Abstract
These experiments sought to identify what form of RNA polymerase transcribes the P1 promoter for the Rhodobacter sphaeroides cytochrome c2 gene (cycA). In vitro, cycA P1 was recognized by an RNA polymerase holoenzyme fraction that transcribes several well-characterized Escherichia coli heat shock (ς32) promoters. The in vivo effects of mutations flanking the transcription initiation site (+1) also suggested that cycA P1 was recognized by an RNA polymerase similar to E. coli Eς32. Function of cycA P1 was not altered by mutations more than 35 bp upstream of position +1 or by alterations downstream of −7. A point mutation at position −34 that is towards the E. coli Eς32 −35 consensus sequence (G34T) increased cycA P1 activity ∼20-fold, while several mutations that reduced or abolished promoter function changed highly conserved bases in presumed −10 or −35 elements. In addition, cycA P1 function was retained in mutant promoters with a spacer region as short as 14 nucleotides. When either wild-type or G34T promoters were incubated with reconstituted RNA polymerase holoenzymes, cycA P1 transcription was observed only with samples containing either a 37-kDa subunit that is a member of the heat shock sigma factor family (Eς37) or a 38-kDa subunit that also allows core RNA polymerase to recognize E. coli heat shock promoters (Eς38) (R. K. Karls, J. Brooks, P. Rossmeissl, J. Luedke, and T. J. Donohue, J. Bacteriol. 180:10–19, 1998).
In order to survive, all cells must continue to generate energy in response to a wide variety of environmental changes and signals. While viability in response to such challenges requires the coordinated induction and repression of numerous genes, relatively little is known about how the expression of energy-generating functions is altered by these stimuli. To analyze the control of proteins that participate in biological energy generation, we are studying transcriptional regulation of the cytochrome c2 gene (cycA) from the facultative phototroph Rhodobacter sphaeroides.
We know that cycA contains several promoters (16, 21, 22). The downstream cycA promoter (cycA P1) provides sufficient cytochrome c2 to support growth, but its output is not altered by conditions that alter levels or demand function of this electron carrier (16, 21, 22). In contrast, activity of the upstream promoter (cycA P2) is increased ∼10-fold under photosynthetic conditions where cytochrome c2 function is required (16, 21, 22). Activity of a third promoter (cycA P3) increased dramatically in cells containing the trans-acting chr-4 mutation (21, 22).
A key unresolved issue is the nature of the RNA polymerase holoenzyme that recognizes each of the three cycA promoters (16, 21, 22). A previous study noted the similarity of DNA sequences upstream of cycA P1 and P2 to promoters recognized by the major eubacterial sigma factor, ς70 (16). Thus, one possibility was that cycA P1 and P2 were each recognized by the previously described R. sphaeroides RNA polymerase holoenzyme (EςD) that transcribes the Escherichia coli ς70-dependent lacUV5 and ColE1 RNA promoters in vitro (10). Alternatively, cycA promoter recognition by minor RNA polymerase holoenzymes (5, 13, 14) could link cytochrome c2 synthesis to conditions that increase or decrease function of alternate sigma factors. This appears to be the case for cycA P3, since the chr-4 mutation appears to inactivate a negative regulator of an R. sphaeroides member of the ςE family of alternative eubacterial sigma factors (18a, 21, 22).
These experiments sought to analyze function of cycA P1. We focused on cycA P1 because sequences within ∼60 bp of the transcription start site (+1) are sufficient for function under both respiratory and photosynthetic conditions (16, 21, 22). In vitro transcription assays indicated that R. sphaeroides EςD (10) did not recognize cycA P1. By analyzing a series of mutant cycA P1 promoters fused to a promoter-less E. coli β-galactosidase (lacZ) gene in vivo, we found that sequences within a presumed RNA polymerase binding site were sufficient for activity. Other promoter mutations identified elements related to the E. coli heat shock (Eς32) consensus sequence that were necessary for cycA P1 function. Reconstituted RNA polymerases were used to demonstrate cycA P1 transcription by a holoenzyme containing either a 37-kDa protein (ς37) related to E. coli ς32 or a 38-kDa protein (ς38) that also recognizes heat shock promoters from enteric bacteria (10, 11). We suggest that one reason why cycA P1 might be transcribed by alternate RNA polymerase is to ensure that cytochrome c2 levels can change when its function in energy generation is required to provide ATP for chaperone activity. Consistent with this suggestion, cycA P1 function is elevated after an increase in temperature when eubacterial chaperones are commonly needed for cells to mount a heat shock response (11).
MATERIALS AND METHODS
Construction of individual cycA transcriptional fusions.
To produce cycA::lacZY′ operon fusions, an E. coli-R. sphaeroides shuttle plasmid (pBM4A) was constructed. In the first step, a promoterless lacZY′ operon from pRS415 (23) was isolated (∼3-kb EcoRI-DraI restriction fragment) and cloned between the EcoRI and HpaI restriction endonuclease sites of pKT231 (1), with lacZY′ and kan transcription in opposite directions, to give pBM12. To place a transcription terminator upstream of lacZY′, the ΩSp cartridge from pHP45Ω (19) was isolated as an EcoRI restriction fragment and cloned into the same site of pUC6S (25) with spc transcription in the same direction as bla, yielding pUC6SΩA. Plasmid pUC6SΩA was then partially digested with SpeI and cloned into the EcoRI site of pBM12 that had been made blunt by treatment with the Klenow fragment of DNA polymerase. The resulting plasmid, pBM4A, has KpnI and StuI restriction endonuclease sites for directional cloning of DNA between the ΩSp cassette and lacZY′ (the direction of spc transcription in pBM4A is opposite that of lacZY′). IncQ plasmids like pBM4A are present at ∼4 to 10 copies per R. sphaeroides cell (4).
A pool of oligonucleotides (spanning from position −76 to +16 relative to the cycA P1 transcription start site) with approximately one random mutation per molecule was generated at the University of Wisconsin Biotechnology Center (Madison, Wis.). To synthesize complementary strands, a 16-mer (C2MUTP2, 5′-CCTCCCAGGCCTTGTA-3′; prepared at the University of Wisconsin Biotechnology Center) was annealed to the oligonucleotide pool (at 37 or 45°C) and extended at 70°C for 1 h with Taq DNA polymerase (Promega Inc., Madison, Wis.). The resultant double-stranded DNA molecules contained KpnI and StuI restriction endonuclease sites at the upstream and downstream ends, respectively, along with flanking sequences (TCCCGG and GGGAGG, respectively) to allow endonucleolytic cleavage and cloning into KpnI- and StuI-digested pBM4A. These plasmids were transformed into E. coli S17-1, conjugated into R. sphaeroides 2.4.1 (3), and screened for LacZ activity on Sistrom’s minimal medium plates (15) containing 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactoside ml−1 and 25 μg (each) of kanamycin and spectinomycin ml−1. For sequencing, R. sphaeroides plasmid DNA was transformed into E. coli DH5α (2), isolated, and extended with Taq DNA polymerase and lacZ-specific primers. E. coli strains were grown at 37°C in L broth (17) with 25 μg (each) of kanamycin and spectinomycin ml−1.
A control reporter plasmid with lacZY′ under control of the wild-type cycA P1 was designated pCM241; derivatives containing mutated cycA P1 fragments were named to describe their lesion. For example, reporter plasmids with cycA P1 deletions are named by the missing bases; 73Δ67 lacks promoter DNA from positions −73 to −67. Plasmids with single point mutations or small insertions upstream of +1 are named for the changed bases (for example, G34T has a G-to-T change at position −34 and 29C28 has a C inserted between coordinates −29 and −28). Reporter plasmids with multiple mutations are named M plus a number (e.g., M1). Several mutations (G34T, 29C28, T12A, and A11C) were introduced by site-directed mutagenesis (17) (oligonucleotides from Genosys Inc., The Woodlands, Tex.). When independent isolates of plasmids containing the same mutation were analyzed, only one set of results is reported, since in every case analyzed, the LacZ levels were indistinguishable (data not shown).
LacZ and primer extension assays.
LacZ activity was measured in exponential-phase cells grown in Sistrom’s minimal medium at 30°C (21). For aerobic cells, 5-ml cultures were grown on a rotary shaker in 125-ml flasks. Duplicate cultures were assayed at least three times for each strain. Primer extension assays (9) used a lacZ-specific primer.
Proteins used for in vitro transcription assays.
Either aerobically grown wild-type or ΔRpoH cells (11) were used to obtain RNA polymerase preparations diminished for EςD by Q-Sepharose chromatography (10). E. coli core RNA polymerase was purchased from Epicentre Technologies, Inc. (Madison, Wis.). Core R. sphaeroides RNA polymerase was obtained from an RpoH null strain (11) by affinity chromatography (10) (∼5 g [wet weight] of cells) on a 0.5-ml bed of resin containing the 4RA2 monoclonal antibody against the α-subunit of E. coli RNA polymerase (provided by N. Thompson and R. Burgess). After the column was washed with 10 volumes of TE (0.1 M Tris [pH 7.9], 0.1 mM EDTA) plus 0.5 M NaCl buffer, core RNA polymerase was eluted with TE supplemented with 0.75 M NH4SO4, 40% 2,3-butanediol (24a). Prior to storage at −20°C, the eluted proteins (1 ml) were dialzyed twice against 500 ml of TE containing 0.1 M NaCl, 50% glycerol, and 1 μM dithiothreitol (DTT).
Proteins to be tested for sigma factor activity were eluted from sodium dodecyl sulfate (SDS)-polyacrylamide gels by slight modifications to existing protocols (7). All steps were performed in polypropylene microcentrifuge tubes. Prior to electrophoresis, RNA polymerase samples were concentrated by a 60-min precipitation in ice-cold 10% trichloroacetic acid. After centrifugation (15 min, 12,000 × g, 4°C), the precipitated proteins were washed twice with 800 μl of 80% acetone–20% 10 mM Tris-Cl (pH 7.9) and once with 80% acetone (with 5 min of centrifugation between washes). Concentrated proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (slab gel dimensions 6 by 8.8 by 1/16 in.) along with molecular weight standards (Bethesda Research Laboratories, Gaithersburg, Md.). After electrophoresis (75 V, 13 to 15 h), the gel was cut into ∼0.5-cm-thick slices starting 1.0 cm below the stacking gel. Gel slices were rinsed twice with 800 μl of 1 mM DTT, overlaid with 400 μl of elution buffer (7), and ground with a conical tipped pestle. After the addition of 200 μl of elution buffer and 1 h of incubation at room temperature with occasional mixing, acrylamide particles were removed by centrifugation and the supernatant (∼450 μl) was removed. Eluted proteins were precipitated with 4 volumes of 80% acetone (10 min, 4°C), harvested by centrifugation, and washed twice (with centrifugation between each wash) in 800 μl of 80% acetone–20% 10 mM Tris-Cl (pH 7.9). After residual acetone was removed by evaporation at room temperature, the precipitated proteins were incubated in 20 μl of dilution buffer containing 6 M guanidine chloride (7) for 1 h at room temperature. To renature the proteins, 50 volumes of dilution buffer were added. Samples were incubated for 12 h at room temperature prior to testing for sigma factor activity or storage at −80°C.
Amino-terminal hexahistidine-tagged R. sphaeroides ςD (His-ςD) was obtained by amplifying the sigA (rpoD) (6) gene and cloning it into pQE30 (Qiagen, Inc., Chatsworth, Calif.). The manufacturer’s protocol was used to isolate R. sphaeroides His-ςD from E. coli. Prior to in vitro transcription reactions, the purified protein was treated with guanidine and renatured (see above). To conform with the nomenclature suggested by Lonetto et al. (13), the protein previously referred to as ς93 (8, 10) will hereafter be called ςD, as DNA sequence analysis of the sigA (rpoD) gene (6) shows that it is most closely related to E. coli RpoD.
In vitro transcription assays.
With the following exceptions, conditions and templates for in vitro transcription assays have been described previously (10). Either RNA polymerase holoenzyme (0.4 pmol plus 4 μl of dilution buffer) or reconstituted enzyme (0.4 pmol of core plus 4 μl of renatured sample to be tested for sigma factor activity) was incubated at room temperature for 1 h and added to the transcription mixture (20 nM plasmid template, 40 mM Tris-Cl [pH 7.9], 2 mM EDTA, 5 mM MgCl2, 100 mM KCl, 1 mM DTT, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 0.05 mM UTP, 1 μCi of [α-32P]UTP) in a final volume of 20 μl. Assays were performed for 20 min at 30°C, Sequenase stop solution (U.S. Biochemical Corp., Cleveland, Ohio) was added, samples were heated to 90°C and then resolved on 6% polyacrylamide–7 M urea gels (10). Transcripts were visualized on dried gels with X-ray film and quantitated using a PhosphorImager and ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.).
The wild-type cycA P1 template (pRKK139) contained DNA from positions −76 to +16 cloned in the KpnI-HincII sites of pRKK137 (16). Site-directed mutagenesis (12) was used to place the G34T mutation in an otherwise wild-type promoter (pRKK138). pRKK137 lacks cycA P1 sequences; it contains the StuI-BamHI multiple cloning sequence from pUC21 (25) cloned in the HincII-BamHI sites of pUC19spf (10).
RESULTS
Transcription of the cycA P1 promoter by an alternate RNA polymerase holoenzyme.
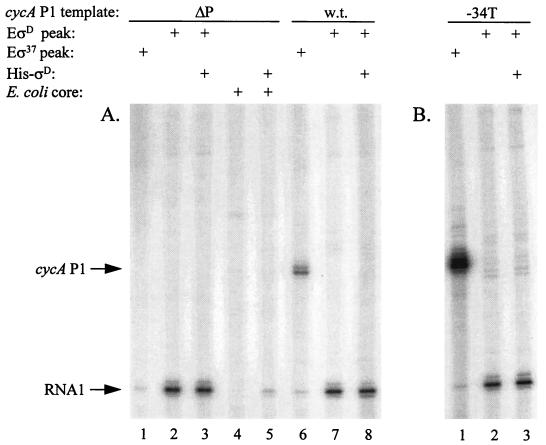
To identify the R. sphaeroides RNA polymerase holoenzyme that recognizes cycA P1, in vitro transcription assays were performed with enzyme samples that recognized different classes of eubacterial promoters (10). A sample enriched in the R. sphaeroides homolog of E. coli Eς70, EςD (8, 10), did not produce a transcript from a cycA P1 template extending from positions −76 to +16 relative to the transcription initiation site (Fig. 1A). However, this enzyme sample produced a Eς70-dependent RNA1 transcript from a promoter near the plasmid DNA replication origin (Fig. 1A). The failure of this EςD sample to transcribe cycA P1 was apparently not due to a limitation for ςD, because a transcript was not observed when an excess of a recombinant R. sphaeroides His-ςD preparation that stimulated RNA1 formation was added to E. coli core RNA polymerase (Fig. 1A).
FIG. 1.
In vitro transcription of cycA P1 templates by different RNA polymerase holoenzymes. (A) Transcription assays were performed as described in Materials and Methods with plasmid templates which lack (ΔP) or contain the wild-type cycA P1 promoter (w.t.) cloned upstream of the spf transcriptional terminator. The RNA polymerases used (+) are indicated above each set of samples. EςD and Eς37 are different R. sphaeroides holoenzymes that can be separated by Q-Sepharose chromatography (10). Where indicated, recombinant R. sphaeroides His-ςD was added to increase the saturation of RNA polymerase preparations. The arrows indicate the positions of RNA1 and cycA P1 transcripts. (B) Analogous assays were performed with a template containing the G34T mutant cycA P1 promoter (−34T).
Instead, cycA P1 was transcribed by an RNA polymerase sample that contained a protein (RpoH or ς37) related to E. coli ς32 (10, 11) (Fig. 1A). Several observations indicate that this transcript was derived from cycA P1. First, it was absent when a control template (pRKK137) that lacks cycA P1 sequences was used (Fig. 1A). The product was also of the expected size after the distance between cycA P1 and a downstream transcription terminator was considered. This set of results suggest cycA P1 is transcribed by an alternate RNA polymerase holoenzyme. Since ς37 was reasonably abundant in the enzyme sample that transcribed cycA P1 (see below; 10), it was possible this promoter was recognized by a holoenzyme, Eς37, that transcribed E. coli ς32-dependent genes (10).
Function of cycA P1 from a reporter gene containing 76 bp of upstream DNA.
To test the previous concept, we sought to analyze the in vivo effects of mutations on cycA P1 function. The normal cycA P1 transcription initiation site (16) was found in primer extension assays with RNA from cells containing wild-type cycA P1 (from positions −76 to +16) fused to lacZ (data not shown). LacZ levels in cells harboring this cycA P1::lacZY′ fusion (pCM241) were ∼10-fold higher than those when a promoter-less version of this plasmid (pBM4A) was present (Fig. 2, top). We present only promoter activity in aerobic cells because control experiments confirmed the prediction (16, 21, 22) that LacZ levels in photosynthetic cultures harboring wild-type or selected mutant cycA P1 reporter plasmids were identical (data not shown). These and other observations verify that the LacZ activity from these low-copy-number plasmids is a valid indicator of cycA P1 function.
FIG. 2.
Upstream or downstream mutations that do or do not alter cycA P1 activity. The sequence of wild-type cycA P1 (plasmid pCM241) is shown at the top with potential −10 and −35 elements set off by spaces. Bases that differ in individual mutant promoters are shown in lowercase letters and in bold type. Small deletions are indicated by dots. Vector sequences contributed by the deletions are underlined twice in panel A. Bases that are identical to those of the wild-type cycA P1 sequence are shown in uppercase letters. The reported β-galactosidase (LacZ) activities are averages of at least three independent cultures and are shown with standard deviations. LacZ activities that are significantly lower than the wild-type activity (∗) or significantly higher than a wild-type reporter gene (‡) are indicated.
Activity of cycA P1 requires sequences between positions −34 and −6.
After analyzing function of an extensive library of mutant cycA P1 promoters, no significant change in LacZ levels was produced from mutant reporter plasmids with either single-base substitutions or a 1-bp deletion downstream of position −7 or point mutations and small deletions between −71 and −39 (data not shown). Thus, it appeared that cycA P1 activity was independent of these sequences.
LacZ levels from a set of mutant cycA P1 promoters with large deletions upstream of position −39 divide them into two groups. Cells containing mutant cycA P1 promoters with deletions ending upstream of −34 produced normal LacZ levels (a subset are shown in Fig. 2A) without altering the transcription initiation site (the primer extension product of the 69Δ49 promoter was indistinguishable from that of a wild-type reporter gene; data not shown). In contrast, LacZ levels from any mutant reporter gene with a deletion that extends downstream of −34 (74Δ32 plus others not shown) were significantly lower than those produced by a wild-type control (Fig. 2A). By comparing LacZ levels produced by cells harboring 41Δ35 or 74Δ32 reporter gene (Fig. 2A), it appeared that sequences downstream of −34 contributed to cycA P1 activity.
Point mutations that alter cycA P1 function map to regions similar to the E. coli ς32 consensus promoter.
Several alterations between positions −34 and −7 significantly alter cycA P1 activity. Although some of these mutant promoters also contain other lesions, the results of our analysis of a large number of point mutations or deletions in these regions suggest that these additional changes do not alter cycA P1 function (see above).
Of all the mutant cycA P1 promoters isolated, only a substitution at position −34 (G34T) that improves the match to the E. coli ς32 consensus −35 element (5) increased its function (∼20-fold [Fig. 2B]). This mutation appears to simply increase cycA P1 function because the G34T transcription initiation site is indistinguishable from a wild-type promoter (in vivo data not shown; see below for in vitro analysis). Other changes at position −34 had either no effect or decreased cycA P1 function (Fig. 2B), so the base preference at this site appears to be T>G≈A>C. Mutations at several other conserved bases in the E. coli ς32 −35 consensus sequence reduced cycA P1 function (Fig. 2B). For example, LacZ levels were reduced by placing an A (T33A) or G (M1) at position −33, placing an A at −32 (G32A), or placing a C at −31 (M3) (Fig. 2B). In contrast, LacZ levels from the sole cycA P1 promoter with a substitution at position −29 (T-to-A change in M9) were indistinguishable from a wild-type reporter plasmid (Fig. 2B). The analogous position is variable in the E. coli ς32 −35 consensus (5), so the failure of the T29A mutation to alter LacZ levels is not surprising.
Several mutations in a region related to the E. coli ς32 −10 consensus sequence also decreased cycA P1 function (Fig. 2C). The reduction in promoter activity caused by mutations at position −12 (T12A) or −11 (A11C or A11T) are expected, since they lowered the match of cycA P1 to conserved positions in the E. coli ς32 consensus −10 element (10). In contrast, a cycA P1 promoter with a C at −9 (M15) or either A (C7A) or T (M18) at −7 had essentially wild-type activity (Fig. 2C). The positions analogous to −9 and −7 are nonconserved in the E. coli ς32 −10 consensus (5), so it is not surprising that these later mutations changes did not significantly alter cycA P1 function.
Other mutations consistent with recognition of cycA P1 by a member of the ς32 family.
Creating a 17-bp spacer (29C28) decreased cycA P1 function slightly, but a mutant promoter with a 15-bp (25ΔT, 16ΔC, or 13ΔC) or 14-bp (M24) spacer had wild-type activity (Fig. 2D). Function of cycA P1 also requires sequences upstream of −10, since changes at −13 (C13A) decreased LacZ levels (Fig. 2D). The E. coli ς32 consensus includes three conserved C residues in an extended −10 element (5), so loss of the C at position −13 is a simple explanation for the lower activity of the C13A and M9 mutant promoters.
Transcription of the G34T promoter by individual R. sphaeroides holoenzymes.
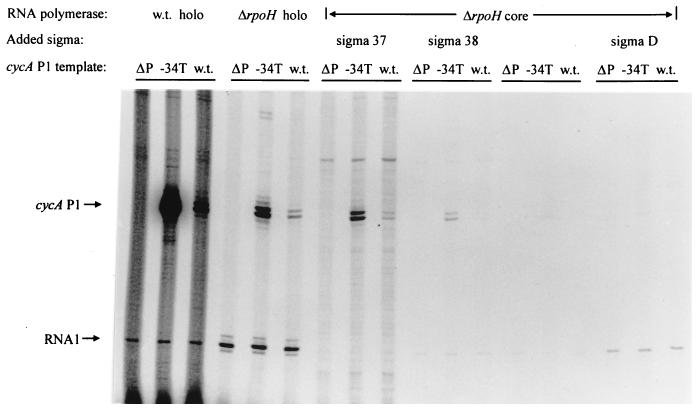
The ability of an Eς37-containing fraction to transcribe cycA P1 in vitro and the in vivo behavior of several mutant promoters suggested that cycA P1 was transcribed by an alternate RNA polymerase holoenzyme. Because it is difficult to obtain homogenous samples of alternate RNA polymerase holoenzymes by chromatography, our Eς37 fraction probably contained additional sigma factors. To assess what holoenzyme transcribed cycA P1, we sought to perform in vitro transcription assays after core RNA polymerase was mixed with potential sigma factors that were isolated from SDS-polyacrylamide gels. These assays initially used the G34T mutant promoter that exhibited increased activity in vivo in order to increase our chances of detecting an in vitro product with reconstituted RNA polymerase holoenzymes. Control experiments showed that the G34T promoter was transcribed by the same R. sphaeroides RNA polymerase holoenzyme preparations that recognized cycA P1 (Fig. 1B). The size of the transcript from the G34T template was also indistinguishable from that produced by wild-type cycA P1. Of equal significance, the approximately eightfold increase in abundance of the G34T product using an RNA polymerase sample from wild-type cells indicates that this mutation alters transcription in a minimal in vitro system (see Fig. 3 for a direct comparison).
FIG. 3.
Transcription of wild-type cycA P1 and G34T templates by individual RNA polymerase holoenzymes. Templates which lack the cycA P1 sequence (ΔP), the wild-type cycA P1 promoter (w.t.), or the G34T (−34T) mutation in the cycA P1 promoter were used as indicated directly above the gel. Above each set of lanes (ΔP, −34T, and w.t.) is indicated the source of RNA polymerase holoenzyme (w.t. holo, holoenzyme preparations from wild-type cells; ΔrpoH holo, analogous material from a ΔRpoH mutant [11]; ΔrpoH core, core RNA polymerase from a ΔRpoH mutant). For assays where core RNA polymerase was reconstituted with potential sigma subunits, ς37 and ςD were obtained from a wild-type RNA polymerase sample (fractions 9 and 19, respectively, in Fig. 4A). ς38 was obtained from the ΔRpoH RNA polymerase sample (fraction 20 in Fig. 4B). The positions of transcripts (arrows) are indicated as follows: RNA1, the EςD-dependent transcript from the oriV promoter present on all templates (10); cycA P1, the specific cycA transcript produced by templates that contain wild-type or G34T mutant promoters.
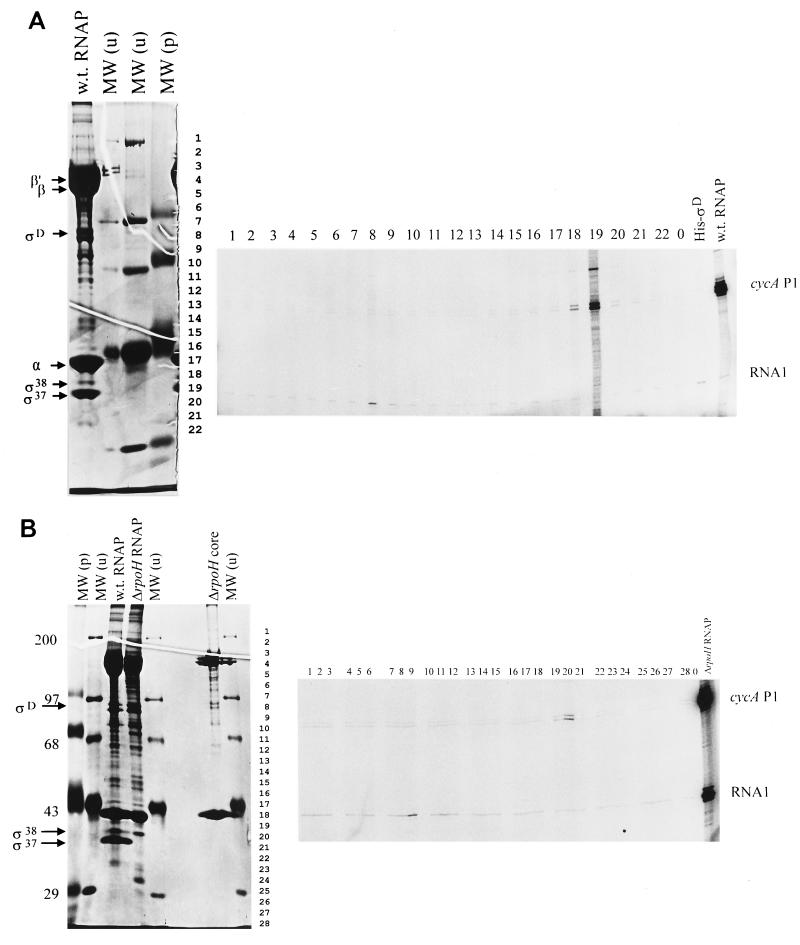
Our core R. sphaeroides RNA polymerase lacked detectable ς37 and contained only small amounts of ςD (Fig. 4A). This explains why little transcription of the G34T, wild-type cycA P1, and the ς70-dependent RNA1 promoter was observed with this sample (Fig. 3). When individual proteins were reconstituted with this core RNA polymerase, the RNA1 transcript was synthesized when either an ∼93-kDa protein (i.e., R. sphaeroides ςD; 10) or recombinant His-ςD was added (Fig. 4A). In contrast, the G34T transcript was generated when fraction 18 or 19 (containing ∼37- or 38-kDa proteins) was added to core RNA polymerase (Fig. 4A). One explanation for synthesis of the cycA P1 transcript in two reconstitution mixtures is the presence of the same sigma factor in both fractions. However, when the abundance of the transcripts in these two reaction mixtures is compared to the levels of the 37-kDa and 38-kDa polypeptides in the RNA polymerase sample used as a source of sigma factors (fractions 18 and 19 [Fig. 4A]), it is also possible that different RNA polymerase holoenzymes (Eς37 and Eς38) transcribe G34T.
FIG. 4.
Transcription of the G34T mutant promoter by reconstituted RNA polymerase holoenzymes. (A) Transcripts produced when the G34T promoter is incubated with different R. sphaeroides RNA polymerase samples. The positions of transcripts are indicated to the right of the rightmost gels as follows: RNA1, the EςD-dependent oriV transcript from the transcription plasmid template (10); cycA P1, the product of the G34T promoter. Reactions in panel A used renatured proteins from a wild-type RNA polymerase sample (shown on the left) as a source of potential sigma factors for addition to core RNA polymerase (shown in panel B) from the ΔRpoH mutant (11). Reactions in panel B used renatured proteins from a RpoH null mutant as a source of potential sigma factors (11) for reconstitution with the same ΔRpoH core RNA polymerase. In panels A and B, numbers above the rightmost gels denote the SDS-polyacrylamide gel fraction that was added to core RNA polymerase as a source of potential sigma factor. Lane 0 contains transcripts produced by core RNA polymerase alone. Lane His-ςD in panel A shows transcripts produced when recombinant R. sphaeroides His-ςD is added to core RNA polymerase. To the left of the leftmost gels in panels A and B is shown the polypeptide composition of RNA polymerase samples used for in vitro transcription reactions. These samples include RNA polymerase samples from wild-type cells (w.t. RNAP in panels A and B) or a ΔRpoH mutant (11) (ΔrpoH RNAP in panel B) and core RNA polymerase from an RpoH null strain (ΔrpoH core in panel B) separated alongside prestained [MW (p)] or unstained [MW (u)] protein molecular weight standards (molecular weights [in thousands] indicated on the left margin). Numbers to the right of the leftmost gel indicate how this gel was sliced to obtain proteins to be tested for sigma factor activity. Individual RNA polymerase subunits are indicated to the left.
A second RNA polymerase holoenzyme transcribes the G34T cycA P1 promoter.
One indication that more than one RNA polymerase holoenzyme transcribes cycA P1 was the ability of RNA polymerase from a strain lacking ς37 (ΔRpoH holo) to recognize both the wild-type and G34T promoters (Fig. 3). To identify the sigma factor within the ΔRpoH enzyme preparation that recognizes these promoters, this holoenzyme was fractionated by SDS-PAGE, reconstituted with core RNA polymerase, and tested for activity with the G34T promoter (Fig. 4B). Of the proteins in the ΔRpoH holoenzyme preparation that were reconstituted with core RNA polymerase, only the ∼38-kDa protein produced a G34T-specific transcript (Fig. 4B). Thus, we conclude that cycA P1 is transcribed by two distinct RNA polymerase holoenzymes, Eς37 and Eς38.
Wild-type cycA P1 is also transcribed by two alternate RNA polymerase holoenzymes.
When wild-type cycA P1 was tested with the same reconstituted holoenzymes, only fractions containing ς37 or ς38 enabled core RNA polymerase to recognize this promoter (Fig. 3). Note that the cycA P1 transcripts are identical in size to those produced from G34T regardless of whether reconstituted Eς37 (generated with the 37-kDa protein from wild-type RNA polymerase preparations) or Eς38 (prepared with the 38-kDa protein from the RpoH mutant; Fig. 3) was used. In addition, the G34T transcript produced by each reconstituted holoenzyme is more abundant than the product of wild-type cycA P1 (∼5-fold more for Eς37 and ∼3-fold more for Eς38; Fig. 3). These results suggest that the G34T mutation increases promoter recognition by both Eς37 and Eς38.
DISCUSSION
Previous studies have shown that cycA P1 function is independent of signals that control genes for other R. sphaeroides energy-generating proteins (16, 21), but it was not known whether its transcription required proteins in addition to RNA polymerase. In addition, the exact cycA P1 promoter was not known, since this region contains several appropriately positioned elements with limited similarity to those recognized by eubacterial RNA polymerase holoenzymes (16). From our in vivo and in vitro analysis of cycA P1 function, we can make several conclusions regarding DNA sequences, RNA polymerase holoenzymes, and proteins required for transcription.
A minimal promoter region is sufficient for cycA P1 function.
The wild-type LacZ levels produced by cycA P1 promoters with mutations upstream of position −34 or downstream of −7 suggested these regions are not target sites for positive or negative regulators. If a transcription factor binds upstream of −34, a significant fraction of the deletions in this region should have altered cycA P1 function either by preventing this interaction or by changing the orientation of this potential regulator and RNA polymerase. Thus, it appears unlikely that the RNA polymerase holoenzymes that recognize cycA P1 require other DNA binding proteins for activity.
Sequences required for cycA P1 function.
In contrast, elements between positions −34 and −7 are required for cycA P1 function, since several mutations reduced transcription. The spectrum of cycA P1 mutations analyzed allows us to rule out a role for previously noted hexamers with similarity to the E. coli ς70 −35 consensus sequence (−36TAGTGA−31 and −30TTGTGT−25) in promoter function (16).
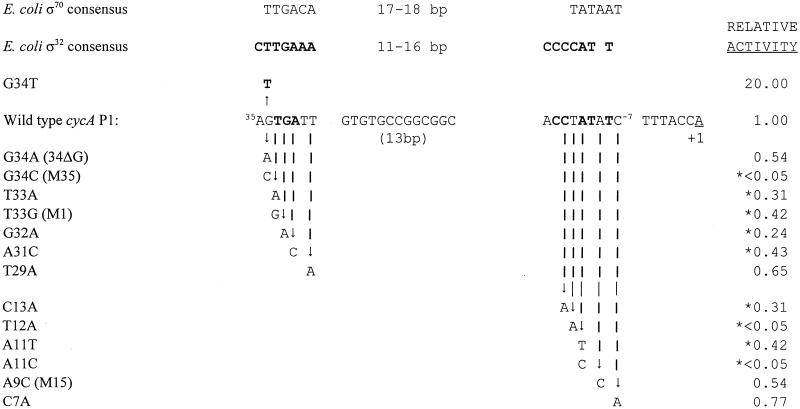
We also found that cycA P1 function was altered by point mutations in elements similar to the −35 (−34GTGATT−29) and −10 (−12TATATC−7) consensus sequences for both E. coli Eς32 (boldfaced bases) and Eς70 (underlined bases). The high degree of amino acid sequence identity in regions 2.4 and 4.2 of both R. sphaeroides ςD (6) and ς37 (11) to their E. coli counterparts predicts that these sigma factors will recognize similar promoter elements. Therefore, conclusions about which holoenzyme transcribes cycA P1 could not have been made solely from an in vivo analysis because our mutant promoter bank did not contain every possible change at all positions. When the in vitro and in vivo results are considered together, we can conclude that cycA P1 is transcribed by alternate R. sphaeroides RNA polymerase holoenzymes that recognize E. coli heat shock promoters (Fig. 5 summarizes data from salient cycA P1 alleles).
FIG. 5.
Effects of point mutations on cycA P1 function. Below the consensus promoters for E. coli Eς70 and Eς32 (top) are listed mutations that increase (in bold type and above the wild-type promoter) or decrease (below the wild-type promoter) cycA P1 activity (normalized to wild-type activity of 1.00; see Fig. 2B to D for primary data). Relative activities were corrected for LacZ levels produced from a control plasmid lacking cycA P1 sequences (pBM4A [Fig. 2]). Asterisks indicate mutations that significantly alter promoter activity. In cases where the cycA P1 mutation is not identical to the name of the mutant promoter, its name is provided in parentheses. Bases shown in bold type in the cycA P1 sequence are identical to the bases in the E. coli Eς70 and Eς32 consensus promoters.
Alternate R. sphaeroides RNA polymerase holoenzymes transcribe cycA P1.
The G34T mutation was particularly useful in demonstrating that alternate RNA polymerase holoenzymes transcribe cycA P1. Even though G34T is a change towards both the ς70 and ς32 −35 consensus sequence (Fig. 5), active preparations of R. sphaeroides EςD failed to transcribe either this promoter or wild-type cycA P1 in vitro. Instead, both wild-type cycA P1 and a G34T template were recognized by R. sphaeroides RNA polymerase holoenzymes containing either a 37-kDa (Eς37) or 38-kDa (Eς38) subunit. R. sphaeroides ς37 is a member of the E. coli ς32 family of alternate sigma factors (10, 11). Recently, Eς38 has also been shown to transcribe E. coli heat shock promoters (11). Eς37 and Eς38 must both contribute to cycA P1 activity in vivo, since its function in steady-state aerobic cells is not abolished by loss of ς37 (11).
Additional suggestions that cycA P1 is recognized by an alternate RNA polymerase holoenzyme came from analyzing how altering the spacer length or sequence changed promoter activity. Spacing of 17 or 18 bp between the −35 and −10 elements is optimal for E. coli Eς70 promoters (18, 20, 24, 26). In contrast, E. coli ς32 promoters have spacer lengths of 12 to 17 bp (10). Thus, function of RNA polymerase holoenzymes with DNA recognition properties similar to Eς32 could explain why decreasing the cycA P1 spacer length to 15 or 14 bp did not affect promoter activity. The reduction in cycA P1 activity caused by changing a conserved C (C13A [Fig. 5]) in a potential extended −10 element similar to E. coli heat shock promoters (5) is also consistent with transcription by R. sphaeroides RNA polymerase holoenzymes (Eς37 and Eς38) related to Eς32.
Potential metabolic significance of cycA P1 transcription by alternate RNA polymerase holoenzymes.
One can envision several reasons why cycA P1 is transcribed by Eς37 and Eς38. Other data indicate that R. sphaeroides Eς37 and Eς38 each transcribe E. coli heat shock genes (11). Thus, in addition to maintaining a pool of cytochrome c2 in steady-state cells, cycA P1 transcription by these alternate RNA polymerase holoenzymes could increase levels of this electron carrier when cells need ATP for cytoplasmic chaperones to assemble or renature macromolecular assemblages (5). Indeed, the modest increase in cycA P1 function that occurs when aerobic wild-type cells are shifted to 42°C is reduced in a ς37 mutant (11).
The increased promoter function caused by the G34T mutation indicates that cycA P1 is not optimal for recognition by either Eς37 or Eς38 (Fig. 5). This, plus the lack of information on how mutations at individual positions in the E. coli ς32 consensus sequence alters its function (5), makes it difficult to decipher consensus promoters for Eς37 and Eς38 and ask if related sequences are present upstream of genes for other R. sphaeroides energy-generating proteins.
In summary, our results indicate that cycA P1 is transcribed by alternate forms of eubacterial RNA polymerase. Both wild-type cycA P1 and a mutant promoter with increased transcription in vivo (G34T) are transcribed in a minimal in vitro system consisting of either reconstituted Eς37 or Eς38. Recognition of cycA P1 by alternate RNA polymerase holoenzymes or its function without additional transcription factors is not in conflict with previous observations that activity of this promoter is increased by the trans-acting chr-4 allele (21, 22). Ongoing experiments suggest that the chr-4 mutation increases function of an R. sphaeroides member of the eubacterial ςE family (18a). By analogy to the situation where E. coli EςE transcribes the ς32 gene (5), it seems likely that increased ςE function caused by the chr-4 mutation simply elevates activity of either Eς37 or Eς38 and thereby raises cycA P1 activity. From our current understanding of cycA P1, it seems clear that a further dissection of its function will increase our understanding of how synthesis of energy-generating electron transport proteins is altered under conditions of metabolic or environmental stress. It could also provide important insights into how gene expression is modulated by regulating function of different RNA polymerase holoenzymes that recognize overlapping promoters.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM37509 to T.J.D. Early in these studies, B.J.M. and R.K.K. were supported by NIH predoctoral training grant GM07215 to UW-Madison.
We thank Nancy Thompson and Richard Burgess for monoclonal antibodies and advice on RNA polymerase reconstitution.
REFERENCES
- 1.Bagdasarian M, Lurz R, Rückert B, Franklin F C H, Bagdasarian M M, Frey J, Timmis K N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 2.Bethesda Research Laboratories. BRL pUC host: Escherichia coli DH5α™ competent cells. Bethesda Res Lab Focus. 1986;8:9–10. [Google Scholar]
- 3.Donohue T J, McEwan A G, Van Doren S, Crofts A R, Kaplan S. Phenotypic and genetic characterization of cytochrome c2 deficient mutants of Rhodobacter sphaeroides. Biochemistry. 1988;27:1918–1924. doi: 10.1021/bi00406a018. [DOI] [PubMed] [Google Scholar]
- 4.Donohue T J, Kaplan S. Genetic techniques in the Rhodospirillaceae. Methods Enzymol. 1991;204:459–485. doi: 10.1016/0076-6879(91)04024-i. [DOI] [PubMed] [Google Scholar]
- 5.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 1382–1399. [Google Scholar]
- 6.Gruber T M, Bryant D A. Molecular systematics of eubacteria using ς70-type sigma factors of group 1 and group 2. J Bacteriol. 1997;179:1734–1747. doi: 10.1128/jb.179.5.1734-1747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hager D A, Burgess R R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980;109:76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- 8.Kansy J, Kaplan S. Purification, characterization, and transcriptional analysis of RNA polymerase from Rhodobacter sphaeroides cells grown chemoheterotrophically and photoheterotrophically. J Biol Chem. 1989;264:13751–13759. [PubMed] [Google Scholar]
- 9.Karls R, Schulz V, Jovanovich S B, Flynn S, Pak A, Reznikoff W S. Pseudorevertants of a lac promoter mutation reveal overlapping nascent promoters. Nucleic Acids Res. 1989;17:3927–3949. doi: 10.1093/nar/17.10.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karls R K, Jin D J, Donohue T J. Transcription properties of RNA polymerase holoenzymes isolated from the purple nonsulfur bacterium Rhodobacter sphaeroides. J Bacteriol. 1993;175:7629–7638. doi: 10.1128/jb.175.23.7629-7638.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karls R K, Brooks J, Rossmiessl P, Luedke J, Donohue T J. Metabolic roles of a Rhodobacter sphaeroides member of the ς32 family. J Bacteriol. 1998;180:10–19. doi: 10.1128/jb.180.1.10-19.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lonetto M, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lonetto M A, Brown K L, Rudd K, Buttner M J. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase ς factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lueking D R, Fraley R T, Kaplan S. Intracytoplasmic membrane synthesis in synchronous cell populations of Rhodopseudomonas sphaeroides. J Biol Chem. 1978;253:451–457. [PubMed] [Google Scholar]
- 16.MacGregor B J, Donohue T J. Evidence for two promoters for the cytochrome c2 gene (cycA) of Rhodobacter sphaeroides. J Bacteriol. 1991;173:3949–3957. doi: 10.1128/jb.173.13.3949-3957.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 18.Mulligan M E, Brosmius J, McClure W R. Characterization in vitro of the effect of spacer length on the activity of Escherichia coli RNA polymerase at the TAC promoter. J Biol Chem. 1985;260:3529–3538. [PubMed] [Google Scholar]
- 18a.Newman, J. D., B. A. Schilke, and T. J. Donohue. Unpublished data.
- 19.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 20.Russell D R, Bennett G N. Construction and analysis of in vivo activity of E. coli promoter mutants that alter the −35 to −10 spacing. Gene. 1982;20:231–243. doi: 10.1016/0378-1119(82)90042-7. [DOI] [PubMed] [Google Scholar]
- 21.Schilke B A, Donohue T J. δ-Aminolevulinate couples cycA transcription to changes in heme availability in Rhodobacter sphaeroides. J Mol Biol. 1992;226:101–115. doi: 10.1016/0022-2836(92)90127-6. [DOI] [PubMed] [Google Scholar]
- 22.Schilke B A, Donohue T J. ChrR positively regulates transcription of the Rhodobacter sphaeroides cytochrome c2 gene. J Bacteriol. 1995;177:1929–1937. doi: 10.1128/jb.177.8.1929-1937.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 24.Stefano J E, Gralla J D. Spacer mutations in the lac ps promoter. Proc Natl Acad Sci USA. 1982;79:1069–1072. doi: 10.1073/pnas.79.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Thompson, N. Personal communication.
- 25.Vieira J, Messing J. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene. 1991;100:189–194. doi: 10.1016/0378-1119(91)90365-i. [DOI] [PubMed] [Google Scholar]
- 26.Warne S E, deHaseth P L. Promoter recognition by Escherichia coli RNA polymerase. Effects of single base pair deletions and insertions in the spacer DNA separating the −10 and −35 regions are dependent on spacer DNA sequence. Biochemistry. 1993;32:6134–6140. doi: 10.1021/bi00075a003. [DOI] [PubMed] [Google Scholar]