Abstract
In the past decade, the frequency of diagnosed fungal infections has risen sharply due to several factors, including the increase in the number of immunosuppressed patients resulting from the AIDS epidemic and treatments during and after organ and bone marrow transplants. Linked with the increase in fungal infections is a recent increase in the frequency with which these infections are recalcitrant to standard antifungal therapy. This review summarizes the factors that contribute to antifungal drug resistance on three levels: (i) clinical factors that result in the inability to successfully treat refractory disease; (ii) cellular factors associated with a resistant fungal strain; and (iii) molecular factors that are ultimately responsible for the resistance phenotype in the cell. Many of the clinical factors that contribute to resistance are associated with the immune status of the patient, with the pharmacology of the drugs, or with the degree or type of fungal infection present. At a cellular level, antifungal drug resistance can be the result of replacement of a susceptible strain with a more resistant strain or species or the alteration of an endogenous strain (by mutation or gene expression) to a resistant phenotype. The molecular mechanisms of resistance that have been identified to date in Candida albicans include overexpression of two types of efflux pumps, overexpression or mutation of the target enzyme, and alteration of other enzymes in the same biosynthetic pathway as the target enzyme. Since the study of antifungal drug resistance is relatively new, other factors that may also contribute to resistance are discussed.
In the 1990s, drug resistance has become an important problem in a variety of infectious diseases including human immunodeficiency virus (HIV) infection, tuberculosis, and other bacterial infections which have profound effects on human health. At the same time, there have been dramatic increases in the incidence of fungal infections, which are probably the result of alterations in immune status associated with the AIDS epidemic, cancer chemotherapy, and organ and bone marrow transplantation. The rise in the incidence fungal infections has exacerbated the need for the next generation of antifungal agents, since many of the currently available drugs have undesirable side effects, are ineffective against new or reemerging fungi, or lead to the rapid development of resistance. Although extremely rare 10 years ago, antifungal drug resistance is quickly becoming a major problem in certain populations, especially those infected with HIV, in whom drug resistance of the agent causing oropharyngeal candidiasis is a major problem (60, 203). For instance, 33% of late-stage AIDS patients in one study had drug-resistant strains of Candida albicans in their oral cavities (101). There are no large-scale surveys of the extent of antifungal drug resistance, which has prompted requests for an international epidemiological survey of this problem (133).
Since the initial studies of antifungal drug resistance in the early 1980s, we have accumulated a body of knowledge concerning the clinical, biochemical, and genetic aspects of this phenomenon. Recently, an elucidation of the molecular aspects of antifungal drug resistance has been initiated. This review will summarize the current knowledge of the clinical and cellular factors that contribute to antifungal drug resistance and will focus on the molecular mechanisms that have recently been described. The elucidation of the molecular mechanisms of drug resistance is still a work in progress. An attempt will be made to highlight research areas that have not yet been investigated.
This review will focus on the pathogenic yeast Candida albicans, since a large body of work on the factors and mechanisms associated with antifungal drug resistance in this organism has recently been reported. When appropriate, antifungal drug resistance in other common medically important fungi, such as Cryptococcus and Aspergillus, will be discussed. This review will not deal extensively with dermatophytes or with medically unimportant fungi. However, it is likely that many of the mechanisms described for C. albicans will be applicable to other fungi, especially since the currently available antifungal drugs are used for a variety of fungal infections.
MECHANISMS OF ACTION OF ANTIFUNGAL DRUGS
With the exception of 5-flucytosine, the antifungal drugs in common usage are directed in some way against ergosterol, the major sterol of the fungal plasma membrane, which is analogous to cholesterol in mammalian cells. Ergosterol in the fungal membrane contributes to a variety of cellular functions. It is important for the fluidity and integrity of the membrane and for the proper function of many membrane-bound enzymes, including chitin synthetase, which is important for proper cell growth and division (77, 204). The mechanisms of action of antifungal drugs discussed below have been reviewed extensively (204).
Polyenes
The polyenes are a class of antifungal drugs that target membranes containing ergosterol. These drugs, which include amphotericin B and nystatin, are amphipathic, having both hydrophobic and hydrophilic sides. The drugs are thought to intercalate into membranes, forming a channel through which cellular components, especially potassium ions, leak and thereby destroying the proton gradient within the membrane (201). The specificity of the drugs for ergosterol-containing membranes is thought to be due to the interactions of the amphotericin B channel with ergosterol in the membrane, although the mechanism of this interaction is unknown. The specificity of amphotericin B for ergosterol-containing membranes may also be associated with phospholipid fatty acids and the ratio of sterol to phospholipids (201). The drugs are less likely to interact with membranes containing cholesterol. It has also been suggested that amphotericin B causes oxidative damage to the fungal plasma membrane (201, 203, 204). However, recent evidence suggests that amphotericin B has an antioxidant effect in vivo, protecting fungal cells against oxidative attack from the host (135).
Ergosterol Biosynthesis Inhibitors
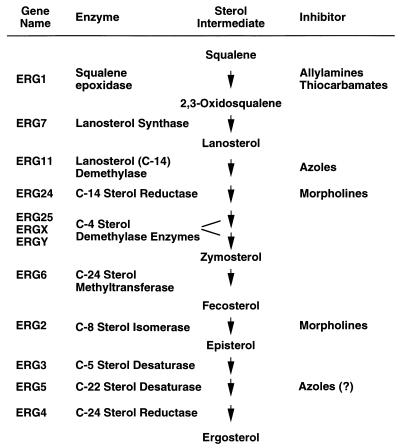
Several inhibitors of the ergosterol biosynthetic pathway have been developed for use against medically important fungi, including allylamines and thiocarbamates, azoles, and morpholines (Table 1). All of these drugs interact with enzymes involved in the synthesis of ergosterol from squalene, which is produced from acetate through acetyl coenzyme A, hydroxymethylglutaryl coenzyme A, and mevalonate. Ergosterol is an important sterol for fungi, since it is the predominant or “bulk” sterol in fungal plasma membranes. In addition, it has an essential “sparking” function in which trace amounts of ergosterol are necessary for the cells to progress through the cell cycle. This sparking function is independent of the bulk sterol in the fungal membranes, since certain sterols can replace the bulk sterol of the membrane without supplying the sparking function for the cell cycle (67). The ergosterol biosynthesis inhibitors disrupt both the bulk sterol of fungal membranes and the sparking function in the cell cycle.
TABLE 1.
Ergosterol biosynthetic pathway from squalene to ergosterola
The table lists the ERG gene designations, the enzyme encoded by the gene, a selected number of sterol intermediates, and the antifungal inhibitors that are directed at specific enzymes. Several sterol intermediates, side paths, and alternate pathways are not listed, for simplification. All of the genes listed have been cloned in S. cerevisiae (27, 103) except ERGX and ERGY. The C-4 sterol demethylase enzymes include three enzyme activities which together remove two methyl groups from C-4 (two arrows). Two of the genes for the C-4 sterol demethylase enzymes, referred to as ERGX and ERGY in the table, have not been cloned and do not yet have gene designations (103). The azole inhibition of ERG5 has recently been suggested (83).
The allylamines (i.e., naftifine and terbinafine) and thiocarbamates (i.e., tolnaftate and tolciclate) inhibit the conversion of squalene to 2,3-oxidosqualene by the enzyme squalene epoxidase (44, 123, 163). This enzyme is the product of the ERG1 gene (gene designations are given in Table 2), which has recently been cloned in C. albicans (Table 1) (161). This class of drugs may inhibit the epoxidase through a naphthalene moiety common to both types of drugs (204). The drugs are noncompetitive inhibitors, and cells accumulate squalene in their presence.
TABLE 2.
Gene nomenclaturea
| Gene | Derivation of name | Organism(s) |
|---|---|---|
| CAD | Cadmium resistance | Saccharomyces |
| CAP | Candida activation protein (transcription factor) | Candida |
| CDR | Candida drug resistance | Candida |
| CFTR | Cystic fibrosis transmembrane conductance receptor | Many systems |
| CYHR | Cycloheximide resistance | Candida |
| ELF | Elongation factor | Candida |
| ERG | Ergosterol biosynthetic enzymesb | Saccharomyces and Candida |
| FCY | Flucytosine resistance | Candida |
| FLR | Fluconazole resistance | Saccharomyces |
| HST6 | Homolog of STE6 | Candida |
| MDR | Multidrug resistance | Many systems |
| MRP | MDR-related protein | Many systems |
| PDR | Pleomorphic drug resistance | Saccharomyces |
| STE6 | Sterile 6 (gene involved in mating − a factor export) | Saccharomyces |
| THR | Threonine biosynthetic enzyme | Saccharomyces and Candida |
| yAP | Yeast activation protein | Saccharomyces |
| YCF | Yeast cadmium factor | Saccharomyces and Candida |
| YEF | Yeast elongation factor | Saccharomyces and Candida |
Gene names are derived from the definition given in the second column. The organism in which the gene has been identified is listed in the third column. In many instances, the gene name is used in more than one species, in many fungi, or in many biological systems. Many gene names represent families of genes in which the gene name is followed by a number which designates a specific gene from the family. For references, see the text.
See Table 1.
The azoles, including both imidazoles (ketoconazole and miconazole) and triazoles (fluconazole, itraconazole, and voriconazole), are directed against lanosterol demethylase in the ergosterol pathway. This enzyme is a cytochrome P-450 enzyme containing a heme moiety in its active site (66, 199). The azoles act through an unhindered nitrogen, which binds to the iron atom of the heme, preventing the activation of oxygen which is necessary for the demethylation of lanosterol (77). In addition to the unhindered nitrogen, a second nitrogen in the azoles is thought to interact directly with the apoprotein of lanosterol demethylase. It is thought that the position of this second nitrogen in relation to the apoprotein may determine the specificity of different azole drugs for the enzyme (66, 199). At high concentrations, the azoles may also interact directly with lipids in the membranes (71, 77). Two other antifungal drug classes, the pyridines (buthiobate and pyrifenox) and the pyrimidines (triarimol and fenarimol), inhibit lanosterol demethylase and are used extensively as antifungal agents in agriculture but are not used in medicine (201). While many fungal species are sensitive to the azoles, Mucor spp. are intrinsically resistant to these drugs (112). Candida krusei and Aspergillus fumigatus are intrinsically resistant to fluconazole and ketoconazole, but both are usually sensitive to itraconazole (112) and voriconazole (118).
The morpholines (i.e., fenpropimorph and amorolfine) inhibit two enzymes in the ergosterol biosynthetic pathway, C-14 sterol reductase and C-8 sterol isomerase. The genes for these two enzymes, ERG24 and ERG2, have not yet been cloned from any medically important fungus, but have been cloned from S. cerevisiae. However, little is known of the interaction of the morpholines with ERG24 or ERG2.
5-Flucytosine
5-Flucytosine (5-FC) has an entirely distinct mode of action from the azoles. 5-FC is taken up into the cell by a cytosine permease and deaminated into 5-fluorouracil (FU) by cytosine deaminase. 5-FC is fungus specific since mammalian cells have little or no cytosine deaminase (203, 204). FU is eventually converted by cellular pyrimidine-processing enzymes into 5-fluoro-dUMP (FdUMP), which is a specific inhibitor of thymidylate synthetase, an essential enzyme for DNA synthesis, and 5-fluoro-UTP (FUTP), which is incorporated into RNA, thus disrupting protein synthesis.
CLINICAL COMPONENTS OF ANTIFUNGAL DRUG RESISTANCE
The AIDS epidemic, improved life-sustaining technologies, and aggressive anticancer therapy have contributed to today’s severely immunosuppressed patient population who survive longer in the immunocompromised state. Mucosal and systemic fungal infections are common in patients lacking intact host defenses, increasing the dependence on antifungal agents for prophylaxis and treatment. Coincident with this increased usage, resistance has been observed. This section will discuss clinical aspects of antifungal resistance: uses of antifungal agents, and the definition and epidemiology of antifungal resistance.
Clinical Uses of Antifungal Agents
Drugs from the polyene class of antifungal agents, specifically amphotericin B, have long been considered the most effective of the systemically administered antifungal agents. The fungicidal antifungal drug amphotericin B has in vitro and in vivo activity against yeasts (Candida spp., Cryptococcus neoformans), molds (Aspergillus spp., Zygomycetes, dematiaceous fungi), and dimorphic fungi. Unfortunately, infusion-related toxicities, the frequent association of renal dysfunction, and the intravenous formulation of amphotericin B have limited the utility of this drug. Similarly, 5-FC, although generally active against Candida spp., C. neoformans, and some molds, has limited clinical utility owing to the frequent association of hematological toxicity and the rapid development of resistance, particularly when it is used as a single agent (discussed in more detail below) (17). The azole antifungal agents, because of their relative safety and ease of delivery, have subsequently become a critical component in the antifungal armamentarium.
Inconsistent levels in blood and high toxicities limited the utility of the first azoles, until ketoconazole was developed and used to treat chronic mucocutaneous candidiasis in the early 1980s (72). Fluconazole, found to be active against C. albicans in vitro and in vivo (162), has since become the drug of choice for the treatment of the most common AIDS-associated opportunistic infection, oropharyngeal candidiasis. Convenient administration and extended activity against non-Candida fungal infections (cryptococcosis, histoplasmosis, and coccidioidomycosis) have made azoles attractive as systemically administered drugs for the therapy and prophylaxis of AIDS-associated opportunistic infections (29, 51). Fluconazole is the preferred azole owing to a high oral bioavailability and good safety profile, although the less expensive ketoconazole is still frequently used for antifungal treatment in other countries (38). A recent study suggests that high doses of fluconazole might be effective therapy for mild blastomycosis in HIV-negative patients (137). Itraconazole and some of the newer azoles such as voriconazole have in vitro activity against various nonyeast fungal infections (molds, dimorphic fungi), but the clinical utility of the newer compounds is still being defined (17, 118).
Over the last several decades, aggressive anticancer therapy has resulted in another patient population that has become increasingly exposed to azole drugs for antifungal treatment and prophylaxis. An increased incidence of invasive Candida infections and attributable mortality have been documented in patients undergoing chemotherapy and bone marrow transplantation (193). Prophylactic fluconazole has been shown to decrease the incidence of local and invasive candidiasis in patients receiving chemotherapy and bone marrow transplantation, and today it is used in some centers for the prevention of candidiasis in neutropenic patients (59, 182, 214).
The role of azole antifungal agents in preventing and treating yeast infections in nonneutropenic patients has been expanding. Fluconazole has been shown to be effective presumptive therapy in surgical intensive care unit patients colonized with Candida spp. (143, 188) and as treatment for candidemia in nonneutropenic patients (156). In addition, azole antifungal agents have been found to be an effective single-dose therapy for vaginal candidiasis (183, 190).
Resistance to each of the antifungal agents has been reported in clinical isolates under specific conditions. However, the increased use of azoles in immunocompromised patients has had the largest effect on the frequency, morbidity, and response to therapy of fungal infections in the hospital and in the outpatient setting. Recently, advances in clinical microbiology have enabled us to detect and define azole resistance. The epidemiology of antifungal resistance, including the incidence, prevalence, and related host risk factors, will be discussed in more detail below.
Definition of Resistance and Role of Susceptibility Testing
Before the development of susceptibility testing of yeasts as outlined by the National Committee for Clinical Laboratory Standards (NCCLS), MIC determinations were inconsistent and varied up to 50,000-fold in different laboratories (49). NCCLS document M27, first published in 1992, has recently been revised (M27-A) and subsequent studies have demonstrated that the interlaboratory reproducibility of MIC determination approximates that of antibacterial testing (159). The macrodilution method has set the groundwork for the development of less cumbersome methods adapted for the clinical laboratory. These methods, relying on microdilution with or without colorimetric indicators or agar diffusion, have been shown to be reproducible and consistent with the standardized macrodilution method according to preliminary studies (141). One complication of the NCCLS protocol is that for certain isolates, the timing of the end point determination can have a major effect (up to 128-fold) on the MIC. A recent study suggests that determining the end point after 24 instead of 48 h ensures that the MICs for these isolates correlate with the in vivo response to azoles (157). In addition, there have been significant advances in susceptibility testing for filamentous fungi (34), as well as in vitro testing of amphotericin B susceptibility. Technical advances of antifungal susceptibility testing have been recently reviewed (141).
Historically, clinical resistance has been defined as persistence or progression of an infection despite appropriate antimicrobial therapy. A successful clinical response to antimicrobial therapy typically not only depends on the susceptibility of the pathogenic organism but also relies heavily on the host immune system, drug penetration and distribution, patient compliance, and absence of a protected or persistent focus of infection (e.g., a catheter or abscess). This is particularly true for fungal infections (Table 3).
TABLE 3.
Factors that may contribute to clinical antifungal drug resistance
| Fungal factors | Drug factors | Host (and other) factors |
|---|---|---|
| Initial MIC | Fungistatic nature of drug | Immune status |
| Cell type | Dosing | Site of infection |
| Yeast/hyphae | Frequency | Severity of infection |
| Switch phenotype | Quantity | Presence of foreign materials (dentures, catheters, prosthetic valves) |
| Serotype | Schedule (intermittent vs continuous) | Abscess formation |
| Genomic stability of strain | Cumulative dose | Patient noncompliance with drug regimen |
| Size of population | Pharmacokinetics | |
| Population “bottlenecks” | Absorption | |
| Biofilms | Distribution | |
| Metabolism | ||
| Drug-drug interactions |
The in vitro resistance of an isolate can be described as either primary or secondary. An organism that is resistant to a drug prior to exposure is described as having primary or intrinsic resistance. Secondary resistance develops in response to exposure to an antimicrobial agent. Both primary and secondary resistance to antifungal agents have been observed.
A correlation of in vitro susceptibility with in vivo response has been observed for mucosal candidal infections in HIV-infected patients. Many groups have noted that the clinical outcome is generally dependent on the in vitro susceptibility of the organism. One group performed fluconazole susceptibility testing on fungal isolates obtained from 87 HIV-infected patients. They observed that persistent oropharyngeal candidiasis correlated with infections caused by yeasts for which the MICs of fluconazole were high (24). Similar trends have been documented by a number of other investigators (9, 94, 108, 126). However, host variables become important when in vivo resistance is compared with in vitro MIC testing, especially in evaluating systemic infections (Table 3). The more immunocompromised the host, the less reliable is the correlation between in vivo resistance and MIC. The ability of C. albicans to form biofilms on surfaces (catheters, teeth, and endothelial cells) has been implicated as a cause of clinical “resistance” despite microbial susceptibility in vitro. One study demonstrated an increased resistance in the cells of the biofilm compared to that in cells growing in suspension (62). The formation of layers of cells may confer protection to organisms in the inner layers, leading to failure of antifungal therapy. Alternatively, infection caused by a resistant organism does not always predict clinical failure. A clinical response to fluconazole occurred in 11 of 13 patients with oral candidiasis even though they were infected with isolates for which the MICs were 32 or 64 μg/ml (155). The correlation between in vivo response and in vitro susceptibility has been reviewed in detail, with most of the data being derived from patients infected with HIV (53).
For aid in clinical interpretation of antifungal susceptibility testing, the NCCLS Subcommittee for Antifungal Susceptibility Testing recently established interpretive breakpoints for testing of fluconazole and itraconazole for Candida infections (158). The breakpoints for fluconazole MICs are as follows: <8 μg/ml, sensitive; 8 to 32 μg/ml, susceptible dose dependent; and ≥64 μg/ml, resistant. The breakpoints for itraconazole MICs are as follows: ≤0.125 μg/ml, sensitive; 0.25 to 0.5 μg/ml, susceptible dose dependent; and ≥1.0 μg/ml, resistant. These breakpoints do not apply for C. krusei, which is intrinsically resistant to most azole drugs. The intermediate levels of resistance are denoted susceptible dose dependent since they are usually treatable with high doses of drug (at least 400 mg of fluconazole per day or doses of itraconazole that maintain the levels in plasma at ≥0.5 μg/ml). Despite the advances in testing and interpretation, the ability of susceptibility testing to guide therapeutic decision making remains controversial. In response to this dilemma, experts in the field recently proposed recommendations for antifungal testing in the clinical laboratory (141). According to these authors, susceptibility testing is not routinely indicated for any fungus. They recommend testing of Candida spp. causing oropharyngeal candidiasis in patients with AIDS who are not responding to azole therapy and in some patients with invasive candidiasis (141).
The establishment of a reproducible method of susceptibility testing not only has resulted in advances in therapeutic decision making, but it also will enable further study of the epidemiology of antifungal resistance in yeasts by allowing comparisons between studies. Recent analyses addressing the prevalence, impact, and microbiological and host risk factors are discussed below and addressed in more detail in two recent reviews (3, 206).
Epidemiology: Prevalence, Risk Factors, and Clinical Implications
Clinical in vitro resistance to all antifungal agents has been reported. Infections caused by azole-resistant Candida spp. have been found in AIDS patients worldwide. Resistance to the polyene antifungal agents and flucytosine have developed in yeasts and a few molds and needs to be taken into consideration during therapeutic decision making.
Polyene resistance.
Reports of amphotericin B resistance are limited, but it appears that severely immunocompromised patients, especially patients with cancer, are at the highest risk. How specific factors such as previous polyene exposure and cytotoxic chemotherapy exposure contribute to the development of amphotericin B resistance has yet to be defined epidemiologically. It appears that amphotericin B resistance in Candida spp. and Cryptococcus neoformans can develop in patients previously exposed to azole antifungal agents. This is thought to be due to an alteration of cellular membrane components.
Several molds (Pseudallescheria boydii, Scopulariopsis, Fusarium), as well as the yeasts Trichosporon beigelii, Candida lusitaniae, and Candida guilliermondii, possess primary resistance to amphotericin B. However, the clinical impact has been limited by the infrequency of invasive infection caused by these resistant organisms and the lack of evidence that such resistance has led to increased numbers of these infections (39, 61, 136, 215). Secondary resistance to amphotericin B has been found in yeasts causing infections in patients with cancer (30, 145, 205). In vitro susceptibility of 29 invasive yeast isolates to amphotericin B was examined in cancer chemotherapy and transplant patients. A significant increase was seen in the average MIC compared to that for colonizing isolates in nonimmunocompromised patients (145). Disseminated infection due to amphotericin B-resistant yeasts has since been reported in other cancer patients undergoing chemotherapy (131), but there have been few reports of amphotericin B resistance developing in other immunocompromised patients. Development of disseminated candidiasis due to amphotericin B-resistant C. albicans has been documented in an immunocompetent trauma patient (30). It was hypothesized that previous exposure to another polyene (nystatin) selected for a resistant strain in this patient. In addition, several strains of fluconazole- and amphotericin B-resistant C. albicans have been found in HIV-infected patients who have received prolonged courses of antifungal prophylaxis with azoles (85). Despite these case reports, secondary resistance to amphotericin B appears to be an infrequent development.
Cryptococcal meningitis requires lifelong suppressive antifungal therapy and recurrence is common despite extended therapy with amphotericin B (43, 91). The in vitro susceptibility of C. neoformans to amphotericin B was tested in isolates from five patients with recurrent meningitis, and no increase in resistance was found in recurrent isolates (25). It was hypothesized that amphotericin B failure occurred because of poor host immune function, poor penetration, and/or inconsistent compliance with medications. Subsequent studies have documented C. neoformans resistance to amphotericin B, although prevalence and significance have not yet been determined (76, 211).
Flucytosine resistance.
Primary resistance to 5-FC is common in certain yeasts and molds. Non-albicans Candida spp., as well as Aspergillus spp., C. neoformans, and the dimorphic fungi, have high rates of 5-FC resistance (47, 178). In addition, secondary resistance is a common development, especially in patients receiving 5-FC monotherapy. There is some indication that the severity of immunosuppression and fungal burden may be important risk factors leading to the development of resistance (47). Because 5-FC resistance develops frequently, the drug should never be used as a single agent to treat either yeast or mold infections. Specific mechanisms of 5-FC resistance in Candida spp. and C. neoformans have been described and are detailed later in this review.
Azole resistance.
As azole antifungal agents have become important in the treatment of mucosal candidiasis in AIDS patients, reports of resistance have increased. In fact, azole resistance has now been found in patients not infected with HIV and, in some situations, in patients not previously exposed to antifungal agents. Several factors contribute to clinical antifungal drug resistance (Table 3).
Several studies have documented that infections due to the intrinsically azole-resistant non-albicans Candida spp. have increased nationwide (128, 149). At one institution, C. albicans comprised 87% of the isolates recovered from blood prior to the use of fluconazole (1987 to 1991) but accounted for only 31% of the isolates in 1992, when fluconazole was used frequently for prophylaxis and treatment (149). Similarly, an increased incidence of non-albicans species with increased azole MICs arose over a 3.5-year study period at another institution (128).
Patients infected with HIV frequently develop oral candidiasis. Candida colonizes the mouths of 64 to 84% of these patients and causes symptomatic disease in up to 46% (109, 116, 167). Reports of azole resistance developing in this setting are numerous (reviewed in reference 160). The prevalence of azole resistance has been estimated to be 21 to 32% in symptomatic patients and up to 14% in asymptomatic patients (109, 116, 155). In a case-control study, advanced immunosuppression and previous exposure to oral azoles were found to be risk factors for the development of resistance (108). In that study, patients with resistant infections had lower mean CD4 counts (11 versus 71/mm3) and a longer duration of antifungal therapy than the matched controls (419 versus 118 days). These results are similar to those found by several other investigators (6, 172). In addition to a long duration of therapy, azole resistance has been reported to occur in association with high total cumulative doses of fluconazole (≥10 g) (120, 212) and with recent exposure to the drug (<1 year prior to the episode) (155). Interestingly, several patients in one study who had received >10 g of fluconazole did not develop resistant candidiasis and several patients with no exposure to fluconazole developed resistant candidiasis (155). The dosing schedule may have an effect on the development of resistance, since patients treated with intermittent therapy were more likely to develop resistance than those treated continuously (64). Still, it is important to be aware that fluconazole resistance has been found in patients treated with daily prophylaxis, weekly prophylaxis, and even episodic single doses (6, 95, 172, 196). These observations emphasize that the development of resistance is complex and relies on multiple host and microbial risk factors.
Both primary resistance and secondary resistance have been documented in HIV-infected patients. Although symptomatic candidiasis develops more commonly due to C. albicans, several groups have documented resistant oral candidiasis caused by non-albicans Candida spp. (108, 155). C. dubliniensis, a new species associated with oral candidiasis in HIV-infected patients, has recently been shown to develop stable fluconazole resistance at a high frequency following exposure to azoles in vitro (124). It has been suggested that C. dubliniensis accounts for a large number of resistant infections in this population, but the prevalence has yet to be determined. Additional fluconazole-resistant species, Candida inconspicua and Candida norvegensis, have recently been reported as pathogens in patients receiving fluconazole (5, 166). Secondary resistance has been confirmed through the study of a patient with AIDS who developed several episodes of thrush caused by a single strain of C. albicans that acquired increasing resistance over time (142, 153). Several groups have suggested that selection for resistant strains occurs with azole treatment (9, 142), but nosocomial transmission (14) and transmission of resistant strains between partners (8, 42) can also occur. The specific mechanisms of acquired azole resistance are discussed in depth later in this review.
Infections due to azole-resistant Candida have also emerged in patients not infected with HIV. Seventeen fluconazole-resistant isolates were found in a collection of 139 strains of C. albicans isolated from patients not infected with HIV at one institution (56). Primary resistance in cancer patients has been well documented, with a change in the spectrum of Candida pathogens to a higher incidence of intrinsically resistant non-albicans species. Several groups have documented an increased incidence of infection due to C. krusei and C. glabrata in cancer and bone marrow transplant patients (226, 227). Recently, the same trend has been noted in surgical intensive care patients treated with fluconazole (55, 164). Whether this is due to selection for more intrinsically resistant species as a result of fluconazole administration or to another risk factor, such as neutropenia, is controversial. Several groups have found no association between fluconazole use and infection with non-albicans species (74). The development of secondary resistance has been confirmed in this setting with recent reports of disseminated disease due to resistant C. albicans developing in patients with leukemia and bone marrow transplantation (114, 131). One patient died from candidiasis after a colonizing isolate developed resistance and disseminated after only 23 days of fluconazole exposure (114). Specific risk factors in this population are not clear and are currently being studied in the bone marrow transplant population.
Azole resistance has been reported as a cause of recurrent vulvovaginal candidiasis (RVVC). The first case of RVVC caused by an azole-resistant C. albicans strain in an HIV-negative woman was reported in 1996 (184). In addition, multiple groups have shown that the incidence of vaginitis due to non-albicans Candida spp. has increased (154, 189). Despite these reports and an increased use of oral azoles for vulvovaginal candidiasis, two groups have recently shown that azole-resistant Candida spp. are rarely the cause of RVVC (46, 107). A longitudinal susceptibility analysis of 177 C. albicans isolates from 50 patients with RVVC did not observe increased MICs to azoles despite long-term exposure (107). The role of azole resistance in recurrent vaginal infections in these immunocompetent hosts will need to be assessed in larger prospective studies.
Although amphotericin B and flucytosine resistance has been reported, azole resistance appears to be emerging as the major problem in patients treated for yeast infections. This may be the result of an increased usage of azoles, which are fungistatic drugs, unlike amphotericin B, which is fungicidal. The importance of selective pressure for resistant strains, induction of secondary resistance, or even nosocomial transmission of resistant strains has not been clearly defined. Genetic typing of isolates in patients with AIDS, recurrent vaginitis, and neutropenia has demonstrated the development of resistance in colonizing strains, suggesting that induction of mutation plays an important role in the development of resistance (146, 175, 186). The remainder of this review discusses specific mechanisms of resistance.
CELLULAR MECHANISMS OF ANTIFUNGAL DRUG RESISTANCE
Fungal disease that is refractory to treatment with antifungal drugs is defined as clinically drug resistant. A common cause of refractory disease is infection with a fungal strain for which the drug MIC is higher than average. Several factors can lead to the presence of a resistant strain in a patient: intrinsic resistance of endogenous strains, replacement with a more resistant species (C. krusei, C. glabrata), replacement with a more resistant strain of C. albicans, genetic alterations that render an endogenous strain resistant, transient gene expression that renders an endogenous strain temporarily resistant, alteration in cell type (yeast/hypha, switch phenotype), size and variability of the population, and population “bottlenecks.” These are discussed in more detail in the following sections.
Change to a More Resistant Species
For each fungal species, there is a distribution of MICs of each antifungal drug. The average MIC of a particular drug can be determined for each species. By using this type of analysis, it is clear that C. krusei is intrinsically more resistant to azole drugs than is C. albicans, with most strains of C. krusei having high azole MICs. In addition, many strains of C. glabrata have azole MICs that are significantly higher than those for most strains of C. albicans (reviewed in references 133 and 160). Recently, Candida norvegensis and Candida inconspicua have also been described as intrinsically resistant species, although these species are uncommon human pathogens (5, 166).
These resistant species may be present by chance in a healthy individual as commensal organisms which can then become a problem when the individual becomes immunocompromised and is administered azole therapy. Alternately, infection with these species can be acquired from the environment or from other individuals. It is thought that in an immunocompetent individual, the endogenous species would prevent these new species from becoming established. However, in an immunocompromised patient, the selective pressure of antifungal drugs may allow the replacement of endogenous species with a more resistant superinfecting species.
The intrinsic resistance of C. krusei and some strains of C. glabrata to azole drugs has resulted in an increased frequency of these species in patient populations that depend on azole drugs for treatment or prophylaxis (see, e.g., references 133, 144, 226, and 227). It is likely that the azole drugs used by these patients suppress the growth of sensitive isolates such as those of C. albicans, and allow the growth of more resistant strains or species.
Change to a More Resistant Strain of C. albicans
In addition to the resistant species such as C. krusei and C. glabrata, there are intrinsically resistant strains of C. albicans that can be part of a commensal growth or can be acquired from the environment or other individuals. Resistant strains of C. albicans can occur as a result of the normal distribution of MICs that all species exhibit, or they can develop resistance through the mechanisms described below.
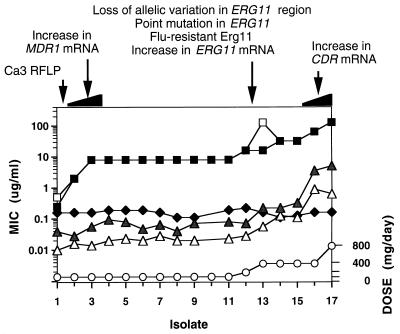
Several studies have investigated the frequency of strain replacement (one strain for another) in a single patient. In one such study (13), strain replacement was seen in the development of resistance in one of four AIDS patients (25%). Sensitive and resistant isolates were obtained from these patients over time, and the isolates were analyzed by pulsed-field gene electrophoresis (molecular karyotyping) and restriction fragment length polymorphism (RFLP). Three of the four patients maintained the same strain, while the sensitive and resistant isolates from the fourth patient were clearly different by RFLP but not by karyotyping. In a second study (120), sensitive and resistant isolates from seven patients were studied by RFLP and karyotyping. In all seven (0% replacement), the sensitive and resistant isolates were the same strain. In a third study (104), multiple sensitive and resistant isolates from three patients were studied by RFLP, karyotyping, and randomly amplified polymorphic DNA. Two of the three patients maintained the same strain (33% replacement). The fourth study used a mixed-linker PCR method to determine that strain replacement accompanied an increase in resistance in only one of five patients (20% replacement) (117). In the final study (142), karyotyping analysis was used to type sensitive and resistant isolates from 10 AIDS patients. Two patients (patients 6 and 20) exhibited sensitive and resistant isolates from the same strain at different times, while several other patients exhibited mixed infections of different strains with different sensitivities at several time points. It is difficult to determine if these mixed infections represent strain replacement over time or mixed infections in which the proportion of sensitive and resistant strains was altered by azole therapy. In one of the two patients from this study, a substrain was selected as resistance developed (225), indicating that the concept of strain replacement is dependent on the technique used to determine strain relatedness. In all of these studies, the sample size is small and so a generalization on the overall frequency of strain replacement is not possible. It is clear that development of resistance within the endogenous strain is much more common than strain replacement. In conclusion, there is a need for a large prospective survey of HIV-infected patients in which the development of resistance and strain identity are closely monitored. Such a study is in progress (139, 155).
The transfer of a strain from one person to another has been documented in several instances. In one case, a strain of C. albicans was transferred orally from one HIV-infected man with oral candidiasis to another (121). In a larger study of 19 sexual partners, at least five pairs had identical or highly related strains of C. albicans (18). In another study, C. albicans strains were identical in women with vaginal candidiasis and their male sexual partner in 8 of 10 cases studied (176). Transfer was not observed in sexual partners without symptoms of disease. None of these studies tested isolates for antifungal susceptibility. Still, the documented strain similarities suggest that transfer of strains can occur, at least in cases in which one of the individuals has disease symptoms.
Genetic Alterations That Render a Strain Resistant
Within a species, intrinsically resistant strains occur as part of the normal distribution of resistance levels (primary resistance). In addition, there is the possibility that a strain can be induced to be resistant by exposure to the drug over long periods (secondary resistance) (160). In these cases, it is hypothesized that the drug itself does not cause resistance but, rather, selects for growth of the more resistant cells in the population. Genetic mutation occurs by chance in all organisms at a low frequency (usually 10−6 to 10−8/gene). However, in a large population of yeast cells under selective drug pressure, specific random mutations that render the cell slightly more resistant will eventually become the dominant strain in the population. Unless the genetic mutations that render a cell resistant also reduce the overall fitness of the cell (133, 202), the mutant strain will persist even in the absence of the selective pressure of the drug.
Given the potential for the transfer of strains between individuals and the genetic stability of resistance, resistant isolates can quickly have a profound impact on the efficacy of the antifungal drug used for treatment, especially within a relatively isolated population with frequent interpersonal contacts, such as HIV-infected populations.
Transient Gene Expression That Renders a Cell Temporarily Resistant
There are recent data suggesting that strains of Candida can become transiently resistant to a drug, a phenomenon called epigenetic resistance. That is, a cell can alter its phenotype, probably through transient gene expression, to become resistant in the presence of the drug, but the resistant phenotype can revert quickly to a susceptible phenotype once the drug pressure is eliminated. Recently, several researchers have demonstrated epigenetic resistance both in vitro (23) and in vivo (67, 203). Given the transient nature of epigenetic resistance, little is known about it at this time.
Alteration in Cell Type
C. albicans has a variety of different cell types which vary in their susceptibility to azoles. C. albicans can be divided into two serotypes, A and B, based on carbohydrate surface markers. The B serotype strains of C. albicans are more sensitive to azoles (specifically ketoconazole) but are more resistant to 5-FC than are the A serotype strains (reviewed in references 132 and 201).
C. albicans is a dimorphic fungus. The yeast-like or blastospore cell types are spherical, budding cells which are associated with commensal growth of C. albicans. Under a variety of conditions, C. albicans can form long, slender hyphal projections, which are often associated with C. albicans pathogenicity (32). Azole drugs can interfere with hyphal production at 0.1 μM, 1/10 the therapeutic concentration of the drug in vivo (132). To date, there have been no studies monitoring the ability of resistant and sensitive clinical isolates of C. albicans to form hyphae. However, it is possible that a resistant strain with the ability to form hyphae even in the presence of azole drugs would be more pathogenic than a sensitive strain that is unable to form hyphae. This possibility needs further study.
In addition to the yeast/hypha transition that can occur in C. albicans, several species of Candida have the ability to alternate between several different forms, referred to as “switch phenotypes” (reviewed in references 185 and 187). These switch phenotypes represent different states of a yeast cell where each state expresses a unique set of genes, cellular structures and cell morphologies. These switch phenotypes are defined by their ability to manifest as different colony morphologies on agar plates. At least two different laboratories have described strains of C. albicans in which two different switch phenotypes exhibit a difference in azole susceptibility (50, 54).
Alterations in the Fungal Population
Antifungal drug resistance is not a constant among the fungal cells in a population. The number of cells that are present in an infection or as a commensal growth will have an effect on antifungal drug resistance, since an increased number of cells will increase the probability that a mutation can occur that confers resistance. Recently, several researchers have observed that there is a great deal of variation in MICs between individual colonies of the same strain from a single patient (75, 179). These differences may reflect genomic instability or variation within a strain. Finally, it is likely that population “bottlenecks” can randomly select for resistance (or susceptibility). A population bottleneck is a set of conditions that randomly selects for a small number of cells from a population. That small subset may contain genetic alterations that will shift the characteristics of a population in a different direction. Such a population “bottleneck” might occur when azole therapy drastically reduces the number of fungal cells in oral candidiasis. In such an example, the azole drug may select for more resistant cells. However, the bottleneck may not necessarily involve drug selection. The same phenomenon might be seen with the removal and/or sterilization of dentures. It is possible that the small subset of fungal cells that survive on the denture or in the oral cavity is more (or less) genetically predisposed to drug resistance.
MOLECULAR MECHANISMS OF AZOLE RESISTANCE
Many different types of mechanisms are known to contribute to a drug-resistant phenotype in eukaryotic cells (Table 4). The most frequent resistance mechanisms include reduction in the import of the drug into the cell; modification or degradation of the drug once it is inside the cell; changes in the interaction of the drug with the target enzyme (binding, activity); changes in other enzymes in the same enzymatic pathway; and an increased efflux of the drug from the cell. Current molecular analysis of antifungal drug resistance has focused on some of these areas, but has not adequately explored other possible mechanisms.
TABLE 4.
Potential molecular mechanisms of antifungal drug resistance
| Alterations in drug import |
| Alterations in intracellular drug processing |
| Modification |
| Degradation |
| Alterations in the target enzyme |
| Point mutations |
| Overexpression |
| Gene amplification |
| Gene conversion or mitotic recombination |
| Alterations in other enzymes in the ergosterol biosynthetic pathway |
| Alterations in efflux pumps |
| ABC transporters |
| Major facilitators |
Matched Sets
It is important in the analysis of the molecular mechanisms of resistance to use a matched set of isolates, that is, sensitive and resistant versions of the same strain, as determined by RFLP or karyotyping. This is required because C. albicans is mostly clonal; that is, it grows vegetatively without sexual reproduction. There is good evidence from Hardy-Weinberg equilibrium experiments (150) and balanced lethal experiments (220) that C. albicans almost never undergoes a sexual cycle. Enzyme analysis shows that C. albicans is not in Hardy-Weinberg equilibrium, which results from a randomly mating population. The disequilibrium in C. albicans indicates that mating is not a common event (150). Most strains of C. albicans contain a large number of balanced lethals (recessive null mutations) in which the cell relies on one functional copy of the gene for activity (220). These recessive lethal mutations could not persist in a population that mated frequently. The clonal nature of C. albicans makes it imperative that matched sets of sensitive and resistant versions of a single strain be characterized when determining the molecular mechanisms of resistance.
RFLP analysis with the repetitive marker Ca3 demonstrates a wide variety of RFLP patterns, suggesting that only rarely are clinical isolates related to each other (105, 177). Specific mutations in a target gene can be the result of allelic variation or selection for a specific resistance phenotype. The difference can be determined only with a matched set of sensitive and resistant isolates from the same strain. To date, few studies have used matched sets of isolates (114, 142, 153, 170, 171, 222, 223, 225).
Drug Import
Defects in drug import are a common mechanism of drug resistance. However, it is important to emphasize the distinction between the import of a drug into a cell and the gradual accumulation of the drug in the cell, which is the result of a balance between import into the cell and efflux of the drug from the cell. Analysis of drug import is difficult at best and requires that mechanisms of import be separated from efflux mechanisms. The standard method for studying drug import is to determine the amount of drug that associates with cells in extremely short periods (less than 10 s). This method has been used extensively in studying drug import in parasitic protozoans (65). The short incubation periods are achieved by mixing the cells and the drug above a layer of mineral oil. After the short incubations, the cells are pelleted through the mineral oil and the amount of drug in the cell pellet is determined. At this time, an analysis of drug import during short periods has not been performed for any medically important fungi.
Studies of fungi have usually used labeled drug to monitor the amount of label that accumulates within the cell over several minutes. These accumulation studies have led to the identification of several fungal efflux mechanisms (see below). One study with C. albicans suggested that accumulation of [3H]ketoconazole required glycolytically derived energy and was controlled by cell viability, environmental pH, and temperature (19). This study also showed that ketoconazole accumulation at low extracellular concentrations was saturable, implying a specific facilitator of import, while accumulation at high concentrations appeared to be by passive diffusion. Curiously, this study found no evidence for export, since the addition of unlabeled drug did not lower the concentration of labeled drug in the cells. Finally, data in this study suggested that other azole drugs and amphotericin B increased the accumulation of labeled ketoconazole in the cell (19). These results are not easily explained by the currently understood molecular mechanisms, which include an energy-dependent efflux pump for fluconazole (see below). The energy requirement for ketoconazole accumulation described in this study is inconsistent with the energy-requiring efflux mechanisms. Elimination of energy levels would inhibit energy-dependent efflux pumps, resulting in increased drug accumulation, the opposite of what is seen with ketoconazole. These results suggest that active transport may be involved in ketoconazole import. It is possible that the ketoconazole and fluconazole are imported by two different mechanisms. Further study is clearly needed.
Drug import may also be affected by the sterol composition of the plasma membrane. Several studies have demonstrated that when the ergosterol component of the membrane is eliminated or reduced in favor of other sterol components such as 14α-methyl sterols, there are concomitant permeability changes in the plasma membrane and a lack of fluidity (204). These changes may lower the capacity of azole drugs to enter the cell.
Modification or Degradation of the Drug
It is important to note that alterations in drug processing (modification or degradation) are important drug resistance mechanisms in a variety of bacterial and eukaryotic systems (20). To date, little analysis of drug modification or degradation within a resistant cell has been performed for the medically important fungi. It has been mentioned that azoles are inert to metabolism by C. albicans (67), although no studies have analyzed the sensitivity of azole drugs to metabolism in resistant strains or non-albicans species.
Modifications of the Ergosterol Biosynthetic Pathway
Another common mechanism of drug resistance is modification of the target enzyme and/or other enzymes in the same biochemical pathway. For azole drugs, that pathway is the ergosterol biosynthetic pathway. Analysis of the sterols of a cell can provide a wealth of information concerning the alterations that have occurred in a resistant strain (Table 1). Modifications in the ergosterol pathway are likely to generate resistance not only to the drug to which the cells are exposed but also to related drugs. This cross-resistance will be discussed in a later section.
A defective lanosterol demethylase (the predominant azole target enzyme) will result in the accumulation of 14α-methyl sterols, especially 14α-methyl fecosterol and the diol 14α-methyl-ergosta-8,24(28)-dien-3β,6α-diol (84). In azole-treated cells, the presence of 14α-methyl sterols can modify the function and fluidity of the plasma membrane. In addition, it appears that cells with 14α-methyl sterols have an increased sensitivity to oxygen-dependent microbicidal systems of the host (180). The diol that accumulates is known to cause growth arrest in Saccharomyces cerevisiae (84, 199) but is thought to be tolerated in C. albicans (11). However, recent studies suggest that accumulation of the diol is correlated with growth arrest even in C. albicans (86). The toxic effects of the diol in S. cerevisiae are eliminated by a mutation in ERG3, which encodes C-5 sterol desaturase (Table 1) (11, 84, 217).
Plasmid complementation of the ERG3 mutation in S. cerevisiae suggests that the ERG3 mutation alone can cause azole resistance (4, 84). Biochemical analysis suggests that ERG3 mutations are responsible for resistance in at least two clinical isolates of C. albicans (86). The sterol composition of Cryptococcus neoformans has been studied when cells are exposed to itraconazole (200). The sterol composition suggests that the drug affects both lanosterol demethylase and the C-4 sterol demethylase enzyme, 3-ketosteroid reductase.
Fungal cell extracts and labeled mevalonate can also be used to monitor the synthesis of sterol intermediates, including squalene, oxidosqualene, lanosterol, desmethyl sterols, and ergosterol (12, 115). The analysis of cell extracts from fungi has been important in monitoring overall sterol synthesis and the level of inhibition of lanosterol demethylase by azole drugs (see, e.g., references 98, 100, and 207). For example, cell extracts from a polyene- and azole-resistant strain of C. albicans (D10) did not contain any detectable lanosterol demethylase activity whereas the revertant strain, D10R, has lanosterol demethylase activity (102). In the absence of drug, strain D10 accumulates 14α-methyl sterols similar to an azole-treated wild-type cell. Strain D10 is also defective in the formation of hyphae, while the revertant forms hyphae at normal rates. This finding suggests that hyphal formation, which is an important virulence factor (32), can be affected by changes in the ergosterol pathway. To date, the genetic mutation in strain D10 has not been identified.
Biochemical analysis of an unrelated azole-resistant strain of C. albicans, the Darlington strain, has shown that lanosterol demethylase activity in this strain does not correlate with resistance (68). Surprisingly, drug accumulation in the Darlington strain is increased compared to that in azole-sensitive strains, suggesting that drug efflux (see below) is altered with resistance (69), although one would expect to see increased efflux in a resistant strain. An analysis of the sterol components of this strain shows an accumulation of fecosterol, suggesting that there may be a defect in ERG2 (not ERG3 as previously reported). The ERG3 gene was recently cloned and expressed in the Darlington strain. Overexpression of the ERG3 gene restored ergosterol as the major sterol of the transformant but did not render the cell susceptible (122).
Cell extracts from fungi other than C. albicans have been used to show that the lanosterol demethylase has become less susceptible in resistant clinical isolates. These include Cryptococcus neoformans (97, 211), A. fumigatus (35), and H. capsulatum (218). The molecular mechanisms for resistance in these isolates were not described. Cell extracts have also been used to characterize a resistant form of lanosterol demethylase from the plant pathogen Ustilago maydis (79).
Molecular Alterations of the ERG11 Gene
As discussed above, the predominant target enzyme of the azole drugs is lanosterol demethylase. The gene encoding this protein is currently designated ERG11 in all fungal species, although it has previously been referred to as ERG16 and CYP51A1 in C. albicans. Several genetic alterations have been identified that are associated with the ERG11 gene of C. albicans, including point mutations in the coding region, overexpression of the gene, gene amplification (which leads to overexpression), and gene conversion or mitotic recombination.
In one study, a point mutation in ERG11 was identified when an azole-resistant clinical isolate was compared with a sensitive isolate from a single strain of C. albicans (223). This point mutation results in the replacement of arginine with lysine at amino acid 467 of the ERG11 gene (abbreviated R467K, where R = Arg and K = Lys). The mutation is positioned near the cysteine which coordinates the fifth position of the iron atom in the heme cofactor. The mutation is thought to cause structural or functional alterations associated with the heme. Recent genetic manipulations suggest that R467K alone is sufficient to cause azole resistance (224). However, in the series of clinical isolates, several alterations occurred simultaneously (223), so it is impossible to determine how much R467K contributes to the overall azole resistance of the isolate.
Point mutations in ERG11 have been developed in laboratory strains that result in azole resistance. The point mutation T315A (the replacement of threonine [T] with alanine [A] at position 315) was constructed in the C. albicans ERG11 gene (100) based on the current understanding of the active site of the enzyme (reviewed in reference 77). The active site represents a pocket positioned on top of the heme cofactor. Substrates or inhibitors enter the active site through a channel that is accessible only with a shift in an α-helix of the apoprotein. Mutations in the active-site pocket, in the channel, and/or in the mobile helix would be predicted to affect the function of the enzyme. The T315A mutation, which is situated in the active-site pocket, was studied in S. cerevisiae, which is more amenable to genetic manipulation. T315A causes a reduction in enzymatic activity and a reduction in azole binding to the active site, resulting in fluconazole resistance. Another point mutation has been identified in the S. cerevisiae ERG11 gene from a laboratory strain that was resistant to azole drugs (73, 228). This mutation, D310G (replacement of aspartic acid [D] with glycine [G] at position 310), is located in the active site of the enzyme in close proximity to the T315A mutation and renders the enzyme inactive. However, the azole resistance of this strain is likely due to an ERG3 suppressor rather than to an inactive ERG11 gene product (84).
A survey of resistant and sensitive clinical isolates has identified seven different point mutations that are associated with azole-resistant isolates (106). However, the matched sensitive isolate was not available from these resistant isolates, which could be used to determine if the point mutations were associated with resistance or just the result of allelic variation. ERG11 genes from a variety of fungal sources have been expressed and manipulated in S. cerevisiae (98, 100, 181) and in Escherichia coli (210). A recent study used functional expression in S. cerevisiae to identify and characterize five ERG11 point mutations from matched sets of sensitive and resistant isolates of C. albicans (168a).
Overexpression of ERG11 has been described in several different clinical isolates (2, 171, 222). In each case, the level of overexpression is not substantial (less than a factor of 5). It is difficult to assess the contribution of ERG11 overexpression to a resistant phenotype, since these limited cases of overexpression have always accompanied other alterations associated with resistance, including the R467K mutation, and overexpression of genes regulating efflux pumps (see below). Overexpression has not been extensively evaluated, so it is difficult to assess its importance. It should be noted that low-level overexpression has also been documented in strains of S. cerevisiae (26, 41, 80, 88). In each case, the effect of overexpression on antifungal susceptibility has been minimal. It remains to be seen if overexpression of ERG11 alone will result in azole resistance.
A common mechanism of resistance in eukaryotic cells is gene amplification. The increase in the number of gene copies usually results in an increase in expression. Overexpression of the ERG11 genes in C. albicans has not been associated with amplification of the ERG11 gene (171, 222). However, in a clinical isolate of C. glabrata, increased levels of lanosterol demethylase were associated with a resistant phenotype (202). These increased levels of the protein were associated with ERG11 gene amplification (113, 203) and correlated with an increase in ERG11 mRNA levels (201). In the same isolate, changes in drug efflux and in ERG7 activity were also observed (203). After 159 subcultures, the gene amplification, increased mRNA levels, enzyme levels, and drug efflux all reverted to normal. However, the revertant retained partial resistance to fluconazole (201). Recently, gene amplification of ERG11 in this isolate has been linked to a chromosome duplication, which results in overexpression of ERG11, as well as altered expression of a variety of other proteins (113).
Another genetic alteration associated with ERG11 has been described in the same clinical isolates in which the R467K mutation was described (223). Most, if not all, strains of C. albicans are diploid, having two alleles of each gene (132). Clinical isolates are usually clonal (105) and contain several sequence differences between the two copies of a gene (allelic differences). While analyzing the R467K mutation, it was observed that all of the allelic differences present in the sensitive isolate were eliminated in the resistant isolates containing R467K. These allelic differences were eliminated from the region of the ERG11 gene including the promoter, coding region, and terminator region, and into the THR1 gene immediately downstream of the ERG11 gene (223). The simplest explanation is that the allelic differences were eliminated by a gene conversion or mitotic recombination involving the ERG11 gene, although other possibilities (gene or chromosome deletion or mating) could not be completely discounted. This gene conversion or mitotic recombination resulted in a cell in which both copies of the ERG11 gene contain the R467K mutation. Genetic evidence suggests that a cell with two copies of the R467K mutation is significantly more resistant to azoles than a cell in which only one allele has the R467K mutation (224). Thus, gene conversion or mitotic recombination, both of which have been found previously in C. albicans, may be an important mechanism in the generation of a resistant phenotype in diploid cells like C. albicans. Gene conversion or mitotic recombination would not be important in haploid organisms such as C. glabrata.
Deletion of the ERG11 gene in C. glabrata has been studied for its effect on enhanced oxidative killing (81). Two ERG11 deletion strains which were fluconazole resistant were more susceptible to killing by H2O2 and by neutrophils than two fluconazole-sensitive strains with intact ERG11 genes. This increased killing (in the absence of drug) may explain why no clinical isolates of any Candida species have yet been identified with a totally nonfunctional ERG11 gene, despite alterations in drug susceptibility due to point mutations in C. albicans.
Alterations in Other ERG Genes
In addition to alterations in the lanosterol demethylase, a common mechanism of resistance is an alteration in other enzymes in the same biosynthetic pathway. The ergosterol biosynthetic pathway beginning with squalene is shown in Table 1. All of the genes listed have been cloned in S. cerevisiae (10, 83, 103). To date, ERG1, ERG2, ERG3, ERG4, ERG7, and ERG11 have been cloned from C. albicans (82, 89, 92, 96, 122, 174), ERG3 and ERG11 have been cloned from C. glabrata (22, 52), and ERG11 has been cloned from C. krusei (22) and C. tropicalis (26). In addition, PRD1, the NADPH-cytochrome P-450 reductase that is important for the function of ERG11, has been cloned from C. albicans (174). Except for the R467K point mutation in ERG11 of C. albicans, no point mutations in the ergosterol pathway have been described from a matched set of azole-resistant and azole-sensitive clinical isolates. Similarly, no gene amplification has been observed for the C. albicans genes to date (171, 222).
The ERG3 and ERG11 genes of C. glabrata have been studied in some detail (52). When the ERG3 gene was deleted from a strain of C. glabrata, the cells remained viable under both aerobic and anaerobic conditions, accumulated the sterol ergosta-7,22-dien-3β-ol, remained susceptible to fluconazole and itraconazole, and became hypersensitive to amphotericin B. When the ERG11 gene was deleted, the cells were viable only under anaerobic conditions, accumulated lanosterol predominantly, became resistant to azoles, and had a reduced susceptibility to amphotericin B. The double mutant (deletion of both ERG3 and ERG11) was aerobically viable, accumulated 14α-methylfecosterol and lanosterol, was resistant to azoles, and had a reduced susceptibility to amphotericin B. The behavior of these mutations for aerobic and anaerobic growth and for sterol accumulation is similar to that described for S. cerevisiae (198). However, the antifungal resistance patterns are not similar, suggesting that the pattern of sterol control and antifungal resistance differs between closely related yeast species (52). This suggests that analysis of the ergosterol biosynthetic pathway in one species may not be applicable to other yeasts.
This analysis has also demonstrated that the ERG3 mRNA level was elevated in ERG11 deletions and that the ERG11 mRNA level was elevated in ERG3 deletions. This suggests that transcriptional control of at least two genes occurred in the ergosterol biosynthetic pathway, perhaps by negative feedback from ergosterol. The lack of ergosterol in the deletion strains may have induced the transcription of ERG3 and/or ERG11. This may be similar to transcriptional control of ERG genes in S. cerevisiae, where ERG11 is regulated by the carbon source, heme, and O2 (aerobic and anaerobic growth) (198).
Sterol analysis of C. albicans clinical isolates has suggested that alterations in ERG3 may be a major cause of azole resistance (85, 86). ERG3 defects prevent the production of the diol that would cause growth arrest (see above). ERG3 defects have been suggested to be a major source of azole resistance in a number of fungi, including the plant pathogen U. maydis (79).
In Cryptococcus neoformans, the azoles appear to directly or indirectly block the C-4 sterol demethylase enzyme complex, which prevents the production of the diol without an ERG3 defect (76). Sterol analysis of fluconazole resistance in C. neoformans suggested that resistant strains may have defects in ERG2 or in ERG3 (211); these strains appear to be cross-resistant to amphotericin B.
In addition to its interactions with lanosterol demethylase, itraconazole interacts with ERG24 in C. neoformans and H. capsulatum, which explains the increased potency of itraconazole against these species (203).
Recently, the ERG5 gene was cloned from S. cerevisiae and found to be another cytochrome P-450 enzyme (83). The azole drugs bind with similar affinity to this enzyme, suggesting that both ERG5 and ERG11 may be responsible for azole tolerance (Table 1).
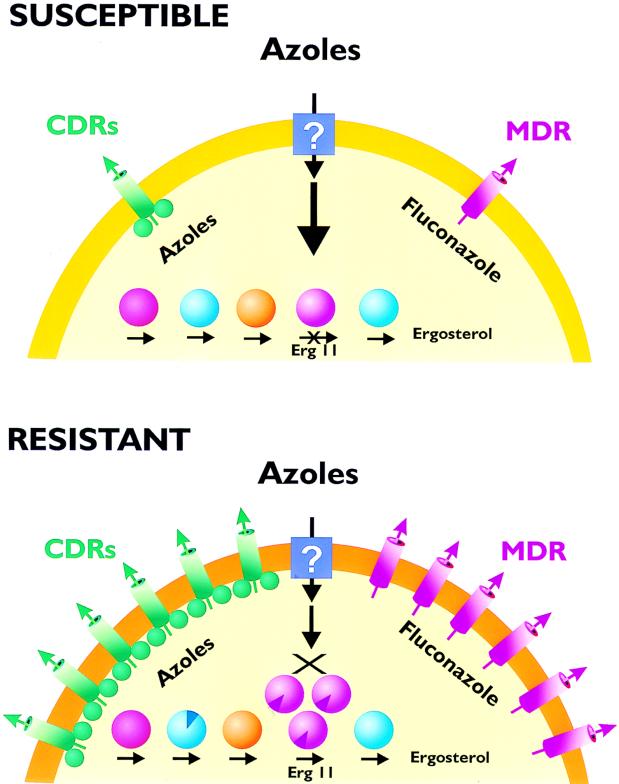
Decreased Accumulation of Drug
As discussed above, little is known about the mechanisms by which a drug enters a fungal cell. However, in recent years, several studies have investigated the accumulation of drugs in cells for which the drug MIC is high. Using radioactively labeled drugs such as fluconazole, these studies have demonstrated that resistant isolates frequently accumulate less drug than do matched sensitive isolates. Several of these studies went on to show that the decreased accumulation is energy dependent.
Reduced accumulation of radiolabeled fluconazole has been documented for a variety of resistant clinical isolates of C. albicans (2, 99, 171, 209). Accumulation in sensitive isolates was shown to be temperature dependent and was not affected by energy inhibitors such as sodium azide or 2-deoxyglucose. In azole-resistant isolates, including a laboratory-induced isolate, fluconazole accumulation was reduced. Intracellular levels of the drug in resistant isolates were then increased by the addition of sodium azide, suggesting that an energy-dependent efflux pump was associated with resistance (2, 171).
Accumulation of fluconazole was studied by using [3H]fluconazole in matched sets of azole-sensitive and azole-resistant isolates of C. glabrata (70, 138, 202). Azole-resistant isolates accumulated significantly less fluconazole than did azole-sensitive isolates from the same strain. Only small changes in ergosterol biosynthesis were detected in the resistant isolates. Metabolic or respiratory inhibitors increased the fluconazole accumulation of azole-resistant isolates, suggesting that energy-dependent efflux pumps were involved. Efflux pump modulators were tested for their effects on fluconazole accumulation. Only benomyl competed with fluconazole under these conditions (138) (benomyl resistance is discussed below).
Reduced drug accumulation has been associated with intrinsic azole resistance to fluconazole, itraconazole, and ketoconazole in C. krusei (111). An itraconazole-resistant isolate of C. krusei did not exhibit any alteration in ergosterol biosynthesis but did show a reduced accumulation of drug compared to a laboratory strain (208). However, a matching sensitive version of the same strain was not available.
Rhodamine 123 (Rh123) is a substrate for a variety of efflux pumps in many systems and has been used to study accumulation in fungal cells. Azole-resistant strains of C. albicans, C. glabrata, and C. krusei accumulate less Rh123 than do azole-sensitive strains (28). Rh123 accumulation is dependent on the growth phase and temperature. Proton uncouplers and the efflux pump modulator reserpine increase accumulation, probably by decreasing the activity of the efflux pumps. In C. glabrata, Rh123 and fluconazole accumulation is competitive, suggesting a common efflux pump, while in C. albicans, the two drugs do not complete (28).
Reduced accumulation has also been suggested as a mechanism of resistance for a C. neoformans laboratory mutant that is resistant to both azoles and polyenes (76). The strain had no detectable change in its sterol pattern. No direct measure of drug accumulation was reported.
For the last several years, there were no reports of itraconazole resistance that were not the result of cross-resistance to fluconazole (201). However, recently, itraconazole resistance in three strains of A. fumigatus was described. One of the strains had a reduced cellular accumulation of labeled itraconazole, suggesting that resistance in this strain is mediated by an efflux pump (35).
Drug accumulation has also been studied for terbinafine in Trichophyton rubrum. Accumulation is 10-fold higher in T. rubrum than in C. albicans, which may explain the high sensitivity of T. rubrum to terbinafine (44).
Two Major Types of Efflux Pumps
Eukaryotic cells contain two types of efflux pumps that are known to contribute to drug resistance: ATP binding cassette (ABC) transporters (ABCT) and major facilitators (MF) (110, 119). Both types of pumps are known to cause drug resistance in other systems. The ABCT are frequently associated with the active efflux of molecules that are toxic to cells and are relatively hydrophobic or lipophilic, as is the case with most azole drugs. The MF have not been studied as extensively as the ABCT but are also associated with relatively hydrophobic molecules such as tetracycline (110).
The ABCT are composed of four protein domains: two membrane-spanning domains (MSD), each consisting of six or seven transmembrane-spanning segments, and two nucleotide binding domains (NBD) (119). The NBD of ABCT bind ATP through an ABC that consists of several conserved peptide motifs, including two Walker domains, a Signature domain and a Center domain. The ATP that is bound to the ABC is used as a source of energy for the ABCT, although the mechanism by which the ATP energy causes transport of the substrate molecule is unknown.
The MF do not contain NBD. They are composed primarily of 12 to 14 transmembrane segments (110, 140). The MF use the proton motive force of the membrane (gradient of H+ across the membrane) as a source of energy. In general, the MF work by antiport; that is, protons are pumped into the cell and substrate molecules are pumped out.
Recently, the complete sequencing of the genome of S. cerevisiae has allowed a determination of the total number of ABCT and MF in the genome of a eukaryotic cell. Thirty ABCT genes that contain an ABC were identified (194). Twenty-three of these genes appear to encode transmembrane segments, which suggests that they could function as drug pumps and are closely related to known ABCT drug pumps. Twenty-eight MF genes were also identified in the S. cerevisiae genome by homology to the five known MF genes of fungi (57). This suggests that S. cerevisiae contains at least 51 different genes encoding efflux pumps that could contribute to the drug resistance levels of the cell. There is good reason to believe that Candida and other fungi would have a similar number of efflux pumps in their genomes (see below).
ABC transporters of Candida.
The 30 ABCT of S. cerevisiae are grouped into six families, based on sequence similarities. Three of these families, PDR5, MRP/CFTR, and MDR, contain ABCT members that are known to cause drug resistance in a variety of systems. To date, 13 efflux pumps have been described in C. albicans. Ten members of the PDR5 family have been described (129, 148, 168, 169, 174). These genes are named CDR (Table 2) and have been linked to azole drug resistance (see below). The HST6 gene of Candida is a member of the MDR gene family and was isolated because it complemented a mutation in STE6, an MDR gene from S. cerevisiae (151). Two groups have identified segments of the Candida YCF1 gene from the MRP/CFTR family (90, 174). The Candida ELF1 gene is a member of the YEF gene family of ABCT (191). This ABCT gene has homology to translation elongation factors and is unlikely to be associated with drug resistance. The YCF1 and ELF1 genes were both isolated by serendipity and have not yet been associated with drug resistance (90, 174, 191). In addition, 14 PCR products with sequence homology to the ABC of ABCT have recently been reported (213).
To date, the only ABCT genes that have been correlated with azole resistance are the CDR genes. CDR1 was first isolated by complementation of the PDR5-mutant strain of S. cerevisiae. The PDR5 mutant of S. cerevisiae is hypersensitive to cycloheximide and chloramphenicol and to azole drugs (148). Complementation of this mutant with CDR1 restored the resistance of the cells to cycloheximide and chloramphenicol, as well as to azole drugs, nystatin, and other unrelated drugs (148, 171). This analysis also indicated that other genes from C. albicans can complement the PDR5 mutant strain of S. cerevisiae. When CDR1 mRNA levels were monitored in azole-resistant clinical isolates of C. albicans, three of five matched sets of sensitive and resistant isolates showed an increase in CDR1 expression (171). In an unrelated study, five azole-resistant strains of C. albicans showed a increased expression of CDR1 compared to azole-sensitive controls (2). In a third study, CDR1 overexpression was correlated with increasing fluconazole MICs in a series of clinical isolates (222).
Genetic deletion of both alleles of CDR1 in C. albicans resulted in a strain that is hypersusceptible to azole drugs, as well as terbinafine (an allylamine), amorolfine (a morpholine), and several metabolic inhibitors (170). The gene disruption did not affect susceptibility to amphotericin B or flucytosine. Recently, it has been shown that overexpression of CDR1 in a CDR1 null mutant resulted in an increased resistance to fluconazole and itraconazole (130).
A second gene, CDR2, has been isolated for its complementation of a PDR5 mutant of S. cerevisiae (169). CDR2 is closely related to CDR1 and confers resistance to azoles, terbinafine and amorolfine, and several metabolic inhibitors. Gene disruption of CDR2 did not cause hypersusceptibility to these agents, but the strain with the double disruption (CDR1 and CDR2) was more hypersusceptible than the strain with the CDR1 single disruption (169). CDR2 overexpression was observed in the resistant isolates from two matched sets of sensitive and resistant clinical isolates. CDR1 overexpression was also observed in the resistant isolates in these pairs, but at a lower level. Overexpression of both genes suggests that transcriptional control may have activated both genes in these isolates. In S. cerevisiae, several transcriptional regulators (PDR3, PDR1, yAP1, yAP2 [CAD1]) control several members of the PDR family of ABCT (110). Recently, the C. albicans homolog of yAP1 was identified (1). This gene, called CAP1, is associated with resistance to azoles, as well as cycloheximide and 4-nitroquinoline-N-oxide (1). In S. cerevisiae, both transcriptional factors CAP1 and yAP1 regulate a new S. cerevisiae MF pump, FLR1, which is associated with resistance to azoles, cycloheximide, and 4-nitroquinoline-N-oxide (1). Finally, in C. albicans strains containing a CDR1 disruption, spontaneous revertants were shown to express CDR2 (169). At least eight other CDR genes have been identified by a variety of molecular and genetic manipulations (129, 152, 168). The relationship between these genes and drug resistance has yet to be established.
Recently, two ABCT were cloned and sequenced from Aspergillus nidulans (33). Transcription of these genes was increased when the cells were grown in the presence of azole drugs and other inhibitory compounds and is correlated with an energy-dependent efflux of azole drugs in this species (36, 37). PCR with degenerate primers has also been used to identify ABCT pumps in both A. fumigatus and A. flavus (195). One of the A. fumigatus pumps confers resistance to the cell wall antifungal agent cilofungin when expressed in S. cerevisiae (195). It is likely that ABCT in these Aspergillus species will be associated with resistance to many classes of antifungal drugs.
Major facilitators in Candida.
To date, the only MF gene that has been cloned in medically important fungi is the MDR1 gene from C. albicans (also referred to as Ben-R) (45), while a related gene, CYHR, has been cloned from the non-pathogenic Candida maltosa (173). The MDR1 gene was originally cloned for its ability to confer both benomyl and methotrexate resistance when transformed into S. cerevisiae (45). This transformant also demonstrates resistance to cycloheximide, benztriazoles, 4-nitroquinoline-N-oxide, and sulfometuron methyl (16). C. albicans is intrinsically resistant to these drugs. In C. albicans, both alleles of the MDR1 gene were deleted by genetic manipulation (58). The cells remained viable but became sensitive to each of the drugs listed except benomyl, suggesting that the gene is important for resistance to most of these drugs and that the cells have more than one mechanism of benomyl resistance.
The C. albicans MDR1 gene was shown to be overexpressed in the fluconazole-resistant isolate in one of five matched sets of clinical isolates (171) and of one of five azole-resistant isolates when compared to azole-sensitive isolates including one matched set (2). In a third study of 17 sequential isolates, overexpression of MDR1 occurred early in this series, correlating with a major increase in resistance (222). The overexpression of MDR1 then continued throughout the series. These experiments suggest a correlation between MDR1 overexpression and azole resistance in clinical isolates.
Genetic manipulations have also demonstrated that major facilitators can be involved in azole resistance. When the MDR1 gene was overexpressed in S. cerevisiae, the cells became resistant to fluconazole but not to ketoconazole or itraconazole (171). The cells were also resistant to benomyl and the other compounds discussed above. When both allelic copies of the MDR1 gene were deleted in a laboratory strain of C. albicans, the cells became more susceptible to 4-nitroquinoline-N-oxide but were unaltered in their susceptibility to other drugs including the azoles (170). If the MDR1 deletion was combined with the CDR1 deletion (discussed above), the cells became more susceptible to cycloheximide or cerulenin than did cells with either single gene deletion (170). The MDR1 gene disruptant had a reduced virulence in a mouse model of systemic candidiasis (15).
Correlation, not cause.
It is important to note that the efflux pumps discussed above, including the CDR genes and the MDR1 gene, show an increased expression that correlates with resistance in clinical isolates of C. albicans. Mutant strains of C. albicans and S. cerevisiae with disruptions in these genes are hypersusceptible to azole and other drugs, and overexpression of these genes in S. cerevisiae can cause an increased resistance. However, there is no definitive proof that overexpression of these genes in C. albicans results in a drug-resistant phenotype.
Possible mechanisms of efflux pump overexpression.
The involvement of the efflux pumps (CDR genes and MDR1) in the azole resistance of clinical isolates has been demonstrated only by increased mRNA levels, and the cause of the increased mRNA levels has not been determined. Increased mRNA levels can be the result of at least three different mechanisms (20). A common mechanism for increased mRNA levels in eukaryotic cells is gene amplification, in which each of many gene copies is transcribed at a normal level so that the total amount of mRNA is increased. There are several mechanisms by which a gene can become amplified (20). Another common mechanism for increased mRNA levels is increased transcription from endogenous copies of the gene, which can be the result of mutations in the promoter of the gene or of alterations in the levels or activity of transcriptional activators that interact with the gene promoter. Finally, mRNA levels will increase if the gene is transcribed at a normal level, but the half-life of the mRNA is increased when it is degraded more slowly. This is usually the result of a mutation in the 3′ untranslated region of the mRNA. To date, there is no data to determine which of these three mechanisms is the most common in clinical isolates.
Summary of Resistance to Azoles
All of the molecular mechanisms described above are summarized in Fig. 1. In a susceptible cell, azoles enter the cell by an unknown mechanism and target lanosterol demethylase, the target enzyme and product of the ERG11 gene, which is part of the ergosterol biosynthetic pathway. Low-level expression of the CDR genes and the MDR1 gene is frequently observed. In a resistant cell, the azoles are blocked from interacting normally with the target enzyme because the enzyme can be modified and/or overexpressed. Mutations in other enzymes in the pathway (such as ERG3) can contribute to resistance. Both ABCT such as the CDR genes and MF such as the MDR1 gene can be overexpressed. The CDR genes appear to remove many azole drugs, while MDR1 appears to be specific for fluconazole.
FIG. 1.
Molecular mechanisms of azole resistance. In a susceptible cell, azole drugs enter the cell through an unknown mechanism, perhaps by passive diffusion. The azoles then inhibit Erg11 (pink circle), blocking the formation of ergosterol. Two types of efflux pumps are expressed at low levels. The CDR proteins are ABCT with both a membrane pore (green tubes) and two ABC domains (green circles). The MDR protein is an MF with a membrane pore (red tubes). In a “model” resistant cell, the azoles also enter the cell through an unknown mechanism. The azole drugs are less effective against Erg11 for two reasons; the enzyme has been modified by specific point mutations (dark slices in pink circles) and the enzyme is overexpressed. Modifications in other enzymes in the ergosterol biosynthetic pathway contribute to azole resistance (dark slices in blue spheres). The sterol components of the plasma membrane are modified (darker orange of membrane). Finally, the azoles are removed from the cell by overexpression of the CDR genes (ABCT) and MDR (MF). The CDR genes are effective against many azole drugs, while MDR appears to be specific for fluconazole. Reprinted from reference 221 with permission of the publisher.
Resistance Is a Gradual Accumulation of Several Alterations
In clinical isolates, it is unlikely that a single mutation will transform a susceptible strain into a highly resistant strain. Resistance usually arises after long periods in the presence of the drug, conditions that may support the gradual, stepwise development of resistance. It is more likely that resistance will gradually evolve over time as the result of several alterations due to continuous selective pressure from the drug. This suggests that several alterations will contribute to the final resistant phenotype. The accumulation of alterations has been documented for one series of 17 clinical isolates (Fig. 2) (222, 223, 225). After the initial selection of a substrain of C. albicans (225), the MDR1 efflux pump was overexpressed (222). Subsequently, three simultaneous changes occurred, all of which involved the ERG11 gene: a point mutation (223), a gene conversion or mitotic recombination of the ERG11 point mutation converting both alleles to the mutant version (223), and overexpression of ERG11 (222). These changes resulted in an enzyme activity that was resistant to fluconazole. Finally, the CDR efflux pumps were overexpressed (222) (Fig. 2). Each of these alterations is likely to contribute in some way to the overall resistance phenotype of the final cell. At this time, there is no indication that the order of these alterations is important. It is likely that each would contribute to the resistance phenotype independent of the timing of the alteration. This gradual increase in resistance associated with specific alterations is an important mechanism of resistance, not only in C. albicans but probably also in other fungi, and it has been shown in a variety of other eukaryotic and prokaryotic systems (20).
FIG. 2.
Time course of the development of fluconazole resistance in a patient with AIDS. Isolates are shown on the x axis in the order in which they were obtained from the patient. The dose that was administered to the patient (○) at the time each isolate was obtained is graphed at the bottom. Doses are given on a linear scale on the right axis in milligrams per day. MICs of fluconazole were determined by macrobroth dilution (□) and microbroth dilution (■). Only two differences were observed between the two methods. MICs of itraconazole (▴), ketoconazole (▵), and amphotericin B (⧫) were determined by E-tests and confirmed by the NCCLS macrobroth dilution method. MICs (in micrograms per milliliter) are given on a logarithmic scale on the left axis. Genetic changes that were identified are summarized above the graph and are described in detail in the text. Reprinted from reference 222 with permission of the publisher.
Cross-Resistance
As described above, clinical isolates that are resistant to one azole are frequently cross-resistant to other azole drugs and can be cross-resistant to polyenes as well. With the current understanding of molecular mechanisms, predictions can be made about the molecular mechanisms that are associated with cross-resistance. Overexpression of the CDR genes is a common mechanism of resistance, and it appears that the CDR genes render a cell resistant to many different azoles while overexpression of MDR1 appears to be specific for fluconazole and is not associated with cross-resistance (169–171). R467K, the one point mutation in ERG11 that is associated with resistance, appears to cause cross-resistance to other azoles but not to amphotericin B (222). It is not clear if specific point mutations in ERG11 will always be associated with cross-resistance. Similarly, there has been insufficient analysis to determine if alterations in other enzymes in the ergosterol pathway will result in cross-resistance.
Unidentified Mechanisms
The work in defining the mechanisms of antifungal drug resistance is still evolving. The mechanisms described above will probably be complemented by descriptions of other, as yet undefined mechanisms such as alterations in drug import or processing. As quickly as new drugs are developed, drug selection will favor new mechanisms that allow the cell to overcome the effects of these drugs.
MOLECULAR MECHANISMS OF NON-AZOLE RESISTANCE
Allylamine and Thiocarbamate Resistance
Allylamine resistance has not been reported for medically important fungi, although resistant strains have been described in S. cerevisiae and the plant pathogen U. maydis (201). Resistance to morpholines or thiocarbamates has not been reported. However, as with any antimicrobial agent, prolonged use will probably select for strain replacement with a resistant isolate or the development of secondary resistance in the original sensitive isolate.
5-Flucytosine Resistance
Primary or intrinsic resistance to 5-FC is a common phenomenon. Estimates suggest that 10% of C. albicans clinical isolates are intrinsically resistant and that 30% will develop secondary resistance (203). The genetics of 5-FC resistance has been investigated in several fungi. In diploid C. albicans, strains with partial resistance appear to be heterozygous for the resistance gene (FCY/fcy). Resistance appears to be a recessive characteristic, and the resistance phenotype (fcy/fcy) can result from further mutation or mitotic recombination of the heterozygote. Mutations in the haploid Cryptococcus neoformans are the most likely cause of 5-FC resistance in these yeasts. Genetic analysis has indicated that there are two genetic loci, FCY1 and FCY2, that can be mutated to produce 5-FC resistance (3).
Primary or intrinsic resistance to 5-FC is usually the result of a defect in cytosine deaminase (203). Secondary resistance to 5-FC in C. albicans is due primarily to a decrease in the activity of the uracil phosphoribosyl transferase (UPRTase), which is involved in the synthesis of FUMP and FdUMP (204, 219). Of the two serotypes of C. albicans, serotype B is more frequently associated with 5-FC resistance (201).
Resistance to 5-FC in C. neoformans can occur and usually results from the loss of UPRTase or cytosine deaminase activities (219). These enzymes make up the pyrimidine salvage pathway. The frequency of 5-FC resistance is lowest in C. albicans, intermediate in C. neoformans, and highest in Aspergillus spp. (201).
Polyene Resistance
Polyene resistance has not been a major clinical problem to date, although polyene-resistant isolates (resistant to nystatin or amphotericin B) have been reported and characterized. Intrinsic resistance to amphotericin B has been reported in Trichosporon beigelii, C. lusitaniae, C. guilliermondii, Pseudallescheria boydii, and certain dematiaceous fungi (3, 111). Amphotericin B-resistant clinical isolates of C. tropicalis have also been found (112), and resistant strains of C. albicans, C. neoformans, Aspergillus nidulans, and A. fennelliae have been constructed in vitro (21, 71, 203). These strains were either exposed to chemical mutagenesis or serially passaged in media containing increasing amounts of polyene. Most polyene-resistant clinical isolates have a greatly reduced ergosterol content in their membranes (201). Based on an analysis of sterol composition, several clinical isolates of C. albicans may be defective in ERG2 or ERG3 (21, 63, 87, 131, 201). Unfortunately, the stability of these mutations or the molecular defects that causes the azole resistance have not been described for any polyene-resistant mutation. One of these strains with an apparent ERG2 defect appears to be unable to form amphotericin B-generated pores in the membrane (63). There is evidence to suggest that alterations in the membrane structure or in the sterol-to-phospholipid ratio in the membrane may be associated with resistance (21, 67).
At least three different types of amphotericin B-resistant mutants of C. neoformans have been isolated, containing alterations in sterol content, alterations that cause cross-resistance to azoles, and alterations that are specific for amphotericin B (78). For most of these isolates, the molecular mechanisms that cause resistance have not been determined. Sterol analysis of one isolate suggested an alteration in ERG2 (87). Environmental isolates of C. neoformans that were serially passaged in a murine model of cryptococcosis became less susceptible to amphotericin B (31). This suggests that the resistance levels of a fungal population can be altered by interactions with the host.
CLINICAL IMPLICATIONS AND FUTURE DIRECTIONS
Dosing
At this time, antifungal drug resistance is clearly becoming a common problem in patients who have a history of antifungal drug use. However, it is not clear what dosing strategies should be used during prophylaxis or treatment to avoid the development of resistance. Azoles, for example, should probably not be used prophylactically for oral or vaginal candidiasis since they may predispose to resistance. However, azole maintenance therapy is be required to prevent the recurrence of cryptococcal meningitis in patients with HIV infection. Several variables need to be considered when trying to minimize the risk for development of resistance, including intermittent versus continuous dosing, the amount of drug administered, the length of treatment, and the immune status of the patient. At this time, none of these factors has been evaluated for its contribution to antifungal resistance, and a large clinical trial by the Mycoses Study Group and the AIDS Clinical Trial Group is under way to evaluate some of these factors for azole use in the HIV-infected population. From experience in other biological systems where drug resistance is a problem, one would predict that continuous treatment with a high dose of drug for as brief a time as possible to effect a cure would result in the least risk for development of resistance, but these factors must be evaluated in clinical trials targeted to specific drugs, infections, and patient populations.
Detection
The NCCLS method for the determination of antifungal susceptibility is currently a standardized technique that can be used to determine the levels of resistance of a yeast strain. The current technique includes both macro- and microdilution broth methods to determine the MIC of a drug. Efforts to make the procedure applicable to a variety of medically important fungi will doubtless improve the value of this technique. However, it will probably remain a time-consuming technique. Quicker methods for the determination of resistance would clearly be welcomed in a clinical laboratory. However, the molecular mechanisms detailed in Fig. 1 suggest that a molecular assay for resistance is unlikely to be simple, because it would have to monitor all of the mechanisms so far described to date and new mechanisms as they are discovered. One alternative that should be explored is to use molecular methods to assess the overall fitness of a cell in the absence and presence of a drug. Such a technique might prove quicker and easier than monitoring all possible mechanisms.
Drug Development
There is a clear need for the next generation of antifungal agents. However, it is important to incorporate what is currently known about antifungal drug resistance into the development of new drugs if those resistance mechanisms are to be avoided. For instance, new and improved azole drugs such as voriconazole should be carefully monitored to determine if they are substrates for the efflux pumps that are already causing resistance to fluconazole (48). This will be difficult since there are probably a large number of efflux pumps in the fungal genome and it is impossible to test each of these pumps at present (see above). New classes of antifungal drugs that are active against azole-resistant isolates, such as the cationic peptide histatin (197), will clearly be important for future treatment strategies.
The broad substrate specificity of the efflux pumps is likely to be a problem even for drugs unrelated to the azoles. Clearly, any new drug should be tested as a substrate for the currently available efflux pumps. However, it is important to keep in mind that these pumps are only a small subset of the efflux pumps encoded in the genome (see above).
One important aspect to keep in mind when examining the molecular mechanisms of antifungal drug resistance is the potential for cross-resistance to the polyenes, especially amphotericin B. Since amphotericin B is the “gold standard” for antifungal drug therapy, it is important to carefully monitor isolates for cross-resistance to this drug. Any therapy or drug that is prone to cause cross-resistance to amphotericin B will have profound negative effects on our ability to treat those fungal infections.
Perhaps the most interesting new antifungal drug target would be the efflux pumps themselves. Any drug that inhibits the activity of these pumps would probably increase the intracellular concentration of drugs such as the azoles, thereby increasing the effectiveness of those drugs. Unfortunately, the currently available inhibitors of these efflux pumps are not effective against fungi, perhaps because the drugs cannot penetrate the fungal cell wall. Clearly, modification of these drugs will be necessary to get the efflux pump inhibitors into the cell.
It is likely that the future of antifungal drug therapy lies in drug combinations. Some drug combinations, such as amphotericin B and 5-FC, are currently in use (93, 216), and others, such as amphotericin B and azoles (165, 192), azoles and 5-FC (40, 127), and azoles and terbinafine (7), have been discussed and are being tested. With the development of new classes of antifungal drug, such as the cell wall inhibitors, and with the knowledge of molecular mechanisms of azole resistance, other potential combinations of drugs are likely to be tested and found effective. One combination that was alluded to above is treatment with azole drugs and an efflux pump inhibitor that is effective against fungal cells.
Other Strategies
The success or failure of antifungal drug therapy is dependent not only on the drug, but also on several other factors (Table 3). Perhaps the best way to improve antifungal drug therapy is to improve the immune status of the host. In many cases, this is not possible, but there are situations in which improving host function may improve outcome. Cytokines such as granulocyte colony-stimulating factor may be useful in the treatment of fungal disease (3, 125). A combination of azoles and cytokines may be an important therapeutic strategy for fungal infections in immunocompromised individuals (147).
Another important strategy in treating fungal disease is the elimination of the disease focus. The removal of fungus-contaminated foreign objects or the surgical removal of abscesses can also reduce the fungal burden and allow the host and antifungal drugs to clear the infection, even with strains that are resistant to standard antifungal therapy.
Finally, new drug delivery systems such as lipid formulations of amphotericin B may have a place in the treatment of antifungal-drug-resistant infections. It is possible that different drug delivery systems will alter the way a yeast cell interacts with a drug and that this will have a negative or positive effect on the development of azole resistance.
CONCLUSIONS
In this review, we have outlined what is currently known about the clinical, cellular, and molecular factors that contribute to antifungal drug resistance. While many contributing factors have been identified (Tables 3 and 4; Fig. 1) (see above), there is only rudimentary information on the mechanism by which these factors contribute to overall drug resistance or the initiating events that trigger any of these factors. Thus, the study of resistance is progressing from a catalog of the basic resistance mechanisms to an understanding of the details of each mechanism and of the steps involved in the activation of these mechanisms. The ongoing developments in the understanding of resistance should assist in the development of diagnostic strategies that identify resistant clinical isolates in patient populations, treatment strategies for resistant fungal infections, and prevention strategies that forestall the development of antifungal drug resistance. Despite all of these efforts, we are faced with the fact that fungi will continually develop new resistance mechanisms to the available antifungal drugs.
ACKNOWLEDGMENTS
We thank many colleagues working on antifungal drug resistance for their contributions to the field, their stimulating and creative discussions, and their advice. We are grateful to them for allowing us access to unpublished work in the writing of this review. We especially thank Steve Kelly and Frank Odds for their comments. We thank the members of our laboratories and the Seattle research community for their comments on the manuscript.
T.C.W. is supported by NIH NIDR grant RO1 DE-11367. K.A.M. is supported by NIH training grant 2T32 A107044-21.
REFERENCES
- 1.Alarco A M, Balan I, Talibi D, Mainville N, Raymond M. AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J Biol Chem. 1997;272:19304–19313. doi: 10.1074/jbc.272.31.19304. [DOI] [PubMed] [Google Scholar]
- 2.Albertson G D, Niimi M, Cannon R D, Jenkinson H F. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob Agents Chemother. 1996;40:2835–2841. doi: 10.1128/aac.40.12.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander B D, Perfect J R. Antifungal resistance trends towards the year 2000: Implications for therapy and new approaches. Drugs. 1997;54:657–678. doi: 10.2165/00003495-199754050-00002. [DOI] [PubMed] [Google Scholar]
- 4.Arthington B A, Bennett L G, Skatrud P L, Guynn C J, Barbuch R J, Ulbright C E, Bard M. Cloning, disruption and sequence of the gene encoding yeast C-5 sterol desaturase. Gene. 1991;102:39–44. doi: 10.1016/0378-1119(91)90535-j. [DOI] [PubMed] [Google Scholar]
- 5.Baily G G, Moore C B, Essayag S M, deWit S, Burnie J P, Denning D W. Candida inconspicua, a fluconazole-resistant pathogen in patients infected with human immunodeficiency virus. Clin Infect Dis. 1997;25:161–163. doi: 10.1086/516894. [DOI] [PubMed] [Google Scholar]
- 6.Baily G G, Perry F M, Denning D W, Mandal B K. Fluconazole-resistant candidosis in an HIV cohort. AIDS. 1994;8:787–792. doi: 10.1097/00002030-199406000-00010. [DOI] [PubMed] [Google Scholar]
- 7.Barchiesi F, DiFrancesco L F, Scalise G. In vitro activities of terbinafine in combination with fluconazole and itraconazole against isolates of Candida albicans with reduced susceptibility to azoles. Antimicrob Agents Chemother. 1997;41:1812–1814. doi: 10.1128/aac.41.8.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barchiesi F, Hollis R J, Del Poeta M, McGough D A, Scalise G, Rinaldi M G, Pfaller M A. Transmission of fluconazole-resistant Candida albicans between patients with AIDS and oropharyngeal candidiasis documented by pulsed-field gel electrophoresis. Clin Infect Dis. 1995;21:561–564. doi: 10.1093/clinids/21.3.561. [DOI] [PubMed] [Google Scholar]
- 9.Barchiesi F, Hollis R J, McGough D A, Scalise G, Rinaldi M G, Pfaller M A. DNA subtypes and fluconazole susceptibilities of Candida albicans isolates from the oral cavities of patients with AIDS. Clin Infect Dis. 1995;20:634–640. doi: 10.1093/clinids/20.3.634. [DOI] [PubMed] [Google Scholar]
- 10.Bard M, Bruner D A, Pierson C A, Lees N D, Biermann B, Frye L, Koegel C, Barbuch R. Cloning and characterization of ERG25, the Saccharomyces cerevisiae gene encoding C-4 sterol methyl oxidase. Proc Natl Acad Sci USA. 1996;93:186–190. doi: 10.1073/pnas.93.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bard M, Lees N D, Turi T, Craft D, Cofrin L, Barbuch R, Koegel C, Loper J C. Sterol synthesis and viability of erg11 (cytochrome P450 lanosterol demethylase) mutations in Saccharomyces cerevisiae and Candida albicans. Lipids. 1993;28:963–967. doi: 10.1007/BF02537115. [DOI] [PubMed] [Google Scholar]
- 12.Barrett-Bee K J, Lane A C, Turner R W. The mode of antifungal action of tolnaftate. J Med Vet Mycol. 1986;24:155–160. doi: 10.1080/02681218680000221. [DOI] [PubMed] [Google Scholar]
- 13.Bart-Delabesse E, Boiron P, Carlotti A, Dupont B. Candida albicans genotyping in studies with patients with AIDS developing resistance to fluconazole. J Clin Microbiol. 1993;31:2933–2937. doi: 10.1128/jcm.31.11.2933-2937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bart-Delabesse E, van Deventer H, Goessens W, Poirot J L, Lioret N, van Belkum A, Dromer F. Contribution of molecular typing methods and antifungal susceptibility testing to the study of a candidemia cluster in a burn care unit. J Clin Microbiol. 1995;33:3278–3283. doi: 10.1128/jcm.33.12.3278-3283.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker J M, Henry L K, Jiang W, Koltin Y. Reduced virulence of Candida albicans mutants affected in multidrug resistance. Infect Immun. 1995;63:4515–518. doi: 10.1128/iai.63.11.4515-4518.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ben-Yaacov R, Knoller S, Caldwell G A, Becker J M, Koltin Y. Candida albicans gene encoding resistance to benomyl and methotrexate is a multidrug resistance gene. Antimicrob Agents Chemother. 1994;38:648–652. doi: 10.1128/aac.38.4.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett J E. Antimicrobial agents: antifungal agents. In: Gilman A G, Rall T W, Nies A S, Taylor P, editors. Goodman and Gilman’s the pharmacological basis of therapeutics. 8th ed. Elmsford, N.Y: Perganon Press, Inc.; 1996. pp. 1175–1190. [Google Scholar]
- 18.Boerlin P, Addo M, Boerlin P F, Durussel C, Pagani J L, Chave J P, Bille J. Transmission of oral Candida albicans strains between HIV-positive patients. Lancet. 1995;345:1052–1053. doi: 10.1016/s0140-6736(95)90793-9. [DOI] [PubMed] [Google Scholar]
- 19.Boiron P, Drouhet E, Dupont B, Improvisi L. Entry of ketoconazole into Candida albicans. Antimicrob Agents Chemother. 1987;31:244–248. doi: 10.1128/aac.31.2.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borst P. Genetic mechanisms of drug resistance. A review. Acta Oncologica. 1991;30:87–105. doi: 10.3109/02841869109091819. [DOI] [PubMed] [Google Scholar]
- 21.Broughton M C, Bard M, Lees N D. Polyene resistance in ergosterol producing strains of Candida albicans. Mycoses. 1991;34:75–83. doi: 10.1111/j.1439-0507.1991.tb00623.x. [DOI] [PubMed] [Google Scholar]
- 22.Burgener-Kairuz P, Zuber J P, Jaunin P, Buchman T G, Bille J, Rossier M. Rapid detection and identification of Candida albicans and Torulopsis (Candida) glabrata in clinical specimens by species-specific nested PCR amplification of a cytochrome P-450 lanosterol-alpha-demethylase (L1A1) gene fragment. J Clin Microbiol. 1994;32:1902–1907. doi: 10.1128/jcm.32.8.1902-1907.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calvet H M, Yeaman M R, Filler S G. Reversible fluconazole resistance in Candida albicans: a potential in vitro model. Antimicrob Agents Chemother. 1997;41:535–539. doi: 10.1128/aac.41.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cameron M L, Schell W A, Bruch S, Bartlett J A, Waskin H A, Perfect J R. Correlation of in vitro fluconazole resistance of Candida isolates in relation to therapy and symptoms of individuals seropositive for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 1993;37:2449–2453. doi: 10.1128/aac.37.11.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casadevall A, Spitzer E D, Webb D, Rinaldi M G. Susceptibilities of serial Cryptococcus neoformans isolates from patients with recurrent cryptococcal meningitis to amphotericin B and fluconazole. Antimicrob Agents Chemother. 1993;37:1383–1386. doi: 10.1128/aac.37.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C, Turi T G, Sanglard D, Loper J C. Isolation of the Candida tropicalis gene for P450 lanosterol demethylase and its expression in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1987;146:1311–1317. doi: 10.1016/0006-291x(87)90792-3. [DOI] [PubMed] [Google Scholar]
- 27.Cherry, J. M., C. Adler, C. Ball, S. Dwight, S. Chervitz, Y. Jia, G. Juvik, S. Weng, and D. Botstein. 1997. Saccharomyces Genome Database. http://genome-www.stanford.edu/Saccharomyces/. [DOI] [PMC free article] [PubMed]
- 28.Clark F S, Parkinson T, Hitchcock C A, Gow N A R. Correlation between rhodamine 123 accumulation and azole sensitivity in Candida species—possible role for drug efflux in drug resistance. Antimicrob Agents Chemother. 1996;40:419–425. doi: 10.1128/aac.40.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Como J A, Dismukes W E. Oral azole drugs as systemic antifungal therapy. N Engl J Med. 1994;330:263–272. doi: 10.1056/NEJM199401273300407. [DOI] [PubMed] [Google Scholar]
- 30.Conly J, Rennie R, Johnson J, Farah S, Hellman L. Disseminated candidiasis due to amphotericin B-resistant Candida albicans. J Infect Dis. 1992;165:761–764. doi: 10.1093/infdis/165.4.761. [DOI] [PubMed] [Google Scholar]
- 31.Currie B, Sanati H, Ibrahim A S, Edwards J E, Jr, Casadevall A, Ghannoum M A. Sterol compositions and susceptibilities to amphotericin B of environmental Cryptococcus neoformans isolates are changed by murine passage. Antimicrob Agents Chemother. 1995;39:1934–1937. doi: 10.1128/aac.39.9.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cutler J E. Putative virulence factors of Candida albicans. Annu Rev Microbiol. 1991;45:187–218. doi: 10.1146/annurev.mi.45.100191.001155. [DOI] [PubMed] [Google Scholar]
- 33.DelSorbo G, Andrade A C, VanNistelrooy J G M, VanKan J A L, Balzi E, DeWaard M A. Multidrug resistance in Aspergillus nidulans involves novel ATP-binding cassette transporters. Mol Gen Genet. 1997;254:417–426. doi: 10.1007/s004380050434. [DOI] [PubMed] [Google Scholar]
- 34.Denning D W, Radford S A, Oakley K L, Hall L, Johnson E M, Warnock D W. Correlation between in-vitro susceptibility testing to itraconazole and in-vivo outcome of Aspergillus fumigatus infection. J Antimicrob Chemother. 1997;40:401–414. doi: 10.1093/jac/40.3.401. [DOI] [PubMed] [Google Scholar]
- 35.Denning D W, Venkateswarlu K, Oakley K L, Anderson M J, Manning N J, Stevens D A, Warnock D W, Kelly S L. Itraconazole resistance in Aspergillus fumigatus. Antimicrob Agents Chemother. 1997;41:1364–1368. doi: 10.1128/aac.41.6.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Waard M A, Van Nistelrooy J G. Induction of fenarimol-efflux activity in Aspergillus nidulans by fungicides inhibiting sterol biosynthesis. J Gen Microbiol. 1981;126:483–489. doi: 10.1099/00221287-126-2-483. [DOI] [PubMed] [Google Scholar]
- 37.De Waard M A, Van Nistelrooy J G M. An energy-dependent efflux mechanism for fenarimol in a wild-type strain and fenarimol-resistant mutants of Aspergillus nidulans. Pestic Biochem Physiol. 1980;13:255–266. [Google Scholar]
- 38.DeWit S, Weerts D, Goossens H, Clumeck N. Comparison of fluconazole and ketoconazole for oropharyngeal candidiasis in AIDS. Lancet. 1989;i:746–748. doi: 10.1016/s0140-6736(89)92572-5. [DOI] [PubMed] [Google Scholar]
- 39.Dick J D, Rosengard B R, Merz W G, Stuart R K, Hutchins G M, Saral R. Fatal disseminated candidiasis due to amphotericin-B-resistant Candida guilliermondii. Ann Intern Med. 1985;102:67–68. doi: 10.7326/0003-4819-102-1-67. [DOI] [PubMed] [Google Scholar]
- 40.Ding J C, Bauer M, Diamond D M, Leal M A, Johnson D, Williams B K, Thomas A M, Najvar L, Graybill J R, Larsen R A. Effect of severity of meningitis on fungicidal activity of flucytosine combined with fluconazole in a murine model of cryptococcal meningitis. Antimicrob Agents Chemother. 1997;41:1589–1593. doi: 10.1128/aac.41.7.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doignon F, Aigle M, Ribereau G P. Resistance to imidazoles and triazoles in Saccharomyces cerevisiae as a new dominant marker. Plasmid. 1993;30:224–233. doi: 10.1006/plas.1993.1054. [DOI] [PubMed] [Google Scholar]
- 42.Dromer F, Improvisi L, Dupont B, Eliaszewicz M, Pialoux G, Fournier S, Feuillie V. Oral transmission of Candida albicans between partners in HIV-infected couples could contribute to dissemination of fluconazole-resistant isolates. AIDS. 1997;11:1095–1101. doi: 10.1097/00002030-199709000-00003. [DOI] [PubMed] [Google Scholar]
- 43.Eng R H, Bishburg E, Smith S M, Kapila R. Cryptococcal infections in patients with acquired immune deficiency syndrome. Am J Med. 1986;81:19–23. doi: 10.1016/0002-9343(86)90176-2. [DOI] [PubMed] [Google Scholar]
- 44.Favre B, Ryder N S. Characterization of squalene epoxidase activity from the dermatophyte Trichophyton rubrum and its inhibition by terbinafine and other antimycotic agents. Antimicrob Agents Chemother. 1996;40:443–447. doi: 10.1128/aac.40.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fling M E, Kopf J, Tamarkin A, Gorman J A, Smith H A, Koltin Y. Analysis of a Candida albicans gene that encodes a novel mechanism for resistance to benomyl and methotrexate. Mol Gen Genet. 1991;227:318–329. doi: 10.1007/BF00259685. [DOI] [PubMed] [Google Scholar]
- 46.Fong I W, Bannatyne R M, Wong P. Lack of in vitro resistance of Candida albicans to ketoconazole, itraconazole and clotrimazole in women treated for recurrent vaginal candidiasis. Genitourin Med. 1993;69:44–46. doi: 10.1136/sti.69.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Francis P, Walsh T J. Evolving role of flucytosine in immunocompromised patients: new insights into safety, pharmacokinetics, and antifungal therapy. Clin Infect Dis. 1992;15:1003–1018. doi: 10.1093/clind/15.6.1003. [DOI] [PubMed] [Google Scholar]
- 48.Fung-Tomc J, White T, Minassian B, Huczko E, Bonner D. Program and Abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. In vitro activity of BMS-207147 (ER-30346) and itraconazole against yeast strains that are resistant or dole-dependent susceptible to fluconazole, abstr. E-69; p. 126. [Google Scholar]
- 49.Galgiani J N, Reiser J, Brass C, Espinel-Ingroff A, Gordon M A, Kerkering T M. Comparison of relative susceptibilities of Candida species to three antifungal agents as determined by unstandardized methods. Antimicrob Agents Chemother. 1987;31:1343–1347. doi: 10.1128/aac.31.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gallagher P J, Bennett D E, Henman M C, Russell R J, Flint S R, Shanley D B, Coleman D C. Reduced azole susceptibility of oral isolates of Candida albicans from HIV-positive patients and a derivative exhibiting colony morphology variation. J Gen Microbiol. 1992;138:1901–1911. doi: 10.1099/00221287-138-9-1901. [DOI] [PubMed] [Google Scholar]
- 51.Gallant J E, Moore R D, Chaisson R E. Prophylaxis for opportunistic infections in patients with HIV infection. Ann Intern Med. 1994;120:932–944. doi: 10.7326/0003-4819-120-11-199406010-00006. [DOI] [PubMed] [Google Scholar]
- 52.Geber A, Hitchcock C A, Swartz J E, Pullen F S, Marsden K E, Kwon-Chung K J, Bennett J E. Deletion of the Candida glabrata ERG3 and ERG11 genes—effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob Agents Chemother. 1995;39:2708–2717. doi: 10.1128/aac.39.12.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghannoum M A, Rex J H, Galgiani J N. Susceptibility testing of fungi—current status of correlation of in vitro data with clinical outcome. J Clin Microbiol. 1996;34:489–495. doi: 10.1128/jcm.34.3.489-495.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghannoum M A, Swairjo I, Soll D R. Variation in lipid and sterol contents in Candida albicans white and opaque phenotypes. J Med Vet Mycol. 1990;28:103–115. [PubMed] [Google Scholar]
- 55.Gleason T G, May A K, Caparelli D, Farr B M, Sawyer R G. Emerging evidence of selection of fluconazole-tolerant fungi in surgical intensive care units. Arch Surg. 1997;132:1197–1201. doi: 10.1001/archsurg.1997.01430350047008. [DOI] [PubMed] [Google Scholar]
- 56.Goff D A, Koletar S L, Buesching W J, Barnishan J, Fass R J. Isolation of fluconazole-resistant Candida albicans from human immunodeficiency virus-negative patients never treated with azoles. Clin Infect Dis. 1995;20:77–83. doi: 10.1093/clinids/20.1.77. [DOI] [PubMed] [Google Scholar]
- 57.Goffeau A, Park J, Paulsen I T, Jonniaux J L, Dinh T, Mordant P, Saier M H. Multidrug-resistant transport proteins in yeast: complete inventory and phylogenetic characterization of yeast open reading frames within the major facilitator superfamily. Yeast. 1997;13:43–54. doi: 10.1002/(SICI)1097-0061(199701)13:1<43::AID-YEA56>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 58.Goldway M, Teff D, Schmidt R, Oppenheim A B, Koltin Y. Multidrug resistance in Candida albicans: disruption of the BENr gene. Antimicrob Agents Chemother. 1995;39:422–426. doi: 10.1128/aac.39.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goodman J L, Winston D J, Greenfield R A, Chandrasekar P H, Fox B, Kaizer H, Shadduck R K, Shea T C, Stiff P, Friedman D J, Powderly W G, Silber J L, Horowitz H, Lichtin A, Wolff S N, Mangan S F, Silver S M, Weisdorf D, Ho W G, Gilbert G, Buell D. A controlled trial of fluconazole to prevent fungal infections in patients undergoing bone marrow transplantation. N Engl J Med. 1992;326:845–851. doi: 10.1056/NEJM199203263261301. [DOI] [PubMed] [Google Scholar]
- 60.Graybill J R. The long and the short of antifungal therapy. Infect Dis Clin North Am. 1988;2:805–825. [PubMed] [Google Scholar]
- 61.Guinet R, Chanas J, Goullier A, Bonnefoy G, Ambroise T P. Fatal septicemia due to amphotericin B-resistant Candida lusitaniae. J Clin Microbiol. 1983;18:443–444. doi: 10.1128/jcm.18.2.443-444.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hawser S P, Douglas L J. Resistance of Candida albicans biofilms to antifungal agents in vitro. Antimicrob Agents Chemother. 1995;39:2128–2131. doi: 10.1128/aac.39.9.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haynes M P, Chong P L, Buckley H R, Pieringer R A. Fluorescence studies on the molecular action of amphotericin B on susceptible and resistant fungal cells. Biochemistry. 1996;35:7983–7992. doi: 10.1021/bi952910c. [DOI] [PubMed] [Google Scholar]
- 64.Heald A E, Cox G M, Schell W A, Bartlett J A, Perfect J R. Oropharyngeal yeast flora and fluconazole resistance in HIV-infected patients receiving long-term continuous versus intermittent fluconazole therapy. AIDS. 1996;10:263–268. doi: 10.1097/00002030-199603000-00004. [DOI] [PubMed] [Google Scholar]
- 65.Hedstrom L, Wang C C. Purine base transport in wild-type and mycophenolic acid-resistant Tritrichomonas foetus. Mol Biochem Parasitol. 1989;35:219–227. doi: 10.1016/0166-6851(89)90208-9. [DOI] [PubMed] [Google Scholar]
- 66.Hitchcock C A. Cytochrome P-450-dependent 14 alpha-sterol demethylase of Candida albicans and its interaction with azole antifungals. Biochem Soc Trans. 1991;19:782–787. doi: 10.1042/bst0190782. [DOI] [PubMed] [Google Scholar]
- 67.Hitchcock C A. Resistance of Candida albicans to azole antifungal agents. Biochem Soc Trans. 1993;21:1039–1047. doi: 10.1042/bst0211039. [DOI] [PubMed] [Google Scholar]
- 68.Hitchcock C A, Barrett-Bee K J, Russell N J. Inhibition of 14 alpha-sterol demethylase activity in Candida albicans Darlington does not correlate with resistance to azole. J Med Vet Mycol. 1987;25:329–333. [PubMed] [Google Scholar]
- 69.Hitchcock C A, Barrett-Bee K J, Russell N J. The lipid composition of azole-sensitive and azole-resistant strains of Candida albicans. J Gen Microbiol. 1986;132:2421–2431. doi: 10.1099/00221287-132-9-2421. [DOI] [PubMed] [Google Scholar]
- 70.Hitchcock C A, Pye G W, Troke P F, Johnson E M, Warnock D W. Fluconazole resistance in Candida glabrata. Antimicrob Agents Chemother. 1993;37:1962–1965. doi: 10.1128/aac.37.9.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hitchcock C A, Russell N J, Barrett B K. Sterols in Candida albicans mutants resistant to polyene or azole antifungals, and of a double mutant C. albicans 6.4. Crit Rev Microbiol. 1987;15:111–115. doi: 10.3109/10408418709104454. [DOI] [PubMed] [Google Scholar]
- 72.Horsburgh C R, Jr, Kirkpatrick C H. Long-term therapy of chronic mucocutaneous candidiasis with ketoconazole: experience with twenty-one patients. Am J Med. 1983;74:23–29. doi: 10.1016/0002-9343(83)90511-9. [DOI] [PubMed] [Google Scholar]
- 73.Ishida N, Aoyama Y, Hatanaka R, Oyama Y, Imajo S, Ishiguro M, Oshima T, Nakazato H, Noguchi T, Maitra U S, Mohan V P, Sprinson D B, Yoshida Y. A single amino acid substitution converts cytochrome P450(14DM) to an inactive form, cytochrome P450SG1: complete primary structures deduced from cloned DNAS. Biochem Biophys Res Commun. 1988;155:317–323. doi: 10.1016/s0006-291x(88)81087-8. [DOI] [PubMed] [Google Scholar]
- 74.Iwen P C, Kelly D M, Reed E C, Hinrichs S H. Invasive infection due to Candida krusei in immunocompromised patients not treated with fluconazole. Clin Infect Dis. 1995;20:342–347. doi: 10.1093/clinids/20.2.342. [DOI] [PubMed] [Google Scholar]
- 75.Johnson E M, Warnock D W, Luker J, Porter S R, Scully C. Emergence of azole drug resistance in Candida species from HIV-infected patients receiving prolonged fluconazole therapy for oral candidosis. J Antimicrob Chemother. 1995;35:103–114. doi: 10.1093/jac/35.1.103. [DOI] [PubMed] [Google Scholar]
- 76.Joseph-Horne T, Hollomon D, Loeffler R S, Kelly S L. Cross-resistance to polyene and azole drugs in Cryptococcus neoformans. Antimicrob Agents Chemother. 1995;39:1526–1529. doi: 10.1128/aac.39.7.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Joseph-Horne T, Hollomon D W. Molecular mechanisms of azole resistance in fungi. FEMS Microbiol Lett. 1997;149:141–149. doi: 10.1111/j.1574-6968.1997.tb10321.x. [DOI] [PubMed] [Google Scholar]
- 78.Joseph-Horne T, Loeffler R S T, Hollomon D W, Kelly S L. Amphotericin B resistant isolates of Cryptococcus neoformans without alteration in sterol biosynthesis. J Med Vet Mycol. 1996;34:223–225. [PubMed] [Google Scholar]
- 79.Joseph-Horne T, Manning N J, Hollomon D, Kelly S L. Defective sterol delta 5(6) desaturase as a cause of azole resistance in Ustilago maydis. FEMS Microbiol Lett. 1995;127:29–34. doi: 10.1111/j.1574-6968.1995.tb07445.x. [DOI] [PubMed] [Google Scholar]
- 80.Kalb V F, Loper J C, Dey C R, Woods C W, Sutter T R. Isolation of a cytochrome P-450 structural gene from Saccharomyces cerevisiae. Gene. 1986;45:237–245. doi: 10.1016/0378-1119(86)90021-1. [DOI] [PubMed] [Google Scholar]
- 81.Kan V L, Geber A, Bennett J E. Enhanced oxidative killing of azole-resistant Candida glabrata strains with ERG11 deletion. Antimicrob Agents Chemother. 1996;40:1717–1719. doi: 10.1128/aac.40.7.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kelly R, Miller S M, Lai M H, Kirsch D R. Cloning and characterization of the 2,3-oxidosqualene cyclase-coding gene of Candida albicans. Gene. 1990;87:177–183. doi: 10.1016/0378-1119(90)90299-7. [DOI] [PubMed] [Google Scholar]
- 83.Kelly S L, Lamb D C, Baldwin B C, Corran A J, Kelly D E. Characterization of Saccharomyces cerevisiae CYP61, sterol delta(22)-desaturase, and inhibition by azole antifungal agents. J Biol Chem. 1997;272:9986–9988. doi: 10.1074/jbc.272.15.9986. [DOI] [PubMed] [Google Scholar]
- 84.Kelly S L, Lamb D C, Corran A J, Baldwin B C, Kelly D E. Mode of action and resistance to azole antifungals associated with the formation of 14 alpha-methylergosta-8,24(28)-dien-3-beta,6-alpha-diol. Biochem Biophys Res Commun. 1995;207:910–915. doi: 10.1006/bbrc.1995.1272. [DOI] [PubMed] [Google Scholar]
- 85.Kelly S L, Lamb D C, Kelly D E, Loeffler J, Einsele H. Resistance to fluconazole and amphotericin in Candida albicans from AIDS patients. Lancet. 1996;348:1523–1524. doi: 10.1016/S0140-6736(05)65949-1. [DOI] [PubMed] [Google Scholar]
- 86.Kelly S L, Lamb D C, Kelly D E, Manning N J, Loeffler J, Hebart H, Schumacher U, Einsele H. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol Delta(5,6) desaturation. FEBS Lett. 1997;400:80–82. doi: 10.1016/s0014-5793(96)01360-9. [DOI] [PubMed] [Google Scholar]
- 87.Kelly S L, Lamb D C, Taylor M, Corran A J, Baldwin B C, Powderly W G. Resistance to amphotericin B associated with defective sterol delta 8→7 isomerase in a Cryptococcus neoformans strain from an AIDS patient. FEMS Microbiol Lett. 1994;122:39–42. doi: 10.1111/j.1574-6968.1994.tb07140.x. [DOI] [PubMed] [Google Scholar]
- 88.Kenna S, Bligh H F, Watson P F, Kelly S L. Genetic and physiological analysis of azole sensitivity in Saccharomyces cerevisiae. J Med Vet Mycol. 1989;27:397–406. doi: 10.1080/02681218980000521. [DOI] [PubMed] [Google Scholar]
- 89.Kirsch D R, Lai M H, O’Sullivan J. Isolation of the gene for cytochrome P450L1A1 (lanosterol 14 alpha-demethylase) from Candida albicans. Gene. 1988;68:229–237. doi: 10.1016/0378-1119(88)90025-x. [DOI] [PubMed] [Google Scholar]
- 90.Kohler, G., and N. Agabian. Personal communication.
- 91.Kovacs J A, Kovacs A A, Polis M, Wright W C, Gill V J, Tuazon C U, Gelmann E P, Lane H C, Longfield R, Overturf G, Macher A M, Fauci A S, Parillo J E, Bennett J E, Masur H. Cryptococcosis in the acquired immunodeficiency syndrome. Ann Intern Med. 1985;103:533–538. doi: 10.7326/0003-4819-103-4-533. [DOI] [PubMed] [Google Scholar]
- 92.Kurtz, M. Personal communication.
- 93.Kwon-Chung K J, Bennett J E. Medical mycology. Philadelphia, Pa: Lea & Febiger; 1992. pp. 81–104. [Google Scholar]
- 94.Lacassin F, Damond F, Chochillon C, Longuet P, Lebras J, Vilde J L, Leport C. Response to fluconazole by 23 patients with human immunodeficiency virus infection and oral candidiasis—pharmacological and mycological factors. Antimicrob Agents Chemother. 1996;40:1961–1963. doi: 10.1128/aac.40.8.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Laguna F, Rodriguez-Tudela J L, Martinez-Suarez J V, Polo R, Valencia E, Diaz-Guerra T M, Dronda F, Pulido F. Patterns of fluconazole susceptibility in isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis due to Candida albicans. Clin Infect Dis. 1997;24:124–130. doi: 10.1093/clinids/24.2.124. [DOI] [PubMed] [Google Scholar]
- 96.Lai M H, Kirsch D R. Nucleotide sequence of cytochrome P450 L1A1 (lanosterol 14 alpha-demethylase) from Candida albicans. Nucleic Acids Res. 1989;17:804. doi: 10.1093/nar/17.2.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lamb D C, Corran A, Baldwin B C, Kwon-Chung J, Kelly S L. Resistant P45051A1 activity in azole antifungal tolerant Cryptococcus neoformans from AIDS patients. FEBS Lett. 1995;368:326–330. doi: 10.1016/0014-5793(95)00684-2. [DOI] [PubMed] [Google Scholar]
- 98.Lamb D C, Kelly D E, Baldwin B C, Gozzo F, Boscott P, Richards W G, Kelly S L. Differential inhibition of Candida albicans CYP51 with azole antifungal stereoisomers. FEMS Microbiol Lett. 1997;149:25–30. doi: 10.1111/j.1574-6968.1997.tb10303.x. [DOI] [PubMed] [Google Scholar]
- 99.Lamb D C, Kelly D E, Manning N J, Kelly S L. Reduced intracellular accumulation of azole antifungal results in resistance in Candida albicans isolate NCPF 3363. FEMS Microbiol Lett. 1997;147:189–193. doi: 10.1111/j.1574-6968.1997.tb10240.x. [DOI] [PubMed] [Google Scholar]
- 100.Lamb D C, Kelly D E, Schunck W H, Shyadehi A Z, Akhtar M, Lowe D J, Baldwin B C, Kelly S L. The mutation T315A in Candida albicans sterol 14 alpha-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J Biol Chem. 1997;272:5682–5688. doi: 10.1074/jbc.272.9.5682. [DOI] [PubMed] [Google Scholar]
- 101.Law D, Moore C B, Wardle H M, Ganguli L A, Keaney M G, Denning D W. High prevalence of antifungal resistance in Candida spp. from patients with AIDS. J Antimicrob Chemother. 1994;34:659–668. doi: 10.1093/jac/34.5.659. [DOI] [PubMed] [Google Scholar]
- 102.Lees N D, Broughton M C, Sanglard D, Bard M. Azole susceptibility and hyphal formation in a cytochrome P-450-deficient mutant of Candida albicans. Antimicrob Agents Chemother. 1990;34:831–836. doi: 10.1128/aac.34.5.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lees N D, Skaggs B, Kirsch D R, Bard M. Cloning of the late genes in the ergosterol biosynthetic pathway of Saccharomyces cerevisiae—a review. Lipids. 1995;30:221–226. doi: 10.1007/BF02537824. [DOI] [PubMed] [Google Scholar]
- 104.Lischewski A, Ruhnke M, Tennagen I, Schonian G, Morschhauser J, Hacker J. Molecular epidemiology of Candida isolates from AIDS patients showing different fluconazole resistance profiles. J Clin Microbiol. 1995;33:769–771. doi: 10.1128/jcm.33.3.769-771.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lockhart S R, Fritch J J, Meier A S, Schroppel K, Srikantha T, Galask R, Soll D R. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C-1 fragment reorganization as demonstrated by DNA fingerprinting and C-1 sequencing. J Clin Microbiol. 1995;33:1501–1509. doi: 10.1128/jcm.33.6.1501-1509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Loffler J, Kelly S L, Hebart H, Schumacher U, LassFlorl C, Einsele H. Molecular analysis of cyp51 from fluconazole-resistant Candida albicans strains. FEMS Microbiol Lett. 1997;151:263–268. doi: 10.1111/j.1574-6968.1997.tb12580.x. [DOI] [PubMed] [Google Scholar]
- 107.Lynch M E, Sobel J D, Fidel P L., Jr Role of antifungal drug resistance in the pathogenesis of recurrent vulvovaginal candidiasis. J Med Vet Mycol. 1996;34:337–339. doi: 10.1080/02681219680000571. [DOI] [PubMed] [Google Scholar]
- 108.Maenza J R, Keruly J C, Moore R D, Chaisson R E, Merz W G, Gallant J E. Risk factors for fluconazole-resistant candidiasis in human immunodeficiency virus-infected patients. J Infect Dis. 1996;173:219–225. doi: 10.1093/infdis/173.1.219. [DOI] [PubMed] [Google Scholar]
- 109.Maenza J R, Merz W G, Romagnoli M J, Keruly J C, Moore R D, Gallant J E. Infection due to fluconazole-resistant Candida in patients with AIDS: prevalence and microbiology. Clin Infect Dis. 1997;24:28–34. doi: 10.1093/clinids/24.1.28. [DOI] [PubMed] [Google Scholar]
- 110.Marger M D, Saier M H., Jr A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- 111.Marichal P, Gorrens J, Coene M C, Le Jeune L, Vanden Bossche H. Origin of differences in susceptibility of Candida krusei to azole antifungal agents. Mycoses. 1995;38:111–117. doi: 10.1111/j.1439-0507.1995.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 112.Marichal P, vanden Bossche H. Mechanisms of resistance to azole antifungals. Acta Biochim Pol. 1995;42:509–516. [PubMed] [Google Scholar]
- 113.Marichal P, Vanden Bossche H, Odds F C, Nobels G, Warnock D W, Timmerman V, Van Broeckhoven C, Fay S, Mose Larsen P. Molecular-biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1997;41:2229–2237. doi: 10.1128/aac.41.10.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Marr K A, White T C, van Burik J H, Bowden R A. Development of a fluconazole-resistant Candida albicans in a patient undergoing bone marrow transplantation. Clin Infect Dis. 1997;25:908–910. doi: 10.1086/515553. [DOI] [PubMed] [Google Scholar]
- 115.Marriott M S. Inhibition of sterol biosynthesis in Candida albicans by imidazole-containing antifungals. J Gen Microbiol. 1980;117:253–255. doi: 10.1099/00221287-117-1-253. [DOI] [PubMed] [Google Scholar]
- 116.Martins M D, LozanoChiu M, Rex J H. Point prevalence of oropharyngeal carriage of fluconazole-resistant Candida in human immunodeficiency virus-infected patients. Clin Infect Dis. 1997;25:843–846. doi: 10.1086/515554. [DOI] [PubMed] [Google Scholar]
- 117.McCullough M, Hume S. A longitudinal study of the change in resistance patterns and genetic relationship of oral Candida albicans from HIV-infected patients. J Med Vet Mycol. 1995;33:33–37. [PubMed] [Google Scholar]
- 118.McGinnis M R, Pasarell L, Sutton D A, Fothergill A W, Cooper C R, Jr, Rinaldi M G. In vitro evaluation of voriconazole against some clinically important fungi. Antimicrob Agents Chemother. 1997;41:1832–1834. doi: 10.1128/aac.41.8.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Michaelis S, Berkower C. Sequence comparison of yeast ATP binding cassette (ABC) proteins. Cold Spring Harbor Symp Quant Biol. 1995;60:291–309. doi: 10.1101/sqb.1995.060.01.034. [DOI] [PubMed] [Google Scholar]
- 120.Millon L, Manteaux A, Reboux G, Drobacheff C, Monod M, Barale T, Michel-Briand Y. Fluconazole-resistant recurrent oral candidiasis in human immunodeficiency virus-positive patients: persistence of Candida albicans strains with the same genotype. J Clin Microbiol. 1994;32:1115–1118. doi: 10.1128/jcm.32.4.1115-1118.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Miyasaki S H, Hicks J B, Greenspan D, Polacheck I, MacPhail L A, White T C, Agabian N, Greenspan J S. The identification and tracking of Candida albicans isolates from oral lesions in HIV-seropositive individuals. J Acquired Immune Defic Syndr. 1992;5:1039–1046. [PubMed] [Google Scholar]
- 122.Miyazaki Y, Geber A, Parkinson T, Hitchcock C, Bennett J E. Abstracts of the 97th General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1997. Complementation of delta 5,6-desaturase did not restore fluconazole susceptibility in Candida albicans Darlington strain, abstr. F-81. [Google Scholar]
- 123.Monk J P, Brogden R N. Naftifine. A review of its antimicrobial activity and therapeutic use in superficial dermatomycoses. Drugs. 1991;42:659–672. doi: 10.2165/00003495-199142040-00008. [DOI] [PubMed] [Google Scholar]
- 124.Moran G P, Sullivan D J, Henman M C, McCreary C E, Harrington B J, Shanley D B, Coleman D C. Antifungal drug susceptibilities of oral Candida dubliniensis isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected subjects and generation of stable fluconazole-resistant derivatives in vitro. Antimicrob Agents Chemother. 1997;41:617–623. doi: 10.1128/aac.41.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Natarajan U, Brummer E, Stevens D A. Effect of granulocyte colony-stimulating factor on the candidacidal activity of polymorphonuclear neutrophils and their collaboration with fluconazole. Antimicrob Agents Chemother. 1997;41:1575–1578. doi: 10.1128/aac.41.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Newman S L, Flanigan T P, Fisher A, Rinaldi M G, Stein M, Vigilante K. Clinically significant mucosal candidiasis resistant to fluconazole treatment in patients with AIDS. Clin Infect Dis. 1994;19:684–686. doi: 10.1093/clinids/19.4.684. [DOI] [PubMed] [Google Scholar]
- 127.Nguyen M H, Najvar L K, Yu C Y, Graybill J R. Combination therapy with fluconazole and flucytosine in the murine model of cryptococcal meningitis. Antimicrob Agents Chemother. 1997;41:1120–1123. doi: 10.1128/aac.41.5.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nguyen M H, Peacock J E, Morris A J, Tanner D C, Nguyen M L, Snydman D R, Wagener M M, Rinaldi M G, Yu V L. The changing face of candidemia—emergence of non-Candida albicans species and antifungal resistance. Am J Med. 1996;100:617–623. doi: 10.1016/s0002-9343(95)00010-0. [DOI] [PubMed] [Google Scholar]
- 129.Niimi M, Cannon R. Multidrug resistance genes in Candida albicans. Jpn J Med Mycol. 1997;38:297–302. [Google Scholar]
- 130.Niimi M, Fischer F J, Piper J, Jenkinson H F, Arisawa M, Cannon R D. Abstracts of the 13th Congress of the International Society for Human and Animal Mycology. 1997. Expression of drug efflux-associated genes and multidrug resistance in Candida albicans, abstr. P456. [Google Scholar]
- 131.Nolte F S, Parkinson T, Falconer D J, Dix S, Williams J, Gilmore C, Geller R, Wingard J R. Isolation and characterization of fluconazole- and amphotericin B-resistant Candida albicans from blood of two patients with leukemia. Antimicrob Agents Chemother. 1997;41:196–199. doi: 10.1128/aac.41.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Odds F C. Candida and candidosis: a review and bibliography. London, United Kingdom: Bailliere Tindall; 1988. [Google Scholar]
- 133.Odds F C. Resistance of yeasts to azole-derivative antifungals. J Antimicrob Chemother. 1993;31:463–471. doi: 10.1093/jac/31.4.463. [DOI] [PubMed] [Google Scholar]
- 134.Reference deleted.
- 135.Osaka K, Ritov V B, Bernardo J F, Branch R A, Kagan V E. Amphotericin B protects cis-parinaric acid against peroxyl radical-induced oxidation: amphotericin B as an antioxidant. Antimicrob Agents Chemother. 1997;41:743–747. doi: 10.1128/aac.41.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pappagianis D, Collins M S, Hector R, Remington J. Development of resistance to amphotericin B in Candida lusitaniae infecting a human. Antimicrob Agents Chemother. 1979;16:123–126. doi: 10.1128/aac.16.2.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pappas P G, Bradsher R W, Kauffman C A, Cloud G A, Thomas C J, Campbell G D, Chapman S W, Newman C, Dismukes W E. Treatment of blastomycosis with higher doses of fluconazole. Clin Infect Dis. 1997;25:200–205. doi: 10.1086/514539. [DOI] [PubMed] [Google Scholar]
- 138.Parkinson T, Falconer D J, Hitchcock C A. Fluconazole resistance due to energy-dependent drug efflux in Candida glabrata. Antimicrob Agents Chemother. 1995;39:1696–1699. doi: 10.1128/aac.39.8.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Patterson, T. F. Personal communication.
- 140.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pfaller M A, Rex J H, Rinaldi M G. Antifungal susceptibility testing: technical advances and potential clinical applications. Clin Infect Dis. 1997;24:776–784. doi: 10.1093/clinids/24.5.776. [DOI] [PubMed] [Google Scholar]
- 142.Pfaller M A, Rhine C J, Redding S W, Smith J, Farinacci G, Fothergill A W, Rinaldi M G. Variations in fluconazole susceptibility and electrophoretic karyotype among oral isolates of Candida albicans from patients with AIDS and oral candidiasis. J Clin Microbiol. 1994;32:59–64. doi: 10.1128/jcm.32.1.59-64.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pittet D, Monod M, Suter P M, Frenk E, Auckenthaler R. Candida colonization and subsequent infections in critically ill surgical patients. Ann Surg. 1994;220:751–758. doi: 10.1097/00000658-199412000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Powderly W G. Mucosal candidiasis caused by non-albicans species of Candida in HIV-positive patients. AIDS. 1992;6:604–605. [PubMed] [Google Scholar]
- 145.Powderly W G, Kobayashi G S, Herzig G P, Medoff G. Amphotericin B-resistant yeast infection in severely immunocompromised patients. Am J Med. 1988;84:826–832. doi: 10.1016/0002-9343(88)90059-9. [DOI] [PubMed] [Google Scholar]
- 146.Powderly W G, Robinson K, Keath E J. Molecular typing of Candida albicans isolated from oral lesions of HIV-infected individuals. AIDS. 1992;6:81–84. doi: 10.1097/00002030-199201000-00010. [DOI] [PubMed] [Google Scholar]
- 147.Poynton C H. Immune modulation by cytokines in the treatment of opportunistic infections. Curr Opin Infect Dis. 1997;10:275–280. [Google Scholar]
- 148.Prasad R, De W P, Goffeau A, Balzi E. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr Genet. 1995;27:320–329. doi: 10.1007/BF00352101. [DOI] [PubMed] [Google Scholar]
- 149.Price M F, LaRocco M T, Gentry L O. Fluconazole susceptibilities of Candida species and distribution of species recovered from blood cultures over a 5-year period. Antimicrob Agents Chemother. 1994;38:1422–1427. doi: 10.1128/aac.38.6.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pujol C, Reynes J, Renaud F, Raymond M, Tibayrenc M, Ayala F J, Janbon F, Malli’e M, Bastide J M. The yeast Candida albicans has a clonal mode of reproduction in a population of infected human immunodeficiency virus-positive patients. Proc Natl Acad Sci USA. 1993;90:9456–9459. doi: 10.1073/pnas.90.20.9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Raymond M, Dignard D, Alarco A-M, Mainville N, Magee B B, Thomas D Y. A Ste6/P-glycoprotein homologue from the asexual yeast, Candida albicans transports the a-factor mating phermone in Saccharomyces cerevisiae. Mol Microbiol. 1998;27:587–598. doi: 10.1046/j.1365-2958.1998.00704.x. [DOI] [PubMed] [Google Scholar]
- 152.Raymond, M., D. Dignard, N. Mainville, and D. Y. Thomas. Personal communication.
- 153.Redding S, Smith J, Farinacci G, Rinaldi M, Fothergill A, Rhine C J, Pfaller M. Resistance of Candida albicans to fluconazole during treatment of oropharyngeal candidiasis in a patient with AIDS: documentation by in vitro susceptibility testing and DNA subtype analysis. Clin Infect Dis. 1994;18:240–242. doi: 10.1093/clinids/18.2.240. [DOI] [PubMed] [Google Scholar]
- 154.Redondo-Lopez V, Lynch M, Schmitt C, Cook R, Sobel J D. Torulopsis glabrata vaginitis: clinical aspects and susceptibility to antifungal agents. Obstet Gynecol. 1990;76:651–655. [PubMed] [Google Scholar]
- 155.Revankar S G, Kirkpatrick W R, McAtee R K, Dib O P, Fothergill A W, Redding S W, Rinaldi M G, Patterson T F. Detection and significance of fluconazole resistance in oropharyngeal candidiasis in Human Immunodeficiency Virus-infected patients. J Infect Dis. 1996;174:821–827. doi: 10.1093/infdis/174.4.821. [DOI] [PubMed] [Google Scholar]
- 156.Rex J H, Bennett J E, Sugar A M, Pappas P G, van der Horst C M, Edwards J E, Washburn R G, Scheld W M, Karchmer A W, Dine A P, Levenstein M J, Webb C D. A randomized trial comparing fluconazole with amphotericin B for the treatment of candidemia in patients without neutropenia. Candidemia Study Group and the National Institute. N Engl J Med. 1994;331:1325–1330. doi: 10.1056/NEJM199411173312001. [DOI] [PubMed] [Google Scholar]
- 157.Rex J H, Nelsom P W, Paetznick V L, Lozano-Chiu M, Espinel-Ingroff A, Anaissie E J. Optimizing the correlation between results of testing in vitro and therapeutic outcome in vivo for fluconazole by testing critical isolates in a murine model of invasive candidiasis. Antimicrob Agents Chemother. 1998;42:129–134. doi: 10.1128/aac.42.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rex J H, Pfaller M A, Galgiani J N, Bartlett M S, Espinel-Ingroff A, Ghannoum M A, Lancaster M, Odds F C, Rinaldi M G, Walsh T J, Barry A L. Development of interpretive breakpoints for antifungal susceptibility testing: conceptual framework and analysis of in vitro/in vivo correlation data for fluconazole, itraconazole, and Candida infections. Clin Infect Dis. 1997;24:235–247. doi: 10.1093/clinids/24.2.235. [DOI] [PubMed] [Google Scholar]
- 159.Rex J H, Pfaller M A, Lancaster M, Odds F C, Bolmstrom A, Rinaldi M G. Quality control guidelines for National Committee for Clinical Laboratory Standards—recommended broth macrodilution testing of ketoconazole and itraconazole. J Clin Microbiol. 1996;34:816–817. doi: 10.1128/jcm.34.4.816-817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Roessner C A, Min C, Hardin S H, Harris Haller L W, McCollum J C, Scott A I. Sequence of the Candida albicans erg7 gene. Gene. 1993;127:149–150. doi: 10.1016/0378-1119(93)90631-c. [DOI] [PubMed] [Google Scholar]
- 162.Rogers T E, Galgiani J N. Activity of fluconazole (UK 49,858) and ketoconazole against Candida albicans in vitro and in vivo. Antimicrob Agents Chemother. 1986;30:418–422. doi: 10.1128/aac.30.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ryder N S. Terbinafine: mode of action and properties of the squalene epoxidase inhibition. Br J Dermatol. 1992;39:2–7. doi: 10.1111/j.1365-2133.1992.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 164.Safran D B, Dawson E. The effect of empiric and prophylactic treatment with fluconazole on yeast isolates in a surgical trauma intensive care unit. Arch Surg. 1997;132:1184–1188. doi: 10.1001/archsurg.1997.01430350034006. [DOI] [PubMed] [Google Scholar]
- 165.Sanati H, Ramos C F, Bayer A S, Ghannoum M A. Combination therapy with amphotericin B and fluconazole against invasive candidiasis in neutropenic-mouse and infective-endocarditis rabbit models. Antimicrob Agents Chemother. 1997;41:1345–1348. doi: 10.1128/aac.41.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sandven P, Nilsen K, Digranes A, Tjade T, Lassen J. Candida norvegensis: a fluconazole-resistant species. Antimicrob Agents Chemother. 1997;41:1375–1376. doi: 10.1128/aac.41.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sangeorzan J A, Bradley S F, He X, Zarins L T, Ridenour G L, Tiballi R N, Kauffman C A. Epidemiology of oral candidiasis in HIV-infected patients: colonization, infection, treatment, and emergence of fluconazole resistance. Am J Med. 1994;97:339–346. doi: 10.1016/0002-9343(94)90300-x. [DOI] [PubMed] [Google Scholar]
- 168.Sanglard, D. Personal communication.
- 168a.Sanglard D, Ischer F, Koymans L, Bille J. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42:241–253. doi: 10.1128/aac.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 170.Sanglard D, Ischer F, Monod M, Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sanglard D, Kuchler K, Ischer F, Pagani J L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sanguineti A, Carmichael J K, Campbell K. Fluconazole-resistant Candida albicans after long-term suppressive therapy. Arch Intern Med. 1993;153:1122–1124. [PubMed] [Google Scholar]
- 173.Sasnauskas K, Jomantiene R, Lebediene E, Lebedys J, Januska A, Janulaitis A. Cloning and sequence analysis of a Candida maltosa gene which confers resistance to cycloheximide. Gene. 1992;116:105–108. doi: 10.1016/0378-1119(92)90636-4. [DOI] [PubMed] [Google Scholar]
- 174.Scherer, S., and Y. Ran. 1996. Candida albicans information: web page. http://alces.med.umn.edu/Candida.html.
- 175.Schmid J, Odds F C, Wiselka M J, Nicholson K G, Soll D R. Genetic similarity and maintenance of Candida albicans strains from a group of AIDS patients, demonstrated by DNA fingerprinting. J Clin Microbiol. 1992;30:935–941. doi: 10.1128/jcm.30.4.935-941.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Schmid J, Rotman M, Reed B, Pierson C L, Soll D R. Genetic similarity of Candida albicans strains from vaginitis patients and their partners. J Clin Microbiol. 1993;31:39–46. doi: 10.1128/jcm.31.1.39-46.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schmid J, Voss E, Soll D R. Computer-assisted methods for assessing strain relatedness in Candida albicans by fingerprinting with the moderately repetitive sequence Ca3. J Clin Microbiol. 1990;28:1236–1243. doi: 10.1128/jcm.28.6.1236-1243.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Schonebeck J, Anséhn S. 5-Fluorocytosine resistance in Candida spp. and Torulopsis glabrata. Sabouraudia. 1973;11:10–20. doi: 10.1080/00362177385190041. [DOI] [PubMed] [Google Scholar]
- 179.Schoofs A, Odds F C, Colebunders R, Ieven M, Wouters L, Goossens H. Isolation of Candida species on media with and without added fluconazole reveals high variability in relative growth susceptibility phenotypes. Antimicrob Agents Chemother. 1997;41:1625–1635. doi: 10.1128/aac.41.8.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shimokawa O, Nakayama H. Increased sensitivity of Candida albicans cells accumulating 14α-methylated sterols to active oxygen: possible relevance to in vivo efficacies of azole antifungal agents. Antimicrob Agents Chemother. 1992;36:1626–1629. doi: 10.1128/aac.36.8.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Shyadehi A Z, Lamb D C, Kelly S L, Kelly D E, Schunck W H, Wright J N, Corina D, Akhtar M. The mechanism of the acyl-carbon bond cleavage reaction catalyzed by recombinant sterol 14-alpha-demethylase of Candida albicans (other names are lanosterol 14-alpha-demethylase, P-450(14dm), and Cyp51) J Biol Chem. 1996;271:12445–12450. doi: 10.1074/jbc.271.21.12445. [DOI] [PubMed] [Google Scholar]
- 182.Slavin M A, Osborne B, Adams R, Levenstein M J, Schoch H G, Feldman A R, Meyers J D, Bowden R A. Efficacy and safety of fluconazole prophylaxis for fungal infections after marrow transplantation—a prospective, randomized, double-blind study. J Infect Dis. 1995;171:1545–1552. doi: 10.1093/infdis/171.6.1545. [DOI] [PubMed] [Google Scholar]
- 183.Sobel J D, Brooker D, Stein G E, Thomason J L, Wermeling D P, Bradley B, Weinstein L. Single oral dose fluconazole compared with conventional clotrimazole topical therapy of Candida vaginitis. Fluconazole Vaginitis Study Group. Am J Obstet Gynecol. 1995;172:1263–1268. doi: 10.1016/0002-9378(95)91490-0. [DOI] [PubMed] [Google Scholar]
- 184.Sobel J D, Vazquez J A. Symptomatic vulvovaginitis due to fluconazole-resistant Candida albicans in a female who was not infected with human immunodeficiency virus. Clin Infect Dis. 1996;22:726–727. doi: 10.1093/clinids/22.4.726. [DOI] [PubMed] [Google Scholar]
- 185.Soll D R. High-frequency switching in Candida albicans. Clin Microbiol Rev. 1992;5:183–203. doi: 10.1128/cmr.5.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Soll D R, Galask R, Isley S, Rao T V, Stone D, Hicks J, Schmid J, Mac K, Hanna C. Switching of Candida albicans during successive episodes of recurrent vaginitis. J Clin Microbiol. 1989;27:681–690. doi: 10.1128/jcm.27.4.681-690.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Soll D R, Morrow B, Srikantha T. High-frequency phenotypic switching in Candida albicans. Trends Genet. 1993;9:61–65. doi: 10.1016/0168-9525(93)90189-O. [DOI] [PubMed] [Google Scholar]
- 188.Solomkin J S. Timing of treatment for nonneutropenic patients colonized with Candida. Am J Surg. 1996;172:44S–48S. doi: 10.1016/s0002-9610(96)00350-9. [DOI] [PubMed] [Google Scholar]
- 189.Spinillo A, Nicola S, Colonna L, Marangoni E, Cavanna C, Michelone G. Frequency and significance of drug resistance in vulvovaginal candidiasis. Gynecol Obstet Invest. 1994;38:130–133. doi: 10.1159/000292465. [DOI] [PubMed] [Google Scholar]
- 190.Stein G E, Mummaw N. Placebo-controlled trial of itraconazole for treatment of acute vaginal candidiasis. Antimicrob Agents Chemother. 1993;37:89–92. doi: 10.1128/aac.37.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sturtevant, J., and R. Calderone. Personal communication.
- 192.Sugar A M. Use of amphotericin B with azole antifungal drugs: what are we doing? Antimicrob Agents Chemother. 1995;39:1907–1912. doi: 10.1128/aac.39.9.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Swerdloff J N, Filler S G, Edwards J E., Jr Severe candidal infections in neutropenic patients. Clin Infect Dis. 1993;2:S457–S467. doi: 10.1093/clinids/17.supplement_2.s457. [DOI] [PubMed] [Google Scholar]
- 194.Taglicht, D., and S. Michaelis. A complete catalogue of Saccharomyces cerevisiae ABC proteins and their relevance to human health and disease. Methods Enzymol., in press. [DOI] [PubMed]
- 195.Tobin M B, Peery R B, Skatrud P L. Genes encoding multiple drug resistance-like proteins in Aspergillus fumigatus and Aspergillus flavus. Gene. 1997;200:1–2. doi: 10.1016/s0378-1119(97)00281-3. [DOI] [PubMed] [Google Scholar]
- 196.Troillet N, Durussel C, Bille J, Glauser M P, Chave J P. Correlation between in vitro susceptibility of Candida albicans and fluconazole-resistant oropharyngeal candidiasis in HIV-infected patients. Eur J Clin Microbiol Infect Dis. 1993;12:911–915. doi: 10.1007/BF01992164. [DOI] [PubMed] [Google Scholar]
- 197.Tsai H, Bobek L A. Studies of the mechanism of human salivary histatin-5 candidacidal activity with histatin-5 variants and azole-sensitive and -resistant Candida species. Antimicrob Agents Chemother. 1997;41:2224–2228. doi: 10.1128/aac.41.10.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Turi T G, Loper J C. Multiple regulatory elements control expression of the gene encoding the Saccharomyces cerevisiae cytochrome P450, lanosterol 14-alpha-demethylase (ERG11) J Biol Chem. 1992;267:2046–2056. [PubMed] [Google Scholar]
- 199.Vanden Bossche H. Ergosterol biosynthesis inhibitors. In: Prasad R, editor. Candida albicans. Berlin, Germany: Springer-Verlag KG; 1991. pp. 239–257. [Google Scholar]
- 200.Vanden Bossche H, Marichal P, Le Jeune L, Coene M C, Gorrens J, Cools W. Effects of itraconazole on cytochrome P-450-dependent sterol 14-alpha-demethylation and reduction of 3-ketosteroids in Cryptococcus neoformans. Antimicrob Agents Chemother. 1993;37:2101–2105. doi: 10.1128/aac.37.10.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Vanden Bossche H, Marichal P, Odds F C. Molecular mechanisms of drug resistance in fungi. Trends Microbiol. 1994;2:393–400. doi: 10.1016/0966-842x(94)90618-1. [DOI] [PubMed] [Google Scholar]
- 202.Vanden Bossche H, Marichal P, Odds F C, Le Jeune L, Coene M C. Characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1992;36:2602–2610. doi: 10.1128/aac.36.12.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Vanden Bossche H, Warnock D W, Dupont B, Kerridge D, Sen G S, Improvisi L, Marichal P, Odds F C, Provost F, Ronin O. Mechanisms and clinical impact of antifungal drug resistance. J Med Vet Mycol. 1994;32:189–202. doi: 10.1080/02681219480000821. [DOI] [PubMed] [Google Scholar]
- 204.Vanden Bossche H, Willemsens G, Marichal P. Anti-Candida drugs—the biochemical basis for their activity. Crit Rev Microbiol. 1987;15:57–72. doi: 10.3109/10408418709104448. [DOI] [PubMed] [Google Scholar]
- 205.Vazquez J A, Lundstrom T, Dembry L, Chandrasekar P, Boikov D, Parri M B, Zervos M J. Invasive Candida guilliermondii infection: in vitro susceptibility studies and molecular analysis. Bone Marrow Transplant. 1995;16:849–853. [PubMed] [Google Scholar]
- 206.Vazquez J A, Sobel J D. Epidemiologic overview of resistance to oral antifungal agents in the immunocompetent host. Opportunistic infection in the immunocompromised host. Excerpta Med Int Congr Ser. 1997;1997:1–11. [Google Scholar]
- 207.Venkateswarlu K, Denning D W, Kelly S L. Inhibition and interaction of cytochrome P450 of Candida krusei with azole antifungal drugs. J Med Vet Mycol. 1997;35:19–25. [PubMed] [Google Scholar]
- 208.Venkateswarlu K, Denning D W, Manning N J, Kelly S L. Reduced accumulation of drug in Candida krusei accounts for itraconazole resistance. Antimicrob Agents Chemother. 1996;40:2443–2446. doi: 10.1128/aac.40.11.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Venkateswarlu K, Denning D W, Manning N J, Kelly S L. Resistance to fluconazole in Candida albicans from AIDS patients correlated with reduced intracellular accumulation of drug. FEMS Microbiol Lett. 1995;131:337–341. doi: 10.1111/j.1574-6968.1995.tb07797.x. [DOI] [PubMed] [Google Scholar]
- 210.Venkateswarlu K, Kelly D E, Kelly S L. Characterization of Saccharomyces cerevisiae CYP51 and a CYP51 fusion protein with NADPH cytochrome P-450 oxidoreductase expressed in Escherichia coli. Antimicrob Agents Chemother. 1997;41:776–780. doi: 10.1128/aac.41.4.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Venkateswarlu K, Taylor M, Manning N J, Rinaldi M G, Kelly S L. Fluconazole tolerance in clinical isolates of Cryptococcus neoformans. Antimicrob Agents Chemother. 1997;41:748–751. doi: 10.1128/aac.41.4.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Vuffray A, Durussel C, Boerlin P, Boerlin P F, Bille J, Glauser M P, Chave J P. Oropharyngeal candidiasis resistant to single-dose therapy with fluconazole in HIV-infected patients. AIDS. 1994;8:708–709. doi: 10.1097/00002030-199405000-00023. [DOI] [PubMed] [Google Scholar]
- 213.Walsh T J, Kasai M, Francesconi A, Landsman D, Chanock S J. New evidence that Candida albicans possesses additional ATP-binding cassette MDR-like genes: implications for antifungal azole resistance. J Med Vet Mycol. 1997;35:133–139. [PubMed] [Google Scholar]
- 214.Walsh T J, Lee J W. Prevention of invasive fungal infections in patients with neoplastic disease. Clin Infect Dis. 1993;2:S468–S480. doi: 10.1093/clinids/17.supplement_2.s468. [DOI] [PubMed] [Google Scholar]
- 215.Walsh T J, Melcher G P, Rinaldi M G, Lecciones J, McGough D A, Kelly P, Lee J, Callender D, Rubin M, Pizzo P A. Trichosporon beigelii, an emerging pathogen resistant to amphotericin B. J Clin Microbiol. 1990;28:1616–1622. doi: 10.1128/jcm.28.7.1616-1622.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Warnock D W. Amphotericin B: an introduction. J Antimicrob Chemother. 1991;28:27–38. doi: 10.1093/jac/28.suppl_b.27. [DOI] [PubMed] [Google Scholar]
- 217.Watson P F, Rose M E, Ellis S W, England H, Kelly S L. Defective sterol C5-6 desaturation and azole resistance: a new hypothesis for the mode of action of azole antifungals. Biochem Biophys Res Commun. 1989;164:1170–1175. doi: 10.1016/0006-291x(89)91792-0. [DOI] [PubMed] [Google Scholar]
- 218.Wheat J, Marichal P, Vanden Bossche H, LeMonte A, Connolly P. Hypothesis on the mechanism of resistance to fluconazole in Histoplasma capsulatum. Antimicrob Agents Chemother. 1997;41:410–414. doi: 10.1128/aac.41.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Whelan W L. The genetic basis of resistance to 5-fluorocytosine in Candida species and Cryptococcus neoformans. Crit Rev Microbiol. 1987;15:45–56. doi: 10.3109/10408418709104447. [DOI] [PubMed] [Google Scholar]
- 220.Whelan W L, Soll D R. Mitotic recombination in Candida albicans: recessive lethal alleles linked to a gene required for methionine biosynthesis. Mol Gen Genet. 1982;187:477–485. doi: 10.1007/BF00332632. [DOI] [PubMed] [Google Scholar]
- 221.White T C. Antifungal drug resistance in Candida albicans. ASM News. 1997;63:427–433. [Google Scholar]
- 222.White T C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from an HIV-infected patient. Antimicrob Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.White T C. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14-alpha-demethylase in Candida albicans. Antimicrob Agents Chemother. 1997;41:1488–1494. doi: 10.1128/aac.41.7.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.White, T. C. Unpublished data.
- 225.White T C, Pfaller M A, Rinaldi R G, Smith J, Redding S W. Stable azole drug resistance associated with a substrain of Candida albicans from an HIV-infected patient. Oral Dis. 1997;3:S102–S109. doi: 10.1111/j.1601-0825.1997.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 226.Wingard J R, Merz W G, Rinaldi M G, Johnson T R, Karp J E, Saral R. Increase in Candida krusei infection among patients with bone marrow transplantation and neutropenia treated prophylactically with fluconazole. N Engl J Med. 1991;325:1274–1277. doi: 10.1056/NEJM199110313251803. [DOI] [PubMed] [Google Scholar]
- 227.Wingard J R, Merz W G, Rinaldi M G, Miller C B, Karp J E, Saral R. Association of Torulopsis glabrata infections with fluconazole prophylaxis in neutropenic bone marrow transplant patients. Antimicrob Agents Chemother. 1993;37:1847–1849. doi: 10.1128/aac.37.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Yoshida Y, Aoyama Y, Nishino T, Katsuki H, Maitra U S, Mohan V P, Sprinson D B. Spectral properties of a novel cytochrome P-450 of a Saccharomyces cerevisiae mutant SG1. A cytochrome P-450 species having a nitrogenous ligand trans to thiolate. Biochem Biophys Res Commun. 1985;127:623–628. doi: 10.1016/s0006-291x(85)80206-0. [DOI] [PubMed] [Google Scholar]