Abstract
Two genera of sepiolid squids—Euprymna, found primarily in shallow, coastal waters of Hawaii and the Western Pacific, and Sepiola, the deeper-, colder-water-dwelling Mediterranean and Atlantic squids—are known to recruit luminous bacteria into light organ symbioses. The light organ symbiont of Euprymna spp. is Vibrio fischeri, but until now, the light organ symbionts of Sepiola spp. have remained inadequately identified. We used a combination of molecular and physiological characteristics to reveal that the light organs of Sepiola affinis and Sepiola robusta contain a mixed population of Vibrio logei and V. fischeri, with V. logei comprising between 63 and 100% of the bacteria in the light organs that we analyzed. V. logei had not previously been known to exist in such symbioses. In addition, this is the first report of two different species of luminous bacteria co-occurring within a single light organ. The luminescence of these symbiotic V. logei strains, as well as that of other isolates of V. logei tested, is reduced when they are grown at temperatures above 20°C, partly due to a limitation in the synthesis of aliphatic aldehyde, a substrate of the luminescence reaction. In contrast, the luminescence of the V. fischeri symbionts is optimal above 24°C and is not enhanced by aldehyde addition. Also, V. fischeri strains were markedly more successful than V. logei at colonizing the light organs of juvenile Euprymna scolopes, especially at 26°C. These findings have important implications for our understanding of the ecological dynamics and evolution of cooperative, and perhaps pathogenic, associations of Vibrio spp. with their animal hosts.
Members of at least two families of squids and seven families of marine fishes are known to harbor a monospecific culture of luminous bacteria in specialized light organs (23, 29). In these cooperative symbioses, the host provides the bacteria with nutrients and a protected environment in exchange for light production, which the host may use for intraspecific communication, prey attraction, or predator avoidance (22). To date, only three species (Vibrio fischeri, Photobacterium leiognathi, and Photobacterium phosphoreum) of the 10 described marine luminous bacteria have been isolated from light organs. P. leiognathi and P. phosphoreum occur primarily as symbionts of marine fishes (14, 28). V. fischeri has been identified as the specific light organ symbiont of at least five species of marine animal hosts, including several bobtail squids in the family Sepiolidae (4, 29) and fishes in the family Monocentridae (10, 31). In all cases, the light organs of marine animals have been reported to contain only a single species of luminous bacteria (23).
Euprymna and Sepiola are two important genera in the family Sepiolidae (19). Euprymna spp. are found primarily in the shallow coastal waters of the Pacific Ocean, in Hawaii (Euprymna scolopes), Australia (Euprymna tasmanica), and the Indo-West Pacific region (Euprymna morsei). The light organ symbiont of these squids has been identified as V. fischeri (6, 18, 24, 30). In contrast, members of the genus Sepiola are found in the Mediterranean Sea, along the Atlantic coastline of Europe, and also in Japan and the Philippines. Until now, the identity of the light organ symbionts of these relatively deeper- and colder-water Sepiola spp. had been reported only for Sepiola atlantica (13).
Preliminary biochemical and microscopic analyses of light organ symbionts isolated from the Mediterranean squids Sepiola affinis and Sepiola robusta suggested that these bacteria were either V. fischeri or the closely related luminous species Vibrio logei. While these two bacteria have proven difficult to differentiate by conventional phenotypic traits (2, 23), a substantial genomic sequence difference does exist among Vibrio spp. in a variable region of the 16S rRNA gene called V1 (36). The length of a PCR-amplified 16S rRNA gene (rDNA) fragment containing this region can be used to categorize different members of the genus Vibrio (33).
We used this PCR product analysis, combined with 16S rDNA sequence information and other phenotypic data, to determine that the predominant light organ symbionts of S. affinis and S. robusta are V. logei. Surprisingly, we found that V. fischeri was a minor but significant component of the symbiont population in some of the light organs, suggesting that there may be a dynamic competition between these two luminous species for dominance. To test this hypothesis, we compared two ecologically relevant activities in representative isolates of V. logei and V. fischeri symbionts: (i) the effect of temperature on growth rate and luminescence potential, and (ii) their ability to colonize the light organs of juvenile E. scolopes squids.
MATERIALS AND METHODS
Collection of specimens of Sepiola spp. and isolation of light organ symbionts.
Specimens of bobtail squids were collected from the upper 50 m of seawater within 1 km of shore near the Laboratorie Arago, Banyuls-sur-Mer, France, between 1 and 15 July 1995. The animals (mantle length, 13 to 24 mm) were identified as S. affinis and S. robusta by using the criteria outlined by Bello (4).
Bacterial symbionts were isolated from three specimens each of freshly collected S. affinis and S. robusta by dissecting an anesthetized animal, removing the light organ, and homogenizing it in sterile seawater. Aliquots of the homogenate were spread on seawater-tryptone-yeast extract (SWT) agar medium (6) and incubated at 25°C overnight. Uniform fields of golden-yellow colonies were observed on all plates, although some light organs yielded two distinct colony size classes. Four to six representative colonies from each light organ (designated either SA for S. affinis symbionts or SR for S. robusta symbionts) were isolated and stored for further study.
PCR amplification of the V1 region of the 16S rRNA gene.
Bacterial cultures of bobtail squid symbionts and reference strains (see Table 1) were streaked on SWT agar and incubated at 20°C overnight. Between 108 and 109 cells from single colonies were inoculated into tubes containing 1.5 ml of distilled water that had been filtered through 0.22-μm-pore-size filters and autoclaved. The cells were lysed by exposure to five cycles of freezing in a dry ice–95% ethanol bath and thawing in a 37°C water bath. Cell debris was pelleted by centrifugation for 5 min. Each supernatant was transferred to a separate tube, and 1 μl of this supernatant was added to 100 μl of the PCR solution (containing 200 μM each deoxynucleoside triphosphate, 10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2, 2.5 U of Taq polymerase, and 0.1 μM each primer). Forward and reverse PCR primers used to specifically amplify V1 (33) were as follows: RU41 (forward) 5′ GATCATGGCTCAGATTGAACG 3′; and RU42 (reverse), 5′ AGGCATTACTCACCCGTCC 3′.
TABLE 1.
Bacterial strains used in this study
| Strain | Source (reference) | Size of V1 regiona | Highest degree of 16S rDNA sequence similarity to: |
|---|---|---|---|
| ATCC 15382 | Pacific cod skin (7) | S | V. logeib |
| ATCC 35077 | Tanner crab lesion (3) | S | V. logeib |
| ATCC 25918 | Seawater (26) | L | V. fischerib |
| ES1114 | E. scolopes light organ (6) | L | V. fischerib |
| MJ1 | Monocentris japonica light organ (31) | L | V. fischerib |
| NCMB 2262 | Atlantic salmon infection (8) | S | V. salmonicidab |
| MdR7 | California coastal seawater (16) | S | V. logei |
| MdRD | California coastal seawater (16) | S | —c |
| SA1 | S. affinis no. 1 (this study) | L | V. fischeri |
| SA2 | S. affinis no. 1 (this study) | L | — |
| SA3 | S. affinis no. 1 (this study) | L | — |
| SA4 | S. affinis no. 1 (this study) | L | — |
| SA5 | S. affinis no. 1 (this study) | S | V. logei |
| SA6 | S. affinis no. 1 (this study) | S | V. logei |
| SA7 | S. affinis no. 2 (this study) | S | — |
| SA8 | S. affinis no. 2 (this study) | S | — |
| SA9 | S. affinis no. 2 (this study) | S | — |
| SA10 | S. affinis no. 2 (this study) | S | — |
| SA11 | S. affinis no. 2 (this study) | S | — |
| SA12 | S. affinis no. 2 (this study) | S | V. logei |
| SA13 | S. affinis no. 3 (this study) | S | — |
| SA14 | S. affinis no. 3 (this study) | S | — |
| SA15 | S. affinis no. 3 (this study) | S | — |
| SA16 | S. affinis no. 3 (this study) | S | — |
| SA17 | S. affinis no. 3 (this study) | S | — |
| SA18 | S. affinis no. 3 (this study) | S | — |
| SR1 | S. robusta no. 1 (this study) | S | — |
| SR2 | S. robusta no. 2 (this study) | S | — |
| SR3 | S. robusta no. 2 (this study) | S | — |
| SR5 | S. robusta no. 2 (this study) | L | V. fischeri |
| SR6 | S. robusta no. 2 (this study) | S | V. logei |
| SR7 | S. robusta no. 3 (this study) | S | — |
| SR8 | S. robusta no. 3 (this study) | S | — |
| SR9 | S. robusta no. 3 (this study) | S | — |
| SR10 | S. robusta no. 3 (this study) | S | — |
S, small (111-bp) V1 region; L, large (121-bp) V1 region.
Taxonomic criteria other than DNA sequence similarity were used to identify these strains (see references).
—, 16S rRNA gene sequencing was not performed for this strain.
These PCR solutions were overlaid with sterile mineral oil and inserted into a thermal cycler for 37 cycles under the following conditions: 94°C, 6 min (first cycle only); 94°C, 1.5 min; 50°C, 1.5 min; 72°C, 2 min; and 72°C, 7 min (final cycle only). Aliquots of each PCR product were then loaded into separate wells of a 3% agarose gel and electrophoretically separated for 5 h at 60 V.
Cloning and sequencing of a portion of the 16S rRNA gene.
Entire 16S rRNA genes from light organ symbionts of S. affinis (strains SA1, SA5, SA6, and SA12), and S. robusta (strains SR5 and SR6) were PCR amplified with the following primers: RU1 (reverse), 5′ CCTTTCCCTCACGGTACTGGT 3′; and RU2 (forward), 5′ TGGCTCAGATTGAACGCTGGCGGC 3′.
PCR products from strains SA1, SA5, SA6, and SR5 were purified by passage through QIAquick columns (Qiagen Inc., Chatsworth, Calif.) and directly sequenced at the Biotechnology-Molecular Biology Instrumentation Facility, University of Hawaii, Manoa. Purified PCR products from strains SA12 and SR6 were ligated into the vector pCRII and cloned in Escherichia coli with the TA cloning kit (Invitrogen Corp., San Diego, Calif.) in accordance with the manufacturer’s instructions. Plasmids containing the desired 2.3-kbp 16S rDNA inserts from strains SA12 and SR6 were then prepared for sequence analysis.
Denatured plasmid DNA (between 300 and 500 ng) was placed into an Eppendorf tube and mixed with 2 μl of 5× sequencing buffer (0.2 M Tris-HCl, pH 7.5; 0.1 M MgCl2; and 0.25 M NaCl) and primer RU2 (10 ng). The tubes were incubated at 37°C for 20 min followed by a 25°C incubation for 10 min to allow the primer to anneal to the 16S rDNA insert. Following this step, the 16S rDNA inserts were sequenced by using the Sequenase version 2.0 sequencing kit (U.S. Biochemicals, Cleveland, Ohio). Sequence data were analyzed with the BLAST algorithm (1) to identify sequences with a high degree of identity to our query sequences.
Biochemical tests for distinguishing V. logei from V. salmonicida.
Because 16S rDNA sequence comparisons were not able to unambiguously differentiate between V. logei and Vibrio salmonicida, we subjected symbionts from Sepiola spp. to three physiological tests that identify biochemical capabilities (lysine decarboxylation, d-galactose fermentation, and nitrate reduction) that differ for these two Vibrio species. The test media were prepared and the results were interpreted in accordance with the Difco Manual (7a), except that each medium was supplemented with marine cations according to the recipe of Farmer and Hickman-Brenner (9). Test organisms were pregrown on SWT agar at 14°C before inoculation, and the reaction tubes were incubated at the same temperature for between 48 and 96 h prior to the recording of results.
Effect of temperature on the growth rates of symbionts of Sepiola spp.
Bacterial strains were inoculated into flasks containing 20 ml of SWT broth to a final optical density (OD) at 600 nm of about 0.01 (approximately 5 × 107 cells/ml). The cultures were then shaken at either 20, 24, or 28°C, and their ODs were measured periodically throughout the logarithmic growth phase.
Effect of temperature and aldehyde on the luminescence of symbiont strains.
Bacterial cultures were inoculated into flasks containing Vibrio harveyi-conditioned broth medium, prepared as described previously (20). This conditioning has been shown to remove an inhibitor of luminescence induction. The cultures were incubated with shaking at either 20, 24, or 26°C. During the logarithmic growth phase, 1.0-ml aliquots of each culture were removed at regular intervals, measured for luminescence, and then exposed to 5 μl of a 0.00005% (vol/vol) decyl aldehyde suspension in water, and the level of luminescence was remeasured.
Colonization of the E. scolopes light organ by symbionts of Sepiola spp.
Bacterial strains to be tested for their ability to colonize juvenile E. scolopes were grown, without shaking, to an OD of between 0.1 and 0.3 in SWT broth at either 21 or 26°C, depending on the temperature at which the corresponding colonization experiment was to be performed. These cultures were diluted to about 2,000 cells per ml in natural seawater to serve as the inocula (17). Newly hatched juveniles of E. scolopes were placed into glass vials containing 5 ml of one of these seawater suspensions of bacteria. The juveniles were exposed to these inocula for 3 h and then washed and placed in 5 ml of natural seawater without added bacteria. The luminescence of the squids was subsequently monitored photometrically in the morning and at dusk for the next 2 to 4 days. After the final luminescence reading was taken, each animal was washed three times in sterile seawater, placed in an Eppendorf tube containing 700 μl of sterile seawater, and homogenized with a pestle. Dilutions of the homogenate were spread in triplicate on SWT agar and incubated at 24°C for between 24 and 48 h. The resulting colonies were checked for luminescence, and estimates of the number of CFU per light organ were calculated.
RESULTS
Collection and identification of light organ symbionts of Mediterranean bobtail squids.
All of the colonies arising from the highest dilutions of Sepiola light organ homogenates had the golden-yellow color characteristic of V. fischeri and V. logei. From the number of CFU per ml of diluted homogenate, it was possible to calculate that each of the six light organs contained between 2 × 107 and 4 × 108 total symbionts. Interestingly, homogenates of two of the six bobtail squids (one specimen of S. affinis and one of S. robusta) gave rise to a mixture of two colony types, based on size and light emission. When observed in the dark, the larger class of colonies produced a relatively brighter bioluminescence. From the one S. affinis specimen, these larger colonies constituted about 37% of the total CFU, while in the S. robusta plating, they accounted for 19%. Five strains were isolated from these two less-abundant colony classes: SA1, SA2, SA3, SA4, and SR5 (Table 1). They and five representatives of the more abundant class, as well as 17 isolates from the other four light organs, were chosen for taxonomic identification.
Differentiation of V. logei and V. fischeri symbionts on the basis of V1 region size.
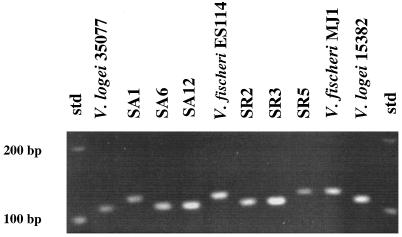
The V. logei type strains ATCC 15382 and ATCC 35077, two luminous bacterial isolates from Southern California seawater (MdR7 and MdRD), and 22 of 27 of the isolates from Sepiola spp. light organs each contained a 111-bp fragment when the portion of its 16S rDNA sequence that spanned the V1 region was PCR amplified (Fig. 1). In contrast, V. fischeri type strain ATCC 25918, the fish symbiont strain MJ1, four light organ isolates from Euprymna spp. (ES114, ES213, EM17, and EM24), and the remaining 5 of the 27 Sepiola light organ symbionts each contained a 121-bp fragment (Fig. 1 and Table 1). These results support the use of this technique as a relatively easy method of distinguishing between V. logei and V. fischeri strains, and they provided the first indication that over 80% of the Sepiola symbionts examined were most closely related to V. logei while the remainder were most closely related to V. fischeri (Table 1).
FIG. 1.
Electrophoretic separation of PCR-amplified 16S rDNA fragments containing the V1 (variable) region. The names of the strains used are indicated above the lanes. std, molecular size standards.
Confirmation of symbiont identification by partial 16S rDNA sequences and biochemical tests.
To further confirm their identity and support the conclusions based on V1 size, partial 16S rDNA sequences from strains SA1, SA5, SA6, SA12, SR5, and SR6 were obtained. When analyzed against the GenBank nucleic acid database, the 16S rDNA sequences (between 250 and 800 bp in length) from SA1 and SR5 each aligned with the highest degrees of identity (90 and 92%, respectively) to V. fischeri 16S rDNA. Similarly, the 16S rDNA sequences of SA5, SA6, SA12, and SR6 aligned with the highest degrees of identity (93, 94, 97, and 99.6%, respectively) to V. logei. Thus, the V1 region size and partial 16S rDNA sequence comparisons were in agreement (Table 1).
During these analyses we discovered that the 16S rDNA sequence from V. salmonicida also aligned with a high (>90%) degree of identity to SA5, SA6, SA12, and SR6 and that V. salmonicida had a 111-bp V1 region. While the data suggested that these four symbionts were most closely related to V. logei, the relatively high degree of similarity to V. salmonicida produced some ambiguity. Thus, we examined the expression of several metabolic activities that differ between these two Vibrio species. Strains of V. logei and V. salmonicida were easily differentiated on the basis of the results of three biochemical tests (Table 2). These data indicate that although V. salmonicida and V. logei strains have very similar 16S rDNA sequences, they exhibit considerable metabolic divergence.
TABLE 2.
Biochemical activities used to distinguish among luminous species
| Strain | Biochemical activitya
|
||
|---|---|---|---|
| Lysine decarboxylation | Nitrate reduction | d-Galactose fermentation | |
| V. logei ATCC 35077 | + | + | + |
| V. fischeri ES114 | + | + | + |
| V. salmonicida NCMB 2262 | − | − | − |
| SA7 | + | + | + |
| SA12 | + | + | + |
| SR3 | + | + | + |
| SR6 | + | + | + |
+, positive; −, negative.
Differential effects of temperature on growth and luminescence of V. logei and V. fischeri.
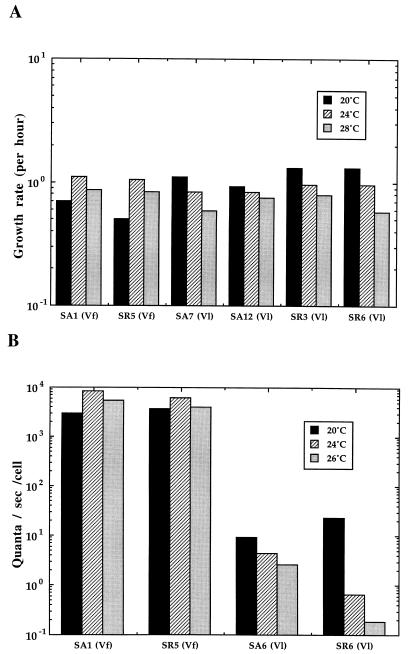
One of the few phenotypic traits that has been reported to be generally useful for distinguishing between V. logei and V. fischeri is the relative psychrophily of the former. Thus, it was not surprising that V. logei strains from either S. affinis or S. robusta grew well on SWT agar plates incubated for 4 days at 4°C whereas V. fischeri strains did not grow at all under those conditions (data not shown). Similarly, V. logei symbionts grew more rapidly in SWT medium at 20°C than their V. fischeri counterparts (Fig. 2A). In contrast, at temperatures above 20°C, the V. fischeri symbionts had a distinct growth advantage over V. logei symbionts (Fig. 2A). All V. logei SA and SR strains that were examined grew in culture with generation times between 1.3 and 2.2 times longer at 28°C than at 20°C; conversely, the generation times of the V. fischeri strains tested (SA1 and SR5) were, respectively, 1.6 and 2.1 times longer at 20°C than at 24°C.
FIG. 2.
(A) Growth rates (per hour) of Mediterranean Sepiola symbionts incubated at three temperatures. (B) Maximum luminescence per cell of exponentially growing Sepiola symbionts exposed to decyl aldehyde.
When grown at 24 and 26°C, V. logei symbionts always emitted a lower level of luminescence per cell than when they were grown at 20°C (Fig. 2B). This effect was due to two temperature-sensitive processes, one that could be relieved by the addition of extraneous aliphatic aldehyde to the cells and another that could not. For instance, exposure of V. logei cells grown at 26°C to decyl aldehyde enhanced their luminescence between 6- and 11-fold (Table 3); however, no enhancement by decyl aldehyde was observed with V. fischeri symbionts from Sepiola spp. Thus, the V. logei symbionts, as well as the ATCC strains (data not shown), appear to have a temperature-sensitive capacity for the synthesis of the natural aldehyde substrate for the luminescence reaction. In addition, even in the presence of excess aldehyde, the maximum luminescence per cell fell as much as 100-fold with increasing growth temperature between 20 and 26°C (Fig. 2B). Once again, this effect was not observed with the V. fischeri strains, whose luminescence per cell generally increased with increasing growth temperature from 20 to 24°C. Thus, both the growth and the luminescence of V. fischeri symbionts appear to be better suited to temperatures at or above 24°C, while those of V. logei symbionts are optimal below 24°C.
TABLE 3.
Effect of temperature and addition of decyl aldehyde on luminescence of representative Sepiola symbionts
| Strain | Avg maximum luminescence at temp (°C) ofa:
|
||
|---|---|---|---|
| 20 | 24 | 26 | |
| V. fischeri | |||
| SA1 | 1.0 | 1.0 | 1.0 |
| SR5 | 1.0 | 1.0 | 1.0 |
| V. logei | |||
| SA6 | 3.8 | 7.4 | 6.6 |
| SR6 | 1.3 | 6.0 | 11.3 |
Average maximum luminescence of log-phase cells grown at each of three temperatures. The values given are the ratios of the specific luminescence (quanta per second per cell) of cells exposed to decyl aldehyde divided by those of cells not exposed to decyl aldehyde.
Colonization of the E. scolopes light organ by Sepiola symbionts.
Newly hatched juveniles of E. scolopes became luminous within 24 h of exposure to the native symbiont V. fischeri ES114 at water temperatures of either 21 or 26°C (Table 4). symbiotic V. fischeri strains from Sepiola spp. also infected the E. scolopes light organ with 100% efficiency at 26°C; however, the average CFU per light organ from squid infected with these strains was lower than that for squid infected with ES114. Interestingly, three of these five V. fischeri strains showed reduced efficiencies of colonization at 21°C (i.e., an average of only 21% of the juveniles were colonized) relative to those at 26°C.
TABLE 4.
Colonization of E. scolopes juveniles by symbionts from Sepiola spp.
| Strain | CFU/squid (105) after 48 h at temp (°C) ofa:
|
|
|---|---|---|
| 21 | 26 | |
| V. fischeri | ||
| ES114 | 2.8 (5/5) | 4.4 (11/11) |
| SA1 | NDb (0/6) | 1.8 (5/5) |
| SA2 | 2.4 (6/6) | 2.2 (5/5) |
| SA3 | 0.2 (1/6) | 0.1 (6/6) |
| SA4 | 0.7 (3/7) | 0.6 (6/6) |
| SR5 | 4.4 (7/7) | 3.1 (7/7) |
| V. logei | ||
| SA5 | 0.9 (2/7) | ND (0/16) |
| SA6 | ND (0/5) | ND (0/6) |
| SA7 | 0.5 (1/11) | 0.3 (1/7) |
| SA12 | ND (0/11) | ND (0/10) |
| SR3 | 0.1 (2/10) | ND (0/7) |
| SR6 | 1.0 (10/10) | 0.1 (3/10) |
Infection assays were performed at either 21 or 26°C. The numbers in parentheses are the number of animals (over the total number tested) that became luminous upon exposure to the bacterial strain.
ND, none detected (<14 CFU/light organ).
In contrast, V. logei strains from Sepiola light organs were even less effective at colonizing juvenile E. scolopes. CFU were not detected in 93% of the light organ homogenates of animals exposed to strain SA5, SA6, SA7, SA12, SR3, and in those six animals that were colonized, it was to a much lower level than that of ES114. Unlike the other V. logei isolates, strain SR6 colonized the E. scolopes light organ with a high efficiency at 21°C; however, both the efficiency and extent of colonization were significantly lower at 26°C. Thus, overall, the V. fischeri symbionts of Sepiola spp. exhibited a relatively increased symbiotic competence at the higher temperature while, in contrast, the V. logei strains showed a relatively decreased symbiotic competence for colonization at the higher temperature.
DISCUSSION
The marine luminous bacteria comprise at least 10 described species in three genera (23). Only three of these luminous species have been previously recognized as participating in specific light organ symbioses with marine animals. We report that a fourth species of luminous bacteria, V. logei, is the predominant light organ symbiont of S. affinis and S. robusta. Surprisingly, in some of these light organs, V. fischeri co-occurs as a less-abundant symbiont. Therefore, in contrast to other known bioluminescent symbioses, the light organs of Sepiola spp. cannot be described as symbiont species specific. While there are relatively few characteristics that can be used to separate strains of V. fischeri and V. logei, the species are nevertheless robust (2), and the symbionts isolated from the light organs of Sepiola species unambiguously group with one or the other of these two species. We must conclude that these symbiotic organs can be inhabited by strains of either V. logei or both species.
Preliminary microscopic and metabolic analyses of light organ symbionts from S. affinis and S. robusta (designated SA and SR strains, respectively) led to the conclusion that these bacteria were strains of either V. fischeri or V. logei. A similar uncertainty exists concerning the light organ symbionts of S. atlantica, which have been reported to be Photobacterium (Vibrio) fischeri, although no information indicating how these strains were distinguished from V. logei was presented (13). Unfortunately, beyond the relative psychrophily of V. logei (Fig. 2A) (2), there are few reported phenotypic differences between these two species, reflecting their close phylogenetic relatedness (15). Nevertheless, Reichelt et al. (27), using total DNA-DNA hybridization, concluded that these two species shared only 37 to 41% DNA sequence identity, prompting them to support the assignment of separate-species status. Recently, it has been reported that a simpler means of differentiating these two species is by the presence or absence of a 10-bp gap in the V1 region of the 16S rDNA sequence (33). PCR amplification of the V1 region of the symbionts from Sepiola spp. indicated that greater than 80% of them were, in fact, V. logei while the remaining strains were V. fischeri (Fig. 1). A mixed colonization population may also exist in the light organs of S. atlantica and perhaps other Sepiola spp. (12). During the course of these analyses, we made the incidental but intriguing discovery that several luminous bacterial isolates from Southern California seawater were also V. logei. Thus, the population of bioluminescent bacteria found in these waters includes this species as well as the previously reported V. fischeri and V. harveyi (32).
Sequences of the 16S rRNA genes from Sepiola symbiont strains SA5, SA6, SA12, and SR6 exhibited the highest degree of identity (>93%) with a V. logei sequence, rather than that of V. fischeri (87%), further supporting the results of the V1 region analyses. Interestingly, the 16S rDNA sequences from these strains also aligned to that of V. salmonicida, a fish pathogen that causes cold-water vibriosis in Norwegian rainbow trout and salmon, with a high degree of identity (≥90%) (8). In addition, the V. salmonicida 16S rRNA gene had the same distinctive 10-bp sequence gap that was found in the V. logei symbionts. Despite these similarities, the presence of three metabolic activities (lysine decarboxylation, nitrate reduction, and d-galactose fermentation) in all of the SA and SR symbiont strains eliminated the possibility that these bacteria were V. salmonicida (Table 2). A similar ambiguity resulting from the high degree of sequence identity between V. logei and V. salmonicida may have arisen in the work of Wiik et al. (35). They described a bacterial strain designated HI651 which, despite deviating metabolically and physiologically from the V. salmonicida type strain NCMB 2262, was designated V. salmonicida on the basis of a partial shared 16S rDNA sequence identity of 99.7%. Fox et al. (11) reported that closely related Bacillus strains could exhibit >99.5% sequence homology yet still belong to distinct species as determined by total DNA-DNA hybridization. Thus, because the 16S rRNA gene evolves slowly relative to the rest of the genome, a considerable degree of metabolic and ecological divergence can occur between two bacterial species whose 16S rDNA sequences do not differ significantly (11). We suggest that the metabolic and physiological traits reported for strain HI651 (35) are actually more consistent with those of V. logei. Because V. logei was not included in the phylogenetic analyses of Wiik et al. (35), this hypothesis remains to be tested.
Our taxonomic studies of Sepiola spp. symbionts also raise fundamental questions about the microecology and dynamics of their mixed-species light organ populations, as well as the evolution of light organ symbioses in general. Specifically, we have addressed the following three questions: (i) is the physiology of V. logei better adapted to cooler ambient conditions than is that of V. fischeri, and, thus, (ii) is temperature an important factor in determining which species of symbiont is predominant in Sepiola spp. light organs; and (iii) is there evidence that V. logei and V. fischeri symbionts of Sepiola spp. have evolved sets of colonization determinants different from those expressed by the native V. fischeri symbiont of E. scolopes?
The luminescence of ATCC strains of V. logei, as well as that of SA and SR V. logei strains, was significantly reduced at warmer temperatures (>20°C). Because luminescence in these strains could be restored by exposing them to decyl aldehyde, it is likely that either the expression of the genes responsible for intrinsic aldehyde synthesis (i.e., luxCDE homologs [23]) or the activity of their gene products is reduced up to 11-fold at warmer growth temperatures. Such a significant temperature dependency was previously observed in the psychrophilic marine bacterium strain WSU (20). While this strain initially was reported to be V. fischeri, its description more closely resembles that of the subsequently established species V. logei. Because strain WSU, as well as the V. logei symbionts of Sepiola, is more dependent on a low ambient temperature for luminescence than are V. fischeri strains (Fig. 2B), comparisons of the components of the lux operons of these two closely related species may provide insight into the physiological control of both the transcription of these genes and the enzymology of bacterial luminescence.
Another indication that the two species exhibit different environmental temperature optima is reflected in the relative colonization effectiveness of symbiotic V. logei and V. fischeri. V. logei strains from Sepiola spp. more often developed a successful symbiosis with a squid host at 21°C than at 26°C, while, in contrast, V. fischeri strains expressed a better capacity to colonize at the higher temperature (Table 4). Because the light organs of Sepiola spp. can contain a mixture of V. logei and V. fischeri (Table 1), symbiosis-competent strains of both species must be found in the Mediterranean Sea. We are tempted to conclude that when Sepiola spp. are infected by a mixture of the two Vibrio species, the identity of the predominant symbiont present in the light organ at any particular time may be determined in part by the ambient water temperature. This conclusion is supported by the observation that the environmental temperature of the Mediterranean Sepiola spp. typically varies between 10 and 22°C both with depth and over the course of the year (5, 25). Thus, one might predict that if the host were to inhabit waters with a temperature in the cooler range, the growth of V. logei would be favored over that of V. fischeri (Fig. 2A). Future experiments, using the native species of Sepiola rather than E. scolopes as the hosts, should serve to directly address this hypothesis.
Both E. scolopes and Sepiola spp. transmit bacterial symbionts horizontally to each new generation of squid (24, 34), and thus the newly hatched squid requires some mechanism(s) for distinguishing the correct symbiont strains from among the myriad other bacteria in seawater. Strains of V. fischeri isolated from the light organs of all Euprymna spp. tested can infect juvenile E. scolopes, suggesting that the symbiosis is not host species specific (24). Sepiola symbionts identified as V. fischeri were also able to colonize the light organ of juvenile E. scolopes, albeit in some cases to a lesser extent than the native symbiont of the squid (Table 4). The fact that V. logei symbionts from Sepiola spp. were capable of colonizing E. scolopes was unexpected because previous attempts either with V. logei strains ATCC 35077 and 15382 or with V. logei isolates from Southern California seawater had been unsuccessful (16, 21). These findings suggest that V. logei strains that have not been isolated from a light organ are missing one or more symbiotic determinants necessary for establishing this association. In addition, the inability of SA and SR V. logei strains to colonize the E. scolopes light organ with as high a success rate as symbiotic V. fischeri strains, particularly at 26°C, suggests that their symbiotic determinants (29) differ sufficiently from those present in the native E. scolopes symbiont to account for the differences in colonization competence (Table 4).
In summary, this study reports five new conclusions. (i) For the first time, V. logei has been demonstrated to be a light organ symbiont, namely of the Mediterranean bobtail squids S. robusta and S. affinis. (ii) The light organs of Sepiola spp. can contain a mixed colonization population consisting of a dominant species (V. logei) and a minor species (V. fischeri). (iii) The light organ symbioses of Mediterranean (Sepiola) and Pacific (Euprymna) squid species, while arising in a common sepiolid ancestor, have evolved independently to associate with different species of Vibrio, perhaps influenced by the environmental temperature of the host. (iv) Luminescence in V. logei is temperature dependent, due in part to an aldehyde substrate limitation. (v) V. logei and V. fischeri symbionts from Sepiola spp. generally exhibit diminished degrees of colonization competence compared to the native V. fischeri symbiont from E. scolopes.
ACKNOWLEDGMENTS
We thank Kyu-Ho Lee for bringing to our attention the PCR approach for differentiating Vibrio species and Michele Nishiguchi and Margaret McFall-Ngai for sharing unpublished data.
This research was supported by grants to E.G.R. from the Office of Naval Research (N00014-93-1-0846) and the National Institutes of Health (RR12294) and to M. McFall-Ngai and E.G.R. from the National Science Foundation (IBN96-01155).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Meyers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bang S S, Baumann P, Nealson K H. Phenotypic characterization of Photobacterium logei (sp. nov.), a species related to P. fischeri. Curr Microbiol. 1978;1:285–288. [Google Scholar]
- 3.Baumann P, Baumann L, Bang S S, Woolkalis M J. Reevaluation of the taxonomy of Vibrio, Beneckea, and Photobacterium: abolition of the genus Beneckea. Curr Microbiol. 1980;4:127–132. [Google Scholar]
- 4.Bello G. A key for the identification of the Mediterranean sepiolids (Mollusca: Cephalopoda) Bull Inst Oceanogr Monaco. 1995;16:41–55. [Google Scholar]
- 5.Bhaud M, Jacques G, Razouls C. Données météorologiques et hydrologiques de la région de Banyuls-sur-Mer, 1965–1966 (point côtier) Vie Milieu. 1967;18:137–151. [Google Scholar]
- 6.Boettcher K J, Ruby E G. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colwell R R. Proposal of a neotype, ATCC 15381, for Vibrio marinus (Russell) Ford 1927 and request for an opinion. Int Bull Bacteriol Nomencl Taxon. 1965;15:165–175. [Google Scholar]
- 7a.Difco Laboratories Inc. Difco manual. 10th ed. Detroit, Mich: Difco Laboratories, Inc.; 1984. [Google Scholar]
- 8.Egidius E, Wiik R, Andersen K, Hoff K A, Hjeltnes B. Vibrio salmonicida sp. nov., a new fish pathogen. Int J Syst Bacteriol. 1986;36:518–520. [Google Scholar]
- 9.Farmer J J, Hickman-Brenner F W. The genera Vibrio and Photobacterium. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1991. pp. 2952–3011. [Google Scholar]
- 10.Fitzgerald J M. Classification of luminous bacteria from the light organ of the Australian pinecone fish, Cleidopus gloriamaris. Arch Microbiol. 1977;112:153–156. [Google Scholar]
- 11.Fox G E, Wisotzkey J D, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 12.Herfurth A H. Beiträge zur kenntnis der bakteriensymbiose der Cephalopoden. Z Morphol Oekol Tiere. 1936;31:561–607. [Google Scholar]
- 13.Herring P J, Clarke M R, von Boletzky S, Ryan K P. The light organs of Sepiola atlantica and Spirula spirula (Mollusca: Cephalopoda): bacterial and intrinsic systems in the order Sepioidea. J Mar Biol Assoc U K. 1981;61:901–916. [Google Scholar]
- 14.Herring P J, Morin J G. Bioluminescence in fishes. In: Herring P J, Morin J G, editors. Bioluminescence in action. New York, N.Y: Academic Press; 1978. pp. 272–293. [Google Scholar]
- 15.Kita-Tsukamoto K, Oyaizu H, Nanba K, Simidu U. Phylogenetic relationships of marine bacteria, mainly members of the family Vibrionaceae, determined on the basis of 16S rRNA sequences. Int J Syst Bacteriol. 1993;43:8–19. doi: 10.1099/00207713-43-1-8. [DOI] [PubMed] [Google Scholar]
- 16.Lee, K.-H., and E. G. Ruby. Unpublished data.
- 17.Lee K-H, Ruby E G. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol. 1994;60:1565–1571. doi: 10.1128/aem.60.5.1565-1571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leisman G, Cohn D, Nealson K H. Bacterial origin of luminescence in marine animals. Science. 1980;208:1271–1273. doi: 10.1126/science.208.4449.1271. [DOI] [PubMed] [Google Scholar]
- 19.Lu C C, Guerra A, Palumbo F, Summers W C. Order Sepiodea Naef, 1916. Smithson Contrib Zool. 1992;513:21–36. [Google Scholar]
- 20.Makemson J C. Control of in vivo luminescence in psychrophilic marine photobacterium. Arch Mikrobiol. 1973;93:347–358. doi: 10.1007/BF00427930. [DOI] [PubMed] [Google Scholar]
- 21.McFall-Ngai M, Ruby E G. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–1493. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- 22.McFall-Ngai M J. Crypsis in the pelagic environment. Am Zool. 1990;30:175–188. [Google Scholar]
- 23.Nealson K H, Hastings J W. The luminous bacteria. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1991. pp. 625–639. [Google Scholar]
- 24.Nishiguchi, M., and M. J. McFall-Ngai. Unpublished data.
- 25.Panhouse M, Jacques G, Neveux J. Hydrologie dans la zone néritique de Banyuls-sur-Mer (Golfe du Lion), 1973. Vie Milieu. 1975;25:77–84. [Google Scholar]
- 26.Reichelt J L, Baumann P. Taxonomy of the marine, luminous bacteria. Arch Mikrobiol. 1973;94:283–330. doi: 10.1007/BF00424970. [DOI] [PubMed] [Google Scholar]
- 27.Reichelt J L, Baumann P, Baumann L. Study of genetic relationships among marine species of the genera Beneckea and Photobacterium by means of in vitro DNA/DNA hybridization. Arch Microbiol. 1976;110:101–120. doi: 10.1007/BF00416975. [DOI] [PubMed] [Google Scholar]
- 28.Reichelt J L, Nealson K H, Hastings J W. The specificity of symbiosis: pony fish and luminescent bacteria. Arch Microbiol. 1977;112:157–161. [Google Scholar]
- 29.Ruby E G. Lessons from a cooperative bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 30.Ruby E G, Asato L M. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- 31.Ruby E G, Nealson K H. Symbiotic associations of Photobacterium fischeri with the marine luminous fish Monocentris japonica: a model of symbiosis based on bacterial studies. Biol Bull. 1976;151:574–586. doi: 10.2307/1540507. [DOI] [PubMed] [Google Scholar]
- 32.Ruby E G, Nealson K H. Seasonal changes in the species composition of luminous bacteria in nearshore seawater. Limnol Oceanogr. 1978;23:530–533. [Google Scholar]
- 33.Soares C A G, Prado R C, Barros C B, Coelho A. Abstracts of the 95th General Meeting of the American Society for Microbiology 1995. Washington, D.C: American Society for Microbiology; 1995. PCR size analysis of 16S rRNA V1 region: a tool for Vibrio taxonomic studies, abstr. R-9; p. 480. [Google Scholar]
- 34.Wei S L, Young R E. Development of symbiotic bacterial luminescence in a nearshore cephalopod, Euprymna scolopes. Mar Biol. 1989;103:541–546. [Google Scholar]
- 35.Wiik R, Stackebrandt E, Valle O, Daae F L, Rødseth O M, Andersen K. Classification of fish-pathogenic vibrios based on comparative 16S rRNA analysis. Int J Syst Bacteriol. 1995;45:421–428. doi: 10.1099/00207713-45-3-421. [DOI] [PubMed] [Google Scholar]
- 36.Zablen L B. Ph.D. thesis. University of Illinois, Urbana; 1976. [Google Scholar]