Abstract
We report the role of a gene (rpoH) from the facultative phototroph Rhodobacter sphaeroides that encodes a protein (ς37) similar to Escherichia coli ς32 and other members of the heat shock family of eubacterial sigma factors. R. sphaeroides ς37 controls genes that function during environmental stress, since an R. sphaeroides ΔRpoH mutant is ∼30-fold more sensitive to the toxic oxyanion tellurite than wild-type cells. However, the ΔRpoH mutant lacks several phenotypes characteristic of E. coli cells lacking ς32. For example, an R. sphaeroides ΔRpoH mutant is not generally defective in phage morphogenesis, since it plates the lytic virus RS1, as well as its wild-type parent. In characterizing the response of R. sphaeroides to heat, we found that its growth temperature profile is different when cells generate energy by aerobic respiration, anaerobic respiration, or photosynthesis. However, growth of the ΔRpoH mutant is comparable to that of a wild-type strain under each of these conditions. The ΔRpoH mutant mounted a heat shock response when aerobically grown cells were shifted from 30 to 42°C, but it exhibited altered induction kinetics of ∼120-, 85-, 75-, and 65-kDa proteins. There was also reduced accumulation of several presumed heat shock transcripts (rpoD PHS, groESL1, etc.) when aerobically grown ΔRpoH cells were placed at 42°C. Under aerobic conditions, it appears that another sigma factor enables the ΔRpoH mutant to mount a heat shock response, since either RNA polymerase preparations from an ΔRpoH mutant, reconstituted Eς37, or a holoenzyme containing a 38-kDa protein (ς38) each transcribed E. coli Eς32-dependent promoters. The lower growth temperature profile of photosynthetic cells is correlated with a difference in heat-inducible gene expression, since neither wild-type cells or the ΔRpoH mutant mount a typical heat shock response after such cultures were shifted from 30 to 37°C.
The heat shock response is a universal phenomenon that allows cells to survive a multitude of environmental stresses (28). This response is characterized by a rapid, transient increase in the rate of synthesis of a highly conserved set of polypeptides known collectively as heat shock proteins (HSPs) (14, 28). While increased expression of HSPs is often seen after a stress, most of these gene products are present at significant levels in steady-state cells to promote protein synthesis, folding, and intracellular localization (28). Molecular chaperone function for HSPs has been demonstrated in key cellular processes such as DNA replication, cell division, or maintenance of active protein conformations during conditions that cause cytoplasmic stress (13, 14, 28, 43).
A molecular picture for what triggers the heat shock response is beginning to emerge. In the enteric bacterium Escherichia coli, an alternative sigma factor (ς32) that recognizes heat shock gene promoters (15) increases in abundance, stability, and activity under conditions that cause the accumulation of misfolded cytoplasmic proteins (38). Use of alternate sigma factors could be a common way to control the eubacterial heat shock response, since proteins related to E. coli ς32 have been identified from a diverse group of proteobacteria (4, 24, 27, 32, 41). These other ς32 family members are believed to function in much the same manner as E. coli ς32, becoming more abundant or active in response to thermal or metabolic stimuli that produce cytoplasmic stress.
While control of the heat shock response by E. coli ς32 is considered the eubacterial paradigm, other ways to regulate procaryotic HSP synthesis exist. An additional alternate sigma factor (ςE) recognizes the E. coli ς32 gene (rpoH) and several other heat shock promoters in response to signals that generate periplasmic stress (11). In several other eubacteria, a cis-active inverted repeat called CIRCE (for controlling inverted repeat of chaperone expression) negatively regulates heat shock gene expression (1, 19, 46). Indeed, recent experiments suggest that individual Bradyrhizobium japonicum heat shock genes are regulated by both members of the ς32 family and CIRCE elements (1, 27).
This work sought to identify the function of a member of the ς32 family from the facultative phototroph Rhodobacter sphaeroides. Previous experiments implicated a 37-kDa R. sphaeroides protein (ς37) that reacted with antibody against E. coli ς32 as a member of the RpoH family (16). The observation that several E. coli heat shock promoters were transcribed by R. sphaeroides RNA polymerase samples that contained this 37-kDa protein supported the provisional designation of ς37 as a member of the ς32 family (16). The recent finding that R. sphaeroides Eς37 transcribes a promoter (cycA P1) for an essential component of the R. sphaeroides photosynthetic apparatus like cytochrome c2 suggested that ς37 might have functions outside its commonly accepted role in HSP synthesis (21). Since little is known about the ability of ς32 family members to recognize genes other than those which encode HSPs, we were particularly interested in asking if Eς37 or its target genes contribute to expression or assembly of proteins that function in biological energy generation by R. sphaeroides.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli (Table 1) strains were grown at 37°C in Luria-Bertani medium (33). R. sphaeroides strains (Table 1) were routinely grown at 30°C in Sistrom’s succinate-based minimal medium A (37). For growth of cells by anaerobic respiration in the dark, dimethyl sulfoxide (DMSO) was used as a terminal electron acceptor in Sistrom’s minimal medium containing 20 mM glucose and 0.2% yeast extract (7). Plating efficiency of phage RS1 was determined with aerobically grown cells (6).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant feature(s) | Source or reference |
|---|---|---|
| R. sphaeroides | ||
| 2.4.1 | Wild type | Laboratory strain |
| RpoH1 | ΔrpoH1::ΩSpr derivative of 2.4.1 | This work |
| RpoH26 | ΔrpoH1::ΩSpr derivative of 2.4.1 with pPJR19 integrated | This work |
| E. coli | ||
| DH5α | Strain used for plasmid maintenance | 3 |
| S17-1 | Conjugative donor for mating | 36 |
| CAG12517 | MC4100 (R40) ΔrpoH30::kan, suppressor tRNA | 44 |
| Plasmids | ||
| pGEM-7Zf(+) | Apr | Promega |
| pSUP202 | Apr Tcr CmrR. sphaeroides suicide plasmid | 36 |
| pHP45Ω | Apr Spr source of 2-kb ΩSpr cartridge | 31 |
| pPJR18 | 4.3-kb EcoRI restriction fragment of rpoH in pGEM-7Zf(+) | This work |
| pPRJ19 | pPJR18 lacking 3-kb SmaI restriction fragment | This work |
| pPJR26 | pPJR19 StyI restriction fragment replaced by ΩSpr cartridge | This work |
| pPJR29 | rpoH region from pPRJ26 in pSUP202 | This work |
Tellurite sensitivity was tested by diluting aerobically grown cells (∼109 cells/ml) sufficiently to produce ∼100 colonies per plate (23). To score for growth phenotypes associated with loss of ς37, solid media were used routinely. Where indicated, growth curves with liquid cultures were performed to test phenotypes associated with the ΔrpoH allele.
Isolation of R. sphaeroides rpoH.
An E. coli ΔRpoH mutant containing the R40 mutation (CAG12517 [44]) was used to isolate an R. sphaeroides gene that recognizes heat shock promoters. While E. coli ΔRpoH strains are unable to grow above 20°C, the R40 mutation allows growth of this strain up to 40°C (17). To identify genes that restore phage growth to CAG12517, this tester strain was infected with a lytic R. sphaeroides NCIB8253 λgt11 library (40) at a multiplicity of infection of 0.05.
Chromosomal mapping and Southern blot analysis.
Restricted R. sphaeroides genomic DNA was separated with a CHEF-DR II apparatus (Bio-Rad Laboratories, Richmond, Calif.) (39). For Southern blot analysis (33), restricted R. sphaeroides DNA was transferred to membranes, hybridized with nick-translated 32P-labeled rpoH probes (Gibco-BRL, Gaithersburg, Md.), washed at moderate stringency (two 5-min washes at room temperature with a solution consisting of 1× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}] and 0.1% sodium dodecyl sulfate [SDS] and two 15-min washes at 45°C with 0.1× SSPE–0.1% SDS), exposed to a PhosphorImaging screen, and visualized with ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.).
DNA sequence analysis.
DNA sequencing used either Taq DNA polymerase and deazanucleotide triphosphates (Promega, Inc., Madison, Wis.) or automated sequencers at the University of Wisconsin Madison Biotechnology Center. A list of plasmids and primers used to sequence the ∼1.5-kb segment of R. sphaeroides DNA in pPJR19 is available upon request. DNA sequences containing rpoH were analyzed with software from the University of Wisconsin Genetics Computer Group, Madison, Wis. (5).
Construction of a ΔRpoH mutant.
To produce an R. sphaeroides ΔRpoH mutant, a StyI restriction fragment internal to rpoH was replaced with a spectinomycin resistance (Spr) gene. Specifically, pPJR19 was digested with StyI, and the ends were made blunt with T4 DNA polymerase and ligated with a 2-kb SmaI restriction fragment carrying an omega (Ω) cartridge encoding Spr (31). The resultant plasmid (pPJR26) was digested with ApaI to produce a restriction fragment containing the interrupted (ΔrpoH1::ΩSpr) gene. After the ends of this restriction fragment were made blunt, it was purified and cloned into pSUP202, a mobilizable suicide vector (36), which had been digested with PstI and EcoRI and treated with T4 DNA polymerase. The resulting plasmid (pPJR29) was used to place a ΔrpoH1::ΩSpr allele in R. sphaeroides 2.4.1 (7). Screening Spr cells (25 μg/ml) for those sensitive to tetracycline (1 μg/ml) identified a strain (RpoH1) where the ΔrpoH1::ΩSpr allele replaced a wild-type gene by homologous recombination (8). One Tcr strain (RpoH26), in which ΔrpoH1::ΩSpr was integrated into the chromosome along with the suicide plasmid, was used to aid genomic mapping of rpoH (see Results).
Rates of protein synthesis.
Rates of protein synthesis were determined before and after log-phase aerobically grown R. sphaeroides cultures (∼109 cells/ml) were shifted from 30 to 42°C. At indicated times, 1-ml samples were removed, labeled with 50 μCi of [35S]Transmet (Amersham Corp., Arlington Heights, Ill.) for 1 min, and chased with a mixture of 1 M cysteine and 1 M methionine for 2 min. At this time, cold trichloroacetic acid was added to a final concentration of 5%, proteins were harvested by centrifugation (13,000 × g, 10 min), the supernatant was aspirated, and the precipitate was washed twice with ice-cold 80% acetone. After evaporation of residual liquid under vacuum, 100 μl of solubilization buffer containing 10% β-mercaptoethanol was added (33). Samples of equal radioactivity were separated by SDS–12.5% polyacrylamide gel electrophoresis prior to analysis and quantitation on a PhosphorImager with ImageQuant software (Molecular Dynamics).
For monitoring the rates of protein synthesis under photosynthetic conditions, cells were grown to mid-exponential phase (∼5 × 108 CFU/ml) at 30°C in 15-ml screw-capped tubes at a light intensity of 10 W/m2. Protein synthesis was monitored as described above.
Primer extension assays.
RNA from cells grown aerobically (45) was used in primer extension assays (2) with these promoter-specific oligonucleotides: rpoD PHS, 5′-CCTCGACCGCCTCCTCGATTTCCT-3′ (3a); groESL1, 5′-CACGGTCATGCAGCGTTTG-3′ (19); rrnB, 5′-AAGACAAAACAAACCGAGACGCCA-3′ (10). Products were separated on denaturing polyacrylamide gels (2) prior to estimation of their levels on a PhosphorImager with ImageQuant software.
In vitro transcription assays.
Conditions for preparation of R. sphaeroides RNA polymerase, reconstitution of core enzyme samples with potential sigma factors, and their use for in vitro transcription assays with plasmid templates have been described previously (21).
Nucleotide sequence accession number.
DNA sequences containing rpoH were deposited with accession no. U82397.
RESULTS
Identification of R. sphaeroides rpoH.
E. coli ΔRpoH mutants are unable to support λ growth because they are limited for the HSPs that are required for phage DNA replication (13). To identify a gene product that supports λ infection, an E. coli ΔRpoH strain was infected with an R. sphaeroides genomic library in the lytic λgt11 vector (40). Phage DNA was purified (33), digested with EcoRI to yield a 4.3-kb restriction fragment, and the DNA was cloned into pGEM-7Zf(+) to yield pPJR18. To confirm that heat shock gene expression was dependent on R. sphaeroides DNA, this plasmid was shown to support 44°C growth and confer λ sensitivity to an E. coli ΔRpoH mutant (data not shown). In addition, a plasmid (pPJR19) containing a smaller, ∼1.5-kb, restriction fragment (Table 1) activated an rpoD PHS::lacZ fusion (data not shown). The following experiments indicate that the responsible R. sphaeroides gene (rpoH) encodes a sigma factor related to proteins in the eubacterial ς32 family.
R. sphaeroides RpoH is a member of the ς32 family.
The R. sphaeroides DNA that activates E. coli heat shock gene expression encodes a 298-amino-acid protein (33.7 kDa) whose putative start codon (coordinate 418 in accession number U82397) begins 10 bases downstream of a potential ribosome binding site (AGAGG). The deduced protein has a high degree of amino acid similarity to eubacterial proteins in the ς32 family (Fig. 1); it displays 68% identity to Caulobacter crescentus ς32 (32, 41), 66% identity to Agrobacterium tumefaciens ς32 (26), 58% identity to B. japonicum ς32 (27), and 40% identity to E. coli ς32 (15, 18). The similarity of R. sphaeroides RpoH to ς32 family members extends throughout the protein, but the considerable conservation in regions 4.2 (−35 recognition) and 2.4 (−10 recognition) explains why Eς37 transcribes E. coli ς32 promoters (16, 21; also see below). The amino acid similarity is particularly striking in a unique and highly conserved 9-amino-acid insertion [Q(R/K)KLFFNLR], designated the RpoH box (26) (Fig. 1), that has been implicated in DnaK-mediated turnover of E. coli ς32 (25). Other features conserved between R. sphaeroides RpoH and related proteins from nonenteric eubacteria include an insertion in region 3.1 and a C-terminal extension (Fig. 1). It appears that the known ς32 family members can provisionally be separated into two groups (Fig. 1), since proteins from the α proteobacteria (R. sphaeroides, C. crescentus, A. tumefaciens, Zymomonas mobilis, and B. japonicum) are more similar to each other and larger than their counterparts from γ proteobacteria (E. coli, Haemophilus influenzae, Pseudomonas aeruginosa, and Citrobacter freundii). The second amino acid residue in the RpoH box also seems to be a distinguishing feature; it is a lysine in proteins from the α proteobacteria and arginine in those from the γ proteobacteria.
FIG. 1.
Alignment of ς32 family members (generated by using the PILEUP program [5]). Identical amino acids are indicated in boldface. The broken lines above the alignment denote conserved regions of eubacterial sigma factors (20). GenBank accession numbers for ς32 family members are as follows: C. crescentus, U39791; A. tumefaciens, D50828; Z. mobilis, D50832; B. japonicum, U55047; H. influenzae, U32713; P. aeruginosa, D50052; C. freundii, X14960; and E. coli, U00039. RpoH box and helix-turn-helix (H-T-H) sequences are underlined. Gaps introduced to maximize alignment are indicated by dots. Asterisks indicate the end of the protein. Abbreviations: Rsph, R. sphaeroides; Ccre, C. crescentus; Atum, A. tumefaciens; Zmob, Z. mobilis; Bjap, B. japonicum; Hinf, H. influenzae; Paer, P. aeruginosa; Cfre, C. freundii; Ecoh, E. coli.
Mapping rpoH.
When wild-type genomic DNA was probed, rpoH mapped to an 1,100-kb AseI restriction fragment (39; data not shown). To better position rpoH, we took advantage of a strain (RpoH26) containing an additional AseI restriction site from the suicide plasmid pPJR29 (Table 1). When RpoH26 DNA is treated with AseI, the ∼1,100-kb AseI restriction fragment was digested in two ∼500-kb fragments (data not shown). This places rpoH near the center of the ∼1,100-kb AseI fragment or at coordinate ∼2250 ± 50 kb on chromosome I.
An R. sphaeroides ΔRpoH strain is not temperature sensitive.
To determine the role of R. sphaeroides ς37, an rpoH insertion-replacement was generated in a wild-type strain. Because we expected a ΔRpoH mutant to be temperature sensitive, like its E. coli counterpart (44), selection for the ΔrpoH1::ΩSpr allele was initially performed at temperatures between 10 and 30°C. To our surprise, cells lacking RpoH (i.e., ones that are both Spr and Tc sensitive) were obtained at all temperatures. To confirm the genotype of these presumptive ΔRpoH mutants, Southern blot analysis with rpoH, suicide plasmid (pSup202), and spc-specific probes was used to show that the ΔrpoH::ΩSpr allele had been incorporated by an even number of crossover events (data not shown).
Respiratory phenotypes associated with the loss of R. sphaeroides ς37.
Both wild-type and ΔRpoH cells grew at temperatures up to 42°C under either aerobic conditions or when DMSO served as an anaerobic electron acceptor (Table 2). Loss of ς37 did not alter the ability of the lytic bacteriophage RS1 (6) to infect cells, since this virus plated at wild-type efficiency on ΔRpoH cells at all temperatures tested (Table 2). Thus, we conclude ς37 is not required for viability, phage replication, or energy generation in the presence or absence of oxygen.
TABLE 2.
Characterization of an R. sphaeroides ΔRpoH mutanta
| Growth condition and parameter | Wild type | ΔRpoH mutant |
|---|---|---|
| Aerobic respiration | ||
| Temp | ||
| 30°C | +b | + |
| 37°C | + | + |
| 42°C | + | + |
| Tellurite | + | (see Fig. 2) |
| Phage growthc | 1.0 | 1.3 |
| Anaerobic respirationd | ||
| Temp | ||
| 30°C | + | + |
| 37°C | + | + |
| 42°C | + | + |
| Photosynthesis | ||
| Temp | ||
| 30°C | + | + |
| 37°C | − | − |
| 42°C | − | − |
| B875c | 225 | 247 |
| B800-850e | 447 | 498 |
| Ratioe | 2.0 | 2.0 |
Unless noted, all phenotypes were scored on solid media.
+, no distinguishable difference between the behavior of wild-type and ΔRpoH cells; −, no growth.
Plating efficiency of phage RS1 under aerobic conditions at 30, 37, or 42°C (6).
With DMSO as an anaerobic electron acceptor.
picomoles of pigment-protein complexes in ∼7 × 108 to 8 × 108 cells grown photosynthetically at 30°C with a light intensity of 10 W/m2 (22). Ratio, B800-850/B875.
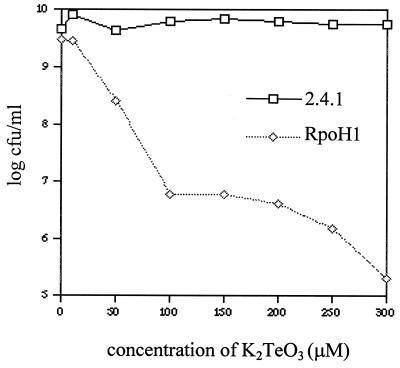
Because resistance to toxic compounds often requires HSPs (28, 43), we compared aerobic sensitivity of wild-type and ΔRpoH cells to the heavy metal oxyanion tellurite. Wild-type cells plate at 100% efficiency at tellurite concentrations up to 300 μM, but the plating efficiency of the ΔRpoH mutant decreases by some 4 orders of magnitude between 10 and 300 μM tellurite (Fig. 2). Tellurite resistance in both strains results in oxyanion reduction, since all colonies exhibited the black phenotype diagnostic of metal deposition (23). At tellurite concentrations greater than 100 μM, plating efficiency of the ΔRpoH mutant plateaus at ∼10−3, suggesting that there is ς37-independent pathway for heavy metal reduction. Finally, the ΔRpoH mutant appears incapable of growth at tellurite concentrations higher than 300 μM, since colonies were not observed when as many as 108 cells are plated.
FIG. 2.
Plating efficiency of aerobically grown R. sphaeroides strains at different tellurite concentrations. Viability of wild-type cells and the ΔRpoH mutant is expressed as the number of CFU per milliliter of original culture at each tellurite concentration.
Protein synthesis after aerobically grown cells are shifted to 42°C.
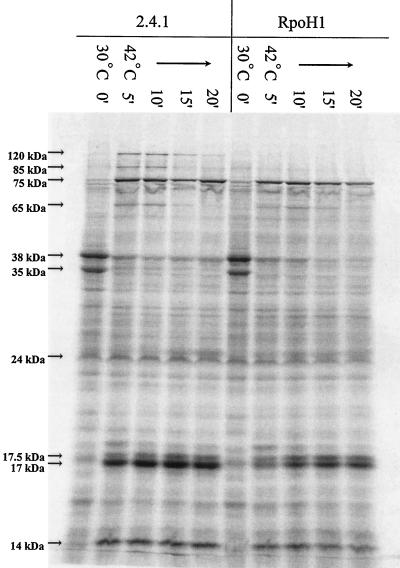
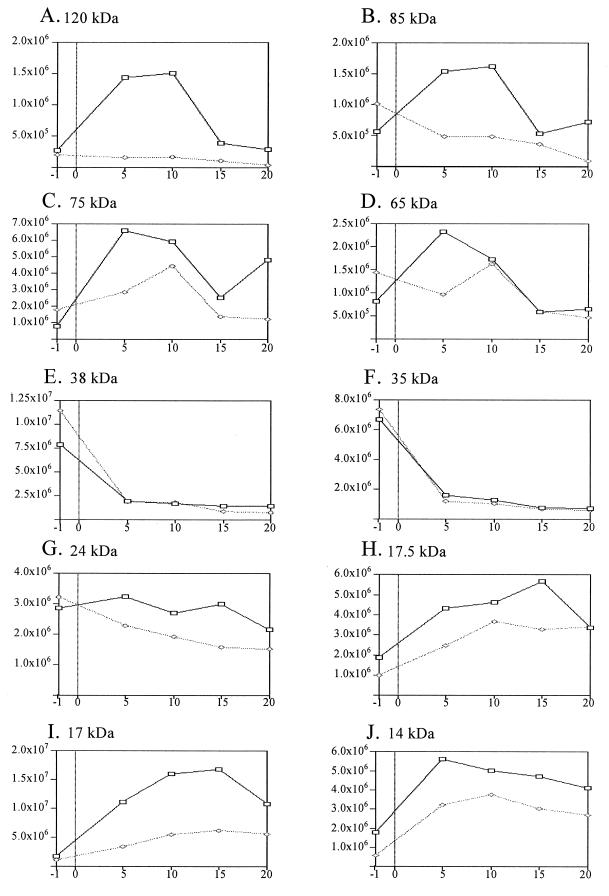
To monitor the ability of cells to mount a heat shock response, protein synthesis was examined before and after aerobically grown cultures were placed at 42°C. In wild-type cells, a pattern of heat-inducible proteins was observed that is characteristic of that seen in other mesophilic eubacteria (14, 43). For example, the synthesis rate of several low-molecular-weight proteins (presumed Hsp10 family members) as well as those of ∼65 (presumed Hsp60), 75 (presumed Hsp70), 85 (presumed Hsp90), and 120 kDa (presumed Clp family member) increased rapidly and transiently after wild-type aerobically grown cells were shifted from 30 to 42°C (Fig. 3). The response observed in wild-type cells can provisionally identify three general classes of heat-responsive proteins. The first class includes three proteins (∼120, 85, and 65 kDa) whose synthesis rate increased within 5 min after a shift to 42°C and then returns to a preshift rate within 20 min (Fig. 4A, B, and D). The synthesis rates of three other proteins (∼75, 17, and 14 kDa) also increased rapidly when wild-type cells were placed at 42°C, but they remained elevated even 20 min after the temperature shift (Fig. 4C, I, and J). The third class includes two proteins (∼38 and 35 kDa) whose synthesis rate decreased rapidly when wild-type cells were shifted to 42°C (Fig. 4E and F).
FIG. 3.
Protein synthesis after aerobically grown wild-type and ΔRpoH cells are shifted from 30 to 42°C. Strains, temperatures, and sampling times (in minutes [indicated by prime symbols]) after the shift are indicated over the gel. The migration of prestained molecular mass standards (Gibco-BRL) was used to estimate the apparent molecular masses of the indicated proteins.
FIG. 4.
Relative synthesis rates of selected proteins before and after aerobically grown cells are shifted to 42°C. The x axes show the minutes after exposure to 42°C (−1 indicates cells sampled 1 min before the temperature shift). The y axes show the relative pixel intensity of individual proteins in Fig. 5. Wild-type (▭) and ΔRpoH ( ) cells are shown.
) cells are shown.
When an aerobically grown culture of the ΔRpoH mutant was shifted to 42°C, the synthesis rates of the 38- and 35-kDa proteins were indistinguishable from those in wild-type cells (Fig. 3 and 4E and F). However, other heat-inducible proteins in wild-type cells behaved differently in the ΔRpoH mutant. For example, there was no detectable increase in the synthesis rate of the heat-inducible 120- and 85-kDa proteins when the ΔRpoH mutant was placed at 42°C (Fig. 4A and B). Within 5 min after the ΔRpoH mutant was incubated at 42°C, there was also smaller increases in the synthesis rates of the heat-inducible 75- and 65-kDa proteins (Fig. 4C and D). By 20 min after the ΔRpoH mutant was placed at 42°C (Fig. 4C), the synthesis rate of the 75-kDa protein approximated that seen in cells grown at 30°C. Despite these differences, heat induction of several potential HSPs in the ΔRpoH mutant is a likely explanation for the aerobic growth of cells lacking ς37 at temperatures up to 42°C (Table 2).
The ΔRpoH mutant contains elevated levels of presumed heat shock transcripts after aerobically grown cells are shifted to 42°C.
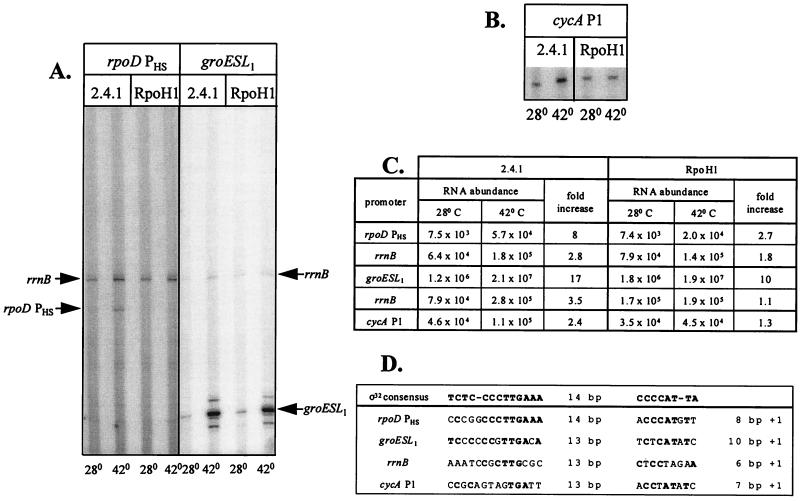
Primer extension assays were performed to test if loss of ς37 altered heat shock promoter function. For this analysis, we chose to monitor several R. sphaeroides promoters, cycA P1, rpoH PHS and groESL1, which have significant similarity to the E. coli ς32 consensus sequence (Fig. 5D). One other, R. sphaeroides rrnB (10, 30), has promoter elements related to the E. coli ς70 and ς32 consensus sequences (Fig. 5D).
FIG. 5.
Transcript levels before and 30 min after aerobically grown wild-type and ΔRpoH cells were shifted to 42°C. (A) Primer extension analysis (∼8 μg of RNA per lane) of the rrnB, rpoD PHS, and groESL1 transcripts at the indicated temperatures. (B) Primer extension analysis (∼8 μg of RNA per lane) of cycA P1-specific transcripts at the indicated temperatures. (C) Transcript abundance (pixel intensity) from panels A and B. Induction ratios denote the increase at 42°C relative to the 30°C level. (D) Comparison of potential R. sphaeroides promoters with the E. coli ς32 consensus sequence; matches are denoted by boldface type.
Levels of cycA P1, rrnB, rpoD PHS, and groESL1-specific transcripts increased ∼2-, 3-, 8- and 17-fold, respectively, 30 min after aerobically grown wild-type cells were placed at 42°C (Fig. 5A to C). If the abundance of these primer extension products is taken as an estimate of promoter function, then loss of ς37 alters their heat inducibility because there were reproducibly smaller increases in the abundance of the rrnB (∼1.1-fold), cycA P1 (∼1.3-fold), rpoD PHS (∼2.7-fold), and groESL1 (∼10-fold) transcripts 30 min after the ΔRpoH mutant was placed at 42°C (Fig. 5A to C). The residual increase in rpoD PHS and groESL1 transcript levels after the ΔRpoH mutant was shifted to 42°C suggests that some other system is increasing promoter function when cells that lack ς37 are placed at a higher temperature.
Heat shock promoters are recognized by multiple R. sphaeroides sigma factors.
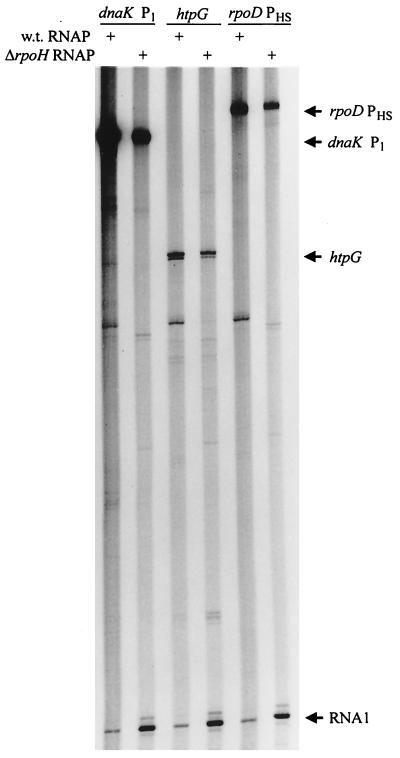
To ask if recognition of heat shock promoters by another RNA polymerase holoenzyme could explain how aerobically grown ΔRpoH cells mount a heat shock response, E. coli dnaK P1, htpG, and rpoD PHS promoters were tested for function in vitro with R. sphaeroides RNA polymerase preparations. Enzyme preparations from wild-type or ΔRpoH cells (21) transcribed all of these E. coli heat shock promoters and produced identically sized transcripts (Fig. 6) which are indistinguishable in size to those generated by E. coli ς32 (16). The lower transcript abundance with equivalent amounts of RNA polymerase from the ΔRpoH mutant probably reflects a reduced level of the cognate holoenzyme in cells that lack ς37.
FIG. 6.
Transcription of E. coli heat shock promoters, dnaK P1, htpG, and rpoD PHS, by mixtures of R. sphaeroides RNA polymerase holoenzymes. Wild-type (w.t.) and ΔrpoH R. sphaeroides RNA polymerase (RNAP) were used (+). The RNA1 transcript is a ς70-dependent product from the origin of DNA replication on all templates (21).
To identify what R. sphaeroides enzyme(s) recognized these E. coli ς32 promoters, RNA polymerase holoenzymes were reconstituted by adding potential sigma factors from the ΔRpoH mutant to a core preparation (21). This analysis identified an ∼38-kDa protein (ς38) that allows transcription of E. coli dnaK P1 when it is added to core RNA polymerase (Fig. 7). This same 38-kDa protein directs transcription of R. sphaeroides cycA P1 when reconstituted with core RNA polymerase subunits (21). Thus, both Eς38 and Eς37 recognize heat shock promoters.
FIG. 7.
R. sphaeroides Eς38 recognizes E. coli dnaK P1. RNA polymerase holoenzymes were reconstituted by adding potential sigma factors (lanes 1 to 28) from the ΔRpoH mutant to a core RNA polymerase preparation from the ΔRpoH mutant (21). The protein in lane 20 was the 38-kDa polypeptide (ς38) that allowed core RNA polymerase to transcribe the cycA P1 heat shock promoter (21).
Loss of ς37 alters the response of photosynthetic cells to increased temperature.
Given the known function of HSPs like GroESL in assembly of photosynthetic functions such as ribulose-1,5-bisphosphate carboxylase (19), we asked if photosynthetic cells were particularly sensitive to the loss of ς37. We were surprised to find that wild-type R. sphaeroides has a lower temperature maximum under photosynthetic conditions, since no growth was observed on solid media at either 37 or 42°C (Table 2). However, loss of ς37 does not make photosynthetic cells temperature sensitive, since the ΔRpoH strain grew normally at temperatures up to 30°C (Fig. 8 and Table 2). Photosynthetic ΔRpoH cells grown at 30°C with moderate light (22) contained levels of light-harvesting bacteriochlorophyll-protein complexes that are indistinguishable from a wild-type strain (Table 2). Thus, loss of ς37 does not have a significant effect on photosynthetic growth or assembly of pigment-protein complexes at 30°C.
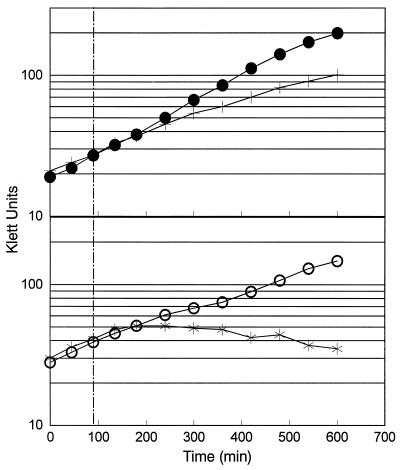
FIG. 8.
Photosynthetic growth of wild-type cells (top) and the ΔRpoH mutant (bottom) at 30°C and after a temperature shift from 30 to 37°C. Photosynthetic growth of wild-type cells (•) and those lacking ς37 (○) at 30°C and the response seen when wild-type cells (+) or the ΔRpoH (✠) mutant are shifted from 30 to 37°C are shown. The broken vertical line indicates when the photosynthetic cultures of wild-type and ΔRpoH cells were placed at 37°C.
During these experiments, we noted a difference in the response of wild-type and ΔRpoH cells to increased temperature under photosynthetic conditions. When a wild-type photosynthetic culture was shifted from 30 to 37°C, there was typically an ∼1.5- to 2.5-fold increase in culture turbidity after the temperature increase (Fig. 8). These turbidity increases cease long before those in a control culture maintained at 30°C. Microscopic examination of wild-type photosynthetic cultures several hours after the shift to 37°C revealed a significant number of paired cells or doublets in the population (data not shown). One explanation for this behavior is that wild-type photosynthetic cultures only complete existing rounds of DNA replication or cell division after the temperature is elevated. Unfortunately, exposure to 37°C under photosynthetic conditions is bactericidal (data not shown), so we were unable to observe increases in CFU after photosynthetic cells were placed at 37°C. In the ΔRpoH mutant, increases in culture turbidity cease rapidly when photosynthetic cultures are shifted from 30 to 37°C (Fig. 8). There are no microscopic indications for the formation of cell doublets after photosynthetic cultures of the ΔRpoH mutant were placed at 37°C (data not shown). Compared with wild-type cells, photosynthetic cells lacking ς37 seem unable to complete previously initiated cell division cycles after exposure to 37°C.
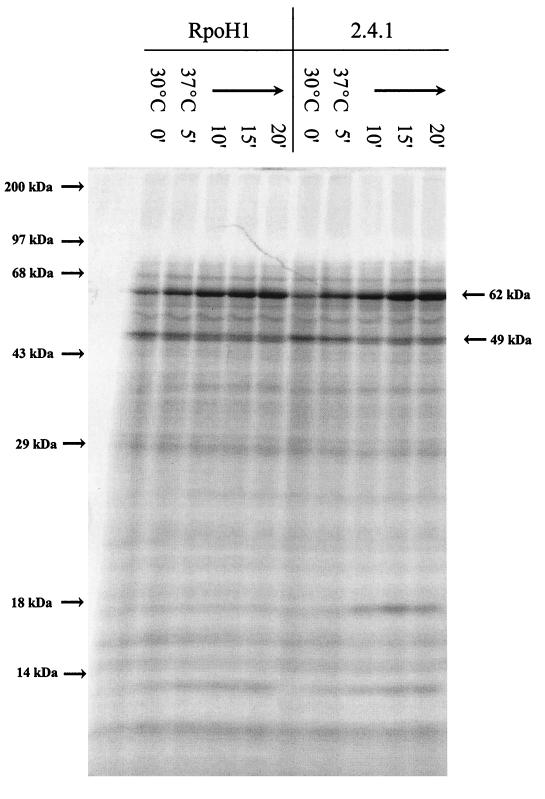
To gain additional insight into potential reasons for this behavior, we monitored bulk protein synthesis before and after photosynthetic cells were shifted from 30 to 37°C. At 30°C, there are significant differences between the protein synthesis pattern of photosynthetic and aerobically grown cultures (compare Fig. 3 to Fig. 9), but the protein synthesis patterns of wild-type and ΔRpoH cells are indistinguishable both before and after photosynthetic cells were incubated at 37°C (Fig. 9). Thus, the different growth response when photosynthetic cultures are shifted to 37°C (Fig. 8) cannot reflect a cessation of translation after the temperature increase. Of equal significance, no detectable heat shock response was observed when photosynthetic wild-type or ΔRpoH cells were shifted to 37°C (compare Fig. 3 and 9); only proteins of ∼62 and 17 kDa exhibited a significant increase in their synthesis rate when photosynthetic cultures of either strain were shifted to 37°C (Fig. 9). Thus, the failure to mount a heat shock response at 37°C under photosynthetic conditions is a likely explanation for why wild-type and ΔRpoH cells are unable to grow at this temperature.
FIG. 9.
Protein synthesis before and after photosynthetic wild-type and ΔRpoH cells are shifted from 30 to 37°C. Temperatures and sampling times (in minutes [indicated by prime symbols]) after the shift are indicated over the gel. The migration of molecular mass standards (Gibco-BRL) indicated to the left of the gel were used to estimate the apparent sizes of the indicated proteins.
DISCUSSION
It was previously suggested that R. sphaeroides Eς37 recognized E. coli heat shock promoters because an RNA polymerase fraction containing a 37-kDa protein transcribed these genes in vitro (16). The amino acid similarity between R. sphaeroides RpoH and proteins in the eubacterial heat shock sigma factor family, its ability to restore bacteriophage λ sensitivity to an E. coli ς32 null mutant, and the heat shock gene expression seen when ς37 is expressed in E. coli cells lacking ς32 indicate that Eς37 recognizes promoters related to the E. coli ς32 consensus sequence. The observation that RNA polymerase preparations from an R. sphaeroides ΔRpoH mutant lack this 37-kDa protein further suggests that rpoH encodes ς37 (21). Additional conclusions and questions generated by this characterization of R. sphaeroides RpoH are presented below.
R. sphaeroides RpoH is essential only under limited conditions.
Growth of cells lacking ς37 at the same temperatures as their wild-type counterparts was surprising when one considers that E. coli ΔRpoH mutants cannot grow at temperatures above 20°C (44). The ability of R. sphaeroides ΔRpoH cells to support infection by the lytic RS1 virus at all temperatures tested also contrasts with the inability of E. coli ΔRpoH mutants to propagate phages such as λ (which served as the basis for our isolation of R. sphaeroides rpoH). Indeed, only two conditions were found where the R. sphaeroides ΔRpoH mutant had a phenotype.
One difference between wild-type and ΔRpoH cells was the increased aerobic sensitivity of cells lacking ς37 to the toxic heavy metal oxyanion tellurite (23). Increased sensitivity of the R. sphaeroides ΔRpoH mutant to tellurite could reflect a limitation of HSPs or other members of a presumed ς37 regulon. While an R. sphaeroides RdxA mutant has a tellurite-sensitive phenotype reminiscent of the ΔRpoH mutant (29), it is not known if this or proteins directly involved in heavy metal reduction are transcribed by Eς37. Alternatively, one promoter for the cytochrome c2 gene (cycA P1) is transcribed by R. sphaeroides Eς37 (21). If cytochrome c2 transferred electrons to the membrane-bound reductase implicated in tellurite reduction (23), then increased cycA P1 activity might be required for heavy metal resistance. This might also explain why tellurite reduction was blocked in cytochrome c2 null mutants (23).
Another phenotype associated with loss of R. sphaeroides ς37 was the rapid cessation in culture turbidity increases when photosynthetic cells were shifted from 30 to 37°C. This behavior of the ΔRpoH mutant does not reflect a total block in macromolecular synthesis, since translation continues after photosynthetic cells are shifted to 37°C. However, the failure of both the ΔRpoH mutant and wild-type cells to mount a heat shock response at 37°C under photosynthetic conditions suggests they are limited for the HSPs needed to progress through the cell cycle. If a component of the photosynthetic apparatus were temperature sensitive, this behavior could reflect a lack of energy for HSP function after such cells are placed at 37°C.
R. sphaeroides mounts a heat shock response in the presence and absence of ς37.
From the universal nature of the heat shock response, it is not surprising that synthesis rates of a number of proteins and the transcript levels from several potential heat shock genes increase after aerobic cells are placed at 42°C (42). Heat induction of several proteins was transient, since the synthesis rate of several putative HSPs (Clp, Hsp90, and Hsp60 homologs) decreased to a new steady state within 20 min after temperature up shift. Other potential HSPs (Hsp70 homologs and several lower-molecular-weight polypeptides) continued to be synthesized at an increased rate even after 20 min at 42°C. Such a persistent induction of HSPs is not seen in well-studied systems like E. coli (14, 43).
When the aerobic heat shock response is analyzed, induction of ∼75-, 65-, and 17-kDa proteins seems partially dependent on ς37, since the magnitude or timing of their synthesis is altered when the ΔRpoH mutant is shifted to 42°C. If the heat-inducible ∼65-kDa protein is a product of the R. sphaeroides groESL1 operon (19), then this reduction probably reflects altered promoter function in cells lacking ς37, since we observed a diminished increase in this transcript 30 min after the ΔRpoH mutant was placed at 42°C. Heat induction of another group of presumed HSPs (∼120 and 85 kDa) could be totally dependent on ς37, since their rate of synthesis is not measurably increased in the ΔRpoH mutant.
The temperature maxima of R. sphaeroides varies with its mode of energy generation.
We were surprised by the selective inability of R. sphaeroides to grow or mount a heat shock response under photosynthetic conditions at temperatures ≥37°C. Photosynthetic temperature maxima of ∼35°C have been reported for many wild-type R. sphaeroides strains (9), yet we have shown that strain 2.4.1 grows and mounts a heat shock response at 42°C when it is generating energy by aerobic or anaerobic respiration. This conditional difference in temperature profile does not reflect an inability to synthesize photosynthetic pigments, since cells using DMSO as an anaerobic electron acceptor at 42°C have colony pigmentation characteristic of the presence of bacteriochlorophyll-protein complexes (data not shown).
R. sphaeroides has two sigma factors which recognize heat shock promoters.
One likely reason why cells lacking ς37 mount a heat shock response is a 38-kDa protein (ς38) that directs core RNA polymerase to recognize ς32 promoters like E. coli dnaK P1 and R. sphaeroides cycA P1 (21). A similar duplication of RpoH function exists in B. japonicum where a second related gene is evident in genomic Southern blots probed with E. coli rpoH (27). At the stringency conditions we employed, no additional sequences related to R. sphaeroides rpoH were observed in genomic Southern blots (data not shown). Until information is available on ς38, its sequence and functional similarity to members of the ς32 family remain open questions.
A B. japonicum ΔrpoH mutation also does not cause temperature sensitivity, presumably because this α proteobacterium contains other related sigma factors (27). Thus, it will be interesting to see if the existence of a second sigma factor that transcribes heat shock genes extends to the other eubacteria from which proteins in the ς32 family have been identified.
The presence of a CIRCE element in the R. sphaeroides groESL1 operon (19) suggests that other mechanisms can contribute to increased heat shock gene expression. The discovery that mutations which increase activity of an R. sphaeroides ςE homolog alters expression of several genes (30a, 34, 35), including one recognized by ς37 (21), reveals additional potential components of this bacterium’s response to environmental or metabolic stress. Experiments are in progress to define the roles and metabolic signals for these alternative R. sphaeroides sigma factors.
ACKNOWLEDGMENTS
These experiments were supported by grant NIH GM37509 to T.J.D. J.B. was supported by NIH Biotechnology predoctoral training grant GM08349 to UW-Madison. P.R. was supported by NIH grant GM36278 and the Fred-Bascom Professorship from the University of Wisconsin Foundation to Carol A. Gross.
REFERENCES
- 1.Babst M, Hennecke H, Fischer H-M. Two different mechanisms are involved in the heat-shock regulation of chaperonin gene expression in Bradyrhizobium japonicum. Mol Microbiol. 1996;19:827–839. doi: 10.1046/j.1365-2958.1996.438968.x. [DOI] [PubMed] [Google Scholar]
- 2.Barber R D, Rott M A, Donohue T J. Characterization of a glutathione-dependent formaldehyde dehydrogenase from Rhodobacter sphaeroides. J Bacteriol. 1996;178:1386–1393. doi: 10.1128/jb.178.5.1386-1393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bethesda Research Laboratories. BRL pUC host: Escherichia coli DH5αTM competent cells. Bethesda Res Lab Focus. 1986;8:9–10. [Google Scholar]
- 3a.Bryant, D., and T. Gruber. Personal communication.
- 4.Daggett Garvin L, Hardies S C. Nucleotide sequence for the htpR gene from Citrobacter freundii. Nucleic Acids Res. 1989;17:4889. doi: 10.1093/nar/17.12.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devereux J R, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donohue T J, Chory J, Goldsand T E, Lynn S P, Kaplan S. Structure and physical map of Rhodopseudomonas sphaeroides bacteriophage RS1 DNA. J Virol. 1985;55:147–157. doi: 10.1128/jvi.55.1.147-157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donohue T J, McEwan A G, Kaplan S. Cloning, DNA sequence, and expression of the Rhodobacter sphaeroides cytochrome c2 gene. J Bacteriol. 1986;168:962–972. doi: 10.1128/jb.168.2.962-972.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donohue T J, McEwan A G, vanDoren S, Crofts A R, Kaplan S. Phenotypic and genetic characterization of cytochrome c2-deficient mutants of Rhodobacter sphaeroides. Biochemistry. 1988;27:1918–1925. doi: 10.1021/bi00406a018. [DOI] [PubMed] [Google Scholar]
- 9.Donohue T J, Kaplan S. Genetic techniques in the Rhodospirillaceae. Methods Enzymol. 1991;204:459–485. doi: 10.1016/0076-6879(91)04024-i. [DOI] [PubMed] [Google Scholar]
- 10.Dryden S C, Kaplan S. Localization and structural analysis of the ribosomal RNA operons of Rhodobacter sphaeroides. Nucleic Acids Res. 1990;24:7267–7277. doi: 10.1093/nar/18.24.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erickson J W, Gross C A. Identification of the ςE subunit of Escherichia coli RNA polymerase: a second alternative sigma factor involved in high-temperature gene expression. Genes Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- 12.Fleischmann R D, Adams M D, White O, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 13.Gething M-J, Sambrook J. Protein folding in the cell. Nature (London) 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 14.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 1382–1399. [Google Scholar]
- 15.Grossman A D, Erickson J W, Gross C A. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell. 1984;38:383–390. doi: 10.1016/0092-8674(84)90493-8. [DOI] [PubMed] [Google Scholar]
- 16.Karls R K, Jin D J, Donohue T J. Transcription properties of RNA polymerase holoenzymes isolated from the purple nonsulfur bacterium Rhodobacter sphaeroides. J Bacteriol. 1993;175:7629–7638. doi: 10.1128/jb.175.23.7629-7638.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusukawa N, Yura T. Heat shock protein GroE of Escherichia coli: key protective roles against thermal stress. Genes Dev. 1988;2:874–882. doi: 10.1101/gad.2.7.874. [DOI] [PubMed] [Google Scholar]
- 18.Landick R, Vaughn V, Lau E T, VanBogelen R A, Erickson J W, Neidhardt F C. Nucleotide sequence of the heat shock regulatory gene of E. coli suggests its protein product may be a transcription factor. Cell. 1984;38:175–182. doi: 10.1016/0092-8674(84)90538-5. [DOI] [PubMed] [Google Scholar]
- 19.Lee W T, Terlesky K C, Tabita F R. Cloning and characterization of two groESL operons of Rhodobacter sphaeroides: transcriptional regulation of the heat-induced groESL operon. J Bacteriol. 1997;179:487–495. doi: 10.1128/jb.179.2.487-495.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lonetto M, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacGregor B J, Karls R K, Donohue T J. Transcription of the Rhodobacter sphaeroides cycA P1 promoter by alternate RNA polymerase holoenzymes. J Bacteriol. 1998;180:1–9. doi: 10.1128/jb.180.1.1-9.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meinhardt S W, Kiley P J, Kaplan S, Crofts A R, Harayama S. Characterization of light-harvesting mutants of Rhodopseudomonas sphaeroides. I. Measurement of the efficiency of energy transfer from light-harvesting complexes to the reaction center. Arch Biochem Biophys. 1984;236:130–139. doi: 10.1016/0003-9861(85)90612-5. [DOI] [PubMed] [Google Scholar]
- 23.Moore M D, Kaplan S. Identification of intrinsic high-level resistance to rare-earth oxides and oxyanions in members of the class Proteobacteria: characterization of tellurite, selenite, and rhodium sesquioxide reduction in Rhodobacter sphaeroides. J Bacteriol. 1992;174:1505–1514. doi: 10.1128/jb.174.5.1505-1514.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naczynski Z M, Mueller C, Kropinski A M. Cloning the gene for the heat shock response positive regulator (ς32 homolog) from Pseudomonas aeruginosa. Can J Microbiol. 1995;41:75–87. doi: 10.1139/m95-010. [DOI] [PubMed] [Google Scholar]
- 25.Nagai H, Yuzawa H, Kanemori M, Yura T. A distinct segment of the ς32 polypeptide is involved in DnaK-mediated negative control of the heat shock response in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:10280–10284. doi: 10.1073/pnas.91.22.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakahigashi K, Yanagi H, Yura T. Isolation and sequence analysis of rpoH genes encoding ς32 homologs from gram negative bacteria: conserved mRNA and protein segments for heat shock regulation. Nucleic Acids Res. 1995;23:4383–4390. [PMC free article] [PubMed] [Google Scholar]
- 27.Narberhaus F, Weiglhofer W, Fischer H-M, Hennecke H. The Bradyrhizobium japonicum rpoH1 gene encoding a ς32-like protein is part of a unique heat shock gene cluster together with groESL1 and three small heat shock genes. J Bacteriol. 1996;178:5337–5346. doi: 10.1128/jb.178.18.5337-5346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neidhardt F C, VanBoglen R A. Heat shock response. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1334–1345. [Google Scholar]
- 29.Neidle E L, Kaplan S. Rhodobacter sphaeroides rdxA, a homolog of Rhizobium meliloti fixG, encodes a membrane protein which may bind cytoplasmic [4Fe-4S] clusters. J Bacteriol. 1992;174:6444–6454. doi: 10.1128/jb.174.20.6444-6454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newlands J T, Gaal T, Mescas J, Gourse R L. Transcription of the Escherichia coli rrnB P1 promoter by the heat shock RNA polymerase (Eς32) in vitro. J Bacteriol. 1993;175:661–668. doi: 10.1128/jb.175.3.661-668.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Newman, J. D., B. A. Schilke, and T. J. Donohue. Unpublished data.
- 31.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 32.Reisenauer A, Mohr C D, Shapiro L. Regulation of a heat shock ς32 homolog in Caulobacter crescentus. J Bacteriol. 1996;178:1919–1927. doi: 10.1128/jb.178.7.1919-1927.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Schilke B A, Donohue T J. δ-Aminolevulinate couples cycA transcription to changes in heme availability in Rhodobacter sphaeroides. J Mol Biol. 1992;226:101–115. doi: 10.1016/0022-2836(92)90127-6. [DOI] [PubMed] [Google Scholar]
- 35.Schilke B A, Donohue T J. ChrR positively regulates transcription of the Rhodobacter sphaeroides cytochrome c2 gene. J Bacteriol. 1995;177:1929–1937. doi: 10.1128/jb.177.8.1929-1937.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 37.Sistrom W R. A requirement for sodium in the growth of Rhodopseudomonas sphaeroides. J Gen Microbiol. 1960;22:778–785. doi: 10.1099/00221287-22-3-778. [DOI] [PubMed] [Google Scholar]
- 38.Straus D B, Walter W A, Gross C A. The heat shock response of Escherichia coli is regulated by changes in the concentration of ς32. Nature. 1987;329:348–351. doi: 10.1038/329348a0. [DOI] [PubMed] [Google Scholar]
- 39.Suwanto A, Kaplan S. Physical and genetic mapping of the Rhodobacter sphaeroides 2.4.1 genome: presence of two unique circular chromosomes. J Bacteriol. 1989;171:5850–5859. doi: 10.1128/jb.171.11.5850-5859.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Usui S, Yu L. Subunit IV (Mr = 14,384) of the cytochrome b-c1 complex from Rhodobacter sphaeroides. J Biol Chem. 1991;266:15644–15649. [PubMed] [Google Scholar]
- 41.Wu J, Newton A. Isolation, identification, and transcriptional specificity of the heat shock sigma factor ς32 from Caulobacter crescentus. J Bacteriol. 1996;178:2094–2101. doi: 10.1128/jb.178.7.2094-2101.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamori T, Yura T. Temperature-induced synthesis of specific proteins in Escherichia coli: evidence for transcriptional control. J Bacteriol. 1980;142:843–851. doi: 10.1128/jb.142.3.843-851.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- 44.Zhou Y, Kusukawa N, Erickson J W, Gross C A, Yura T. Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor ς32. J Bacteriol. 1988;170:3640–3649. doi: 10.1128/jb.170.8.3640-3649.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu Y S, Kaplan S. Effects of light, oxygen, and substrates on steady-state levels of mRNA coding for ribulose-1,5-bisphosphate carboxylase and light-harvesting and reaction center polypeptides in Rhodopseudomonas sphaeroides. J Bacteriol. 1985;162:925–932. doi: 10.1128/jb.162.3.925-932.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuber U, Schumann W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J Bacteriol. 1994;176:1359–1363. doi: 10.1128/jb.176.5.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]