Abstract
The psd gene of Bacillus subtilis Marburg, encoding phosphatidylserine decarboxylase, has been cloned and sequenced. It encodes a polypeptide of 263 amino acid residues (deduced molecular weight of 29,689) and is located just downstream of pss, the structural gene for phosphatidylserine synthase that catalyzes the preceding reaction in phosphatidylethanolamine synthesis (M. Okada, H. Matsuzaki, I. Shibuya, and K. Matsumoto, J. Bacteriol. 176:7456–7461, 1994). Introduction of a plasmid containing the psd gene into temperature-sensitive Escherichia coli psd-2 mutant cells allowed growth at otherwise restrictive temperature. Phosphatidylserine was not detected in the psd-2 mutant cells harboring the plasmid; it accumulated in the mutant up to 29% of the total phospholipids without the plasmid. An enzyme activity that catalyzes decarboxylation of 14C-labeled phosphatidylserine to form phosphatidylethanolamine was detected in E. coli psd-2 cells harboring a Bacillus psd plasmid. E. coli cells harboring the psd plasmid, the expression of which was under the control of the T7φ10 promoter, produced proteins of 32 and 29 kDa upon induction. A pulse-labeling experiment suggested that the 32-kDa protein is the primary translation product and is processed into the 29-kDa protein. The psd gene, together with pss, was located by Southern hybridization to the 238- to 306-kb SfiI-NotI fragment of the chromosome. A B. subtilis strain harboring an interrupted psd allele, psd1::neo, was constructed. The null psd mutant contained no phosphatidylethanolamine and accumulated phosphatidylserine. It grew well without supplementation of divalent cations which are essential for the E. coli pssA null mutant lacking phosphatidylethanolamine. In both the B. subtilis null pss and psd mutants, glucosyldiacylglycerol content increased two- to fourfold. The results suggest that the lack of phosphatidylethanolamine in the B. subtilis membrane may be compensated for by the increases in the contents of glucosyldiacylglycerols by an unknown mechanism.
Biosynthesis of membrane lipids in gram-positive Bacillus species has not been studied extensively, and it has been assumed to follow the standard pathways established in Escherichia coli (7, 29, 39, 40). As for the synthesis of phosphatidylethanolamine in Bacillus species, two enzymes involved, CDP-diacylglycerol-dependent phosphatidylserine synthase and phosphatidylserine decarboxylase, are associated with membranes (11, 12, 26, 40), whereas upon disruption of the cell, the phosphatidylserine synthase of E. coli is mainly associated with ribosomes, not with the membrane (31, 42) because of its hydrophilic and basic nature (6). This property of the E. coli synthase appears to be related to the regulation mechanism for phosphatidylethanolamine synthesis (30, 46). In Bacillus subtilis, with the membrane-localized enzymes, the phosphatidylethanolamine synthesis should be regulated in a very different way (7, 32); however, no data on the biosynthetic pathway and regulation have been available, despite the wealth of its genetic information.
In order to get pertinent information on the physiological roles of phosphatidylethanolamine and the possible regulatory mechanism of its synthesis in Bacilli, as well as for use as a heterologous gene probe for elucidation of the regulatory mechanism in E. coli, we have cloned and characterized the structural gene pss for the phosphatidylserine synthase of B. subtilis Marburg (38, 43). During the course of cloning the pss gene, a reading frame showing a homology with known phosphatidylserine decarboxylase sequences was found downstream of the pss gene. This report describes the characterization of the B. subtilis gene for phosphatidylserine decarboxylase, psd, and its translation product. By constructing the strain bearing an interrupted chromosomal allele of psd or pss, we further examined the effect of phosphatidylethanolamine deprivation from the B. subtilis membrane.
MATERIALS AND METHODS
Bacterial strains and plasmids.
B. subtilis Marburg and E. coli K-12 strains and plasmids used in this study are listed in Table 1. For structures of plasmids, see Fig. 1. The reference B. subtilis strains for Southern hybridization mapping are described in the following section.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| B. subtilis Marburg | ||
| 160 | trpB160 | H. Saito |
| SDB01 | 160 psd1::neo | This work |
| SDB02 | 160 Δpss10::spc | This work |
| E. coli | ||
| OS2101 | pssA1 pyrD34 thyA33 galK35 str | 36 |
| EH150 | psd-2 | 15 |
| JM109(DE3) | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB)/[F′ proAB+ lacIqlacZΔM15 traD36] [λ lacUV5-T7 gene 1] | 47 |
| XL1-Blue | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 lac/[F′::Tn10 proAB+ lacIqlacZΔM15 traD36] | 4 |
| Plasmids | ||
| pWSK29 | pSC101 harboring a multicloning site and T7φ10 and lac promoters, Ampr | 51 |
| pBM201 | pWSK29 harboring a 4.6-kb fragment of the B. subtilis chromosome | 38 |
| pBM102 | pBM201 lacking 346 bases from the 5′ end of the 4.6-kb insert | 38 |
| pBM22 | pWSK29 harboring a 1,244-base fragment of the B. subtilis chromosome | 38 |
| pBM213 | pWSK29 harboring an EcoRI-AvaII fragment of the B. subtilis chromosome | 38 |
| pBM61 | pBM201 lacking 730 bases from the 5′ end of the insert | 38 |
| pBM106 | pBM201 lacking 1,375 bases from the 5′ end of the insert | This work |
| pBM281 | pBM106 lacking the 3′ HindIII fragment of the insert | This work |
| pBM912 | pBM201 lacking 1,497 bases from the 5′ end of the insert | This work |
| pBM903 | pBM201 lacking 1,584 bases from the 5′ end of the insert | This work |
| pBM2 | pWSK29 carrying the 2,035-base PstI fragment of pBM201 | 38 |
| pBEST502 | pGEM4 (Promega) neo | 20 |
| pYH01 | pBM102 harboring psd1::neo; neo was inserted at the unique PstI site in psd | This work |
| pDH1726 | pSB119 (pUC) spc | 14 |
| pSF01 | pBM2 harboring Δpss10::spc, MunI-HindIII region of pss was replaced with spc | This work |
| pDH88his | pDH88 harboring spac promoter (18), lacI, and B. subtilis hisA | 23 |
| pLUCK100 | pDH88his harboring the pss-psd locus under the spac promoter | This work |
FIG. 1.
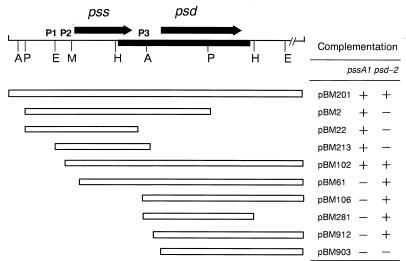
Physical map of the B. subtilis pss and psd gene loci and complementation of pssA1 and psd-2 mutations. The physical map of the 4.6-kbp fragment of B. subtilis chromosome cloned on pWSK29, designated pBM201, is presented. The solid line in the map indicates the regions sequenced. The horizontal arrows show the direction and extent of the reading frames of the pss and psd genes. P1, P2, and P3 indicate putative promoter sites. Horizontal bars indicate the regions subcloned on the respective plasmids; their complementation abilities are also listed (see Table 1 and Materials and Methods for more details). Except for pBM22, the T7φ10 promoter of pWSK29 was placed on the left ends of the subcloned fragments. Complementation of temperature-sensitive mutations of OS2101 (pssA1) and EH150 (psd) was examined at 42°C on NBY plates. A, AvaII; E, EcoRI; H, HindIII; P, PstI; M, MunI.
Media and bacterial growth.
Luria-Bertani broth contained 1% tryptone (Difco, Detroit, Mich.), 0.5% yeast extract (Difco), and 1% NaCl. NBY medium (35) contained 0.8% nutrient broth (Difco), 0.5% polypeptone (Dainihon Seiyaku, Tokyo), 0.2% yeast extract, and 0.1% NaCl and was adjusted to pH 7.2 with NaOH. TY broth, which contained 1% tryptone, 0.5% yeast extract, and 0.5% NaCl; synthetic media CI and CII for development of competence (2); and Penassay broth (Difco) were used for B. subtilis. When required, the following supplements were added to the media (per liter): 50 mg of thymine, 5 mg of thiamine-hydrochloride, 50 mg of ampicillin (Sigma), 20 mg of neomycin (Wako Pure Chem.), and 50 mg of spectinomycin (Sigma). Growth of bacteria was monitored by measuring turbidity with a Klett-Summerson photoelectric colorimeter (no. 54 filter). For solid media, 1.5% agar (Difco) was included.
DNA preparation and manipulations.
For construction, isolation, and identification of recombinant DNA, the methods described by Sambrook et al. (45) were used. Plasmid DNAs were prepared from XL1-Blue cells harboring the respective plasmids by the alkaline lysis method. Fragments of DNA were recovered from agarose gels with a GENECLEAN II kit from BIO 101 (La Jolla, Calif.). Restriction endonuclease digestion, filling of cohesive ends, and ligation were performed with enzymes from Takara Shuzo (Kyoto), Nippon Gene (Tokyo), and New England Biolabs (Beverly, Mass.) under the conditions recommended by the suppliers. Competent cells of strains OS2101 and XL1-Blue were prepared by the standard CaCl2 method.
DNA sequencing.
Subclones for DNA sequencing were obtained by cloning the defined restriction fragments and overlapping deletion fragments made by using exonuclease III and Sl nuclease into pWSK29 vector (51). Single-stranded DNA templates were prepared from phage M13KO7-infected XL1-Blue cells harboring each of the subclone plasmids (45). Chain termination reactions were performed as described in the manual supplied by the United States Biochemical Corporation (Cleveland, Ohio) with Sequenase version 2.0 DNA polymerase and a fluorescein-labeled 21-mer M13 universal primer (Yuki Gosei Kogyo, Tokyo, Japan). Nucleotide mixtures containing 7-deaza-dGTP and 7-deaza-dATP (Pharmacia) were used to prevent formation of intrastrand secondary structures. Reaction products were analyzed with an automated laser fluorescent DNA sequencer, DSQ-1 (Shimadzu, Kyoto). To verify the sequence data, both strands were sequenced. The DNA sequence and the deduced amino acid sequence were examined with sequence analysis programs of GENETYX software (Software Development Co., Tokyo, Japan).
Analysis of plasmid-encoded protein.
Detection and molecular weight determination of phosphatidylserine decarboxylase were performed by the T7 RNA polymerase-promoter system (10). Strain JM109(DE3) harboring pBM281 was grown in LB medium supplemented with 100 μg of ampicillin per ml, and at 70 Klett units (about 2 × 108 cells per ml), isopropyl-β-d-thiogalactopyranoside (Takara Shuzo) was added to a final concentration of 5 mM, and after 20 min of incubation, this was followed by an addition of rifampin (Sigma, St. Louis, Mo. [final concentration of 100 μg per ml]). After a further 12 min of incubation, 50 μCi of l-[35S]methionine (1,152 Ci/mmol [American Radiolabeled Chemicals, Inc., St. Louis, Mo.]) was added, and the mixture was incubated for 10, 50, and 100 min. The cells were then collected and solubilized in a sample buffer (3% sodium dodecyl sulfate, 5% mercaptoethanol) at 100°C for 3 min. Labeled proteins were subjected to sodium dodecyl sulfate–12.5% polyacrylamide gel electrophoresis, and they were visualized and quantified with the BAS 1000 Mac bioimaging analyzer (Fuji Photo Film, Tokyo, Japan).
Phosphatidylserine decarboxylase assay.
Cells grown to late log phase for 6 h at 43°C in LB medium were collected and suspended in a potassium phosphate buffer (100 mM [pH 7.4]) and disrupted by sonication with a Branson sonifier followed by fractionation into supernatant and crude membrane fractions by centrifugation for 1 h at 100,000 × g after removal of cell debris. The tightly packed pellet was suspended in 100 mM potassium phosphate buffer (pH 7.4) and used as the crude membrane fraction. The enzyme activities in the membrane and supernatant fractions were assayed essentially by the method of Hawrot and Kennedy (15). The assay mixture (0.1 ml) contained 0.1 M potassium phosphate buffer (pH 7.0), 0.1% Triton X-100 (wt/vol), 0.2 mM l-phosphatidyl[U-14C]serine (800 dpm/nmol), and the enzyme fraction (40 ng). After incubation at 30°C for 30 min, the reaction was terminated by the addition of methanol containing 0.1 N HCl. Chloroform-soluble materials were extracted and dried, and the lipids were then redissolved in chloroform-methanol (2:1 [vol/vol]), applied to a silica gel plate (no. 60; Merck, Darmstadt, Germany), and developed in chloroform-methanol-acetic acid (65:25:10 [vol/vol/vol]). The positions of the radioactive phospholipids were determined and quantified with the bioimaging analyzer.
Lipid analysis.
Membrane lipids were labeled with 0.5 μCi of [1-14C]acetic acid per ml (57.2 mCi/mmol [Amersham]) for at least six generations of cultivation of the mutant cells in Penassay broth (5 ml) or cultivated in LB broth with no radioisotope and then were harvested in the late exponential phase. Lipids were extracted by the method of Bligh and Dyer (1), and halves of the lipid fractions were separated by two-dimensional thin-layer chromatography on silica gel (no. 60; Merck, Darmstadt), first (x dimension) with chloroform-methanol-water (65:25:4 [vol/vol/vol]) and then (y dimension) with chloroform-methanol-acetic acid (65:25:10 [vol/vol/vol]). Spots for 14C-labelled lipid were visualized and quantified with the BAS 1000 bioimaging analyzer. Phospholipids were visualized by uniformly spraying Dittmer-Lester reagent (8), and spots were quantified with a high-speed thin-layer chromatography (TLC) scanner (model CS-920; Shimadzu, Kyoto). Glycolipids were visualized with a spray of orcinol-sulfuric acid mixture, and lipid species having free amino groups were detected with a ninhydrin spray. Molar percentages of each component were calculated.
Southern hybridization mapping of psd.
The procedures for labelling of pBM102 (pss-psd) and pBM281 (psd) probes with digoxigenin (Boehringer Mannheim), blotting, hybridization, and identification of hybridized restriction fragments of reference genomes were described previously (21). For the hybridization with NotI-digested fragments, reference genome DNAs from the CU741, BEST4041, BEST4087, and BEST4133 strains were used, and for the hybridization with SfiI-digested fragments, reference genome DNAs from the OA101, BEST3015, BEST3028, and BEST3055 strains (21) were used.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have appeared in the GSDB, DDBJ, EMBL, and NCBI nucleotide databases under accession no. D38022.
RESULTS AND DISCUSSION
Cloning and sequencing of the structural gene for phosphatidylserine decarboxylase of B. subtilis.
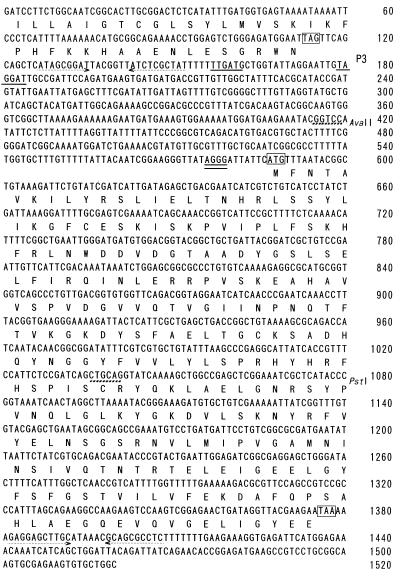
During the course of the cloning of a B. subtilis structural gene for phosphatidylserine synthase, pss (38), a reading frame which showed homology with phosphatidylserine decarboxylases of E. coli, Saccharomyces cerevisiae, and CHO-K1 cells was found 461 nucleotides downstream of the coding frame of the pss gene (Fig. 1). We therefore sequenced the downstream region to the end of the reading frame. The open reading frame, starting at nucleotide 587 and extending up to the stop codon at 1,375 (Fig. 2), encoded a polypeptide of 263 amino acid residues with a deduced molecular weight of 29,689. Six base pairs upstream of the initiation codon was a putative 5′-AGGG-3′ ribosome binding site. A potential promoter consensus sequence (13) starting at nucleotide 1,157 was found and designated as P3, and those with lower homology scores were also found downstream of P3. Immediately upstream of P3, a possible ρ-independent terminator signal of pss was observed, and the reading frame was also followed by a possible ρ-independent terminator signal.
FIG. 2.
Nucleotide sequence of the B. subtilis psd locus. Numbering begins at nucleotide 1,001 of the sequence presented previously (38), and the newly sequenced region in this work starts from nucleotide 422. Possible promoter P3 is underlined. A putative ribosome binding site is double underlined, and the start and stop codons are boxed. The dotted arrows indicate inverted repeats of a possible ρ-independent terminator. The PstI site used for the disruption of psd is shown by bold dashed lines. From nucleotide 106 to nucleotide 591 (overlapping with the end of pss and the start of psd), an open reading frame predicting a polypeptide of 162 amino acid residues is possible. The deduced sequence has partial similarity (31%) to the product of the E. coli dedA gene, the function of which is presently unknown (GenBank accession no. S53037).
We then examined whether this reading frame could complement the temperature sensitivity of the E. coli EH150 (psd-2) strain which has a thermosensitive phosphatidylserine decarboxylase activity (16). EH150 cells harboring pBM201 did grow at 42°C (Fig. 1). Subclones of 3′ end deletions lacking this reading frame region could not support growth. The subclone of the 5′ end deletion-containing plasmid pBM912, which lacked up to nucleotide 497, supported growth. However, the plasmid pBM903, which had a deletion up to nucleotide 586, 1 base upstream of the initiation codon, did not support the growth, probably because it lacked the putative ribosome binding site of the reading frame. These results indicated that this reading frame could complement the temperature-sensitive phosphatidylserine decarboxylase activity of the E. coli EH150 (psd-2) strain. Since this reading frame was verified to code for B. subtilis phosphatidylserine decarboxylase by the functional assay and the gene disruption described below, we designate the gene as psd.
Analysis of the B. subtilis psd gene product.
The deduced primary sequence of the product of B. subtilis psd gene was compared with those of the phosphatidylserine decarboxylases of E. coli (27), S. cerevisiae (5, 48), and CHO cells (24). The B. subtilis product, a 263-amino-acid residue protein, was the smallest among the known enzymes; the numbers of residues of E. coli, S. cerevisiae, and CHO cells are 322, 500, and 370, respectively. The B. subtilis sequence shared 27% identical amino acids with those of E. coli and CHO cells and 29% identical residues with that of S. cerevisiae. When the related amino acid substitutes were included, the similarity values were 43, 40, and 41% with the products from E. coli, CHO cells, and S. cerevisiae, respectively. There were several conserved segments among these four products. The similarity at the carboxyl-terminal end of the B. subtilis product included the conserved segment GX3GXFX2GST(V/I)(V/I)X2F, corresponding to the posttranslational processing site in E. coli decarboxylase, which is cleaved into the β subunit of 28,579 Da and the small pyruvoyl-containing α subunit of 7,332 Da (27). Putative processing of the product of B. subtilis psd (29,689 Da) would result in a pyruvoyl-containing subunit of 3,700 Da and a 25,973-Da subunit. Hydropathy analysis (25) of the sequence indicated that the conserved segment for the processing is hydrophobic, with the remainder primarily hydrophilic (data not shown), as in the cases of the E. coli (27), CHO cell, and S. cerevisiae (5) counterparts.
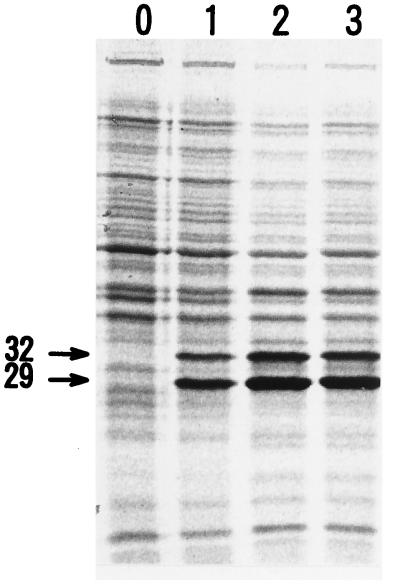
The product of the psd gene was then examined by the T7 RNA polymerase-promoter system. An autoradiogram of 35S-labeled proteins after induction with the addition of isopropyl-β-d-thiogalactopyranoside indicated that E. coli JM109(DE3) cells harboring pBM281 produced a large amount of 32- and 29-kDa proteins (Fig. 3). Prolonged labeling led to a decrease in the proportion of 32-kDa protein with a concomitant increase in 29-kDa protein; when the labeling was for 10 min, the proportion of 29- versus 32-kDa protein was 2.2, and with 100 min of labeling, the ratio was 3.5. After 4 h of incubation, a ratio of 10 was observed (Coomassie brilliant blue staining [data not shown]). The molecular weights of these induced proteins coincided with the predicted molecular weights for the primary product and its processed large subunit, respectively. Therefore, the results suggested that the translated product of B. subtilis psd was processed in E. coli cells into a pyruvoyl-containing small subunit and the remaining large subunit, as in the case of E. coli (27). The majority of the overproduced primary product was observed in the membrane fraction, and a large amount of the processed 29-kDA protein was observed in the 100,000 × g supernatant fraction of the sonically disrupted cells (data not shown), as in the case of the E. coli counterpart (49). This loose association of the overproduced decarboxylase with the E. coli membrane might be correlated with its primarily hydrophilic nature (Fig. 4).
FIG. 3.
Processing of the product of the B. subtilis psd gene. Cells of JM109(DE3) transformed with pMB281 were inoculated into LB medium supplemented with 100 μg of ampicillin per ml, and at 70 Klett units, isopropyl-β-d-galactopyranoside was added to a final concentration of 5 mM. After 20 min of incubation, this addition was followed by addition of rifampin (the final concentration of 100 μg per ml). After a further 12 min of incubation, 50 μCi of l-[35S]methionine per ml was added, and this mixture was then incubated for 10, 50, and 100 min (lanes, 1, 2, and 3, respectively; lane 0 is the transformant harboring the pWSK29 control). Cells were then collected and suspended in the sample buffer and subjected to sodium dodecyl sulfate–12.5% polyacrylamide gel electrophoresis. Labeled proteins were visualized with the Bio-Image Analyzer BAS 1000 Mac BAS System.
FIG. 4.
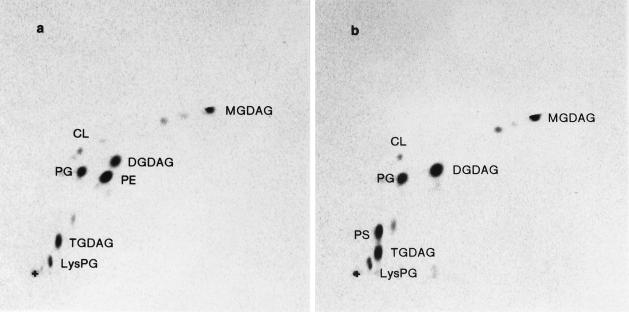
Autoradiograms demonstrating the absence of phosphatidylethanolamine (PE) and accumulation of phosphatidylserine (PS) in a B. subtilis psd disruptant. Cells of the B. subtilis Marburg wild type (a) and psd1::neo mutant (b) were labelled with 0.5 μCi of [1-14C]acetic acid per ml for six generations of cultivation in Penassay broth (5 ml), lipids were extracted, and halves of the lipid fractions were separated by two-dimensional TLC, which was carried out as described in the text. Plates were developed first (x dimension) with chloroform-methanol-water (65:25:4 [vol/vol/vol]) and then (y dimension) with chloroform-methanol-acetic acid (65:25:10 [vol/vol/vol]). +, origins of chromatography. CL, cardiolipin; PG, phosphatidylglycerol; LysPG, lysylphosphatidylglycerol; MGDAG, DGDAG, and TGDAG, mono-, di-, and triglucosyldiacylglycerols, respectively.
Expression of the B. subtilis psd gene in E. coli.
When strain EH150 (psd-2) was kept at 42°C for 6 h, phosphatidylserine accumulated to 29% of the total phospholipids, with the concomitant decrease in the phosphatidylethanolamine level to 51%, as previously described (15). After introduction of pBM281 containing the Bacillus psd gene, the cells of EH150 regained normal growth and recovered the phosphatidylethanolamine content to the wild-type level of 73%, with the decrease of phosphatidylserine to below the detectable limit. The phosphatidylserine decarboxylase activity of EH150 cells harboring pBM281 was then examined. The cells of strain EH150 harboring pBM281 were cultivated at 43°C for 6 h to inactivate the thermosensitive host phosphatidylserine decarboxylase (16, 17). The membrane fraction of the cells harboring pBM281 showed higher phosphatidylserine decarboxylase activity (8.5 nmol/min/mg) than that from cells without the plasmid (0.2 nmol/min/mg). This value was about two times higher than that of the wild-type E. coli cells, and the activity in the soluble fraction was 7% of that of the membrane fraction. This result indicated that the cloned DNA fragment really contained the structural gene for phosphatidylserine decarboxylase of B. subtilis and that the enzyme activity of the B. subtilis decarboxylase was detectable under the conditions described for the E. coli counterpart (9).
It should be noted that the amino acid sequences of the four decarboxylases are highly homologous and that the genes of S. cerevisiae (5) and B. subtilis were able to complement the temperature-sensitive defect of the E. coli (psd-2) mutant. Therefore, the primary product of B. subtilis psd probably undergoes a similar processing to become an active phosphatidylserine decarboxylase like the E. coli product, the processing to the active form is expected to be an autocatalytic process (27, 28), as for the prohistidine decarboxylase (50).
Chromosomal locus of pss-psd genes.
To determine the locus of psd gene on the SfiI and NotI restriction map of the B. subtilis chromosome (21), Southern hybridization analysis was conducted. The procedures for blotting and hybridization with digoxigenin-labeled probe and identification of hybridized restriction fragment were performed as described previously (21). Plasmid pBM102, which contained both the pss and psd genes, hybridized with the 22N fragment of the NotI digest. In the case of the SfiI digest, the BS fragment hybridized with the probe. Experiments with gels of different running conditions and with the probe (pBM281) containing the psd gene alone were consistent with this assignment. Accordingly, both pss and psd genes were unambiguously localized within the overlapping segment (68 kbp) of the 22N and BS fragments: the 238- to 306-kb region on the 4,188-kb physical map (19).
A B. subtilis mutant that showed temperature-sensitive net synthesis of phosphatidylethanolamine was reported (29). Because the in vitro phosphatidylethanolamine synthesis with the membrane of this mutant was no more temperature sensitive than the wild-type preparation, the mutation was considered to affect the synthesis indirectly. The mutation was linked by transformation to aroD, which is located at around 2,600 kb on the physical map (19), and, therefore, it is not in the pss or psd gene region. In E. coli, pss and psd genes are located at different loci on the chromosome; pss and psd were mapped at 49 min (37, 41) and at 83 min (15), respectively. The result of the Southern mapping with the probes of pss and psd genes showing no sign of multiple signals of hybridization indicated an unequivocal close linkage of the two genes, in accordance with the data from nucleotide sequencing. The close linkage may suggest an organized control of phosphatidylserine synthase and decarboxylase, constituting an operon in which the both genes are transcribed into a single mRNA. Although the presence of a putative ρ-independent terminator signal and a promoter sequence between the two genes (Fig. 2) might oppose an organization in an operon, there are many examples of complex operons having sets of promoters and terminators for dynamic and fine control. In E. coli, all phospholipid enzymes appear to be synthesized constitutively, and lipid synthesis is considered to be regulated by far more complex mechanisms than those presently known (46). Studies of transcription of B. subtilis cells to define the nature of the linkage of the pss and psd genes will answer this question.
Disruption of the psd gene and its effect on lipid composition.
To construct a strain harboring an interrupted chromosomal allele of psd, we first prepared a B. subtilis strain, 160, transformed with a covering plasmid, pLUCK100, which has a spac promoter-controllable pss-psd gene combination. This strain, 160/pLUCK100, and the parental strain, 160, were then transformed with the DNA fragment of the psd gene region from plasmid pYH01, in which the gene was interrupted with the neo (20) gene at its unique PstI site (Fig. 2). From both recipient strains, neomycin-resistant transformants appeared at frequencies comparable to those with chromosomal markers. Unexpectedly, all of the neomycin-resistant transformants of strain 160 without the covering plasmid required no divalent cation, which is essential for E. coli pssA null mutants (6, 44). Analysis of the chromosomal DNA of the neomycin-resistant transformants by the PCR method using primers for the psd gene produced an amplified DNA product corresponding to the size of the neo-interrupted psd gene, and no product corresponding to the intact psd gene was detected, indicating that the chromosomal psd gene was correctly disrupted by homologous recombination with the insertion of neo as constructed on plasmid pYH01. The resistant strain, designated as SDB01 (psd1::neo), showed the same growth rates as the wild-type strain in LB, TY, and NBY media without supplementation with divalent cations and synthetic medium CI. In all cases, no difference in final cell densities was observed.
The composition of lipids from the psd disruptant (SDB01) was compared with that of the wild type after labelling with [1-14C]acetic acid followed by two-dimensional TLC on a silica gel plate (Fig. 4). No significant radiolabel was detected in the region corresponding to phosphatidylethanolamine, and a concomitant accumulation of phosphatidylserine (corresponding to 40% of the amount of phosphatidylethanolamine of the wild type) was observed (Table 2). The absence of phosphatidylethanolamine in B. subtilis cells, therefore, does not have any adverse effect on their growth, which is quite different from the case of E. coli phosphatidylethanolamine-deficient mutants, which require divalent cations (6, 44). Phosphatidylethanolamine, therefore, seems dispensable for the growth of B. subtilis cells. This is compatible with the previous report that phosphatidylethanolamine was practically absent from the membrane of B. subtilis cells that had been cultured in a chemostat under Mg2+-limited conditions at pH 8.0 (33). Phosphatidylethanolamine is a major component of the membrane of E. coli, and the different phenotypes of the disruptants of E. coli and B. subtilis could be due to the difference in the membrane. The B. subtilis null pss mutant SDB02 (Δpss10::spc) constructed in the same way lacked both phosphatidylethanolamine and phosphatidylserine in the membrane, yet grew normally without further addition of divalent cations. This indicated that the accumulated phosphatidylserine in place of phosphatidylethanolamine in the psd disruptant had little effect for the growth of the disruptant. In the psd-disrupted strain, apparent increases in monoglucosyldiacylglycerol (2-fold), diglucosyldiacylglycerol (3.1-fold), and triglucosyldiacylglycerol (1.6-fold) were observed. Similar increases in diglucosyldiacylglycerol (3.1-fold) and triglucosyldiacylglycerol (4.1-fold), but not in monoglucosyldiacylglycerol, were observed in the lipid fraction of the pss null mutant strain (Table 2). This is consistent with the previous observation that the membrane levels of phosphatidylethanolamine and diglucosyldiacylglycerol changed inversely in response to changes in the culture conditions (34). The need for phosphatidylethanolamine in the B. subtilis membrane may be satisfied by increases in the contents of glucosyldiacylglycerols. A careful inspection of the proportion of the lipids in Table 2 revealed that the sums of glucosyldiacylglycerols plus phosphatidylethanolamine (including phosphatidylserine) of the three strains were quite equal; wild-type, psd null, and pss null mutant lipids were 78.8, 80.1, and 79.7% of the total lipids, respectively. In B. subtilis, glucosyldiacylglycerols are believed to be synthesized by transfer of glucose from UDP-glucose to diacylglycerol, which is produced by dephosphorylation from phosphatidic acid according to the reactions detected in other organisms (7). Therefore, it is possible that the regulated cellular level (3) of CDP-diacylglycerol results in the accumulation of phosphatidic acid in the absence of phosphatidylserine synthesis, which in turn accelerates the synthesis of diacylglycerol, the putative substrate for glucosyldiacylglycerols. However, the reasons for the apparently specific increase of glucosyldiacylglycerols but not of phosphatidylglycerol derivatives in the two null mutants, as well as for the decrease of monoglucosyldiacylglycerol in the pss null mutant, are unknown.
TABLE 2.
Lipid composition of B. subtilis psd1::neo and Δpss10::spc mutants
| Strain | % of 14C-labelled total lipida
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| MGDAG | DGDAG | TGDAG | CL | PG | LysPG | PE | PS | Others | |
| 160 (wild type) | 8.0 | 12.6 | 8.9 | 0.8 | 16.2 | 2.4 | 49.3 | <0.1 | 1.8 |
| SDB01 (psd1::neo) | 15.8 | 29.0 | 14.3 | 0.6 | 13.1 | 2.7 | <0.1 | 21.0 | 2.2 |
| SDB02 (Δpss10::spc) | 3.8 | 39.5 | 36.4 | 3.1 | 10.5 | 2.9 | <0.1 | <0.1 | 3.9 |
Values were calculated from the PSL unit of the BAS 1000 bioimaging analyzer. MGDAG, DGDAG, and TGDAG, mono-, di-, and triglucosyldiacylglycerols, respectively; CL, cardiolipin; PG, phosphatidylglycerol; LysPG, lysylphosphatidylglycerol; PE, phosphatidylethanolamine; PS, phosphatidylserine.
The complete absence of phosphatidylethanolamine in B. subtilis psd-disrupted and pss null mutant cells implied that phosphatidylethanolamine is synthesized solely through phosphatidylserine. Thus, it is highly probable that both gram-negative and -positive bacteria adopt not the Kennedy pathway (22) but the de novo phosphatidylserine pathway to form phosphatidylethanolamine.
ACKNOWLEDGMENTS
We thank W. Dowhan for providing us with E. coli EH150 and S. Kushner and P. Stragier for vector plasmids. Thanks are also due to S. Fuchizawa and K. Komori for construction of B. subtilis SDB02.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan.
REFERENCES
- 1.Ames G F. Lipids of Salmonella typhimurium and Escherichia coli: structure and metabolism. J Bacteriol. 1968;95:833–843. doi: 10.1128/jb.95.3.833-843.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostopoulos C, Crawford I. Transformation studies on the linkage of markers in the tryptophan pathway in Bacillus subtilis. Proc Natl Acad Sci USA. 1961;47:378–390. doi: 10.1073/pnas.47.3.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell R M, Cronan J E., Jr Mutants of Escherichia coli defective in membrane phospholipid synthesis. Phenotypic suppression of sn-glycerol-3-phosphate acyltransferase Km mutants by loss of feedback inhibition of the biosynthetic sn-glycero l-3-phosphate dehydrogenase. J Biol Chem. 1975;250:7153–7158. [PubMed] [Google Scholar]
- 4.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 5.Clancey C J, Chang S C, Dowhan W. Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant. J Biol Chem. 1993;268:24580–24590. [PubMed] [Google Scholar]
- 6.DeChavigny A, Heacock P N, Dowhan W. Sequence and inactivation of the pss gene of Escherichia coli. Phosphatidylethanolamine may not be essential for cell viability. J Biol Chem. 1991;266:5323–5332. [PubMed] [Google Scholar]
- 7.de Mendoza D, Grau R, Cronan J E., Jr . Biosynthesis and function of membrane lipids. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 411–421. [Google Scholar]
- 8.Dittmer J C, Lester R L. A simple, specific spray for the detection of phospholipids on thin layer chromatograms. J Lipid Res. 1964;5:126–127. [PubMed] [Google Scholar]
- 9.Dowhan W, Wickner W T, Kennedy E P. Purification and properties of phosphatidylserine decarboxylase from Escherichia coli. J Biol Chem. 1974;249:3079–3084. [PubMed] [Google Scholar]
- 10.Dubendorff J W, Studier F W. Controlling basal expression in an inducible T7 expression system by blocking the target T7 promoter with lac repressor. J Mol Biol. 1991;219:45–59. doi: 10.1016/0022-2836(91)90856-2. [DOI] [PubMed] [Google Scholar]
- 11.Dutt A, Dowhan W. Characterization of a membrane-associated cytidine diphosphate-diacylglycerol-dependent phosphatidylserine synthase in bacilli. J Bacteriol. 1981;147:535–542. doi: 10.1128/jb.147.2.535-542.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutt A, Dowhan W. Purification and characterization of a membrane-associated phosphatidylserine synthase from Bacillus licheniformis. Biochemistry. 1985;24:1073–1079. doi: 10.1021/bi00326a001. [DOI] [PubMed] [Google Scholar]
- 13.Graves M C, Rabinowitz J M. In vivo and in vitro transcription of the Clostridium pasteurianum ferredoxin gene: evidence for “extended” promoter elements in gram-positive organisms. J Biol Chem. 1986;261:11409–11415. [PubMed] [Google Scholar]
- 14.Guerout-Fleury A M, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 15.Hawrot E, Kennedy E P. Biogenesis of membrane lipids: mutants of Escherichia coli with temperature-sensitive phosphatidylserine decarboxylase. Proc Natl Acad Sci USA. 1975;72:1112–1116. doi: 10.1073/pnas.72.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawrot E, Kennedy E P. Conditional lethal phosphatidylserine decarboxylase mutants of Escherichia coli. Mol Gen Genet. 1976;148:271–279. doi: 10.1007/BF00332901. [DOI] [PubMed] [Google Scholar]
- 17.Hawrot E, Kennedy E P. Phospholipid composition and membrane function in phosphatidylserine decarboxylase mutants of Escherichia coli. J Biol Chem. 1978;253:8213–8220. [PubMed] [Google Scholar]
- 18.Henner D J. Inducible expression of regulatory genes in Bacillus subtilis. Methods Enzymol. 1993;68:243–255. doi: 10.1016/0076-6879(90)85022-g. [DOI] [PubMed] [Google Scholar]
- 19.Itaya M. Physical map of the Bacillus subtilis 168 chromosome. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 463–471. [Google Scholar]
- 20.Itaya M, Kondo K, Tanaka T. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis. Nucleic Acids Res. 1989;17:4410. doi: 10.1093/nar/17.11.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itaya M, Tanaka T. Complete physical map of the Bacillus subtilis 168 chromosome constructed by a gene-directed mutagenesis method. J Mol Biol. 1991;220:631–648. doi: 10.1016/0022-2836(91)90106-g. [DOI] [PubMed] [Google Scholar]
- 22.Kennedy E P, Weiss S B. The function of cytidine coenzymes in the biosynthesis of phospholipids. J Biol Chem. 1956;222:193–214. [PubMed] [Google Scholar]
- 23.Kishi T, Miura K, Matsumoto K, Hirokawa H. Complementation assay of primer protein: gene expression systems of plasmid vectors support the infection of suppressor sensitive mutant phages M2 and φ29. Jpn J Genet. 1993;68:243–255. doi: 10.1266/jjg.68.243. [DOI] [PubMed] [Google Scholar]
- 24.Kuge O, Nishijima M, Akamatsu Y. A cloned gene encoding phosphatidylserine decarboxylase complements the phosphatidylserine biosynthetic defect of a chinese hamster ovary cell mutant. J Biol Chem. 1991;266:6370–6376. [PubMed] [Google Scholar]
- 25.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 26.Langley K E, Kennedy E P. Energetics of rapid transmembrane movement and of compositional asymmetry of phosphatidylethanolamine in membranes of Bacillus megaterium. Proc Natl Acad Sci USA. 1979;76:6245–6249. doi: 10.1073/pnas.76.12.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q-X, Dowhan W. Structural characterization of Escherichia coli phosphatidylserine decarboxylase. J Biol Chem. 1988;263:11516–11522. [PubMed] [Google Scholar]
- 28.Li Q-X, Dowhan W. Studies on the mechanism of formation of the pyruvate prosthetic group of phosphatidylserine decarboxylase from Escherichia coli. J Biol Chem. 1990;265:4111–4115. [PubMed] [Google Scholar]
- 29.Lindgren V, Holmgren E, Rutberg L. Bacillus subtilis mutant with temperature-sensitive net synthesis of phosphatidylethanolamine. J Bacteriol. 1977;132:473–484. doi: 10.1128/jb.132.2.473-484.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Louie K, Chen Y-C, Dowhan W. Substrate-induced membrane association of phosphatidylserine synthase from Escherichia coli. J Bacteriol. 1986;165:805–812. doi: 10.1128/jb.165.3.805-812.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Louie K, Dowhan W. Investigations on the association of phosphatidyl serine synthase with the ribosomal component from Escherichia coli. J Biol Chem. 1980;255:1124–1127. [PubMed] [Google Scholar]
- 32.Matsumoto K. Phosphatidylserine synthase from bacteria. Biochim Biophys Acta. 1997;1348(Special Issue):214–227. doi: 10.1016/s0005-2760(97)00110-0. [DOI] [PubMed] [Google Scholar]
- 33.Minnikin D E, Abdolrahimzadeh H. Effect of pH on the proportions of polar lipids, in chemostat cultures of Bacillus subtilis. J Bacteriol. 1974;120:999–1003. doi: 10.1128/jb.120.3.999-1003.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minnikin D E, Abdolrahimzadeh H, Baddiley J. Variation of polar lipid composition of Bacillus subtilis (Marburg) with different growth conditions. FEBS Lett. 1972;27:16–18. doi: 10.1016/0014-5793(72)80398-3. [DOI] [PubMed] [Google Scholar]
- 35.Ohta A, Obara T, Asami Y, Shibuya I. Molecular cloning of the cls gene responsible for cardiolipin synthesis in Escherichia coli and phenotypic consequences of its amplification. J Bacteriol. 1985;163:506–514. doi: 10.1128/jb.163.2.506-514.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohta A, Shibuya I. Membrane phospholipid synthesis and phenotypic correlation of an Escherichia coli pss mutant. J Bacteriol. 1977;132:434–443. doi: 10.1128/jb.132.2.434-443.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohta A, Shibuya I, Maruo B. Escherichia coli mutants with temperature-sensitive phosphatidylserine synthetase: genetic analysis. Agric Biol Chem. 1975;39:2443–2445. [Google Scholar]
- 38.Okada M, Matsuzaki H, Shibuya I, Matsumoto K. Cloning, sequencing, and expression in Escherichia coli of the Bacillus subtilis gene for phosphatidylserine synthase. J Bacteriol. 1994;176:7456–7461. doi: 10.1128/jb.176.24.7456-7461.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Op den Kamp J A F, Redai I, van Deenen L L M. Phospholipid composition of Bacillus subtilis. J Bacteriol. 1969;99:298–303. doi: 10.1128/jb.99.1.298-303.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patterson P H, Lennarz W J. Studies on the membrane of bacilli. I. Phospholipid biosynthesis. J Biol Chem. 1971;246:1062–1072. [PubMed] [Google Scholar]
- 41.Raetz C R H. Phosphatidylserine synthetase mutants of Escherichia coli. Genetic mapping and membrane phospholipid composition. J Biol Chem. 1976;251:3242–3249. [PubMed] [Google Scholar]
- 42.Raetz C R H, Kennedy E P. The association of phosphatidylserine synthetase with ribosomes in extracts of Escherichia coli. J Biol Chem. 1972;247:2008–2014. [PubMed] [Google Scholar]
- 43.Saha S K, Furukawa Y, Matsuzaki H, Shibuya I, Matsumoto K. Directed mutagenesis, Ser-56 to Pro of Bacillus subtilis phosphatidylserine synthase drastically lowers enzymatic activity and relieves amplification toxicity in Escherichia coli. Biosci Biotechnol Biochem. 1996;60:630–633. doi: 10.1271/bbb.60.630. [DOI] [PubMed] [Google Scholar]
- 44.Saha S K, Nishijima S, Matsuzaki H, Shibuya I, Matsumoto K. A regulatory mechanism for the balanced synthesis of membrane phospholipid species in Escherichia coli. Biosci Biotechnol Biochem. 1996;60:111–116. doi: 10.1271/bbb.60.111. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 46.Shibuya I. Metabolic regulations and biological functions of phospholipids in Escherichia coli. Prog Lipid Res. 1992;31:245–299. doi: 10.1016/0163-7827(92)90010-g. [DOI] [PubMed] [Google Scholar]
- 47.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 48.Trotter P J, Pedretti J, Voelker D R. Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J Biol Chem. 1993;268:21416–21424. [PubMed] [Google Scholar]
- 49.Tyhach R J, Hawrot E, Satre M, Kennedy E P. Increased synthesis of phosphatidylserine decarboxylase in a strain of Escherichia coli bearing a hybrid plasmid. Altered association of enzyme with the membrane. J Biol Chem. 1979;254:627–633. [PubMed] [Google Scholar]
- 50.van Poelji P D, Snell E S. Pyruvoyl-dependent enzymes. Annu Rev Biochem. 1990;59:29–59. doi: 10.1146/annurev.bi.59.070190.000333. [DOI] [PubMed] [Google Scholar]
- 51.Wang R F, Kushner S. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]