Abstract
The European Cooperation in Science and Technology (COST) is an intergovernmental organization dedicated to funding and coordinating scientific and technological research in Europe, fostering collaboration among researchers and institutions across countries. Recently, COST Action funded the "Genome Editing to treat Human Diseases" (GenE-HumDi) network, uniting various stakeholders such as pharmaceutical companies, academic institutions, regulatory agencies, biotech firms, and patient advocacy groups. GenE-HumDi’s primary objective is to expedite the application of genome editing for therapeutic purposes in treating human diseases. To achieve this goal, GenE-HumDi is organized in several working groups, each focusing on specific aspects. These groups aim to enhance genome editing technologies, assess delivery systems, address safety concerns, promote clinical translation, and develop regulatory guidelines. The network seeks to establish standard procedures and guidelines for these areas to standardize scientific practices and facilitate knowledge sharing. Furthermore, GenE-HumDi aims to communicate its findings to the public in accessible yet rigorous language, emphasizing genome editing’s potential to revolutionize the treatment of many human diseases. The inaugural GenE-HumDi meeting, held in Granada, Spain, in March 2023, featured presentations from experts in the field, discussing recent breakthroughs in delivery methods, safety measures, clinical translation, and regulatory aspects related to gene editing.
Keywords: MT: RNA/DNA Editing, European Cooperation in Science and Technology, COST, GenE-HumDi, genome editing, base editors, delivery systems, regulatory guidelines
Graphical abstract
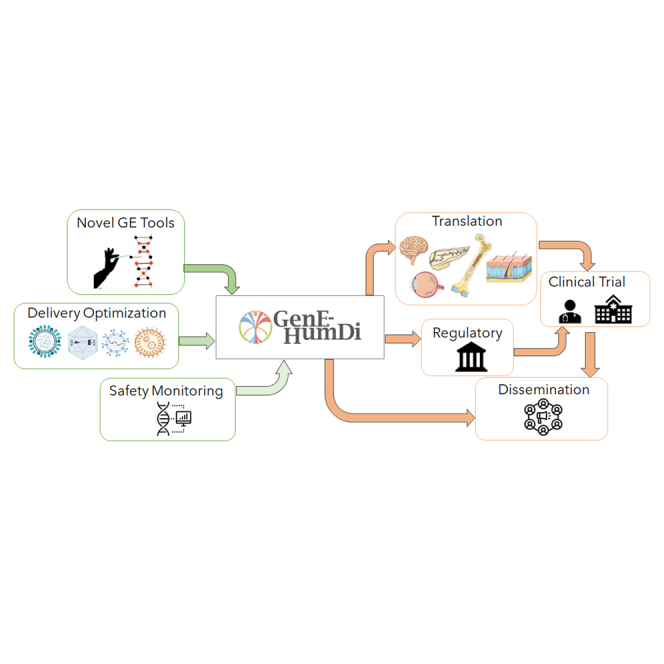
The COST Action GenE-HumDi network unites experts, pharmaceutical industry, academia, and regulatory authorities to accelerate genome editing for treating human diseases. GenE-HumDi’s mission includes refining technology, ensuring safety, and promoting accessibility.
Introduction
Genome editors are transformative technologies that can address genetic causes of human diseases. Tremendous progress has been made across different classes of genome editors, from meganucleases, zinc finger nucleases (ZFNs), transcription activator-like endonucleases (TALENs), and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR). The simplicity with which CRISPR genome editors can be programmed by a short-associated guide RNA (gRNA) has stimulated their broad application in life sciences research and clinical development. Exciting additions to the CRISPR editing toolbox include base editors (BE) that can precisely install certain point mutations and prime editors (PE) that can copy information in a prime editing gRNA (pegRNA) into a nicked DNA target. Each of these editors is now being rapidly developed into genome editing (GE) medicines, by both companies and academic groups. The first GE medicine, exa-cel, for treatment of sickle-cell disease (SCD) or transfusion-dependent beta-thalassemia is expected to be approved by the Food and Drug Administration (FDA) in December 2023.
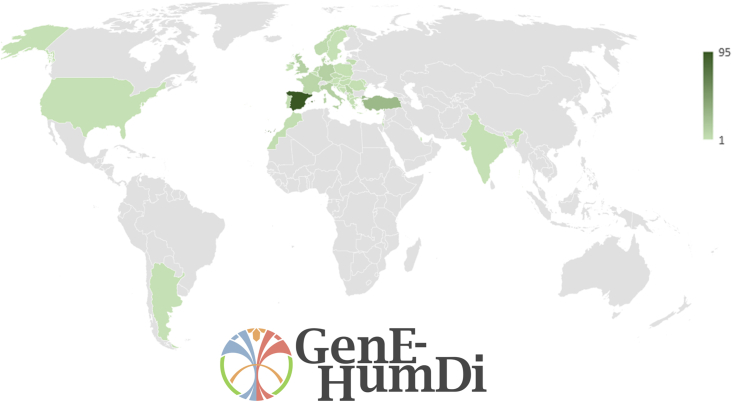
In the context of advancing scientific and technological innovation, the COST (European Cooperation in Science and Technology, cost.eu) provides a platform for researchers and experts to collaborate and exchange knowledge and expertise across different fields and disciplines. COST’s central mission is to promote cross-border collaboration and networking among researchers and institutions spanning various European and neighboring countries with the aim of advancing scientific and technological innovation in Europe. These initiatives involve a series of calls for projects, where researchers and institutions can submit proposals for scientific and technological projects related to the initiative’s theme. The funding is provided to selected projects, which are expected to collaborate and network within the framework of the initiative. In particular, the COST Action Genome Editing to treat Human Diseases (GenE-HumDi), with 269 participants distributed across 38 countries (Figure 1) is focused on exploring the use of GE to treat diseases that affect humans.
Figure 1.
The map illustrates the global reach of GenE-HumDi COST Action, with 269 participants distributed across 38 countries
Each participant is actively involved in at least one of the consortium’s working groups.
Specifically, the GenE-HumDi Action aims to promote collaboration between academic institutions, pharmaceutical companies, hospitals, regulators, and patient advocacy associations to accelerate the translation of GE technologies into effective treatments for human diseases. During its 4 years of operation, the GenE-HumDi network (https://www.genehumdi.eu/) aims to regularly discuss the establishment of standard operating procedures and guidelines focusing on key areas for the accelerated translation of GE into the clinic: (1) design of improved GE tools and their delivery; (2) assessment of the safety of GE platforms; (3) creation of a roadmap for the translation of GE from bench to bedside; (4) mapping of industry engagement and intellectual property; and (5) evaluation and promotion of regulatory guidelines for GE clinical translation and commercialization. The implementation of GenE-HumDi, spanning 26 member countries of COST, is overseen by a management committee composed of national experts, as well as a chair, Dr Karim Benbdellah, and a vice-chair, Dr Alessia Cavazza. The initiative has been structured into seven working groups (WGs). A dedicated WG, named "Improvement of GE technologies" and led by Dr Rasmus O. Bak, has been established to consolidate information on the efficiency and specificity of various existing GE tools. A "Delivery Strategies" WG led by Dr Yonglun Luo has been established with the aim of evaluating the optimal delivery methods of different GE tools for each cell type, animal model, and route of administration. The third WG, headed by Dr Ayal Hendel, focuses on “safety issues” related to GE. Its main objective is to outline and standardize current methodologies for assessing the cytotoxicity and genotoxicity of different GE platforms, including off-target and on-target effects, unintended recombination events, and cell population biases. A fourth WG, “Translation into the Clinic” led by Dr Alessia Cavazza, is dedicated to the multifaceted translational aspects of GE for clinical practice. The primary objective of this WG is to evaluate the status of GE clinical translation in Europe for rare inherited diseases, cancers, and infectious diseases, as well as to develop a roadmap for protocol adoption and manufacturing of GE-based medicines. The “Technological Transfer and Industry” WG headed by Monika Paule aims to draw guidelines to provide cost-effective GMP-grade sourcing of GE-based medicines and promote intellectual property management between collaborating partners. The WG focused on “Regulatory Issues,” headed by Prof Toni Cathomen, aims to ensure the regulatory adequacy of preclinical models and methods used to assess the efficacy and safety of various in vivo and ex vivo GE tools. This WG group also aims to promote the development of regulatory guidelines for the translation of GE into clinical practice. Finally, a "Dissemination" WG led by Dr Lluis Montoliu focuses on the promulgation of the results stemming from our Action and the integration of research and data analysis to decrease knowledge fragmentation among partners and to communicate the relevant advances in this field to a wider non-scientific audience. Eighty-seven scientists belonging to the Gene-HumDi Action from both academia and industry from 29 different countries gathered at the Center for Genomics and Oncological Research (GENYO) for the inaugural meeting of the Action (13–15 March 2023, Granada, Spain), to review the state-of-the-art of the GE field and debate on its future directions. The following proceedings summarize key considerations and highlights from the meeting, which revolved around the seven main pillars of the Action.
Improving gene editing technologies
Development of GE technologies moves at an exceptionally high pace. Fueled by the advent of the CRISPR-Cas technology, the community has seen over the past 10 years several new GE tools being developed and existing tools being refined. This has shaped major research areas in (1) conventional DNA editing methods relying on DNA double-strand breaks (DSBs), (2) DSB-free gene editing tools such as BE and PE, (3) tools that enable site-specific integration of large genetic payloads, and (4) genetic engineering tools that do not change the DNA code but instead edit RNA or manipulate epigenetic and/or transcriptional status of a gene. Various GE technologies have been developed and, while CRISPR-Cas-based tools dominate the field, it is important to recognize alternative tools that have been evaluated in clinical studies, such as ZFNs and TALENs, as well as less embraced strategies for editing the genome.
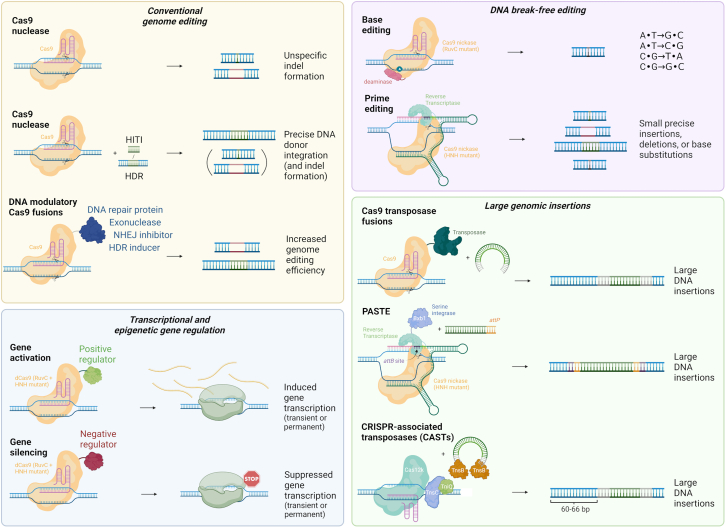
CRISPR-Cas was originally developed to introduce site-specific DNA breaks to edit genomic sequences, but as shown in Figure 2 it has been reconfigured and repurposed for a wide range of applications. Rasmus Bak (Aarhus University, Aarhus, Denmark) and Julian Grünewald (Technical University of Munich, Munich, Germany) presented some of the recent efforts made in developing and applying new tools for genetic engineering, including BE and PE, as well as CRISPR-Cas-based transcriptional modulators.1,2 These tools allow researchers to modify DNA in a more precise and controlled manner, with the potential to correct disease-causing mutations and without the need to introduce DSBs at the target site. PE can create new DNA sequences by inserting, deleting, or replacing specific genomic sequences. CRISPR-Cas transcriptional modulators, on the other hand, allow researchers to turn genes on or off without changing the DNA sequence.3
Figure 2.
Gene editing technologies based on CRISPR-Cas systems
The CRISPR-Cas toolbox contains multiple versions of Cas enzymes combined with other proteins to manipulate genomic DNA. For conventional genome editing, Cas9 nucleases are used to create DNA double-strand breaks (DSBs) that facilitate insertions or deletions (indels) of base pairs at the target site introduced by DSB correction via the non-homologous end-joining (NHEJ) pathway, leading to disruption of target DNA sequences. For precise edits, Cas9 nucleases are supplemented with a DNA template for its integration into the desired target locus by either homology-directed repair (HDR) or by homology-independent targeted integration (HITI). These approaches are accompanied by the simultaneous introduction of undesired indels, as such other approaches have fused different DNA modulatory proteins to Cas9 to alter the indel spectrum or to affect the HDR:indel ratio to favor HDR. The DNA break-free base and prime editors (BE and PE, respectively) display high product purity of the editing outcome and highly decrease the risks associated with DNA DSBs, including induction of gross chromosomal aberrations. BE mediates single-base substitutions, while PE can create small precise insertions, deletions, or base substitutions. To induce insertion of large DNA regions, some systems utilize Cas9 fused to transposases, serine integrases (PASTE) or CRISPR-associated transposases (CASTs) to insert large donor DNA templates. PASTE inserts an attB site into the desired genomic location by prime editing, followed by the integration of the donor DNA via the serine integrase (e.g., BxbI) acting on the flanking attP site. The CAST system uses CRISPR-associated transposases to insert transposon DNA engineered to carry the desired cargo. To manipulate the transcriptional status of a gene, a nuclease-deactivated version of Cas9 (dCas9) is employed that maintains the ability to bind a specific DNA target. When fused to transcriptional activators or repressors, target genes can be dialed up or down. By instead using fusion proteins that regulate the epigenetic status of a gene, inherited epigenetic marks can lead to permanent modulation of transcription.
During his presentation, Giedrius Gasiunas (CasZyme, Vilnius, Lithuania) discussed the challenges associated with using the Cas9 proteins for GE applications. The requirement for a protospacer adjacent motif (PAM) to bind to a target, the lack of specificity, and size limitations for its viral delivery are significant obstacles for Cas9-based GE. To overcome these challenges, Gasiunas and his team explored the natural variation of Cas enzymes to develop RNA-guided tools with diverse and potentially beneficial properties.4 Through biochemical screens of Cas9 orthologues of the type II family and Cas12 proteins from the novel type V family, the team found a wide range of activities both in vitro and in-cellula, making them an attractive alternative to traditional Cas9 enzymes.5 Similarly, Lluis Montoliu (CNB-CSIC, Madrid, Spain) presented a collaborative effort leading to the resurrection of some CRISPR-associated nucleases from tens to thousands of millions of years ago, obtained through statistical inference applying a maximum likelihood approach, while searching for novel Cas nucleases from bacteria that might have not interacted with human beings and, as such, were not known by our immune system. He presented the corresponding validation experiments in human cells showing that these ancestral Cas nucleases were suitable for GE applications, with a comparable efficiency to today’s nucleases.6 Marc Güell (Pompeu Fabra University, Barcelona, Spain) discussed the limitations of the CRISPR technology when employed to generate large genomic changes and the need for improved methods. To address this issue, Güell and colleagues combined the CRISPR system with the gene-transfer capacity of transposases to create novel gene writers that can efficiently introduce large genomic changes with precise control.7 This new system, known as Find and Cut-and Transfer (FiCAT) enables researchers to evolve and optimize CRISPR enzymes for a wide range of applications, including gene regulation and epigenetic modifications. Duško Lainšček (National Institute of Chemistry, Slovenia) presented his work on augmenting the action of the CRISPR-Cas9 system by noncovalent tethering of Cas9 protein to the exonuclease III via coiled-coil forming heterodimeric peptides (CCExo).8 Indeed, bringing the exonuclease III into proximity of the Cas9-induced DSB results in additional DNA recessions with final larger deletions and increased rate of gene alterations. CCExo showed robust increased GE action determined for several different genes in various cell lines, as well in human primary cells and in somatic adult cells in vivo, with no additional undesired DNA cleavage observed by circularization for in vitro reporting of cleavage effects by sequencing (CIRCLE-seq). As such, this system could be applied to treat different diseases, which was exemplified by the speaker by using CCExo for targeting the BCR-ABL1 fusion chromosome, a main cause of chronic myelogenous leukemia, in patient’s cells and in xenograft animal model.
Strategies to deliver GE tools
Unlike the rapid and continuous development of GE technologies, approaches to deliver the GE cargos specifically and efficiently into the target cells and tissues are evolving at a much slower pace and now represent one of the major limiting factors in advancing CRISPR therapeutic applications. Although smaller or split Cas proteins were engineered to make them compatible with the low immunogenic and pathogenic adeno-associated viral (AAV)9 vectors, concerns are not fully addressed regarding the unintended off-target effects due to relatively high and long-term AAV-mediated CRISPR expression, as well as CRISPR-independent adverse effects associated with viral vector genome integration into host cells. Besides, it is vital to select the right delivery method to achieve a better GE outcome in terms of efficiency, efficacy, and specificity depending on the GE strategies and applications chosen.
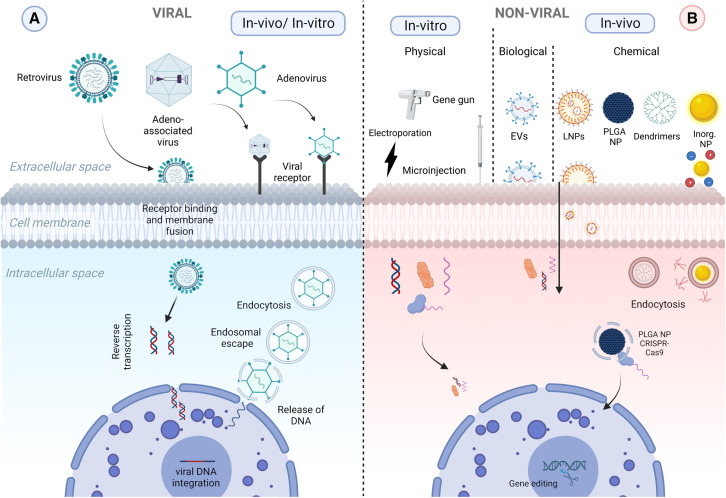
The overarching goal of the “Delivery Strategies” WG3 and session is to critically evaluate and identify the most effective ex vivo and in vivo delivery systems for GE applications (illustrated in Figure 3). Specifically, the different speakers assessed the suitability of various delivery methods for delivering genetic material to different cell types and animal models, with the aim of advancing the field of GE and promoting the development of novel therapeutic approaches, based either on viral and non-viral delivery systems. As excellently illustrated by Virginia Arechavala-Gomeza (Biobizkaia Health Research Institute, Bilbao, Spain), delivery of nucleic acid therapeutics has been the subject of much interest since early antisense oligonucleotide therapies were developed with mixed results10,11: while therapies targeting tissues such as the eye or the central nervous system provided patients with life-changing therapeutic options,12 others targeting the muscle showed limited clinical benefit.13 Researchers in the field soon realized that delivery was the main hurdle stopping these new drugs from reaching their full potential.14 The issues related to the delivery of these compounds can be divided in two areas: access to target tissue and escape from the endosomes.15 Most nucleic acid therapeutics, and in particular GE tools, are too large and too charged to bypass these barriers on their own, hence there is an urgent need for alternative and more efficient delivery systems.
Figure 3.
Gene editing delivery systems
Schematic illustration of the varieties of the tools to deliver genome editing components, classified into two categories based on the different constituents and cellular entry mechanisms: viral (A) and non-viral methods (B). In the first category, the most widely used viruses for delivery of GE tools are retroviruses, adeno-associated viruses, and adenoviruses, where entry mechanisms of the gene editing components into the target cell are virus specific. Viral methods can be used for both in vitro and in vivo applications. The non-viral delivery methods can be further split into three subgroups: physical methods utilized for in vitro gene editing (gene gun, electroporation and microinjection), and biological (extracellular vesicles, EVs) or chemical (lipid nanoparticles, LNPs; Poly lactic-co-glycolic acid nanoparticles, PLGA NPs; dendrimers and inorganic nanoparticles) methods for in vivo gene editing.
Rosario Sanchez (GENYO-UGR, Granada, Spain) presented on the use of non-viral nanoparticles as delivery systems for GE tools in cancer immunotherapy. They developed a multifunctional nanodevice capable of efficient delivery of CRISPR-Cas9 and GE tools to T cells, enhancing their ability to recognize and target cancer cells. Additionally, the potential of polymeric and plasmonic nanoparticles as delivery systems for mRNA-based therapeutics was discussed. Poly(lactic-co-glycolic acid) PLGA nanoparticles offered protection and improved cellular uptake of mRNA, while plasmonic nanoparticles increased mRNA concentration and uptake through photothermal effects.16,17,18 The presentation also compared PLGA nanoparticles and electroporation to deliver CRISPR-Cas9, highlighting their advantages and limitations in terms of effectiveness, specificity, and safety. Although PLGA nanoparticles showed lower editing efficiency compared with nucleofection, further improvements in encapsulation methods were suggested.
On the other hand, dendrimers represent a special family of polymers that are emerging as promising vectors for nucleic acid delivery by virtue of their well-defined dendritic structure and cooperative multivalency.19,20,21,22,23,24 Ling Peng (Aix-Marseille Université, Marseille, France) discussed the potential application as well as limitations of amphiphilic dendrimers to improve endosome release in nucleic acid delivery. These conjugates can mimic lipid vectors and exploit membrane-fusion-mediated delivery, while simultaneously retaining the multivalent properties of polymer vectors that allow endocytosis-based delivery benefiting from the proton-sponge effect. In the future, researchers may investigate the potential of using amphiphilic dendrimers as delivery vehicles for various types of RNA therapeutics, such as antisense oligonucleotides for gene silencing, small activating RNA for gene activation, and mRNA and single guide RNA for GE.
Another emerging promising approach that is now widely utilized in clinical settings for the delivery of GE reagents involves the use of synthetic lipid nanoparticles (LNPs). By encapsulating specific gRNA and Cas9 mRNA, LNPs provide a DNA-free CRISPR delivery system that can be readily taken up by cells through endocytosis.25 Julie Lund Petersen (Aarhus University, Aarhus, Denmark) discussed the formulation and utilization of LNPs for the encapsulation of CRISPR RNA. Notably, their research demonstrated the efficacy of LNPs in delivering GFP mRNA, as evidenced by the high levels of GFP expression observed in vitro in cultured cells and in vivo following subretinal and hippocampal injections in mice.
Safety issues related to GE therapeutic applications
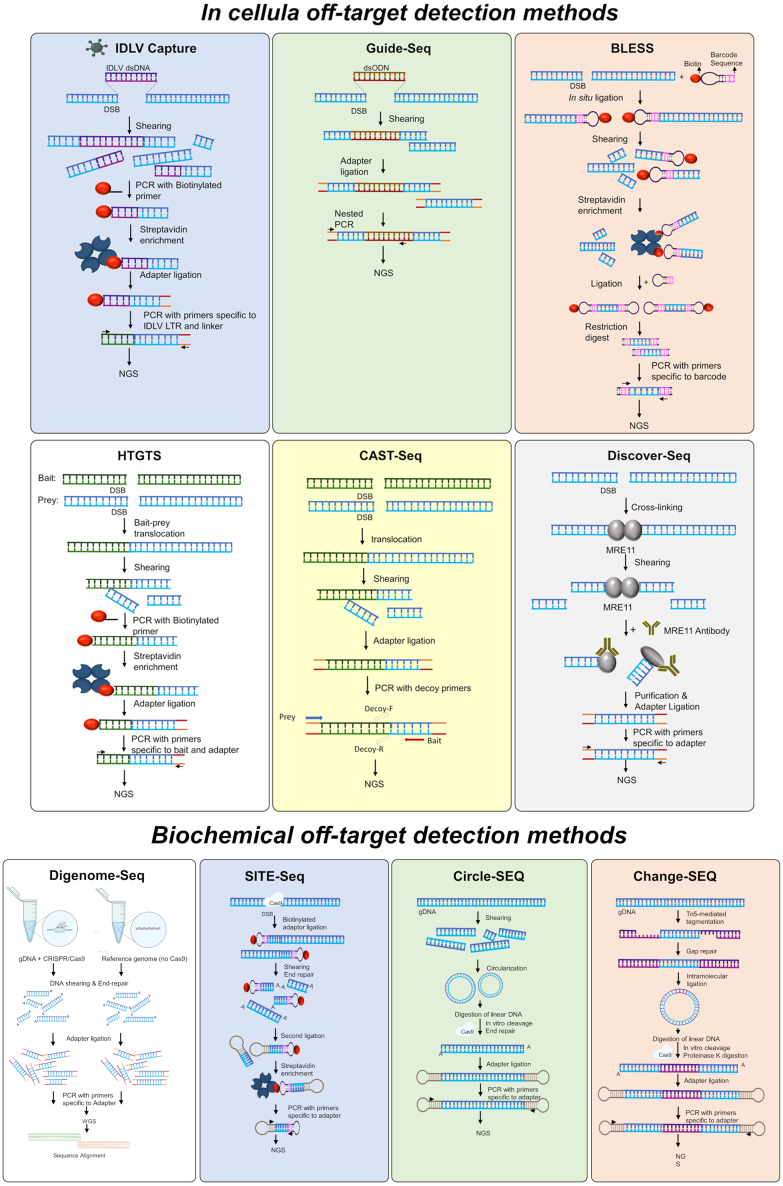
Despite the revolutionary nature of CRISPR-Cas9 GE, concerns about its promiscuous nuclease activity and unintended off-target effects have been raised. The off-target activity may lead to the introduction of unintended insertions/deletions (indels) or structural variations, posing significant safety concerns. One approach to mitigate off-target genotoxicity involves the development of more specific nucleases, such as alternative Cas variants.26 Another approach involves utilizing chemically modified gRNAs, which have been shown to increase both the efficiency and specificity of the system.27 Last, extensive research is dedicated to the development of sensitive assays and tools for the prediction and detection of off-target sites. Strategies aimed at enhancing sensitivity include the utilization of Cas9 overexpression systems, advancements in sequencing methods, and the development of improved computational algorithms. The insights gained from the off-target detection methods could in turn aid in the selection and refinement of more precise editing systems. Apart from off-target genotoxicity, additional toxicity concerns arise from the foreign DNA that may be used to deliver any of the editing components, be it by non-viral or viral vectors. The presence of foreign nucleic acids triggers DNA damage response (DDR) pathways, leading to cell-cycle arrest, transcriptional blockage, reduced proliferation, and potentially apoptosis.28,29 Ensuring safety in the various aspects described is of utmost importance, particularly as Cas9-based therapeutic GE has entered clinical trials. Several talks focused on establishing a standardized protocol for the measurement and evaluation of on- and off-target activity, which included generating big data to develop models that better predict off-target activity. The speakers compared the prediction of cell-based, cell-free, and in silico off-target identification methods and highlighted the need for a combination of experimental methods to assess the safety of gRNAs (reviewed in Figure 4). Shengdar Q. Tsai (St. Jude Children’s Research Hospital, Memphis, USA) the keynote speaker of the meeting, discussed several cell-based and biochemical methods for understanding off-target effects, including integration-deficient lentiviral capture, high-throughput genome-wide translocation sequencing, and genome-wide unbiased identification of DSBs enabled by sequencing (GUIDE-seq), and circularization for high-throughput analysis of nucleases genome-wide effects (CHANGE-seq) among others.30,31 He highlighted the importance of using sensitive and unbiased methods for defining the potential safety and genotoxicity of editors through the lens of developing SCD GE therapies. Ayal Hendel (Bar-Ilan University, Ramat Gan, Israel) addressed the safety concerns involved in the development of a homology-directed repair (HDR)-based correction strategy in CD34+ hematopoietic stem and progenitor cells (HSPCs). To assess potential off-target effects, the Hendel group implemented a two-step approach involving the identification of off-target sites using GUIDE-seq in a Cas9 overexpression system, and the quantification of their activity through rhAmpSeq. While high off-target activity was observed in an HEK-293 Cas9 cell line, the use of ribonucleoprotein (RNP) complexes and high-fidelity (Hi-Fi) Cas9 in CD34+ HSPCs significantly reduced toxicity.32 Hendel also discussed viral vector-related safety aspects, noting that viral HDR donors triggered a DDR proportional to the AAV multiplicity of infection used.33 Furthermore, higher viral doses led to extended vector genome presence, decreased cell yields, and positive selection of unedited cells. However, by reducing the viral vector dosage, significant improvements were observed in HSPC survival, allowing successful T cell differentiation of corrected SCID-RAG2 patient HSPCs in vitro.34 These findings highlight that minimizing the viral vector dose to strike a delicate balance between non-toxicity and optimal editing efficiency is essential for upholding the highest standards of safety. Marcello Maresca (Astrazeneca, Molndal, Sweden) discussed the limitations of current strategies for mapping CRISPR-Cas9 off-target effects, which can impact their sensitivity. His team has developed a new off-target assessment workflow using duplex sequencing, which can increase the sensitivity of CRISPR-Cas9 mutation detection by one order of magnitude and the reduction of false positive and negative rates, which enabled the identification of previously missed off-target mutations associated with wild-type SpCas9 treatment in an in vivo humanized PCSK9 mouse model of hypercholesterolemia. In addition to this innovative technique, Maresca also discussed the development and features of a novel, precise Cas9 nuclease, known as PsCas9. This new nuclease shows high intrinsic specificity and is considered a promising alternative to SpCas9 for both research and clinical purposes in the field of GE. Maresca believes that using a highly sensitive off-target assay in conjunction with PsCas9 could provide more accurate GE treatments and safety assessment.35
Figure 4.
Sensitive assays and tools for prediction and detection of off-target sites
Schematic overview of in-cellula off-target detection methods. Upper panel, from left to right: integrase-deficient lentivirus capture (IDLV Capture); genome-wide, unbiased identification of DSBs enabled by sequencing (Guide-Seq); direct in situ breaks labeling, enrichment on streptavidin and next-generation sequencing (BLESS). Lower panel, from left to right: high-throughput, genome-wide translocation sequencing (HTGTS); chromosomal aberrations analysis by single targeted linker-mediated PCR sequencing (CAST-seq); discovery of in situ Cas off-targets and verification by sequencing (Discover-seq). DSB, double-stand break; NGS, next-generation sequencing; dsODN, double-strand oligo DNA; LTR, long terminal repeat. Schematic overview of biochemical off-target detection methods, from left to right Digenome-seq, Site-seq, Circle-seq and CHANGE-seq.
Jan Gorodkin (University of Copenhagen, Copenhagen, Denmark) highlighted the challenge of balancing on-target efficiency and minimizing off-target effects when selecting gRNAs for a given genomic region, and emphasized the need for advanced computational tools to analyze and predict gRNA activity, as well as the importance of experimental validation to confirm the predicted results. In this regard, Gorodkin’s team developed the CRISPR on/off framework, a computational methodology based on a deep learning-based predictor and a binding energy model, which aims to facilitate the selection of optimal CRISPR-Cas9 gRNAs for GE.36,37,38,39,40 Its web interface (http://rth.dk/resources/crispr) provides easy access to the tools and allows users to input genomic regions of interest for gRNA design. The deep learning-based predictor uses a convolutional neural network to predict on-target efficiency, while the binding energy model predicts off-target effects based on the binding energies of the gRNA and potential off-target.36 Stefan Seemann (University of Copenhagen, Copenhagen, Denmark) presented a tool called CRISPRroots, which combines off-target predictions, variant calling, and differential expression analysis of RNA-seq data to evaluate successful on-target editing while also identifying any predicted off-target effects that could be contributing to the observed gene expression changes.37 The tool uses a combination of off-target predictions, variant calling, and differential expression analysis of RNA-seq data to evaluate successful on-target effects while also identifying potential off-target effects in genes with altered expression. The method provides an unbiased analysis of somatic variants and differentially expressed genes linked to predicted on- and off-targets, allowing researchers to validate on-target edits and prioritize potential off-targets for experimental validation.37 Giandomenico Turchiano (University College London, London, UK) discussed the comprehensive characterization of genome stability in ex vivo gene-edited primary cells. He presented various methods for evaluating mutations, including computer-based prediction of off-targets, in vitro and in-cellula DSB in DNA, in vitro base editor off-targets, and chromosomal aberrations. Turchiano emphasized the importance of using a combination of techniques to gain a comprehensive understanding of genome stability. He introduced the MEGA approach, which utilizes multiplexed droplet digital PCR (ddPCR) for mutation analysis, enabling the simultaneous detection and tracking of different types of mutations induced by designer nucleases. MEGA provided insights into DNA repair dynamics, quantified the presence of episomal AAV DNA, and proposed RNP thresholds to optimize HDR efficiency. Additionally, Turchiano discussed the association of low-frequency indels with other aberrations and highlighted the findings from using the CAST-seq technology, revealing large deletions, inversions, and translocations at on-target sites, including unbalanced translocations and homology-mediated events. On a similar note, Toni Cathomen (Medical Center-University of Freiburg, Freiburg, Germany) discussed the concept of sensitivity and specificity when it comes to off-target analysis. He explained that CAST-seq has a high sensitivity, with a limit of detection of approximately one event in 10,000 cells. making CAST-seq analysis more sensitive than traditional translocation assays41 Importantly, the linear range for CAST-seq analysis is 0.01%–1%, allowing it to detect changes in the frequency of events within this range. In collaboration with AstraZeneca Cathomen’s team tested CAST-seq in vivo edited samples using a CRISPR-Cas9 system targeting Pcsk9 with a “promiscuous” gRNA.42 They established that out of the 99 off-target sites identified by CAST-seq, 90 could be tested by NGS and 88 of them were true off-target sites as further validated by using rhAmpSeq and CRISPECTOR.43
As noted by Manuel Gonçalves (Leiden University Medical Center, Leiden, Netherlands), achieving precise GE using programmable nucleases remains challenging mostly due to the prevalent repair of DSBs by the non-homologous end-joining (NHEJ) repair pathway rather than HDR. Besides local- and chromosome-wide generation of complex structural variants,44 NHEJ can yield target protein imbalances and loss of cell fitness, which limit the effectiveness of DSB-dependent GE approaches.45 In addition, on-target DSBs are known to trigger the activation of P53, which hinders the effectiveness of GE in therapeutically relevant DNA damage sensitive stem cells.46,47 While the use of high-specificity programmable nucleases can dramatically reduce the occurrence of off-target DNA cleavage, they are not capable of eliminating the unintended effects caused by on-target DSB formation, with a high risk of affecting the function or fitness of edited cells. In this context, the Gonçalves’ team is currently exploring HDR-based gene knock-in techniques that rely on conventional or high-specificity CRISPR-Cas9 nickases, as single-stranded DNA breaks (SSBs), or nicks, are not canonical NHEJ substrates.45,48,49 This research builds on earlier findings from his laboratory showing that although SSBs are per se poor HDR stimuli, simultaneous formation of SSBs at chromosomal sites and matching donor DNA constructs elicits HDR-mediated gene knock-ins while avoiding P53-dependent DDR activation.50,51 Indeed, fostering otherwise inefficient SSB-dependent HDR such as by in trans paired nicking (ITPN) approaches allows for seamless chromosomal editing, including at multi-copy or essential genomic tracts.45,49 Moreover, ITPN could also be suitable for allele-specific editing,52,53 one-step biallelic editing,48,54 or knocking-in whole transgenes at safe harbor loci.45,49
Translation of GE therapeutic platforms into the clinic
As recently pointed out,55 there are >70 GE clinical trials (with <10 sponsored by EU countries; clinicaltrialsregister.eu) currently ongoing or in the recruiting phase around the globe, highlighting the incredible pace at which the field has advanced in the past 10 years, since the development of CRISPR as a GE tool. Of these trials, almost half were related to genetically modified T cell-based immunotherapies, 25% to viral infections and 35% to monogenic disorders, mostly affecting the hematopoietic system. One of the most exciting milestones in the field is likely represented by the imminent approval by US and European regulatory agencies of the first CRISPR-based medicine to treat SCD and β-thalassemia (exa-cel). Clinical successes like exa-cel are bringing the potential of a GE-based treatment closer to patients; however, more therapeutic opportunities are needed and further obstacles need to be addressed for a smooth transition from preclinical research to clinical applications. These include the urgent need to establish standardized protocols and procedures for GE manufacturing, delivery, and monitoring in clinical trials, as well as clearing key regulatory aspects of GE research and, critically, establishing a pricing framework for these innovative therapeutics. The talks delivered during the “Translation into the clinic” WG session aimed at discussing the state-of-the-art of GE clinical translation for different diseases (depicted in Figure 5) and setting the stage for the development of a roadmap that could guide future research and investment in this promising field.
Figure 5.
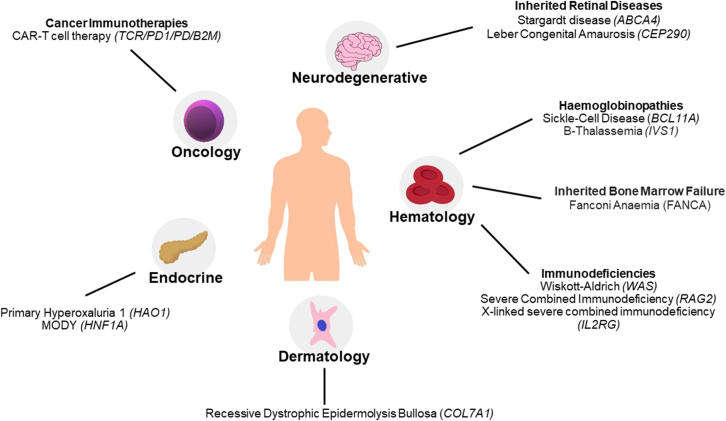
Applications of gene editing in the treatment of inherited rare diseases, cancer, and infectious diseases
Summary diagram of conditions treatable by gene editing that are currently being investigated by GenE-HumDi COST Action members. Specific gene targets are listed in italics per each disease.
A series of presentations illustrated the preclinical and clinical development of GE strategies to treat genetic blood disorders, which are among the most advanced GE therapeutic applications now entering the clinic. The groups of Alessia Cavazza (University College London, London, UK) and Ayal Hendel (Bar-Ilan University, Ramat Gan, Israel) are devising HDR-based CRISPR-Cas GE platforms to treat various primary immunodeficiencies, such as Wiskott-Aldrich syndrome, syndrome RAG2-severe combined immunodeficiency (RAG2-SCID), and X-linked SCID SCID-X1, that rely on the use of AAV6 donor vectors to introduce the corrective gene into its endogenous locus in HSPCs.34,56 Cavazza highlighted the challenges that need to be addressed when translating advanced therapies from bench to bedside, and in particular the plethora of technologies that are required to thoroughly assess correct manufacturing and safety of a GE product in late preclinical studies, pinpointing the importance of establishing standardized protocols and guidelines to streamline the access of these treatments to patients. The development of therapeutic strategies to treat such ultrarare genetic diseases entails a series of further complications, including the availability of patient samples and the need to implement newborn screening programs for early diagnosis of the disease to ensure therapeutic benefits from these GE strategies. In this regard, Hendel’s lab has established a disease modeling system for several forms of SCIDs in primary human HSPCs using a multiplexed HDR platform based on CRISPR-Cas9-AAV6, thus allowing to easily assess the efficacy of a therapeutic approach while sparing precious patient samples. Hemoglobinopathies, such as SCD and β thalassemia, are the most frequent monogenic diseases worldwide affecting the production of the adult hemoglobin β-chain. The current curative treatment for this condition is allogeneic HSPC transplantation; however, some of its limitations have pushed scientists to find alternative therapeutic options. CRISPR-Cas has emerged as a powerful tool to treat hemoglobinopathies and many approaches have been developed in the past few years, some of which have already reached the clinical stage. Shengdar Q. Tsai (St. Jude Children’s Research Hospital, Memphis, TN, USA) provided an overview of the encouraging outcomes of current clinical trials for exa-cel57 and shared his team’s efforts to test an autologous genome-edited HSPC therapy for SCD at St. Jude Children’s Research Hospital.58 Annarita Miccio (Imagine Institute, Paris, France) proposed to use BE as an alternative approach to correct prevalent β-thalassemic mutations, such as IVS1-110 G>A.59 This approach offers the potential to improve the efficacy and safety of autologous gene-edited HSPC transplantation, which is currently limited by the safety concerns raised by unwanted on- and off-target events due to the cleavage of the Cas9 nuclease. Cells corrected via BE showed increased levels of adult hemoglobin production, leading to an improvement in the β-thalassemic phenotype. A similar approach shown by Annarita Miccio is the use of BEs to generate mutations in the −200 region of the fetal globin promoters that create a KLF1 activator binding site, with the aim of inducing the expression of fetal hemoglobin as a universal treatment for β-hemoglobinopathies. She reported that correction of patient HSPCs using BE is safe and leads to reactivation of fetal hemoglobin at higher levels compared with a Cas9-nuclease approach, while avoiding the generation of indels and large genomic rearrangements.60 With shared interest in hemoglobinopathies, mutation-specific repair and fetal hemoglobin induction, Carsten W. Lederer (The Cyprus Institute of Neurology & Genetics, Nicosia, Cyprus) also introduced the concept of tag-activated microRNA (miRNA)-mediated endogene deactivation (TAMED) as a potentially general therapy and research approach.61 TAMED draws on targeted insertion of miRNA recognition site (MRS) tags and on endogenous miRNA expression to achieve lineage-specific silencing of tagged endogenes. For the abundant erythroid miR-451a and the use case of BCL11A downregulation based on tagging with miR-451a cognate MRSs, Lederer’s team established proof of principle for TAMED, while concluding that therapeutically relevant efficiencies and wider application will depend on improved donor chemistries.62,63
Potential therapies employing nucleases and BE have been developed for other blood disorders, such as Fanconi anemia (FA), a rare inherited bone marrow failure syndrome. The main obstacle in treating this condition is the reduced number of HSPCs found in these patients; however, once corrected, these cells can ameliorate the diseases as shown in transplantation studies. Paula Rio (CIEMAT, Madrid, Spain) is optimizing gene editing strategies to correct HSPCs from FA patients. The team exploited BE to introduce a silent mutation in a safe harbor locus in HSPCs and observed editing efficiencies up to 80%, while preserving the clonogenic and long-term repopulating ability of these cells.64 They employed a parallel approach using a gRNA to generate a therapeutic SNP in primary FA HSPCs achieving a 42%–64% frequency of correction and a proliferative advantage of corrected cells. Overall, the findings demonstrate that various GE strategies can be used to correct mutations in FA patients and provide promising insights for the future clinical application of gene editing in FA.
Cancer immunotherapy is one of the most promising recent breakthroughs in medicine and aims at driving the patient’s own immune system to fight tumor cells. Within this field, cell-based immunotherapy using T cells engineered with chimeric antigen receptors (CARs) has gained momentum after the approval of several CAR-T cell-based medicines for the treatment of CD19+ B cell malignancies by both the FDA and EMA.65 GE is being utilized to optimize existing technologies, such as for example to manufacture “off-the-shelf” CAR-T cell products or to increase their safety, sensitivity and longevity through gene knockout/knockin.66 As such, GE applications in the context of cell-based immunotherapy has been the focus of many talks during the meeting. Karim Benabdellah (GENYO, Granada, Spain) presented the CARAML-EXO project, which aims to enhance CAR-T cell therapy for acute myeloid leukemia. The project aims to generate safe and effective universal CAR-T lymphocytes by knocking out the T cell receptor (TCR) and PD1 genes, and establishing a suitable combination of TCR/PD/PD1 knockout CAR-T subpopulations. Juan R. Rodriguez-Madoz’s team (CIMA Universidad de Navarra, Navarra, Spain) has developed a novel approach using CRISPR-based GE technologies and virus-free gene-transfer strategies with Sleeping Beauty transposons. Their aim is to generate allogeneic CAR-T cells that are depleted of human leukocyte antigen class I (HLA-I) and TCR complexes. This one-step generation of edited CAR-T cells has been optimized for large-scale production, enabling their potential use in clinical settings. Finally, Noelia Maldonado-Pérez from Francisco Martin’s group (GENYO, Granada, Spain) investigated the efficacy and safety of generating off-the-shelf TCR knockout (KO) ARI CAR-T cells (ARI CAR-T cells is the first academic CAR-T cell product authorized by the the Spanish Agency of Medicines and Medical Devices [AEMPS] under the “hospital exemption” approval pathway). Although KO leads to on-target large deletions that should be monitored as a potential safety issue, the data presented by Maldonado-Pérez suggest that disrupting the TCR proves to be a viable strategy for producing functional allogeneic ARI-0001 CAR-T cells with a similar phenotype and antitumor efficacy.67
Despite the potential clinical efficacy of GE combined with CAR-T cell therapy in hematologic diseases, challenges such as tumor heterogeneity, immune evasion, limited trafficking and persistence, adverse effects, immunosuppression, and manufacturing issues restrict their potential in treating solid tumor diseases or viral infections in immunosuppressed patients. Michael Schmueck-Henneresse’s team (Charité-Universitätsmedizin Berlin, Germany) aims to overcome some of these challenges by understanding how T cells coordinate an effective immune memory against virus-infected or tumor cells and how this can be specifically modified for therapeutic purposes. They use CRISPR-Cas-based gene modifications to enhance T cell migration to solid tumors through transgenic expression of chimeric receptors as a targeted adaptation of homing chemokine systems for CAR-T cell products. Both Michael Schmueck-Henneresse and Gal Cafri (Sheba Medical Center, Ramat Gan, Israel) also discussed the use of effector T cells for the treatment of tumors and viral infections in vulnerable patients, as well as the clinical use of regulatory T cells after transplantation. On a similar note, Cristina Maccalli (Sidra Medicine, Doha, Qatar) employs genomic and immunological characterization of cancer stem cells to identify the mechanisms of resistance to T cell-mediated immune responses in solid tumors, increasing cell-based immunotherapy efficacy by selecting high-affinity, antigen-specific TCRs.
A plethora of therapeutic GE applications for a variety of other diseases were also discussed. For example, Laura Torrella (CIMA, University of Navarra, Spain) is exploring the potential of GE to treat a rare metabolic disorder that can lead to end-stage renal failure and caused by mutations in the Hao1 gene.68 Torrella developed GE strategies using CRISPR-Cas9 nickases delivered by AAV to the liver of a PH1 mouse model to treat PH1, showing reduced oxalate accumulation and prevention of renal damage. Jose Bonafont (DanausGT, Madrid, Spain) presented a GE-based curative approach for recessive dystrophic epidermolysis bullosa (RDEB), a rare skin genetic disorder caused by mutations in the COL7A1 gene, which encodes the type VII collagen protein (C7). Jose Bonafont used the CRISPR-Cas9 system to edit the genome of patients with RDEB. Specifically, two gRNAs were used to direct the Cas9 enzyme to cut the DNA at two specific sites within the COL7A1 gene, with the aim to delete the exon-bearing mutation and restore the production of functional type VII collagen.69 He also presented an HDR-based protocol to precisely correct the mutation by using CRISPR-Cas9 in combination with AAV6-donor template delivery.70
Last, two speakers addressed the use of CRISPR-based disease modeling as a powerful tool for understanding disease mechanism, identifying potential therapeutic targets, and testing new treatment approaches. Alejandro Garanto (Radboud University Medical Center, Nijmegen, the Netherlands) is working on the development of molecular therapies for inherited retinal diseases, with a special focus on Stargardt disease. The team has used GE tools in different ways: (1) to correct or include variants to generate isogenic controls and verify pathogenicity upon differentiation to human retinal models; and (2) to develop a novel therapeutic strategy to target pathogenic variants in ABCA4. Since the eye is a model organ for therapeutics, the overall goal is to halt or slow down the progression of the disease to improve the quality of life of the patients. Neli Kachamakova-Trojanowska (Malopolska Center of Biotechnology, Jagiellonian University, Krakow, Poland) employed hiPSCs to study the causative molecular mechanisms of HNF1A-onset diabetes of the young (HNF1-MODY) which leads to increased risk for cardiovascular diseases and retinopathy.71 Kachamakova-Trojanowska’s team used the CRISPR-Cas9 system to introduce mutations in the HNF1A gene in a healthy donor hiPSC line. This approach allowed them to show that endothelial cells derived from hiPSCs with mono- or biallelic mutations in the HNF1A gene exhibited increased vascular permeability in comparison with the respective control cells,72 which could contribute to the endothelial dysfunction observed in patients with HNF1A-MODY.
Regulatory issues
The field of somatic GE trials is rapidly advancing in Europe and the United States, with a focus on cancer treatment and monogenic conditions. While the potential benefits of these therapies are apparent, the understanding of associated risks is still evolving. The WG on Regulatory Issues aims to facilitate the transition of these therapies from the lab to clinical trials by providing recommendations for evaluating their safety, quality, and addressing potential risks. During the first year of the Gene-HumDI Action, members of the Spanish Agency of Medicines and Medical Devices (AEMPS) and Andalusian Network for the design and translation of Advanced Therapies (RAdyTA) have joined our network and more representatives from national regulatory bodies of many EU countries will be recruited in the following years to ensure a thorough discussion of important regulatory aspects necessary for the successful translation of GE into clinical applications. Toni Cathomen (Medical Center-University of Freiburg, Freiburg, Germany) focused on the regulatory considerations for safety analyses of genetically engineered cell products in clinical applications. Cathomen emphasized that on-target and off-target aberrations are an inevitable side effect of engineering the genome. Regulatory agencies therefore require comprehensive off-target analyses in non-clinical studies to identify and mitigate the risk of genotoxicity. Incorporating regulatory requirements into the tests used for off-target analyses helps ensure that GE tools meet safety standards and increase the likelihood of regulatory approval for clinical use. Establishing common standards for quality control, data processing, performance parameters, and reference materials are hence crucial for accurate, reliable, and reproducible off-target analyses across different developers and platforms. Harmonizing these standards and parameters is essential for meeting regulatory guidelines and promoting the safe and effective use of GE technologies in clinical applications. María José del Pino (AEMPS, Spain) and Gloria Carmona (RAdyTA) explained the different types of Advanced Therapy Medicinal Products (ATMPs) in the European and Spanish regulatory context, and where currently treatments based on GE are framed challenges related to ATMP regulation and translation into GMP facilities. They emphasized that Spain in general, and Andalusia in particular, is a good host country and region, respectively, for the evaluation and implementation of clinical trials based on ATMPs, with multidisciplinary teams that have good experience in the evaluation and authorization of ATMP products. María José del Pino discussed the importance of scientific and regulatory advice in obtaining clinical trial authorization and marketing authorization for ATMPs and gene therapy. The assessment and counseling process for each product is conducted on a case-by-case basis, considering the unique characteristics of each ATMP, therapeutic area, and disease. There are three main pathways for assessment and counseling: (1) through the innovation office of AEMPS for non-profit institutions, academia, universities, start-ups, biotechnology, and pharmaceutical companies; (2) through EMA and its working groups, such as the Innovation Task Force or the Scientific Advice Working Party, as well as programs like STARS (Strengthening Regulatory Science); and (3) through the EU-Innovation Network (EU-IN), led by the Head of Medicines Agencies and the EMA, which addresses emerging topics requiring action by the European Medicines regulatory network. In particular, the EU-IN published the Horizon Scanning Genome Editing Report in 2021, providing guidance on regulatory considerations for the coming years.
Conclusions
The GenE-HumDi network, funded by COST, represents a significant milestone in advancing the translation of GE technologies into effective treatments for human diseases. By fostering collaboration among diverse stakeholders, including researchers, pharmaceutical companies, regulatory agencies, and patient advocacy associations, GenE-HumDi aims to reduce knowledge fragmentation by harmonizing the state of the art of the GE field and facilitate the development of standardized procedures and guidelines. The network’s first meeting showcased the recent breakthroughs and novel developments in the GE field, including delivery strategies, safety considerations, clinical translation, and regulatory aspects. With the recent forthcoming approvals of gene editing medicines by European and American regulatory authorities, the field is rapidly progressing toward bringing innovative therapies closer to patients. However, challenges remain, including the need for standardized protocols related to safety, regulatory clarity, and establishment of pricing frameworks. The GenE-HumDi network is strongly committed to contribute significantly to overcoming these obstacles and driving the future of GE as a valuable alternative for treating a broad range of human diseases.
Acknowledgments
This publication is based upon work from COST Action Gene Editing for the treatment of Human Diseases, CA21113 (https://www.genehumdi.eu) supported by COST (European Cooperation of Science and Technology). The study was also supported by the Consejería de Universidad, Investigación e Innovación under Plan Andaluz de Investigación, Desarrollo e Innovación (PAIDI 2020) (ProyExcel_00875) de la junta de Andalucía, and by Consejería de Salud y familia de la Junta de Andalucia/FEDER (PECART-0027-2020), K.B. held Nicolas Monardes contract from Consejería de Salud y Consumo de la Junta de Andalucía.
Author contributions
A.C.: Conceptualization, data curation, Network administration, investigation, visualization, writing—original draft, writing—review and editing. A.H., R.O.B., P.R., M.G., D.L., V.A., L.P., F.Z.H., J.H., F.G.O., C.G.M., C.L., K.M., G.G., N.K., M.A.F.V.G.; A.G., L.M., M.M, J.G., M.L., R.S., J.R.R.M., N.M.P., L.T., M.S.H., C.M., J.G., G.C., A.M., F.M., G.T., T.C., Y.L., T.Q.S.: validation, review editing. K.B.: Conceptualization, funding acquisition, investigation, Network administration, resources, supervision, visualization, writing—review.
Declaration of interests
P.R. has licensed the PGK:FANCAWpre∗ LV medicinal product and receives funding and equity from Rocket Pharmaceuticals, Inc., patents and royalties, research & consulting funding. D.L. is an inventor on a patent National Institute of Chemistry filed (WO/2021/032759 patent application, European patent application EP 3783104, China patent application CN 114269930 with National Phase entry EP2020756868). R.O.B. holds patents related to CRISPR-Cas genome editing and has equity in Graphite Bio and is consultant for UNIKUM Tx. G.G. holds patents related to CRISPR-Cas genome editing, is an employee of CasZyme, and has equity in CasZyme. S.Q.T. is a co-inventor on patents for GUIDE-seq, CHANGE-seq, and other genome editing technologies and a member of the scientific advisory boards of Prime Medicine and Ensoma. T.C. is a co-inventor on patents for CAST-seq, Abnoba-Seq, and other genome editing technologies, and a member of the scientific advisory boards of Cimeo Therapeutics, Excision BioTherapeutics, and GenCC. A.C. and G.T. are inventors on a patent for MEGA (WO/2023/079285), G.T. is also co-inventor on a patent for CAST-seq.
Contributor Information
Alessia Cavazza, Email: a.cavazza@ucl.ac.uk.
Ayal Hendel, Email: ayal.hendel@biu.ac.il.
Rasmus O. Bak, Email: bak@biomed.au.dk.
Yonglun Luo, Email: alun@biomed.au.dk.
Shengdar Q. Tsai, Email: Shengdar.Tsai@STJUDE.ORG.
Karim Benabdellah, Email: karim.benabdel@genyo.es.
References
- 1.Gao Z., Ravendran S., Mikkelsen N.S., Haldrup J., Cai H., Ding X., Paludan S.R., Thomsen M.K., Mikkelsen J.G., Bak R.O. A truncated reverse transcriptase enhances prime editing by split AAV vectors. Mol. Ther. 2022;30:2942–2951. doi: 10.1016/j.ymthe.2022.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan Q., Gao X. Multiplex base- and prime-editing with drive-and-process CRISPR arrays. Nat. Commun. 2022;13:2771. doi: 10.1038/s41467-022-30514-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jensen T.I., Mikkelsen N.S., Gao Z., Foßelteder J., Pabst G., Axelgaard E., Laustsen A., König S., Reinisch A., Bak R.O. Targeted regulation of transcription in primary cells using CRISPRa and CRISPRi. Genome Res. 2021;31:2120–2130. doi: 10.1101/gr.275607.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gasiunas G., Young J.K., Karvelis T., Kazlauskas D., Urbaitis T., Jasnauskaite M., Grusyte M.M., Paulraj S., Wang P.H., Hou Z., et al. A catalogue of biochemically diverse CRISPR-Cas9 orthologs. Nat. Commun. 2020;11:5512. doi: 10.1038/s41467-020-19344-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urbaitis T., Gasiunas G., Young J.K., Hou Z., Paulraj S., Godliauskaite E., Juskeviciene M.M., Stitilyte M., Jasnauskaite M., Mabuchi M., et al. A new family of CRISPR-type V nucleases with C-rich PAM recognition. EMBO Rep. 2022;23 doi: 10.15252/embr.202255481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alonso-Lerma B., Jabalera Y., Samperio S., Morin M., Fernandez A., Hille L.T., Silverstein R.A., Quesada-Ganuza A., Reifs A., Fernández-Peñalver S., et al. Evolution of CRISPR-associated endonucleases as inferred from resurrected proteins. Nat. Microbiol. 2023;8:77–90. doi: 10.1038/s41564-022-01265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pallarès-Masmitjà M., Ivančić D., Mir-Pedrol J., Jaraba-Wallace J., Tagliani T., Oliva B., Rahmeh A., Sánchez-Mejías A., Güell M. Find and cut-and-transfer (FiCAT) mammalian genome engineering. Nat. Commun. 2021;12:7071. doi: 10.1038/s41467-021-27183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lainšček D., Forstnerič V., Mikolič V., Malenšek Š., Pečan P., Benčina M., Sever M., Podgornik H., Jerala R. Coiled-coil heterodimer-based recruitment of an exonuclease to CRISPR/Cas for enhanced gene editing. Nat. Commun. 2022;13:3604. doi: 10.1038/s41467-022-31386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis J.R., Wang X., Witte I.P., Huang T.P., Levy J.M., Raguram A., Banskota S., Seidah N.G., Musunuru K., Liu D.R. Efficient in vivo base editing via single adeno-associated viruses with size-optimized genomes encoding compact adenine base editors. Nat. Biomed. Eng. 2022;6:1272–1283. doi: 10.1038/s41551-022-00911-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinali M., Arechavala-Gomeza V., Feng L., Cirak S., Hunt D., Adkin C., Guglieri M., Ashton E., Abbs S., Nihoyannopoulos P., et al. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8:918–928. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cirak S., Arechavala-Gomeza V., Guglieri M., Feng L., Torelli S., Anthony K., Abbs S., Garralda M.E., Bourke J., Wells D.J., et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiriboga C.A., Swoboda K.J., Darras B.T., Iannaccone S.T., Montes J., De Vivo D.C., Norris D.A., Bennett C.F., Bishop K.M. Results from a phase 1 study of nusinersen (ISIS-SMN(Rx)) in children with spinal muscular atrophy. Neurology. 2016;86:890–897. doi: 10.1212/WNL.0000000000002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendell J.R., Khan N., Sha N., Eliopoulos H., McDonald C.M., Goemans N., Mercuri E., Lowes L.P., Alfano L.N., Eteplirsen Study Group Comparison of Long-term Ambulatory Function in Patients with Duchenne Muscular Dystrophy Treated with Eteplirsen and Matched Natural History Controls. J. Neuromuscul. Dis. 2021;8:469–479. doi: 10.3233/JND-200548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godfrey C., Desviat L.R., Smedsrød B., Piétri-Rouxel F., Denti M.A., Disterer P., Lorain S., Nogales-Gadea G., Sardone V., Anwar R., et al. Delivery is key: lessons learnt from developing splice-switching antisense therapies. EMBO Mol. Med. 2017;9:545–557. doi: 10.15252/emmm.201607199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dowdy S.F., Setten R.L., Cui X.S., Jadhav S.G. Delivery of RNA Therapeutics: The Great Endosomal Escape. Nucleic Acid Ther. 2022;32:361–368. doi: 10.1089/nat.2022.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu E., Liu F. PLGA-based drug delivery systems in treating bone tumors. Front. Bioeng. Biotechnol. 2023;11 doi: 10.3389/fbioe.2023.1199343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Na Y., Zhang N., Zhong X., Gu J., Yan C., Yin S., Lei X., Zhao J., Geng F. Polylactic-co-glycolic acid-based nanoparticles modified with peptides and other linkers cross the blood-brain barrier for targeted drug delivery. Nanomedicine. 2023;18:125–143. doi: 10.2217/nnm-2022-0287. [DOI] [PubMed] [Google Scholar]
- 18.Abbasi R., Shineh G., Mobaraki M., Doughty S., Tayebi L. Structural parameters of nanoparticles affecting their toxicity for biomedical applications: a review. J. Nano Res. 2023;25:43. doi: 10.1007/s11051-023-05690-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J., Zhu D., Liu X., Peng L. Amphiphilic Dendrimer Vectors for RNA Delivery: State-of-the-Art and Future Perspective. Acc. Mater. Res. 2022;3:484–497. doi: 10.1021/accountsmr.1c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyu Z., Ding L., Tintaru A., Peng L. Self-Assembling Supramolecular Dendrimers for Biomedical Applications: Lessons Learned from Poly(amidoamine) Dendrimers. Acc. Chem. Res. 2020;53:2936–2949. doi: 10.1021/acs.accounts.0c00589. [DOI] [PubMed] [Google Scholar]
- 21.Xiong Y., Ke R., Zhang Q., Lan W., Yuan W., Chan K.N.I., Roussel T., Jiang Y., Wu J., Liu S., et al. Small Activating RNA Modulation of the G Protein-Coupled Receptor for Cancer Treatment. Adv. Sci. 2022;9 doi: 10.1002/advs.202200562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J., Zhu D., Lian B., Shi K., Chen P., Li Y., Lin W., Ding L., Long Q., Wang Y., et al. Cargo-selective and adaptive delivery of nucleic acid therapeutics by bola-amphiphilic dendrimers. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2220787120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y., Lyu Z., Ralahy B., Liu J., Roussel T., Ding L., Tang J., Kosta A., Giorgio S., Tomasini R., et al. Dendrimer nanosystems for adaptive tumor-assisted drug delivery via extracellular vesicle hijacking. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2215308120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J., Ellert-Miklaszewska A., Garofalo S., Dey A.K., Tang J., Jiang Y., Clément F., Marche P.N., Liu X., Kaminska B., et al. Synthesis and use of an amphiphilic dendrimer for siRNA delivery into primary immune cells. Nat. Protoc. 2021;16:327–351. doi: 10.1038/s41596-020-00418-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Hees M., Slott S., Hansen A.H., Kim H.S., Ji H.P., Astakhova K. New approaches to moderate CRISPR-Cas9 activity: Addressing issues of cellular uptake and endosomal escape. Mol. Ther. 2022;30:32–46. doi: 10.1016/j.ymthe.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cebrian-Serrano A., Davies B. CRISPR-Cas orthologues and variants: optimizing the repertoire, specificity and delivery of genome engineering tools. Mamm. Genome. 2017;28:247–261. doi: 10.1007/s00335-017-9697-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen D., Rosenberg M., Hendel A. Using Synthetically Engineered Guide RNAs to Enhance CRISPR Genome Editing Systems in Mammalian Cells. Front. Genome Ed. 2020;2 doi: 10.3389/fgeed.2020.617910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allen D., Kalter N., Rosenberg M., Hendel A. Homology-Directed-Repair-Based Genome Editing in HSPCs for the Treatment of Inborn Errors of Immunity and Blood Disorders. Pharmaceutics. 2023;15 doi: 10.3390/pharmaceutics15051329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dorset S.R., Bak R.O. The p53 challenge of hematopoietic stem cell gene editing. Mol. Ther. Methods Clin. Dev. 2023;30:83–89. doi: 10.1016/j.omtm.2023.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P., et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazzarotto C.R., Malinin N.L., Li Y., Zhang R., Yang Y., Lee G., Cowley E., He Y., Lan X., Jividen K., et al. CHANGE-seq reveals genetic and epigenetic effects on CRISPR-Cas9 genome-wide activity. Nat. Biotechnol. 2020;38:1317–1327. doi: 10.1038/s41587-020-0555-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapiro J., Tovin A., Iancu O., Allen D., Hendel A. Chemical Modification of Guide RNAs for Improved CRISPR Activity in CD34+ Human Hematopoietic Stem and Progenitor Cells. Methods Mol. Biol. 2021;2162:37–48. doi: 10.1007/978-1-0716-0687-2_3. [DOI] [PubMed] [Google Scholar]
- 33.Allen D., Weiss L.E., Saguy A., Rosenberg M., Iancu O., Matalon O., Lee C., Beider K., Nagler A., Shechtman Y., Hendel A. High-Throughput Imaging of CRISPR- and Recombinant Adeno-Associated Virus-Induced DNA Damage Response in Human Hematopoietic Stem and Progenitor Cells. CRISPR J. 2022;5:80–94. doi: 10.1089/crispr.2021.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iancu O., Allen D., Knop O., Zehavi Y., Breier D., Arbiv A., Lev A., Lee Y.N., Beider K., Nagler A., et al. Multiplex HDR for disease and correction modeling of SCID by CRISPR genome editing in human HSPCs. Mol. Ther. Nucleic Acids. 2023;31:105–121. doi: 10.1016/j.omtn.2022.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bestas B., Wimberger S., Degtev D., Madsen A., Rottner A.K., Karlsson F., Naumenko S., Callahan M., Touza J.L., Francescatto M., et al. A Type II-B Cas9 nuclease with minimized off-targets and reduced chromosomal translocations in vivo. Nat. Commun. 2023;14:5474. doi: 10.1038/s41467-023-41240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anthon C., Corsi G.I., Gorodkin J. CRISPRon/off: CRISPR/Cas9 on- and off-target gRNA design. Bioinformatics. 2022;38:5437–5439. doi: 10.1093/bioinformatics/btac697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corsi G.I., Gadekar V.P., Gorodkin J., Seemann S.E. CRISPRroots: on- and off-target assessment of RNA-seq data in CRISPR-Cas9 edited cells. Nucleic Acids Res. 2022;50:e20. doi: 10.1093/nar/gkab1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corsi G.I., Qu K., Alkan F., Pan X., Luo Y., Gorodkin J. CRISPR/Cas9 gRNA activity depends on free energy changes and on the target PAM context. Nat. Commun. 2022;13:3006. doi: 10.1038/s41467-022-30515-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan X., Qu K., Yuan H., Xiang X., Anthon C., Pashkova L., Liang X., Han P., Corsi G.I., Xu F., et al. Massively targeted evaluation of therapeutic CRISPR off-targets in cells. Nat. Commun. 2022;13:4049. doi: 10.1038/s41467-022-31543-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiang X., Corsi G.I., Anthon C., Qu K., Pan X., Liang X., Han P., Dong Z., Liu L., Zhong J., et al. Enhancing CRISPR-Cas9 gRNA efficiency prediction by data integration and deep learning. Nat. Commun. 2021;12:3238. doi: 10.1038/s41467-021-23576-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turchiano G., Andrieux G., Klermund J., Blattner G., Pennucci V., El Gaz M., Monaco G., Poddar S., Mussolino C., Cornu T.I., et al. Quantitative evaluation of chromosomal rearrangements in gene-edited human stem cells by CAST-Seq. Cell Stem Cell. 2021;28:1136–1147.e5. doi: 10.1016/j.stem.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 42.Akcakaya P., Bobbin M.L., Guo J.A., Malagon-Lopez J., Clement K., Garcia S.P., Fellows M.D., Porritt M.J., Firth M.A., Carreras A., et al. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature. 2018;561:416–419. doi: 10.1038/s41586-018-0500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amit I., Iancu O., Levy-Jurgenson A., Kurgan G., McNeill M.S., Rettig G.R., Allen D., Breier D., Ben Haim N., Wang Y., et al. CRISPECTOR provides accurate estimation of genome editing translocation and off-target activity from comparative NGS data. Nat. Commun. 2021;12:3042. doi: 10.1038/s41467-021-22417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X., Tasca F., Wang Q., Liu J., Janssen J.M., Brescia M.D., Bellin M., Szuhai K., Kenrick J., Frock R.L., Gonçalves M.A.F.V. Expanding the editable genome and CRISPR-Cas9 versatility using DNA cutting-free gene targeting based on in trans paired nicking. Nucleic Acids Res. 2020;48:974–995. doi: 10.1093/nar/gkz1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ihry R.J., Worringer K.A., Salick M.R., Frias E., Ho D., Theriault K., Kommineni S., Chen J., Sondey M., Ye C., et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 2018;24:939–946. doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- 47.Schiroli G., Conti A., Ferrari S., Della Volpe L., Jacob A., Albano L., Beretta S., Calabria A., Vavassori V., Gasparini P., et al. Precise Gene Editing Preserves Hematopoietic Stem Cell Function following Transient p53-Mediated DNA Damage Response. Cell Stem Cell. 2019;24:551–565.e8. doi: 10.1016/j.stem.2019.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X., Janssen J.M., Liu J., Maggio I., 't Jong A.E.J., Mikkers H.M.M., Gonçalves M.A.F.V. In trans paired nicking triggers seamless genome editing without double-stranded DNA cutting. Nat. Commun. 2017;8:657. doi: 10.1038/s41467-017-00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q., Liu J., Janssen J.M., Gonçalves M.A.F.V. Precise homology-directed installation of large genomic edits in human cells with cleaving and nicking high-specificity Cas9 variants. Nucleic Acids Res. 2023;51:3465–3484. doi: 10.1093/nar/gkad165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Nierop G.P., de Vries A.A.F., Holkers M., Vrijsen K.R., Gonçalves M.A.F.V. Stimulation of homology-directed gene targeting at an endogenous human locus by a nicking endonuclease. Nucleic Acids Res. 2009;37:5725–5736. doi: 10.1093/nar/gkp643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonçalves M.A.F.V., van Nierop G.P., Holkers M., de Vries A.A.F. Concerted nicking of donor and chromosomal acceptor DNA promotes homology-directed gene targeting in human cells. Nucleic Acids Res. 2012;40:3443–3455. doi: 10.1093/nar/gkr1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bollen Y., Hageman J.H., van Leenen P., Derks L.L.M., Ponsioen B., Buissant des Amorie J.R., Verlaan-Klink I., van den Bos M., Terstappen L.W.M.M., van Boxtel R., Snippert H.J.G. Efficient and error-free fluorescent gene tagging in human organoids without double-strand DNA cleavage. PLoS Biol. 2022;20 doi: 10.1371/journal.pbio.3001527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fortschegger K., Husa A.M., Schinnerl D., Nebral K., Strehl S. Expression of RUNX1-JAK2 in Human Induced Pluripotent Stem Cell-Derived Hematopoietic Cells Activates the JAK-STAT and MYC Pathways. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22147576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bollen Y., Stelloo E., van Leenen P., van den Bos M., Ponsioen B., Lu B., van Roosmalen M.J., Bolhaqueiro A.C.F., Kimberley C., Mossner M., et al. Reconstructing single-cell karyotype alterations in colorectal cancer identifies punctuated and gradual diversification patterns. Nat. Genet. 2021;53:1187–1195. doi: 10.1038/s41588-021-00891-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eshka S.F.A., Bahador M., Gordan M.M., Karbasi S., Tabar Z.M., Basiri M. A systematic review of gene editing clinical trials. medRxiv. 2022 doi: 10.1101/2022.11.24.22282599. Preprint at. [DOI] [Google Scholar]
- 56.Rai R., Romito M., Rivers E., Turchiano G., Blattner G., Vetharoy W., Ladon D., Andrieux G., Zhang F., Zinicola M., et al. Targeted gene correction of human hematopoietic stem cells for the treatment of Wiskott - Aldrich Syndrome. Nat. Commun. 2020;11:4034. doi: 10.1038/s41467-020-17626-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frangoul H., Ho T.W., Corbacioglu S. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and beta-Thalassemia. Reply. N. Engl. J. Med. 2021;384:e91. doi: 10.1056/NEJMc2103481. [DOI] [PubMed] [Google Scholar]
- 58.Traxler E.A., Yao Y., Wang Y.D., Woodard K.J., Kurita R., Nakamura Y., Hughes J.R., Hardison R.C., Blobel G.A., Li C., Weiss M.J. A genome-editing strategy to treat beta-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat. Med. 2016;22:987–990. doi: 10.1038/nm.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hardouin G., Antoniou P., Martinucci P., Felix T., Manceau S., Joseph L., Masson C., Scaramuzza S., Ferrari G., Cavazzana M., Miccio A. Adenine base editor-mediated correction of the common and severe IVS1-110 (G>A) beta-thalassemia mutation. Blood. 2023;141:1169–1179. doi: 10.1182/blood.2022016629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Antoniou P., Hardouin G., Martinucci P., Frati G., Felix T., Chalumeau A., Fontana L., Martin J., Masson C., Brusson M., et al. Base-editing-mediated dissection of a gamma-globin cis-regulatory element for the therapeutic reactivation of fetal hemoglobin expression. Nat. Commun. 2022;13:6618. doi: 10.1038/s41467-022-34493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Papasavva P.L., Patsali P., Loucari C.C., Kurita R., Nakamura Y., Kleanthous M., Lederer C.W. CRISPR Editing Enables Consequential Tag-Activated MicroRNA-Mediated Endogene Deactivation. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23031082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kountouris P., Stephanou C., Lederer C.W., Traeger-Synodinos J., Bento C., Harteveld C.L., Fylaktou E., Koopmann T.T., Halim-Fikri H., Michailidou K., et al. Adapting the ACMG/AMP variant classification framework: A perspective from the ClinGen Hemoglobinopathy Variant Curation Expert Panel. Hum. Mutat. 2022;43:1089–1096. doi: 10.1002/humu.24280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kountouris P., Stephanou C., Archer N., Bonifazi F., Giannuzzi V., Kuo K.H.M., Maggio A., Makani J., Mañú-Pereira M.D.M., Michailidou K., et al. The International Hemoglobinopathy Research Network (INHERENT): An international initiative to study the role of genetic modifiers in hemoglobinopathies. Am. J. Hematol. 2021;96:E416–E420. doi: 10.1002/ajh.26323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siegner S.M., Ugalde L., Clemens A., Garcia-Garcia L., Bueren J.A., Rio P., Karasu M.E., Corn J.E. Adenine base editing efficiently restores the function of Fanconi anemia hematopoietic stem and progenitor cells. Nat. Commun. 2022;13:6900. doi: 10.1038/s41467-022-34479-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kochenderfer J.N., Dudley M.E., Feldman S.A., Wilson W.H., Spaner D.E., Maric I., Stetler-Stevenson M., Phan G.Q., Hughes M.S., Sherry R.M., et al. B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood. 2012;119:2709–2720. doi: 10.1182/blood-2011-10-384388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jo S., Das S., Williams A., Chretien A.S., Pagliardini T., Le Roy A., Fernandez J.P., Le Clerre D., Jahangiri B., Chion-Sotinel I., et al. Endowing universal CAR T-cell with immune-evasive properties using TALEN-gene editing. Nat. Commun. 2022;13:3453. doi: 10.1038/s41467-022-30896-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maldonado-Pérez N., Tristán-Manzano M., Justicia-Lirio P., Martínez-Planes E., Muñoz P., Pavlovic K., Cortijo-Gutiérrez M., Blanco-Benítez C., Castella M., Juan M., et al. Efficacy and safety of universal (TCRKO) ARI-0001 CAR-T cells for the treatment of B-cell lymphoma. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1011858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Groothoff J.W., Metry E., Deesker L., Garrelfs S., Acquaviva C., Almardini R., Beck B.B., Boyer O., Cerkauskiene R., Ferraro P.M., et al. Clinical practice recommendations for primary hyperoxaluria: an expert consensus statement from ERKNet and OxalEurope. Nat. Rev. Nephrol. 2023;19:194–211. doi: 10.1038/s41581-022-00661-1. [DOI] [PubMed] [Google Scholar]
- 69.Bonafont J., Mencía Á., García M., Torres R., Rodríguez S., Carretero M., Chacón-Solano E., Modamio-Høybjør S., Marinas L., León C., et al. Clinically Relevant Correction of Recessive Dystrophic Epidermolysis Bullosa by Dual sgRNA CRISPR/Cas9-Mediated Gene Editing. Mol. Ther. 2019;27:986–998. doi: 10.1016/j.ymthe.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bonafont J., Mencía A., Chacón-Solano E., Srifa W., Vaidyanathan S., Romano R., Garcia M., Hervás-Salcedo R., Ugalde L., Duarte B., et al. Correction of recessive dystrophic epidermolysis bullosa by homology-directed repair-mediated genome editing. Mol. Ther. 2021;29:2008–2018. doi: 10.1016/j.ymthe.2021.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Szopa M., Wolkow J., Matejko B., Skupien J., Klupa T., Wybrańska I., Trznadel-Morawska I., Kiec-Wilk B., Borowiec M., Malecki M.T. Prevalence of Retinopathy in Adult Patients with GCK-MODY and HNF1A-MODY. Exp. Clin. Endocrinol. Diabetes. 2015;123:524–528. doi: 10.1055/s-0035-1559605. [DOI] [PubMed] [Google Scholar]
- 72.Kachamakova-Trojanowska N., Stepniewski J., Dulak J. Human iPSCs-Derived Endothelial Cells with Mutation in HNF1A as a Model of Maturity-Onset Diabetes of the Young. Cells. 2019;8 doi: 10.3390/cells8111440. [DOI] [PMC free article] [PubMed] [Google Scholar]