Abstract
Ralstonia solanacearum is the causal agent of bacterial wilt of many agriculturally important crops. Exopolysaccharide synthesized by products of the epsI operon is the major virulence factor for R. solanacearum. Expression of epsI has been demonstrated to be under the control of several proteins, including several two-component regulators. Overexpression of EpsR was found previously to reduce the amount of synthesis specifically from the epsI promoter. Here we present data that a single chromosomal copy of epsR activates the epsI promoter, suggesting that EpsR is a concentration-dependent effector of epsI gene expression. Furthermore, the ability of EpsR to modulate epsI expression is dependent on the phosphorylation state of EpsR. Gel mobility shift assays suggest that EpsR can specifically bind the epsI promoter and that this binding requires a phosphorylated form of EpsR.
Many prokaryotes produce extracellular polysaccharides that have important roles in pathogenesis (8). In the plant pathogen Ralstonia solanacearum, extracellular polysaccharides (EPS) are major virulence factors required to cause the agriculturally important disease bacterial wilt (15). Although the exact role of EPS has not been demonstrated, it may interfere with water transport in the plant by plugging the xylem vessels, leading to wilt (20).
Several genes involved in EPS production in R. solanacearum have been identified. Structural gene clusters include opsI (7, 22), opsII (29), rgnII (10), and epsI (10). The opsI and opsII gene clusters are important for both EPS and lipopolysaccharide syntheses since mutations in them affect the production of both macromolecules. The opsI cluster consists of at least seven genes, some of which are important for nucleotide sugar synthesis (7, 22). The rgnII cluster is largely uncharacterized since it is required for EPS production only in culture, not in plants (10). Mutational analyses of epsI suggest that it encodes proteins responsible for synthesis of the acidic component of EPS, which is absolutely required for R. solanacearum infection of plants (10, 23, 32).
Sequence analysis revealed that epsI encodes polypeptides transcribed from a single promoter (18). Regulation of epsI is complex, involving at least seven proteins, including the highly basic XpsR, which likely affects expression of epsI directly (17). The two-component regulatory systems VsrB-VsrC (19) and VsrA-VsrD (37) have been shown genetically to positively regulate epsI and xpsR expression, respectively. The LysR-like transcriptional regulator PhcA has been demonstrated to positively regulate xpsR expression (4, 5, 17). Finally, EpsR overexpressed in plasmids of four to six copies per cell can specifically reduce synthesis from the epsI promoter, decreasing EPS production from colonies (16, 21, 29). EpsR has sequence similarity to effector proteins of two-component regulatory systems (21). A thorough Tn5::lacZ mutagenesis of R. solanacearum selecting for EPS-defective strains resulted in the identification and lacZ tagging of 12 complementation groups of EPS genes (29). Overexpression of EpsR affected the expression of only epsI::lacZ genes, suggesting a specific interaction between EpsR and the epsI promoter. In this work, we provide evidence that the chromosomal copy of epsR encodes a positive regulator of epsI. An epsR insertional mutation also reduced the virulence of R. solanacearum. We show that a mutation at the putative phosphorylation site renders EpsR unable to regulate synthesis of epsI and prevents an EpsR-dependent gel mobility shift of the epsI promoter.
MATERIALS AND METHODS
Growth and maintenance of bacterial strains.
The strains and plasmids used in the experiments described herein are listed in Table 1. R. solanacearum was routinely cultured in CPG medium (per liter, 10 g of tryptone, 5 g of glucose, 1 g of Casamino Acids, 1 g of yeast extract, and 15 g of agar as appropriate) at 30°C. Antibiotics, where used, were at the following concentrations: kanamycin, 50 μg/ml; tetracycline, 15 μg/ml; ampicillin, 100 μg/ml; streptomycin, 25 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source |
|---|---|---|
| Escherichia coli DH5α | F−endA1 hrdR17 (Rk− mK+) recA1 | 14 |
| Ralstonia solanaceaum | ||
| K60 | Wild type | 26 |
| S49 | EPS−; epsI::lacZ fusion; Kanr | 29 |
| S70 | EPS−; epsI::lacZ fusion; Kanr | 29 |
| S90 | EPS−; rgnII::lacZ fusion; Kanr | 29 |
| S112 | EPS−; vsrB::lacZ or vsrC::lacZ fusion; Kanr | 29 |
| S50 | EPS−; ops::lacZ fusion; Kanr | 29 |
| K60::epsRΩ | EPS+; EpsR− | This work |
| S49::epsRΩ | EPS−; epsI::lacZ fusion; epsR Kanr Smr | This work |
| S70::epsRΩ | EPS−; epsI::lacZ fusion; epsR Kanr Smr | This work |
| S90::epsRΩ | EPS−; rgnII::lacZ fusion; epsR Kanr Smr | This work |
| S112::epsRΩ | EPS−; vsrB::lacZ or vsrC::lacZ fusion epsR Kanr Smr | This work |
| S50::epsRΩ | EPS−; opsG::lacZ fusion; epsR Kanr Smr | This work |
| S49::epsRD-A 1 | EPS−; epsI::lacZ fusion; epsR EpsRD-A+ Kanr Smr | This work |
| S49::epsRD-A 10 | EPS−; epsI::lacZ fusion; epsR EpsRD-A+ Kanr Smr | This work |
| Plasmids | ||
| pLAFR3 | Tcr | 25 |
| pGL10 | Kanr | Y. Brun |
| pGL10lacZ | Promoterless lacZ gene from mini-Tn5::lacZ cloned into pGL10; Kanr | This work |
| pGL10Ω | Ω fragment from pHP45Ω cloned into the BglII site of pGL10; Kanr Smr | This work |
| pGepsI::lacZ | Nucleotides 1 to 260 of the epsI promoter cloned into the EcoRI and BamHI sites of pGL10lacZ; Kanr Smr | This work |
| pGepsR::lacZ | Nucleotides 25 to 365 of epsR cloned into the EcoRI and KpnI sites of pGL10lacZ; Kanr Smr | This work |
| pGopsG::lacZ | 360-bp fragment containing the opsG promoter cloned into the EcoRI site of pGL10lacZ; Kanr Smr | This work |
| Mini-Tn5lacZ2 | Ampr Kanr | 28 |
| pHP45Ω | Smr Ampr | Y. Brun; 34 |
| pepsRΩ | Ω cassette cloned into the SmaI site of epsR contained on pET11epsR | This work |
| pTAepsI | Nucleotides 1 to 260 of epsI cloned into PCRII as a PCR product; Kanr Smr | This work |
| pKLS44 | Ampr | 21 |
| pET11epsR | Ampr | 21 |
| pGepsR | Nucleotides 25 to 1290 of epsR cloned into pGL10 | This work |
| pepsR | Nucleotides 25 to 1142 of epsR cloned into pLAFR3; Tetr | This work |
| pepsRD-A | Nucleotides 25 to 1142 of epsRD-A cloned into EcoRI and HindIII sites of pLAFR3; Tetr | This work |
| pBepsRD-A | Nucleotides 25 to 1142 of epsRD-A cloned in pBSKS | This work |
| pepsRD-AΩ | Nucleotides 286 to 1142 of epsRD-A cloned in pBSKS | This work |
| pKL4 | Tcr | 16 |
| pL112 | Tcr | 29 |
| pL90 | Tcr | 29 |
Ampr, ampicillin resistance; Smr, streptomycin resistance; Tcr, tetracycline resistance; Kanr, kanamycin resistance.
Molecular techniques.
Plasmids were isolated from Escherichia coli by using Qiagen (Chatsworth, Calif.) columns. Chromosomal DNA isolation and Southern hybridizations were done as previously described (21). To transform R. solanacearum, 2-ml cultures at an optical density at 600 nm (OD600) of approximately 1.0 were washed three times with sterile water and finally resuspended in 100 μl of water. The cells were then electroporated with 0.5 μg of plasmid with a Gene Pulser (Bio-Rad, Hercules, Calif.) set at 25 μF with a field strength of 6,000 V/cm. After electroporation, the cells were incubated for 3 h in CPG broth before being plated onto selective medium.
Site-directed mutagenesis.
The aspartate at residue 47 of EpsR was changed to alanine by amplifying epsR in two halves by using PCR with pGepsR as a template. The 5′ half of epsR was generated with the following primers: PUC19PC (5′ GCCTGCAGGTCGACTCTAG 3′), which hybridizes with sequence in the plasmid vector of pGepsR, and epsRD-A5′ (5′ CAGCGGCTGCGTGGGCGGCAAG 3′), which hybridizes to nucleotides (nt) 486 to 508 of epsR (21). The 3′ half of epsR was amplified by using epsRD-A3′, which hybridizes to nt 500 to 523 and contains an AlwNI site (5′ CAGCCGCTGAACTGGCCGTGATC 3′), and epsR3′ EcoRI, which hybridizes to nt 1125 to 1142 and contains an EcoRI site (5′ GAATTCCCGCGACGCGACAGCGCG 3′). The PCR products encoded by the 5′ and 3′ halves of epsR were individually cloned into PCRII (Novagen, Milwaukee, Wis.), creating TAepsRD-A5′ and TAepsRD-A3′ respectively. Inserts from TAepsRD-A5′ and TAepsRD-A3′ were released with HindIII-AlwNI and EcoRI-AlwNI digestions, respectively. The fragments were ligated to pBSKS+ linearized with EcoRI and HindIII to reconstitute epsR encoding the amino acid change at residue 47, creating pBepsRD-A. Clones which contained epsRD-A mutations were verified by screening for the presence of the AlwNI site. Since ampicillin selection is ineffective in R. solanacearum K60, we cloned the Ω fragment (encoding streptomycin resistance) into the HindIII site in the polylinker region of pBepsRD-A, creating pepsRD-AΩ. This plasmid was then transformed by electroporation into the epsI mutant S49, selecting for streptomycin resistance. To investigate the effect of overexpression of EpsRD-A in R. solanacearum, the EcoRI-HindIII fragment from pepsRD-A was cloned into the EcoRI and HindIII sites of pLAFR3, creating pepsRD-A.
In vivo labeling and Western blot analysis.
Strains grown in 2 ml of CPG broth to an OD600 of 0.5 were washed three times in low-phosphate medium (M9 medium containing 30 μM Na2HPO4 and KH2PO4) (36) and then resuspended in 2 ml of low-phosphate medium and incubated at 30°C for an additional 4 h. One-half millicurie of orthophosphate (200 mCi/mmol; Amersham, Arlington Heights, Ill.) was added to 1-ml aliquots of cells. Orthophosphate can freely diffuse into bacterial cells, be incorporated into nucleotides, and subsequently be used as a substrate for phosphorylation. After a 20-min incubation at 30°C, the samples were washed twice with sterile water and resuspended in 60 μl of 1× Laemmli loading dye (27). Ten microliters of each sample was loaded on a sodium dodecyl sulfate–10% polyacrylamide gel. After electrophoresis, the gel was wrapped in plastic wrap and exposed to X-ray film for 1 h at −80°C. After autoradiography, the gel was washed extensively with 1× Western transfer buffer (10% methanol, 200 mM glycine, 25 mM Tris [pH 8.3]). The gel was blotted onto a nitrocellulose membrane and probed with anti-EpsR antibodies as previously described (21).
Gel mobility shift.
DNA probes used for gel mobility shift assays contained a 240-bp fragment (nt −140 to +100) of the epsI promoter. The 240-bp fragment was produced by digesting pTAepsI with EcoRI and then end labeling with Klenow polymerase, [α-32P]dATP (3,000 Ci/mmol), and dTTP. In competition experiments, a 320-bp fragment containing the opsG promoter was made by PCR as previously described (21). epsI promoter used in competition experiments was also synthesized by PCR by using K60 chromosomal DNA as the template and the following primers: epsI 5′, which hybridizes to nt 1 to 20 (5′ GAATTCTCTGTCGAATTGGG) and epsI 3′, which hybridizes to nt 218 to 238 (5′ GGATCCGCTTACGAACATGAATGCG 3′) (18). Protein extracts were made from 50 ml of R. solanacearum culture grown to an OD600 of approximately 1.0. The cells were harvested, washed once in extraction buffer (50 mM Tris-HCl [pH 7.9], 10 mM EDTA, 10% glycerol, 10 mM KCl, 1 mM dithiothreitol, 0.5 mM sodium pyrophosphate, 0.4 mg of phenylmethylsulfonyl fluoride per ml), and finally resuspended in 1 ml of extraction buffer. Lysates were prepared by sonication with three 10-s bursts with a model 50 Sonic Dismembrator (Fisher Scientific, Pittsburgh, Pa.) set to 30% output. Protein concentrations were determined by the Bradford assay with bovine serum albumin as the standard (3). DNA binding reaction components, including 50-μg protein extracts, competitor DNAs (when appropriate), and binding buffer [50 mM Tris-HCl (pH 7.9), 5 mM MgCl2, 30 mM KCl, 12% glycerol, 0.5 μg of poly(dI-dC) per ml, 1 mM dithiothreitol] were incubated for 15 min at room temperature before the addition of 5 nM end-labeled epsI promoter. Reaction mixtures were incubated for a further 20 min at room temperature before electrophoresis in a 3.5% (wt/vol) polyacrylamide (Tris-glycine [pH 7.9]) native gel at 70 V for 12 h at 4°C.
RESULTS
EpsR can act as both a positive and negative regulator of epsI expression.
We previously determined that overexpression of EpsR in plasmids of four to six copies per cell resulted in decreased expression from the epsI promoter (29). To examine the effect of inactivation of the chromosomal copy of epsR, we inserted an Ω cassette carrying streptomycin resistance into the unique SmaI site in epsR contained in PET11epsR, creating pepsRΩ. This plasmid contains a ColEI replication origin which is not utilized in R. solanacearum. Exchange of epsR::Ω with the chromosomal copy of epsR was made in the wild type, K60, and the EPS mutants S49, S50, S70, and S112 by homologous recombination. The homologous integration of epsR::Ω was checked by Southern blotting by using labeled epsR DNA as a probe (data not shown). Mutations in epsR were obtained in several strains, S70 and S49, which have epsI::lacZ fusions, S50, which has an opsI::lacZ fusion, S112, which has a vsrB::lacZ or vsrC::lacZ fusion, and finally S90, which contains a rgnII::lacZ fusion (29).
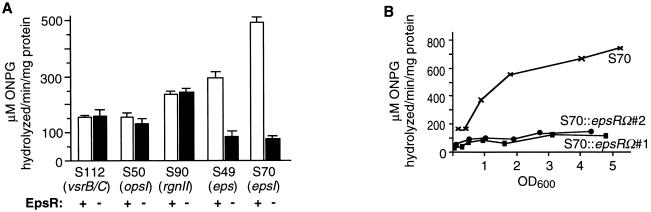
Expression of EPS genes in the absence of EpsR was assayed by measuring β-galactosidase activity. In two S70 and two S49 strains with independently derived epsRΩ insertional mutations, epsI expression was reduced four- to sixfold (Fig. 1A). Since overexpression of EpsR repressed epsI expression (29), we were surprised to find that an epsRΩ mutation also reduced epsI expression. Expression of other genes involved in EPS biosynthesis, including opsI, rgnII, and vsrB or vsrC, were unaffected by the inactivation of epsR (Fig. 1A), consistent with the previous report that EpsR specifically affected the epsI promoter. Since epsI expression increases with culture density (29), we assayed epsI expression during growth of the culture in strains which contain an epsRΩ insertional mutation. Samples of S70 (epsR+) and two independently derived S70::epsRΩ mutant strains were collected at the indicated optical densities and assayed for β-galactosidase activity. Strains lacking EpsR had a reduced, but not a complete lack of, β-galactosidase activity throughout the growth of the culture, with the greatest difference from the activity of strain 70 at the higher optical densities (Fig. 1B). These data suggest that EpsR acts as positive regulator of epsI synthesis when present in one copy and that the negative effect on epsI expression was due to overexpression by multicopy plasmids.
FIG. 1.
(A) Effect of inactivation of EpsR on expression from different EPS genes. Cells used were grown to an OD600 of 1.5 and then assayed as described previously (29, 30). The bars represent an average of three independent trials. (B) Effect of the epsRΩ mutation on expression from the epsI promoter at different cell densities. Cells used for the assay were collected at the indicated optical densities and then frozen at −70°C until use. β-Galactosidase activities (micromoles of ONPG [o-nitrophenyl-β-d-galactopyranoside] hydrolyzed per minute per milligram of protein) were determined and plotted.
In planta analysis of EpsR.
The effect of single and multiple copies of epsR on the ability of R. solanacearum to kill eggplant seedlings was assayed as previously described (7, 21, 22, 40). At least 10 plants were inoculated with either the wild type, K60, K60/pKL4 (overexpressing EpsR), or K60::epsRΩ (epsR insertional mutant), and the results of two independently performed assays are presented (Table 2). The wild type, K60, caused death in a majority of the plants by 11 days postinoculation (Table 2). Overexpression of EpsR slowed the wilting process, with only approximately half of the plants killed at 11 days postinoculation (Table 2). However, at 14 days postinoculation, more than half of the plants inoculated with K60/pKL4 died, possibly due to the loss of pKL4, since there is no antibiotic selection in the plant. Strains with an epsRΩ insertional mutation showed reduced wilting, with only approximately half of the inoculated plants killed at 11 days postinoculation. This result is consistent with reduction but not abolition of epsI expression due to the lack of EpsR. As expected for a stable genetic change, no significant increase in plant death occurred by 14 days postinoculation.
TABLE 2.
Effect of epsRΩ mutation and overexpression on the ability of R. solanacearum to kill eggplant seedlings
| Strain and trial no. | No. of seedlings killed/no. inoculateda
|
|
|---|---|---|
| Day 11 | Day 14 | |
| K60 | ||
| Trial 1 | 12/13 | 12/13 |
| Trial 2 | 9/10 | 10/10 |
| K60/PKL4 | ||
| Trial 1 | 6/13 | 9/13 |
| Trial 2 | 5/13 | 8/13 |
| K60::epsRΩ | ||
| Trial 1 | 6/13 | 7/13 |
| Trial 2 | 7/12 | 7/12 |
Inoculated seedlings were scored for death at 11 and 14 days postinoculation.
Regulation of EPS genes.
To facilitate a more convenient analysis of the effects of epsR, we fused the promoters of epsI, opsG, and epsR to pGL10::lacZ, which contains a promoterless lacZ gene. The epsI promoter contained nt −140 to +120 in plasmid pGepsI::lacZ, the opsG promoter contained nt −360 to +1 in pGopsG::lacZ, and the epsR promoter contained nt −280 to +40 in pGepsR::lacZ. Promoter activities when EpsR is overexpressed or absent were assayed by measuring β-galactosidase activity. Transformation of strain K60 with pGepsI::lacZ and pKL4 resulted in a reduction of epsI expression to 15% of that of K60 transformed with pGepsI::lacZ and pLAFR3 (Table 3). Furthermore, β-galactosidase activity resulting from strains transformed with pGopsG::lacZ was unchanged, as expected, whether EpsR was overexpressed or absent (Table 3). The epsI and opsG promoters on plasmids, thus, mimicked the phenotype of their chromosomal counterparts with regards to EpsR regulation and demonstrated the efficacy of the system (29). Strains transformed with pGepsR::lacZ had approximately 150 U of β-galactosidase activity, demonstrating that the epsR promoter is expressed when contained on a plasmid (Table 3). However, the expression of the epsR promoter was not affected when strain K60 harbored both pGepsR::lacZ and pKL4 or when K60 contained pGepsR::lacZ and an epsRΩ insertion mutation (Table 3). Therefore, EpsR does not regulate its own expression.
TABLE 3.
Activity of the epsI, opsG, and epsR promoters in the presence or absence of EpsR
| Strain | EpsRa | Plasmids | β-Galacto- sidase activity (Miller Ub) | Relative activityc |
|---|---|---|---|---|
| K60 | + | pGepsI::lacZ + pLAFR3 | 337.3 | 1.0 |
| K60 | +++ | pGepsI::lacZ + pKL4 | 50.4 | 0.2 |
| K60 | + | pGL10::lacZ + pLAFR3 | 36.5 | 0.1 |
| K60 | + | pGopsG::lacZ + pLAFR3 | 533.4 | 1.0 |
| K60::epsRΩ | − | pGopsG::lacZ + pLAFR3 | 612.9 | 1.2 |
| K60 | +++ | pGopsG::lacZ + pKL4 | 600.9 | 1.1 |
| K60 | + | pGepsR::lacZ + pLAFR3 | 177.8 | 1.0 |
| K60::epsRΩ | − | pGepsR::lacZ + pLAFR3 | 169.7 | 0.9 |
| K60 | +++ | pGepsR::lacZ + pKL4 | 157.5 | 0.9 |
+, produced from one copy of epsR; +++, produced from pKL4; −, not produced.
β-Galactosidase activity is expressed in Miller units (micromoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per milligram of protein [30]). Values shown are the average of two independent trials.
Activity of each promoter in the presence of wild-type levels of EpsR is adjusted to 1.0.
EpsR is phosphorylated in vivo.
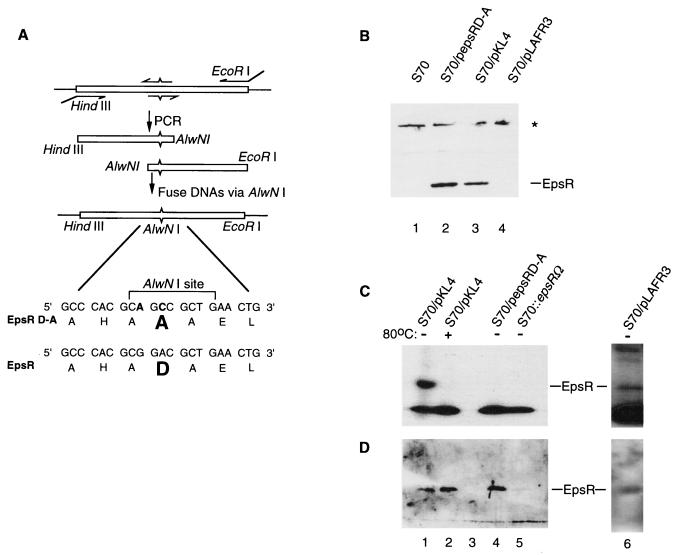
Most effector proteins of two-component regulatory systems are modified at a conserved aspartic acid by phosphorylation, which can modulate a variety of the protein’s activities (33). EpsR contains an aspartate (D) at amino acid 47, which is the most likely site of phosphorylation (Fig. 2A). We changed D47 to an alanine by using the scheme diagrammed in Fig. 2A. The 5′ and 3′ halves of epsR were amplified by PCR by using primers which changed codon 47 to an alanine while adding a unique AlwNI site (Fig. 2A). The alanine codon created is one which is commonly used in R. solanacearum (6). DNA carrying the mutant version of epsR was cloned into pLAFR3, creating pepsRD-A.
FIG. 2.
(A) A schematic diagram of site-directed mutagenesis of the conserved aspartate (D47) in epsR is shown. Oligonucleotide primers are indicated with arrows. The internal pair of primers contains changes in the epsR sequence, indicated by ^ or ˇ. Changes made in the epsR sequence are shown at the bottom of the figure, and the encoded amino acids are in the standard one-letter code. The original aspartate in the EpsR sequence and the alanine in the EpsRD-A sequence are shown in bold type. (B) Western blot analysis shows that EpsRD-A is expressed in R. solanacearum (S70) when cells are transformed with pepsRD-A. The star designates nonspecific recognition of protein by the anti-EpsR antibodies. (C) EpsR (lane 1), but not EpsRD-A (lane 3), is phosphorylated. A longer exposure of this gel (lane 6) reveals the phosphorylation of chromosomally derived EpsR. (D) After autoradiography, the gel in Fig. 3C was subjected to Western blot analysis with anti-EpsR serum (21). The results show that EpsR and EpsRD-A are present in cell lysates in similar quantities.
Western blot analysis using antibodies directed against EpsR detected a protein of the size predicted for EpsR in extracts of strain S70 containing pKL4, the EpsR-overexpressing plasmid (Fig. 2B). A protein of similar size and abundance is produced in S70 transformed with pepsRD-A, demonstrating that the D-A mutation did not noticeably affect the stability of the resultant protein. This protein band was not readily apparent in lysates made from strains which contained only the chromosomal copy of epsR (Fig. 2B). However, a longer exposure revealed the lower level of EpsR made from this strain (data not shown) (6).
To determine if EpsR is phosphorylated in vivo, radiolabeled orthophosphate was added to cells adapted to growth in low-phosphate medium. Lysates made from S70/pKL4 produced a band of the same size as that of EpsR (Fig. 2C). The covalent bond formed between aspartate and the phosphate group in response regulators is inherently unstable (38). In lysates heated to 80°C for 15 min prior to electrophoresis, the band corresponding to EpsR was no longer detectable. Moreover, this band was absent in epsR insertion mutant strains and was apparent in strains which contained the chromosomal copy of epsR only after prolonged exposure (Fig. 2C, lane 6), suggesting that EpsR is phosphorylated in the absence of overexpression. No band corresponding to EpsRD-A can be detected in lysates made from cells which contain pepsRD-A (Fig. 2C). The same gel was stripped of labeled phosphate, transferred to a nitrocellulose membrane, and probed with anti-EpsR antibodies to show that EpsRD-A was being expressed in the labeled cells (Fig. 2D). EpsRD-A was present at levels comparable to those of wild-type EpsR (compare lanes 4 and 2). Therefore, D47 of EpsR is critical for phosphorylation.
EpsRD-A cannot modulate expression of epsI.
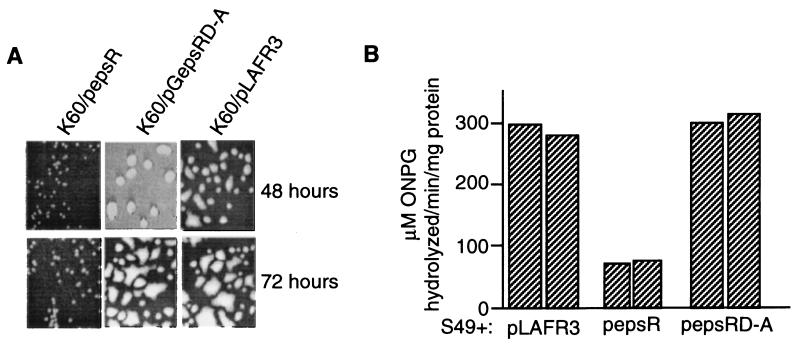
To examine the effect of EpsRD-A on EPS production, K60 was transformed with pepsRD-A. The wild type, K60, became visibly mucoid approximately 48 h after plating (Fig. 3A). Overexpression of EpsRD-A did not grossly affect the abundance of EPS produced by K60, in contrast to overexpression of wild-type EpsR from plasmid pepsR, which visibly reduced the amount of EPS produced by K60 colonies (Fig. 3A). The plasmid pepsR contains a PCR-derived clone of epsR and demonstrates that EpsR is sufficient to reduce production of EPS. As previously reported, the effect of EpsR lessens as the culture ages, since colonies transformed for 72 h are noticeably more mucoid than those transformed for 48 h (21).
FIG. 3.
(A) Colony morphology of K60 transformed for 48 and 72 h with pepsR, pGepsRD-A, and pLAFR3. (B) Overexpression of EpsRD-A does not affect expression of the epsI promoter as measured in strain S49, containing the epsI::lacZ fusion. Cells containing the plasmids indicated at the bottom of the graph were grown to an OD600 of 1.2 and assayed for β-galactosidase activity (see legend to Fig. 1 for clarification of values) as described in Materials and Methods.
The effect of EpsRD-A on the expression of the epsI promoter was measured in strain S49. Consistent with previous results, wild-type EpsR showed epsI expression reduced fivefold in comparison to that of cells which contained pLAFR3. Overexpression of EpsRD-A had no effect on epsI expression (Fig. 3B). Therefore, phosphorylation of EpsR is required for repression of epsI expression.
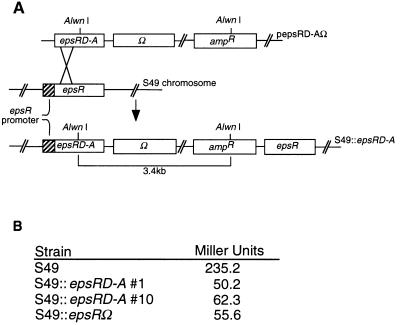
The effect of one copy of epsRD-A on epsI expression was examined. To replace wild-type epsR with epsRD-A, S49 was transformed with pepsRD-AΩ, which contained a promoterless epsRD-A gene (Fig. 4A). Streptomycin- and ampicillin-resistant colonies were selected. Southern blotting confirmed that 2 of 20 isolates (strains 1 and 10) examined had recombination events which occurred before codon 47 of epsR. The resulting strain should express only the epsRD-A allele, since the other epsR copy is left without a functional promoter (Fig. 4A). Promoter activity in strains which contain one copy of epsRD-A is reduced between four- and sixfold, based on measurements of two independently isolated epsRD-A integrants (Fig. 4B). Since this is approximately the same reduction of expression of epsI observed in strain S49 lacking a functional copy of epsR, phosphorylation is required for activation of epsI expression.
FIG. 4.
(A) Schematic diagram for construction of S49 chromosomes with one copy of epsRD-A. The desired integration event and the resulting chromosome are shown. (B) Expression from the epsI promoter is reduced in S49 strains which contain epsRD-A.
EpsR participates in a complex with the epsI promoter.
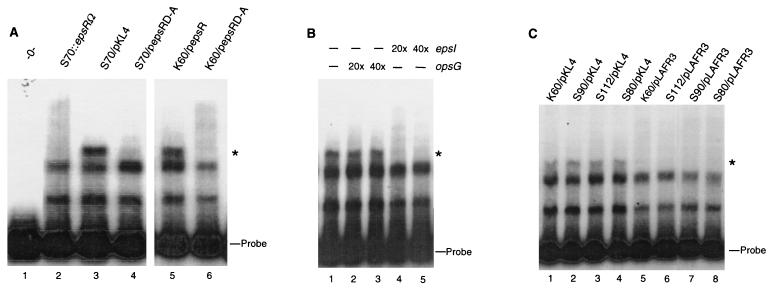
All of the results described above suggest that phosphorylation plays a critical role in the activity of EpsR. We used a gel mobility shift assay to examine the interaction of EpsR with the epsI promoter region and the role of phosphorylation in this interaction. Clarified extracts made from strains K60 and S70 overexpressing wild-type EpsR from either pKL4 or pepsR were incubated with a labeled 240-bp probe containing a functional epsI promoter. Three predominant bands were observed in a gel mobility shift assay, two of which were present in all extracts, including S70::epsRΩ, which does not contain a functional copy of epsR, and the third band, with the slowest mobility, which was present only in extracts from cells overexpressing EpsR (Fig. 5A, compare lane 2 to lanes 3 and 5). This band likely contains EpsR, while the two lower bands are not specific to EpsR, since they appear in all reactions, including ones which used lysates lacking EpsR (Fig. 5A, lane 1). In addition, extracts made from K60 and from S70, which overexpresses EpsRD-A, did not give rise to the specific band (Fig. 5A, lanes 4 and 6). These results strongly suggest that EpsR expressed in either wild-type R. solanacearum or a mutant lacking the acidic exopolysaccharide can form a complex with the epsI promoter. Furthermore, phosphorylation of EpsR is required for complex formation with the epsI promoter.
FIG. 5.
(A) Gel mobility shift assay shows that EpsR mediates a gel shift of the epsI promoter. Strains used to make each extract are indicated above the lanes. The EpsR-dependent shifted band is designated with a star. S70::epsRΩ containing pLAFR3 serves as a negative control since it lacks a functional copy of epsR. (B) Extracts overexpressing EpsR specifically shift the epsI promoter. A molar excess of nonradiolabeled DNA containing the opsG (nt −310 to +10) or epsI (nt −140 to +100) promoters was added as indicated above the autoradiogram. (C) The EpsR-mediated gel shift is detected in extracts made from strains S90 (xpsR), S112 (vsrB or vsrC), and S80 (phcA) overexpressing EpsR from pKL4 (29). Free probe is indicated to the right of each gel.
To further determine whether the unique shifted band represents a complex specific to epsI, we performed reactions with cell extracts preincubated with a 20- or 40-fold molar excess of nonradiolabeled epsI and opsG promoter sequences (Fig. 5B). Unlabeled epsI promoter abolished the band corresponding to the EpsR-induced complex, while preincubation with the opsG promoter region had no effect on the integrity of this band, suggesting that the proteins in the shifted complex bind the epsI promoter specifically (Fig. 5B). The lack of an EpsR-specific gel shift with extracts containing wild-type levels of EpsR may be due to a lack of sensitivity in the gel mobility shift assay or that the shifted complex we observed contains EpsR in a multimeric form.
To address the possibility that any of the known epsI regulators, XpsR, VsrB or VsrC, and PhcA, play a role in the EpsR-associated gel shift, we made clarified extracts from mutant strains S90 (xpsR), S112 (vsrB or vsrC), and S80 (phcA) containing pKL4 and tested them for the ability to form the EpsR-associated shifted complex. The EpsR-specific gel shift of the epsI promoter was still detected in these mutant backgrounds at levels similar to those of strain K60 containing pKL4, suggesting that PhcA, XpsR, and VsrB or VsrC are not required for the EpsR-induced gel shift of the epsI promoter (Fig. 5C).
DISCUSSION
An acidic form of EPS produced by the epsI gene cluster is a major virulence factor of the bacterial wilt pathogen, R. solanacearum. Disruption of the epsI structural genes will reduce or eliminate the ability of R. solanacearum to kill plants. Three signal transduction systems, VsrA-VsrD (37), VsrB-VsrC (19), and PhcA (4, 5, 17), positively affect epsI expression. EpsR has been reported to be a negative regulator of EPS when present in multicopy plasmids, decreasing EPS production and expression from the epsI promoter (16, 21, 29). In this work, we have extended the characterization of EpsR and found that a single copy of epsR in the R. solanacearum chromosome is required for wild-type levels of epsI expression and virulence. Thus, the level of EpsR protein in the cell can result in different phenotypes. Furthermore, both the repressive and stimulatory effects of EpsR and the formation of an EpsR-specific complex with the epsI promoter require an aspartate residue, which is important for the phosphorylation of EpsR.
After visual inspection of colonies with an epsR mutation, it was previously reported that one copy of EpsR was not affecting EPS expression (21). However, when expression from the epsI promoter was examined with the more sensitive and quantitative lacZ promoter fusions, expression was reduced in strains with an epsRΩ mutation. This observation, coupled with the fact that epsRΩ mutant strains are reproducibly affected in virulence, suggests that the chromosomal copy of epsR is positively regulating epsI synthesis, and only when overexpressed from multicopy plasmids can EpsR act as a repressor of epsI. We note that epsI expression and virulence are not abolished in epsRΩ mutant strains, suggesting that EpsR is contributing to, but not essential for, expression of epsI.
Together, our results suggest that EpsR directly regulates epsI expression. First, EpsR does not affect the expression of any of the other regulators of epsI. Second, overexpression of EpsR and the absence of the chromosomal copy of epsR affected expression from only epsI. Third, multicopy plasmids carrying the known positive regulators of epsI did not alleviate EpsR-mediated repression of epsI expression, suggesting that EpsR does not simply titrate one of these regulators (data not shown) (6). Our data does not eliminate the possibility that overexpression of EpsR interferes with the ability of another positive regulator to associate with the epsI promoter.
It is possible, although unlikely, that EpsR does not interact directly with the epsI promoter. Since our binding assays were done with crude cell extracts, we cannot rule out the possibility that overexpression of EpsR may in some way stimulate another protein to bind the epsI promoter, which in turn accounts for the observed epsI gel shift in extracts overexpressing EpsR. However, the EpsR-specific gel shift of the epsI promoter was observed with lysates putatively lacking PhcA, XpsR, and VsrB or VsrC, suggesting that these proteins are not required for the EpsR-induced gel shift of the epsI promoter. The C terminus of EpsR is homologous to the DNA-binding domains of other response regulators which have been shown to bind DNA (21). EpsR is highly basic (predicted pI of 8.5), which is consistent with the idea that EpsR has a natural affinity for DNA. All of these results are consistent with the working model that the activity of EpsR is mediated through formation of a complex with the epsI promoter.
EpsR-specific gel shift of the epsI promoter requires phosphorylation of EpsR. For R. solanacearum, this is the first direct demonstration of transcriptional regulation by protein modification. Most response regulators are thought to be phosphorylated at a conserved aspartate found in the N terminus. Several proteins have been shown experimentally to be phosphorylated, which in turn regulates many different aspects of the protein’s activity, including dimerization, DNA binding, and transcriptional regulation (2, 11–13, 24, 33, 39). EpsR has extensive homology with response regulators in the OmpR class; however, its phosphorylation domain is unique. EpsR is missing two conserved aspartate residues at amino acid positions 13 and 14, which are present in nearly all response regulators. In NarL, these residues are thought to form an acid pocket which facilitates phosphorylation (1). This suggests that the mechanism of phosphorylation for EpsR may be different from that of proteins such as NarL and NarP. Although EpsR is missing these residues, D47 is clearly required for phosphorylation, likely serving as the phosphate acceptor. We were unable to detect an EpsR-mediated gel shift of the epsI promoter by using extracts made from E. coli overexpressing EpsR. Coincident with this, preliminary results indicate that EpsR is not phosphorylated in E. coli, providing further proof that phosphorylation is required for the EpsR-mediated gel shift observed when R. solanacearum extracts are used (data not shown) (6). For E. coli strains transformed with either pKL4 or pepsR, we were unable to identify a phosphorylated band corresponding to EpsR. The identity of the kinase is not known; however, a functional homolog appears to be one that is absent in E. coli.
Approximately 1% of the nonessential genes in R. solanacearum are directly or indirectly involved in EPS production (29). At present, all regulators of EPS production affect expression from the epsI promoter. In E. coli, the epsI promoter is not expressed when carried on the plasmid pGepsI::lacZ, although other promoters involved in EPS production (opsG) are expressed (6). Since expression of epsI in R. solanacearum is dependent on several regulatory proteins, it is not surprising that this gene is not expressed in E. coli. Determining the signals required for activation of epsI, how each of the regulators interacts with each of the other regulators, and what conditions in planta might result in overexpression of EpsR are potential topics of future study.
The paradigm for differential regulation of a promoter is the lambda phage protein, which regulates its own expression by binding to different operator sequences located in its promoter (35). Several other bacterial proteins have been shown to act as both activators and repressors of transcription. In E. coli, the response regulators NarL and NarP can both positively and negatively affect transcription by the location of their DNA binding sites with respect to the transcriptional start site (9). The flagellar genes of Caulobacter crescentus are also both positively and negatively regulated by the response regulator FlbD (31). Should EpsR levels be regulated in the cell, then EpsR may interact specifically with the epsI promoter at multiple sites and with differing affinities. While it is possible that the repressive effect of EpsR is due to artificial overexpression, an abundance of EpsR retains the ability to specifically interact with the epsI promoter. Perhaps, with normal cellular levels of EpsR, a high-affinity binding site will be recognized, leading to activation of epsI. However, in our in vitro experiments, we are unable to detect an EpsR-mediated gel shift of the epsI promoter by using extracts containing wild-type levels of EpsR.
ACKNOWLEDGMENTS
We thank E. O’Reilly and C. Bauer for helpful comments on the manuscript and Y. Brun for plasmids, pGL10 and pHP45gΩ.
We thank Indiana University for funds to carry out this research. M.C. gratefully acknowledges the Konetzka fellowship.
REFERENCES
- 1.Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus P R P, Dickerson R E. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 2.Boucher P E, Menozzi F D, Locht C. The modular architecture of bacterial response regulators. J Mol Biol. 1994;241:363–377. doi: 10.1006/jmbi.1994.1513. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Brumbley S M, Carney B F, Denny T P. Phenotype conversion in Pseudomonas solanacearum due to spontaneous inactivation of PhcA, a putative LysR transcriptional regulator. J Bacteriol. 1993;175:5477–5487. doi: 10.1128/jb.175.17.5477-5487.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brumbley S M, Denny T P. Cloning of wild-type Pseudomonas solanacearum phcA, a gene that when mutated alters expression of multiple traits that contribute to virulence. J Bacteriol. 1990;172:5677–5685. doi: 10.1128/jb.172.10.5677-5685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman, M., and C. Kao. Unpublished data.
- 7.Cook D, Sequeira L. Genetic and biochemical characterization of a gene cluster from Pseudomonas solanacearum required for extracellular polysaccharide production and for virulence. J Bacteriol. 1991;173:1654–1662. doi: 10.1128/jb.173.5.1654-1662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coplin D L, Cook D. Molecular genetics of extracellular polysaccharide biosynthesis in vascular phytopathogenic bacteria. Mol Plant-Microbe Interact. 1990;3:271–279. doi: 10.1094/mpmi-3-271. [DOI] [PubMed] [Google Scholar]
- 9.Darwin A, Stewart V. Nitrate and nitrite regulation of the Fnr-dependent aeg-46.5 promoter of Escherichia coli K-12 is mediated by competition between homologous response regulators (NarL and NarP) for a common DNA-binding site. J Mol Biol. 1995;251:15–29. doi: 10.1006/jmbi.1995.0412. [DOI] [PubMed] [Google Scholar]
- 10.Denny T, Baek S. Genetic evidence that extracellular polysaccharide is a virulence factor of Pseudomonas solanacearum. Mol Plant-Microbe Interact. 1990;4:198–206. [Google Scholar]
- 11.Egan S M, Stewart V. Mutational analysis of nitrate regulatory gene narL in Escherichia coli K-12. J Bacteriol. 1991;173:4424–4432. doi: 10.1128/jb.173.14.4424-4432.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiedler U, Weiss V. A common switch in activation of the response regulators NtrC and PhoB: phosphorylation induces dimerization of the receiver modules. EMBO J. 1995;14:3696–3705. doi: 10.1002/j.1460-2075.1995.tb00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross R, Aricó B, Pappuoli R. Families of bacterial signal-transducing proteins. Mol Microbiol. 1989;3:1661–1667. doi: 10.1111/j.1365-2958.1989.tb00152.x. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 15.Hayward A C. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol. 1991;29:65–108. doi: 10.1146/annurev.py.29.090191.000433. [DOI] [PubMed] [Google Scholar]
- 16.Huang H, Sequeira L. Identification of a locus that regulates multiple functions in Pseudomonas solanacearum. J Bacteriol. 1990;172:4728–4731. doi: 10.1128/jb.172.8.4728-4731.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J, Carney B F, Denny T P, Weissinger A K, Schell M A. A complex network regulates expression of eps and other virulence genes of Pseudomonas solanacearum. J Bacteriol. 1995;177:1259–1267. doi: 10.1128/jb.177.5.1259-1267.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Schell M. Molecular characterization of the eps gene cluster of Pseudomonas solanacearum and its transcriptional regulation at a single promoter. Mol Microbiol. 1995;16:977–989. doi: 10.1111/j.1365-2958.1995.tb02323.x. [DOI] [PubMed] [Google Scholar]
- 19.Huang J, Denny T, Schell M A. VsrB, a regulator of virulence genes of Pseudomonas solanacearum, is homologous to sensors of the two-component regulator family. J Bacteriol. 1993;175:6169–6178. doi: 10.1128/jb.175.19.6169-6178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussain A, Kelman A. Relation of slime production to mechanism of wilting and pathogenicity of Pseudomonas solanacearum. Phytopathology. 1958;48:155–165. [Google Scholar]
- 21.Kao C C, Gosti F, Huang H, Sequeira L. Characterization of a negative regulator of exopolysaccharide production by the plant-pathogenic bacterium Pseudomonas solanacearum. Mol Plant-Microbe Interact. 1994;7:121–130. doi: 10.1094/mpmi-7-0121. [DOI] [PubMed] [Google Scholar]
- 22.Kao C C, Sequeira L. A gene cluster required for coordinated biosynthesis of lipopolysaccharide and extracellular polysaccharide also affects the virulence of Pseudomonas solanacearum. J Bacteriol. 1991;173:7841–7848. doi: 10.1128/jb.173.24.7841-7847.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao C C, Barlow L, Sequeira L. Extracellular polysaccharide is required for wild-type virulence of Pseudomonas solanacearum. J Bacteriol. 1992;174:1068–1071. doi: 10.1128/jb.174.3.1068-1071.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karimova G, Bellalou J, Ullman A. Phosphorylation-dependent binding of BvgA to the upstream region of the cyaA gene of Bordetella pertussis. Mol Microbiol. 1996;20:489–496. doi: 10.1046/j.1365-2958.1996.5231057.x. [DOI] [PubMed] [Google Scholar]
- 25.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 26.Kelman A. The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on a tetrazolium medium. Phytopathology. 1954;44:693–695. [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Lorenzo V, Herrero M, Jakubzik V, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McWilliams R, Chapman M, Kowalczuk K M, Hersberger D, Sun J S, Kao C. Complementation analysis of Pseudomonas solanacearum extracellular polysaccharide mutants and identification of genes responsive to EpsR. Mol Plant-Microbe Interact. 1995;8:837–844. doi: 10.1094/mpmi-8-0837. [DOI] [PubMed] [Google Scholar]
- 30.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 31.Mullin D A, Vanway S M, Blankenship C A, Mullin A H. FlbD has a DNA-binding activity near its carboxy terminus that recognizes ftr sequences involved in positive and negative regulation of flagellar gene transcription in Caulobacter crescentus. J Bacteriol. 1994;176:5971–5981. doi: 10.1128/jb.176.19.5971-5981.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orgambide G, Montrozier H, Servin P, Roussel J, Trigalet-Demery D, Trigalet A. High heterogeneity of exopolysaccharides of Pseudomonas solanacearum strain GM1000 and complete structure of the major polysaccharide. J Biol Chem. 1991;266:8312–8321. [PubMed] [Google Scholar]
- 33.Parkinson J S, Kofoid E C. Communicating modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 34.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 35.Ptashne M. A genetic switch. 2nd ed. Cambridge, Mass: Blackwell Scientific Publications; 1992. . Press, New York, N.Y. [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Schell M A, Denny T P, Huang J. VsrA, a second two-component sensor regulating virulence genes of Pseudomonas solanacearum. Mol Microbiol. 1993;11:489–500. doi: 10.1111/j.1365-2958.1994.tb00330.x. [DOI] [PubMed] [Google Scholar]
- 38.Stock J B, Surette M G, McCleary W R, Stock A M. Signal transduction in bacterial chemotaxis. J Biol Chem. 1992;267:19753–19756. [PubMed] [Google Scholar]
- 39.Wingrove J A, Mangan E K, Gober J W. Spatial and temporal phosphorylation of a transcriptional activator regulates pole-specific gene expression in Caulobacter. Genes Dev. 1993;7:1979–1992. doi: 10.1101/gad.7.10.1979. [DOI] [PubMed] [Google Scholar]
- 40.Xu P, Leong S, Sequeira L. Molecular cloning of genes that specify virulence in Pseudomonas solanacearum. J Bacteriol. 1988;170:617–622. doi: 10.1128/jb.170.2.617-622.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]