Abstract
Background
Salmonella enterica are important foodborne pathogens and the third leading cause of death among diarrheal infections worldwide. This cross-sectional study investigated the frequency of antibiotic-resistant Salmonella enterica in commercial and smallholder farm environments in the Ashanti Region of Ghana. A total of 1490 environmental samples, comprising 800 (53.7%) soil (from poultry, pigs, sheep, goats and cattle farms), 409 (27.4%) pooled poultry fecal and 281 (18.9%) dust (from poultry farms) samples, were collected from 30 commercial and 64 smallholder farms. All samples were processed using standard culture methods. Isolates were identified by biochemical methods and confirmed using the VITEK 2 System. Antibiotic susceptibility testing was carried out by disk diffusion following the EUCAST guidelines. Serotyping was performed using the Kauffman White Le Minor Scheme.
Results
The overall Salmonella frequency was 6.0% (n/N = 90/1490); the frequency varied according to the type of sample collected and included: 8.9% for dust (n/N = 25/281), 6.5% for soil (n/N = 52/800) and 3.2% for pooled poultry fecal samples (n/N = 13/409). Salmonella was also recovered from commercial farm environments (8.6%, n/N = 68/793) than from smallholder farms (3.2%, n/N = 22/697) (PR = 2.7, CI: 1.7 – 4.4). Thirty-four different Salmonella serovars were identified, the two most common being Rubislaw (27.8%, n/N = 25/90) and Tamale (12.2%, n/N = 11/90). Serovar diversity was highest in strains from soil samples (70.6%, n/N = 24/34) compared to those found in the dust (35.2%, n/N = 12/34) and in fecal samples (29.4%, n/N = 10/34). Salmonella frequency was much higher in the rainy season (8.4%, n/N = 85/1007) than in the dry season (1.0%, n/N = 5/483) (PR = 8.4, 95% CI: 3.3 – 20.0). Approximately 14.4% (n/N = 13/90) of the isolates were resistant to at least one of the tested antimicrobials, with 84.6% (n/N = 11/13) being resistant to multiple antibiotics. All Salmonella Kentucky (n = 5) were resistant to ciprofloxacin.
Conclusion
This study showed that farm environments represent an important reservoir for antibiotic-resistant Salmonella, which warrants monitoring and good husbandry practices, especially in commercial farms during the rainy season, to control the spread of this pathogen.
Keywords: Salmonella enterica, Environmental reservoirs, Antibiotic resistance, Farms, Rural Ghana
Background
Recent data suggest that non-typhoidal Salmonella invasive disease causes between 46,400 and 123,000 annual deaths worldwide, mostly in sub-Saharan Africa [1]. So far, more than 2,600 Salmonella serovars have been described. Salmonella transmission in humans is typically of zoonotic origin, and the majority of infections in both humans and animals are caused by subspecies 1 of Salmonella enterica [2]. Nevertheless, Salmonella enterica has not been widely studied in sub-Saharan Africa [3]. Even though anthroponotic transmission has been suggested, it is yet to be confirmed [4].
Antimicrobial resistance (AMR) leading to difficult-to-treat Salmonella infections in humans is a global health concern [5]. Multidrug-resistant (MDR) Salmonella and emerging fluoroquinolone resistance are rising worldwide [6]. In Ghana, resistant Salmonella have been reported from humans [7, 8], farm animals [9, 10], water sources [11], fresh milk [12] and vegetables [13]. Despite Ghana having a national action plan for antimicrobial use [14], poultry and livestock farmers continue to overuse antimicrobials such as oxytetracycline, neomycin and tylosin for growth promotion, prophylaxis and infection treatment [15], fostering the development of AMR.
Salmonella are highly adaptive bacteria that can persist in the environment for prolonged periods [16]. The presence of Salmonella in soil may originate from fecal droppings, organic fertilizers or dust from farms [17]; hence environmental niches such as soil and dust might present possible transmission reservoirs for infections. So far, only a few studies have been conducted on Salmonella in soil and dust, and data for Africa is limited [18, 19]. The majority of Salmonella studies conducted in Ghana focused on commercial farm animals [9, 10], slaughterhouses [20, 21] and markets [13, 22]. In this study, we examined the frequency and antibiotic resistance of Salmonella enterica isolated from environmental samples (dust and soil) and animal feces collected from commercial and smallholder farm environments in two communities in the Ashanti Region of Ghana.
Results
Salmonella frequencies in environmental samples from rural and semi-urban communities
A total of 1490 environmental samples, comprising 800 (53.7%) soil, 409 (27.4%) pooled fecal and 281 (18.9%) samples from dust, were collected from 30 commercial and 64 smallholder farms in the rural Agogo and semi-urban Ejisu communities of Ghana. The overall Salmonella frequency was 6.0% (n/N = 90/1490). Frequencies from the two study sites: rural (4.9%, n/N = 37/750) and semi-urban (7.2%, n/N = 53/740), were similar (PR = 0.7, 95% CI: 0.5 – 1.0). Overall Salmonella frequencies varied according to the type of sample collected: 8.9% (n/N = 25/281) for dust, 6.5% (n/N = 52/800) for soil and 3.2% (n/N = 13/409) for fecal samples. The highest frequency recorded by commercial farms was observed in samples from dust (20.8%, n/N = 15/72) followed by soil (13.1%, n/N = 34/260) samples. Out of the 40 smallholders and 15 commercial farms sampled from rural Agogo, 12 (30%) and 6 (40%) were positive for Salmonella, respectively. Also, in the semi-urban community, more commercial farms (86.7%, n/N = 13/15) were positive for Salmonella than smallholder farms (8.3%, n/N = 2/24) (PR = 10.4, 95% CI: 2.7 – 39.8). Table 1 details Salmonella frequencies detected from the various sample types and study sites.
Table 1.
Salmonella frequencies detected from various sample types from rural (Agogo) and semi-urban (Ejisu) communities
| Sample type | Rural, % (n/N) | Semi-urban, % (n/N) | Total, % (n/N) | ||
|---|---|---|---|---|---|
| Commercial | Smallholder | Commercial | Smallholder | ||
| Soil | 4.3 (8/186) | 4.2 (9/214) | 13.1 (34/260) | 0.7 (1/140) | 6.5 (52/800) |
| Fecal | 1.0 (1/104) | 9.5 (10/105) | 2.0 (2/100) | 0 (0/100) | 3.2 (13/409) |
| Dust | 11.3 (8 /71) | 1.4 (1/70) | 20.8 (15/72) | 1.5 (1/68) | 8.9 (25/281) |
| Total | 4.7 (17/361) | 5.1 (20/389) | 11.8 (51/432) | 0.6 (2/308) | 6.0 (90/1490) |
n positive sample, N total sample size
Factors associated with Salmonella frequencies
Table 2 shows a Poisson regression analysis of possible factors associated with Salmonella frequencies. From the adjusted prevalence ratios, more Salmonella were recovered from commercial farms (8.6%, n/N = 68/793) than from smallholder farms (3.2%, n/N = 22/697) (PR = 2.6, CI: 1.6 – 4.3). Also, Salmonella was 2.7 (95% CI: 1.4 – 5.5) and 1.9 (95% CI: 1.1 – 3.6) times more likely to be isolated from dust and soil than from fecal samples. In the farm environment, Salmonella isolation rates in soil within ≤ 5 m around the pen and > 5 m away from the pen were similar (PR = 1.3, 95% CI: 0.6 – 2.4).
Table 2.
Poisson regression analysis of possible factors associated with Salmonella frequency
| Variable | Crude ratios PR (95% CI) |
Adjusted ratios PR (95% CI) |
|---|---|---|
| All Salmonella isolates | ||
| Commercial vs. smallholder farms | 2.7 (1.7 – 4.4) | 2.6 (1.6 – 4.3) |
| Ejisu (semi-urban) vs. Agogo (rural) | 1.4 (0.9 – 2.2) | 1.3 (0.8 – 2.0) |
| Dust vs. fecal | 2.8 (1.4 – 5.6) | 2.7 (1.4 – 5.5) |
| Dust vs. soil | 1.3 (0.8 – 2.1) | 1.4 (0.8 – 2.3) |
| Soil vs fecal | 2.0 (1.1 – 3.9) | 1.9 (1.1 – 3.6) |
| Soil Salmonella isolates | ||
| Poultry vs. Other livestock | 0.5 (0.3 – 0.8) | 0.5 (0.3 – 0.9) |
| ≤ 5 m near pen vs. > 5 m away from pen | 1.4 (0.8 – 2.4) | 1.3 (0.6 – 2.4) |
Salmonella serovar distribution
All Salmonella isolated from environmental samples belonged to the Salmonella enterica species. The vast majority (96.7%, n/N = 87/90) belonged to the subspecies enterica, 2.2% (n/N = 2/90) belonged to the subspecies diarizonae and 1.1% (n/N = 1/90) were subspecies salamae (Table 3). The diarizonae and salamae subspecies were only found in the soil. A total of 34 different Salmonella serovars were identified in this study. Serovar diversity was highest in strains from soil samples (70.6%, n/N = 24/34) compared to those found in the dust (35.2%, n/N = 12/34) and in fecal samples (29.4%, n/N = 10/34). Also, the serovar diversity from smallholder farms (77.2%, n/N = 17/22) was broader than what was found in commercial farms (29.4%, n/N = 20/68) (PR = 2.3, 95% CI: 1.6 – 3.2). Overall, the two most common serovars were Rubislaw (27.8%, n/N = 25/90) and Tamale (12.2%, n/N = 11/90). Most serovar Tamale (72.7%, n/N = 8/11) and Rubislaw (68.0%, n/N = 17/25) were identified from dust and soil, respectively (Table 3).
Table 3.
Distribution of Salmonella serovars isolated from environmental samples
| subsp. | serovar | Soil, N = 52 % (n) |
Dust, N = 25 % (n) |
Fecal, N = 13 % (n) |
Total, N = 90 % (n) |
|---|---|---|---|---|---|
| enterica | Rubislaw | 32.7 (17) | 24.0 (6) | 15.4 (2) | 27.8 (25) |
| Tamale | 1.9 (1) | 32.0 (8) | 15.4 (2) | 12.2 (11) | |
| Kentucky | 5.8 (3) | 8.0 (2) | - | 5.6 (5) | |
| Bochum | 5.8 (3) | 4.0 (1) | - | 4.4 (4) | |
| Yovokome | 3.8 (2) | - | 15.4 (2) | 4.4 (4) | |
| Agona | 5.8 (3) | - | - | 3.3 (3) | |
| Reading | 3.8 (2) | - | 7.7 (1) | 3.3 (3) | |
| Westphalia | 3.8 (2) | - | - | 2.2 (2) | |
| Montevideo | 3.8 (2) | - | - | 2.2 (2) | |
| Chester | 1.9 (1) | 4.0 (1) | - | 2.2 (2) | |
| Epinay | 1.9 (1) | 4.0 (1) | - | 2.2 (2) | |
| Ilala | 1.9 (1) | 4.0 (1) | - | 2.2 (2) | |
| Agama | 1.9 (1) | - | - | 1.1 (1) | |
| Alachua | 1.9 (1) | - | - | 1.1 (1) | |
| Duisburg | 1.9 (1) | - | - | 1.1 (1) | |
| Give | 1.9 (1) | - | - | 1.1 (1) | |
| Honelis | 1.9 (1) | - | - | 1.1 (1) | |
| Poona | 1.9 (1) | - | - | 1.1 (1) | |
| Mundonobo | 1.9 (1) | - | - | 1.1 (1) | |
| Saphra | 1.9 (1) | - | - | 1.1 (1) | |
| Serologically Rough | 1.9 (1) | - | - | 1.1 (1) | |
| Typhimurium | - | 4.0 (1) | - | 1.1 (1) | |
| Durban | - | 4.0 (1) | - | 1.1 (1) | |
| Elisabethville | - | 4.0 (1) | - | 1.1 (1) | |
| Redhill | - | 4.0 (1) | - | 1.1 (1) | |
| Konongo | - | 4.0 (1) | - | 1.1 (1) | |
| Aschersleben | - | - | 7.7 (1) | 1.1 (1) | |
| Gaminara | - | - | 7.7 (1) | 1.1 (1) | |
| Wagenia | - | - | 7.7 (1) | 1.1 (1) | |
| Wien | - | - | 7.7 (1) | 1.1 (1) | |
| Lexington | - | - | 7.7 (1) | 1.1 (1) | |
| Unidentified | 3.8 (2) | - | 7.7 (1) | 3.3 (3) | |
| diarizonae | Ssp. IIIb | 3.8 (2) | - | - | 2.2 (2) |
| salamae | Ssp. II | 1.9 (1) | - | - | 1.1 (1) |
| Total | 100 (52) | 100 (25) | 100 (13) | 100 (90) |
Seasonal variation of Salmonella frequency
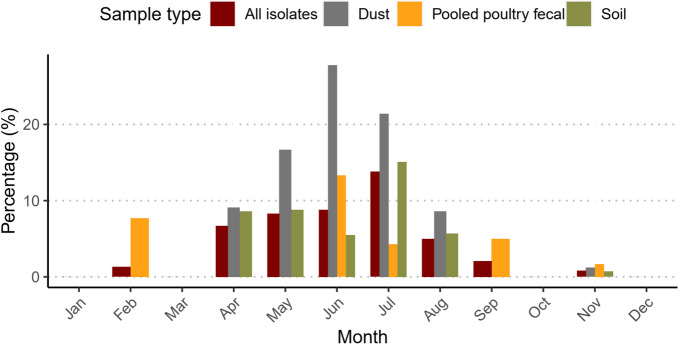
Figure 1 shows the monthly isolation rate of Salmonella from dust, fecal and soil during the sampling period. In January, March, October and December, no Salmonella were isolated. The highest Salmonella frequency from dust (23.6%, n/N = 17/74), soil (12.3%, n/N = 15/122) and fecal (7.8%, n/N = 9/115) samples were recorded between June and July. Furthermore, the overall Salmonella frequency was much higher in the rainy season (8.4%, n/N = 85/1007) than in the dry season (1.0%, n/N = 5/483) (PR = 8.4, 95% CI: 3.3 – 20.0).
Fig. 1.
Seasonal frequencies of Salmonella isolated from dust, pooled fecal and soil samples collected from farm environments
Antimicrobial resistance
Among all the antibiotics tested, the highest rate of resistance was to ciprofloxacin (12.2%, n/N = 11/90), whereas all Salmonella were susceptible to cefotaxime, cefoxitin, ceftazidime and meropenem. In terms of serovars and associated resistance, all Salmonella Kentucky (n = 5) were resistant to ciprofloxacin. Ciprofloxacin resistance was also found in the serovars Chester (18.1%, n/N = 2/11), Ilala (18.1%, n/N = 2/11), Montevideo (9.0%, n/N = 1/11), and Epinay (9.0%, n/N = 1/11). None of the strains was MDR (resistant to chloramphenicol, ampicillin, and trimethoprim-sulfamethoxazole), but 14.4% (n/N = 13/90) of the Salmonella were resistant to at least one tested antibiotic, and 12.2% (n/N = 11/90) were resistant to multiple antibiotics. All of the 11 multiple antibiotic resistant isolates were either from the soil (54.5%, n/N = 6/11) or dust (45.4%, n/N = 5/11). Multiple antibiotic resistance was seen in the serovars Epinay, Montevideo, Chester, Ilala, and Kentucky. Table 4 further shows the observed antibiotic resistance in Salmonella serovar isolated from the different sample types.
Table 4.
Antibiotic resistance patterns in different Salmonella serovars isolated from environmental samples
| Sample type | Serovar | Resistance, % (n/N) | |||
|---|---|---|---|---|---|
| Ampicillin | Ciprofloxacin | Trimethoprim-sulfamethoxazole | Chloramphenicol | ||
| Soil | Kentucky | 100 (3/3) | 100 (3/3) | 33.3 (1/3) | 0 (0/3) |
| Soil | Montevideo | 50 (1/2) | 50 (1/2) | 0 (0/2) | 0 (0/2) |
| Soil | Chester | 0 (0/1) | 100 (1/1) | 100 (1/1) | 0 (0/1) |
| Soil | Ilala | 0 (0/1) | 100 (1/1) | 100 (1/1) | 0 (0/1) |
| Dust | Kentucky | 100 (2/2) | 100 (2/2) | 50 (1/2) | 0 (0/1) |
| Dust | Epinay | 100 (1/1) | 100 (1/1) | 100 (1/1) | 0 (0/1) |
| Dust | Chester | 0 (0/1) | 100 (1/1) | 100 (1/1) | 0 (0/1) |
| Dust | Ilala | 0 (0/1) | 100 (1/1) | 100 (1/1) | 0 (0/1) |
| Fecal | Lexington | 0 (0/1) | 0 (0/1) | 0 (0/1) | 100 (1/1) |
Discussion
This cross-sectional study investigated the frequency and antimicrobial resistance of Salmonella enterica isolated from soil, dust and fecal samples collected from commercial and smallholder farms in two communities in the Ashanti Region of Ghana. The overall Salmonella frequency observed in this study was 6%. This raises concerns for public health, particularly for workers and individuals residing near these farms, as Salmonella is a zoonotic pathogen with the potential to cause foodborne illness in humans. The current Salmonella frequency detected is similar to what was reported from studies conducted in Nairobi (2.6–5.9%) [23] and Nigeria (10%) [24]. However, it is far less than the 44% reported by a similar study from commercial poultry farms and markets in Ghana [9]. This disparity could be due to the differences in the types of environmental samples analyzed and the geographical locations. In commercial farms, we observed the highest Salmonella frequency in the dust. In agreement with the current findings, several studies have also associated dust generated in farms with Salmonella transmission in farm animals and sporadic human outbreaks [25, 26].
In humans, S. Enteritidis and S. Typhimurium have been reported as the most common serovars isolated from clinical samples [27]. Our study observed a very low prevalence of S. Typhimurium and no S. Enteritidis. This could be attributed to the fact that S. Typhimurium is more frequently associated with human socio-demographic features rather than environmental reservoirs [28]. Interestingly, our study found no S. Enteritidis, while a similar previous study conducted in Kumasi, Ghana, reported a 10.6% prevalence of this serovar [9]. This suggests the need for further research to confirm whether the Salmonella serovar distribution in the farm environments in Ghana is changing. Nonetheless, we identified other Salmonella enterica serovars, potentially capable of causing human infections but known to inhabit various environmental sources, including soil, water, plants, and animals. The current study observed a diverse serovar distribution in the environment, mostly isolated from soil and dust. This observation is not unusual since earlier findings have reported diverse serovars in environmental samples [29, 30]. The most common serovars identified in this study were S. Rubislaw and S. Tamale. In agreement with our findings, S. Rubislaw has been recovered in Ghana from dust and poultry feces [9] and drinking water sources [31]. In Australia, the first recorded instance of Salmonella Rubislaw gastroenteritis in humans was linked to the terrarium of a pet lizard [32].
In the current study, the isolation rate for Salmonella was much higher in the rainy than in the dry season. This finding is similar to reports from studies conducted in Uganda [33] and Southern Ethiopia [34] that investigated the frequency of Salmonella in different environmental samples collected during the rainy and dry seasons. In the rainy season, there is increased availability of contaminated water sources and potential flooding, which create favorable conditions for bacterial growth and survival. Also, in most areas in Africa, the spread of Salmonella is aided by inadequate sanitation and poor drainage systems [35]. In contrast, temperate climates record high occurrences of Salmonella during the summer months [36] because during warm temperatures, delayed refrigeration of food products creates ideal conditions for Salmonella to grow.
This study recorded higher antimicrobial resistance in commercial farms than in smallholder farms. This is not surprising because a recent study in the same area reported higher usage of antimicrobials in commercial farms than on the smallholder farm level [15]. Also, a significant level of multiple antibiotic resistance, especially resistance to fluoroquinolones, was observed. This is particularly concerning because fluoroquinolones are amongst the most important drugs for treating a wide range of infections in the country. Our study did not detect MDR; only one isolate was resistant to chloramphenicol. Contrary, other earlier studies done in Ghana have reported a high rate of MDR Salmonella in humans [11] and poultry [9]. But in agreement with the current finding, recent similar studies conducted in Nairobi and Ghana have recorded 100% [37] and 91% [20] susceptibility to chloramphenicol, respectively. This high Salmonella susceptibility to chloramphenicol is reassuring since it is commonly used in low-income countries to treat Salmonellosis [38]. Nonetheless, high resistance was observed for ampicillin and trimethoprim-sulfamethoxazole, which are likewise recommended for treating salmonellosis [38]. The high trimethoprim-sulfamethoxazole resistance observed probably reflects farmers' high use of sulfonamides in animal farming [39].
There were a few limitations to this study. Sampling was done in only two districts, so our results might not represent other geographical regions of Ghana. Dust and fecal samples were collected from only poultry farms, while soil samples were collected from poultry and livestock farms that raised pigs, goats, sheep, or cattle. Hence, caution must be taken when comparing dust and fecal isolates to isolates from the soil. Also, the Salmonella isolated from dust and soil may not be coming from the farm animals alone since feces from reptiles, rodents, and other non-farm animals may be part of the dust and soil collected from commercial and smallholder farms. Despite the aforementioned limitations, our study provides enhanced insights into the types of Salmonella serovars found in dust, soil, and animal feces from commercial and smallholder farms.
Conclusion
In conclusion, this study reports on diverse Salmonella serovars circulating in environmental samples (dust, soil and feces) from commercial and smallholder farms in Ghana. The data shows that ecological niches might present a transmission reservoir for antibiotic-resistant Salmonella. Hence, it is important to monitor such niches for surveillance purposes, especially commercial farms during the rainy seasons, to enable the implementation of control strategies. Last but not least, these findings warrant encouraging good husbandry practices, such as farms having concrete floors and periodic dust removal from environments.
Methods
Study site and sample collection
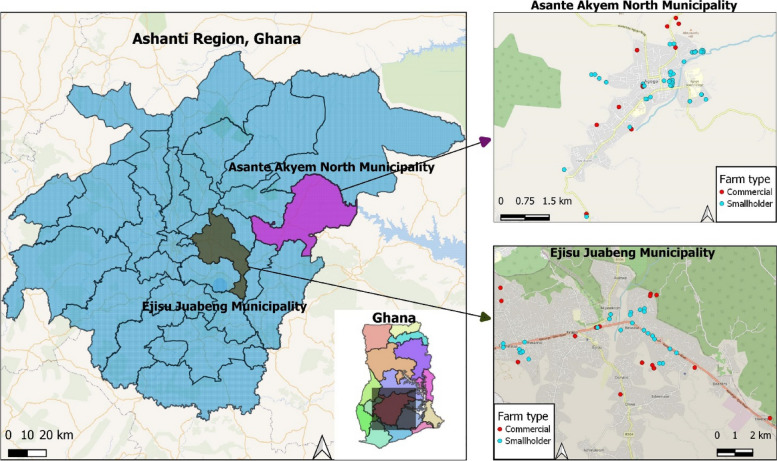
This study was carried out in the Asante Akyem North Municipality, a rural community and the Ejisu Juabeng Municipality, a semi-urban community located in the Ashanti region of Ghana (Fig. 2). Ghana has a tropical climate with two main seasons. The rainy season lasts from April to October, and the dry season lasts from November to March. Sampling was done between April 2019 and November 2020. Commercial and smallholder farms were selected using snowball sampling. Following the farmers' consent to the study, soil, dust and pooled fecal samples were collected from the farm environments. Soil samples around the animal pen and farm environments were collected from poultry and livestock (pigs, sheep, goats and cattle) farms.
Fig. 2.
Location of commercial and smallholder farms sampled in the Asante Akyem North Municipality and the Ejisu Juabeng Municipality of Ghana. (The authors created this map using QGIS software)
In contrast, dust and fecal samples were collected from poultry farms only. 1490 samples were collected from 30 commercial and 64 smallholder farms. Approximately 53.2% (n/N = 793/1490) of the samples were from commercial farms, and 46.8% (n/N = 697/1490) were from smallholder farms. For the environmental soil samples, 630 were collected from poultry farms, while 67, 67, 18, and 17 were collected from farms that kept pigs, sheep, goats, and cattle. Additionally, 409 pooled poultry fecal samples were collected, and 281 dust samples were obtained from poultry farms. A farm was considered commercial if it had at least 500 caged poultry and/or any quantity of caged livestock with an intensive housing system. In contrast, smallholder farms (small-scale agriculture) were households with free-roaming poultry (mainly indigenous breeds) and/or livestock with shelter provided by basic or temporary roofing. Farms with multiple pen houses were visited more than once; however, each was sampled only once.
Soil samples were collected from commercial and smallholder farms at 0-5 cm depth, with a core sampler measuring 5 cm in diameter and length. From each farm, two categories of soil samples were taken; soil from within ≤ 5 m around the pen structure and more than 5 m away from the pen structure but within the farm environment (including the working area, walkways, and part of the household where free-range animals roam and humans reside). Pooled fecal samples were collected from poultry farms using a pair of socks (strips mobs nurse cap, Hubei Zhiyue Non-woven products Co. Ltd) soaked in 0.9% normal saline. Socks were worn over a farm boot and used to take ten steps in a figure-of-eight-like pattern around the pen perimeter, as described by Andoh et al. [9]. Dust samples were collected from each poultry pen house using sterile socks moistened with 0.9% normal saline. The socks were used to clean the farms' fences, doors, and feeding/water troughs. All samples were placed in labelled sterile plastic containers and transported in a cool box (4–8 °C) to the Kumasi Center for Collaborative Research (KCCR) within 1–4 h for further processing.
Salmonella culture and identification
Fecal, dust and soil (5 g) samples were pre-enriched in buffered peptone water (Oxiod) and incubated at 35–37℃ for 18–24 h in a normal atmosphere. This was further enriched in selenite broth (Difco, BD) and then incubated at 35–37℃ for 18–24 h in a normal atmosphere. The enrichment broth was then cultured on Xylose Lysine Deoxycholate agar (XLD) (Difco, BD) and incubated at 35–37℃ for 18–24 h in a normal atmosphere. Suspected Salmonella colonies were presumptively identified using a Salmonella latex test (Oxoid) and the analytical profile index test (API 20E, bioMérieux, Marcy l’Etoile, France). All Salmonella were confirmed using the automated VITEK 2 System and serotyped following the White-Kaufmann Le Minor scheme [40] at the National Reference Centre for Salmonella and Other Bacterial Enteric Pathogens at the Robert Koch Institute (RKI), Germany.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using the Kirby Bauer disk diffusion method and interpreted following the European Committee on Antimicrobial Susceptibility Testing (EUCAST, version 12.0) guidelines. Salmonella Typhimurium ATCC 14028 was used as a reference strain for quality control. Confirmed Salmonella strains were tested against ampicillin, cefotaxime, cefoxitin, ceftazidime, chloramphenicol, ciprofloxacin, meropenem and trimethoprim-sulfamethoxazole. Isolates resistant to chloramphenicol, ampicillin, and trimethoprim-sulfamethoxazole were considered MDR [41]. Multiple antibiotic resistance was defined as resistance to three or more antimicrobials of different substance classes.
Statistical analysis
Categorical variables were described using absolute frequencies and their corresponding percentages. Association between two categorical variables were shown using prevalence ratios and their corresponding 95% confidence intervals (CI). Crude and adjusted prevalence ratios (PR) and their respective 95% CIs were calculated in bivariate and multivariate analyses, respectively, using Poisson regression to show possible factors associated with Salmonella frequency. Because of the exploratory nature of this study, p-values were not calculated. All analyses were conducted using R statistical software (version 4.1.1), and the epiR (2.0.19) package was applied to calculate PRs. The ggplot2 package was used to plot bar charts. QGIS software, version 3.24.0 [42], was used to draw a map showing the sampled farms' geographical location.
Acknowledgements
The authors acknowledge the field and technical support of Abdul Seidu Razak and Cynthia Adu Kyerewaa from the Kumasi Centre for Collaborative Research in Tropical Medicine (KCCR). We would also like to thank Susanne Kulbe, Marita Wahnfried, Bettina Fürholzner and the Robert Koch Institute for contributing to this project. We thank all the commercial and smallholder farmers for their time and cooperation.
Abbreviations
- AMR
Antimicrobial resistance
- EUCAST
European Committee on Antimicrobial Susceptibility Testing
- MDR
Multidrug-resistant
- API
Analytical profile index test
- KCCR
Kumasi Center for Collaborative Research
- XLD
Xylose Lysine Deoxycholate agar
Authors’ contributions
Linda Aurelia Ofori: LAO; Dennis Fosu: DF; Seth Ofori: SO; Charity Wiafe Akenten: CWA; Antje Flieger: AF; Sandra Simon: SS; Anna Jaeger: AJ;, Maike Lamshöft: ML; Juergen May: JM; Kwasi Obiri-Danso: KOD; Richard Phillips: RP; Daniel Haile Chercos: DHC; Ellis Kobina Paintsil: EKP; Denise Dekker: DD. Conceptualization: L.A.O., D.F. and D.D.; methodology: D.F., A.J. and M.L., C.W.A., S.O. S.S. and E.K.P.; validation: L.A.O., K.O.D. R.P., S.S., A.F. and D.D.; formal analysis: E.K.P., and D.H.C.; data curation: L.A.O., C.W.A., and S.O.; original draft preparation: D.F and E.K.P; writing, review and editing: L.A.O., D.F, E.K.P., K.O.D., C.W.A., R.P., A.J. M.L., S.O., A.F., S.S. and D.D.; supervision: L.A.O., D.D. and J.M.; funding acquisition: S.S., A.F., D.D. and J.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by "The German Research Foundation (DFG) within the project “Genetic Adaptation of non-typhoidal Salmonella within Human and Animal Reservoirs in sub-Saharan Africa” (DFG; grant number 649070). The funding body played no role in the study's design and collection, analysis, data interpretation, and manuscript writing.
Availability of data and materials
The data used to support the findings of this study are available from the corresponding author upon request.
Declarations
Ethics approval and consent to participants
The owner of each farm was informed of the study purpose, and oral permission was obtained before sampling.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Parisi A, Stanaway JD, Sarkar K, Crump JA. The global burden of non-typhoidal Salmonella invasive disease: a systematic analysis for the global burden of disease study 2017. Int J Infect Dis. 2020;101:341. doi: 10.1016/j.ijid.2020.09.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanner JR, Kingsley RA. Evolution of Salmonella within hosts. Trends Microbiol. 2018;26(12):986–998. doi: 10.1016/j.tim.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tack B, Vanaenrode J, Verbakel JY, Toelen J, Jacobs J. Invasive non-typhoidal Salmonella infections in sub-saharan Africa: a systematic review on antimicrobial resistance and treatment. BMC Med. 2020;18(1):212. doi: 10.1186/s12916-020-01652-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silva C, Calva E, Maloy S. One health and food-borne disease: salmonella transmission between humans, animals, and plants. Microbiol Spectr. 2014;2(1):2.1.08. doi: 10.1128/microbiolspec.oh-0020-2013. [DOI] [PubMed] [Google Scholar]
- 5.Ehrhardt K, Becker AL, Grassl GA. Determinants of persistent Salmonella infections. Curr Opin Immunol. 2023;82(102306):102306. doi: 10.1016/j.coi.2023.102306. [DOI] [PubMed] [Google Scholar]
- 6.Thong K-L. Characterization of drug resistant Salmonella enterica serotype typhimurium by antibiograms, plasmids, integrons, resistance genes and PFGE. J Microbiol Biotechnol. 2010;20(6):1042–1052. doi: 10.4014/jmb.0910.10028. [DOI] [PubMed] [Google Scholar]
- 7.Aldrich C, Hartman H, Feasey N, Chattaway MA, Dekker D, Al-Emran HM, Larkin L, McCormick J, Sarpong N, Le Hello S, Adu-Sarkodie Y, Panzner U, Park SE, Im J, Marks F, May J, Dallman TJ, Eibach D. Emergence of phylogenetically diverse and fluoroquinolone-resistant Salmonella Enteritidis as a cause of invasive nontyphoidal Salmonella disease in Ghana. PLoS Negl Trop Dis. 2019;13(6):e0007485. doi: 10.1371/journal.pntd.0007485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andoh LA, Ahmed S, Olsen JE, Obiri-Danso K, Newman MJ, Opintan JA, Dalsgaard A. Prevalence and characterization of Salmonella among humans in Ghana. Trop Med Health. 2017;45(1):1–11. doi: 10.1186/s41182-017-0043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andoh LA, Dalsgaard A, Obiri-Danso K, Newman MJ, Barco L, Olsen JE. Prevalence and antimicrobial resistance of Salmonella serovars isolated from poultry in Ghana. Epidemiol Infect. 2016;144(15):3288–3299. doi: 10.1017/s0950268816001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ekli R, Adzitey F, Huda N. Prevalence of resistant Salmonella spp. isolated from raw meat and liver of cattle in the Wa Municipality of Ghana. IOP Conf Ser Earth Environ Sci. 2019;287(1):012006. doi: 10.1088/1755-1315/287/1/012006. [DOI] [Google Scholar]
- 11.Dekker D, Krumkamp R, Eibach D, Sarpong N, Boahen KG, Frimpong M, Fechtner E, Poppert S, Hagen RM, Schwarz NG, Adu-Sarkodie Y, Owusu-Dabo E, Im J, Marks F, Frickmann H, May J. Characterization of Salmonella enterica from invasive bloodstream infections and water sources in rural Ghana. BMC Infect Dis. 2018;18(1):47. doi: 10.1186/s12879-018-2957-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunadu APH, Holmes M, Miller EL, Grant AJ. Microbiological quality and antimicrobial resistance characterization of Salmonella spp. in fresh milk value chains in Ghana. Int J Food Microbiol. 2018;277:41–49. doi: 10.1016/j.ijfoodmicro.2018.04.025. [DOI] [PubMed] [Google Scholar]
- 13.Adzitey F. Antibiotic resistance of Escherichia coli and Salmonella enterica isolated from cabbage and lettuce samples in Tamale metropolis of Ghana. Int J Food Contam. 2018;5(1). 10.1186/s40550-018-0068-z.
- 14.Ghana Ministry of Health, Ministry of Food and Agriculture, Ministry of Environment, Science, Technology and Innovation, Ministry of Fisheries and Aquaculture Development. Ghana national action plan for antimicrobial use and resistance. 2017–2021. Available online: http://www.moh.gov.gh/wpcontent/uploads/2018/04/NAP_FINAL_PDF_A4_19.3.2018-SIGNED-1.pdf. Accessed 22 May 2023.
- 15.Paintsil EK, Ofori LA, Akenten CW, Fosu D, Ofori S, Lamshöft M, May J, Danso KO, Krumkamp R, Dekker D. Antimicrobial usage in commercial and domestic poultry farming in two communities in the Ashanti region of Ghana. Antibiot (Basel Switzerland) 2021;10(7):800. doi: 10.3390/antibiotics10070800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schierstaedt J, Jechalke S, Nesme J, Neuhaus K, Sørensen SJ, Grosch R, Smalla K, Schikora A. Salmonella persistence in soil depends on reciprocal interactions with indigenous microorganisms. Environ Microbiol. 2020;22(7):2639–2652. doi: 10.1111/1462-2920.14972. [DOI] [PubMed] [Google Scholar]
- 17.Semenov AM, Kuprianov AA, Bruggen AHC, Semenov AM, Kuprianov AA. Transfer of enteric pathogens to successive habitats as part of microbial cycles linked references are available on JSTOR for this article. Transfer of enteric pathogens to successive habitats as part of microbial cycles’. 2022;60:239–49. 10.1007/s00248-0.
- 18.Čučak D, Babić O, Tamaš I, Simeunović J, Karaman M, Kovač D, Novaković M, Markov S, Knežević P, Stojanov I, Obradović V, Radnović D. Prevalence, antibiotic resistance and diversity of Salmonella isolates from soils and sediments in Serbia. Int J Environ Res. 2018;12(6):829–841. doi: 10.1007/s41742-018-0138-3. [DOI] [Google Scholar]
- 19.Gu G, Strawn LK, Zheng J, Reed EA, Rideout SL. Diversity and dynamics of Salmonella enterica in water sources, poultry litters, and field soils amended with poultry litter in a major agricultural area of Virginia. Front Microbiol. 2019;10:2868. doi: 10.3389/fmicb.2019.02868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parry-Hanson Kunadu A, Otwey RY, Mosi L. Microbiological quality and Salmonella prevalence, serovar distribution and antimicrobial resistance associated with informal raw chicken processing in Accra, Ghana. Food Control. 2020;118(107440):107440. doi: 10.1016/j.foodcont.2020.107440. [DOI] [Google Scholar]
- 21.Adzitey F, Teye GA, Amoako DG. Prevalence, phylogenomic insights, and phenotypic characterization of Salmonella enterica isolated from meats in the Tamale metropolis of Ghana. Food Sci Nutr. 2020;8(7):3647–3655. doi: 10.1002/fsn3.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dekker D, Eibach D, Boahen KG, Akenten CW, Pfeifer Y, Zautner AE, Mertens E, Krumkamp R, Jaeger A, Flieger A, Owusu-Dabo E, May J. Fluoroquinolone-resistant salmonella enterica, campylobacter spp., and arcobacter butzleri from local and imported poultry meat in Kumasi, Ghana. Foodborne Pathog Dis. 2019;16(5):352–358. doi: 10.1089/fpd.2018.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nyabundi D, Onkoba N, Kimathi R, Nyachieo A, Juma G, Kinyanjui P, Kamau J. Molecular characterization and antibiotic resistance profiles of Salmonella isolated from fecal matter of domestic animals and animal products in Nairobi. Trop Dis Travel Med Vaccines. 2017;3(1):2. doi: 10.1186/s40794-016-0045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agada G. Prevalence and antibiotic resistance profile of Salmonella isolates from commercial poultry and poultry farm-handlers in Jos, Plateau state, Nigeria. Br Microbiol Res J. 2014;4(4):462–479. doi: 10.9734/bmrj/2014/5872. [DOI] [Google Scholar]
- 25.Mitchell BW, Buhr RJ, Berrang ME, Bailey JS, Cox NA. Reducing airborne pathogens, dust and Salmonella transmission in experimental hatching cabinets using an electrostatic space charge system. Poult Sci. 2002;81(1):49–55. doi: 10.1093/ps/81.1.49. [DOI] [PubMed] [Google Scholar]
- 26.Pal A, Bailey MA, Talorico AA, Krehling JT, Macklin KS, Price SB, Buhr RJ, Bourassa DV. Impact of poultry litter Salmonella levels and moisture on transfer of Salmonella through associated in vitro generated dust. Poult Sci. 2021;100(8):101236. doi: 10.1016/j.psj.2021.101236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park SE, Pham DT, Pak GD, Panzner U, Espinoza LMC, von Kalckreuth V, Im J, Mogeni OD, Schütt-Gerowitt H, Crump JA, Breiman RF, Adu-Sarkodie Y, Owusu-Dabo E, Rakotozandrindrainy R, Bassiahi Soura A, Aseffa A, Gasmelseed N, Sooka A, Keddy KH, et al. The genomic epidemiology of multi-drug resistant invasive non-typhoidal Salmonella in selected sub-Saharan African countries. BMJ Global Health. 2021;6(8):e005659. doi: 10.1136/bmjgh-2021-005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson KMJ, Mor SM, Ward MP, Walsh MG. Divergent geography of Salmonella Wangata and Salmonella Typhimurium epidemiology in New South Wales, Australia. One Health (Amsterdam Netherlands) 2019;7(100092):100092. doi: 10.1016/j.onehlt.2019.100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson KMJ, Hill-Cawthorne GA, Ward MP, Mor SM. Diversity of Salmonella serotypes from humans, food, domestic animals and wildlife in New South Wales, Australia. BMC Infect Dis. 2018;18(1):623. doi: 10.1186/s12879-018-3563-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Somda NS, Bonkoungou IJO, Sambe-Ba B, Drabo MS, Wane AA, Sawadogo-Lingani H, Savadogo A. Diversity and antimicrobial drug resistance of non-typhoid Salmonella serotypes isolated in lettuce, irrigation water and clinical samples in Burkina Faso. J Agric Food Res. 2021;5(100167):100167. doi: 10.1016/j.jafr.2021.100167. [DOI] [Google Scholar]
- 31.Dekker D, Krumkamp R, Sarpong N, Frickmann H, Boahen K, Frimpong M, Asare R, Larbi R, Hagen R, Poppert S, Rabsch W, Marks F, Adu-Sarkodie Y, May J. Drinking water from dug wells in rural Ghana — Salmonella contamination, environmental factors, and genotypes. Int J Environ Res Public Health. 2015;12(4):3535–3546. doi: 10.3390/ijerph120403535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moffatt CRM, Lafferty AR, Khan S, Krsteski R, Valcanis M, Powling J, Veitch M. Salmonella Rubislaw gastroenteritis linked to a pet lizard. Med J Australia. 2010;193(1):54–55. doi: 10.5694/j.1326-5377.2010.tb03743.x. [DOI] [PubMed] [Google Scholar]
- 33.Ball TA, Monte DF, Aidara-Kane A, Matheu J, Ru H, Thakur S, Ejobi F, Fedorka-Cray PJ. International lineages of Salmonella enterica serovars isolated from chicken farms, Wakiso District, Uganda. In bioRxiv. 2019. 10.1101/707372. [DOI] [PMC free article] [PubMed]
- 34.Gebeyehu A, Taye M, Abebe R. Isolation, molecular detection and antimicrobial susceptibility profile of Salmonella from raw cow milk collected from dairy farms and households in southern Ethiopia. BMC Microbiol. 2022;22(1):84. doi: 10.1186/s12866-022-02504-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adesegun O, Adeyemi O, Ehioghae O, Rabor D, Binuyo T, Alafin B, Nnagha O, Idowu A, Osonuga A. Current trends in the epidemiology and management of enteric fever in Africa: a literature review. Asian Pac J Trop Med. 2020;13(5):204. doi: 10.4103/1995-7645.283515. [DOI] [Google Scholar]
- 36.Smith BA, Meadows S, Meyers R, Parmley EJ, Fazil A. Seasonality and zoonotic foodborne pathogens in Canada: relationships between climate and Campylobacter, E. coli and Salmonella in meat products. Epidemiol Infect. 2019;147(e190):e190. doi: 10.1017/S0950268819000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nyandjou YMC, Yakubu SE, Abdullahi IO, Machido DA. Multidrug resistance patterns and multiple antibiotic resistance index of Salmonella species isolated from waste dumps in Zaria Metropolis, Nigeria. J Appl Sci Environ Manage. 2019;23(1):41–46. [Google Scholar]
- 38.Crump JA, Medalla FM, Joyce KW, Krueger AL, Hoekstra RM, Whichard JM, Barzilay EJ. Antimicrobial resistance among invasive Nontyphoidal Salmonella enterica isolates in the United States: National Antimicrobial Resistance Monitoring System, 1996 to 2007. Antimicrob Agents Chemother. 2011;55(3):1148–1154. doi: 10.1128/aac.01333-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth N, Käsbohrer A, Mayrhofer S, Zitz U, Hofacre C, Domig KJ. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: a global overview. Poult Sci. 2019;98(4):1791–1804. doi: 10.3382/ps/pey539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grimont PA, Weill F-X. Antigenic Formulae of the Salmonella Serovars. 9th. Paris, France: WHO Collaborating Center for Reference and Research on Salmonella, Institut Pasteur. 2007. http://www.scacm.org/free/Antigenic%20Formulae%20of%20the%20Salmonella%20Serovars%202007%209th%20edition.pdf.
- 41.Rowe B, Ward LR, Threlfall EJ. Multidrug-resistant Salmonella typhi: a worldwide epidemic. Clin Infect Diseases: Official Publication Infect Dis Soc Am. 1997;24(Suppl 1):S106–109. doi: 10.1093/clinids/24.supplement_1.s106. [DOI] [PubMed] [Google Scholar]
- 42.QGIS Development Team . QGIS geographic information system. Zurich: Open Source Geospatial Foundation Project; 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.