Abstract
Predicting how biological communities respond to disturbance requires understanding the forces that govern their assembly. We propose using human skin piercings as a model system for studying community assembly after rapid environmental change. Local skin sterilization provides a ‘clean slate’ within the novel ecological niche created by the piercing. Stochastic assembly processes can dominate skin microbiomes due to the influence of environmental exposure on local dispersal, but deterministic processes might play a greater role within occluded skin piercings if piercing habitats impose strong selection pressures on colonizing species. Here we explore the human ear-piercing microbiome and demonstrate that community assembly is predominantly stochastic but becomes significantly more deterministic with time, producing increasingly diverse and ecologically complex communities. We also observed changes in two dominant and medically relevant antagonists (Cutibacterium acnes and Staphylococcus epidermidis), consistent with competitive exclusion induced by a transition from sebaceous to moist environments. By exploiting this common yet uniquely human practice, we show that skin piercings are not just culturally significant but also represent ecosystem engineering on the human body. The novel habitats and communities that skin piercings produce may provide general insights into biological responses to environmental disturbances with implications for both ecosystem and human health.
Keywords: 16S, bacteria, biodiversity, ecological niche, environmental change, skin microbiome
1. Introduction
How communities of coexisting species originate and are maintained is known as community assembly, and these processes determine which species thrive and which perish [1,2]. Similar ecological conditions across environments might result in community convergence because deterministic niche selection can promote analogous community profiles [3]. Community divergence may be driven by changing ecological pressures, but stochastic processes such as the order and timing of migration and random extirpation of populations can also play significant roles [4]. Initial colonizers may exert priority effects where the arrival of one species affects the subsequent colonization and/or establishment of a different species and produces historical contingency, in which chance effects can have long lasting consequences for community structure [4,5]. These priority effects can be pronounced during ecological succession as communities shift to a stable state after perturbation [6,7]. Understanding the mechanisms that underlie community assembly will ultimately lead to better predictions of community as well as individual species responses to environmental change.
Community assembly processes of human microbiomes have gained recent interest due to an increasing awareness of their ecological and medical significance [5,8,9]. Human microbiomes refer to the collective microorganisms and genes found within or on human beings [10]. Such microbiota consist largely of bacteria but also viruses, archaea, and microscopic eukaryotes like protists and fungi, and different communities inhabit various parts of the human body [10,11]. As the largest organ, the skin represents a diverse ecosystem of habitats for microbes that are constantly exposed to changing external conditions [12]. Although a dearth of nutrients results in the skin containing relatively low biodiversity [13], distinct core taxa can be found on ecologically dissimilar parts of the skin [14,15]. Two of the most common and abundant genera present on human skin include Cutibacterium (formerly Propionibacterium) and Staphylococcus, which dominate at sebaceous and moist sites, respectively [13,16]. Functionally, the human skin microbiome plays an important role in maintaining cutaneous health [17]. Pathogenic species such as Staphylococcus pyogenes, Staphylococcus aureus, and group A Streptococcus are known to cause skin infections while commensal species like Cutibacterium acnes and Staphylococcus epidermidis can protect from pathogens through regulation of the immune system [18]. Shifts in the human skin microbiome have been associated with many disease states like atopic dermatitis [19], psoriasis [20], acne vulgaris [21] and chronic wounds [22], but studies on the community dynamics of healthy skin microbiomes, especially after extrinsic perturbation, are less common (but see [23–26]). These studies show that many common activities such as using cosmetics and topical creams, sun exposure, direct contact sport, mineral bathing, and moving homes constitute rapid environmental disturbances for the human skin microbiome. These activities have potentially significant and unintentional impacts on microbial community assembly with differential ecological and evolutionary responses according to scale and functional contexts of specific taxonomic groups. Thus, the human skin microbiome is highly dynamic and the underlying assembly processes can have important health implications, such as in understanding ecological succession of microbial communities during wound recovery or dermatological disorders [27,28]. Community assembly processes of human skin microbiomes are also readily apparent due to relatively simple taxonomic compositions and short bacterial generation times.
One uniquely human activity that might affect the skin microbiome is skin piercing. Skin piercing has been present in human societies at least as far back as Ötzi the Iceman, who lived nearly 5000 years ago and was found to have pierced earlobes [29]. Skin piercings have been used to express both individual and group identities and served significant roles in traditional customs and rites of passage in various cultures around the world [30]. In addition to its anthropological and sociological importance, here we propose human skin piercings as a model for studying biological community assembly processes after rapid environmental change. Piercing practices typically begin with surface sterilization of the skin, which we hypothesize functions as a major environmental disturbance to the local skin microbiome. The piercing of the skin then reshapes the skin's physical topology, which is immediately followed by insertion of surgical stainless-steel studs for usually at least two weeks. This is expected to produce a novel ecological niche that differs from the previously unpierced skin in many ways such as surface area, temperature, acidity, humidity, and environmental exposure. This drastic environmental shift should fundamentally alter the ecological and evolutionary forces acting on the piercing microbiome.
Here, we hypothesize that human ear-piercing microbiomes (1) become more diverse and ecologically complex because the novel piercing environment offers increased protection, stability, and nutrients, (2) will exhibit less historical contingency because ecological succession will result in the deterministic assembly of an equilibrium community structure, and (3) reflect a transition of the skin environment from sebaceous to moist through increased moisture retention, resulting in a reduction of Cutibacterium and an increase in Staphylococcus. We tested these hypotheses by sampling the microbiomes of human ear-piercings over a two-week time period. Longitudinal samples of skin microbiomes from adjacent unsterilized and unpierced skin were collected simultaneously as controls for temporal variation. Other than previous clinical investigations of piercing infections, to our knowledge, this study represents the first investigation of the human piercing microbiome.
2. Methods
(a) . Human research ethics approval
Protocols for study participant recruitment, data security, sample collection, and associated procedures were approved by the McGill University Research Ethics Board Office (REB-1 no. 70-0617).
(b) . Sample collection
From October 2019 to March 2020, we recruited 28 individuals who were receiving earlobe piercings at Tattoo Lounge MTL in Montreal, Quebec, Canada and received their written, informed consent to participate in the study (electronic supplementary material, figure S3). Following standard ear-piercing protocols, we sterilized the earlobe skin area to be pierced with a benzalkonium chloride antiseptic towelette (Jedmon Products) immediately before piercing. We pierced earlobes using a sterilized bevelled hollow needle (Ruthless/Precision) and then inserted a 5/16 surgical steel grade (316L) piercing labret stud composed of chromium, nickel, and molybdenum. Both needle and stud were dipped in a water-based lubricant jelly (Personelle, Jean Coutu) to minimize friction and then cleaned off afterwards using a cotton-tipped swab. We collected skin swab samples using the DNA/RNA Shield Collection Tube w/Swab – DX (Zymo Research), which was used to preserve nucleic acids within samples at ambient temperatures. The piercer collected samples from the earlobe to be pierced and an adjacent unsterilized part of the ear farther up the ear but still part of the earlobe skin to serve as a temporal control. Samples were collected both before and after the piercing event (defined as a three-part process that includes (A) skin sterilization followed by (B) skin piercing and then (C) insertion of the metal stud). Study participants were then instructed to self-sample both the piercing and the adjacent skin control while wearing gloves over the following two weeks at specified timepoints: 12 h, 1 day, 3 days, one week, and two weeks. Additionally, environmental controls were collected by the piercer before the piercing and by the participant at the one- and two -week timepoints by waving a swab in the air for 30 s. In total, we collected 17 samples from each participant.
(c) . DNA extraction and amplicon sequencing
We extracted DNA from swabs using the DNeasy PowerSoil kit (QIAGEN) and then purified using the OneStep PCR Inhibitor Removal kit (Zymo Research). Skin swab samples and environmental controls were processed with a DNA extraction negative control included within each batch of 24 extractions. This work was carried out in a laboratory facility designed to handle low-copy and highly degraded environmental DNA samples through mitigation of contamination factors (e.g. no exposure to PCR products, regular deep cleaning, and strict usage protocols limited to trained personnel). The V1–V3 region of the 16S rRNA gene was PCR amplified using the primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 518R (5′-ATTACCGCGGCTGCTGG-3′) [31]. Library preparation, quality control, and high throughput sequencing with Illumina MiSeq v2/v3 were conducted at Génome Québec and the McGill Genome Centre (Montreal, Quebec, Canada).
(d) . Data processing
We processed raw sequences using the QIIME2 bioinformatics pipeline [32]. We trimmed primer sequences using cutadapt before generating amplicon sequence variants (ASVs) using DADA2 [33]. We identified contaminant ASVs using environmental and DNA extraction negative controls for each sequencing batch with the prevalence-based method at a classification threshold of p* = 0.5 within decontam [34]. We considered the unpierced control of each individual to be experimentally valid only if it exhibited no significant differences from the microbiome of the skin to be pierced prior to piercing. Thus, we defined statistical outlier individuals as having an absolute difference in ASV richness between sample and control prior to piercing that was greater than 1.5 times the interquartile range across all individuals [35]. We removed a total of 1047 contaminant ASVs and two statistical outlier individuals resulting in 10 915 ASVs across 392 samples with a mean sequencing depth of 27 817 reads per sample. We aligned ASVs using MAFFT and built phylogenetic trees using FastTree 2 based on Jukes–Cantor distances. For taxonomic assignment, we used the 27F/518R 16S rRNA primers to in silico extract the target V1–V3 amplicon from the SILVA 132 database [36]. We trained a naïve Bayes classifier using scikit-learn on the extracted database and then used it to taxonomically assign ASVs from domain down to species. We accepted assignments if classification confidence was at least 0.7 [37].
(e) . Statistical analyses
We normalized ASV counts via total sum scaling (TSS), and calculated biodiversity indices, principal coordinate analysis (PCoA), and PERMANOVA (999 permutations) using the R ‘phyloseq’ and ‘vegan’ packages implemented within MicrobiomeAnalyst 2.0 [38–40]. We did not rarefy data to maximize the amount of data analysed and the number of participants included in the study [41]. We measured alpha and beta diversities using Chao1 and Bray–Curtis dissimilarity, respectively. We calculated betadisper separately using the R ‘vegan’ package version 2.6-2 and used ‘ggstatsplot’ version 0.10.0 for plotting within RStudio Desktop version 2022.12.0 + 353 and R version 4.2.2 [39,42,43]. We built ASV co-occurrence networks using random matrix theory (RMT)-based Spearman's rank correlation through the Molecular Ecological Network Analysis (MENA) pipeline implemented within the Integrated Network Analysis Pipeline (iNAP) [44]. We first filtered data by retaining only ASVs present in greater than 15% of samples and then log transformed the filtered data before calculating similarity matrices allowing a single timepoint lag for time-dependent interactions. We visualized co-occurrence networks using Cytoscape version 3.9.1 keeping only nodes with valid genus-level taxonomic assignments and edges with a p-value < 0.05. We used the ‘iCAMP’ R package v. 1.5.12 [45] to calculate pNST [46] and infer community assembly mechanisms by phylogenetic bin-based null model analysis. We used bootstrapping tests with a resampling size of 1000 to assess significant pairwise differences between timepoints. We classified core microbiome community taxa based on a minimum of 5% relative abundance across at least 50% of all samples.
3. Results and discussion
(a) . Human piercings increase diversity and ecological complexity
Sudden events that cause drastic environmental change for human skin can lead to fundamental shifts in skin microbiomes. For example, human birth involves moving from an environment that is liquid and mostly sterile to one exposed to air and microbial colonization, which contributes to increased skin microbiome diversity and differentiation for human infants through their first year of life [47]. Analogously, we found that skin piercings were strongly associated with a significant increase in ASV richness (i.e. number of unique DNA sequences) at the piercing site over two weeks (figure 1a). By contrast, ASV richness of the unpierced controls remained stable over the same time period (figure 1b). Skin piercings likely represent the creation of hospitable niches for certain bacteria that thrive in areas of greater occlusion, moisture, and nutrient retention. Occlusion increases skin pH and produces moisture through transepidermal water loss, which supports bacterial growth and survival [48]. Piercing studs may physically trap and accumulate debris including sweat, sebum, and pieces of stratum corneum that serve as primary nutrient sources for most human skin microbiome members [49]. Undisturbed occluded skin microbiomes also exhibit the greatest longitudinal stability, presumably due to physical protection from external perturbation [14].
Figure 1.

Human piercings affect skin microbiome diversities. Alpha diversity of the (a) piercing microbiome increased significantly by two weeks after piercing, but (b) unpierced controls did not. Large red dots indicate medians, box and whiskers show the minimum, maximum, median, and 25th and 75th percentiles, and violin plots represent probability densities. Pairwise Wilcoxon signed-rank test V statistics and p-values are shown above each plot. (c) PCoA of Bray–Curtis dissimilarities before piercing (teal) and two weeks after (rose) show a significant increase in dispersion over time (betadisper, ANOVA, F = 4.9053, p = 0.03101). (d) No significant changes were observed in the unpierced controls (betadisper, ANOVA, F = 0.0189, p = 0.8911; PERMANOVA, F = 1.0692, p = 0.321).
Piercings were also associated with a significant increase in dispersion of beta diversity (i.e. increased dissimilarity in community composition between piercing microbiomes) by two weeks after piercing (figure 1c) whereas beta diversity did not change between timepoints in the unpierced controls (figure 1d). Thus, the piercing environment may have caused greater variance in community structure over time, which is consistent with the increase in alpha diversity and supports the idea that piercings produce novel ecological niches. The greatest source of temporal variation in beta diversity of both piercing microbiomes and unpierced controls, however, was differences between individuals (electronic supplementary material, Appendix, figure S1). Metadata of study participants collected through questionnaire surveys revealed no significant relationships between the skin piercing microbiome and host factors and behaviors including hygiene, travel, and physical activity (electronic supplementary material, Appendix, Methods). However, some behavioural differences across individuals, such as heterogeneity during self-sampling (e.g. pressure applied to swabs), may not have been captured by our questionnaire and these effects may have masked the importance of more nuanced factors on the skin piercing microbiome.
Because beta diversity did not change significantly until the two-week timepoint, the two-week sampling period of this study may have been insufficient to fully capture the ecological succession process. Human skin microbiome communities can be surprisingly stable even at highly exposed and perturbed body sites like the face and palm as well as in the long-term for up to two years [50,51]. Other recent longitudinal studies on chronic and acute skin disturbances such as diabetic foot ulcers [52], burn wounds [53], and chlorhexidine disinfectants [54] have demonstrated that post-disturbance community structure remains quite stable from 3 to 56 days later despite other significant ecological impacts.
Network analyses revealed co-occurrence and exclusion patterns of human skin microbiomes driven largely by body sites representing distinct microbial habitats [55]. Environmental factors (e.g. elevation [56] and urban living [57]), skin physiology (e.g. ageing [58] and skin sensitivity [59]), and skin products (e.g. lotion [60]) can also affect various properties of skin microbiome network topology. Correlational analyses have been widely used to infer real-world biotic interactions from amplicon sequencing data but suffer from producing spurious and indirect associations, especially for rare ASVs in zero-inflated data typical of skin microbiomes [61–63]. Time-series experiments can help address these issues by making it possible to infer directionality and time-dependency of interactions, which are often asymmetrical [64]. To examine how piercings may have impacted the ecological interactions within skin microbiomes, we constructed co-occurrence networks via the MENA pipeline. Although there was little difference in the absolute number of nodes between piercing and unpierced networks, the number of edges was consistently higher in the piercing network, suggesting a greater number of biotic interactions among microbiome members (figure 2a and b). This increase in ecological complexity is potentially associated with more available resources [65], which we predicted to occur due to accumulation of nutrients in the occluded piercing environment. The relationship between ecological complexity and resilience to environmental disturbance can be either positive [66] or negative [67] depending on the interdependency of interactions, and complexity can have significant implications for microbial ecosystem functioning [68]. Ecological complexity is also sensitive to shifting selection pressures [69], which is further evidence suggesting that the piercing environment represents a novel ecological niche.
Figure 2.

Piercing microbiomes exhibit a greater proportion of positive and direct ecological interactions. Molecular ecological networks of the (a) piercing microbiome (orange) contained 35 nodes and 121 edges, and (b) unpierced controls (blue) contained 37 nodes and 90 edges. Each node represents an individual ASV labelled by its genus identity (only nodes with genus identities shown), and edges represent correlation-inferred interactions. Concurrent interactions are shown as solid lines and time-dependent interactions are shown as dotted lines. The piercing and unpierced networks contain 69% and 79% time-dependent interactions, respectively. Positive interactions are coloured in green and negative interactions are coloured in red. The positive-to-negative links (P/N) ratio is 1.42 for the piercing network and 0.88 for the unpierced network.
(b) . Stochasticity and determinism during community assembly in piercing microbiomes
To directly assess the relative contribution of deterministic and stochastic processes in the community assembly of the piercing microbiome, we used community assembly mechanisms by phylogenetic-bin-based null model analysis (iCAMP). iCAMP employs the beta net relatedness index (βNRI) and taxonomic beta diversities with the modified Raup–Crick (RC) metric to partition deterministic processes into either heterogeneous selection or homogeneous selection, and stochastic processes into homogenizing dispersal, dispersal limitation, or drift [45]. In contrast to heterogeneous selection, homogeneous selection occurs when environmental conditions are stable and consistently exerting similar selection pressures over space and/or time [70]. Homogeneous selection typically leads to greater phylogenetic relatedness because related communities are often ecologically similar whereas heterogeneous selection produces greater phylogenetic dissimilarity. The iCAMP analysis indicated that stochasticity was dominant in both piercing microbiomes and unpierced controls (figure 3a), specifically through dispersal limitation (relative importance of 73.1% in piercing, 76.8% in unpierced) with minimal contributions from drift (0.25% in piercing, 0.43% in unpierced) and none from homogenizing dispersal (figure 3b). Deterministic assembly processes were largely accounted for by homogeneous selection (24.3% in piercing, 19.8% in unpierced) with minor contributions from heterogeneous selection (2.3% in piercing, 2.9% in unpierced) (figure 3b). We found that two weeks after piercing, the relative contribution of dispersal limitation decreases while homogeneous selection increases (figure 3b). Stochasticity between the to-be pierced and unpierced control skin differed, with high variation observed in both. This may suggest substantial variation in the proportion of stochastic versus deterministic assembly processes even at very short distances between adjacent skin of the same body part. However, alpha diversity between pierced samples and unpierced controls does exhibit strong correspondence, providing support for the validity of using adjacent unpierced skin as controls (electronic supplementary material, Appendix, figure S1A and C). The dominance of stochasticity and its decrease over time in piercing microbiomes was also supported by phylogenetic normalized stochasticity ratio (pNST) analyses (electronic supplementary material, Appendix, figure S2), which is based on beta mean nearest taxon distance (βMNTD) [46]. These results suggest that community assembly of the piercing microbiome becomes more deterministic with time, consistent with the hypothesis that piercings produce a novel yet consistent and stable microhabitat that leads to homogeneous selection pressures.
Figure 3.
Community assembly of piercing microbiomes became more deterministic. (a) Relative contribution of stochastic assembly processes between piercing microbiomes (orange) and unpierced controls (blue). Stochasticity significantly decreased by two weeks after piercing (pairwise bootstrap, p = 0.0352). Box and whiskers show the minimum, maximum, median and 25th and 75th percentiles. (b) Relative contribution of deterministic (open markers) and stochastic (closed markers) processes to community assembly in piercing microbiomes (orange) and unpierced controls (blue). Deterministic processes include homogeneous selection (open square) and heterogeneous selection (open diamond), and stochastic processes include dispersal limitation (solid circle), homogenizing dispersal (solid square), and drift (solid triangle). Error bars indicate standard deviations around the mean.
To better understand the difference in temporal dynamics between piercing microbiomes and unpierced controls, we next explored time-lagged correlations within ecological networks, which can be indicative of time-dependent interactions such as priority effects. Both piercing microbiomes and unpierced controls were comprised of significantly more time-dependent interactions, with 69% in the piercing (one-sided one-proportion Z test, p = 9.66×10−6) and 79% in the unpierced (one-sided one-proportion Z test, p = 2.11 × 10−8) networks (figure 2a and b). The lower proportion of time-dependent interactions in the piercing network could be caused by an increase in deterministic selection forces (e.g. environmental filtering) of the newly created environmental niches within piercings. An increase in determinism reduces the relative importance of stochastic processes like historically contingent dispersal [71], although the difference in proportions between piercing and unpierced networks was insignificant (one-sided two-proportion Z test, Z = −1.54, p = 0.062, 95% CI [−1, 0.004]). Another potential and non-mutually exclusive explanation could be that environmental disturbance from piercings compresses the spatio-temporal niche of the microbiome by increasing species abundances which leads to greater species interactions that are not time-lagged. Evidence for this mechanism was recently discovered when anthropogenic landscape modification was found to increase co-occurrence of wildlife species [72].
Network correlations can be either positive or negative reflecting the nature of potential ecological interactions. The positive-to-negative links (P/N) ratio has been proposed as a marker for differentiating healthy and diseased microbiome networks by detecting shifts in the balance between facilitative and inhibitive interactions [73]. Here, the P/N ratio was able to distinguish piercing (P/N = 1.42) from unpierced (P/N = 0.88) networks (figure 2a and b), with more positive than negative edges in the piercing network, whereas the opposite was true of the unpierced network. The proportion of positive edges was significantly greater in the piercing than unpierced network (one-sided two-proportion Z test, p = 0.0418, 95% CI [0.007, 1]). Positive network associations may represent facultative and obligatory commensalisms or mutualisms between taxa, but they can also reflect the co-occurrence of taxa with high niche overlap that are ecologically or functionally similar (i.e. environmental filtering) [74]. There is evidence that, during secondary succession (i.e. post-disturbance recolonization), a general shift to positive interactions may help a community respond to environmental stress through neighbourhood habitat amelioration, where one species changes the environment in a way that facilitates the growth and survival of another species [75]. Positive biotic interactions and environmental filtering are not mutually exclusive because positively interacting taxa that share similar niches would increase positive network associations through both mechanisms. Regardless, both are deterministic processes [76]. Thus, contrasting P/N ratios of piercing and unpierced networks suggests that piercings are strongly associated with a deterministic ecological shift for the local skin microbiome.
(c) . Piercings cause a shift towards moist skin microbiomes
While piercing infections are common medical complications [77] and a variety of specific pathologies have been identified [78,79], the community composition and temporal dynamics of uninfected human piercing microbiomes have yet to be characterized. Because piercings can potentially trap moisture by mitigating evaporation, we predicted that the piercing microbiome should develop to resemble skin microbiomes found in moist areas such as the nose, armpit, or groin. We found that the two-week phylum-level community composition of the piercing microbiome was dominated by Actinobacteriota (Actinomycetota) and Firmicutes (Bacillota), followed by Proteobacteria (Pseudomonadota) with relatively few Bacteroidota (Bacteroidetes). Actinobacteriota was largely represented by the families Propionibacteriaceae and Corynebacteriaceae, specifically the genera Cutibacterium and Corynebacterium, respectively. Firmicutes was mainly comprised of Staphylococcaceae and Streptococcaceae at the family level and Staphyloccocus and Streptococcus at the genus level, respectively. Although we could not assign a species identity to a large proportion of ASVs, just two species, namely Cutibacterium acnes and Staphylococcus epidermidis, emerged as core taxa of the piercing microbiome given their relative abundance and wide prevalence (figure 4a). These two species encompassed more than half of the community in 58% of our samples, with an average of 44% C. acnes and 8.6% S. epidermidis. Corynebacterium was the third most dominant genus at 6.6%, but the most prominent Corynebacterium ASV could not be classified to species-level and all genus-level Corynebacterium ASVs failed to meet the core taxa criteria. Following C. acnes and S. epidermidis across time confirms that they experience dramatic longitudinal shifts in the expected directions: a significant decrease in the relative abundance of C. acnes and a significant increase in relative abundance of S. epidermidis (figure 4b and c). These findings are consistent with a moist piercing environment because Cutibacterium species are known to be dominant members of sebaceous skin microbiomes, including specifically the external auditory canal, while Staphylococcus is mainly associated with moist skin [80]. These significant longitudinal changes in C. acnes and S. epidermidis were not observed in the unpierced controls (figure 4d and e).
Figure 4.
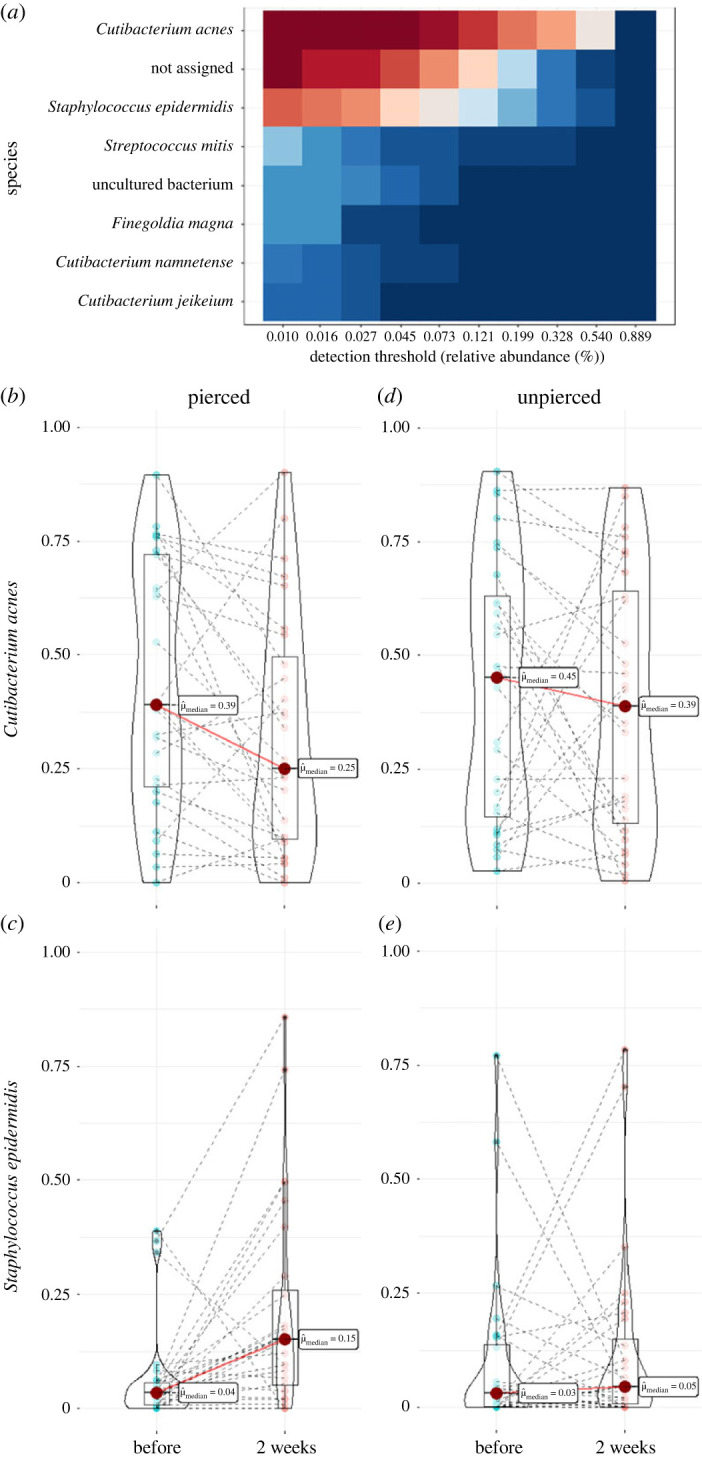
Two species, Cutibacterium acnes and Staphyloccocus epidermidis, dominate the piercing microbiome and shift in opposite directions over time in terms of prevalence and relative abundance. (a) Species-level heat maps of piercing microbiomes at the two-week timepoint. Red and teal respectively represent more and less prevalence at increasing relative abundance thresholds. (b) C. acnes decreased significantly in relative abundance (Wilcoxon signed-rank test, V = 309, p = 0.02), and (c) S. epidermidis increased significantly in relative abundance (Wilcoxon signed-ranked test, V = 66, p = 1.88 × 10−3) in the piercing microbiome two weeks after piercing. No significant changes in relative abundance were observed for (d) C. acnes (Wilcoxon signed-rank test, V = 234, p = 0.49) or (e) S. epidermidis (Wilcoxon signed-rank test, V = 159, p = 0.48) in unpierced controls by the two-week timepoint. Large red dots indicate medians, box and whiskers show the minimum, maximum, median and 25th and 75th percentiles, and violin plots represent probability densities.
Beyond associations with distinct skin ecologies, the two core taxa, C. acnes and S. epidermidis, are well-known commensals and opportunistic pathogens of human skin as well as direct antagonists. Both C. acnes and S. epidermidis are common members of skin microbiomes that help maintain skin homeostasis through competitive exclusion of potential pathogens, production of antibacterial bacteriocins, and priming of the skin's innate Toll-like receptor (TLR) immune system [18,81]. Against each other, however, they compete using a variety of methods including the production of antimicrobial short chain fatty acids [82], bacteriocins and polymorphic toxins [83], and electricity [84]. The strong antagonism between C. acnes and S. epidermidis may help explain the observed change in the piercing microbiome. If the novel piercing environment directly affects a single species, through either greater selection against C. acnes or increased relative fitness of S. epidermidis, it should induce an opposite trajectory of the corresponding species. Numerous skin diseases like acnes, dermatitis, rosacea, and psoriasis have been associated with lower relative abundance and/or a loss in taxonomic diversity of Cutibacterium [85]. However, over-colonization of C. acnes can lead to microcomedone formation and acne if S. epidermidis fails to control its proliferation [86]. An imbalance between C. acnes and S. epidermidis has also been shown to activate skin inflammation through the production of cytokines [87]. While both species are known opportunistic pathogens given the right environmental context [88], S. epidermidis represents the most common source of infections on indwelling medical devices such as central venous catheters and joint prostheses [89]. A major similarity between internal medical implants and external skin piercings is the insertion of foreign metal objects into the human body, which involves direct contact with the microbiota living in and on human skin. We hypothesize that the novel piercing ecological niche is more advantageous to S. epidermidis due to increased moisture, decreased sebum, and/or the new metal surface area that may support biofilm growth. The growing population of S. epidermidis can then reduce C. acnes abundance through antagonistic interactions, but further studies will be needed to confirm this hypothesis. Such ecological relationships between specific dominant species could potentially be exploited to inform pre- and probiotic treatments to prevent and control skin infections through competition or direct antagonisms with pathogenic microbiota [90]. Thus, skin piercings may serve as a model for understanding environmental disturbances by shedding light on the ecological dynamics of specific, medically relevant species.
(d) . Piercings as a model for studying biological responses to environmental change
Our study provides the first glimpse into the bacterial communities inhabiting human ear-piercings. We show that the piercing process—skin sterilization, piercing of the skin, and insertion of a metal stud—has a demonstrable impact on the ecology of the local skin microbiome. Despite sterilization serving as a major environmental disturbance that kills many resident species, we found that, over time, the new piercing environment was significantly associated with greater biodiversity and ecological complexity, with fundamental differences in the nature of biotic interactions compared to exposed earlobe skin. The assembly of piercing microbiomes, however, remained dominated by stochastic dispersal typical of other skin microbiomes. Piercing microbiomes did not converge towards a single community structure but rather composition varied widely across individuals. Despite this, deterministic homogeneous selection did become more important with time, indicating some level of environmental filtering in the piercing environment. Piercing microbiomes showed less historical contingency than unpierced controls, consistent with greater contemporary selection. Similar to the microbiome of belly buttons [91], piercing microbiome communities are diverse but contain a few predictably dominant taxa. Specifically, we identified two major species, C. acnes and S. epidermidis, that show a change consistent with their known competitive antagonism and habitat associations, suggesting that piercings are moist environments. Studying how these two medically significant core species respond to rapidly changing environmental conditions within their natural communities may provide novel avenues to understanding their pathogenicity, of which interspecies interactions are known to play a major role [92,93]. Ecological disturbance experiments in natural ecosystems have traditionally been labour-intensive and difficult to replicate [94]. By significantly altering the composition and ecology of the resident human microbiome, skin piercings could serve as a model for insights into the response of microbiomes to environmental disturbance as well as community assembly processes more generally. As human beings, we have practised the art of skin piercing for cultural, religious, and personal expression across diverse societies for thousands of years. Here we reveal that skin piercings also represent a form of ecosystem self-engineering of the ecological landscape that is the human skin.
Acknowledgements
We thank R. Odgers for assistance with sample collection, and M. Farrell and A. Kamino for helping to coordinate the study at Tattoo Lounge MTL. We also thank S. Nason for designing recruitment materials, L. Bennett for laboratory work, Å. Lind for logistics, and J. Shapiro, F.-J. Lapointe, and A.C. Gerstein for helpful feedback.
Contributor Information
Charles C. Y. Xu, Email: charles.cong.xu@gmail.com.
Rowan D. H. Barrett, Email: rowan.barrett@mcgill.ca.
Ethics
Protocols for study participant recruitment, data security, sample collection, and associated procedures were approved by the McGill University Research Ethics Board Office (REB-1 no. 70-0617).
Data accessibility
Data and materials availability: raw sequencing data are available on NCBI Sequence Read Archive under BioProject PRJNA956301. Data and code are available on Zenodo: https://doi.org/10.5281/zenodo.7832626 [95].
Supplementary material is available online [96].
Declaration of AI use
We have not used AI-assisted technologies in creating this article.
Authors' contributions
C.C.Y.X.: conceptualization, data curation, formal analysis, investigation, methodology, project administration, supervision, validation, visualization, writing—original draft, writing—review and editing; J.L.: data curation, investigation, validation; A.A.: data curation, methodology, validation; É.M.W.: investigation, methodology, resources, supervision, validation; R.D.H.B.: conceptualization, data curation, funding acquisition, methodology, project administration, resources, supervision, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
The authors declare that they have no competing interests.
Funding
C.C.Y.X. was partially funded by a Vanier Canada Graduate Scholarship. This work was funded by the Natural Sciences and Engineering Research Council of Canada (RGPIN-2019-04549); and a Canada Research Chair awarded to R.D.H.B.
References
- 1.Nemergut DR, et al. 2013. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342-356. ( 10.1128/MMBR.00051-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fukami T. 2009. Community assembly dynamics in space. In Community ecology: processes, models and applications (eds Verhoef HA, Morin PJ), pp. 45-54. Oxford, UK: Oxford University Press. [Google Scholar]
- 3.Clements FE. 1936. Nature and structure of the climax. J. Ecol. 24, 252-284. ( 10.2307/2256278) [DOI] [Google Scholar]
- 4.Fukami T. 2015. Historical contingency in community assembly: integrating niches, species pools, and priority effects. Annu. Rev. Ecol. Evol. Syst. 46, 1-23. ( 10.1146/annurev-ecolsys-110411-160340) [DOI] [Google Scholar]
- 5.Debray R, Herbert RA, Jaffe AL, Crits-Christoph A, Power ME, Koskella B. 2022. Priority effects in microbiome assembly. Nat. Rev. Microbiol. 20, 109-121. ( 10.1038/s41579-021-00604-w) [DOI] [PubMed] [Google Scholar]
- 6.Zhou J, et al. 2014. Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc. Natl Acad. Sci. USA 111, e836-e845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang C, HilleRisLambers J. 2016. Integrating succession and community assembly perspectives. F1000Res. 5, 2294. ( 10.12688/f1000research.8973.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martino C, Dilmore AH, Burcham ZM, Metcalf JL, Jeste D, Knight R. et al. 2022. Microbiota succession throughout life from the cradle to the grave. Nat. Rev. Microbiol. 20, 707-720. ( 10.1038/s41579-022-00768-z) [DOI] [PubMed] [Google Scholar]
- 9.Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK. 2020. Plant–microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607-621. ( 10.1038/s41579-020-0412-1) [DOI] [PubMed] [Google Scholar]
- 10.Marchesi JR, Ravel J. 2015. The vocabulary of microbiome research: a proposal. Microbiome 3, 1-3. ( 10.1186/s40168-015-0094-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. 2018. Current understanding of the human microbiome. Nat. Med. 24, 392-400. ( 10.1038/nm.4517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grice EA, Segre JA. 2011. The skin microbiome. Nat. Rev. Microbiol. 9, 244-253. ( 10.1038/nrmicro2537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byrd AL, Belkaid Y, Segre JA. 2018. The human skin microbiome. Nat. Rev. Microbiol. 16, 143-155. ( 10.1038/nrmicro.2017.157) [DOI] [PubMed] [Google Scholar]
- 14.Grice EA, et al. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190-1192. ( 10.1126/science.1171700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326, 1694-1697. ( 10.1126/science.1177486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong HH, Segre JA. 2012. Skin microbiome: looking back to move forward. J. Invest. Dermatol. 132, 933-939. ( 10.1038/jid.2011.417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schommer NN, Gallo RL. 2013. Structure and function of the human skin microbiome. Trends Microbiol. 21, 660-668. ( 10.1016/j.tim.2013.10.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanford JA, Gallo RL. 2013. Functions of the skin microbiota in health and disease. Semin. Immunol. 25, 370-377. ( 10.1016/j.smim.2013.09.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakatsuji T, Gallo RL. 2019. The role of the skin microbiome in atopic dermatitis. Ann. Allergy Asthma Immunol. 122, 263-269. ( 10.1016/j.anai.2018.12.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tett A, et al. 2017. Unexplored diversity and strain-level structure of the skin microbiome associated with psoriasis. npj Biofilms Microbiomes 3, 14. ( 10.1038/s41522-017-0022-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu H, Li H. 2019. Acne, the skin microbiome, and antibiotic treatment. Am. J. Clin. Dermatol. 20, 335-344. ( 10.1007/s40257-018-00417-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomic-Canic M, Burgess JL, O'Neill KE, Strbo N, Pastar I. 2020. Skin microbiota and its interplay with wound healing. Am. J. Clin. Dermatol. 21, 36-43. ( 10.1007/s40257-020-00536-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meadow JF, Bateman AC, Herkert KM, O'Connor TK, Green JL. 2013. Significant changes in the skin microbiome mediated by the sport of roller derby. PeerJ 1, e53. ( 10.7717/peerj.53) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moskovicz V, Gross A, Mizrahi B. 2020. Extrinsic factors shaping the skin microbiome. Microorganisms 8, 1023. ( 10.3390/microorganisms8071023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brandwein M, et al. 2018. Temporal stability of the healthy human skin microbiome following dead sea climatotherapy. Acta Derm. Venerol. 98, 256-261. ( 10.2340/00015555-2769) [DOI] [PubMed] [Google Scholar]
- 26.Lax S, et al. 2014. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048-1052. ( 10.1126/science.1254529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musthaq S, Mazuy A, Jakus J. 2018. The microbiome in dermatology. Clin. Dermatol. 36, 390-398. ( 10.1016/j.clindermatol.2018.03.012) [DOI] [PubMed] [Google Scholar]
- 28.White EK, Grice EA. 2023. The wound microbiome. Cold Spring Harb. Perspect. Biol. 15, a041218. ( 10.1101/cshperspect.a041218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rauf D. 2019. The culture of body piercing, 1st edn. New York, NY: Rosen Publishing. [Google Scholar]
- 30.Rush JA. 2005. Spiritual tattoo: a cultural history of tattooing, piercing, scarification, branding, and implants. Berkeley, CA: Frog, Ltd. Distributed by North Atlantic Books. [Google Scholar]
- 31.Meisel JS, Hannigan GD, Tyldsley AS, Sanmiguel AJ, Hodkinson BP, Zheng Q, Grice EA. 2016. Skin microbiome surveys are strongly influenced by experimental design. J. Invest. Dermatol. 136, 947-956. ( 10.1016/j.jid.2016.01.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolyen E, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852-857. ( 10.1038/s41587-019-0209-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Callahan BJ, Mcmurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581-583. ( 10.1038/nmeth.3869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. 2018. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 1-4. ( 10.1186/s40168-018-0605-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rousseeuw PJ, Hubert M. 2011. Robust statistics for outlier detection. WIREs Data Mining Knowl. Discov. 1, 73-79. ( 10.1002/widm.2) [DOI] [Google Scholar]
- 36.Yilmaz P, et al. 2014. The SILVA and ‘All-species Living Tree Project (LTP)’ taxonomic frameworks. Nucl. Acids Res. 42, D643-D648. ( 10.1093/nar/gkt1209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziemski M, Wisanwanichthan T, Bokulich NA, Kaehler BD. 2021. Beating naive Bayes at taxonomic classification of 16S rRNA gene sequences. Front. Microbiol. 12, 644487. ( 10.3389/fmicb.2021.644487) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chong J, Liu P, Zhou G, Xia J. 2020. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 15, 799-821. ( 10.1038/s41596-019-0264-1) [DOI] [PubMed] [Google Scholar]
- 39.Oksanen J, et al. 2023. vegan: Community ecology package. R package version 2.6-4. See https://CRAN.R-project.org/package=vegan. [Google Scholar]
- 40.McMurdie PJ, Holme S. 2013. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217. ( 10.1371/journal.pone.0061217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 10, e1003531. ( 10.1371/journal.pcbi.1003531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Posit Team. 2023. RStudio: integrated development environment for R. Boston, MA: Posit Software, PBC. See http://www.posit.co/. [Google Scholar]
- 43.R Core Team. 2023. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. See https://www.R-project.org/. [Google Scholar]
- 44.Feng K, et al. 2022. iNAP: an integrated network analysis pipeline for microbiome studies. iMeta 1, e13. ( 10.1002/imt2.13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ning D, et al. 2020. A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nat. Commun. 11, 4717. ( 10.1038/s41467-020-18560-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ning D, Deng Y, Tiedje JM, Zhou J. 2019. A general framework for quantitatively assessing ecological stochasticity. Proc. Natl Acad. Sci. USA 116, 16 892-16 898. ( 10.1073/pnas.1904623116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Capone KA, Dowd SE, Stamatas GN, Nikolovski J. 2011. Diversity of the human skin microbiome early in life. J. Invest. Dermatol. 131, 2026-2032. ( 10.1038/jid.2011.168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aly R, Shirley C, Cunico B, Maibach HI. 1978. Effect of prolonged occlusion on the microbial flora, pH, carbon dioxide and transepidermal water loss on human skin. J. Invest. Dermatol. 71, 378-381. ( 10.1111/1523-1747.ep12556778) [DOI] [PubMed] [Google Scholar]
- 49.Scharschmidt TC, Fischbach MA. 2013. What lives on our skin: ecology, genomics and therapeutic opportunities of the skin microbiome. Drug Discov. Today Dis. Mech. 10, e83-e89. ( 10.1016/j.ddmec.2012.12.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oh J, Byrd AL, Park M, Kong HH, Segre JA. 2016. Temporal stability of the human skin microbiome. Cell 165, 854-866. ( 10.1016/j.cell.2016.04.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hillebrand GG, Dimitriu P, Malik K, Park Y, Qu D, Mohn WW, Kong R. 2021. Temporal variation of the facial skin microbiome: a 2-year longitudinal study in healthy adults. Plast. Reconstr. Surg. 147, 50S-61S. ( 10.1097/PRS.0000000000007621) [DOI] [PubMed] [Google Scholar]
- 52.MacDonald A, Brodell JD, Daiss JL, Schwarz EM, Oh I. 2019. Evidence of differential microbiomes in healing versus non-healing diabetic foot ulcers prior to and following foot salvage therapy. J. Orthop. Res. 37, 1596-1603. ( 10.1002/jor.24279) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lima KM, Davis RR, Liu SY, Greenhalgh DG, Tran NK. 2021. Longitudinal profiling of the burn patient cutaneous and gastrointestinal microbiota: a pilot study. Sci. Rep. 11, 10667. ( 10.1038/s41598-021-89822-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mougeot J-LC, Beckman MF, Bahrani Mougeot F, Horton JM. 2022. Cutaneous microbiome profiles following chlorhexidine treatment in a 72-hour daily follow-up paired design: a pilot study. Microbiol. Spectr. 10, e01753-21. ( 10.1128/spectrum.01753-21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C. 2012. Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol. 8, e1002606. ( 10.1371/journal.pcbi.1002606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, et al. 2019. Elevation is associated with human skin microbiomes. Microorganisms 7, 611. ( 10.3390/microorganisms7120611) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim H-J, et al. 2018. Fragile skin microbiomes in megacities are assembled by a predominantly niche-based process. Sci. Adv. 4, e1701581. ( 10.1126/sciadv.1701581) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim H-J, et al. 2019. Segregation of age-related skin microbiome characteristics by functionality. Sci. Rep. 9, 16748. ( 10.1038/s41598-019-53266-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keum HL, Kim H, Kim H-J, Park T, Kim S, An S, Sul WJ. 2020. Structures of the skin microbiome and mycobiome depending on skin sensitivity. Microorganisms 8, 1032. ( 10.3390/microorganisms8071032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murphy B, et al. 2022. Alteration of barrier properties, stratum corneum ceramides and microbiome composition in response to lotion application on cosmetic dry skin. Sci. Rep. 12, 5223. ( 10.1038/s41598-022-09231-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carr A, Diener C, Baliga NS, Gibbons SM. 2019. Use and abuse of correlation analyses in microbial ecology. ISME J. 13, 2647-2655. ( 10.1038/s41396-019-0459-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirano H, Takemoto K. 2019. Difficulty in inferring microbial community structure based on co-occurrence network approaches. BMC Bioinform. 20, 329. ( 10.1186/s12859-019-2915-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Layeghifard M, Hwang DM, Guttman DS. 2017. Disentangling interactions in the microbiome: a network perspective. Trends Microbiol. 25, 217-228. ( 10.1016/j.tim.2016.11.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ai D, Li X, Liu G, Liang X, Xia LC. 2019. Constructing the microbial association network from large-scale time series data using granger causality. Genes 10, 216. ( 10.3390/genes10030216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo J, et al. 2020. Soil fungal assemblage complexity is dependent on soil fertility and dominated by deterministic processes. New Phytol. 226, 232-243. ( 10.1111/nph.16345) [DOI] [PubMed] [Google Scholar]
- 66.Santolini M, Barabási A-L. 2018. Predicting perturbation patterns from the topology of biological networks. Proc. Natl Acad. Sci. USA 115, e6375-e6383. ( 10.1073/pnas.1720589115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y, Zhang R, Zheng Q, Deng Y, Van Nostrand JD, Zhou J, Jiao N. 2016. Bacterioplankton community resilience to ocean acidification: evidence from microbial network analysis. ICES Mar. Sci. Symp. 73, 865-875. ( 10.1093/icesjms/fsv187) [DOI] [Google Scholar]
- 68.Wagg C, Schlaeppi K, Banerjee S, Kuramae EE, van der Heijden MGA. 2019. Fungal–bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 10, 4841. ( 10.1038/s41467-019-12798-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiong C, et al. 2021. Host selection shapes crop microbiome assembly and network complexity. New Phytol. 229, 1091-1104. ( 10.1111/nph.16890) [DOI] [PubMed] [Google Scholar]
- 70.Stegen JC, Lin X, Fredrickson JK, Konopka AE. 2015. Estimating and mapping ecological processes influencing microbial community assembly. Front. Microbiol. 6, 370. ( 10.3389/fmicb.2015.00370) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou J, Ning D. 2017. Stochastic community assembly: does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 81, e00002-17. ( 10.1128/MMBR.00002-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gilbert NA, Stenglein JL, Pauli JN, Zuckerberg B. 2022. Human disturbance compresses the spatiotemporal niche. Proc. Natl Acad. Sci. USA 119, e2206339119. ( 10.1073/pnas.2206339119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma (Sam) Z. 2018. The P/N (positive-to-negative links) ratio in complex networks—a promising in silico biomarker for detecting changes occurring in the human microbiome. Microb. Ecol. 75, 1063-1073. ( 10.1007/s00248-017-1079-7) [DOI] [PubMed] [Google Scholar]
- 74.Hernandez DJ, David AS, Menges ES, Searcy CA, Afkhami ME. 2021. Environmental stress destabilizes microbial networks. ISME J. 15, 1722-1734. ( 10.1038/s41396-020-00882-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bertness MD, Callaway R. 1994. Positive interactions in communities. Trends Ecol. Evol. 9, 191-193. ( 10.1016/0169-5347(94)90088-4) [DOI] [PubMed] [Google Scholar]
- 76.Chase JM, Myers JA. 2011. Disentangling the importance of ecological niches from stochastic processes across scales. Phil. Trans. R. Soc. B 366, 2351-2363. ( 10.1098/rstb.2011.0063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stirn A. 2003. Body piercing: medical consequences and psychological motivations. Lancet 361, 1205-1215. ( 10.1016/S0140-6736(03)12955-8) [DOI] [PubMed] [Google Scholar]
- 78.Messahel A, Musgrove B. 2009. Infective complications of tattooing and skin piercing. J. Infect. Public Health 2, 7-13. ( 10.1016/j.jiph.2009.01.006) [DOI] [PubMed] [Google Scholar]
- 79.Tweeten SSM, Rickman LS. 1998. Infectious complications of body piercing. Clin. Infect. Dis. 26, 735-740. ( 10.1086/514586) [DOI] [PubMed] [Google Scholar]
- 80.Fournière M, Latire T, Souak D, Feuilloley MGJ, Bedoux G. 2020. Staphylococcus epidermidis and Cutibacterium acnes: two major sentinels of skin microbiota and the influence of cosmetics. Microorganisms 8, 1752. ( 10.3390/microorganisms8111752) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cogen AL, Nizet V, Gallo RL. 2008. Skin microbiota: a source of disease or defence? Br. J. Dermatol. 158, 442-455. ( 10.1111/j.1365-2133.2008.08437.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakamura K, O'Neill AM, Williams MR, Cau L, Nakatsuji T, Horswill AR, Gallo RL. et al. 2020. Short chain fatty acids produced by Cutibacterium acnes inhibit biofilm formation by Staphylococcus epidermidis. Sci. Rep. 10, 21237. ( 10.1038/s41598-020-77790-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Christensen GJM, Scholz CF, Enghild J, Rohde H, Kilian M, Thürmer A, Brzuszkiewicz E, Lomholt HB, Brüggemann H. 2016. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. BMC Genomics 17, 152. ( 10.1186/s12864-016-2489-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marito S, Keshari S, Traisaeng S, My DTT, Balasubramaniam A, Adi P, Hsieh M-F, Herr DR, Huang C-M. 2021. Electricity-producing Staphylococcus epidermidis counteracts Cutibacterium acnes. Sci. Rep. 11, 12001. ( 10.1038/s41598-021-91398-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rozas M, Hart De Ruijter A, Fabrega MJ, Zorgani A, Guell M, Paetzold B, Brillet F. 2021. From dysbiosis to healthy skin: major contributions of Cutibacterium acnes to skin homeostasis. Microorganisms 9, 628. ( 10.3390/microorganisms9030628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Claudel J-P, Auffret N, Leccia M-T, Poli F, Corvec S, Dréno B. 2019. Staphylococcus epidermidis: a potential new player in the physiopathology of acne? Dermatology 235, 287-294. ( 10.1159/000499858) [DOI] [PubMed] [Google Scholar]
- 87.Dagnelie M-A, Corvec S, Timon-David E, Khammari A, Dréno B. 2022. Cutibacterium acnes and Staphylococcus epidermidis: the unmissable modulators of skin inflammatory response. Exp. Dermatol. 31, 406-412. ( 10.1111/exd.14467) [DOI] [PubMed] [Google Scholar]
- 88.Niazi SA, Clarke D, Do T, Gilbert SC, Mannocci F, Beighton D. 2010. Propionibacterium acnes and Staphylococcus epidermidis isolated from refractory endodontic lesions are opportunistic pathogens. J. Clin. Microbiol. 48, 3859-3869. ( 10.1128/JCM.01326-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Uçkay I, Pittet D, Vaudaux P, Sax H, Lew D, Waldvogel F. 2009. Foreign body infections due to Staphylococcus epidermidis. Ann. Med. 41, 109-119. ( 10.1080/07853890802337045) [DOI] [PubMed] [Google Scholar]
- 90.Maguire M, Maguire G. 2017. The role of microbiota, and probiotics and prebiotics in skin health. Arch. Dermatol. Res. 309, 411-421. ( 10.1007/s00403-017-1750-3) [DOI] [PubMed] [Google Scholar]
- 91.Hulcr J, Latimer AM, Henley JB, Rountree NR, Fierer N, Lucky A, Lowman MD, Dunn RR. 2012. A jungle in there: bacteria in belly buttons are highly diverse, but predictable. PLoS ONE 7, e47712. ( 10.1371/journal.pone.0047712) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sabaté Brescó M, Harris LG, Thompson K, Stanic B, Morgenstern M, O'mahony L, Richards RG, Moriarty TF. 2017. Pathogenic mechanisms and host interactions in Staphylococcus epidermidis device-related infection. Front. Microbiol. 8, 1401. ( 10.3389/fmicb.2017.01401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ramasamy S, Barnard E, Dawson TL, Li H. 2019. The role of the skin microbiota in acne pathophysiology. Br. J. Dermatol. 181, 691-699. ( 10.1111/bjd.18230) [DOI] [PubMed] [Google Scholar]
- 94.Simberloff DS, Wilson EO. 1970. Experimental zoogeography of islands: a two-year record of colonization. Ecology 51, 934-937. ( 10.2307/1933995) [DOI] [Google Scholar]
- 95.Xu C. 2023. Code for: piercing microbiome. Zenodo. ( 10.5281/zenodo.7832626) [DOI]
- 96.Xu CCY, Lemoine J, Albert A, Whirter ÉM, Barrett RDH. 2023. Community assembly of the human piercing microbiome. Figshare. ( 10.6084/m9.figshare.c.6927518) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Xu C. 2023. Code for: piercing microbiome. Zenodo. ( 10.5281/zenodo.7832626) [DOI]
- Xu CCY, Lemoine J, Albert A, Whirter ÉM, Barrett RDH. 2023. Community assembly of the human piercing microbiome. Figshare. ( 10.6084/m9.figshare.c.6927518) [DOI] [PMC free article] [PubMed]
Data Availability Statement
Data and materials availability: raw sequencing data are available on NCBI Sequence Read Archive under BioProject PRJNA956301. Data and code are available on Zenodo: https://doi.org/10.5281/zenodo.7832626 [95].
Supplementary material is available online [96].



