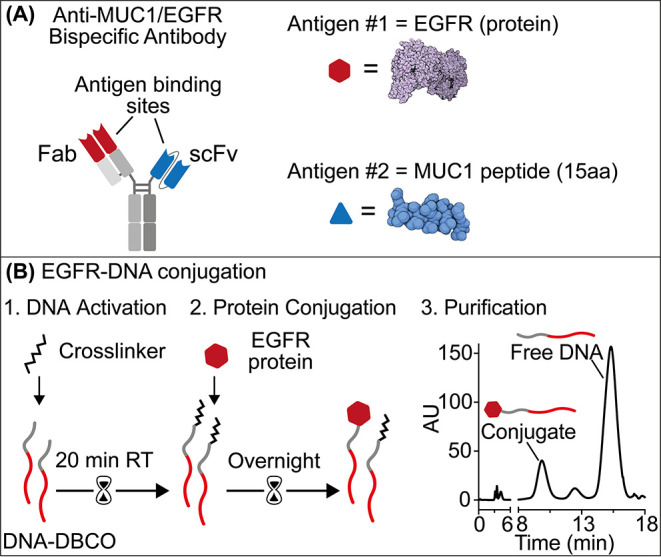
(A) General schematic of the bispecific Anti-MUC1/EGFR antibody. (B) Schematic representation of the reaction for conjugation of EGFR to a DNA strand and purification.
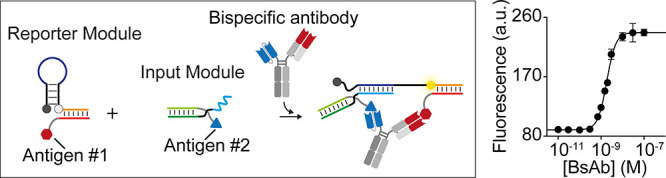
Using the antigen-conjugated nucleic acid strands described above, we set out to demonstrate two possible strategies for BsAb detection. In the first one we adapted an approach recently described by our group for monoclonal antibody detection based on antibody-induced colocalization of antigen-conjugated nucleic acid strands.37 The system comprises two modules: a reporter module and an input module. The reporter module is a duplex DNA obtained by hybridization of a fluorophore/quencher-modified hairpin DNA strand flanking a 15-base single strand and the EGFR-conjugated DNA strand described above (Figure 2A). The input module is instead a duplex of a DNA strand with a portion complementary to the loop of the hairpin DNA strand and the MUC1 peptide-conjugated PNA strand (Figure 2A). Bivalent binding of BsAb to the antigen-conjugated strands colocalizes the reporter and input modules, thus increasing their local concentration and enabling their hybridization (Figure 2A). Such antibody-induced hybridization triggers the opening of the stem-loop structure and enhances the fluorescence as a function of the antibody concentration.
Figure 2.
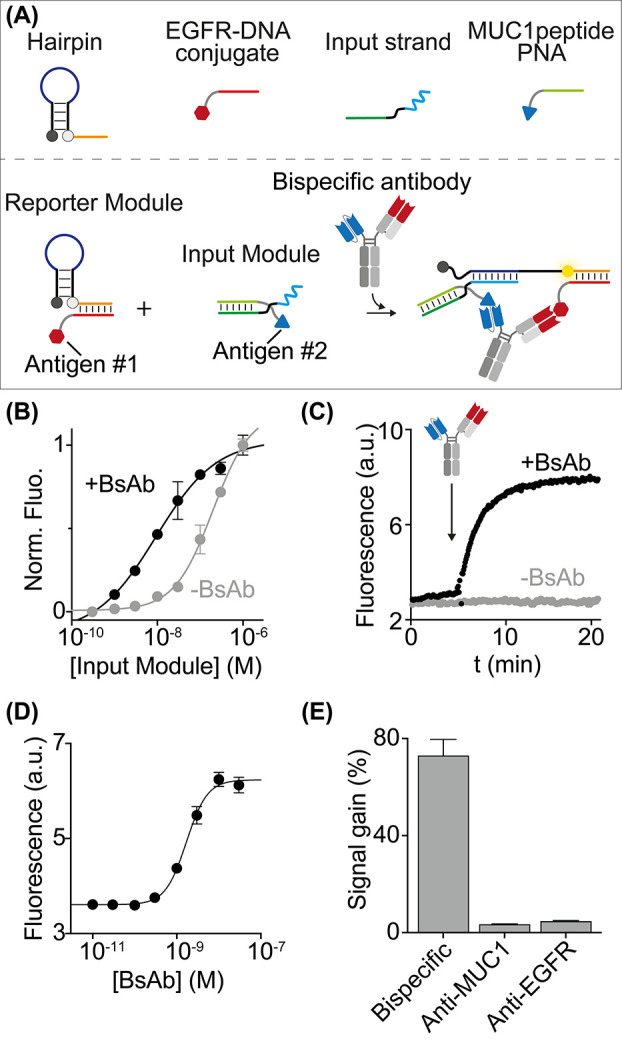
(A) Schematic of the antigen-conjugated nucleic acid strands platform. (B) Dose–response curve (fit eq (1)) obtained by adding an increasing concentration of the input module to a fixed (10 nM) concentration of the reporter module in the absence and presence of a saturating concentration (100 nM) of BsAb. (C) Fluorescence kinetic traces obtained in the absence (gray) and presence (black) of BsAb (100 nM) at a fixed concentration of reporter module (10 nM) and input module (30 nM). (D) Dose–response curve (fit eq (1)) at increasing concentration of BsAb. (E) Signal gain values (fit eq (3)) obtained at a saturating concentration (100 nM) of the BsAb and the two related monoclonal antibodies. Experiments were performed in 20 μL of 10 mM Na2HPO4, 137 mM NaCl, and 2.7 mM KCl at pH 7.4 at 25 °C. Experimental values in this and the following figures represent averages of three separate measurements, and error bars reflect standard deviations.
To optimize the platform’s performance for BsAb detection and maximize its signal response, we first generated binding curves by adding increasing concentrations of the input module at a fixed concentration (10 nM) of the reporter module in the absence and presence of a saturating concentration of BsAb (100 nM). In the presence of the target antibody, we observe an increase in binding affinity (K1/2 (−BsAb) = 195 ± 2 nM and K1/2 (+BsAb) = 9.4 ± 0.4 nM), supporting the hypothesis that antibody-induced colocalization is critical for the sensing mechanism (Figure 2B). We find that a concentration of 30 nM of the input module leads to the largest difference in fluorescence signal between the absence and presence of BsAb (Figure S1). Using the optimized concentrations of the reporter module (i.e., 30 nM) and the input module (i.e., 10 nM), we tested the platform to detect the BsAb. Since it is a direct approach, no washing or multiple reaction steps are required, and we observe signal saturation after addition of the BsAb after approximately 30 min (Figure 2C). The platform proves to be sensitive (K1/2 = 1.7 ± 0.4 nM) with a calculated detection limit (defined here as the antibody concentration that gives a signal equal to the blank value plus three standard deviation) of 0.6 nM (Figure 2D). The presence of the two related monoclonal antibodies causes only a minimal signal (2.7% for Anti-MUC1 and 3.3% for Anti-EGFR) (Figure 2E).
Comparable results in terms of sensitivity and specificity were obtained with platforms designed for the detection of the two monoclonal antibodies using the same recognition element in the two modules (Figures S2 & S3).
To demonstrate the potential application of our sensing platform at the point-of-care, we adapted the antibody detection measurements to the format of a plate reader (Figure 3A). Using the plate reader format, the platform confirmed sensitive detection of BsAb directly in a 50% plasma sample (K1/2 = 1.5 ± 0.2 nM; limit of detection (LOD) = 0.4 nM) (Figure 3B). The analytical performance of our platform was evaluated by spiking different matrix samples (buffer solution, 10% plasma, and 50% plasma) with five known BsAb concentrations (0.7, 1, 1.5, 2, 3 nM; n = 3 for each concentration) during intrarun experiments. A BIAS% (or systematic error), defined as the difference between the expected result and the true value, less than +15% was obtained with these experiments. A good correlation (within ±20% error) between spiked and measured BsAb concentration in the linear range was also observed (Figures 3C & S4). The CV% (Percent Coefficient of Variation), defined as the agreement between independent measurements and the precision obtained by our method, was <3% (see also Supporting Information for analytical characterization). Finally, the lowest tested BsAb concentration determined with acceptable accuracy and precision, defined as the Low Limit of Quantification (LLOQ), was 0.7 nM.
Figure 3.
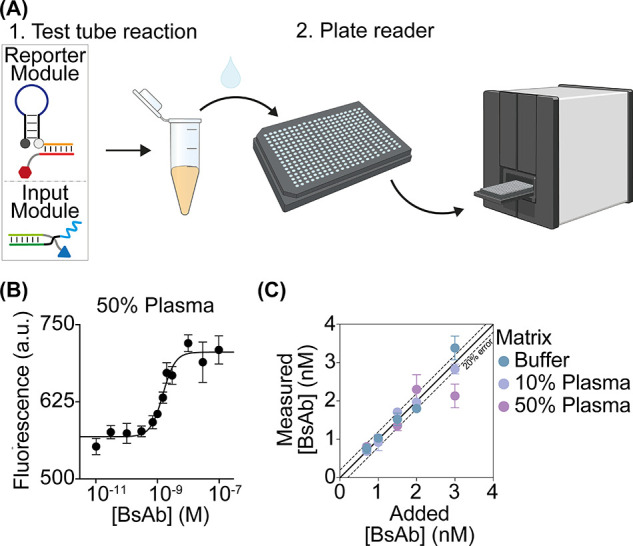
(A) Schematic representation of the plate reader platform for BsAb detection. (B) Fluorescence signals in a 50% plasma solution spiked with increasing concentrations of BsAb (fit eq (1)). (C) Correlation between added (0.7, 1, 1.5, 2, 3 nM) and measured BsAb concentrations (fit eq (4)) in different matrix samples (buffer solution, 10% plasma, and 50% plasma). Experiments were performed in a 20 μL solution containing 10 mM Na2HPO4, 137 mM NaCl, and 2.7 mM KCl, pH 7.4, containing the reporter module (10 nM), the input module (30 nM), and the BsAb at the indicated concentration.
We also evaluated the stability of the method by calculating the percent stability (STAB%), defined as the ratio between the measured concentration and the added concentration after storage of the platform under different conditions. Short-term (or benchtop) stability was evaluated by testing the BsAb concentrations of the linear range after the platform components were stored at room temperature (RT) for 4 h. Instead, freeze–thaw stability was evaluated by testing these concentrations after the components were stored at −80 °C for at least 12 h and then thawed three times. The results showed that the method is stable with an STAB% between 80 and 120%.
Because antigen-conjugated synthetic nucleic acid strands are programmable, they can be used to detect BsAb via a variety of mechanisms. To demonstrate this, we employed a second strategy for the detection of the same bispecific target antibody. The approach employs an antibody-induced strand displacement reaction previously demonstrated for monospecific antibodies.38,39 Specifically, this strategy uses a target duplex labeled with a fluorophore/quencher pair and two unmodified scaffold strands (split #1 and #2, Figure 4A) that can hybridize to the antigen-conjugated DNA strands. The scaffold strands consist of three sections: (i) a complementary sequence to the MUC1 peptide-PNA or EGFR-DNA conjugates; (ii) a stem-forming section (black); and (iii) a toehold or invasion sequence required to activate the strand displacement reaction. Binding of BsAb to the two recognition elements induces colocalization of the bimolecular complexes (split #1/EGFR-DNA conjugate and split #2/MUC1 peptide-PNA conjugate) and promotes hybridization between the stem-forming portions leading to activation of the strand displacement reaction. This induces the release of the fluorophore-labeled reporter strand and a subsequent increase in the measured fluorescence signal. This strategy could, in principle, allow one to reduce possible non-specific interactions in absence of the target antibody, enabling a better optimization of the signal-to-noise ratio.
Figure 4.
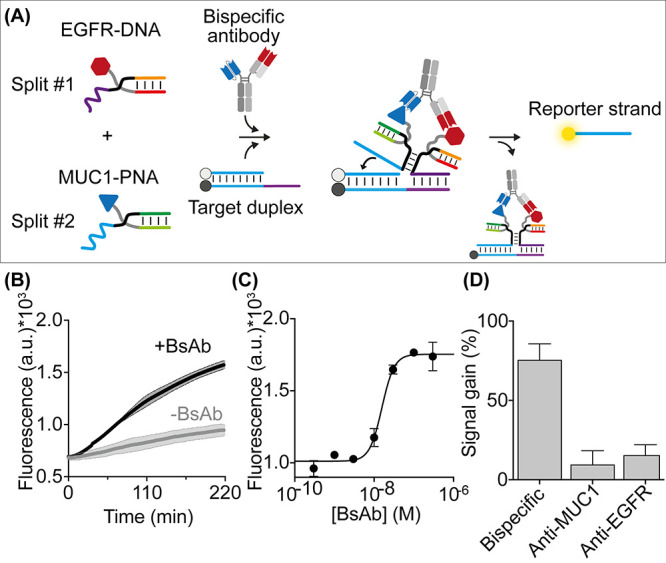
(A) Antibody-responsive strand displacement reaction for the detection of BsAb. (B) Fluorescence kinetic traces obtained in the absence (gray) and presence (black) of BsAb. (C) Fluorescence values in a 50% plasma solution supplemented with increasing BsAb concentrations (fit eq (1)). (D) Signal gain values (fit eq (3)) at a saturating concentration (100 nM) of BsAb and the two related monoclonal antibodies. Experiments were performed in 20 μL of 50 mM Na2HPO4, 150 mM NaCl, pH 7.0 containing the DNA target duplex (60 nM), split #1 + #2 (both at 100 nM), EGFR-DNA conjugate and MUC1-PNA conjugate (both at 120 nM), and BsAb at the indicated concentration.
As a first step toward the characterization of the Ab-induced colocalization, we designed a bivalent DNA strand that acts as an Ab mimic and binds the first portion of Split #1 and #2, inducing a similar colocalization to that expected from the binding of a bivalent antibody (Figure S5). We then used our platform to detect the BsAb in a 50% plasma solution. The presence of the antibody efficiently induces a strand displacement reaction in a concentration-dependent manner (K1/2 = 15 ± 1 nM; LOD = 8 nM) (Figure 4B,C). The overall reaction efficiency increases with BsAb concentration until it saturates at about 100 nM. BsAb detection is highly specific, as no significant fluorescence signals are observed at saturating concentrations of the related monoclonal antibodies (Anti-MUC1 and Anti-EGFR antibodies) (Figure 4D).
Conclusions
In the present study, we have demonstrated two different sensing platforms that employ antigen-conjugated nucleic acid strands for the detection of a bispecific antibody against the tumor-related proteins Mucin1 and EGFR. The systems we developed can efficiently detect the target BsAb with high sensitivity, specificity (no significant activation with monospecific antibodies was observed), and good selectivity in complex sample matrices (plasma). They also present advantageous features in comparison with standard methods such as ELISA that make them suitable for point-of-care applications. In particular, both platforms developed in this work, as also other DNA-based systems for antibodies detection reported recently by our and other research groups,35 are rapid and cost-effective and can be easily adapted to detect other therapeutic bispecific antibodies.40 For a more detailed comparison between our approaches, other DNA-based sensors, and ELISA we refer to our recent Perspective published in this journal.35
The development of similar DNA-based point-of-care methods for the detection of therapeutic bispecific antibodies would improve the characterization and monitoring of immunotherapies, thereby increasing their efficacy. A possible limitation of the approaches we described in this work is that, like the majority of analytical systems that do not rely on an amplification step, they cannot achieve the sensitivity of other amplification-based antibody detection methods such as ELISA.33 For example, in this work we achieve sensitivities in the low nanomolar level, which is in the same order of the plasma level expected in patients treated with therapeutic antibodies.41 But for applications in which the level of the target is expected to be below the nanomolar sensitivity reached here, an additional amplification step should be added. This could be achieved through, for example, the use of enzymes or nonenzymatic reaction cascades.42,43
Acknowledgments
The work was supported by the Innovative Analytics Technologies project funded by Merck (to F.R.).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssensors.3c01717.
Reagents and materials, oligonucleotide sequences, experimental details; Kinetic traces, binding curves, and specificity experiments with Anti-MUC1 and Anti-EGFR monoclonal antibodies; Binding curves in buffer, 10% and 50% plasma samples with Bispecific antibody; Binding curve with Ab mimic strand (PDF)
Author Present Address
⊥ Boehringer Ingelheim Pharma GmbH & Co. KG, Birkendorfer Str. 65, 88400, Biberach an der Riß, Germany
Author Contributions
∥ These authors contributed equally. DM and SB designed and performed all experiments. SB, SR, FR, LB, SWF, CS, SM, and CK wrote the manuscript. SR and FR supervised the research.
The authors declare the following competing financial interest(s): The work was supported by the Innovative Analytics Technologies project funded by Merck.
Supplementary Material
References
- Beck A.; Wurch T.; Bailly C.; Corvaia N. Strategies and Challenges for the next Generation of Therapeutic Antibodies. Nat. Rev. Immunol. 2010, 10 (5), 345–352. 10.1038/nri2747. [DOI] [PubMed] [Google Scholar]
- Leavy O. Therapeutic Antibodies: Past, Present and Future. Nat. Rev. Immunol. 2010, 10 (5), 297–297. 10.1038/nri2763. [DOI] [PubMed] [Google Scholar]
- Lu R.-M.; Hwang Y.-C.; Liu I.-J.; Lee C.-C.; Tsai H.-Z.; Li H.-J.; Wu H.-C. Development of Therapeutic Antibodies for the Treatment of Diseases. J. Biomed. Sci. 2020, 27 (1), 1. 10.1186/s12929-019-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyiadzis M.; Foon K. A. Approved Monoclonal Antibodies for Cancer Therapy. Expert Opin. Biol. Ther. 2008, 8 (8), 1151–1158. 10.1517/14712598.8.8.1151. [DOI] [PubMed] [Google Scholar]
- Jahanshahlu L.; Rezaei N. Monoclonal Antibody as a Potential Anti-COVID-19. Biomed. Pharmacother. 2020, 129, 110337. 10.1016/j.biopha.2020.110337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A. C.; Carter P. J. Therapeutic Antibodies for Autoimmunity and Inflammation. Nat. Rev. Immunol. 2010, 10 (5), 301–316. 10.1038/nri2761. [DOI] [PubMed] [Google Scholar]
- Melero I.; Hervas-Stubbs S.; Glennie M.; Pardoll D. M.; Chen L. Immunostimulatory Monoclonal Antibodies for Cancer Therapy. Nat. Rev. Cancer 2007, 7 (2), 95–106. 10.1038/nrc2051. [DOI] [PubMed] [Google Scholar]
- Norman R. A.; Ambrosetti F.; Bonvin A. M. J. J.; Colwell L. J.; Kelm S.; Kumar S.; Krawczyk K. Computational Approaches to Therapeutic Antibody Design: Established Methods and Emerging Trends. Brief. Bioinform. 2020, 21 (5), 1549–1567. 10.1093/bib/bbz095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presta L. G. Molecular Engineering and Design of Therapeutic Antibodies. Curr. Opin. Immunol. 2008, 20 (4), 460–470. 10.1016/j.coi.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Tir N.; Heistinger L.; Grünwald-Gruber C.; Jakob L. A.; Dickgiesser S.; Rasche N.; Mattanovich D. From Strain Engineering to Process Development: Monoclonal Antibody Production with an Unnatural Amino Acid in Pichia Pastoris. Microb. Cell Factories 2022, 21 (1), 157. 10.1186/s12934-022-01882-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. K.; Maynard J. A. Engineering Therapeutic Antibodies to Combat Infectious Diseases. Curr. Opin. Chem. Eng. 2018, 19, 131–141. 10.1016/j.coche.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne H.; Conroy P. J.; Whisstock J. C.; O’Kennedy R. J. A Tale of Two Specificities: Bispecific Antibodies for Therapeutic and Diagnostic Applications. Trends Biotechnol. 2013, 31 (11), 621–632. 10.1016/j.tibtech.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlén E.; Veitonmäki N.; Norlén P. Bispecific Antibodies in Cancer Immunotherapy. Ther. Adv. Vaccines Immunother. 2018, 6 (1), 3–17. 10.1177/2515135518763280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk A.-L.; Mebrahtu A.; Ko B.-K.; Lundqvist M.; Karlander M.; Lee H.-J.; Frejd F. Y.; Kim K.-T.; Lee J.-S.; Rockberg J. Bispecific Antibody Molecule Inhibits Tumor Cell Proliferation More Efficiently Than the Two-Molecule Combination. Drugs RD 2021, 21 (2), 157–168. 10.1007/s40268-021-00339-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein C.; Cuello A. C. Hybrid Hybridomas and Their Use in Immunohistochemistry. Nature 1983, 305 (5934), 537–540. 10.1038/305537a0. [DOI] [PubMed] [Google Scholar]
- Labrijn A. F.; Janmaat M. L.; Reichert J. M.; Parren P. W. H. I. Bispecific Antibodies: A Mechanistic Review of the Pipeline. Nat. Rev. Drug Discovery 2019, 18 (8), 585–608. 10.1038/s41573-019-0028-1. [DOI] [PubMed] [Google Scholar]
- Cao M.; Parthemore C.; Jiao Y.; Korman S.; Aspelund M.; Hunter A.; Kilby G.; Chen X. Characterization and Monitoring of a Novel Light-Heavy-Light Chain Mispair in a Therapeutic Bispecific Antibody. J. Pharm. Sci. 2021, 110 (8), 2904–2915. 10.1016/j.xphs.2021.04.010. [DOI] [PubMed] [Google Scholar]
- Wang C.; Vemulapalli B.; Cao M.; Gadre D.; Wang J.; Hunter A.; Wang X.; Liu D. A Systematic Approach for Analysis and Characterization of Mispairing in Bispecific Antibodies with Asymmetric Architecture. mAbs 2018, 10 (8), 1226–1235. 10.1080/19420862.2018.1511198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann U.; Kontermann R. E. The Making of Bispecific Antibodies. mAbs 2017, 9 (2), 182–212. 10.1080/19420862.2016.1268307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-L.; Cui Y.; Liu X.; Liu G.; Dong X.; Tang L.; Hung Y.; Wang C.; Feng M.-Q. A Bispecific Antibody Targeting HER2 and PD-L1 Inhibits Tumor Growth with Superior Efficacy. J. Biol. Chem. 2021, 297 (6), 101420. 10.1016/j.jbc.2021.101420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.; van Duijnhoven S. M. J.; Sijts A. J. A. M.; van Elsas A. Bispecific Antibodies Targeting Dual Tumor-Associated Antigens in Cancer Therapy. J. Cancer Res. Clin. Oncol. 2020, 146 (12), 3111–3122. 10.1007/s00432-020-03404-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Chen K.; Lei Q.; Ma P.; Yuan A. Q.; Zhao Y.; Jiang Y.; Fang H.; Xing S.; Fang Y.; Jiang N.; Miao H.; Zhang M.; Sun S.; Yu Z.; Tao W.; Zhu Q.; Nie Y.; Li N. The State of the Art of Bispecific Antibodies for Treating Human Malignancies. EMBO Mol. Med. 2021, 13 (9), e14291. 10.15252/emmm.202114291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann M. D.; Bivi N.; Calderon B.; Shimizu T.; Delafontaine B.; Liu Z. T.; Szpurka A. M.; Copeland V.; Hodi F. S.; Rottey S.; Aftimos P.; Piao Y.; Gandhi L.; Galvao V. R.; Leow C. C.; Doi T. Safety and Immunogenicity of LY3415244, a Bispecific Antibody Against TIM-3 and PD-L1, in Patients With Advanced Solid Tumors. Clin. Cancer Res. 2021, 27 (10), 2773–2781. 10.1158/1078-0432.CCR-20-3716. [DOI] [PubMed] [Google Scholar]
- Weiner L. M.; Murray J. C.; Shuptrine C. W. Antibody-Based Immunotherapy of Cancer. Cell 2012, 148 (6), 1081–1084. 10.1016/j.cell.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine D. J.; Dane E. L. Enhancing Cancer Immunotherapy with Nanomedicine. Nat. Rev. Immunol. 2020, 20 (5), 321–334. 10.1038/s41577-019-0269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centanni M.; Moes D. J. A. R.; Trocóniz I. F.; Ciccolini J.; van Hasselt J. G. C. Clinical Pharmacokinetics and Pharmacodynamics of Immune Checkpoint Inhibitors. Clin. Pharmacokinet. 2019, 58 (7), 835–857. 10.1007/s40262-019-00748-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. Y.; Schaefer G.; Lesaca I.; Lee C. V.; Wong P. Y.; Jiang G. Two-in-One” Approach for Bioassay Selection for Dual Specificity Antibodies. J. Immunol. Methods 2017, 448, 74–79. 10.1016/j.jim.2017.05.011. [DOI] [PubMed] [Google Scholar]
- Meschendoerfer W.; Gassner C.; Lipsmeier F.; Regula J. T.; Moelleken J. SPR-Based Assays Enable the Full Functional Analysis of Bispecific Molecules. J. Pharm. Biomed. Anal. 2017, 132, 141–147. 10.1016/j.jpba.2016.09.028. [DOI] [PubMed] [Google Scholar]
- Ljungars A.; Schiött T.; Mattson U.; Steppa J.; Hambe B.; Semmrich M.; Ohlin M.; Tornberg U.-C.; Mattsson M. A Bispecific IgG Format Containing Four Independent Antigen Binding Sites. Sci. Rep. 2020, 10 (1), 1546. 10.1038/s41598-020-58150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian E. A.; Hussmann G. P.; Babu M.; Prophet M.; Mazor Y.; Chen W.; Grigoriadou C.; Lin S. A Single Homogeneous Assay for Simultaneous Measurement of Bispecific Antibody Target Binding. J. Immunol. Methods 2021, 496, 113099. 10.1016/j.jim.2021.113099. [DOI] [PubMed] [Google Scholar]
- Gassner C.; Lipsmeier F.; Metzger P.; Beck H.; Schnueriger A.; Regula J. T.; Moelleken J. Development and Validation of a Novel SPR-Based Assay Principle for Bispecific Molecules. J. Pharm. Biomed. Anal. 2015, 102, 144–149. 10.1016/j.jpba.2014.09.007. [DOI] [PubMed] [Google Scholar]
- Pei M.; Wang Y.; Tang L.; Wu W.; Wang C.; Chen Y.-L. Dual-Target Bridging ELISA for Bispecific Antibodies. Bio-Protoc 2022, 12 (19), e4522. 10.21769/BioProtoc.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C.; Zhu H.; Deng L.; Meng X.; Li K.; Xu W.; Zhao L.; Liu Y.; Zhu Z.; Huang H. Bispecific Antibody Simultaneously Targeting PD1 and HER2 Inhibits Tumor Growth via Direct Tumor Cell Killing in Combination with PD1/PDL1 Blockade and HER2 Inhibition. Acta Pharmacol. Sin. 2022, 43 (3), 672–680. 10.1038/s41401-021-00683-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak S.; Marszal A.; Matscheko N.; Rant U. Kinetic Analysis of Ternary and Binary Binding Modes of the Bispecific Antibody Emicizumab. mAbs 2023, 15 (1), 2149053. 10.1080/19420862.2022.2149053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo S.; Bracaglia S.; Sorrentino D.; Ricci F. Synthetic Antigen-Conjugated DNA Systems for Antibody Detection and Characterization. ACS Sens 2023, 8, 2415. 10.1021/acssensors.3c00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Sáez N.; Peregrina J. M.; Corzana F. Principles of Mucin Structure: Implications for the Rational Design of Cancer Vaccines Derived from MUC1-Glycopeptides. Chem. Soc. Rev. 2017, 46 (23), 7154–7175. 10.1039/C6CS00858E. [DOI] [PubMed] [Google Scholar]
- Porchetta A.; Ippodrino R.; Marini B.; Caruso A.; Caccuri F.; Ricci F. Programmable Nucleic Acid Nanoswitches for the Rapid, Single-Step Detection of Antibodies in Bodily Fluids. J. Am. Chem. Soc. 2018, 140 (3), 947–953. 10.1021/jacs.7b09347. [DOI] [PubMed] [Google Scholar]
- Bracaglia S.; Ranallo S.; Plaxco K. W.; Ricci F. Programmable, Multiplexed DNA Circuits Supporting Clinically Relevant, Electrochemical Antibody Detection. ACS Sens. 2021, 6 (6), 2442–2448. 10.1021/acssensors.1c00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo S.; Sorrentino D.; Ricci F. Orthogonal Regulation of DNA Nanostructure Self-Assembly and Disassembly Using Antibodies. Nat. Commun. 2019, 10 (1), 5509. 10.1038/s41467-019-13104-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H. Bispecific Antibodies and Antibody-Drug Conjugates for Cancer Therapy: Technological Considerations. Biomolecules 2020, 10 (3), 360. 10.3390/biom10030360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekrul I.; Pfrepper C.; Calatzis G.; Giebl A.; Siegemund A.; Grützner S.; Spannagl M. Approximation of Emicizumab Plasma Levels in Emergency Situations. A Practical Approach. Haemophilia 2021, 27 (2), e214–e220. 10.1111/hae.14264. [DOI] [PubMed] [Google Scholar]
- Tsai C.; Robinson P. V.; Spencer C. A.; Bertozzi C. R. Ultrasensitive Antibody Detection by Agglutination-PCR (ADAP). ACS Cent. Sci. 2016, 2 (3), 139–147. 10.1021/acscentsci.5b00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N.; Liu L.; Xiang M.-H.; Liu J.-W.; Yu R.-Q.; Jiang J.-H. Proximity-Induced Hybridization Chain Assembly with Small-Molecule Linked DNA for Single-Step Amplified Detection of Antibodies. Chem. Commun. 2019, 55 (30), 4387–4390. 10.1039/C9CC01654F. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


