Abstract
In this study, metagenomic sequencing technology was employed to analyze the ITS1 region sequence of the ITS rDNA gene of endophytic fungi and 16S sequence of endophytic bacteria in tea leaves with varying degrees of infection by tea blister blight disease as well as healthy tea leaves. Subsequently, a comparative analysis was conducted on the endophytic microbial diversity and the community structure in tea leaves. The findings of this investigation reveal a shift in the dominant endophytic fungal genera from Ascomycota to Basidiomycota as the disease progressed. Furthermore, a negative correlation was observed between Exobasidium and Talaromyce, with Talaromyce exhibiting potential as an antagonist against the disease. Meanwhile, our findings reveal that Proteobacteria, Firmicutes, and Actinobacteria were the three most abundant bacteria phyla in tea leaves. As the disease progressed, there was an increase in the relative abundance of Actinobacteria, while Variovorax, Sphingomonas, and Pseudomonas were found to have higher abundance in later stages. The diversity analysis results indicated that the endophytic microbial diversity and the community structure in tea leaves in the diseased group were lower than those in the healthy control group. In general, blister blight disease altered the community structure of endophytic microorganisms in tea leaves, resulting in a few species with high abundance. The study lays a foundation for investigating the pathogenic mechanism of tea blister disease and establishing a theoretical basis for controlling diseases in tea trees.
Keywords: Blister blight disease, Endophytic microbial, Diversity, Community structure, Tea leaves
Introduction
Endophytes are non-pathogenic organisms that live in plant tissues (Rahman et al. 2018). Most plant endophytes are harmless although a few of them are opportunistic pathogen (Bové, 2006). The presence of endophytic microorganisms in plants represents a valuable and innovative microbial resource, as extensive research has demonstrated their indispensable role in promoting plant growth and health (Niu et al. 2022; Kunpeng et al. 2022; Sun et al. 2022; Sharma and Kumar 2021; Gupta et al. 2020). Endophytes possess the ability to enhance plant adaptability by modulating community structure, thereby conferring resistance against abiotic and biological stresses (Nadira et al. 2021; Liu et al. 2022). At present, there are numerous studies on endophytic microorganisms of tea leaves (Mu et al. 2022; Tibpromma et al. 2022; Jia et al. 2022; Hazarika et al. 2021; Kabir et al. 2023). Nevertheless, the majority of these studies are limited to utilizing traditional isolation and culture methods in order to obtain a restricted number of culturable endophytic microorganisms and explore their functions, such as disease resistance, insect resistance, and drought resistance. However, research has shown that pure culture methods only enable the detection of a minimal fraction of uncultured microorganisms (Liu et al. 2022). In the context of sequencing technology evolution, high-throughput sequencing enables direct detection of endophytes in host tissues. This technology offers a more comprehensive and intuitive analysis of endophyte community composition and diversity in plant tissues due to its large sequencing base and high depth, providing a foundation for investigating pathogenic antagonistic microorganisms or compound microbial agents (Tamošiūnė et al. 2019).
Tea blister blight disease, also known as tea gall and leaf swelling disease, is a highly significant fungal infection that affects tea plants in numerous countries including China, India, Sri Lanka, Malaysia, Thailand, and Japan (Pandey et al. 2021; Barman et al. 2020; Rachmad et al. 2022; Sen et al. 2020; Zhang et al. 2023). Numerous studies have demonstrated that tea tree leaves, when infected with tea blister blight disease, can disrupt the electron transport pathway of the respiratory chain by secreting pathogenic factors that damage mitochondria (Chakraborty et al. 2007). Additionally, it has been observed that leaf growth, physiological parameters, and photosynthetic activity significantly decrease during the infection process of blister blight disease (Premkumar et al. 2008). These findings indicate that pathogen infection not only destroys host tissue structure but also disrupts normal and biochemical processes. Moreover, tea blister blight disease can substantially reduce tea polyphenols and catechins, thereby negatively impacting both yield and quality of tea leaves (Zhou et al 2023). However, there has been no research conducted on the impact of blister blight disease on the endophytic microbial community of tea leaves.
This study aims to collect healthy tea plant leaves and leaves infected with blister blight disease at different stages (early, middle, and late) of disease progression, followed by metagenomic sequencing of endophytic fungi and bacteria using high-throughput sequencing technology. By exploring the relationship between them, this study intends to provide a theoretical basis for preventing and controlling tea diseases, as well as laying the foundation for further exploration of the underlying pathology.
Materials and methods
Materials collection
In September 2022, tea leaf samples were collected from five tea gardens situated in Jinji Village, Suiyang Town, Fenggang County, Guizhou Province (28°19′95.8''N, 107°02′75.4"E). The sampled tea trees belong to the Longjing 43 cultivar and are grown at an average altitude of 866 m with an annual precipitation of 1160.00 mm and an annual mean temperature of 15.10 °C. Each replicate was selected from a distinct location, and within each location, four groups of tea leaves samples were collected from tea tree affected by blister blight disease at different stages (early, middle, and late) of disease progression as well as healthy tea leaves. The healthy group was designated as CB1, while the disease groups at early stage (lesion diameter of 3–6 mm, Fig. 1A), middle stage (expansion of the disease results in a smooth and concave leaf surface, with bright front sides and grayish or dark backsides that exhibit noticeable thickening, Fig. 1B), and late stage (exhaustion of cellular nutrition within the lesion tissue leads to a host resistance reaction, resulting in the limited expansion of necrotic, brown, and ulcerated lesions, Fig. 1C) of disease progression were labeled CB2, CB3, and CB4, respectively. Each group consisted of three replicates resulting in a total of 12 samples. The sampling technique employed was the five-point method which first determines the midpoint of the diagonal line as the central sampling point, and then selects four points on the diagonal line with the same distance from the central sample point as the sample point, and each sample includes five sub-samples (Maki et al. 2023). The collected samples were pre-treated under sterile conditions using a three-step disinfection process (75% alcohol-5% sodium hypochlorite-75% alcohol) and subsequently stored at − 80 °C (Liu et al. 2022).
Fig. 1.
Collected tea leaves which show different stages of disease symptoms of blister blight disease. A disease groups at early stage; B disease groups at middle stage; C disease groups at late stage
DNA extraction, PCR amplification, and high-throughput sequencing
The sterilized samples were pulverized using a mixed grinder (MM 400, Retsch, Germany), followed by total community DNA extraction with the Qiagen DNeasy Plant Mini kit (Qiagen, Redwood, CA, USA). Polymerase chain reactions (PCR) were performed using fungal primers (ITS1F and ITS2R) and bacterial primers (799F and 1193R). To enhance the efficiency of PCR and prevent amplification of mitochondrial and plastid templates, two types of peptide nucleic acid clamps (mPNA and pPNA) for blocking mitochondrial and plastid DNA were introduced into the PCR mixture (Lundberg et al. 2013). A 50 μL PCR system, consisting of 25 μL of a 2 × PCR mixture, 2 μL each of primers (5 μM), 2.5 μL pPNA (5 μM), 2.5 μL mPNA (5 μM), 2 ng template DNA and 16 μL ddH2O, was utilized with the thermal cycle conditions of PCR as follows: 3 min at 94 °C (initial denaturation), 30 cycles of 15 s at 94 °C, 10 s at 75 °C, 10 s at 55 °C, 30 s at 68 °C, and the final extension at 72 °C for 10 min. The amplicons were purified using a gel extraction kit (OMEGA bio-tek, Doraville, GA, USA) and the DNA concentration was quantified by Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The amplicons that had been purified were combined in equal molar concentrations and subjected to paired-end sequencing on the Illumina Miseq-PE 250 sequencing platform (Illumina Inc., San Diego, CA). The sequence data have been deposited in NCBI and assigned a BioProject ID PRJNA979171 (http://www.ncbi.nlm.nih.gov/bioproject/979171).
Sequence analyses
The Illumina Miseq-PE 250 platform (Illumina Inc., San Diego, CA) was utilized for sequencing the samples, and subsequently processed using QIIME2 (Bokulich et al. 2018). The primer fragments were eliminated using qiime cutadapt trim-pairing, followed by the removal of sequences lacking matching primers. Subsequently, the data underwent rigorous quality control, de-noising, splicing, and chimera removal via qiime dada2 noise-paired. The amplicon sequence variants (ASVs) and the corresponding tables were merged while singleton ASVs were excluded. To ensure consistency, R language scripts were utilized to perform length distribution statistics on high-quality sequences in all samples. The QIIME2 package incorporates the classification-sklearn algorithm, and default parameters are employed to classify each ASV's characteristic sequence based on the UNITE database (https://unite.ut.ee/) and Greengenes database (http://greengenes.secondgenome.com/) using the Naive Bayes classifier for annotation, respectively. Finally, the community structure of samples at various taxonomic levels was visualized using R 3.6.0.
Statistical analysis
Alpha diversity and beta diversity indices are commonly utilized to assess species richness within and among habitats (Whittaker 1972). The unflattened ASV table was utilized to compute the alpha diversity index via QIIME2. Post hoc analyses including Kruskal–Wallis rank-sum test and Dunn's test were performed to confirm statistical significance of differences observed. Data visualization was conducted using R script. Beta diversity was calculated utilizing Bray–Curtis distance metric and assessed through QIIME2 (Martino et al. 2019). A principal coordinate analysis (PCoA) was conducted on the distance matrix, followed by a clustering hierarchy analysis using the UPGMA algorithm and Bray–Curtis distance matrix. The Wayne diagram was generated based on the ASV abundance table, which recorded the presence or absence of each ASV in different sample groups. The petal diagram was then constructed for visualization purposes. Correlation analysis was performed using igraph with reference to the ASV abundance table, and the results were presented visually.
Results
Analysis of microbial community composition
After undergoing quality filtering and removal of singleton and chimeric sequences, the ITS datasets of four groups (CB1, CB2, CB3, and CB4) exhibited a range of 81,138 to 131,449 reads per sample (148–339 bp in length) with an average of 113,576 reads and comprising 653 ASVs. The samples' 16S rRNA reads ranged from 145,318 to 163,353 per sample (156–384 bp in length), averaging at 156,098 reads assigned to a total of 741 ASVs.
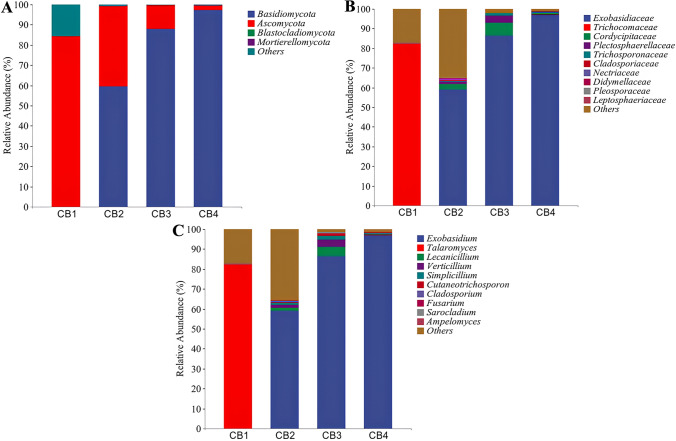
For fungi, Fig. 2A illustrates that the top four most abundant phyla were Ascomycota, Basidiomycota, Mortierellomycota, and Blastocladiomycota. Notably, during the CB1 period, Ascomycota was found to be dominant in healthy tea leaves with a relative abundance of 84.32%. With disease progression, the relative abundance of Ascomycota gradually declined from 84.32% to 2.33%, while Basidiomycota emerged as the dominant phylum during CB2–CB4 period and rapidly increased in relative abundance from 59.53% to 97.17%. Meanwhile, as illustrated in Fig. 2B, the top 10 most abundant families comprised Exobasidiaceae, Cordycipitaceae, Plectosphaerellaceae, Trichosporonaceae, Cladosporiaceae, Nectriaceae, Didymellaceae, Pleosporaceae, and Leptosphaeriaceae. In the CB1 group, the highest abundance was Trichocomaceae, accounting for 82.06%. In the CB2–CB4 group, the highest abundance was Exobasidiaceae, accounting for 58.92%, 86.43%, and 96.97%, respectively. Furthermore, as shown in Fig. 2C, the top 10 most abundant genera were Exobasidium, Talaromyces, Lecanicillium, Verticillium, Simplicillium, Cutaneotrichosporon, Cladosporium, Fusarium, Sarocladium, and Ampelomyces. In the CB1 group, Talaromyces was the dominant fungi, accounting for 82.06%. In the CB2–CB4 groups, Exobasidium was the dominant genus, accounting for 58.92%, 86.43%, and 96.97%, respectively.
Fig. 2.
In the graph, the horizontal coordinate is the relative abundance of endophytic bacteria at the phylum, family, and genus levels affected by blister blight disease at different stages (early, middle, and late) of disease progression as well as healthy ones, and the vertical coordinate is the relative abundance of each taxon at a specific classification level. A Relative abundance of endophytic fungi at the phylum level; B Relative abundance of endophytic fungi at the family level; and C Relative abundance of endophytic fungi at the genus level
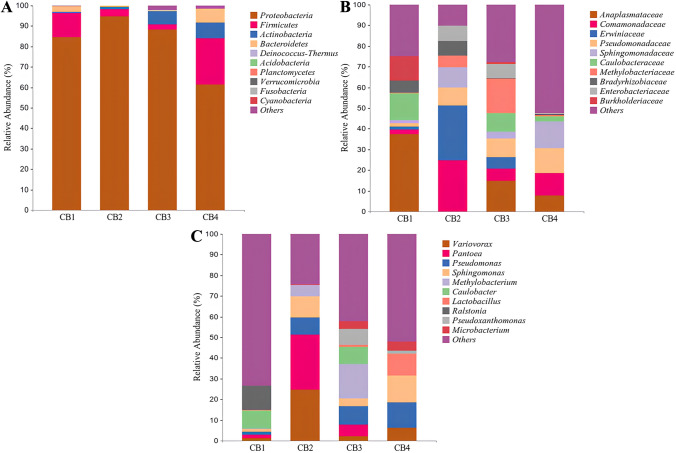
For bacteria, at the phylum level (Fig. 3A), the top 10 most abundant phyla in the four groups are Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes, Deinococcus-Thermus, Acidobacteria, Planctomycetes, Verrucomicrobia, Fusobacteria, and Cyanobacteria. In the CB1–CB4 groups, Proteobacteria was found to be the most abundant phylum, comprising 84.49%, 94.62%, 88.29%, and 61.27% of the total bacterial community, respectively. Meanwhile, at the family level (Fig. 3B), the top 10 most abundant families comprised Anaplasmataceae, Caulobacteraceae, Burkholderiaceae, Bradyrhizobiaceae, Comamonadaceae, Pseudomonadaceae, Erwiniaceae, Sphingomonadaceae, Methylobacteriaceae, and Enterobacteriaceae. During the CB1 period, Anaplasmataceae exhibited a higher relative abundance of 37.40%, making it the dominant phylum of endophytic bacteria in healthy tea leaves. As the disease progressed, the relative abundance of Anaplasmataceae gradually decreased from 31.40% to 7.66%. In addition, at the genus level (Fig. 3C), the top 10 most abundant genera in the four groups were Variovorax, Pantoea, Pseudomonas, Sphingomonas, Methylobacterium, Caulobacter, Lactobacillus, Ralstonia, Pseudoxanthomonas, and Microbacterium. In the CB1 group, Ralstonia and Caulobacter were the dominant bacteria, accounting for 11.72% and 8.59%, respectively. In the CB2 group, Pantoea and Variovorax were the dominant genera, accounting for 26.45% and 24.76%, respectively. In the CB3 group, Methylobacterium was the dominant genus, accounting for 16.72%. In the CB4 group, Sphingomonas and Pseudomonas were the dominant genera, accounting for 12.96% and 12.17%, respectively. It is noteworthy that Variovorax, Pseudomonas, and Sphingomonas are present in all four stages of the disease, with their relative abundance gradually increasing as the disease progresses.
Fig. 3.
In the graph, the horizontal coordinate is the relative abundance of endophytic bacteria at the phylum, family, and genus levels affected by blister blight disease at different stages (early, middle, and late) of disease progression as well as healthy ones, and the vertical coordinate is the relative abundance of each taxon at a specific classification level. A Relative abundance of endophytic bacteria at the phylum level; B Relative abundance of endophytic bacteria at the family level; and C Relative abundance of endophytic bacteria at the genus level
Alpha diversity analysis
Alpha diversity, also known as within-habitat diversity, refers to the richness, diversity, and evenness of species in locally homogeneous habitats (Chao 1984; Shannon 2001; Simpson 1949). To assess microbial community alpha diversity, in this study, Chao1 and Observed species indices were utilized to quantify richness, while Shannon and Simpson indices were employed to measure diversity. According to the results of Alpha diversity index analysis (Table 1), blister blight disease has been found to reduce the richness and the diversity of endophytic microorganisms. The impact on the diversity of endophytic fungi was most pronounced during the early stage, exhibiting significantly higher diversity compared to later stages of infection.
Table 1.
Alpha diversity indices of bacteria and fungi in the tea leaves at different stages of disease progression
| Classify | Fungi | Bacteria | ||||||
|---|---|---|---|---|---|---|---|---|
| Chao1 | Observed species | Shannon | Simpson | Chao1 | Observed species | Shannon | Simpson | |
| CB1 | 54.38 ± 8.10a | 53.63 ± 7.97a | 1.16 ± 0.28b | 0.33 ± 0.16ab | 46.45 ± 15.96a | 39.70 ± 13.26a | 4.07 ± 0.41a | 0.91 ± 0.02a |
| CB2 | 70.05 ± 20.87a | 69.33 ± 20.62a | 1.74 ± 0.35a | 0.55 ± 0.03a | 55.82 ± 16.55a | 38.40 ± 11.26a | 3.60 ± 0.76a | 0.83 ± 0.10a |
| CB3 | 41.28 ± 6.12a | 40.03 ± 6.25a | 1.00 ± 0.33b | 0.25 ± 0.08bc | 38.41 ± 5.75a | 37.47 ± 5.41a | 4.38 ± 0.22a | 0.93 ± 0.01a |
| CB4 | 55.29 ± 19.89a | 54.60 ± 19.97a | 0.33 ± 0.25c | 0.06 ± 0.05c | 45.35 ± 12.05a | 35.93 ± 13.00a | 3.81 ± 1.11a | 0.86 ± 0.09a |
Beta diversity analysis
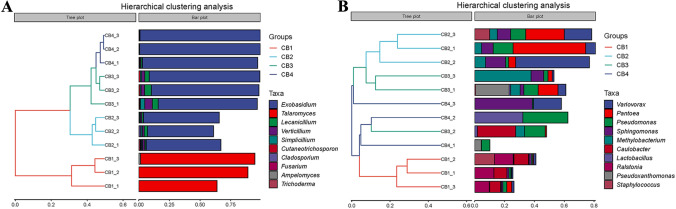
The beta diversity index is a widely used metric for quantifying the dissimilarity in community composition among different habitats, as indicated by differences in sample composition (Lozupone et al. 2007). In this study, the beta diversity of the samples was computed utilizing QIIME software and the outcomes of this analysis are presented in Fig. 4. Inspection of Fig. 4A reveals that the four samples can be classified into two major branches. CB1 groups are clustered together in one large branch, while CB2–CB4 groups form another large branch. These findings suggest that the incidence of blister blight disease significantly altered the community structure of endophytic fungi in tea leaves, resulting in a noticeable clustering between sample groups. Figure 4B illustrates the beta diversity outcomes of endophytic bacteria, revealing a significant impact of blister blight disease on early-stage endophytic bacteria in tea leaves.
Fig. 4.
Beta diversity analysis of endophytic fungi and bacteria in tea leaves affected by blister blight disease at different stages (early, middle, and late) of disease progression as well as healthy ones. In the figure, the panel on the left is a hierarchical clustering tree, where samples are grouped according to their similarity. The shorter the branch length between samples, the more similar the two samples are. The panel on the right (drawn by default) is a stacked bar chart of the top 10 genera in abundance. A Hierarchical clustering tree with branch support of endophytic fungi; B Hierarchical clustering tree with branch support of endophytic bacteria
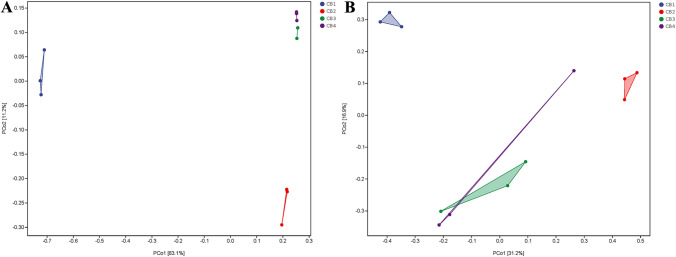
PCoA is a technique utilized to expand low-dimensional spaces by projecting sample distance matrices while maintaining the original distance relationship as closely as possible (Ramette 2007). The PCoA analysis results presented in Fig. 5 indicate that PCoA1 accounts for 83.1% of the functional variation in fungi and 31.2% in bacteria. In line with the results on endophytic microbial community structure, fungal and bacterial spectra derived from CB1 samples exhibited significant dissimilarities compared to those of other samples (acquired from the upper left quadrant of the PCoA plots).
Fig. 5.
Each dot in the diagram represents a sample, and different colored dots indicate different groupings. The percentages in the brackets represent the percentage of the sample difference data (distance matrix) that can be explained by the corresponding axis. A PCoA analysis of endophytic fungi communities in tea leaves affected by blister blight disease at different stages (early, middle, and late) of disease progression as well as healthy ones; B PCoA analysis of endophytic bacteria communities in tea leaves affected by blister blight disease at different stages (early, middle, and late) of disease progression as well as healthy ones
Analyses of the network structure
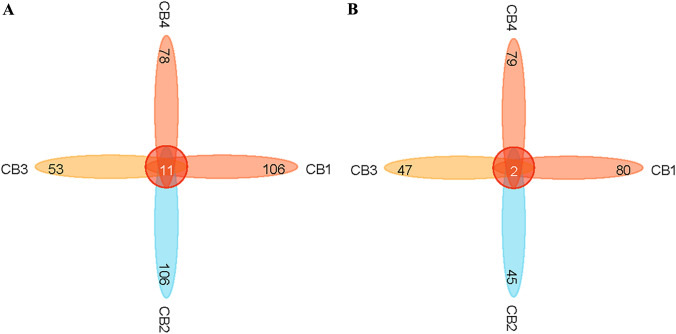
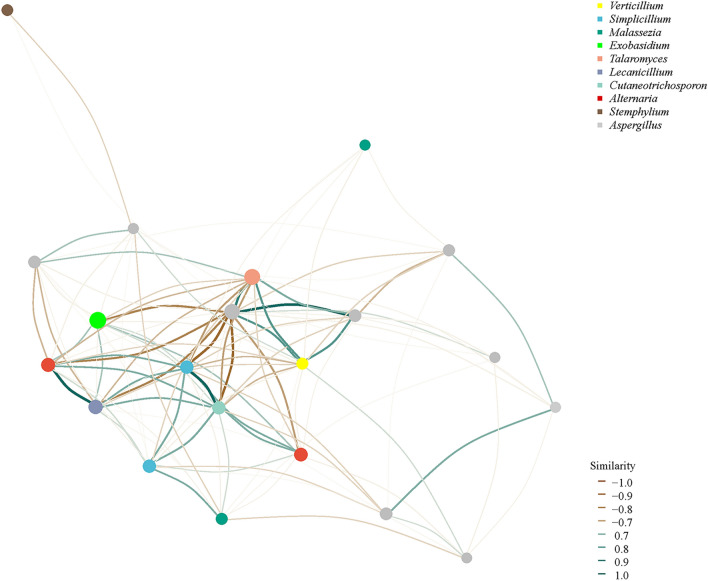
The abundance and the similarity of ASVs in four samples were analyzed. For fungi, Fig. 6A showed that CB1 and CB2 had 117 fungi species and 106 endemic species, CB3 had 64 fungi species and 53 endemic species, and CB4 had 89 fungi species and 78 endemic species. Regarding bacterial aspects, Fig. 6B illustrates that CB1, CB2, CB3, and CB4 contained a total of 82, 47, 49, and 81 bacterial species, including 80, 45, 47, and 79 endemic ones. The results indicate a significant decline in both the total number of species and the number of endemic species within CB3 and CB4 groups as blister blight disease progresses, compared to the healthy period of CB1, indicating a reduction in the diversity of endophytic microbial diversity in infected tea leaves. Furthermore, an induced subgraph function of igraph was utilized to conduct network analysis based on ASV abundance in order to explore the correlation among endophytic microbial members in tea leaves (Gustavsen et al. 2019). Figure 7 illustrates a positive correlation between Exbasidium and Simplicillium, Cutaneotrichosporon, Verticillium, and Alternaria, while showing a negative correlation with Talaromyces and Stemphylium. However, the limited diversity of common microorganisms prevented endophytic bacteria from establishing microbial networks.
Fig. 6.
ASV level of endophytic bacteria distribution in tea leaves affected by blister blight disease at different stages (early, middle, and late) of disease progression as well as healthy ones. In the diagram, each block represents a group, the overlapping area between the blocks indicates the ASV shared between the corresponding groups, and the number of each block indicates the number of ASVs contained in that block. A Venn diagram shows the number of ASVs shared and unique fungi species; B Venn diagram shows the number of ASVs shared and unique bacterial species
Fig. 7.
Correlation network analysis of fungi diversity in tea leaves affected by blister blight disease at different stages (early, middle, and late) of disease progression as well as healthy ones. The size of annotation nodes for the top 10 species, distinguished by different colors, is proportional to their abundance (The unit is log2(CPM/n). The presence of an edge between two connected nodes indicates a correlation between them
Discussion
Blister blight disease is a devastating affliction of tea leaves caused by Exobasidium vexans Masee (E. vexans), which colonizes the foliage and has a severe impact on tea production. During its early stages, E. vexans was identified as an obligate parasitic fungus (Jayaswall et al. 2016; Chaliha et al. 2020; Chandra et al. 2017). Although it has been demonstrated that E. vexans can thrive on potato dextrose agar medium supplemented with natural active substances, carbon sources, tea extracts, and calcium carbonate, a comprehensive investigation into the biology of the isolated strains remains unreported (Chaliha et al. 2017). Therefore, there is debate as to whether E. vexans can be artificially cultured, which has hindered research on the pathogenesis of blister blight disease and its prevention and control. To circumvent this bottleneck issue, amplicon sequencing technology was employed to analyze the microbial community present in blister blight disease tissue. This approach allows a comprehensive and accurate depiction of pathogenic microorganism interactions, which has significant implications for pathogen identification and disease prevention and control.
In this study, it was observed that the microbial diversity of endophytic bacteria was significantly lower in diseased leaves compared to healthy ones. The three most abundant phyla of endophytic bacteria were Proteobacteria, Firmicutes, and Actinobacteria, which were found across all four sample groups. Previous studies have demonstrated the efficient utilization of plant secretions by Proteobacteria and Firmicutes, which play a crucial role in promoting plant growth (Trivedi et al. 2020). It is noteworthy that the relative abundance of Actinobacteria increases as the disease progresses. Actinobacteria, a Gram-positive bacterium, produces various metabolites, such as antibiotics and antagonistic substances, whose efficacy has been demonstrated in different soil- and ocean-derived strains (Rateb et al. 2018; Lu et al. 2020). The results of the study indicate that Variovorax, a genus classified under Actinobacteria, exhibited higher relative abundance in susceptible leaves compared to healthy ones. This genus not only promotes plant root growth within complex microbial communities but also exerts an antagonistic effect on plant pathogens (Finkel et al. 2020; Hong et al. 2017). Additionally, the relative abundances of Sphingomonas and Pseudomonas were significantly higher in susceptible leaves than in healthy ones. Both Sphingomonas and Pseudomonas have been shown to exhibit antagonistic effects against pathogens (Dong et al. 2022; Yu et al. 2022; Hadian et al. 2023). Pseudomonas has the ability to simultaneously produce multiple antibiotics in response to diverse plant pathogenic microorganisms (Biessy et al. 2021; Aghdam et al. 2022; Lv et al. 2023). This study has concluded that tea plants can stimulate the defense mechanisms of endophytic microorganisms in tea leaves following an attack by blister blight disease. This response enhances the abundance of certain endophytic bacteria with potential disease resistance, which may aid in increasing the resistance of tea leaves to invasion by blister blight disease.
The study revealed that endophytic fungi belonging to the phyla Ascomycota and Basidiomycota were the primary dominant species. The genus Talaromyce was found to be predominant in healthy tea leaves, while the genus Exobasidium exhibited dominance in susceptible tea leaves. Talaromyce, known for its chitinase secretion and competitive edge, serves as a potent biological agent against plant pathogens. For instance, Talaromyces flavus is frequently utilized as a biological antagonist to suppress the proliferation of pathogens (Bahramian et al. 2016; Shabani et al. 2024). Therefore, the findings of this study hold significant implications for tea plant disease management, as Talaromyce has the potential to serve as a valuable biological control resource in the event of pathogen outbreak.
Our findings indicate that Exobasidium is the predominant genus present in diseased tea leaves. Studies have shown that Exobasidium harbors E. reticulatum and E. vexans that are known to cause significant damage to tea plants (Kerr and Rodrigo 1967). Interestingly, our study revealed that Exobasidium had a relative abundance of 0.18% in healthy tea leaves, indicating its active colonization within the plant prior to the onset of blister blight disease. Additionally, we have observed an increase in the relative abundance of E. vexans in tea leaves following infection by blister blight disease, which may be attributed to the disruption of the host's endophytic microbial communities. Consequently, fungi that play an antagonistic role could lose their competitiveness, allowing for rapid niche occupation by the pathogen.
Notably, we have observed significant differences in the community structure of endophytic fungi between healthy and diseased leaves. Specifically, an increase in the relative abundance of Exobasidium in diseased leaves corresponded with a decrease in the relative abundance of Talaromyce. At the same time, Talaromyces is also often used as a biocontrol bacterium in the prevention and control of other diseases (Huong et al. 2022; Dethoup et al. 2022; Di et al. 2023). This suggests potential niche competition between these two fungal species and highlights the possibility for Talaromyce to be developed as a biocontrol agent for blister blight disease. In addition, plant diseases can also result from a synergistic effect between different pathogenic microorganisms on a single host plant, leading to an escalation in disease severity (Ruiz-Bedoya et al. 2023). For example, previous research conducted by Barman et al. has demonstrated that the domesticated tea clone TV17 exhibits symptoms resembling blister blight disease when co-infected with Pestalotiopsis spp. and Nigrospora spp. under controlled greenhouse conditions (Barman et al. 2020). Therefore, to investigate the relationship between Exobasidium and other endophytic fungi, we constructed a microbial correlation network and discovered a positive correlation between Exobasidium and Alternaria. Alternaria is a genus of fungi widely distributed in the natural environment that causes several important crop diseases, which can significantly impact crop production and lead to substantial losses (Xu et al. 2019; Xin et al. 2021). Previous studies have shown that Alternaria tenuis and Alternaria longipes, two major pathogens causing leaf mold disease in tea leaves, belong to the genus Alternaria (Yin et al. 2021; Yin et al. 2021). Whether Exobasidium and Alternaria exhibit a synergistic effect in the pathogenesis of blister blight disease requires further experimental verification. Meanwhile, we have discovered a negative correlation between Exobasidium and Talaromyce. Consequently, we have observed a significant increase in the relative abundance of Exobasidium and a corresponding decrease in the relative abundance of Talaromyce in tea leaves afflicted with blister blight disease. This implies the possibility of an antagonistic relationship or niche overlap, which aligns with our prior conjecture.
In summary, this study has revealed that the community structure of endophytic fungi undergoes changes in response to blister blight disease-induced damage on tea leaves. The highest richness of endophytic fungi was detected during the early stage of infection, while the lowest richness was observed during the middle and late stages of infection. Furthermore, the diversity of endophytic fungi experienced a temporary increase during the initial phase of infection before declining continuously and reaching its lowest point in later stages. In contrast, the richness and the diversity of the endophytic bacterial community varied across different periods, yet no statistically significant differences were observed. The beta analysis also indicated a more profound impact on the composition of the endophytic fungal community compared to that of the endophytic bacterial community. Furthermore, the identification of endophytic microbial species in infected leaves at different stages demonstrated potential niche competition between Talaromyce in endophytic fungi and pathogenic bacteria causing blister blight disease. This competition was negatively correlated with Exobasidium, Talaromyce, and Stemphylium in the construction of a complex network. These findings are highly significant for studying compound microbial agents. Additionally, the relative abundance of endophytic bacteria provides insights into the potential resistance of Variovorax, Sphingomonas, and Pseudomonas to invasion by blister blight disease. Further verification of these findings could clarify the antagonistic abilities of these bacterial species.
Moreover, Chen et al. (2023) discovered that Cladosporium was the predominant bacterium in the vulnerable site of blister blight disease and exhibited greater viability than E. vexans on PDA plates. Consequently, this bacterium hindered the isolation of E. vexans, which proved challenging to culture in vitro. Conversely, this study identified an increase in Exobasidium within the endophytic fungal community of tea leaves as disease progression occurred. The proportion of Exobasidium increased to 96.97% in the later stage of infection, indicating that it is feasible to obtain E. vexans by isolating endophytic fungi from susceptible tissues.
Conclusion
This study utilized metagenomic sequencing technology to uncover the response of endophytic microbial communities in tea leaves to the threat of tea blister blight disease. The results indicate that the abundance and the diversity of endophytic fungi initially increase and then decrease with the progression of the disease. On the other hand, the abundance and the diversity of endophytic bacterial communities show some differences at different stages, but the differences are not significant due to the activation of defense mechanisms of endophytic bacteria by the infection of blister blight disease. Moreover, based on the analysis of community structures in samples from different time periods, a negative correlation was observed between Talaromyce and pathogenic fungi during the disease development. In addition, the relative abundance of endophytic bacteria provides insights into the potential resistance of Variovorax, Sphingomonas, and Pseudomonas against the invasion of blister blight disease. These findings hold significant implications for researching composite microbial preparations.
Funding
This work was supported by the Science and Technology Support Program of Guizhou, China (Nos. [2023]085 and [2020]1Y001) and the National Natural Science Foundation of China (No. 32160077).
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Aghdam NMN, Baghaee-Ravari S, Shiri A. Antimicrobial capacity of Pseudomonas brassicacearum strain EnPb against potato soft rot agent. Eur J Plant Pathol. 2022;165:215–231. doi: 10.1007/s10658-022-02600-z. [DOI] [Google Scholar]
- Bahramian D, Naraghi L, Heydari A. Effectiveness of the chemical stabilizers of Talaromyces flavus in biological control of tomato and greenhouse cucumber vascular wilt disease. J Plant Prot Res. 2016;56(3):291–297. doi: 10.1515/jppr-2016-0045. [DOI] [Google Scholar]
- Barman A, Nath A, Thakur D. Identification and characterization of fungi associated with blister blight lesions of tea (Camellia sinensis L. Kuntze) isolated from Meghalaya, India: Microbiol Res; 2020. [DOI] [PubMed] [Google Scholar]
- Biessy A, Novinscak A, St-Onge R, Léger G, Zboralski A, Filion M. Inhibition of three potato pathogens by phenazine-producing Pseudomonas spp. is associated with multiple biocontrol-related traits. mSphere. 2021 doi: 10.1128/mSphere.00427-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Caporaso JG. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bové JM (2006) Huanglongbing: A destructive, newly emerging, century-old disease of citrus. J Plant Pathol 88(1):7–37. https://www.jstor.org/stable/41998278
- Chakraborty BN, Sharma M. Serological detection and immunogold localization of cross-reactive antigens shared by Camellia sinensis and Exobasidium vexans. J Appl Microbiol. 2007;103:1669–1680. doi: 10.1111/j.1365-2672.2007.03459.x. [DOI] [PubMed] [Google Scholar]
- Chaliha C, Kalita E, Verma PK. Optimizing in vitro culture conditions for the biotrophic fungi Exobasidium vexans through response surface methodology. Indian J Microbiol. 2020;60:167–174. doi: 10.1007/s12088-019-00846-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S, Chakraborty N, Panda K, Acharya K. Chitosan-induced immunity in Camellia sinensis (L.) O. Kuntze against blister blight disease is mediated by nitric-oxide. Plant Physiol Biochem. 2017;115:298–307. doi: 10.1016/j.plaphy.2017.04.008. [DOI] [PubMed] [Google Scholar]
- Chao A (1984) Nonparametric estimation of the number of classes in a population. Scand J Stat 11(4): 265–270. https://www.jstor.org/stable/4615964
- Chen YY, Zhou B, Li JL, Tang H, Cui YY, Wu WH, Liu JY, Tang JC. Blister blight lesions of tea (Camellia sinensis L. Kuntze) leaves: Microbial diversity analysis and identification of the disease fungi. Chin Agric Sci Bull. 2023;39(6):116–123. [Google Scholar]
- Dethoup T, Klaram R, Pankaew T, Jantasorn A. Impact of fungicides and plant extracts on biocontrol agents and side-effects of Trichoderma spp. on rice growth. Eur J Plant Pathol. 2022;164:567–582. doi: 10.1007/s10658-022-02581-z. [DOI] [Google Scholar]
- Di C, Han Z, Chai C, Sun J, Wu F, Pan K. Improvement of Straw Changed Soil Microbial Flora Composition and Suppressed Chinese Cabbage (Brassica rapa L. ssp. pekinensis) Clubroot Disease. Agronomy. 2023 doi: 10.3390/agronomy13071688. [DOI] [Google Scholar]
- Dong LP, Rong ZZ, Yunzeng Z, Jianping X, Hongkai W, Zhengyi W, Hongye L. The phyllosphere microbiome shifts toward combating melanose pathogen. Microbiome. 2022 doi: 10.1186/s40168-022-01234-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel OM, Salas-González I, Castrillo G, Conway JM, Law TF, Teixeira PJPL, Wilson ED, Fitzpatrick CR, Jones CD, Dangl JL. A single bacterial genus maintains root growth in a complex microbiome. Nature. 2020;587:103–108. doi: 10.1038/s41586-020-2778-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Chaturvedi P, Kulkarni MG, Staden JV. A critical review on exploiting the pharmaceutical potential of plant endophytic fungi. Biotechnol Adv. 2020 doi: 10.1016/j.biotechadv.2019.107462. [DOI] [PubMed] [Google Scholar]
- Gustavsen JA, Pai S, Isserlin R, Demchak B, Pico AR. RCy3: Network biology using cytoscape from within R. Research. 2019 doi: 10.1101/793166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadian S, Supronienė S, Kulaitienė J, Hasanzadeh N. Effect of epiphytic bacteria from citrus against green mold post-harvest diseases of citrus. Horticulturae. 2023;9(7):764. doi: 10.3390/horticulturae9070764. [DOI] [Google Scholar]
- Hazarika SN, Saikia K, Borah A, Thakur D. Prospecting endophytic bacteria endowed with plant growth promoting potential isolated from Camellia sinensis. Front Microbiol. 2021 doi: 10.3389/fmicb.2021.738058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CE, Jo SH, Jo IH, Jeong H, Park JM. Draft genome sequence of the endophytic bacterium Variovorax paradoxus KB5, which has antagonistic activity against a phytopathogen, Pseudomonas syringae pv. tomato DC3000. Gen Announc. 2017 doi: 10.1128/genomea.00950-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huong NTM, Hoai PTT, Thao PTH, Huong TT, Chinh VD. Growth stimulation, phosphate resolution, and resistance to fungal pathogens of some endogenous fungal strains in the rhizospheres of medicinal plants in vietnam. Molecules. 2022;27(16):5051. doi: 10.3390/molecules27165051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaswall K, Mahajan P, Singh G, Parmar R, Seth R, Raina A, Swarnkar MK, Singh AK, Sharma SR, RK, Transcriptome analysis reveals candidate genes involved in blister blight defense in tea (Camellia sinensis (L) Kuntze) Sci Rep. 2016;6:30412. doi: 10.1038/srep30412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HY, Xi ZG, Ma JQ, Li YF, Hao CH, LU MQ, Zhang ZZ, Deng WW, Endophytic bacteria from the leaves of two types of albino tea plants, indicating the plant growth promoting properties. Plant Growth Regul. 2022;96:331–343. doi: 10.1007/s10725-021-00779-5. [DOI] [Google Scholar]
- Kabir MH, Unban K, Kodchasee P, Govindarajan RK, Lumyong S, Suwannarach N, Wongputtisin P, Shetty K, Khanongnuch C. Endophytic bacteria isolated from tea leaves (Camellia sinensis var. assamica) enhanced plant-growth-promoting activity. Agriculture. 2023 doi: 10.3390/agriculture13030533. [DOI] [Google Scholar]
- Kerr A, Rodrigo WRF. Epidemiology of tea blister blight (Exobasidium vexans): IV Disease Forecasting. Trans Brit Mycol Soc. 1967;50(4):609–614. doi: 10.1016/S0007-1536(67)80092-5. [DOI] [Google Scholar]
- Liu SJ, Moon CD, Zheng N, Huws S, Zhao SG, Wang JQ. Opportunities and challenges of using metagenomic data to bring uncultured microbes into cultivation. Microbiome. 2022;10:76. doi: 10.1186/s40168-022-01272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73(5):1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SL, Wang JM, Sheng RL, Fang YW, Guo RH. Novel bioactive polyketides isolated from marine actinomycetes: An update review from 2013 to 2019. Chem Biodivers. 2020 doi: 10.1002/cbdv.202000562. [DOI] [PubMed] [Google Scholar]
- Lundberg D, Yourstone S, Mieczkowski P, Jones CD, Dangl JL. Practical innovations for high-throughput amplicon sequencing. Nat Methods. 2013;10:999–1002. doi: 10.1038/nmeth.2634. [DOI] [PubMed] [Google Scholar]
- Lv NN, Tao CY, Ou YN, Wang JB, Deng XH, Liu HJ, Shen ZZ, Li R, Shen QR. Root-associated antagonistic Pseudomonas spp. contribute to soil suppressiveness against banana Fusarium wilt disease of banana. Microbiol Spectr. 2023 doi: 10.1128/spectrum.03525-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki KA, Wolff B, Varuzza L, Green SJ, Barb JJ. Multi-amplicon microbiome data analysis pipelines for mixed orientation sequences using QIIME2: Assessing reference database, variable region and pre-processing bias in classification of mock bacterial community samples. PLoS ONE. 2023 doi: 10.1371/journal.pone.0280293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu TC, Liu RY, Zhang XG, Li Z, Xia JW. Diaporthe cha sp. nov on Camellia sinensis in China. Nova Hedwigia. 2022;115(3–4):473–485. doi: 10.1127/nova_hedwigia/2022/0713. [DOI] [Google Scholar]
- Martino C, Morton JT, Marotz CA, Thompson LR, Tripathi A, Knight R, Zengler K. A novel sparse compositional technique reveals microbial perturbations. mSystems. 2019 doi: 10.1128/msystems.00016-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu S, Gao Y, Zi HX, Liu Y, Liu XM, Xiong XQ, Yao QQ, Qin ZW, Chen N, Guo L, Yang YZ, Qin P, Lin JZ, Zhu YH. The osmolyte-producing endophyte Streptomyces albidoflavus OsiLf-2 induces drought and salt tolerance in rice via a multi-level mechanism. Crop J. 2022;10:375–386. doi: 10.1016/j.cj.2021.06.008. [DOI] [Google Scholar]
- Pandey AK, Sinniah GD, Babu A, Tanti A. How the global tea industry copes with fungal diseases-challenges and opportunities. Plant Dis. 2021;105(7):1868–1879. doi: 10.1094/PDIS-09-20-1945-FE. [DOI] [PubMed] [Google Scholar]
- Premkumar R, Ponmurugan P, Manian S. Growth and photosynthetic and biochemical responses of tea cultivars to blister blight infection. Photosynthetica. 2008;46(1):135–138. doi: 10.1007/s11099-008-0021-0. [DOI] [Google Scholar]
- Rachmad FD, Priyatmojo A, Widiastuti A. Genetic diversity analysis of Exobasidium vexans causing tea blister blight in Wonosobo, Central Java, Indonesia using RAPD markers. Arch Phytopathol Plant Prot. 2022;55(10):1234–1249. doi: 10.1080/03235408.2022.2086026. [DOI] [Google Scholar]
- Rahman SFSA, Singh E, Pieterse CMJ, Schenk PM. Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 2018;267:102–111. doi: 10.1016/j.plantsci.2017.11.012. [DOI] [PubMed] [Google Scholar]
- Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol. 2007;62(2):142–160. doi: 10.1111/j.1574-6941.2007.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rateb ME, Ebel R, Jaspars M. Natural product diversity of actinobacteria in the Atacama Desert. Antonie Van Leeuwenhoek. 2018;111:1467–1477. doi: 10.1007/s10482-018-1030-z. [DOI] [PubMed] [Google Scholar]
- Ruiz-Bedoya T, Wang PW, Desveaux D, Guttman DS. Cooperative virulence via the collective action of secreted pathogen effectors. Nat Microbiol. 2023 doi: 10.1038/s41564-023-01328-8. [DOI] [PubMed] [Google Scholar]
- Sen S, Rai M, Das D, Chandra S, Acharya K. Blister blight a threatened problem in tea industry: A review. J King Saud Univ Sci. 2020;32(8):3265–3272. doi: 10.1016/j.jksus.2020.09.008. [DOI] [Google Scholar]
- Shabani MH, Naraghi L, Maleki M, Negahban M. Evaluation of the efficacy of different concentrations of nano-capsules containing Talaromyces flavus with two forms of powder and suspension in reducing the incidence of cotton Verticillium wilt. Braz J Biol. 2024 doi: 10.1590/1519-6984.262480. [DOI] [PubMed] [Google Scholar]
- Simpson EH. Measurement of diversity. Nature. 1949 doi: 10.1038/163688a0. [DOI] [Google Scholar]
- Shannon CE. A mathematical theory of communication. GetMobile-Mob Compu Commun Rev. 2001;27(1):23–27. doi: 10.1145/3599184.3599192. [DOI] [Google Scholar]
- Sharma P, Kumar S. Bioremediation of heavy metals from industrial effluents by endophytes and their metabolic activity: Recent advances. Bioresource Technol. 2021;339:125589. doi: 10.1016/j.biortech.2021.125589. [DOI] [PubMed] [Google Scholar]
- Sun K, Lu F, Huang PW, Tang MJ, Xu FJ, Zhang W, Zhou JY, Zhao P, Jia Y, Dai CC. Root endophyte differentially regulates plant response to NO3- and NH4+ nutrition by modulating N fluxes at the plant-fungal interface. Plant Cell Environ. 2022;45(6):1813–1828. doi: 10.1111/pce.14304. [DOI] [PubMed] [Google Scholar]
- Tamošiūnė I, Andriūnaitė E, Stanys V, Baniulis D (2019) Exploring diversity of bacterial endophyte communities using advanced sequencing technology. In: Kumar, V., Prasad, R., Kumar, M., Choudhary, D. (eds) Microbiome in Plant Health and Disease. Springer, Singapore. 10.1007/978-981-13-8495-0_20
- Tibpromma S, Karunarathna SC, Bhat JD, Suwannarach N, Stephenson SL, Elgorban AM, Al-Rejaie S, Xu J, Mortimer PE. Using culture-dependent and molecular techniques to identify endophytic fungi associated with tea leaves (Camellia spp) in Yunnan Province. China: Diversity; 2022. [Google Scholar]
- Trivedi P, Leach JE, Tringe SG, Sa TM, Singh BK. Plant-microbiome interactions: from community assembly to plant health. Nat Rev Microbiol. 2020;18:607–621. doi: 10.1038/s41579-020-0412-1. [DOI] [PubMed] [Google Scholar]
- Whittaker RH. Evolution and measurement of species diversity. Taxon. 1972;21(2–3):213–251. doi: 10.2307/1218190. [DOI] [Google Scholar]
- Xin JJ, Liu Y, Li HY, Chen SM, Jiang JF, Song AP, Fang WM, Chen FD. CmMLO17 and its partner CmKIC potentially support Alternaria alternata growth in Chrysanthemum morifolium. Hortic Res. 2021;8:101. doi: 10.1038/s41438-021-00534-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Song N, Song N, Ma L, Wu JS. IRE1-bZIP60 pathway is required for Nicotiana attenuata resistance to fungal pathogen Alternaria alternata. Front Plant Sci. 2019;10:263. doi: 10.3389/fpls.2019.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin QX, An XL, Wu X, Dharmasena DSP, Li DX, Jiang SL, Wang Y, Wang DL, Chen Z (2021) First report of Alternaria longipes causing leaf spot on tea in China. 105(12): 4167. 10.1094/PDIS-07-20-1583-PDN [DOI] [PubMed]
- Yu HR, Yan FF, Wang YL, Tong XY, Chen D, Ye Q, Piao RZ, Zhao HY. Antagonistic effects of Sphingomonas and Pseudomonas aeruginosa on 4 kinds of pathogenic bacteria of Giaseng. Asian Agric Res. 2022;14(05):31–35. doi: 10.19601/j.enki.issn1943-9903.2022.05.008. [DOI] [Google Scholar]
- Zhang J, Wang ZB, Guo HW, Sun XL, Xiao Q. Research progresses on blister blight in tea plant. Acta Phytopathol Sin. 2023 doi: 10.13926/j.cnki.apps.001016(inChinese). [DOI] [Google Scholar]
- Zhou XL, Hu LH, Huy HN, Thanh TL, Zhou CB, Mei X, Buensanteai K. The changes of metabolites, quality components and antioxidant activity of tea (Camellia sinensis L.) infected with Exobasidium vexans by applying UPLC-MS/MS based widely targeted metabolome and biochemical analysis. Phytopathology. 2023 doi: 10.1094/PHYTO-03-23-0105-R. [DOI] [PubMed] [Google Scholar]