Abstract
Fusarium wilt disease of banana, caused by the notorious soil-borne pathogen Fusarium oxysporum f. sp. cubense Tropical Race 4 (Foc TR4), is extremely difficult to manage. Manipulation of soil pH or application of synthetic iron chelators can suppress the disease through iron starvation, which inhibits the germination of pathogen propagules called chlamydospores. However, the effect of iron starvation on chlamydospore germination is largely unknown. In this study, scanning electron microscopy was used to assemble the developmental sequence of chlamydospore germination and to assess the effect of iron starvation and pH in vitro. Germination occurs in three distinct phenotypic transitions (swelling, polarized growth, outgrowth). Outgrowth, characterized by formation of a single protrusion (germ tube), occurred at 2 to 3 h, and a maximum value of 69.3% to 76.7% outgrowth was observed at 8 to 10 h after germination induction. Germination exhibited plasticity with pH as over 60% of the chlamydospores formed a germ tube between pH 3 and pH 11. Iron-starved chlamydospores exhibited polarized-growth arrest, characterized by the inability to form a germ tube. Gene expression analysis of rnr1 and rnr2, which encode the iron-dependent enzyme ribonucleotide reductase, showed that rnr2 was upregulated (p < 0.0001) in iron-starved chlamydospores compared to the control. Collectively, these findings suggest that iron and extracellular pH are crucial for chlamydospore germination in Foc TR4. Moreover, inhibition of germination by iron starvation may be linked to a different mechanism, rather than repression of the function of ribonucleotide reductase, the enzyme that controls growth by regulation of DNA synthesis.
Keywords: Germ tube, Outgrowth, Polarized, Ribonucleotide reductase, Quiescence
Introduction
Chlamydospores are hardy thick-walled asexual spores produced by diverse fungi, including Fusarium oxysporum f. sp. cubense (Foc), the notorious root-infecting pathogen causing Fusarium wilt disease of banana (Viljoen et al. 2020). Fusarium oxysporum f. sp. cubense Tropical Race 4 (Foc TR4), is considered the most virulent and devastating race of Foc. Foc TR4 is spreading inexorably and poses a threat to the food security and income of nearly 400 million people that depend on banana (Zheng et al. 2018; Viljoen et al. 2020; van Westerhoven et al. 2022). Chlamydospores are the primary source of Foc inoculum and can remain quiescent in infested soils for decades. When suitable conditions are encountered, chlamydospores in soil undergo a revival cellular process called germination (Pegg et al. 2019; Were et al. 2022a). Chlamydospore germination is a crucial step in the infection of host roots and development of Fusarium wilt (Pegg et al. 2019). Fusarium wilt can be suppressed by targeting chlamydospore germination through the manipulation of soil pH to 7.0 or close thereto, and by reduction of the bioavailability of iron through application of iron chelators (Peng et al. 1999; Dita et al. 2018; Segura-Mena et al. 2021). However, the mechanisms of inhibition and the distinct role of iron and pH on chlamydospore germination are largely unknown.
Quiescence in fungal spores is a non-proliferative cellular state of reversible cell cycle arrest (Rittershaus et al. 2013; Blatzer and Latgé 2021). Quiescence is characterized by reduced metabolic activity and accumulation of storage molecules, such as trehalose (Wyatt et al. 2013; Hayer et al. 2014). The transition from quiescence to germination generally occurs in three distinct phenotypic transitions: dormancy to swelling (isotropic increase in size), swelling to polarized growth, and the emergence of a germ tube (outgrowth) (Sephton-Clark and Voelz 2018). Outgrowth marks the end of the germination process. It is considered the first step in the formation of a fungal colony and a key developmental stage in the life cycle of fungi (Sephton-Clark and Voelz 2018). Spore germination has been widely investigated in fungal pathogens (Hayer et al. 2014; Turgeman et al. 2016). However, spore germination in Foc TR4 is largely unknown. Moreover, most studies on spore germination in Foc have been conducted using conidia (Li et al. 2011; Meldrum et al. 2013; Deng et al. 2015). However, conidia of Fusarium oxysporum may not be appropriate substitutes for chlamydospores. This is because both types of spores are typically formed under distinct environmental conditions, perform distinct functions in the fungus’s life cycle, and exhibit different degrees of persistence in soil (Ohara and Tsuge 2004; Ohara Leslie and Summerell 2006).
Spore germination is marked by heightened metabolic activity during which biomolecules are synthesised to rebuild hyphae from the disintegrating spore (Deng et al. 2015; Sephton-Clark and Voelz 2018; Balotf et al. 2021). For example, outgrowth in filamentous fungi occurs after nuclear division which, in turn, is dependent on DNA synthesis (Cohen et al. 2019; Greene et al. 2020; Steenwyk 2021). The synthesis of DNA requires ribonucleotide reductase (RNR), the iron-dependent enzyme which converts ribonucleotides to deoxyribonucleotides (dNTPs) which are the precursors for DNA synthesis and repair (Greene et al. 2020; Steenwyk 2021). In F. oxysporum, RNR has two non-identical homodimeric subunits, a large subunit Rnr1 and a small one-Rnr2, which are encoded by the genes rnr1 and rnr2, respectively (Cohen et al. 2019). The Rnr1 subunit contains the catalytic site and allosteric sites that control enzyme activity and specificity (Greene et al. 2020). On the other hand, the Rnr2 subunit contains a non-heme iron centre, which generates a tyrosyl free radical that is essential for catalysis.
Iron acquisition is crucial for the survival and growth of invading microorganisms. In Fusarium oxysporum, iron homeostasis is regulated by the basic leucine zipper (bZIP) transcription factor, HapX (López-Berges et al. 2012a, b). Under conditions of iron starvation, HapX is activated, resulting in the synthesis of low-molecular-weight (< 1 kDa) secondary metabolites called siderophores, that are essential for iron scavenging and storage (Hider and Kong 2010). Although several siderophores, including ferrichrome, fusigen, and fusarinine, have been identified in Foc (Anke et al. 1973; Beckmann et al. 2013; Were et al. 2022a, b), the mechanisms of siderophore-mediated iron transport remain largely unknown.
Iron is an indispensable micronutrient for all eukaryotic organisms and an essential cofactor of RNR (Furukawa et al. 1992; Colombo et al. 2014; Fukada et al. 2019). Given the significance of iron, we hypothesized that iron is fundamental for outgrowth in Foc TR4 and that iron scarcity leads to selective optimization of RNR function by enhanced expression of rnr1 and rnr2 genes. Accordingly, the specific objectives were to assess (i) the effect of iron starvation on outgrowth and the expression of rnr1 and rnr2 genes, and (ii) the effect of extracellular pH on outgrowth.
Materials and methods
Fungal strain, culture conditions and production of chlamydospores
Fusarium oxysporum f. sp. cubense Tropical Race 4 (TR4) VCG 01213/16 was obtained as a frozen stock on 30% (v/v) glycerol from the Department of Plant Pathology Stellenbosch University, South Africa. The isolate was preserved at −80 °C and revived by culturing on potato dextrose agar (PDA) at 28 °C for 5 days. All chemicals, unless otherwise stated, were purchased from Carl Roth (Karlsruhe, Germany).
Chlamydospore germination time-course assay
Chlamydospores were produced from the culture and purified as previously described (Goyal et al. 1973; Were et al. 2022b). A time-course experiment was conducted to determine the developmental sequence of germinating chlamydospores of Foc TR4. The experiment was conducted in 24-well culture plates (Costar, Cambridge, MA, USA) using Barz broth (Were et al. 2022b). Samples for phenotypic analysis of germination were retrieved after 30 min and thereafter every hour for duration of 10 h post induction of germination (h.p.i). Samples were retrieved in 2-mL tubes, chilled on ice, and centrifuged at 13,000 r.p.m. for 10 min at 4 °C (Eppendorf 5810R Centrifuge, Eppendorf, Hamburg, Germany). Chlamydospores were washed with ice-cold cell wash buffer (PBS-T: 137 mM, NaCl; 2.7 mM, KCl; 2 mM, KH2PO4; 0.005% v/v, Triton X-100; pH 7.4) by vortexing at maximum speed for 1 min, followed by centrifugation at 13,000 r.p.m. for 10 min at 4 °C. This procedure was repeated using sterile deionized water.
A developmental sequence of germinating chlamydospores was assembled by bright-field microscopy using a LeicaDM750 microscope equipped with a Leica ICC50 HD camera (Leica, Heerbrugg, Switzerland). Germinated, polarized, and round chlamydospores were counted using a haemocytometer and a Zeiss Axioskop microscope (Carl Zeiss, Jena, Germany). Micrographs were captured using a LeicaDM750 microscope, converted to 8-bit grayscale or 24-bit RGB, and annotated using CorelDraw 12.0 (Corel, Ottawa, Canada). This procedure allowed for the monitoring of the progression of chlamydospore germination.
Scanning electron microscopy (SEM)
Scanning electron microscopy (SEM) was used to obtain ultra-structural details of germinating chlamydospores. Chlamydospore specimens for SEM analysis were prepared as described previously (Were et al. 2022b). Specimens were visualized with a Zeiss Merlin scanning electron microscope (Carl Zeiss) with a Gemini-type field emission gun electron column (FEG-SEM) equipped with two Oxford Instruments X-MaxN 150 SDDs. Typical imaging conditions were magnification of 1–3 × 104, a working distance of 5–10 mm, 2–3 kV, a beam current 100–200 pA and using an In Lens secondary electron detector. Micrographs of specimens were captured in TIF format using a pixel averaging noise reduction algorithm and SmartSEM software (Carl Zeiss).
Cellular metabolic activity and germination in iron-starved chlamydospores
The effect of iron-starvation on chlamydospore germination was assessed by determining the percent germination and cellular metabolic activity of iron-starved chlamydospores. Iron starvation was induced using 2,2′-dipyridyl, a synthetic lipophilic iron chelator that sequesters both extracellular and intracellular Fe2+ and Fe3+ pools (Breuer et al. 1995; Romeo et al. 2001; Asai et al. 2022). A stock solution of 2,2′-dipyridyl (20 mM) was prepared in dimethyl sulfoxide (DMSO), filter-sterilized using a 0.22 µm syringe filter (Sartorius, Göttingen, Germany), and stored in the dark at 4 °C.
Cellular metabolic activity was determined using the Alamar Blue kit (Bio-Rad, Hercules, CA, USA), that measures activity of the mitochondrial respiratory chain as a readout of cell viability. The assay was conducted in Barz medium supplemented with different concentrations of 2,2′-dipyridyl [1, 10, 100, 200 µM and control (10% v/v DMSO)] by following the manufacturer’s protocol (Bio-Rad, Hercules, CA, USA). Absorbance was measured using a microplate reader (Tecan, Maennedorf, Switzerland). Absorbances were normalized to the DMSO control, where cell viability was set to 100%. The effect of iron-starvation on chlamydospore germination was conducted in 24-well plates as described earlier using Barz supplemented with 2,2′-dipyridyl [1, 10, 100, 200 µM and control (10% v/v DMSO)]. Plates were incubated for ten hours and germinated chlamydospores were determined as described earlier.
To further establish the effect of iron starvation on the chlamydospore germination process, a two-step experiment (iron starvation and iron repletion) was conducted. First, in the iron starvation experiment, chlamydospores (103 per well) were incubated in a 24-well culture plate containing 900 µL of Barz broth per well supplemented with 2,2′-dipyridyl (100 µM). After 5 h, rounded, polarized, and outgrown chlamydospores was determined as described earlier. In the second, iron-replete experiment, chlamydospores from the iron-starvation experiment were washed as described earlier to remove 2,2′-dipyridyl and further incubated for 5 h in fresh Barz medium. Afterward, rounded, polarized, and outgrown chlamydospores was determined as described earlier.
RNA extraction and cDNA synthesis
To analyse the expression of RNR genes (rnr1 and rnr2), chlamydospore samples were collected from both iron-starvation and repletion experiments. The Chlamydospores were obtained by pooling suspensions from two wells and then transferred into 2-mL tubes. The samples were washed as described earlier. Then, samples were stabilized by immediately suspending in 500 µL of RNAlater RNA (Qiagen, Hilden, Germany) and stored at −80 °C until RNA extraction. Samples were retrieved and thawed on ice prior to extraction of total RNA. Total RNA was isolated from chlamydospores using the RNeasy kit following the manufacture’s protocol (Qiagen). RNA samples were subsequently reverse transcribed to cDNA using a QuantiTect Reverse Transcription Kit following the manufacture’s protocol (Qiagen).
Quantitative RT-PCR
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was performed on a StepOne Real-Time PCR system (Applied BioSystems, CA, USA) using Power SYBR Green PCR Master Mix (Applied Biosystems) and gene-specific primer pairs (Table 1) (Cohen et al. 2019). All samples were amplified in triplicate using the following thermal cycling conditions: 95 °C for 20 s, 40 cycles of priming at 54 °C for 20 s, and elongation at 72 °C for 20 s. Cycle thresholds were obtained using the StepOne software (v2.0, Applied BioSystems, CA, USA). Relative gene expression levels were normalized to that of the Fusarium oxysporum actin-lateral binding (ALb) gene using the ΔΔCt method (Yuan et al. 2006).
Table 1.
Primers used for analysis of the expression of ribonucleotide reductase genes (rnr1 and rnr2) and actin-lateral binding (ALb) gene in chlamydospores of Fusarium oxysporum f. sp. cubense Tropical Race 4 (Cohen et al. 2019)
| Target gene | Primer name | Sequence (5ʹ → 3ʹ) |
|---|---|---|
| rnr1 (F. oxysporum ribonucleotide reductase large subunit) | rnr1-F | CATCAAGGCTGATGTTGAGG |
| rnr1-R | CTTGACACCCAACTCTTCTTCC | |
| rnr2 (F. oxysporum ribonucleotide reductase small subunit) | rnr2-F | TACTTTGGATCCGGAAGCTG |
| rnr2-R | TTTTCATCCACCCTGAGTCC | |
| ALb (F. oxysporum actin-lateral binding protein) | ALb-F | GGTTTCCCTTCAGCCTTTTC |
| ALb-R | CGGAGCTGGTTCATTTTCTC |
Statistical analyses
Data analysis and visualisation were performed using R (v.4.0.2; R Development Core Team 2020). Prior to statistical analysis, data was checked for normality and homoscedasticity (Kozak and Piepho 2018). Homoscedasticity of the data was verified using Levene’s test, whereas the normality the data was verified using the Shapiro–Wilk test and diagnostic plots (histograms and Q-Q plots). Comparisons were made between treatments and the control. Statistical significance was declared if p < 0.05. Significant differences between treatments and the control were tested using analysis of variance (ANOVA) and subsequent post hoc analysis using Tukey’s Honest Significant Difference (Tukey HSD) test. All data are expressed as mean ± standard error of the mean (Kozak and Piepho 2020).
Results
Germination of Foc TR4 chlamydospores is asynchronous
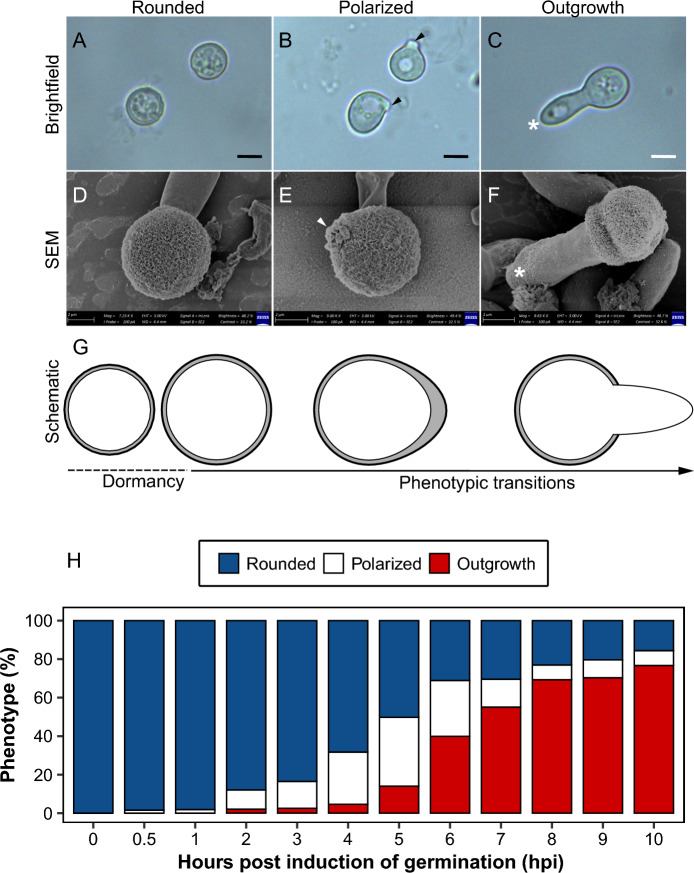
Chlamydospores appeared round and enveloped in a double layered wall: a thin inner wall and a thick outer wall, when examined with brightfield microscopy (Fig. 1A). The outer wall was composed of distinctly fibrillar material, when examined by SEM (Fig. 1D–F). Germination was characterized by three distinct phenotypic transitions: swelling, to polarized growth, to outgrowth (Fig. 1A–H). Swelling occurred within 0.5 to 1 h post induction of germination (h.p.i), during which the rounded chlamydospores appeared larger than the dormant chlamydospores and possessed a highly granulated cytoplasm (Fig. 1A, G). At 2 to 3 h.p.i, deposition and concentration of cellular material was observed at a single specific point on the chlamydospore inner wall (Fig. 1B). This cellular material, whose molecular identity was not determined, appeared as a bright spot towards which growth was directed, thereby defining the spore front and domain. This domain was maintained, resulting in a polarized chlamydospore with a pear-shaped form (Fig. 1B, E). Outgrowth, the most conspicuous transition, was observed starting at 2 to 3 h.p.i and a maximum value of 69.3% to 76.7% chlamydospores produced a single protrusion (germ tube) between 8 to 10 h.p.i (Fig. 1C, F and H). At this point, chlamydospores were considered to be fully germinated.
Fig. 1.
Bright-field and scanning electron microscopy micrographs showing the transitions during germination of chlamydospores of Fusarium oxysporum f. sp. cubense Tropical Race 4 (Foc TR4): rounded (A, D), polarized growth (B, E), and outgrowth (C, F). The stages of germination are further illustrated in the schematic (G). The site of polarity establishment (indicated by arrow heads) on polarized chlamydospores, is the point where the germ tube (indicated by white asterisk) emerges during outgrowth. Scale bar = 20 µm. Phenotypic changes during the process of germination of chlamydospores in Foc TR4 (H)
Notably, the germ tube emerged from the chlamydospore by the extension of the inner spore wall after local rapture of the outer wall at the site of polarity establishment (Fig. 1B). The germ tube progressively grew into hyphae that extended along a polar axis (Fig. 1C). Importantly, swelling, polarized growth, and outgrowth occurred subsequently, but not simultaneously, as evidenced by the rapid remarkable transition that was observed in some chlamydospores. This observation suggests that chlamydospore germination in Foc TR4 is an asynchronous process (Fig. 1H).
Iron is essential for metabolic activity and outgrowth in chlamydospores
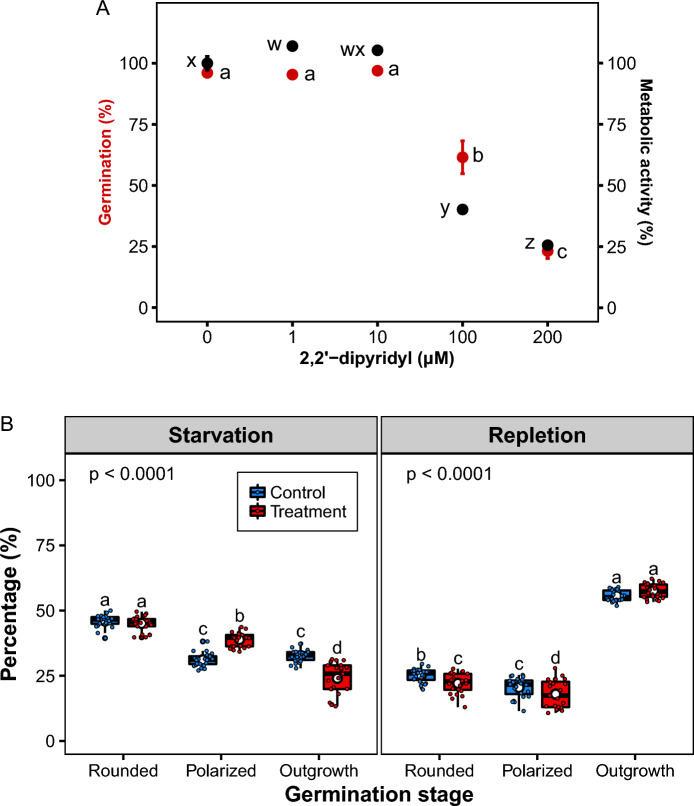
At low concentration (1 and 10 µM) of 2,2′-dipyridyl, cellular metabolic activity of chlamydospores was stimulated, but decreased (p < 0.0001) as the concentration of 2,2′-dipyridyl increased (Fig. 2A). Similarly, chlamydospore germination decreased (p < 0.0001) as the concentration of 2,2′-dipyridyl increased (Fig. 2B). Iron-starved chlamydospores underwent through swelling but halted their developmental progression, thereby accumulating at polarized growth stage. The percentage of polarized chlamydospores in the treatment was 38.90 ± 0.54% compared to 31.11 ± 0.52 in the control (Fig. 2B). This polarized growth-arrest was characterized by the reduced inability to form a germ tube. Only 24.01 ± 1.24% of chlamydospores switched to outgrowth in the presence of 2,2′-dipyridyl, compared to 32.70 ± 0.49% in the control (p < 0.0001).
Fig. 2.
Effect of iron starvation on cellular metabolic activity and germination on chlamydospores of Fusarium oxysporum f. sp. cubense Tropical Race 4 (A, B). Boxplots show the upper and lower quartile, median (bold horizontal bar), mean (white circle), and whiskers (vertical lines)
Chlamydospores, which exhibited polarized growth-arrest, contained perceptibly smaller and fewer cytoplasmic inclusions, indicating defects in replication or differentiation of cellular organelles. In marked contrast, control chlamydospores exhibited no discernible change in cytoplasmic inclusions and progressed normally through germination, resulting in the formation of a germ tube (outgrowth). Chlamydospores exhibiting polarized growth-arrest were rescued by iron repletion, resulting in restoration of germination (p < 0.0001) (Fig. 2B).
Expression of the rnr2 gene is up-regulated in iron-starved chlamydospores
Expression of the rnr2 gene was up-regulated in iron-starved chlamydospores, as compared with the control (p < 0.0001) (Fig. 3A). In contrast, expression of the rnr1 gene did not differ between iron-starved chlamydospores and the control (p > 0.05) (Fig. 3A).
Fig. 3.
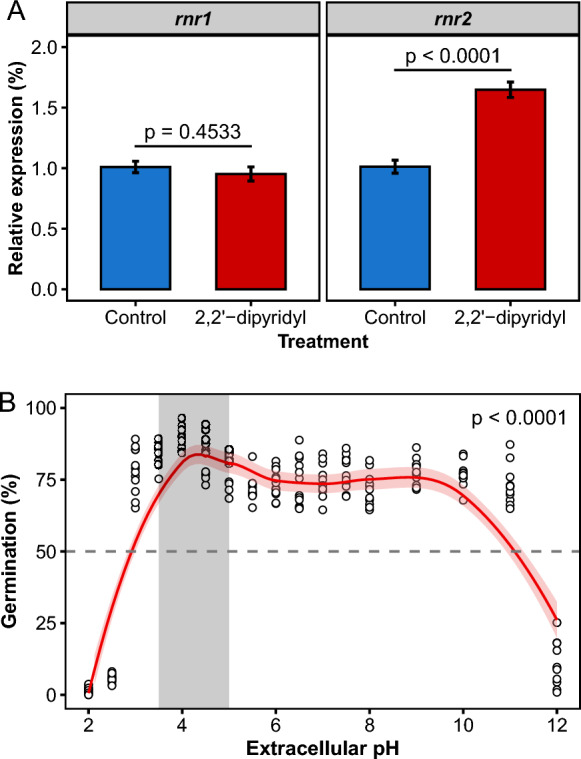
Effect of iron starvation on the expression of genes rnr1 and rnr2 that encode the Rnr1 and Rnr2 subunits, respectively, of the ribonucleotide reductase enzyme in chlamydospores of Fusarium oxysporum f. sp. cubense Tropical Race 4 (Foc TR4) (A). Effect of pH on chlamydospore germination in Foc TR4 (B). The dotted line denotes the 50% germination of chlamydospores
Chlamydospore germination exhibits plasticity with extracellular pH
Chlamydospore germination was influenced by extracellular pH (p < 0.0001) (Fig. 3B). Germination was apparent over a wide pH range and exhibited double maxima; a primary peak (optimal) on the acid side from pH 3.5 to 4.5, where 75.3% to 94.3% of chlamydospores formed a germ tube. A secondary peak was noted close to neutrality between pH 8 and 9.5, where 67.8% to 84% of chlamydospores formed a germ tube (Fig. 3B). An increase of 0.5 pH units from pH 2.5 to 3.0 induced a 14-fold increase in germination (p < 0.0001). Notably, chlamydospores appeared small in Barz broth below pH 3.0 compared to chlamydospores above pH 10.0. Moreover, disintegration of incipient germ tubes was frequently observed below pH 3.0. Above pH 10.0, however, the produced germ tubes were often large with a small terminal globular structure, resembling a newly formed chlamydospore.
Discussion
Chlamydospore germination is crucial in the infection of banana roots by the notorious fungal pathogen Foc TR4 and severity of Fusarium wilt disease (Dita et al. 2018; Pegg et al. 2019). Fusarium wilt can be suppressed by targeting chlamydospore germination through iron starvation caused by application of iron chelators or manipulation of soil pH, but the underlying mechanism of inhibition remains elusive (Peng et al. 1999; Segura-Mena et al. 2021). The findings of this study demonstrate that iron-deficient chlamydospores are unable to form a germ tube.
Consistent with previous work (Plante and Labbé 2019), we found that iron starvation results in polarized growth arrest and a concomitant decrease in cellular metabolic activity. Our data suggest that chlamydospore germination is a developmental process that requires iron. Spore germination in Foc TR4 entails resumption of metabolism and de novo synthesis of macromolecules to rebuild hyphae from the disintegrating spore (Deng et al. 2015). Macromolecular biosynthesis and energy generation require a unique enzymatic repertoire and bioavailability of cofactors, such as iron (Philpott et al. 2012). Polarized-growth arrest observed in Foc TR4 chlamydospores may reflect attenuation of cell cycle progression arising from a paucity of dNTPs available for DNA synthesis in iron-starved chlamydospores (Renton and Jeitner 1996; Cohen et al. 2019; Greene et al. 2020). DNA synthesis immediately precedes mitosis and subsequently nuclear division, which is indispensable for outgrowth in many fungi (Fukada et al. 2019). Nuclear division and migration of the daughter nucleus into the germ tube are contingent on mitosis control (Renton and Jeitner 1996; Cohen et al. 2019; Greene et al. 2020). Iron is required for assembly and activity of ribonucleotide reductase, an enzyme that catalyses the rate-limiting step in the production of dNTPs, the precursors for DNA synthesis (Cohen et al. 2019; Greene et al. 2020). Thus, the enhanced expression of rnr2 suggests that iron scarcity leads to selective optimization of RNR function at the expense of other non-essential iron-dependent processes, to allow for DNA synthesis and repair. Similar findings have also been reported in Saccharomyces cerevisiae (Sanvisens et al. 2011).
Germination of Foc TR4 chlamydospores occurred over a wide pH range, which is consistent with studies in F. oxysporum (Cruz et al. 2019), Harpophora maydis (Degani and Goldblat 2014), and Rhizopus delemar (Turgeman et al. 2016). Maximum germination observed in the acidic and alkaline pH may denote different mechanism by which pH modulates germination. Specifically, the isoelectric point of protoplasmic proteins at this pH is at a low point, but increases under acidic and alkaline conditions (Caracuel et al. 2003; Turgeman et al. 2016). Moreover, interference with cellular processes, including protein synthesis, protein folding and therefore enzyme activity, cell wall remodelling and reduced availability of nutrients, are likely involved (Gaitanaki et al. 1990; Caracuel et al. 2003; Turgeman et al. 2016). Under field conditions, however, extreme acidic and alkaline conditions rarely occur. Nevertheless, from a biological perspective, our findings suggest a robust adaptation of Foc TR4 to ambient pH. Thus, from a virulence perspective, this might reflect the potential of Foc TR4 to infect plants in diverse soils.
The pH of the environment plays a crucial role in the infection process of F. oxysporum for instance, through the regulation of nutrient bioavailability and modulation of virulence factors such as fusaric acid (Gielkens et al. 1999; Fernandes et al. 2017; López-Díaz et al. 2018; Palmieri et al. 2023). Fusarium oxysporum relies on the highly conserved zinc finger transcription factor PacC and the PalH/Rim signalling pathway to sense and respond to extracellular pH (Caracuel et al. 2003). In planta, F. oxysporum secretes an array of effector proteins, which promote host colonization. For instance, a functional homolog of the plant regulatory peptide RALF (rapid alkalinization factor) efficiently induces host alkalinisation which, in turn stimulates MAPK-driven invasive growth (Fernandes et al. 2017; Mariscal et al. 2022).
Chlamydospore germination in Foc TR4 was characterized by the developmental sequence of three discrete morphological changes transitioning to swelling, polarized growth, and outgrowth. These transitions did not occur uniformly in a chlamydospore population, suggesting that the process of chlamydospore germination in Foc TR4 is asynchronous. Germination asynchrony may be a strategy for biological bet-hedging. It is a strategy employed by organisms to mitigate risk from unpredictable environments. Biological bet-hedging is particularly crucial for fungi that inhabit soils, which frequently experiences disturbances and fluctuations in temperature, humidity, and nutrient availability. (Stelkens et al. 2016), or may denote heterogeneity within a spore population (Wyatt et al. 2013). Asynchronous germination has also been reported in Penicillium marneffei (Zuber et al. 2003), Saccharomyces paradoxus (Stelkens et al. 2016), and Cryptococcus neoformans (Ortiz et al. 2021).
Phenotypic transitions observed in germinating chlamydospores of Foc TR4 are reminiscent of filamentous fungi, but differ in the timing between transitions (Sephton-Clark and Voelz 2018). This difference could reflect strain variability of the pathogen or distinct experimental systems. Chlamydospore swelling at the onset of germination suggests an increase in spore volume, which is typical of germinating fungal spores (Sharma et al. 2016; Hayer et al. 2014). Increase in spore volume results from hydration of the spore and is a fundamental step in the germination process (Hayer et al. 2014; Sephton-Clark and Voelz 2018). Hydration of the spore can be passive, following hydrolysis of trehalose to glucose (Sharma et al. 2016) or ATP-driven via aquaporins (Turgeman et al. 2016).
Polarized growth in filamentous fungi is well established (Ghose et al. 2021). A polar site is defined inside the cell (polarity establishment), from where growth is directed along a polar axis (polarity maintenance) (Ghose et al. 2021). Polarized growth is regulated by spatio-temporal activation of Rho-family guanosine triphosphatases (GTPases) and requires specialized GTP-binding proteins, called septins. Septins localize in a cortical region and serve as positional landmarks for activation and recruitment of the polarity machinery (Bonazzi et al. 2014; Ghose et al. 2021; Hall and Wallace 2022). This defines the cell’s front, thereby creating a polarity domain and protein complex, termed the polarisome (Ghose et al. 2021; Mishra et al. 2022). The polarisome is sustained by a constant supply of exocytic vesicles containing precursors for cell surface expansion and growth of the cell tip (Mishra et al. 2022). Vesicles aggregate at the cell tip and together with cytoskeleton components form the Spitzenkörper, a highly dynamic and pleomorphic complex that regulates hyphal growth and morphogenesis (Bonazzi et al. 2014; Ghose et al. 2021; Mishra et al. 2022). Hence, for now, it may be only speculated that the bright spot observed in polarized chlamydospores represents the polarisome, but further confirmative analysis will be necessary.
Conclusion
Our results suggest that the process of chlamydospore germination in Foc TR4 is developmentally orchestrated and iron-dependent. Our findings highlight the role of iron and pH in the process of chlamydospore germination and suggest that in soil, disease suppression by manipulation of soil pH may act via other mechanisms besides the alteration of iron bioavailability. Although iron is crucial, the role of other elements like copper in chlamydospore germination and virulence of Foc TR4 cannot be ignored, especially given the impact of pH on their bioavailability. Therefore, it is necessary to investigate the importance and function of copper in these processes.
Acknowledgements
We thank Dr. Diane Morstert for providing the Foc TR4 isolate used in this study and Prof. Dr. Lydia-Marie Joubert for her support with SEM. The technical assistance of Carolin Stahl, Julia Asch, and Ms. Sharney Abrahams is gratefully acknowledged. We thank Dr. Walter Ocimati for the many fruitful discussions.
Authors contribution
EW conceived the idea; EW, AV, and FR developed the methodology; FR and AV sourced the funding for this study; EW conducted the experiments, data curation and analysis and drafted the original manuscript; FR and AV revised and gave further inputs to the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported with funding from the German Academic Exchange Service (DAAD) through the Food Security Center (FSC) of the University of Hohenheim (Stuttgart, Germany).
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Anke H, Anke T, Diekmann H (1973) Biosynthesis of sideramines in fungi. Fusigen synthetase from extracts of Fusarium cubense. FEBS Lett 36:323-325. 10.1016/0014-5793(73)80401-6 [DOI] [PubMed]
- Asai Y, Hiratsuka T, Ueda M, Kawamura Y, Asamizu S, Onaka H, Arioka M, Nishimura S, Yoshida M. Differential biosynthesis and roles of two ferrichrome-type siderophores, ASP2397/AS2488053 and ferricrocin, in Acremonium persicinum. ACS Chem Biol. 2022;17:207–216. doi: 10.1021/acschembio.1c00867. [DOI] [PubMed] [Google Scholar]
- Balotf S, Tegg RS, Nichols DS, Wilson CR. Spore germination of the obligate biotroph Spongospora subterranea: Transcriptome analysis reveals germination associated genes. Front Microbiol. 2021;12:1557. doi: 10.3389/fmicb.2021.691877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann N, Schafferer L, Schrettl M, Binder U, Talasz H, Lindner H, Haas H (2013) Characterization of the link between ornithine, arginine, polyamine and siderophore metabolism in Aspergillus fumigatus. PloS One 8:e67426. 10.1371/journal.pone.0067426 [DOI] [PMC free article] [PubMed]
- Blatzer M, Latgé JP. Fungal spores are future-proofed. Nat Microbiol. 2021;6:979–980. doi: 10.1038/s41564-021-00959-z. [DOI] [PubMed] [Google Scholar]
- Bonazzi D, Julien JD, Romao M, Seddiki R, Piel M, Boudaoud A, Minc N. Symmetry breaking in spore germination relies on an interplay between polar cap stability and spore wall mechanics. Dev Cell. 2014;28:534–546. doi: 10.1016/j.devcel.2014.01.023. [DOI] [PubMed] [Google Scholar]
- Breuer W, Epsztejn S, Cabantchik ZI. Iron Acquired from transferrin by K562 Cells Is delivered into a cytoplasmic pool of chelatable iron (II)(∗) J Biol Chem. 1995;270:24209–24215. doi: 10.1074/jbc.270.41.24209. [DOI] [PubMed] [Google Scholar]
- Caracuel Z, Roncero MI, Espeso EA, González-Verdejo CI, García-Maceira FI, Di Pietro A. The pH signalling transcription factor PacC controls virulence in the plant pathogen Fusarium oxysporum. Mol Microbiol. 2003;48:765–779. doi: 10.1046/j.1365-2958.2003.03465.x. [DOI] [PubMed] [Google Scholar]
- Cohen R, Milo S, Sharma S, Savidor A, Covo S. Ribonucleotide reductase from Fusarium oxysporum does not respond to DNA replication stress. DNA Repair. 2019;83:102674. doi: 10.1016/j.dnarep.2019.102674. [DOI] [PubMed] [Google Scholar]
- Colombo C, Palumbo G, He JZ, Pinton R, Cesco S. Review on iron availability in soil: interaction of Fe minerals, plants, and microbes. J Soils Sediments. 2014;14:538–548. doi: 10.1007/s11368-013-0814-z. [DOI] [Google Scholar]
- Cruz DR, Leandro LF, Munkvold GP (2019) Effects of temperature and pH on Fusarium oxysporum and soybean seedling disease. Plant Dis 103:3234–3243. 10.1094/PDIS-11-18-1952-RE [DOI] [PubMed]
- Degani O, Goldblat Y (2014) Ambient stresses regulate the development of the maize late wilt causing agent, Harpophora maydis. Agric Sci 2014
- Deng GM, Yang QS, He WD, Li CY, Yang J, Zuo CW, Gao J, Sheng O, Lu SY, Zhang S, Yi GJ. Proteomic analysis of conidia germination in Fusarium oxysporum f. sp. cubense tropical race 4 reveals new targets in ergosterol biosynthesis pathway for controlling Fusarium wilt of banana. Appl Microbiol Biotechnol. 2015;99:7189–7207. doi: 10.1007/s00253-015-6768-x. [DOI] [PubMed] [Google Scholar]
- Dita M, Barquero M, Heck D, Mizubuti ES, Staver CP (2018) Fusarium wilt of banana: current knowledge on epidemiology and research needs toward sustainable disease management. Front Plant Sci 9:1468. 10.3389/fpls.2018.01468 [DOI] [PMC free article] [PubMed]
- Fernandes TR, Segorbe D, Prusky D, Di Pietro A. How alkalinization drives fungal pathogenicity. PLoS Pathog. 2017;13:e1006621. doi: 10.1371/journal.ppat.1006621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada F, Kodama S, Nishiuchi T, Kajikawa N, Kubo Y (2019) Plant pathogenic fungi Colletotrichum and Magnaporthe share a common G1 phase monitoring strategy for proper appressorium development. New Phytol 222:1909–1923. 10.1111/nph.15728 [DOI] [PubMed]
- Furukawa T, Naitoh Y, Kohno H, Tokunaga R, Taketani S (1992) Iron deprivation decreases ribonucleotide reductase activity and DNA synthesis. Life Sci 50:2059–2065. 10.1016/0024-3205(92)90572-7 [DOI] [PubMed]
- Gaitanaki CJ, Sugden PH, Fuller SJ. Stimulation of protein synthesis by raised extracellular pH in cardiac myocytes and perfused hearts. FEBS Lett. 1990;260:42–44. doi: 10.1016/0014-5793(90)80061-M. [DOI] [PubMed] [Google Scholar]
- Ghose D, Jacobs K, Ramirez S, Elston T, Lew D. Chemotactic movement of a polarity site enables yeast cells to find their mates. Proc Natl Acad Sci USA. 2021 doi: 10.1073/pnas.2025445118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal JP, Maraite H, Meyer JA. Abundant production of chlamydospores by Fusarium oxysporum f. sp. melonis in soil extract with glucose. Neth J Plant Pathol. 1973;79:162–164. doi: 10.1007/BF01976712. [DOI] [Google Scholar]
- Greene BL, Kang G, Cui C, Bennati M, Nocera DG, Drennan CL, Stubbe J. Ribonucleotide reductases: structure, chemistry, and metabolism suggest new therapeutic targets. Ann Rev Biochem. 2020;89:45–75. doi: 10.1146/annurev-biochem-013118-111843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RA, Wallace EW. Post-transcriptional control of fungal cell wall synthesis. Cell Surf. 2022;8:100074. doi: 10.1016/j.tcsw.2022.100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayer K, Stratford M, Archer DB. Germination of Aspergillus niger conidia is triggered by nitrogen compounds related to L-amino acids. Appl Environ Microbiol. 2014;80:6046–6053. doi: 10.1128/AEM.01078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hider RC, Kong X. Chemistry and biology of siderophores. Nat Prod Rep. 2010;27:637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- Kozak M, Piepho HP. What’s normal anyway? Residual plots are more telling than significance tests when checking ANOVA assumptions. J Agron Crop Sci. 2018;204:86–98. doi: 10.1111/jac.12220. [DOI] [Google Scholar]
- Kozak M, Piepho HP. Analyzing designed experiments: should we report standard deviations or standard errors of the mean or standard errors of the difference or what? Exp Agric. 2020;56:312–319. doi: 10.1111/jac.12220. [DOI] [Google Scholar]
- Leslie JF, Summerell BA. The Fusarium laboratory manual. Iowa: Blackwell Publishing; 2006. [Google Scholar]
- Li C, Chen S, Zuo C, Sun Q, Ye Q, Yi G, Huang B. The use of GFP-transformed isolates to study infection of banana with Fusarium oxysporum f. sp. cubense race 4. Eur J Plant Pathol. 2011;131:327–340. doi: 10.1007/s10658-011-9811-5. [DOI] [Google Scholar]
- López-Berges MS, Capilla J, Turrà D, Schafferer L, Matthijs S, Jöchl C, Cornelis P, Guarro J, Haas H, Di Pietro A. HapX-mediated iron homeostasis is essential for rhizosphere competence and virulence of the soilborne pathogen Fusarium oxysporum. Plant Cell. 2012;24:3805–3822. doi: 10.1105/tpc.112.098624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Berges MS, Capilla J, Turra D, Schafferer L, Matthijs S, Jochl C, Cornelis P, Guarro J, Haas H, Di Pietro A. HapX-mediated iron homeostasis is essential for rhizosphere competence and virulence of the soilborne pathogen Fusarium oxysporum. Plant Cell. 2012;24:3805–3822. doi: 10.1105/tpc.112.098624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R, Minc N, Peter M. Cells under pressure: how yeast cells respond to mechanical forces. Trends Microbiol. 2022 doi: 10.1016/j.tim.2021.11.006. [DOI] [PubMed] [Google Scholar]
- Ohara T, Tsuge T. FoSTUA, encoding a basic helix-loop-helix protein, differentially regulates development of three kinds of asexual spores, macroconidia, microconidia, and chlamydospores, in the fungal plant pathogen Fusarium oxysporum. Eukaryot Cell. 2004;3:1412–1422. doi: 10.1128/EC.3.6.1412-1422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz SC, Huang M, Hull CM. Discovery of fungus-specific targets and inhibitors using chemical phenotyping of pathogenic spore germination. Mbio. 2021;12:e01672–e1721. doi: 10.1128/mBio.01672-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri D, Segorbe D, López-Berges MS, De Curtis F, Lima G, Di Pietro A, Turrà D. Alkaline pH, Low Iron Availability, Poor Nitrogen Sources and CWI MAPK Signaling Are Associated with Increased Fusaric Acid Production in Fusarium oxysporum. Toxins. 2023;15:50. doi: 10.3390/toxins15010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg KG, Coates LM, O’Neill WT, Turner DW (2019) The epidemiology of Fusarium wilt of banana. Front Plant Sci 10:1395. 10.3389/fpls.2019.01395 [DOI] [PMC free article] [PubMed]
- Peng HX, Sivasithamparam K, Turner DW. Chlamydospore germination and Fusarium wilt of banana plantlets in suppressive and conducive soils are affected by physical and chemical factors. Soil Biol Biochem. 1999;31:1363–1374. doi: 10.1016/S0038-0717(99)00045-0. [DOI] [Google Scholar]
- Philpott CC, Leidgens S, Frey AG. Metabolic remodeling in iron-deficient fungi. Biochim Biophys Acta - Mol Cell Res. 2012;1823:1509–1520. doi: 10.1016/j.bbamcr.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante S, Labbé S (2019) Spore germination requires ferrichrome biosynthesis and the siderophore transporter Str1 in Schizosaccharomyces pombe. Genetics 211:893–911. 10.1534/genetics.118.301843 [DOI] [PMC free article] [PubMed]
- Renton FJ, Jeitner TM. Cell cycle-dependent inhibition of the proliferation of human neural tumor cell lines by iron chelators. Biochem Pharmacol. 1996;51:1553–1561. doi: 10.1016/0006-2952(96)00099-8. [DOI] [PubMed] [Google Scholar]
- Rittershaus ES, Baek SH, Sassetti CM. The normalcy of dormancy: common themes in microbial quiescence. Cell Host Microbe. 2013;13:643–651. doi: 10.1016/j.chom.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo AM, Christen L, Niles EG, Kosman DJ. Intracellular chelation of Fe by bipyridyl inhibits DNA virus replication: ribonucleotide reductase maturation as a probe of intracellular Fe pools. J Biol Chem. 2001;276:24301–24308. doi: 10.1074/jbc.M010806200. [DOI] [PubMed] [Google Scholar]
- Sanvisens N, Bañó MC, Huang M, Puig S. Regulation of ribonucleotide reductase in response to iron deficiency. Mol Cell. 2011;44:759–769. doi: 10.1016/j.molcel.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura-Mena RA, Stoorvogel JJ, García-Bastidas F, Salacinas-Niez M, Kema GH, Sandoval JA. Evaluating the potential of soil management to reduce the effect of Fusarium oxysporum f. sp. cubense in banana (Musa AAA) Euro J Plant Pathol. 2021;160:441–455. doi: 10.1007/s10658-021-02255-2. [DOI] [Google Scholar]
- Sharma M, Sengupta A, Ghosh R, Agarwal G, Tarafdar A, Nagavardhini A, Pande S, Varshney RK. Genome wide transcriptome profiling of Fusarium oxysporum f. sp. ciceris conidial germination reveals new insights into infection-related genes. Sci Rep. 2016;6:1–11. doi: 10.1038/srep37353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk JL. Evolutionary divergence in DNA damage responses among fungi. Mbio. 2021;12:e03348–e3420. doi: 10.1128/mBio.03348-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelkens RB, Miller EL, Greig D. Asynchronous spore germination in isogenic natural isolates of Saccharomyces paradoxus. FEMS Yeast Res. 2016;16:fow012. doi: 10.1093/femsyr/fow012. [DOI] [PubMed] [Google Scholar]
- Turgeman T, Shatil-Cohen A, Moshelion M, Teper-Bamnolker P, Skory CD, Lichter A, Eshel D. The role of aquaporins in pH-dependent germination of Rhizopus delemar spores. PLoS ONE. 2016;11:e0150543. doi: 10.1371/journal.pone.0150543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Westerhoven AC, Meijer HJ, Seidl MF, Kema GH. Uncontained spread of Fusarium wilt of banana threatens African food security. Plos Pathog. 2022;18:e1010769. doi: 10.1371/journal.ppat.1010769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Were E, Viljoen A, Rasche F. Back to the roots: understanding banana below-ground interactions is crucial for effective management of Fusarium wilt. Plant Pathol. 2022 doi: 10.1111/ppa.13641. [DOI] [Google Scholar]
- Were E, Schöne J, Viljoen A, Rasche F. Phenolics mediate suppression of Fusarium oxysporum f. sp. cubense TR4 by legume root exudates. Rhizosphere. 2022;21:100459. doi: 10.1016/j.rhisph.2021.100459. [DOI] [Google Scholar]
- Wyatt TT, Wösten HA, Dijksterhuis J. Fungal spores for dispersion in space and time. Adv Appl Microbiol. 2013;85:43–91. doi: 10.1016/B978-0-12-407672-3.00002-2. [DOI] [PubMed] [Google Scholar]
- Yuan JS, Reed A, Chen F, Stewart CN. Statistical analysis of real-time PCR data. BMC Bioinform. 2006;7:1–12. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S-J, García-Bastidas FA, Li X, Zeng L, Bai T, Xu S, Yin K, Li H, Fu G, Yu Y, Yang L, Nguyen HC, Douangboupha B, Khaing AA, Drenth A, Seidl MF, Meijer HJG, Kema GHJ. New geographical insights of the latest expansion of Fusarium oxysporum f. sp. cubense Tropical Race 4 into the greater Mekong subregion. Front Plant Sci. 2018;2018:457. doi: 10.3389/fpls.2018.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber S, Hynes MJ, Andrianopoulos A. The G-protein α-subunit GasC plays a major role in germination in the dimorphic fungus Penicillium marneffei. Genetics. 2003;164:487–499. doi: 10.1093/genetics/164.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Díaz C, Rahjoo V, Sulyok M, Ghionna V, Martín-Vicente A, Capilla J, Di Pietro A, López-Berges MS (2018) Fusaric acid contributes to virulence of Fusarium oxysporum on plant and mammalian hosts. Mol Plant Pathol 19: 440–453 [DOI] [PMC free article] [PubMed]
- Mariscal, M, Fernandes, TR, Di Pietro A (2022) Role of pH in the Control of Fungal MAPK Signalling and Pathogenicity. In Plant Relationships: Fungal-Plant Interactions (pp. 227–238). Cham: Springer International Publishing. 10.1007/978-3-031-16503-0_9
- Meldrum RA, Daly AM, Tran-Nguyen LT, Aitken EA (2013) The effect of surface sterilants on spore germination of Fusarium oxysporum f. sp. cubense tropical race 4. Crop Prot. 54: 194-198
- R Development Core Team R (2020) A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna, Austria https://www.R-project.org/.
- Sephton-Clark PC, Voelz K (2018) Spore germination of pathogenic filamentous fungi. Pages 117–157: In Advances in applied microbiology. Academic Press, London 10.1016/bs.aambs.2017.10.002 [DOI] [PubMed]
- Viljoen A, Ma LJ, Molina AB (2020) Fusarium wilt (Panama disease) and monoculture banana production: Resurgence of a century-old disease. Pages 159–184 In: Emerging plant diseases and global food security. J. B. Ristaino and A. Records eds. APS Press St. Paul, MN, USA. 10.1094/9780890546383.008