Abstract
Mycobacterium tuberculosis RNA polymerase is 1,000-fold more sensitive to rifampin than Escherichia coli RNA polymerase. Chimeric E. coli RNA polymerase in which the β-subunit segment encompassing rifampin regions I and II (amino acids [aa] 463 through 590) was replaced with the corresponding region from M. tuberculosis (aa 382 through 509) did not show an increased sensitivity to the antibiotic. Thus, the difference in amino acid sequence between the rifampin regions I and II of the two species does not account for the difference in rifampin sensitivity of the two polymerases.
Rifampin (RIF) is a broad-spectrum antibiotic that is used in the therapy of many infectious diseases, in particular, tuberculosis. The cellular target of RIF is RNA polymerase (RNAP) (7). RIF blocks the initiation of transcription by preventing the synthesis of RNAs larger than dinucleotides. At the same time, RIF has no effect on the formation of the first phosphodiester bond and does not inhibit RNA elongation. These facts led to the proposal that RIF sterically blocks the path of the nascent RNA during initiation (5, 9, 15, 25).
An analysis of the three-dimensional structure of Thermus aquaticus RNAP in a complex with RIF revealed that the antibiotic binds near the RNAP active site at a protein pocket formed by the β subunit (2). RIF overlaps with the position of the third RNA nucleotide in the elongation complex (11). These data strongly supported the initial hypothesis on the steric mechanism of RIF action.
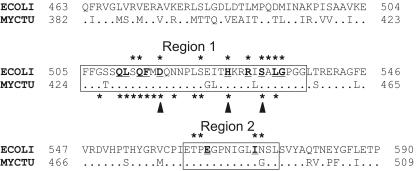
All known mutations leading to RIF resistance in bacteria (Rifr mutations) map in four regions in the RNAP β subunit. In total, 23 positions of the β subunit have been implicated in RIF resistance in different bacteria (Fig. 1) (2, 3, 8, 10, 12-14, 17, 18, 20, 22, 23). The majority of the mutations are found in the β-subunit regions I and II (positions 505 to 537 and 562 to 572 in Escherichia coli numbering); in addition, there are two mutations at the 687 (region III) and 146 positions of the β subunit. Most mutations found in clinical isolates of Mycobacterium tuberculosis are localized in three positions of the β-subunit region I (Fig. 1), while no mutations in regions II and III were described (18, 21, 24).
FIG. 1.
Alignment of the β-subunit RIF regions of E. coli (ECOLI) and M. tuberculosis (MYCTU) RNAPs. Dots in the sequence of M. tuberculosis represent amino acids that are identical to those of E. coli. Numbers on both sides of the sequences indicate the positions of amino acids starting from the N terminus of the protein. The first and the second RIF regions are boxed. Positions of the Rifr mutations are marked with asterisks; mutations in E. coli and M. tuberculosis RNAPs are shown above and below the alignment, respectively. Amino acids that directly interact with RIF in the structure of T. aquaticus RNAP (2) are bold and underlined. Three positions of the first RIF region which most frequently mutate in M. tuberculosis are indicated by arrowheads under the sequence.
All amino acids changed by Rifr mutations are spatially grouped around the RIF binding pocket (2, 26). Twelve amino acid residues of RNAP are involved in direct interactions with RIF, and substitutions of 11 of them were shown to lead to RIF resistance (Fig. 1). Twelve additional amino acids changed by Rifr mutations surround the RIF pocket but do not make direct contact with the antibiotic. The effect of these substitutions is likely to come from local changes of the structure of the RIF pocket weakening the binding of the antibiotic.
RNAPs of various bacteria possess different levels of RIF sensitivity. M. tuberculosis RNAP was shown to be about 1,000-fold more sensitive to RIF than the RNAP from E. coli. It also formed much more stable complexes with the antibiotic (6). The formation of stable RIF-RNAP complexes probably explains the bactericidal activity of RIF and may be one of the factors explaining the efficacy of RIF in the tuberculosis therapy. It was reasonable to suppose that the different RIF sensitivities of E. coli and M. tuberculosis RNAPs result from the differences in the β-subunit regions involved in the binding of RIF. To test this hypothesis, we constructed two chimeric E. coli RNAPs in which either the RIF region I alone (amino acids [aa] 505 through 537) or a larger β-subunit segment encompassing RIF regions I and II and a portion of flanking sequences (aa 463 through 590) were replaced with the corresponding sequences from M. tuberculosis (Mtu[I] RNAP and Mtu[I-II] RNAP, respectively) (Fig. 1).
The mutant rpoB genes encoding the chimeric β subunits were generated by standard PCR mutagenesis methods. To produce the Mtu[I] gene, recognition sites of two restriction endonucleases, EcoRI and SmaI, were introduced by two-stage PCRs at both sides of the RIF region I (codons 504 and 539) in the plasmid containing E. coli rpoB (pMKSe2) (22). In each case, the first PCR was done with a reverse mutagenic primer containing the sequence of the restriction site (5′-CGAAGAATTCTTTCACTGCTGC and 5′-GCACGTTCCCGGGTCAGACC for EcoRI and SmaI sites [underlined], respectively) and direct primer 5′-GCTGGCTAAGCTGAGCC beginning at codon 321 of the rpoB gene. This product was used as a megaprimer in the second PCRs, with reverse primer 5′-GGAGAGCGCAGCTTCACCC beginning at codon 863. The resulting PCR product was treated with ClaI and BspEI and cloned into the same sites of the E. coli rpoB gene (located at codons 433 and 846). Then, the M. tuberculosis rpoB segment comprising RIF region I (codons 424 to 556) was amplified from the I376 cosmid with primers containing EcoRI and SmaI sites (5′-CAAGGAATTCTTCGGCACCAGCCAGC and 5′-CGCTCCCGGGTCAGACCGCCTGGCCCCAGC for the EcoRI site and the SmaI site [underlined], respectively) and cloned into the E. coli plasmid.
The Mtu[I-II] rpoB gene was generated in essentially the same way. In the first stage, the recognition sites of MfeI and MluI restriction endonucleases were introduced by two-stage PCRs at codons 463 and 590 of the rpoB gene in the pMKSe2 plasmid by using reverse mutagenic primers 5′-GGCCAACGCGCAATTGGTTTTCCGC and 5′-CACTTTACGATACGCGTTCTCAGGG (with the MfeI site and the MluI site, respectively, underlined). Then, the corresponding M. tuberculosis rpoB segment (codons 382 through 509) was amplified with primers 5′-GCTGATCCAAAACCAATTGCGCGTCGGC and 5′-CACCTTGCGGTACGCGTTTTCGATG (with the MfeI site and the MluI site, respectively, underlined) and cloned into the MfeI and MluI sites of the E. coli rpoB gene.
The RNA polymerases containing the chimeric β subunits were reconstituted in vitro from individual subunits (1). Two control RNAPs were reconstituted in parallel. The first one was wild-type RIF-sensitive (Rifs) RNAP, and the second contained a β subunit with a point Rifr mutation at position 531 (S531F). The transcription activities and RIF sensitivities of RNAPs were studied with an in vitro transcription test using a DNA template containing a T7 A1 promoter followed by a λ tR2 terminator (19). The transcription of this template results in the synthesis of two major products corresponding to a full-length 130-nucleotide RNA transcribed to the end of the DNA fragment and a shorter 106-nucleotide product terminated at the tR2 sequence. The reaction mixture contained reconstituted core RNAP, σ subunit, a promoter DNA fragment (50 nM), and a mixture of nucleotides (25 μM concentrations of ATP, CTP, and GTP and 5 μM of UTP with the addition of [α-32P]UTP). RIF was added 3 min prior to the addition of the nucleotides. Transcription was performed for 10 min at 37°C. The measured specific activities of the Rifr, Mtu[I], and Mtu[I-II] RNAPs relative to the Rifs enzyme were 105, 75, and 45%. Most probably, the lower activity of the Mtu[I-II] enzyme is explained by some defects in RNAP reconstitution caused by the mutation. At the same time, the mutation did not have a dramatic effect on the interactions of RNAP with promoters, elongation, and termination of RNA synthesis (Fig. 2 and data not shown).
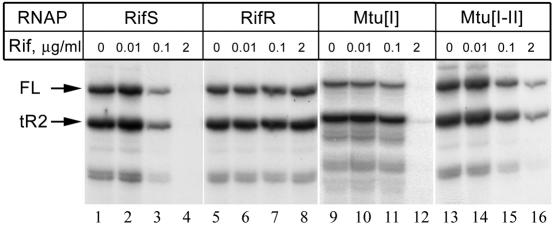
FIG. 2.
The effect of RIF on the activity of RNAPs. The concentrations of RNAP in the reaction were 10 nM in the cases of the Rifs (RifS), Rifr (RifR), and Mtu[I] RNAPs and 20 nM in the case of the Mtu[I-II] enzyme. Full-length and terminated RNA transcripts are marked FL and tR2, respectively.
In agreement with published data, the activity of the wild-type Rifs RNAP was completely inhibited at 2 μg of RIF/ml, while the activity of the control Rifr RNAP was not affected at this concentration of antibiotic (Fig. 2). Contrary to expectations, neither chimeric RNAP possessed an increased RIF sensitivity in comparison with the E. coli Rifs RNAP. Moreover, the Mtu[I-II] chimera happened to be even more RIF resistant than the wild-type E. coli Rifs enzyme (Fig. 2, compare lanes 4 and 16).
The sequences forming the RIF pocket are highly conserved among bacteria; the first and the second RIF regions of M. tuberculosis differ from the E. coli sequence at only five positions (Fig. 1). In addition, the substituted segment of the E. coli β subunit (aa 463 through 590) differs from the sequence of M. tuberculosis at 4 positions between the two RIF regions and at 27 positions at both sides of the segment (these parts of the protein are weakly conserved and do not contain any known Rifr mutations) (Fig. 1). Our results demonstrate that all these differences do not account for the different RIF sensitivities of E. coli and M. tuberculosis RNAPs. At the same time, the changes in the nonconserved regions surrounding the RIF pocket are apparently responsible for the increased RIF resistance of the Mtu[I-II] chimeric RNAP.
It should be noted that none of the five amino acids that differ in the RIF regions of the two RNAPs makes direct contacts with the antibiotic in the complex of RIF with T. aquaticus RNAP (2). Only one of these amino acids (position 573 in E. coli) was shown to be replaced by the Rifr mutation (Fig. 1). The low conservation of these positions and the absence of Rifr mutations support the idea that these amino acids are not involved in RIF binding. In contrast, all 12 amino acids that directly interact with RIF (including positions in M. tuberculosis which are most frequently changed by Rifr mutations) are identical in E. coli and M. tuberculosis RNAPs (Fig. 1). Thus, the increased RIF sensitivity of M. tuberculosis RNAP must be explained by the differences in other RNAP regions which may indirectly result in structural changes of the RIF pocket increasing the affinity of RNAP to the antibiotic.
Previously, RNAP from thermophilic T. aquaticus strains was shown to be about 100-fold less sensitive to RIF than E. coli RNAP (2, 4). As in the case with the polymerase from M. tuberculosis, all 12 amino acids of T. aquaticus RNAP that directly interact with RIF are identical to those in the E. coli enzyme. This fact led to the proposal that the increased RIF resistance of T. aquaticus RNAP also results from changes in the protein regions outside of the RIF binding pocket (16). These data clearly illustrate that the sensitivities of enzymes (and in particular, RNAP) to antibiotics are determined not only by the protein regions that are directly involved in antibiotic binding but also by other parts of the protein that can indirectly affect the structure of the binding site. Most probably, the structure of the RIF pocket may be affected by the nearby protein regions, in particular, the N-terminal region of the β subunit, where a single Rifr mutation (position 146 in E. coli) has been found (8, 13). M. tuberculosis RNAP differs from the E. coli enzyme at the adjacent position 145 (where Ile is substituted for Val). This substitution may somehow improve RIF binding to M. tuberculosis RNAP. Another possibility is that the different RIF sensitivities of the E. coli and M. tuberculosis enzymes are explained by multiple amino acid changes dispersed over several regions of RNAP. The identification of these regions is an important subject for further studies.
It should be noted that no mutations in the rifampin regions of the β subunit have been found in 5 to 10% of RIF-resistant clinical isolates of M. tuberculosis. It was proposed that in these cases, Rifr mutations may be present in other regions of the β subunit or in other subunits of M. tuberculosis RNAP (8, 18, 24). Our results indicate that these mutations may be localized in the RNAP regions responsible for its high affinity to RIF. Thus, new molecular mechanisms underlying the RIF resistance in M. tuberculosis and other bacteria may be discovered in the future. These considerations must be taken into account when developing new strategies for antituberculosis therapy.
Acknowledgments
The work was supported by Russian Foundation for Basic Research grant 02-04-48525 and by National Institutes of Health grant GM30717 to A. Goldfarb.
We thank N. Kurepina for the I376 cosmid containing the M. tuberculosis H37Rv rpoB gene and S. Borukhov for help in protein preparation.
REFERENCES
- 1.Borukhov, S., and A. Goldfarb. 1993. Recombinant Escherichia coli RNA polymerase: purification of individually overexpressed subunits and in vitro assembly. Protein Expr. Purif. 4:503-511. [DOI] [PubMed] [Google Scholar]
- 2.Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901-912. [DOI] [PubMed] [Google Scholar]
- 3.Cole, S. T. 1996. Rifamycin resistance in mycobacteria. Res. Microbiol. 147:48-52. [DOI] [PubMed] [Google Scholar]
- 4.Fabry, M., J. Sumegi, and P. Venetianer. 1976. Purification and properties of the RNA polymerase of an extremely thermophilic bacterium: Thermus aquaticus T2. Biochim. Biophys. Acta 435:228-235. [DOI] [PubMed] [Google Scholar]
- 5.Handschin, J. C., and W. Wehrli. 1976. On the kinetics of the rifampicin-RNA-polymerase complex. Differences between crude and purified enzyme fractions. Eur. J. Biochem. 66:309-317. [DOI] [PubMed] [Google Scholar]
- 6.Harshey, R. M., and T. Ramakrishnan. 1976. Purification and properties of DNA-dependent RNA polymerase from Mycobacterium tuberculosis H37RV. Biochim. Biophys. Acta 432:49-59. [DOI] [PubMed] [Google Scholar]
- 7.Hartmann, G., K. O. Honikel, F. Knusel, and J. Nuesch. 1967. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim. Biophys. Acta 145:843-844. [DOI] [PubMed] [Google Scholar]
- 8.Heep, M., B. Brandstatter, U. Rieger, N. Lehn, E. Richter, S. Rusch-Gerdes, and S. Niemann. 2001. Frequency of rpoB mutations inside and outside the cluster I region in rifampin-resistant clinical Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 39:107-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinkle, D. C., W. F. Mangel, and M. J. Chamberlin. 1972. Studies of the binding of Escherichia coli RNA polymerase to DNA. IV. The effect of rifampicin on binding and on RNA chain initiation. J. Mol. Biol. 70:209-220. [DOI] [PubMed] [Google Scholar]
- 10.Jin, D. J., and C. A. Gross. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J. Mol. Biol. 202:45-58. [DOI] [PubMed] [Google Scholar]
- 11.Korzheva, N., A. Mustaev, M. Kozlov, A. Malhotra, V. Nikiforov, A. Goldfarb, and S. A. Darst. 2000. A structural model of transcription elongation. Science 289:619-625. [DOI] [PubMed] [Google Scholar]
- 12.Lisitsyn, N. A., S. O. Gur'ev, E. D. Sverdlov, E. P. Moiseeva, and V. G. Nikiforov. 1984. Nucleotide substitutions in the rpoB gene leading to rifampicin resistance of E. coli RNA polymerase. Bioorg. Khim. 10:127-128. (In Russian.) [PubMed] [Google Scholar]
- 13.Lisitsyn, N. A., E. D. Sverdlov, E. P. Moiseyeva, O. N. Danilevskaya, and V. G. Nikiforov. 1984. Mutation to rifampicin resistance at the beginning of the RNA polymerase beta subunit gene in Escherichia coli. Mol. Gen. Genet. 196:173-174. [DOI] [PubMed] [Google Scholar]
- 14.Mariam, D. H., Y. Mengistu, S. E. Hoffner, and D. I. Andersson. 2004. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 48:1289-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClure, W. R., and C. L. Cech. 1978. On the mechanism of rifampicin inhibition of RNA synthesis. J. Biol. Chem. 253:8949-8956. [PubMed] [Google Scholar]
- 16.Minakhin, L., S. Nechaev, E. A. Campbell, and K. Severinov. 2001. Recombinant Thermus aquaticus RNA polymerase, a new tool for structure-based analysis of transcription. J. Bacteriol. 183:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morlock, G. P., B. B. Plikaytis, and J. T. Crawford. 2000. Characterization of spontaneous, in vitro-selected, rifampin-resistant mutants of Mycobacterium tuberculosis strain H37Rv. Antimicrob. Agents Chemother. 44:3298-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musser, J. M. 1995. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev. 8:496-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nudler, E., M. Kashlev, V. Nikiforov, and A. Goldfarb. 1995. Coupling between transcription termination and RNA polymerase inchworming. Cell 81:351-357. [DOI] [PubMed] [Google Scholar]
- 20.Ovchinnikov, Y. A., G. S. Monastyrskaya, S. O. Guriev, N. F. Kalinina, E. D. Sverdlov, A. I. Gragerov, I. A. Bass, I. F. Kiver, E. P. Moiseyeva, V. N. Igumnov, S. Z. Mindlin, V. G. Nikiforov, and R. B. Khesin. 1983. RNA polymerase rifampicin resistance mutations in Escherichia coli: sequence changes and dominance. Mol. Gen. Genet. 190:344-348. [DOI] [PubMed] [Google Scholar]
- 21.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuberc. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 22.Severinov, K., M. Soushko, A. Goldfarb, and V. Nikiforov. 1993. Rifampicin region revisited. New rifampicin-resistant and streptolydigin-resistant mutants in the beta subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 268:14820-14825. [PubMed] [Google Scholar]
- 23.Severinov, K., M. Soushko, A. Goldfarb, and V. Nikiforov. 1994. RifR mutations in the beginning of the Escherichia coli rpoB gene. Mol. Gen. Genet. 244:120-126. [DOI] [PubMed] [Google Scholar]
- 24.Williams, D. L., C. Waguespack, K. Eisenach, J. T. Crawford, F. Portaels, M. Salfinger, C. M. Nolan, C. Abe, V. Sticht-Groh, and T. P. Gillis. 1994. Characterization of rifampin-resistance in pathogenic mycobacteria. Antimicrob. Agents Chemother. 38:2380-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yarbrough, L. R., F. Y. Wu, and C. W. Wu. 1976. Molecular mechanism of the rifampicin-RNA polymerase interaction. Biochemistry 15:2669-2676. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, G., E. A. Campbell, L. Minakhin, C. Richter, K. Severinov, and S. A. Darst. 1999. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell 98:811-824. [DOI] [PubMed] [Google Scholar]