Abstract
An efflux system, CmeABC, in Campylobacter jejuni was previously described, and a second efflux system, CmeDEF, has now been identified. The substrates of CmeDEF include ampicillin, ethidium bromide, acridine, sodium dodecyl sulfate (SDS), deoxycholate, triclosan, and cetrimide, but not ciprofloxacin or erythromycin. C. jejuni NCTC11168 and two efflux pump knockout strains, cmeB::Kanr and cmeF::Kanr, were exposed to 0.5 to 1 μg of ciprofloxacin/ml in agar plates. All mutants arising from NCTC11168 were resistant to ciprofloxacin but not to other agents and contained a mutation resulting in the replacement of threonine 86 with isoleucine in the quinolone resistance-determining region of GyrA. Mutants with two distinct phenotypes were selected from the efflux pump knockout strains. Mutants with the first phenotype were resistant to ciprofloxacin only and had the same substitution within GyrA as the NCTC11168-derived mutants. Irrespective of the parent strain, mutants with the second phenotype were resistant to ciprofloxacin, chloramphenicol, tetracycline, ethidium bromide, acridine orange, and SDS and had no mutation in gyrA. These mutants expressed levels of the efflux pump genes cmeB and cmeF and the major outer membrane protein gene porA similar to those expressed by the respective parent strains. No mutations were detected in cmeF or cmeB. Accumulation assays revealed that the mutants accumulated lower concentrations of drug. These data suggest the involvement of a non-CmeB or -CmeF efflux pump or reduced uptake conferring multiple-antibiotic resistance, which can be selected after exposure to a fluoroquinolone.
Over the past 5 years, Campylobacter spp. have been the most common cause of bacterial human gastroenteritis worldwide (1, 2). Infections are usually treated with fluid replenishment. However, in cases of persistent enteritis or extraintestinal infections, a fluoroquinolone or erythromycin is usually the antibiotic of choice (1, 2). In recent years, there has been a worldwide increase in the incidence of both human and veterinary isolates of Campylobacter resistant to ciprofloxacin (8, 10, 17, 22, 28). Data indicate that this resistance can be selected for by fluoroquinolones both in vitro and in vivo (26, 27). Fluoroquinolone resistance is usually due to substitutions in the quinolone resistance-determining region (QRDR) of GyrA (28). In Campylobacter spp., substitution of threonine 86 for isoleucine in GyrA is most frequently associated with resistance, although other substitutions have also been documented (18, 28). As elsewhere in the world, ciprofloxacin-resistant Campylobacter spp. have been isolated in England and Wales (17, 32). Studies in our laboratory have shown that the majority (∼90%) of such isolates possess a gyrA mutation (28). Between the years 2000 and 2001, ∼15% (9,000 cases) of the annual number of Campylobacter spp. reported in England and Wales were ciprofloxacin resistant. Our studies estimate that 15% (1,350) of these cases have multiple-antibiotic resistance (MAR) (unpublished data).
Fluoroquinolone resistance can also be conferred by efflux pumps, which limit the access of antibiotics to their targets by actively pumping out these molecules, thereby preventing the intracellular accumulation necessary for lethality (13). Since efflux pumps have broad substrate specificity, they confer MAR (13). MAR bacteria tend to be resistant to at least three classes of structurally unrelated antibiotics. Efflux pumps of the resistance-nodulation-division (RND) family appear to be one of the most widespread antibiotic resistance mechanisms in gram-negative bacteria (4, 24, 29). Efflux was first postulated as a mechanism of multiple-antibiotic resistance in Campylobacter spp. 8 years ago (5). In 2002, the efflux system CmeABC in Campylobacter jejuni was described for NCTC11168 (30), in which CmeA was the putative membrane fusion protein, CmeB was the putative efflux pump, and CmeC was the putative outer membrane channel protein. This was subsequently confirmed independently by Lin et al. with a poultry isolate (15). CmeB can confer fluoroquinolone resistance and multiple-antibiotic resistance, including antibiotics, detergents, dyes, disinfectants, and bile (15, 30). In other gram-negative bacteria, such as Escherichia coli, fluoroquinolone resistance due to efflux can occur synergistically with substitutions in GyrA (25). Recently, Lin et al. (16) demonstrated that CmeB and CmeC are essential for bile tolerance and colonization of chickens by C. jejuni.
A second efflux pump, CmeF, linked to a putative outer membrane protein, CmeD, by a putative membrane fusion protein, CmeE, is described in this report.
It has been found for Staphylococcus aureus that in the presence of an efflux pump inhibitor the number of norfloxacin-resistant mutants selected in vitro is reduced (19, 20). It was hypothesized that C. jejuni lacking an efflux pump(s) would give rise to fewer ciprofloxacin-resistant mutants than a wild-type strain. It was also hypothesized that mutants derived from a cmeB::Kanr strain would compensate for the loss of CmeB by overexpressing cmeF, and vice versa. Therefore, the aims of this study were (i) to characterize a second RND efflux pump, cmeF, in C. jejuni and (ii) to use ciprofloxacin to select for mutants from NCTC11168, cmeB::Kanr, and cmeF::Kanr strains to determine whether disruption of efflux pump genes affected the frequency of selecting ciprofloxacin-resistant mutants and to characterize any mutants selected.
MATERIALS AND METHODS
Bacterial strains and growth media.
C. jejuni NCTC11168 (26), cmeB::Kanr (30) and cmeF::Kanr strains, and a mutant of NCTC11168 overexpressing cmeB were used throughout this study (Table 1). All strains were routinely cultivated under microaerophilic conditions (5% O2, 7.5% CO2, 85% N2) on Mueller-Hinton (MH) agar plates supplemented with 5% sterile horse blood.
TABLE 1.
Strains used in this study
| Strain no. and group | Description | Source or reference |
|---|---|---|
| P270 | C. jejuni NCTC11168; amenable to genetic manipulation | 26 |
| P1031 | Ciprofloxacin-resistant mutant selected from P270 | This study |
| P1038 | cmeB::Kanr | 30 |
| P1035 | MAR mutant selected from P1038 | This study |
| P1040 | Ciprofloxacin-resistant mutant selected from P1038 | This study |
| P1043 | cmeF::Kanr | This study |
| P1036 | MAR mutant selected from P1043 | This study |
| P1045 | Ciprofloxacin-resistant mutant selected from P1043 | This study |
| P1048 | CmeB overexpressing mutant selected from P270 | This study |
Identification and characterization of CmeF.
The efflux pump CmeF was identified and characterized exactly as previously described for CmeB (3, 30). A 1-kb fragment was amplified within the coding region of cmeF using primers cmeF-a and cmeF-b (Table 2), and the PCR product was cloned into the pGEM-T Easy vector (Promega). A 100-bp deletion and a unique BglII site were created within the insert by inverse PCR (3, 12, 30) with primers cmeF-c and cmeF-d (Table 2). In order to disrupt the gene, a 1.4-kb BamHI restriction fragment containing a kanamycin cassette was then inserted within the BglII site of the cmeF insert. The construct was then used to transform the genome sequence of C. jejuni strain NCTC11168 and allowed to replace the gene on the chromosome by homologous recombination (3, 12, 30).
TABLE 2.
Oligonucleotide primers used in this study
| Gene and primer | Nucleotide sequence | Source or reference |
|---|---|---|
| 16S rRNA gene | ||
| cj16S1 | 5′-GAGAGGCAGATGGAATTG-3′ | This study |
| cj16S2 | 5′-CCTACCTCCTATACCTGG-3′ | This study |
| cmeB | This study | |
| cmeB-1 | 5′-CCTACCTCCTATACCTGG-3′ | This study |
| cmeB-2 | 5′-TTGAACTTGTGCCGCTGG-3′ | |
| cmeF | ||
| cmeF-1 | 5′-GCACTAATGCACGCATTG-3′ | This study |
| cmeF-2 | 5′-TGCACCGCTCCTAAGGAA-3′ | This study |
| cmeF-a | 5′-TTATCCTAGCCACTCTTG-3′ | This study |
| cmeF-b | 5′-CCTATAGAAGCTCCGCCA-3′ | This study |
| cmeF-d | 5′-CGC(AGATCT)ATGCGTGCATTAGTGCCT-3′ | This study |
| cmeF-c | 5′-CGC(AGATCT)TTAGCGTAGGAAGCATGG-3′ | This study |
| gyrA | ||
| cjgyrA-1 | 5′-GCCTGACGCAAGAGATGGTT-3′ | 27 |
| cjgyrA-2 | 5′-TCAGTATAACGCATCGCAGC-3′ | 27 |
| gyrB | ||
| cjgyrB-1 | 5′-ATGGCAGCTAGAGGAAGAGA-3′ | 27 |
| cjgyrB-2 | 5′-GTGATCCATCAACATCCGCA-3′ | 27 |
| porA | ||
| porA-1 | 5′-TAATGCTGCTGATGGTGG-3′ | This study |
| porA-2 | 5′-AAGCACCTTCAAGTGTCC-3′ | This study |
tBLASTN and SWISS-PROT analysis of CmeB and CmeF.
Amino acid sequence homology of CmeB and CmeF were determined using the tBLASTN algorithm. Their putative tertiary structures were compared to the AcrB crystal structure (21) using Swiss-Model software (http://www.expasy.ch).
Selection of ciprofloxacin-resistant and MAR laboratory mutants.
Mutants with decreased susceptibility to ciprofloxacin were selected from NCTC11168, cmeB::Kanr, and cmeF::Kanr strains using the method described by Griggs et al. (11). Briefly, the three parent strains were grown overnight in antibiotic-free MH broth, concentrated by centrifugation, and resuspended in sterile MH broth to give a range of inocula (105 to 109 CFU/ml). Replicate (n = 5) agar plates containing ciprofloxacin at two and four times the MICs for the respective parent strains were inoculated with 100 μl of each suspension and incubated for 2 to 5 days under microaerophilic conditions. Ten colonies resembling the morphology of the original strain were randomly chosen from each selecting plate and subcultured onto antibiotic-free media. These were examined for susceptibility and resistance mechanisms.
A cmeB-overexpressing mutant was selected by serial passage (increasing the antibiotic concentration with each passage) of NCTC11168 on replicate (n = 8) MH agar plates containing cefotaxime at 0.5, 2, and 4 times the MIC. Colonies from the antibiotic-containing plates were emulsified into saline and plated onto fresh antibiotic-free agar plates. The viable-cell count was determined by counting the number of colonies on the antibiotic-free plates. The mutation frequency was determined by dividing the number of colonies on the antibiotic-containing plates by the viable-cell count.
Growth kinetics.
Cultures (50 ml) were inoculated with 2 ml of an overnight broth and grown under microaerophilic conditions over a period of 24 h in MH broth, and 1-ml aliquots were withdrawn at 2-h intervals. Cell growth was measured spectrophotometrically by determining the optical density at 600 nm (OD600).
Determination of the MIC.
The MICs of the antibiotics ampicillin, chloramphenicol, ciprofloxacin, erythromycin, kanamycin, and tetracycline; the dyes acridine orange and ethidium bromide; the detergents sodium dodecyl sulfate (SDS) and sodium deoxycholate; and the disinfectants cetrimide and triclosan were determined by the agar doubling dilution method as recommended by the NCCLS (23).
Accumulation of ciprofloxacin and ethidium bromide.
The concentration of ciprofloxacin accumulated was measured as previously described (30). For ethidium bromide, cells were cultured to mid-log phase (OD600, 0.4 to 0.5), harvested, and washed in sodium phosphate buffer (pH 7.0) to an OD600 of 0.2 (10 ml). After the cells were preincubated for 15 min at 37°C, a 2-ml time zero aliquot was withdrawn, and ethidium bromide was added to a final concentration of 2 μg/ml. Further aliquots were withdrawn at 5, 10, 15, and 20 min after the addition of ethidium bromide. The aliquots were stored on ice until the fluorescence was determined. Fluorescence was measured at an excitation of 530 nm and an emission of 600 nm. Accumulation experiments were performed with or without the protonophore carbonyl cyanide-m-chlorophenyl hydrazone (CCCP) added to a final concentration of 150 μM after withdrawal of the 5-min aliquot. All experiments were performed in duplicate on at least three separate occasions. The data were analyzed by Student's t test, and a P value of ≤0.05 was considered significant.
Sequence analysis of the QRDR of gyrA or gyrB.
Mutation detection by denaturing high-performance liquid chromatography and DNA sequencing were performed as described previously (7). The primers used are listed in Table 2.
Expression of cmeB, cmeF, and porA.
Expression of cmeB, cmeF, and porA (which encodes the major outer membrane protein [MOMP]) was measured by comparative reverse transcription-PCR exactly as described previously (6, 31). The primers used are listed in Table 2. porA expression was confirmed by SDS-polyacrylamide gel electrophoresis (9).
RESULTS
Identification and characterization of CmeF.
A second efflux system, corresponding to regions Cj1031, -1032, and -1033 in the C. jejuni NCTC11168 genome sequence, was identified by homology to E. coli AcrB, AcrD, and AcrF and C. jejuniCmeB. The inner membrane efflux pump was designated CmeF (Cj1033), CmeD (Cj1031) the outer membrane protein, and CmeE (Cj1032) the membrane fusion protein. Like CmeB, CmeF was large (3,000 bp) and had a predicted 12-transmembrane (TM) helical domain structure (30). CmeF had a moderate amino acid similarity to the E. coli RND efflux pump proteins AcrB, AcrD, and AcrF (26.9, 27.1, and 28.7% identity, respectively). CmeB had higher homology with the E. coli proteins (42.2, 43.6, and 42.2%, respectively) (30). CmeF showed only 25% amino acid identity with CmeB. P1043 (cmeF::Kanr) was more susceptible to ampicillin, ethidium bromide, acridine, SDS, deoxycholate, triclosan, and cetrimide (Table 3). In contrast, P1038 (cmeB::Kanr) was also more susceptible to ciprofloxacin, erythromycin, and tetracycline (Table 3) (30). Interestingly, and as found for P1038 (cmeB::Kanr), P1043 (cmeF::Kanr) was also more susceptible to bile (Table 3). Although the two efflux pumps are RND-type pumps and are predicted to have similar basic tripartite structures, these data suggest that they have different substrate-binding sites and conductance properties.
TABLE 3.
Phenotypes of selected mutants
| Strain | Phenotypea | MIC (μg/ml)b
|
Accumulation
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amp | Cip | Chl | Ery | Tet | EtBr | AO | SDS | Deox | Bile | Cet | Tri | Cipc | EtBrd | ||
| P270 | WT | 4 | 0.25 | 1 | 0.25 | 0.25 | 4 | 16 | 8 | 16 | 64 | 8 | 4 | 15.9 ± 1 | 14.6 ± 1 |
| P1031 | Cipr | 2 | 32 | 1 | 0.25 | 0.25 | 4 | 16 | 8 | 16 | 64 | 8 | 4 | NDe | ND |
| P1048 | CmeB+ | 32 | 8 | 16 | 4 | 8 | 32 | 64 | 64 | 64 | 128 | 64 | 32 | 8.3 ± 2 | 7.4 |
| P1038 | CmeB− | 1 | 0.06 | 0.5 | 0.06 | 0.06 | 2 | 8 | 2 | 4 | 16 | 2 | 1 | 24.4 ± 2 | 21.7 ± 2 |
| P1040 | Cipr | 1 | 16 | 1 | 0.06 | 0.06 | 2 | 8 | 2 | 4 | 16 | 2 | 1 | ND | ND |
| P1035 | MAR | 1 | 4 | 4 | 0.06 | 0.06 | 8 | 32 | 8 | 4 | 16 | 2 | 1 | 19.8 ± 2 | 17.98 ± 2 |
| P1043 | CmeF− | 0.5 | 0.25 | 1 | 0.25 | 0.25 | 1 | 4 | 4 | 8 | 32 | 2 | 1 | 17 ± 1 | 23.2 ± 1 |
| P1045 | Cipr | 0.5 | 32 | 2 | 0.25 | 0.25 | 1 | 4 | 4 | 8 | 32 | 2 | 1 | ND | ND |
| P1036 | MAR | 0.5 | 4 | 4 | 0.25 | 0.25 | 8 | 32 | 8 | 8 | 32 | 2 | 1 | 13.2 ± 2 | 19.1 ± 1 |
WT, wild type.
Amp, ampicillin; Cip, ciprofloxacin; Chl, chloramphenicol; Ery, erythromycin; Kan, kanamycin; Tet, tetracycline; EtBr, ethidium bromide; AO, acridine orange; Deox, sodium deoxycholate; Cet, cetrimide; Tri, triclosan.
Micrograms of ciprofloxacin per nanogram of dry cells after 5 min of exposure (± standard deviation).
Fluorescence units of ethidium bromide after 5 min of exposure (± standard deviation).
ND, not determined.
Protein structure of CmeF.
TBLASTN comparisons revealed that the amino acid sequence of CmeB was >40% identical to that of AcrB in both the TM and non-TM domain regions (21). However, CmeF was <30% identical to AcrB and CmeB throughout the TM and non-TM regions (data not shown). Swiss-Model analysis also revealed that the CmeB tertiary structure was similar to that of AcrB, whereas the structure of CmeF could not be modeled due to its low structural similarity to AcrB.
Antibiotic susceptibility of ciprofloxacin-resistant mutants and mutations in the QRDR of gyrA or gyrB.
Mutants were selected on agar containing ciprofloxacin from cmeB::Kanr (P1038) and cmeF::Kanr (P1043) strains as readily as from NCTC11168 (P270), with similar mutation frequencies of 2.2 × 10−9, 1.7 × 10−9, and 2.5 × 10−9, respectively. Growth curves showed that P1038 (cmeB::Kanr), P1043 (cmeF::Kanr), and P1048 and the mutants were able to grow as well as P270 (wild type) under normal laboratory conditions (data not shown).
All mutants (n = 8) selected from NCTC11168 (P270) had an ∼100-fold decrease in susceptibility to ciprofloxacin. These mutants also had a mutation resulting in the replacement of threonine at position 86 with isoleucine in GyrA (e.g., P1031). Two mutant phenotypes were selected from each strain (P1038 [cmeB::Kanr] and P1043 [cmeF::Kanr]). The first phenotype, illustrated by strains P1040 and P1045, was similar to that of the NCTC11168-derived mutants and had an ∼100-fold decrease in susceptibility to ciprofloxacin, a Thr86-Ile GyrA substitution, and no mutations in gyrB. The second mutant phenotype, illustrated by P1035 and P1036, had an ∼16- to 30-fold decrease in susceptibility to ciprofloxacin and decreased susceptibility to chloramphenicol, ethidium bromide, acridine orange, and SDS. Susceptibilities to erythromycin, tetracycline, deoxycholate, and bile were unchanged. There were no mutations in gyrA or gyrB. The cefotaxime-selected cmeB-overexpressing mutant, P1048, was highly resistant to multiple agents and very resistant to bile (Table 3).
On three separate occasions, disruption of cmeF in P1035 and disruption of cmeB in P1036 were attempted by using a chloramphenicol resistance cassette (33). No double mutants were obtained. However, disruption of cmeF in NCTC11168 with this cassette in parallel gave rise to cmeF::Chlorr mutants (data not shown). DNA sequencing of cmeF in P1035 and cmeB in P1036 revealed no mutations in either entire structural gene.
Accumulation of ciprofloxacin and ethidium bromide.
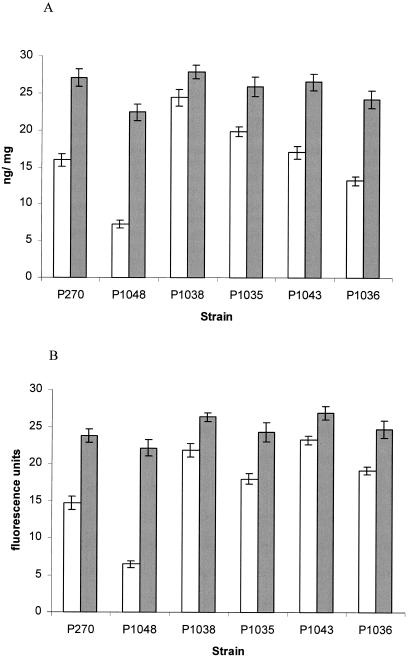
Accumulation of ciprofloxacin by P1043 (cmeF::Kanr) was no different than that of the parent strain (P270) (Table 3). However, accumulation of ethidium bromide increased 1.6-fold compared to that of the wild type (P270) (P = 0.0048) (Fig. 1B). CCCP increased the accumulation of ethidium bromide by P1043 by 16% and that of P270 by 48% (data not shown). Compared with the wild-type parent strain (P270), the concentration of ciprofloxacin and ethidium bromide accumulated was also reduced in P1048 (48 and 50%, respectively; P = 0.0016 and 0.0017, respectively) (Table 3). CCCP increased the accumulation of ciprofloxacin and ethidium bromide by P1048 to 79.7 and 82.0%, respectively, of that of the wild type (data not shown).
FIG. 1.
Accumulation of ciprofloxacin (A) and ethidium bromide (B) without (open bars) or with (shaded bars) CCCP by P270 (NCTC11168), P1048 (cmeB++ [overexpressed]), P1038 (cmeB::Kanr), P1035 (cmeB::Kanr MAR mutant), P1043 (cmeF::Kanr), and P1036 (cmeF::Kanr MAR mutant). The values are from five independent experiments expressed as means ± standard deviations and were obtained at the 15-min time point.
Accumulation assays of the MAR mutants derived from the insertionally inactivated mutants revealed that they accumulated less ciprofloxacin and ethidium bromide than their respective parent strains. P1035 accumulated 19% less ciprofloxacin and 11% less ethidium bromide than its parent strain, P1038 (P = 0.0043) (Table 3). CCCP increased this concentration in P1035 by 7.1 and 7.9%, respectively (data not shown). However, P1035 still accumulated higher concentrations of ciprofloxacin and ethidium bromide than NCTC11168 (P270) (Table 3). Similarly, P1036 accumulated 23% less ciprofloxacin and 18% less ethidium bromide than P1043 (P = 0.0049 and 0.0042). CCCP increased this concentration by 5 and 8%, respectively (Fig. 1). P1036 accumulated 17% less ciprofloxacin than NCTC11168, whereas it accumulated 24% more ethidium bromide.
Expression of cmeB, cmeF, and porA.
There was no significant difference between the expression levels of cmeB by P1043 and P1036. Similarly, cmeF expression by P1038 and P1036 was not significantly different from that of P270 or the ciprofloxacin-resistant mutants (P1038, P1040, and P1045). None of the strains showed any difference in the level of expression of porA (data not shown). Only the cefotaxime-selected MAR mutant, P1048, overexpressed cmeB (2.4-fold).
DISCUSSION
A second RND-type efflux system, CmeDEF, in C. jejuni is described in this study. This system has different substrate-binding properties and is thus functionally distinct from the previously described CmeABC. However, these systems have some overlap in their substrate profiles.
More important is the observation that lack of CmeF also increased sensitivity to bile. This phenomenon was previously observed with two different strains lacking CmeB (15, 30), and recently it has been shown that CmeB is essential for colonization of chickens (16). As no up-regulation of cmeB or cmeF was detected in the cmeF::Kanr or cmeB::Kanr strain, respectively, it is intriguing that C. jejuni should have two proteins involved in bile resistance but that lack of either protein confers susceptibility. These data suggest that the two pumps have low binding specificity for bile.
The hypothesis that ciprofloxacin-resistant mutants would be selected less frequently from the knockout mutants was not supported, as there was no significant difference among the three strains in the frequency of mutation to resistance. In addition, the hypothesis that any selected mutants would compensate for the lack of CmeB by overexpressing cmeF, and vice versa, was also not supported. The latter data provide further evidence that cmeB and cmeF are expressed independently.
Ciprofloxacin selected for mutants with the threonine 86-to-isoleucine substitution in the QRDR of GyrA of NCTC11168 and the cmeB::Kanr and cmeF::Kanr strains. However, ciprofloxacin also selected for MAR mutants derived from P1038 (cmeB::Kanr) and P1043 (cmeF::Kanr), whose substrate profiles were different from that of the cefotaxime-selected MAR cmeB-overexpressing mutant (P1048). Compared with P1048, the ciprofloxacin-selected MAR mutants, P1035 and P1036, had more limited substrate profiles. Data from accumulation experiments suggest that these mutants either take up less of these agents or have enhanced efflux. However, this appears not to be mediated by CmeB, CmeF, or PorA/MOMP, the major outer membrane protein (porin) (14). In addition, resistance to bile was not restored in the MAR mutants.
Despite repeated attempts, deletion of cmeF in P1035, or of cmeB in P1036, was unsuccessful, suggesting that one RND pump is essential for viability. Although double mutants could not be selected, DNA sequencing of cmeF and cmeB in each mutant revealed no mutations and ruled out substrate specificity alterations or substrate efficiency changes in CmeB or CmeF as a cause of multiple-antibiotic resistance. Ciprofloxacin remains the first- or second-line antibiotic for treatment of Campylobacter sp. infections. Therefore, the use of fluoroquinolones may select for resistance not only to these agents, but to other antibiotics. These data have shown that ciprofloxacin can select for resistance to fluoroquinolones. However, this study suggests that fluoroquinolones can also select for MAR mutants. Other work in our laboratory has shown that MAR in C. jejuni cannot be fully accounted for by the known mechanisms (31); therefore, it is possible that multiple-antibiotic resistance may be mediated by another efflux pump. As the C. jejuni genome contains at least eight other non-RND efflux pumps that could be involved in the transport of antibiotics, further investigation of these is now warranted.
Acknowledgments
We are grateful to the Department of Environment, Food and Rural Affairs (formerly The Ministry for Agriculture, Fisheries and Food) for project grant CTA9903.
L.J.V.P. is a recipient of the Bristol Myers Squibb Nonrestricted Grant in Infectious Diseases.
REFERENCES
- 1.Aarestrup, F. M., and J. Engberg. 2001. Antimicrobial resistance of thermophilic Campylobacter. Vet. Res. 32:311-321. [DOI] [PubMed] [Google Scholar]
- 2.Allos, B. M. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32:1201-1206. [DOI] [PubMed] [Google Scholar]
- 3.Arnoud, H., M. Van Vliet, A. C. Wood, J. Henderson, K. Wooldridge, and J. M. Ketley. 1998. Genetic manipulation of enteric Campylobacter species. Methods Microbiol. 27:407-421. [Google Scholar]
- 4.Borges-Wamsley, M. I., and A. R. Walmsley. 2001. The structure and function of drug pumps. Trends Microbiol. 9:71-79. [DOI] [PubMed] [Google Scholar]
- 5.Charvalos, E., Y. Tselentis, M. M Hamzehpour, T. Kohler, and J. C. Pechere. 1995. Evidence for an efflux pump in multidrug-resistant Campylobacter jejuni. Antimicrob. Agents Chemother. 39:2019-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eaves, D. J. V., V. Ricci, and L. J. V. Piddock. 2004. Expression of acrB, acrF, acrB, marA and soxS in Salmonella enterica serovar Typhimirium: role in multiple antibiotic resistance. Antimicrob. Agents Chemother. 48:1145-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaves, D. J. V., E. Liebana, M. J. Woodward, and L. J. V. Piddock. 2002. Detection of gyrA mutations in quinolone-resistant Salmonella enterica by denaturing high-performance liquid chromatography. J. Clin. Microbiol. 40:4121-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enberg, J., F. M. Aarestrup, D. E. Taylor, P. Gerner-Smidt, and I. Nachamkin. 2001. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7:24-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett, M., Y. F. Jin, V. Ricci, and L. J. V. Piddock. 1996. Contribution of individual mechanisms to fluoroquinolone resistance in Escherichia coli strains isolated from humans and animals. Antimicrob. Agents Chemother. 40:2380-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaunt, P. N., and L. J. V. Piddock. 1996. Ciprofloxacin resistant Campylobacter spp. in humans: an epidemiological and laboratory study. J. Antimicrob. Chemother. 37:47-57. [DOI] [PubMed] [Google Scholar]
- 11.Griggs, D. J., H. Marona, and L. J. V. Piddock. 2003. Selection of moxifloxacin-resistant Staphylococcus aureus compared with five other fluoroquinolones. J. Antimicrob. Chemother. 51:1403-1407. [DOI] [PubMed] [Google Scholar]
- 12.Jagannathan, A., C. Constantinidou, and C. Penn. 2001. Roles of rpoN, fliA, and flgR in expression of flagella in Campylobacter jejuni. J. Bacteriol. 183:2937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohler, T., J. C. Pechere, and P. Plesiat. 1999. Bacterial antibiotic efflux systems of medical importance. Cell. Mol. Life Sci. 56:771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Labesse, G., E. S. Garnotel, S. Bonnel, G. Pumas, J. M. Pages, and J. M. Bola. 2001. MOMP, a divergent porin from Campylobacter: cloning and primary structural characterization. Biochem. Biophys. Res. Commun. 280:380-387. [DOI] [PubMed] [Google Scholar]
- 15.Lin, J., L. O. Michel, and Q. Zhang. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46:2124-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, J., O. Sahin, L. O. Michel, and Q. Zhang. 2003. Critical role of the multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect. Immun. 8:4250-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livermore, D. M., D. James, M. Reacher, C. Graham, T. Nichols, P. Stephens, A. P. Johnson, and R. C. George. 2002. Trends in fluoroquinolone (ciprofloxacin) resistance in Enterobacteriaceae from bacteremias, England and Wales, 1990-1999. Emerg. Infect. Dis. 8:473-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo, N., O. Shahin, J. Lin, L. O. Michel, and Q. Zhang. 2003. In vivo selection of Campylobacter isolates with high-level fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob. Agents Chemother. 47:390-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markham, P., E. Westhause, K. Klyachko, M. E. Johnson, and A. A. Neyfakh. 1999. Multiple novel inhibitors of the NorA multidrug transporter of Staphylococcus aureus. Antimicrob. Agents Chemother. 43:2404-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markham, P. N., and A. A. Neyfakh. 1996. Inhibition of the multidrug transporter NorA prevents emergence of norfloxacin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 406:2673-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 22.Nachamkin, I., H. Ung, and M. Li. 2003. Increasing fluoroquinolone resistance in Campylobacter jejuni, Pennsylvania, USA, 1982-2001. Emerg. Infect. Dis. 8:1501-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.NCCLS. 2002. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 24.Nikaido, H., and H. I. Zgurskaya. 2001. AcrAB and related multidrug efflux pumps in Escherichia coli. J. Mol. Microbiol. Biotechnol. 3:215-218. [PubMed] [Google Scholar]
- 25.Oethinger, M., W. V. Kern, A. S. Jellen-Ritter, L. M. McMurry, and S. B. Levy. 2000. Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the AcrAB efflux pump. Antimicrob. Agents Chemother. 44:10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karleyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. Van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 27.Payot, S., A Cloeckaert, and E. Chalus-Dancla. 2002. Selection and characterization of fluoroquinolone resistant mutants of Campylobacter jejuni using enrofloxacin. Microb. Drug Resist. 84:335-343. [DOI] [PubMed] [Google Scholar]
- 28.Piddock, L. J. V., V. Ricci, L. Pumbwe, M. J. Everett, and D. J. G. Griggs. 2003. Fluoroquinolone resistance in Campylobacter species from man and animals: detection of mutations in topoisomerase genes. J. Antimicrob. Chemother. 51:19-26. [DOI] [PubMed] [Google Scholar]
- 29.Poole, K. 2001. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 3:255-264. [PubMed] [Google Scholar]
- 30.Pumbwe, L., and L. J. V. Piddock. 2002. Identification and molecular characterisation of CmeB, a Campylobacter jejuni multidrug efflux pump. FEMS Microbiol. Lett. 206:185-189. [DOI] [PubMed] [Google Scholar]
- 31.Pumbwe, L., L. P. Randall, M. J. Woodward, and L. J. V. Piddock. 2004. Expression of the efflux pump genes cmeB, cmeF and the porin gene porA in multiple antibiotic resistant isolates of Campylobacter jejuni. J. Antimicrob. Chemother. 54:341-347. [DOI] [PubMed] [Google Scholar]
- 32.Thwaites, R. T., and J. A. Frost. 1999. Drug resistant Campylobacter jejuni, C. coli and C. lari isolated from humans in North West England and Wales in 1997. J. Clin. Pathol. 52:812-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression and cloning vector construction. Gene 94:23-28. [DOI] [PubMed] [Google Scholar]