Abstract
A series of triclosan azo-adducts were synthesized to investigate their structure-activity relationship against Mycobacterium tuberculosis and non-tuberculous mycobacteria. The series' most potent compound was four and sixteen times more active than triclosan and rifabutin against drug-resistant Mycobacterium abscessus, respectively, while being less cytotoxic to human macrophages than triclosan on day one. Additionally, one of the azo-adducts was twice as efficient against M. tuberculosis as triclosan and twice as effective against Mycobacterium marinum as isoniazid. Furthermore, the synthesized azo-adducts were equally effective against M. abscessus strains overexpressing InhA, suggesting that these compounds work through a distinct mechanism.
Keywords: Anti-mycobacterial, Cytotoxicity, Multi drug resistance, Overexpressing InhA strain, Triclosan
1. Introduction
Tuberculosis (TB) remains a major global public health threat. It is an infectious disease that mainly affects the lungs and is transmitted through the air by respiratory secretions [1]. TB is caused by a group of closely related bacterial species called Mycobacterium tuberculosis complex. Today, M. tuberculosis (Mtb) and Mycobacterium bovis is the key cause of TB in humans and other animal species. Mycobacterium africanum and Mycobacteruim microti are the other members of the complex that are not known to cause TB in humans [2]. Despite being a preventable and curable disease, an estimated 10.6 million people contracted TB in 2021, and 1.6 million died globally. In 2021, a total of 1.9 million TB cases were reported in India, up from 1.6 million cases in 2020, representing a 19 % increase over the previous year [3]. Drug-susceptible TB can be successfully treated using a combination of three or four drugs. Anti-TB first-line drugs include isoniazid (INH) (1), ethambutol (2) and rifampicin (RIF) (3), while second-line drugs include ethionamide (6), ciprofloxacin (4), cycloserine (5), and para-amino salicylic acid (7) (Fig. 1) [4].
Fig. 1.
Drugs used in the treatment of TB (1–7) and structure of TCS in inset (8).
Mycolic acids are important components of the bacterial cell wall that contribute to drug tolerance, virulence, and cell viability [5]. The type II fatty acid synthase (FAS-II) multienzyme system is the primary target of drugs used to prevent mycolic acid formation in mycobacterium [6]. The catalase-peroxidase KatG of Mtb activates the prodrug INH. When it binds to InhA, an enoyl-acyl carrier protein reductase, it forms an adduct that inhibits the synthesis of mycolic acids [[7], [8], [9], [10]]. However, mutations in the KatG gene can induce resistance to INH. The emergence of multidrug-resistant TB (MDR-TB), extensively drug-resistant TB (XDR-TB), and totally drug-resistant TB (TDR-TB) has exacerbated the situation [11,12]. MDR-TB and XDR-TB strains are primarily resistant to the most effective drugs (INH; RIF), bringing the infectious disease back into scientific focus [13,14]. Non-tubercular mycobacterial (NTM) infections have become a serious and growing threat [15,16]. NTM infections have grown dramatically in many affluent countries, surpassing TB as a public health concern. NTM infections have frequently been identified as the primary cause of pulmonary infections, notably in people with cystic fibrosis and other lung disorders [[17], [18], [19], [20]]. M. abscessus (Mabs) has recently emerged as one of the most difficult NTM pathogens to eradicate, as these mycobacteria are intrinsically resistant not only to traditional antituberculars, including INH and RIF, but also to the vast majority of currently available antibiotics [20,21].
Triclosan (TCS) (8) was initially believed to be a nonspecific bacterial membrane disruptor until it was discovered to be an InhA inhibitor, a significant and appealing anti-TB target [22]. Unlike INH, this scaffold does not require KatG bioactivation and, thus, has the potential to help overcome the prevailing resistance mechanism and be used to treat resistant bacterial infections [23]. Ring-A is a critical moiety in TCS for inhibiting InhA, whereas chlorine atoms have no effect (Fig. 1). The ether oxygen bond is also important because it improves TCS's interaction with InhA, resulting in enhanced biological activity. This ether oxygen also aids in the formation of the stable InhA-TCS-NAD+ complex [22]. TCS is found in the blood, urine, breast milk, and amniotic fluid and accumulates in fatty tissues. Toxicity studies in mammals show that TCS in consumer products is not expected to cause adverse health effects as it is not mutagenic, carcinogenic, or developmental toxicant [23,24].
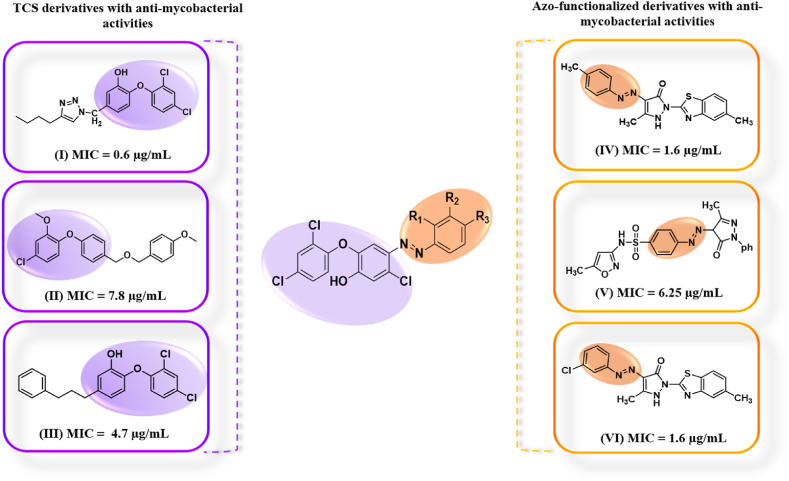
Azo dyes are another important family of organic compounds that have piqued the interest of researchers due to their wide range of applications in a variety of domains such as food, medicines, paper, cosmetics, textile, and leather industries. Heterocyclic azo dyes and azo-dye-based hybrids have a wide range of pharmacological effects, including antibacterial, antifungal, antituberculosis, anticancer, antioxidant, and anti-inflammatory activities [25,26]. A recent study revealed the anti-mycobacterial activities of azo dyes based on conventional diazo-coupling reaction between aniline derivatives with 5-methyl-2-(6-methyl-1,3-benzothiazol-2-yl)-2,4-dihydro-3H-pyrazol-3-one (MIC = 1.6 μg/mL) (Ⅳ,Ⅵ), sulfamethoxazole (MIC = 6.25 μg/mL) (Ⅴ) at lower temperature, with the most active compound being comparable to the standard drug streptomycin (Fig. 2) [27,28]. TCS derivatives are being synthesized and investigated as potential agents against TB. The addition of 3-phenylpropyl (Ⅲ) and (4-Butyl-1H-1,2,3-triazol-1-yl)methyl (Ⅰ) groups enhances the anti-tubercular activity, resulting in MIC values of 4.7 μg/mL and 0.6 μg/mL, respectively [23]. Conversely, linking 1-methoxy-4-((methoxy)methyl)benzene (Ⅱ) to TCS leads to reduced activity (MIC = 7.8 μg/mL) compared to TCS alone [29] (Fig. 2).
Fig. 2.
Schematic diagram showing the design strategy for choosing the molecular fragments (Triclosan highlighted in purple and azo functionality in orange) in the synthesis of TCS azo-adducts.
We previously showed that isatin-mono- and bis-INH hybrids had anti-TB potential, with the majority of the compounds exhibiting strong activity against Mtb in vitro and in macrophages [30]. We were able to ascertain their mode of action through the overexpression of InhA, cross-resistance determination, selection of resistant mutants, and biochemical investigation. The work was later expanded to include the synthesis of promising quinoline-INH hybrids as anti-mycobacterial compounds [31]. The current study describes the synthesis of TCS-based azo adducts (Fig. 2), along with an evaluation of their anti-mycobacterial activity.
2. Results and discussion
2.1. Synthetic chemistry
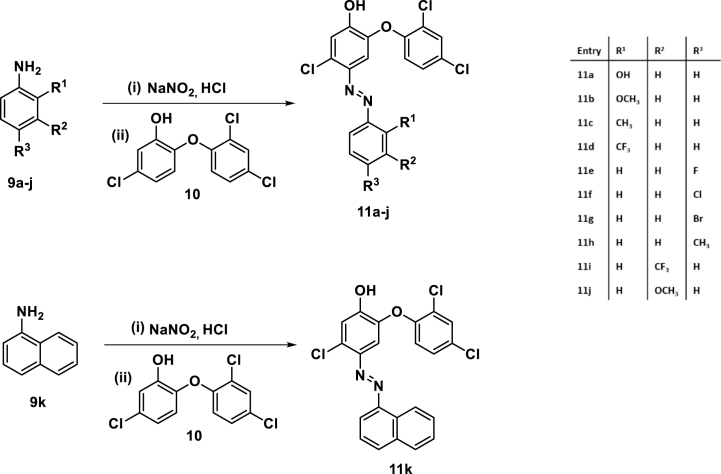
The desired TCS-based azo dyes were obtained through an initial diazotization of substituted anilines in conc. HCl at temperatures below 5 οC, followed by the addition of sodium nitrite. This was then added to a TCS solution in 35 % NaOH over 15 min at 0 οC. The reaction was stirred at room temperature for 2 h before the precipitates were filtered, washed with ice-cold water, and dried under reduced pressure (<b>Scheme 1</b>). The isolated crude products were then purified through column chromatography using a mixture of ethyl acetate:hexane (5:95) as eluent.
Scheme 1.
Synthesis of TCS azo-adducts 11a-k.
The structures of the synthesized TCS-aniline hybrids were assigned based on the spectral and analytical evidence. For instance, compound 11b exhibited a molecular ion peak at m/z C19H13Cl3N2O3 [M]+ 421.9982, [M+2]+ 423.9953 and [M+4]+ 425.9945 in the HRMS spectrum. Its 1H NMR exhibited the presence of a singlet at δ 3.78 corresponding to methoxy (-OCH3) group of anisidine core, a doublet at δ 6.88 because of aromatic ring proton at ortho position to ether linkage and a multiplet at δ 7.33–7.27 because of TCS ring protons.
2.2. Biological activities
2.2.1. Anti-mycobacterial and cytotoxic activities
The synthesized scaffolds were evaluated for their anti-mycobacterial activities toward Mtb, M. marinum (Mmar) and Mabs strains and the results are enlisted in Table 1 with INH, TCS, Rifabutin (RFB) and NITD-916 [32,33] used as standards. A careful inspection of the activity results revealed an intriguing structure-activity relationship (SAR) between the synthesized compounds and the tested strains. The majority of the compounds displayed activity against Mtb that was comparable or less active than TCS, except for 11e (R3 =4-F), 11f (R3 =4-Cl), and 11i (R2 =3-CF3), which had two folds the activity of TCS.
Table 1.
MIC99 (μg/mL) values determined in 7H9 broth supplemented with OADC of TCS analogs against M. tuberculosis mc26230, M. marinum M strain and in CaMHB at 30 °C against the rough variant of M. abscessus CIP104536T (3 independent experiments).
| Entry | |||||||
|---|---|---|---|---|---|---|---|
| M. tuberculosis | M. tuberculosis pMV261:inhAMTB | M. abscessus | M. abscessus pMV261:inhAMAB | M. abscessus pMV261:inhAMAB (G96V) | M. abscessus pMV261:inhAMAB (G96S) | M. marinum | |
| 11a | 25 | 50 | 3.125 | 3.125 | 3.125 | 3.125 | 25 |
| 11b | 25 | 25 | 0.78 | 0.78 | 0.78 | 0.78 | 25 |
| 11c | >100 | >100 | 25 | 25 | 25 | 25 | >100 |
| 11d | 12.5 | 25 | 12.5 | 12.5 | 12.5 | 12.5 | 12.5 |
| 11e | 6.25 | 12.5 | 1.56 | 1.56 | 1.56 | 1.56 | 12.5 |
| 11f | 6.25 | 6.25 | 1.56 | 1.56 | 1.56 | 1.56 | 6.25 |
| 11g | 25 | 25 | 6.25 | 6.25 | 6.25 | 6.25 | 25 |
| 11h | 12.5 | 25 | 12.5 | 12.5 | 12.5 | 12.5 | 12.5 |
| 11i | 6.25 | 6.25 | 3.125 | 3.125 | 3.125 | 3.125 | 12.5 |
| 11j | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| 11k | 12.5 | 12.5 | 3.125 | 3.125 | 3.125 | 3.125 | 12.5 |
| TCS | 12.5 | 25 | 3.125 | 3.125 | 3.125 | 3.125 | 6.25 |
| NITD-916 | <0.195 | 1.56 | 1.56 | 3.125 | 25 | 50 | 3.125 |
| INH | 0.05 | 0.8 | ND | ND | ND | ND | 3.125 |
| RFB | ND | ND | 12.5 | 12.5 | 12.5 | 12.5 | ND |
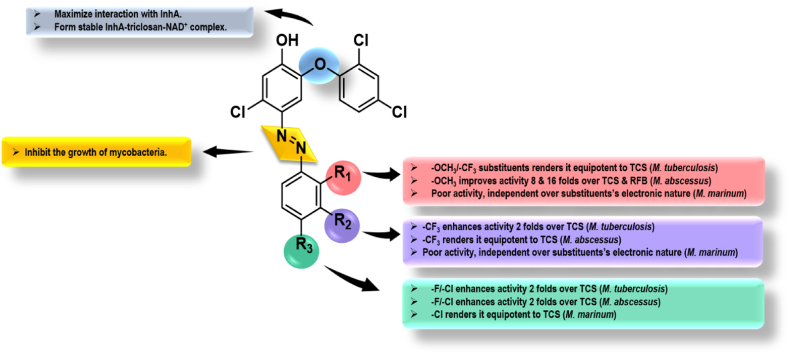
The compounds with polar donating hydroxy/methoxy substituents on the aromatic ring proved to be inactive as evidenced by 11j. The activity data against Mabs indicated a variety of compounds that were more active than the standard, TCS and RFB. The compound 11b (R1 = 2-OCH3) was shown to be the most active in the series, with a MIC99 of 0.78 μg/mL, four-fold more active than TCS. The presence of an inductively electron donating or electron withdrawing substituent at the ortho position of an aryl ring lowered the anti-mycobacterial activity as evident from 11c and 11d. The inclusion of an electron-withdrawing substituent at the para position resulted in TCS-azo adducts that were two and eight-fold more potent than TCS and RFB, respectively. At the meta position, a similar preference for electron-withdrawing substituents was noted, with 11i (R2 = 3-CF3) being equipotent to TCS, however the inclusion of a polar donating group proved to be deleterious, as seen in the case of 11j (R2 = 3-OCH3). The anti-mycobacterial activity of synthesized TCS-azo compounds against Mmar, a slow-growing mycobacterial species capable of infecting fish and causing skin lesions in humans, revealed that most of the compounds were two-four fold less potent than TCS and two-eight fold less potent than INH, regardless of the nature of the aromatic ring substituent. The graphical representation of SAR is provided in Fig. 3.
Fig. 3.
Graphical representation of SAR.
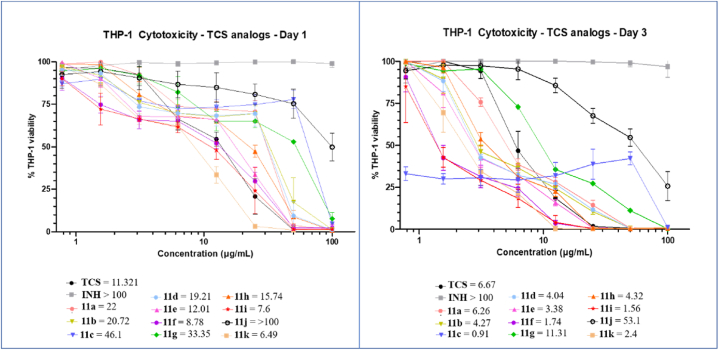
Crucial determinants in INH resistance include increased InhA expression or mutations that reduce the enzyme's affinity for binding to NADH. However, it is unclear if the newly synthesized azo-adducts share the exact inhibitory mechanism as INH by targeting InhA or follow a distinct approach. The inhA gene was cloned into pMV261 under the control of the strong hsp60 promoter [34] and the resulting plasmid was introduced into Mtb [35] to determine whether overexpression of the enoyl ACP reductase affects the MIC values of 11a-k in Middlebrook 7H9 supplemented with oleic acid-albumin-dextrose-catalase (OADC) enrichment. As expected, overexpression of InhAMTB in Mtb resulted in high levels of resistance to INH and NITD-916 in agreement with previous studies [32,34] while only a moderate two-fold resistance level was noticed with TCS, consistent with a previous report [36]. The level of resistance to one subset of the TCS analogs (compounds 11a, 11d, 11e, 11h) was also low (two-fold increase) in the strain carrying pMV261:inhAMBT while the MIC of the other compounds did not vary between the wild-type strain and the strain overexpressing InhA, suggesting that the lethal effects of these compounds may also stem from disrupting other mycobacterial targets. In addition, TCS and its azo adducts were equally active against the parental Mabs and the Mabs strains overexpressing either InhAMAB, InhAMAB (G96V) or InhAMAB (G96S) (the latter strains being highly resistant to the InhA inhibitor NITD-916) [33]. This may suggest that, in Mabs, TCS and its azo adducts inhibit other targets than InhA, a possibility supported by the fact that we were unable to select TCS-resistant spontaneous mutant on plates containing increasing TCS concentration (data not shown). However, further studies are required to figure out whether TCS inhibits various targets in this species and determine their precise mechanisms of action in this species. The scaffolds were next evaluated on human THP-1 macrophages on days 1 and 3 to establish their cytotoxicity levels (Fig. 4).
Fig. 4.
Cytotoxicity assay of TCS, INH and 11a-k after 1 and 3 days in THP-1 cells. CC50 values are indicated for each compound.
The selectivity index (SI) was calculated against the tested strains, defined as the ratio of cytotoxicity to inhibitory activities (Table 2). Evidently, on day one, most synthesized compounds were either less toxic or had equivalent cytotoxicity to TCS and displayed higher SI values against Mtb than TCS. The most active molecule 11b on Mabs shows a SI of 26.56 11a, 11e, 11f, and 11g had slightly higher SI than TCS against Mabs at day 1, with 11b being approximately seven-fold more selective. The SI values are comparable or lower than TCS in Mmar (Table 2). On day 3, most of the compounds showed comparable cytotoxicity and selectivity to TCS. In general, the TCS azo-adducts show very modest SI values against the 3 mycobacterial species.
Table 2.
In vitro cytotoxicity evaluation of 11a-k in human THP-1 macrophages, exposed for 1 day or 3 days with increasing concentrations of each compound and calculated selectivity indices (SI).
| Entry | Cytotoxicity CC50 (Day-1) | Selectivity index |
Cytotoxicity CC50 (Day-3) | Selectivity index |
||||
|---|---|---|---|---|---|---|---|---|
| M. tuberculosis | M. abscessus | M. marinum | M. tuberculosis | M. abscessus | M. marinum | |||
| 11a | 22 ± 0.12 | 0.88 | 7.04 | 0.88 | 6.26 ± 0.22 | 0.25 | 2.00 | 0.25 |
| 11b | 20.72 ± 2.6 | 0.83 | 26.56 | 0.82 | 4.27 ± 1.07 | 0.34 | 5.47 | 0.17 |
| 11c | 46.1 ± 1.3 | – | 1.84 | – | 0.91 ± 0.3 | – | 0.03 | – |
| 11d | 19.21 ± 0.3 | 1.53 | 1.53 | 1.53 | 4.04 ± 0.9 | 0.32 | 0.32 | 0.32 |
| 11e | 12.01 ± 0.25 | 1.92 | 7.70 | 0.96 | 3.38 ± 0.45 | 0.54 | 2.16 | 0.27 |
| 11f | 8.78 ± 0.37 | 1.40 | 5.62 | 1.40 | 1.74 ± 0.38 | 0.27 | 1.11 | 0.27 |
| 11g | 33.35 ± 1.71 | 1.33 | 5.33 | 1.33 | 11.31 ± 1.7 | 0.45 | 1.80 | 0.45 |
| 11h | 15.74 ± 0.8 | 1.25 | 1.26 | 1.25 | 4.32 ± 0.36 | 0.34 | 0.17 | 0.34 |
| 11i | 7.6 ± 1.3 | 1.21 | 2.43 | 0.60 | 1.56 ± 0.25 | 0.24 | 0.49 | 0.12 |
| 11j | >100 | – | – | – | 53.1 ± 7.5 | – | – | – |
| 11k | 6.49 ± 0.31 | 0.51 | 2.08 | 0.51 | 2.4 ± 0.24 | 0.19 | 0.38 | 0.19 |
| TCS | 11.321 ± 0.11 | 0.90 | 3.62 | 1.81 | 6.67 ± 0.67 | 0.53 | 2.13 | 1.06 |
| INH | >100 | – | – | – | >100 | – | – | – |
In order to better apprehend the mode of action of the newly synthesized TCS azo-adducts and their potential functional connection with INH, we conducted experiments using spontaneously INH-resistant strains of Mtb. Our selection of strains included three mutants of Mtb mc26230 harboring mutations in the catalase-peroxydase KatG (Table 3). These mutants (designated 16dR5X, 16fR5X and 16gR20X)) had previously shown resistance to INH and quinoline-INH hybrids [31]. Since resistance to INH primarily arises from mutations in katG, we investigated the potential implication of KatG in the mechanism of action of the TCS azo-adducts by MIC determination on the KatG mutant strains. As shown in Table 3, there were no changes in the MIC values for TCS azo-adducts and TCS, in contrast to INH. These findings strongly suggests that, unlike INH, the synthesized compounds do not necessitate KatG-mediated biotransformation.
Table 3.
MIC (μg/mL) values determined in 7H9 broth supplemented with OADC of TCS analogs against M. tuberculosis mc26230 WT and three M. tuberculosis mc26230 KatG mutants selected for their resistance to INH and quinoline-INH hybrids. (2 independent experiments).
| M. tuberculosis | M. tuberculosis 16dR5X | M. tuberculosis 16fR5X | M. tuberculosis 16gR20X | |
|---|---|---|---|---|
| 11a | 25 | 25 | 25 | 25 |
| 11b | 25 | 25 | 25 | 25 |
| 11c | >100 | >100 | >100 | >100 |
| 11d | 12.5 | 12.5 | 12.5 | 12.5 |
| 11e | 6.25 | 6.25 | 6.25 | 6.25 |
| 11f | 6.25 | 6.25 | 6.25 | 6.25 |
| 11g | 25 | 25 | 25 | 25 |
| 11h | 12.5 | 12.5 | 12.5 | 12.5 |
| 11i | 6.25 | 6.25 | 6.25 | 6.25 |
| 11j | >100 | >100 | >100 | >100 |
| 11k | 12.5 | 12.5 | 12.5 | 12.5 |
| TCS | 12.5 | 12.5 | 12.5 | 12.5 |
| INH | 0.05 | >3.2 | >3.2 | >3.2 |
Furthermore, the predicted physicochemical properties and absorption, distribution, metabolism, elimination, and toxicity (ADMET) descriptors revealed that the majority of the synthesized compounds had good drug-like properties (Table S1 in the supplemental material) (http://www.swiss adme.ch/; https://biosig.lab.uq.edu.au/pkcsm/prediction). The TCS azo-adducts are predicted to exhibit decreased blood-brain barrier permeability (logBB ranging from −0.05 to −0.37), reducing their ability to induce neurotoxicity or psychotropic adverse effects. The data analysis showed that (a) all molecules have a molecular weight between 407 and 472, which is within the recognised limit of 500, (b) a logP between 5.53 and 6.79, showing that the compounds are lipophilic, and (c) conform to hydrogen bond donor and acceptor properties. The majority of the compounds demonstrated high solubility, intestinal absorption, and P-glycoprotein I and II inhibitory activity. Additionally, the compounds do not act as substrates for P-glycoproteins, a key ATP-dependent efflux pump with broad substrate specificity. The majority of the compounds inhibit most CYP450 isoforms, which play a significant role in metabolism, increasing the chance of drug clearance and lowering the risk of drug-drug interactions and other severe side effects.
2.2.2. In silico studies
Molecular docking can provide useful information about the interactions of ligands with their targets. This technique now forms an indispensable part of structure-based drug development as the precision and speed of molecular docking approaches have been remarkably improved [37]. The enoyl ACP reductase InhA is one of the proteins that has been reported to play a significant part against drug-sensitive and drug-resistant Mtb strains [38] and is involved in the mycolic acid biosynthesis. It is responsible for catalyzing trans double bond reduction in the FAS-II pathway, which is associated with a carbonyl group of an intermediate that is covalently linked to an ACP [39].
To substantiate the experimental anti-Mtb and anti-Mabs results, molecular docking studies of one of the most potent compounds and the least potent compounds, (11f and 11b against Mtb) and (11b and 11c against Mabs), were performed along with TCS (control drug) in the binding pocket of InhAMTB and InhAMAB by employing an induced fit docking protocol. This docking protocol combines Glide and Prime refinement modules to predict ligand binding modes by allowing flexibility of the receptor at the same time to create a reliable model of the ligand-receptor complex. The reported crystal structures of InhAMTB (PDB ID: 1P45) with TCS enclosed and InhAMAB (PDB ID: 7U0M) were retrieved from the RCSB data bank.
2.2.3. InhAMTB
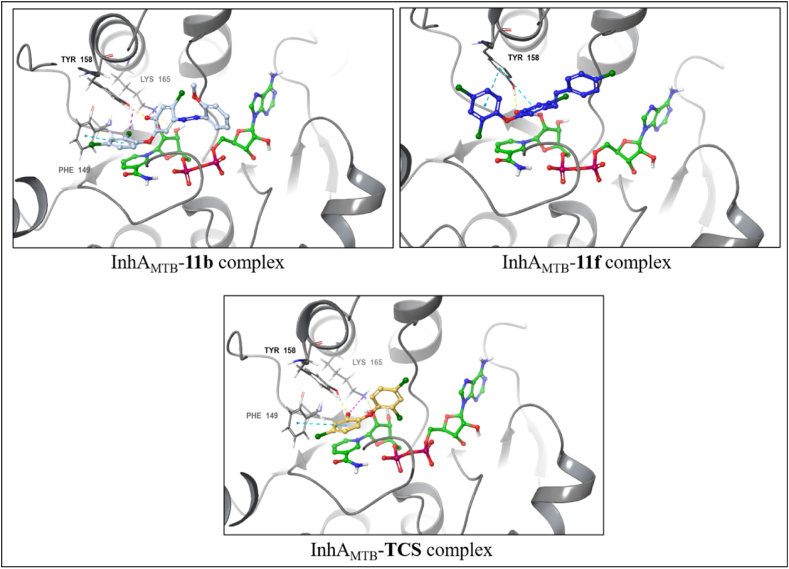
The docking data obtained revealed a similar trend as IC50 values of evaluated compounds. Compound 11f exhibiting potent activity against Mtb with an MIC of 6.25 μg/mL showed the highest docking score (most negative value) of −11.078 and binding free energy of −68.57 kcal/mol (Table 4), corroborating their anti-TB activity. InhAMTB-11f complex is characterized by hydrogen bond interaction via hydroxy group and Tyr158. The complex was further stabilized by two π-π stacking interactions with that same amino acid residue with two phenolic groups (Fig. 5).
Table 4.
Induced fit docking (IFD) and MM-GBSA (ΔG bind) calculations of selected compounds on InhAMTB and InhAMAB.
| Target | Ligand | MIC99 (μg/mL) | Docking score (kcal/mol) | Glide Emodel | Glide energy | MM-GBSA ΔG bind (kcal/mol) |
|---|---|---|---|---|---|---|
| InhAMTB | 11b | 25 | −9.215 | −91.212 | −57.541 | −52.00 |
| 11f | 6.25 | −11.078 | −92.364 | −57.319 | −68.57 | |
| TCS | 12.5 | −7.258 | −69.400 | −48.512 | −36.40 | |
| InhAMAB | 11b | 0.78 | −9.893 | −93.578 | −62.647 | −87.59 |
| 11c | 25 | −7.728 | −66.141 | −50.845 | −76.76 | |
| TCS | 3.125 | −8.032 | −50.294 | −37.764 | −72.99 |
Fig. 5.
Predicted binding modes of compounds 11b, 11f, and TCS at the active site of InhAMTB (PBD ID:1P45). Protein-ligand interactions are shown as dashed lines: hydrogen bonds (yellow), halogen bonds (purple), π-π stacking (light-blue), and salt bridges (magenta).
On the other hand, the docking score of −9.215 and binding free energy of −52.00 kcal/mol of compound 11b revealed its reduced potency against Mtb. InhAMTB-11b complex comprises of hydrogen bond interaction between hydroxy group with Tyr158 as well as π-π stacking interaction between 2,4-dichlorophenyl ring and Phe149. In addition to these interactions, a salt bridge between the hydroxy group and Lys165 and halogen bond between the chlorine atom in the second position with Tyr158, respectively. TCS (docking score = −7.258 and binding free energy = −36.40 kcal/mol) maintained hydrogen bond and salt bridge interaction with Tyr158 and Lys165. In addition, π-π stacking interaction was observed through Phe149.
2.2.4. InhAMAB
Compound 11b, showing the highest potency against Mabs showed the highest docking score of −9.893 and binding free energy of −87.59 kcal/mol (Table 4). The docked complex is characterized by hydrogen bond interaction hydroxy group and Tyr158. In addition, two more interactions were observed, viz salt bridge interaction via hydroxy group with Lys165 and halogen bond interaction via one chlorine atom of 2,4-dichlorophenyl ring with Thr196. The reduced potency of compound 11c was endorsed by its lower docking score of −7.728 and binding free energy of −76.76 kcal/mol. InhAMAB-11c complex featured π-π stacking interaction between 2,4-dichlorophenyl ring and Phe149 as well as establishing two halogen bond interactions with Met98 via the chlorine atom of the centre phenyl ring (Fig. 6). Lastly, TCS having a docking score of −8.032 and binding free energy of −72.99 kcal/mol furnished similar interactions as in InhAMAB-11c complex regarding Tyr158 and Phe149 amino acid residues; moreover, one halogen bond interaction with Thr196.
Fig. 6.
Predicted binding modes of compounds 11b, 11c and TCS at the active site of InhAMAB (PBD ID: 7U0M). Protein-ligand interactions are shown as dashed lines: hydrogen bonds (yellow), halogen bonds (purple), π-π stacking (light-blue), salt bridges (magenta).
3. Conclusion
In summary, a new series of TCS azo-adducts were synthesized and evaluated against TB and non-TB mycobacteria. SAR demonstrated a dependence of activity on both the position and electronic nature of aryl ring substituents. When evaluated against Mtb, Mabs, and Mmar, compound 11b revealed to be equipotent, four times more potent, and four times less potent, respectively, to TCS and sixteen folds more potent than RFB against Mabs. The cytotoxicity profile showed that 11b was less cytotoxic on human macrophages than TCS on day 1 and had a selectivity index of 26.56 on Mabs, which was better than TCS's SI of 3.62. In addition, the azo-adduct 11f was twice as potent against Mtb as TCS and twice as effective against Mmar as INH. Future medicinal chemistry programs are needed to improve the SI values fo TCS azo-derivatives prior to test them in pre-clinical animal models. Finally, the in silico computed docking scores and binding free energies of representative compounds supported the experimentally observed activity trends involving key interactions responsible for their host-guest relationship. However, the primary in vivo target of these compounds remains to be established.
4. Experimental
4.1. General informations
The reactions were performed by employing standard protocols and techniques. Melting points were recorded using open capillaries and Stuart digital melting-point apparatus (SMP10) and uncorrected. JEOL (400 MHz) and Bruker (500 MHz) spectrometers were used to record 1H NMR spectra, and JEOL (100 MHz) and Bruker (125 MHz) spectrometers for 13C NMR spectra with DMSO‑d6 as a solvent. The chemical shifts (δ) were expressed in parts per million (ppm) and coupling constants (J values) were specified in hertz (Hz). Splitting patterns are designated as s: singlet, d: doublet, t: triplet, m: multiplet, dd: double of the doublet ddd: doublet of doublet of doublet. Using ESI as the source, mass spectral data were assembled on Bruker high-resolution mass spectrometer (micrOTOF QII) equipment.
4.2. General procedures for the synthesis of conjugates 11a-k
Added substituted aniline (10 mmol) and concentrated HCl (2 mL, 20 mmol) to 15 mL of water. Cooled down the resulting solution to 2 °C. Added a solution of NaNO2 (0.8 g, 10 mmol) in 10 mL of water dropwise to the resulting solution, maintaining the temperature below 5 °C. The resulting mixture was added slowly to a 15 mL of an aqueous solution of TCS (1.16 g, 4 mmol), containing NaOH 35 %, until homogenization. Maintain the reaction mixture at 0 °C. Stir the reaction mixture for 15 min. Allow the reaction mixture to stay at room temperature for 2 h. Filter off the resulting precipitate. Wash the resulting precipitate with water. Dry the resulting precipitate.
4.2.1. (E)-5-Chloro-2-(2,4-dichlorophenoxy)-4-((2-hydroxyphenyl)diazenyl)phenol (11a)
Pale yellow (yield 84 %); m.p.: >200 °C; 1H NMR (500 MHz, DMSO‑d6) δ 13.32 (s, 1H, –OH, exchangable with D2O), 9.42 (s, 1H, –OH, exchangable with D2O), 7.45 (t, J = 6.8 Hz, 1H, ArH), 7.34 (t, J = 7.4 Hz, 1H, ArH), 7.24 (t, J = 7.4 Hz, 1H, ArH), 7.19 (d, J = 7.3 Hz, 1H, ArH), 7.06–6.80 (m, 1H, ArH). 13C NMR (125 MHz DMSO‑d6) δ 155.13, 154.11, 152.57, 141.97, 137.20, 134.70, 133.61, 133.51, 131.44, 129.53, 126.62, 122.45, 121.88, 120.30, 119.21, 118.24, 117.73, 115.71. HRMS calcd for C18H11Cl3N2O3 [M]+ 407.9835, [M+2]+ 409.9806 and [M+4]+ 411.9776 and found 407.9822, 409.9820, and 411.9762.
4.2.2. (Z)-5-Chloro-2-(2,4-dichlorophenoxy)-4-((2-methoxyphenyl)diazenyl)phenol (11b)
Yellow (yield 88 %); m.p.: 110–112 °C; 1H NMR (400 MHz, DMSO‑d6) δ 7.69 (d, J = 2.5 Hz, 1H, H5), 7.44 (t, J = 7.8 Hz, 1H, ArH), 7.39 (dt, J = 7.9, 1.4 Hz, 1H, H4), 7.35 (s, 1H, H2), 7.33–7.28 (m, 2H, ArH + H3), 7.22 (s, 1H, H1), 7.08 (ddd, J = 7.9, 2.6, 1.3 Hz, 1H, ArH), 6.88 (d, J = 8.9 Hz, 1H, ArH), 3.78 (s, 3H, –OCH3). 13C NMR (100 MHz, DMSO‑d6) δ 160.55, 153.74, 152.09, 142.80, 140.74, 133.16, 130.80, 130.40, 129.01, 127.71, 123.96, 119.22, 118.46, 117.99, 116.33, 109.68, 107.27, 79.68, 55.87. HRMS calcd for C19H13Cl3N2O3 [M]+ 421.9992, [M+2]+ 423.9962 and [M+4]+ 425.9933 and found 421.9982, 423.9953 and 425.9945.
4.2.3. (E)-5-Chloro-2-(2,4-dichlorophenoxy)-4-(o-tolyldiazenyl)phenol (11c)
Orange yellow (yield 85 %); m.p.: 111–113 °C; 1H NMR (500 MHz, DMSO‑d6) δ 7.72 (d, J = 1.6 Hz, 1H, H5), 7.55 (d, J = 8.0 Hz, 1H, H6), 7.40 (d, J = 7.0 Hz, 3H, ArH), 7.36–7.28 (m, 2H, H1+H4), 7.20 (s, 1H, H2), 6.91 (d, J = 8.8 Hz, 1H, H3), 2.58 (s, 3H, –CH3). 13C NMR (125 MHz, DMSO‑d6) δ 154.36, 152.09, 150.43, 142.76, 140.84, 137.88, 132.99, 131.93, 131.62, 130.23, 128.83, 127.45, 127.11, 123.77, 119.01, 118.39, 116.00, 109.94, 17.60. HRMS calcd for C19H13Cl3N2O2 [M]+ 406.0043, [M+2]+ 408.0013 and [M+4]+ 409.9984 and found 408.0029, 409.0002 and 410.0011.
4.2.4. (E)-5-Chloro-2-(2,4-dichlorophenoxy)-4-((2-(trifluoromethyl)phenyl)diazenyl)phenol (11d)
Dark orange (yield 87 %); m.p.: 148–150 °C; 1H NMR (500 MHz, DMSO‑d6) δ 7.92 (d, J = 7.8 Hz, 1H, H6), 7.83 (t, J = 7.6 Hz, 1H, H7), 7.77 (m, 2H, H5+H9), 7.72 (t, J = 7.5 Hz, 1H, H8), 7.40 (dd, J = 8.8, 2.3 Hz, 1H, H4), 7.27 (s, 1H, H1), 7.22 (s, 1H, H2), 7.07 (d, J = 8.8 Hz, 1H, H3). 13C NMR (100 MHz, DMSO‑d6) δ 154.56, 151.25, 148.99, 143.86, 140.83, 134.25, 133.59, 131.83, 130.48, 129.17, 128.67, 125.79, 124.92, 120.86, 118.41, 116.69, 108.27. HRMS calcd for C19H10Cl3F3N2O2 [M]+459.9760, [M+2]+ 461.9730and [M+4]+ 463.9701 and found 459.9751, 461.9722 and 463.9715.
4.2.5. (E)-5-Chloro-2-(2,4-dichlorophenoxy)-4-((4-fluorophenyl)diazenyl)phenol (11e)
Pale yellow (yield 84 %); m.p.: 149–151 °C; 1H NMR (400 MHz, DMSO‑d6) δ 7.83 (dd, J = 8.9, 5.4 Hz, 2H, ArH), 7.66–7.64 (m, 1H, H5), 7.37–7.31 (m, 3H, H1+2 × ArH), 7.27 (dd, J = 8.9, 2.6 Hz, 1H, H4), 7.19 (s, 1H, H2), 6.84 (d, J = 8.9 Hz, 1H, H3). 13C NMR (125 MHz, DMSO‑d6) δ 164.69, 162.71, 152.36, 149.49, 143.33, 138.64, 133.90, 130.10, 128.74, 127.02, 124.96, 124.89, 123.50, 118.81, 118.73, 116.86, 116.68, 109.61. HRMS calcd for C18H10Cl3FN2O2 [M]+ 409.9792, [M+2]+ 411.9762 and [M+4]+ 413.9733 and found 409.9779, 411.9751 and 413.9721.
4.2.6. (E)-5-Chloro-4-((4-chlorophenyl)diazenyl)-2-(2,4-dichlorophenoxy)phenol (11f)
Yellowish green (yield 89 %); m.p.: 147–149 °C; 1H NMR (400 MHz, DMSO‑d6) δ 7.73 (d, J = 8.7 Hz, 2H, H6+H9), 7.62 (d, J = 2.5 Hz, 1H, H5), 7.53 (d, J = 8.7 Hz, 2H, H7+H8), 7.41 (s, 1H, H1), 7.24 (dd, J = 8.9, 2.6 Hz, 1H, H4), 6.85 (s, 1H, H2), 6.78 (d, J = 8.9 Hz, 1H, H3). 13C NMR (100 MHz, DMSO‑d6) δ 152.57, 151.57, 143.83, 137.27, 137.20, 135.03, 134.58, 129.98, 129.86, 128.64, 126.60, 124.11, 123.21, 119.23, 118.51, 109.67. HRMS calcd for C18H10Cl4N2O2 [M]+ 425.9496, [M+2]+ 427.9467 and [M+4]+ 429.9437 and found 425.9482, 427.9476 and 429.9449.
4.2.7. (E)-4-((4-bromophenyl)diazenyl)-5-chloro-2-(2,4-dichlorophenoxy)phenol (11g)
Pale yellow (yield 87 %); m.p.: 147–149 °C; 1H NMR (400 MHz, DMSO‑d6) δ 7.67 (m, 4H, ArH), 7.64 (d, J = 2.5 Hz, 1H, H5), 7.39 (s, 1H, H1), 7.26 (dd, J = 8.8, 2.5 Hz, 1H, H4), 6.95 (s, 1H, H2), 6.80 (d, J = 8.8 Hz, 1H, H3). 13C NMR (125 MHz, DMSO‑d6) δ 153.88, 151.89, 151.28, 142.88, 140.76, 133.22, 133.02, 130.36, 128.96, 127.81, 125.28, 124.92, 124.05, 119.38, 118.39, 109.43. HRMS calcd for C18H10BrCl3N2O2 [M]+ 469.8991, [M+2]+ 471.8971 and [M+4]+ 473.8941 and found 469.8972, 471.8982 and 473.8953.
4.2.8. (E)-5-Chloro-2-(2,4-dichlorophenoxy)-4-(p-tolyldiazenyl)phenol (11h)
Yellowish green (yield 86 %); m.p.: 147–149 °C; 1H NMR (400 MHz, DMSO‑d6) δ 8.27 (s, 1H, –OH, exchangable with D2O), 7.70 (m, 3H, H5+2ArH), 7.33 (m, 2H, H1+ArH), 7.33–7.28 (m, 2H, ArH + H4), 7.21 (s, 1H, H2), 6.88 (d, J = 8.9 Hz, 1H, H3), 2.34 (s, 3H, –CH3). 13C NMR (125 MHz, DMSO‑d6) δ 157.34, 155.19, 152.23, 141.66, 139.74, 135.78, 132.59, 131.83, 130.69, 130.53 128.65, 127.55, 126.01, 124.47, 120.11, 119.49, 118.10, 107.24, 18.60. HRMS calcd for C19H13Cl3N2O2 [M]+ 406.0043, [M+2]+ 408.0013 and [M+4]+ 409.9984 and found 406.0054, 408.0025 and 410.0013.
4.2.9. (E)-5-Chloro-2-(2,4-dichlorophenoxy)-4-((3-(trifluoromethyl)phenyl)diazenyl)phenol (11i)
Yellowish brown (yield 87 %); m.p.: 111–112 °C; 1H NMR (500 MHz, DMSO‑d6) δ 8.09 (m, 2H, H8 +H9), 7.89 (d, J = 7.6 Hz, 1H, H6), 7.81 (t, J = 7.8 Hz, 1H, H7), 7.73 (s, 1H, H5), 7.46 (s, 1H, H1), 7.34 (d, J = 8.8 Hz, 1H, H4), 7.22 (s, 1H, H2), 6.92 (d, J = 8.8 Hz, 1H, H3). 13C NMR (100 MHz, DMSO‑d6) δ 152.58, 152.04, 143.10, 139.92, 134.11, 131.34, 130.81, 130.55, 130.30, 128.91, 127.55, 126.78, 125.38, 123.84, 119.15, 118.62, 109.69. HRMS calcd for C19H10Cl3F3N2O2 [M]+ 459.9760, [M+2]+ 461.9730 and [M+4]+ 463.9701 and found 459.9772, 461.9719 and 463.9722.
4.2.10. (E)-5-Chloro-2-(2,4-dichlorophenoxy)-4-((3-methoxyphenyl)diazenyl)phenol (11j)
Dark brown (yield 84 %); m.p.: 161–163 °C; 1H NMR (500 MHz, DMSO‑d6) δ 7.73 (d, J = 2.0 Hz, 1H, H5), 7.50–7.43 (m, 2H, ArH), 7.39 (s, 1H, H1), 7.37–7.33 (m, 2H, ArH), 7.27 (s, 1H, H2), 7.12 (d, J = 7.5 Hz, 1H, H4), 6.93 (d, J = 8.8 Hz, 1H, H3), 3.83 (s, 1H, –OCH3). 13C NMR (125 MHz, DMSO‑d6) δ 160.49, 153.68, 152.00, 142.75, 140.77, 133.03, 130.74, 130.34, 128.95, 127.70, 123.95, 119.21, 118.38, 117.94, 116.29, 116.11, 109.58, 107.22, 55.82. HRMS calcd for C19H13Cl3N2O3 [M]+ 421.9992, [M+2]+ 423.9962 and [M+4]+ 425.9933 and found 421.9979, 423.9949 and 425.9948.
4.2.11. (E)-5-Chloro-2-(2,4-dichlorophenoxy)-4-(naphthalen-1-yldiazenyl)phenol (11k)
Dark brown (yield 81 %); m.p.: 115–117 °C; 1H NMR (400 MHz, DMSO‑d6) δ 8.76–8.72 (m, 1H, ArH), 8.09 (d, J = 8.1 Hz, 1H, ArH), 8.00 (dd, J = 6.1, 3.3 Hz, 1H, ArH), 7.75 (dd, J = 7.5, 0.9 Hz, 1H, ArH), 7.71 (d, J = 2.6 Hz, 1H, H5), 7.65 (s, 1H, H1), 7.61 (ddd, J = 5.8, 5.4, 2.1 Hz, 3H, ArH), 7.30 (dd, J = 8.9, 2.6 Hz, 1H, H4), 7.26 (s, 1H, H2), 6.90 (d, J = 8.9 Hz, 1H, H3). 13C NMR (125 MHz DMSO‑d6) δ 154.00, 152.19, 147.22, 142.67, 141.53, 134.43, 133.30, 132.11, 130.77, 130.26, 128.85, 128.56, 127.80, 127.48, 127.22, 126.34, 123.76, 123.35, 118.89, 118.42, 112.94, 110.49. C22H13Cl3N2O2 [M]+ 442.0043, [M+2]+ 444.0013 and [M+4]+ 445.9984 and found 442.0031, 444.0026 and 446.0012.
4.3. Docking protocols
The selected compounds were drawn using ChemDraw software and then imported into Schrödinger suite_2021-2 to prepare their ligands for docking using the LigPrep module [40], while their protonation and ionization states were determined using Epik [41]. The optimization of the representative compound (11b) showed the lower energy of its E-configuration (21.2381 kcal/mol) over its Z-conunterpart (44.7232 kcal/mol) and thus were preferred for the ligand preparation studies. The crystal structures of the target proteins, InhAMTB (PDB ID: 1P45) and InhAMAB (PDB ID: 7U0M) were obtained from RCSB Protein Data Bank and prepared using Protein Preparation Wizard [42] by adding hydrogen atoms and missing residues, assigning bond orders, and removing water molecules. Optimization of the hydrogen bond network was done using an OPLS4 force field and restrained minimization to RMSD of 0.30 Å.
The prepared ligands were docked into the active site of the prepared protein using Induced Fit Docking (IFD) [43] using the standard parameters. Briefly, the receptor's grid box was defined as the centroid of the workspace ligand and the ligand size was set at ≤ 20 Å with a conformational sampling at a 2.5 kcal/mol energy window while the protein structure was subjected to a brief constrained refinement (RMSD ≤0.18 Å). Thereafter, structures within 30 kcal mol−1 were submitted for glide redocking with extra precision mode and the best receptor-ligand complex was redocked to improve the final pose. Lastly, the best models (based on docking score, IFD score, glide emodel, and glide energy) were selected to calculate ligands' binding free energy (ΔG bind) using Prime Molecular Mechanics-Generalized Borne Surface Area (MM-GBSA) protocol [44].
4.4. Strains and culture conditions
The pantothenate-auxotrophic Mtb mc26230 strain and its derivatives were maintained in Middlebrook 7H9 broth (BD Difco) supplemented with 10 % oleic-albumin-dextrose-catalase enrichment (OADC), 0.025 % tyloxapol (Sigma-Aldrich) and 109 μM pantothenic acid at 37 °C without agitation. Strain mc26230 carrying pMV261:inhAMTB was grown in the presence of 25 μg/mL kanamycin.
Mmar M strain and Mabs CIP104536T reference rough strain were grown in 7H9 broth supplemented with OADC enrichment and 0.025 % tyloxapol. Mabs harbouring pMV261:inhAMTB was grown as reported previously [33].
4.5. Drug susceptibility testing
Minimal Inhibitory Concentration (MIC) is defined as the lowest compound concentration inhibiting 99 % of bacterial growth. Cultures were grown in 7H9 broth supplemented with 10 % OADC, 0.025 % tyloxapol at 37 °C for Mtb (supplemented with 109 μM pantothenic acid), at 30 °C for Mmar and in CaMHB for Mabs, according to the CLSI guidelines [45]. MIC determination was done using the broth dilution method. A log-phase culture was diluted to an OD600 = 0.05 (Mtb) or with an inoculum of 5.106 CFU/mL (Mabs and Mmar) in adequate broth. The bacterial suspension was seeded in 100 μL volumes in all wells of a 96-well plate, except for the first column, to which 198 μL was added. Compounds were directly added to the first-column wells and a serial two-fold serial dilution was then carried out starting from the first column. After 7 days (Mtb) or 4–5 days (Mabs and Mmar), MICs were recorded by visual inspection. TCS, INH, RFB and NITD-916 were included as reference drug.
4.6. Cytotoxicity assay
Human THP-1 monocytes were grown in RPMI medium supplemented with 10 % Fetal Bovine Serum (Sigma-Aldrich) (RPMIFBS) and incubated at 37 °C with 5 % CO2. Cells were differentiated with 20 ng/mL Phorbol Myristate Acetate in 96-well plate (2.104 cells/well) for 48 h and exposed to decreasing concentrations of 11a-k compounds, INH or TCS (ranging from 100 μg/mL to 0.78 μg/mL). Following incubation (24 or 72 h), 10 % v/v resazurin was added to each well and left to incubate for few hours at 37 °C with 5 % CO2. Data were measured using a fluorescent plate reader (excitation 540 nm, emission 590 nm).
Data availability statement
Data associated with this study is provided in the Electronic Supplementary Information file.
CRediT authorship contribution statement
Shekhar: Writing – original draft, Methodology, Writing – original draft, Methodology. Matthéo Alcaraz: Writing – original draft, Methodology. Pule Seboletswe: Software. Neha Manhas: Software. Laurent Kremer: Writing – review & editing, Writing – original draft, Methodology. Parvesh Singh: Writing – review & editing, Writing – original draft, Software. Vipan Kumar: Writing – review & editing, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Shekhar reports financial support was provided by University Grants Commission, India. Vipan Kumar reports financial support was provided by Council for Scientific and Industrial Research, India. Mattheo Alcaraz reports financial support was provided by French National Research Agency, France. Associate Editor, Heliyon (Pharmaceutical Sciences Section) Vipan Kumar If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
UGC(NFSC) has been acknowledged for providing fellowship to Shekhar (Ref. No. 1758/CSIR-UGC NET JUNE 2019). VK thanks Council of Scientific and Industrial Research (CSIR) for providing financial support (Grant No. 02(0400)/21/EMR-II). PS gratefully acknowledges the Centre for High-Performance Computing (CHPC), Cape Town (South Africa), for the supercomputing facilities. We acknowledge the Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation for funding the PhD of MA. This project has been funded by the French National Research Agency ANR-19-CE15-0012-01 (SUNLIVE).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e22182.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Who . World Health Organization; 2022. Global Tuberculosis Report. 25-05-2023) [Google Scholar]
- 2.Sandhu G. Tuberculosis: current situation, challenges and overview of its control programs in India. J. Global Infect. Dis. 2011;3:143–150. doi: 10.4103/0974-777X.81691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welfare F. 2022. INDIA TB REPORT 2022 Coming Together to End TB Altogether. [Google Scholar]
- 4.Dheda K., Gumbo T., Maartens G., Dooley K.E., Mcnerney R., Murray M., Furin J., Nardell E.A., Nuermberger E., Mcilleron H., Fennelly K.P., Goemaere E., Jaramillo E., Low M., Jara C.M., Padayatchi N., Warren R.M. The Lancet Respiratory Medicine Commission the epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. Lancet Respir. Med. Comm. 2017;2600:291–360. doi: 10.1016/S2213-2600(17)30079-6. [DOI] [PubMed] [Google Scholar]
- 5.Bhatt A., Molle V., Besra G.S., Jacobs W.R., Kremer L., I U.D.L., Biosciences I.F.R. MicroReview the Mycobacterium tuberculosis FAS-II condensing enzymes : their role in mycolic acid biosynthesis , acid-fastness , pathogenesis and in future drug development. Mol. Microbiol. 2007;64:1442–1454. doi: 10.1111/j.1365-2958.2007.05761.x. [DOI] [PubMed] [Google Scholar]
- 6.Li M., Huang Q., Zhang W., Cao Y., Wang Z., Zhao Z., Zhang X., Zhang J. A novel acyl-AcpM-binding protein confers intrinsic sensitivity to fatty acid synthase type II inhibitors in Mycobacterium smegmatis. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.846722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Lett. to Nat. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 8.Johnsson K., Froland W.A., Schultz P.G., Mycobacte W.K. Overexpression , purification , and characterization of the catalase-peroxidase KatG from Mycobacterium tuberculosis. J. Biol. Chem. 1997;272:2834–2840. doi: 10.1074/jbc.272.5.2834. [DOI] [PubMed] [Google Scholar]
- 9.Rozwarski D.A., Grant G.A., Barton D.H.R., Jr W.R.J., Sacchettini J.C. Modif icaf ion of the NADH of the lsoniazid Target (In h A) from Mycobacterium tuberculosis. Science. 1998;279(80):98–102. doi: 10.1126/science.279.5347.98. [DOI] [PubMed] [Google Scholar]
- 10.Wilming M., Johnsson K. Spontaneous Formation of the bioactive form of the tuberculosis drug isoniazid. Communications. 1999;5:2588–2590. doi: 10.1002/(sici)1521-3773(19990903)38:17<2588::aid-anie2588>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 11.Lafeuille E., Veziris N., Sougakoff W., Roure F., Le Dû D., Dournon N., Caumes E., Jarlier V., Aubry A., Robert J., Bernard C. XDR-tuberculosis in France: community transmission due to non-compliance with isolation precautions. Med. Maladies Infect. 2016;46:52–55. doi: 10.1016/j.medmal.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Cole S., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., Iii, Tekaia F. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/24206. [DOI] [PubMed] [Google Scholar]
- 13.Bemer-Melchior P., Bryskier A., Drugeon H.B. Comparison of the in vitro activities of rifapentine and rifampicin against Mycobacterium tuberculosis complex. J. Antimicrob. Chemother. 2000;46:571–575. doi: 10.1093/jac/46.4.571. [DOI] [PubMed] [Google Scholar]
- 14.Jain A., Mondal R. Extensively drug-resistant tuberculosis: current challenges and threats. FEMS Immunol. Med. Microbiol. 2008;53:145–150. doi: 10.1111/j.1574-695X.2008.00400.x. [DOI] [PubMed] [Google Scholar]
- 15.Aksamit T.R., Philley J.V., Griffith D.E. Nontuberculous mycobacterial (NTM) lung disease: the top ten essentials. Respir. Med. 2014;108:417–425. doi: 10.1016/j.rmed.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 16.Cook J.L. Nontuberculous mycobacteria: opportunistic environmental pathogens for predisposed hosts. Br. Med. Bull. 2010;96:45–59. doi: 10.1093/bmb/ldq035. [DOI] [PubMed] [Google Scholar]
- 17.Cowman S., Van Ingen J., Griffith D., Loebinger M.R. State of the Art disease Non-tuberculous mycobacterial pulmonary disease. Eur. Respir. J. 2019;54 doi: 10.1183/13993003.00250-2019. [DOI] [PubMed] [Google Scholar]
- 18.Johnson M.M., Odell J.A. Nontuberculous mycobacterial pulmonary infections. J. Thorac. Dis. 2014;6:210–220. doi: 10.3978/j.issn.2072-1439.2013.12.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jo B.E., Gilljam M., Lindblad A., Ridell M., Wold A.E., Welinder-olsson C. Molecular epidemiology of Mycobacterium abscessus , with focus on cystic fibrosis. J. Clin. Microbiol. 2007;45:1497–1504. doi: 10.1128/JCM.02592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansen M.D., Herrmann J.L., Kremer L. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat. Rev. Microbiol. 2020;18:392–407. doi: 10.1038/s41579-020-0331-1. [DOI] [PubMed] [Google Scholar]
- 21.Schim van der Loeff I., Owens S. Non-tuberculous mycobacterial infection in children. Paediatr. Child Health. 2021;31:102–109. doi: 10.1016/j.paed.2020.12.002. [DOI] [Google Scholar]
- 22.Parikh S.L., Xiao G., Tonge P.J. Accelerated publications inhibition of InhA , the enoyl reductase from Mycobacterium tuberculosis , by. Biochemistry. 2000;39:7645–7650. doi: 10.1021/bi0008940. [DOI] [PubMed] [Google Scholar]
- 23.Vosátka R., Krátký M., Vinšová J. Triclosan and its derivatives as antimycobacterial active agents. Eur. J. Pharm. Sci. 2018;114:318–331. doi: 10.1016/j.ejps.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 24.Dann A.B., Hontela A. Triclosan: environmental exposure, toxicity and mechanisms of action. J. Appl. Toxicol. 2011;31:285–311. doi: 10.1002/jat.1660. [DOI] [PubMed] [Google Scholar]
- 25.Yazdanbakhsh M.R., Youse H., Mamaghani M., Moradi E.O., Rassa M., Pouramir H., Bagheri M. Synthesis , spectral characterization and antimicrobial activity of some new azo dyes derived from 4 , 6-dihydroxypyrimidine. J. Mol. Liq. 2012;169:21–26. doi: 10.1016/j.molliq.2012.03.003. [DOI] [Google Scholar]
- 26.Keshavayya J. Synthesis , structural investigations and in vitro biological evaluation of N , N -dimethyl aniline derivatives based azo dyes as potential pharmacological agents. J. Mol. Struct. 2019;1186:404–412. doi: 10.1016/j.molstruc.2019.03.042. [DOI] [Google Scholar]
- 27.Mallikarjuna N.M., Keshavayya J. Journal of King Saud University – science Synthesis , spectroscopic characterization and pharmacological studies on novel sulfamethaxazole based azo dyes. J. King Saud Univ. Sci. 2020;32:251–259. doi: 10.1016/j.jksus.2018.04.033. [DOI] [Google Scholar]
- 28.Maliyappa M.R., Keshavayya J., Mallikarjuna N.M., Pushpavathi I. Novel substituted aniline based heterocyclic dispersed azo dyes coupling with 5-methyl-2-(6-methyl-1, 3-benzothiazol-2-yl)-2, 4-dihydro-3H-pyrazol-3-one: synthesis, structural, computational and biological studies. J. Mol. Struct. 2020;1205 doi: 10.1016/j.molstruc.2019.127576. [DOI] [Google Scholar]
- 29.Chetty S., Armstrong T., Sharma Kharkwal S., Drewe W.C., De Matteis C.I., Evangelopoulos D., Bhakta S., Thomas N.R. New inha inhibitors based on expanded triclosan and di-triclosan analogues to develop a new treatment for tuberculosis. Pharmaceuticals. 2021;14 doi: 10.3390/ph14040361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johansen M.D., Shalini, Kumar S., Raynaud C., Quan D.H., Britton W.J., Hansbro P.M., Kumar V., Kremer L. Biological and biochemical evaluation of isatin-isoniazid hybrids as bactericidal candidates against mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2021;65 doi: 10.1128/AAC.00011-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alcaraz M., Sharma B., Roquet-Banères F., Conde C., Cochard T., Biet F., Kumar V., Kremer L. Designing quinoline-isoniazid hybrids as potent anti-tubercular agents inhibiting mycolic acid biosynthesis. Eur. J. Med. Chem. 2022;239 doi: 10.1016/j.ejmech.2022.114531. [DOI] [PubMed] [Google Scholar]
- 32.Manjunatha U.H., Rao S.P.S., Kondreddi R.R., Noble C.G., Camacho L.R., Tan B.H., Ng S.H., Ng P.S., Ma N.L., Lakshminarayana S.B., Herve M., Barnes S.W., Yu W., Kuhen K., Blasco F., Beer D., Walker J.R., Tonge P.J., Glynne R., Smith P.W., Diagana T.T. Direct inhibitors of InhA are active against. Mycobacterium tuberculosis, Sci. Transl. Med. 2015;7:269ra3. doi: 10.1126/scitranslmed.3010597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alcaraz M., Roquet-Banères F., Leon-Icaza S.A., Abendroth J., Boudehen Y.M., Cougoule C., Edwards T.E., Kremer L. Efficacy and mode of action of a direct inhibitor of Mycobacterium abscessus InhA. ACS Infect. Dis. 2022;8:2171–2186. doi: 10.1021/acsinfecdis.2c00314. [DOI] [PubMed] [Google Scholar]
- 34.Larsen M.H., Vilchèze C., Besra G.S., Parsons L., Heifets L., Hazbon M.H., Alland D., Sacchettini J.C., Jacobs W.R. Overexpression of inhA , but not kasA , confers resistance to isoniazid and ethionamide in Mycobacterium smegmatis , M. bovis BCG and M. Tuberculosis. 2002;2:453–466. doi: 10.1046/j.1365-2958.2002.03162.x. [DOI] [PubMed] [Google Scholar]
- 35.Stover C.K., de Ia Cruz V.F., Fuerst T.R., Burlein J.E., Benson L., Bennett L.T., Bansal G.P., Young J.F., Lee M.H., Hatfull G.F., Snappert S.B., Barlettat R.G., Jacobs Jrt W.R., Bloomt B.R. New use of BCG for recombinant vaccines, N. R. Eta. Proc. Natn. Acad Sci. US.A. 1989;13:1483–1488. doi: 10.1038/351456a0. [DOI] [Google Scholar]
- 36.Stec J., Vilchèze C., Lun S., Perryman A.L., Wang X., Freundlich J.S., Bishai W., Jacobs W.R., Kozikowski A.P. Biological evaluation of potent triclosan-derived inhibitors of the enoyl-acyl carrier protein reductase InhA in drug-sensitive and drug-resistant strains of mycobacterium tuberculosis. ChemMedChem. 2014;9:2528–2537. doi: 10.1002/cmdc.201402255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al Sheikh Ali A., Khan D., Naqvi A., Al-Blewi F.F., Rezki N., Aouad M.R., Hagar M. Design, synthesis, molecular modeling, anticancer studies, and density functional theory calculations of 4-(1,2,4-Triazol-3-ylsulfanylmethyl)-1,2,3-triazole derivatives. ACS Omega. 2021;6:301–316. doi: 10.1021/acsomega.0c04595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dhameliya T.M., Bhakhar K.A., Gajjar N.D., Patel K.A., Devani A.A., Hirani R.V. Recent advancements and developments in search of anti-tuberculosis agents: a quinquennial update and future directions. J. Mol. Struct. 2022;1248 doi: 10.1016/j.molstruc.2021.131473. [DOI] [Google Scholar]
- 39.Prasad M.S., Bhole R.P., Khedekar P.B., Chikhale R.V. Mycobacterium enoyl acyl carrier protein reductase (InhA): a key target for antitubercular drug discovery. Bioorg. Chem. 2021;115 doi: 10.1016/j.bioorg.2021.105242. [DOI] [PubMed] [Google Scholar]
- 40.Cinu T.A., Sidhartha S.K., Indira B., Varadaraj B.G., Vishnu P.S., Shenoy G.G. Design, synthesis and evaluation of antitubercular activity of Triclosan analogues. Arab. J. Chem. 2019;12:3316–3323. doi: 10.1016/j.arabjc.2015.09.003. [DOI] [Google Scholar]
- 41.Armstrong T., Lamont M., Lanne A., Alderwick L.J., Thomas N.R. Inhibition of Mycobacterium tuberculosis InhA: design, synthesis and evaluation of new di-triclosan derivatives. Bioorganic Med. Chem. 2020;28 doi: 10.1016/j.bmc.2020.115744. [DOI] [PubMed] [Google Scholar]
- 42.Khade A.B., Eshwara V.K., Boshoff H.I.M., Arora K., Tiwari A., Bhat P., Tiwari M., Shenoy G.G. Design, synthesis, biological evaluation and molecular dynamic simulation studies of diphenyl ether derivatives as antitubercular and antibacterial agents. ChemistrySelect. 2020;5:201–210. doi: 10.1002/slct.201903305. [DOI] [Google Scholar]
- 43.Schrödinger Release 2021-2: Induced Fit Docking Protocol; Glide, Schrödinger, LLC, New York, NY. Prime, Schrödinger, LLC; New York, NY: 2021. 2021. [Google Scholar]
- 44.Schrödinger Release 2021-2: Prime-MMGBSA. Schrödinger, LLC; New York, NY: 2021. [Google Scholar]
- 45.Woods G.L. 2011. Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study is provided in the Electronic Supplementary Information file.