Abstract
A recombinant vaccinia virus, expressing the NS3-to-NS5 region of the N clone of hepatitis C virus (HCV), was generated and utilized both in a gel-based assay and in an enzyme-linked immunosorbent assay (ELISA) to evaluate the pyrrolidine-5,5-trans-lactams, a series of inhibitors of the HCV NS3/4A protease. The absolute levels of processed, mature HCV nonstructural proteins in this system were found to decrease in the presence of the trans-lactams. Monitoring of this reduction enabled end points and 50% inhibitory concentrations to be calculated in order to rank the active compounds according to potency. These compounds had no effect on the transcription or translation of the NS3-5 polyprotein at concentrations shown to inhibit NS3/4A protease, and they were shown to be specific inhibitors of this protease. The ELISA, originally developed using the vaccinia virus expression system, was modified to utilize Huh-7 cells containing an HCV replicon. Results with this assay correlated well with those obtained with the recombinant vaccinia virus assays. These results demonstrate the utility of these assays for the characterization of NS3/4A protease inhibitors. In addition, inhibitors of other viral targets, such as polymerase and helicase, can be evaluated in the context of the replicon ELISA.
It is estimated that at least 170 million people worldwide have been infected by the hepatitis C virus (HCV). Most of these individuals will have a chronic infection that may eventually lead to fibrosis, cirrhosis, and liver failure or hepatocellular carcinoma (21). This severe, end-stage liver disease is currently the major indication for liver transplantation in developed countries, and consequently, HCV infection is rapidly becoming a leading health issue. The current treatment of choice for chronic hepatitis C is a combination of pegylated alpha interferon and ribavirin. The outcome of therapy, however, depends upon several factors, including the viral load and the HCV genotype. Individuals with genotype 1, the most prevalent genotype in Europe, the United States, and Japan, are generally more refractory to current treatment than individuals infected with genotype 2 or 3. Fewer than 50% of patients with genotype 1 demonstrate a sustained virological response to this therapy. The development of more-effective anti-HCV therapies is, therefore, a priority.
Although non-A, non-B hepatitis was known for many years, HCV itself was first identified in 1989 (11) and, since then, has been classified as a member of the Hepacivirus genus within the family Flaviviridae (30). This family also includes the flaviviruses, such as yellow fever virus and dengue virus, and the animal pestiviruses, such as bovine viral diarrhea virus. HCV is a small, enveloped, single-stranded RNA virus with a genome of ∼9,600 nucleotides which consists of a 5′ nontranslated region, a single large open reading frame (ORF), and a short 3′ nontranslated region. The ORF encodes a single large polyprotein of ∼3,010 amino acids which is processed, co- and posttranslationally, by host signal peptidases and virally encoded proteases into 10 proteins. The structural proteins consist of the core protein, the envelope glycoproteins, E1 and E2, and possibly p7. The nonstructural (NS) proteins comprise the NS2/3 protease, NS3 serine protease/helicase, NS4A cofactor for protease and helicase, NS4B, NS5A, and the RNA-dependent RNA polymerase, NS5B. There is possibly an 11th protein arising from a frameshift event in the core protein region (9). The molecular virology and biochemical properties of these proteins have been extensively reviewed (13, 26).
The downstream cleavage events in the nonstructural region are carried out by the serine protease, which is located in the N-terminal third of the NS3 protein along with the NS4A cofactor (12, 34). The NS3/4A protease is a trypsin-like serine protease which has been shown to be essential for replication of the virus (18) and is consequently a valid target for therapeutic intervention (25).
Unlike most other members of the Flaviviridae, HCV cannot be efficiently cultured in vitro. Consequently, the evaluation of potential antiviral compounds in a biologically relevant context is extremely difficult. Cell-based assays complement enzymatic assays, because they can determine whether potential inhibitors are able to penetrate the cell and act in an appropriate cellular environment. A number of these systems with the potential to identify inhibitors of NS3/4A protease have been described (19). Some include the use of chimeras of either Sindbis virus or poliovirus containing the HCV NS3 protease, in which the production of infectious virus is dependent on the activity of the NS3 protease (10, 17). Other systems utilize reporter genes such as secreted alkaline phosphatase, the secretion of which is dependent on cleavage by NS3 (22, 29). Recent reviews have described several classes of inhibitors of HCV NS3/4A protease and their development (14, 25, 26, 28). For example, RNA aptamers which bind specifically to NS3/4A and inhibit protease activity have been generated (16). Other classes include substrate-based inhibitors, such as boronic acid, phosphonates, α-ketoamides, hydrazinoureas, and product-based peptidic inhibitors, and non-substrate based inhibitors, such as thiazolidines, benzanilides, phenanthrenequinone, and nitrobenzamide derivatives. BILN 2061, a protease inhibitor designed by using a substrate-based approach, has recently been reported to be effective in reducing HCV loads in patients after 2 days (20).
Here we describe the development of cell-based assay systems which extend and complement enzyme-based assays for the evaluation of potential antiviral compounds, specifically the pyrrolidine-5,5-trans-lactams (referred to below as trans-lactams), which are inhibitors of NS3/4A protease (1, 2, 3, 4, 5, 31, 32). The gel-based assay, utilizing a recombinant vaccinia virus expressing NS3 to NS5 (rVV NS3-5), has the advantage of allowing direct visualization of the inhibition of NS3/4A processing. This was developed further into an enzyme-linked immunosorbent assay (ELISA), and when the replicon system (6, 23) subsequently became available, we developed a replicon ELISA. Perhaps the greatest advantage of the replicon ELISA is that it can be used to assess the effects of inhibition of protease in the most relevant cell-based system to date. Moreover, it can be used to screen potential inhibitors of other viral targets, including polymerase and helicase, as well as replication complex inhibitors.
MATERIALS AND METHODS
Cell culture.
The African green monkey kidney epithelial cell line BT7H, human TK−143 (HTK−) cells, and the human hepatoma replicon cell line Huh-7 (clone 5-15) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, nonessential amino acids, 2 mM glutamine, 100 IU of penicillin/ml, and 100 μg of streptomycin/ml. BT7H and Huh-7 clone 5-15 cells were also cultured in the presence of 500 μg of G418 (Geneticin; Life Technologies)/ml, while the HTK− cells had 20 μg of 5-bromo-2′-deoxyuridine/ml in the maintenance medium. BT7H cells are derived from the BSC-1 cell line and constitutively express bacteriophage T7 RNA polymerase. The Huh-7 clone 5-15 was obtained from R. Bartenschlager (University of Heidelberg, Heidelberg, Germany) and is a stable cell line which expresses an HCV subgenomic replicon (23). All cells were maintained in a 5% CO2 incubator at 37°C.
Recombinant vaccinia virus.
The vaccinia virus transfer vector pTM3, kindly supplied by B. Moss (27), was used to generate a recombinant vaccinia virus, rVV NS3-5, expressing a polyprotein comprising the nonstructural proteins NS3 to NS5B. A BsrGI-ClaI fragment from the HCV N (type 1b) clone (8) was first subcloned into a modified pGEM vector, and the remainder of the NS3 protein was cloned in by PCR. The resulting EcoRI-BamHI fragment was then transferred into pTM3, and the resulting vector was designated pTM-NS3-5. This transfer vector contains a T7 RNA polymerase promoter upstream of the encephalomyocarditis virus internal ribosome entry site (IRES), followed by the NS3-5 ORF. Recombinant vaccinia virus was generated by homologous recombination into the thymidine kinase gene of wild-type vaccinia virus DNA. Recombinant virus was selected on HTK− cells grown in a medium containing 30 μg of hypoxanthine/ml, 500 μg of xanthine/ml, and 5 μg of mycophenolic acid/ml (15). Stocks of recombinant vaccinia virus were grown and amplified on HTK− cell monolayers, and titers of infectious virus were determined by a plaque assay on HTK− cells.
Gel-based assay.
BT7H cells were seeded into 6-well plates and grown for 18 h under 5% CO2 at 37°C until confluent. Cells were infected with rVV NS3-5 at a multiplicity of infection of 0.5 to 1.0 particle-forming units per cell for 1 h at 37°C. The virus was replaced with medium with or without the compound dissolved in dimethyl sulfoxide (final concentration, 0.25%). Plates were incubated for a further 18 h under 5% CO2 at 37°C. Metabolic labeling was carried out as follows. Cells were washed with phosphate-buffered saline (PBS) and then starved in a methionine-free medium for 1 h. The medium was removed, and cells were incubated for a further 2 h in a methionine-free medium supplemented with 150 kBq of [35S]methionine (NEN-Dupont)/well. Where compounds were added, they were included at all stages of the procedure. Cells were washed once with PBS, harvested in ice-cold PBS, and pelleted in a centrifuge. Cell pellets were resuspended in polyacrylamide gel electrophoresis (PAGE) loading buffer, boiled for 10 min, and analyzed by sodium dodecyl sulfate (SDS)-PAGE and autoradiography.
Western blot analysis.
Cells were harvested and resuspended in PBS and PAGE loading buffer. Samples were run on SDS-10% polyacrylamide gels, and the proteins were transferred to nitrocellulose membranes. Membranes were blocked in 10% (wt/vol) skim milk powder-0.05% Tween 20 in PBS and then probed with an antibody in the same buffer. After incubation with a horseradish peroxidase (HRP)-conjugated anti-species antibody, development was carried out using 3,3′-diaminobenzidine tetrahydrochloride (Sigma Fast DAB) or enhanced chemiluminescence (Amersham Pharmacia Biotech).
Immunoprecipitation of nonstructural proteins.
rVV NS3-5-infected cells were labeled with [35S]methionine as for the gel-based assay. Cells were harvested and boiled in disruption buffer (2% SDS, 2% mercaptoethanol, 10% glycerol, 125 mM Tris HCl [pH 6.8]). Samples were immunoprecipitated for 18 h at 4°C with an antibody in radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS in PBS). Immune complexes were bound to stapylococcal protein A (Pansorbin; Calbiochem) and washed three times in RIPA buffer. Samples were analyzed by SDS-PAGE and autoradiography.
Northern blot analysis.
Cytoplasmic RNA was extracted from rVV NS3-5-infected BT7H cells by using the QIAGEN RNeasy kit as instructed by the manufacturer. A 40-mer oligonucleotide probe derived from the NS5B region of the N clone (5′-TCCTCAACACGGATGTCATTCTCAGTGACCGTTGAGTCAA-3′) was labeled by using the BrightStar psoralen-biotin kit (Ambion). Cytoplasmic RNA samples were transferred to nylon membranes (Hybond-N+; Amersham Pharmacia Biotech) and hybridized to the biotinylated probe for 18 h at 42°C. Membranes were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS, followed by 0.1× SSC-0.1% SDS at 42°C. Detection was carried out using the BrightStar BioDetect kit (Ambion).
ELISA.
Preformed, confluent BT7H cells in 96-well plates were infected for 1 h with rVV NS3-5 at a multiplicity of infection of ∼0.1 to 0.5. The virus was removed, and doubling dilutions of a compound in DMEM, ranging from 100 to 0.8 μM, were added at a final volume of 200 μl per well in 0.25% dimethyl sulfoxide. Plates were incubated for 18 h under 5% CO2 at 37°C. Cells were washed twice in PBS and then fixed and permeabilized in a 1:1 mixture of methanol and acetone. Cells were washed once more in PBS, blocked for 1 h in 2% (wt/vol) skim milk-0.05%(vol/vol) Tween 20 in PBS, and then washed once with PBS-0.05% (vol/vol) Tween 20 (PBST). Cells were probed for 2 h with a murine monoclonal antibody against NS protein (anti-C100 [Biogenesis, Poole, United Kingdom] or anti-NS4A and anti-NS5A [Virostat, Portland, Maine]) and then washed three times in PBST. Cells were incubated with an HRP-conjugated secondary antibody for a further 1 h and were then washed five times with PBST. Development was carried out using ο-phenylenediamine dihydrochloride-urea hydrogen peroxide (Sigma Fast OPD) and was terminated with 20% sulfuric acid, and plates were read spectrophotometrically at 490 nm.
The replicon ELISA utilized Huh-7 clone 5-15 cells at a concentration of 4 × 104 per well, added simultaneously with the compound. Plates were incubated for 3 days at 37°C under 5% CO2; thereafter, the cells were fixed and processed as for the rVV NS3-5 ELISA.
Selection of the primary antibody for ELISA.
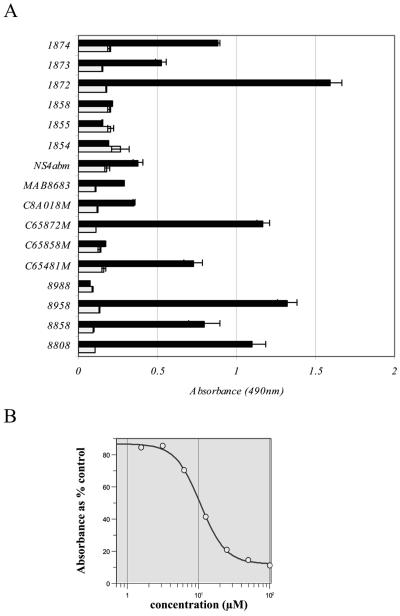
A number of commercially available murine monoclonal immunoglobulin G antibodies, raised against HCV NS proteins, were assayed at 4 μg/ml for binding to the proteins expressed by rVV NS3-5. The signal was compared to that from an uninfected BT7H cell control to determine suitability for ELISA. Antibodies C65481M (anti-NS5), C65858M (anti-NS4A), C65872M (anti-NS4) and C8A018M (anti-NS4) were obtained from Biodesign, Oxford, United Kingdom; 8808 (anti-NS5), 8858 (anti-NS5), 8958 (anti-C100), and 8988 (anti-NS4) were from Biogenesis; MAB8683 (anti-NS4) was from Chemicon, Harrow, United Kingdom; and 1854, 1855, 1858, 1872 (all anti-NS4A), 1873 (anti-NS5), and 1874 (anti-NS5A) were from Virostat.
Vero cytotoxicity assay.
Vero (African green monkey kidney) cells were seeded into 96-well plates at a density of 4,000 per well in 75 μl of growth medium consisting of DMEM supplemented with 5% fetal calf serum and 1% penicillin and streptomycin. Cells were allowed to settle and adhere for 1 h at 37°C. Drug dilutions were made in growth medium ranging from 500 to 31 μM, and 75 μl was added to duplicate wells. Plates were incubated for 4 days under 5% CO2 at 37°C. Twenty microliters of a 5-mg/ml solution of MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide] in PBS was then added to each well. After a 2-h incubation at 37°C, the supernatants were aspirated from the wells and replaced with 125 μl of acidified isopropanol containing 0.5% SDS per well. The plates were maintained on a shaking incubator for 20 min and were then read spectrophotometrically at 570 nm. The mean absorbance values of replicate test wells were expressed as percentages of the mean cell control well values and were then plotted against drug concentration to allow the calculation of the CC50 (the concentration that is cytotoxic to 50% of the cell population).
Biotinylated-probe analysis to determine the potency of trans-lactams against NS3/4A in vaccinia virus-infected cells.
rVV NS3-5 infected whole-cell extracts or replicon cell extracts were preincubated with different concentrations of compound 1q (Fig. 1) for 1 h prior to the addition of 5 μM biotinylated trans-lactam probe (compound 6) for 45 min. Samples were run under denaturing conditions on a 10% NuPAGE gel (Invitrogen) and were blotted onto nitrocellulose membranes. Blots were probed with HRP-neutravidin (Bio-Rad) and visualized by using Super Signal Femto reagent (Pierce) prior to imaging on a Proexpress imager (Perkin-Elmer Biosystems).
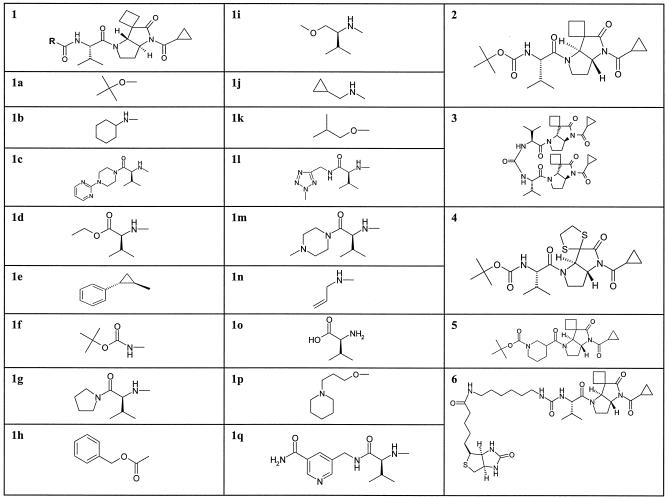
FIG. 1.
Structures of pyrrolidine-5,5-trans-lactam compounds. Structure 1 is the parent for compounds 1a to 1q. The position of the R group in structure 1 is indicated on the left. R groups only are shown for compounds 1a to 1q. The complete structures of compounds 2 to 6 are given separately.
Structures of pyrrolidine-5,5-trans-lactam compounds.
The structures of the compounds used in these studies are shown in Fig. 1.
RESULTS
Gel-based assay.
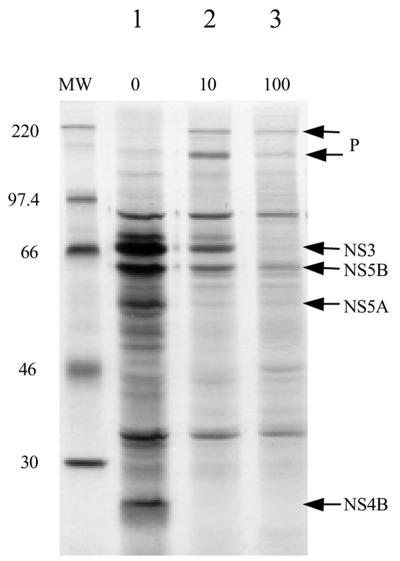
A recombinant vaccinia virus, rVV NS3-5, expressing the NS proteins of HCV (NS3 through NS5B) under the control of the encephalomyocarditis virus IRES, was generated from the genotype 1b N clone (8). Infection of BT7H cells with this virus resulted in rapid inhibition of host cell protein synthesis, and the majority of proteins observed after metabolic labeling were HCV derived (Fig. 2, lane 1). Proteins were identified by their relative molecular weight and reaction with antibodies. When a trans-lactam (compound 1a), active against the HCV NS3/4A protease, was added to the system, there was a disappearance of fully processed proteins and a simultaneous appearance of high-molecular-weight proteins. There is not a stoichiometric relationship between the loss of mature proteins and the appearance of high-molecular-weight proteins, because the small amount of detectable high-molecular-weight protein does not proportionally account for the large decrease in the amount of mature proteins. This is probably due to increased degradation by the cell, since the NS3-5 polyprotein, with a nonfunctional NS3/4A protease, is found only at very low levels (V. Chung, unpublished data). These high-molecular-weight proteins were not seen when the trans-lactam was added to the same cells infected with wild-type vaccinia virus (data not shown). Western blot analysis and immunoprecipitation with specific anti-HCV antibodies against NS3 and NS5B confirmed that these high-molecular-weight proteins were derived from the recombinant HCV NS3-5 polyprotein and represent uncleaved or partially cleaved polyprotein (data not shown).
FIG. 2.
Metabolic labeling of vaccinia virus-infected BT7H cells in the presence and absence of compound 1a. BT7H cells were infected with rVV NS3-5. Compound 1a was added at 10 or 100 μM throughout the assay, and cells were metabolically labeled with [35S]methionine at 19 to 21 h postinfection (see Materials and Methods). Cell lysates were analyzed by SDS-PAGE and autoradiography. Fully processed nonstructural proteins are indicated by arrows. P, precursor proteins. Due to the low molecular weight of NS4A, it is not visible on these gels. MW, molecular weight marker (in thousands).
Specificity.
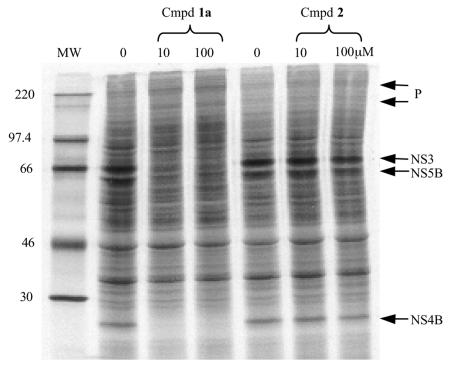
The specificity of this inhibition was investigated by using enantiomers, compounds 1a and 2, one of which had biochemical activity against NS3/4A protease while the other was inactive. The active enantiomer, compound 1a, inhibited NS3/4A-dependent processing, while the inactive enantiomer, compound 2, showed no effect at 100 μM (Fig. 3). In addition, a trans-lactam compound active against another protease, elastase (24), was shown to be inactive in this assay (data not shown).
FIG. 3.
Enantiomers, compounds 1a and 2, exhibit different activities in the gel-based assay. Compound (Cmpd) 1a was active at 10 μM, as demonstrated by the loss of fully processed nonstructural proteins NS3, NS4B, and NS5B, while its enantiomer, compound 2, was inactive at 100 μM. P, precursor proteins. MW, molecular weight marker (in thousands).
Effects of trans-lactams on transcription and translation.
Since the high-molecular-weight polyproteins in this system were less abundant than the processed products, we looked at possible effects on transcription and translation. To investigate the effect on transcription, the potency of a pair of enantiomers, compounds 1a and 2, was examined. Cells were infected with rVV NS3-5 in the presence or absence of the active or inactive enantiomer. These compounds showed no effect on the transcription by the T7 polymerase of mRNA coding for the NS3-5 polyprotein. This was exemplified by measuring RNA levels by Northern hybridization using a probe to the NS5B region of the genome (Fig. 4). Another recombinant vaccinia virus expressing the structural proteins of HCV under the control of the same promoter and with the same IRES background was used to infect cells in the presence of an active trans-lactam compound in order to ensure that there was no general effect on translation of recombinant proteins in this system. There was no effect on the expression of HCV core protein at drug concentrations that inhibited NS3/4A protease (data not shown).
FIG. 4.
Effects of trans-lactams on transcription. Cytoplasmic RNA was extracted from BT7H cells that either were left uninfected (U) or were infected with rVV NS3-5 or wild-type vaccinia virus (WT). Cells infected with rVV NS3-5 were also treated with compound 1a or 2 at the concentrations shown. Northern blot analysis was carried out using a biotinylated oligonucleotide probe to the NS5B region. All lanes contain 0.2 μg of cytoplasmic RNA, except for the positive control (P), which contains 10 ng of the NS3-5 RNA transcript.
Protein expression in the presence of trans-lactams.
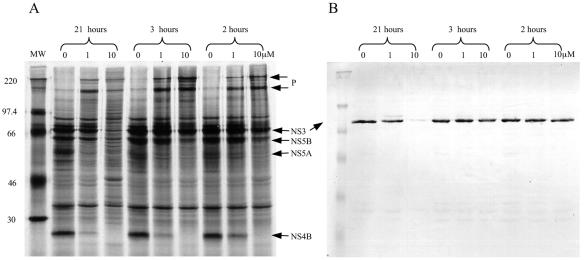
The recombinant vaccinia virus system described here expresses the NS3-5 polyprotein continuously. However, the assay pulse-labels only proteins that are newly synthesized 19 to 21 h postinfection. To estimate the amounts of protein synthesized at different time points after infection, an experiment was carried out in which an active trans-lactam was added to the cells only during the 1-h methionine starvation and/or the 2-h labeling stage (Fig. 5A). Western blot analysis of parallel samples was used to visualize the total amount of NS3 protein present in the cells (Fig. 5B). Incubation of infected cells with the trans-lactam for 21 h resulted in the depletion of most, if not all, of the NS3 in the system, whereas the protein was still evident after 3 h of incubation with the trans-lactam. However, inhibition of newly synthesized NS3 was still possible in 2 h despite the high levels of preexisting NS3 in the system. This is best illustrated by the decrease in the amount of NS4B (Fig. 5A).
FIG. 5.
Protein expression in the presence of a trans-lactam. Compound 1b was added, at the concentrations indicated, to BT7H cells at different stages during the labeling process. The 21-h samples were continuously exposed to the compound. The 3-h sample was exposed to the compound from 18 to 21 h postinfection, during both the methionine starvation and the labeling phase. The 2-h sample was exposed to the compound from 19 to 21 h postinfection, during the metabolic labeling phase only. SDS-PAGE was carried out on the labeled samples. Western blot analysis with an in-house anti-NS3 polyclonal antibody was performed on parallel samples to assess the total amount of NS3 in the system. (A) Autoradiograph showing metabolically labeled proteins. MW, molecular weight marker (in thousands). P, precursor proteins. (B) Western blot analysis with the anti-NS3 antibody.
trans-lactam potency can be ranked by using the gel-based assay.
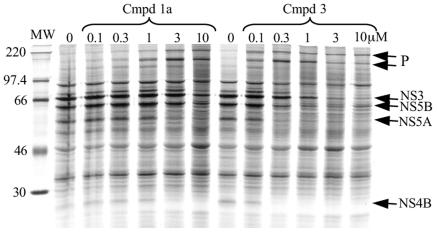
The potencies of a number of trans-lactam compounds have been evaluated by using the gel-based assay. Dose responses resulting in an end point were obtained, enabling ranking of compounds according to potency. The end point is defined as the lowest concentration at which NS4B is no longer visible and there is a simultaneous increase in the amount of high-molecular-weight precursor proteins. This is illustrated in Fig. 6, where the two trans-lactams used, compounds 1a and 3, had end points of 3 and 0.3 μM, respectively.
FIG. 6.
trans-lactam compounds can be ranked according to potency in the gel-based assay. Dose responses of compounds 1a and 3 were carried out at the concentrations indicated, from which end points can be determined. The end point is defined as the lowest concentration at which NS4B is no longer visible—in this case, 3 μM for compound 1a and 0.3 μM for compound 3. MW, molecular weight marker (in thousands).
Adaptation of the gel-based vaccinia virus assay to an ELISA format.
Not only was NS3-dependent processing inhibited in the presence of trans-lactams, but also the absolute levels of processed proteins decreased. Based on this observation, the gel-based assay was modified to an ELISA format. This format had the advantages of being nonisotopic, higher throughput, more quantitative, and less labor-intensive as well as less subjective. This assay involved infecting preformed, confluent BT7H monolayers with rVV NS3-5 in 96-well plates. Compounds were added after the removal of the vaccinia virus inoculum, and after a 24-h incubation, cells were fixed and permeabilized and the ELISA was performed.
A panel of commercially available murine monoclonal antibodies recognizing different nonstructural proteins of HCV was assessed for binding to the proteins expressed in rVV NS3-5-infected BT7H cells (Fig. 7A). Signals were compared to those obtained from uninfected BT7H cells, and suitable antibodies were titrated to determine appropriate working dilutions. By using this format, 50% inhibitory concentrations (IC50) were determined for a number of compounds. An example of an IC50 curve for compound 1a is shown in Fig. 7B.
FIG. 7.
Selection of the primary antibody for the rVV NS3-5 ELISA. (A) Commercially available murine monoclonal antibodies against HCV nonstructural proteins were assayed at 4 μg/ml against rVV NS3-5-infected BT7H cells (dark bars) and uninfected BT7H cells (light bars). Each bar represents the mean result for four (rVV NS3-5-infected cells) or three (uninfected cells) samples within the same experiment ± the standard error of the mean. Antibodies against the C-100 antigen (8958) and against NS5 (8808 and 1872) were selected for use in the ELISA (see Materials and Methods). (B) An example of an IC50 curve for compound 1a using antibody 8958.
A panel of trans-lactams that had been ranked for potency by the gel-based assay was titrated in the rVV NS3-5 ELISA by using two monoclonal antibodies of different specificities (anti-C100 antibody 8958 and anti-NS5 antibody 8808). Both assays, as well as both antibodies, ranked the compounds in essentially the same order (Table 1), confirming the correlation between the inhibition of cleavage and the absolute levels of NS proteins. Furthermore, cytotoxicity data obtained by a Vero cell assay confirmed that the potency of the compounds was not related to any cytotoxic effect.
TABLE 1.
Comparison of trans-lactam potency in rVV NS3-5 gel-based assay and ELISA using antibodies of different specificitiesa
| Compound IDb | Gel-based assay end pointc (μM) | ELISA IC50 (μM) ± SEd
|
Vero cell CC50 (μM) | Selectivity indexg | |
|---|---|---|---|---|---|
| Anti-C100 antibodye | Anti-NS5A antibodyf | ||||
| 1c | <0.3 | <1.6 | 0.4 ± 0.1 | 124 | >78 |
| 1d | 1 | 1.2 ± 0.7 | 0.3 ± 0.2 | 144 | 120 |
| 1e | <0.3 | 2.1 ± 0.1 | NT | 76 | 36 |
| 1f | 1 | 4.6 ± 0.4 | 5.2 ± 0.6 | 78 | 17 |
| 1g | 1 | 5.9 ± 0.6 | 5.4 ± 0.3 | 270 | 46 |
| 1h | 1 | NT | 6.1 ± 0.6 | 95 | 16h |
| 1i | 3 | 7.8 ± 0.7 | NT | 382 | 49 |
| 1j | 3 | 9.0 ± 1.4 | NT | 320 | 36 |
| 1k | 1 | 9.1 ± 0.7 | 8.0 ± 0.6 | 290 | 32 |
| 1a | 3 | 11.8 ± 0.7 | 8.3 ± 0.4 | 92 | 8 |
| 1l | 3 | NT | 13.4 ± 0.9 | 279 | 21h |
| 1m | 3 | NT | 15.1 ± 0.7 | >500 | >33h |
| 1n | 3-10 | NT | 20.3 ± 2.6 | 331 | 16h |
| 1o | 3-10 | 67.9 | >100 | 500 | 7 |
| 4 | 3 | >100 | >100 | >500 | NC |
| 1p | 10 | >100 | 23.4 ± 2.2 | 531 | NC |
| 1q | >10 | >100 | 50.7 ± 13.7 | >500 | NC |
Results are ranked according to data from the ELISA using the anti-C100 antibody.
ID, identification number.
Defined as the lowest concentration at which NS4B is no longer visible.
Standard errors were derived from a GraFit software program used to calculate IC50. Compounds were tested once. NT, not tested.
8958, from Biogenesis (see Materials and Methods).
8808, from Biogenesis (see Materials and Methods).
Calculated by dividing the Vero cell CC50 by the ELISA IC50 (anti-C100 antibody). NC, not calculated.
Calculated by using the anti-NS5A antibody result.
The HCV replicon-based ELISA.
With the advent of an HCV replicon system, consisting of Huh-7 cells stably transfected with an HCV minigenome expressing NS3-5 (23), the ELISA was further modified to utilize these cells. A suspension of 4 × 104 Huh-7 clone 5-15 cells per well was added to dilutions of the compound in 96-well plates, and the plates were incubated for 3 days. Thereafter, the cells were fixed and permeabilized, and an ELISA was performed as for the vaccinia virus ELISA using the anti-C100 monoclonal antibody 8958. IC50 were calculated from plots of percent inhibition against compound concentration using the GraFit software package (Erithacus Software). The potencies of a panel of trans-lactams were determined by both the rVV NS3-5 ELISA and the replicon ELISA and were compared. The ranking of IC50 in the two assays was in general agreement (Table 2). The biochemical potencies of these compounds were also measured by a fluorogenic assay and expressed by the kinetic parameter kobs/I (1). This assay was run in a continuous-readout mode at three concentrations to generate a more robust kinetic measure of potency than that obtained using a chromogenic assay, where more-potent compounds were acting as active-site titrants (1). In this case, the kobs/I values were calculated by dividing the observed rate constant (kobs) by the molar concentration used; generally, the larger the value, the greater the potency of the compound.
TABLE 2.
Comparison of trans-lactam potencies determined by the rVV NS3-5 and replicon ELISAsa
| Compound IDb | IC50 (μM) by the:
|
Biochemical potency (kobs/I [M−1 s−1])c | |
|---|---|---|---|
| Vaccinia virus ELISA | Replicon ELISA | ||
| 1d | 0.3 | <0.78 | 2,292 |
| 1c | 0.4 | 0.45 | 7,760 |
| 1i | 7.8 | 1.1 | 362 |
| 1j | 9 | 2.8 | 853 |
| 1n | 20.3 | 6.4 | 543 |
| 1a | 8.3 | 9.0 | 300 |
| 4 | >100 | 97 | 671 |
| 5 | 75 | >100 | 112 |
| 2 | >100 | >100 | 0 |
Results are ranked according to the data from the replicon ELISA.
ID, identification number.
Measured by a fluorogenic assay as described by Andrews et al. (1).
Use of biotinylated probes for the detection of HCV NS3 in vaccinia virus-infected cells.
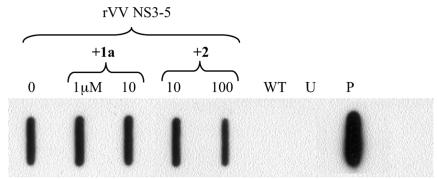
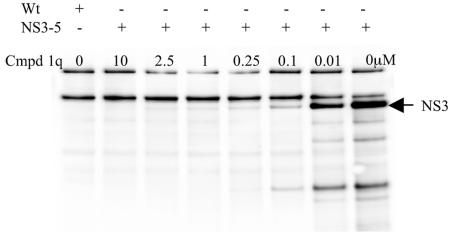
When NS3/4A protease is incubated with a trans-lactam, active NS3/4A is covalently modified by the trans-lactam, which acylates the active-site serine (N. M. Gray, unpublished data). This covalent modification can be exploited for the detection of active NS3/4A protease in crude mixtures by preincubation of the test trans-lactam prior to the modification of the remaining active NS3/4A by biotinylated trans-lactam. A 1-h preincubation of rVV NS3-5-infected whole-cell extracts with a biochemically active, unlabeled trans-lactam, compound 1q, prior to the addition of a biotin-labeled trans-lactam probe (compound 6) results in the titration of activity against NS3 (Fig. 8). The result shows the chemiluminescent signal from the NS3 band decreasing with increasing concentrations of the test trans-lactam, compound 1q. This method can be used to obtain a quantitative IC50 result for these compounds: in this case, 0.1 μM for compound 1q. The same biotinylated trans-lactam was used to show that the protease present in the replicon cell line, 5-15, was active.
FIG. 8.
Use of a biotinylated trans-lactam probe for the detection of HCV NS3 in vaccinia virus-infected cells. Extracts of rVV NS3-5-infected cells were preincubated with compound (Cmpd) 1q, at the concentrations indicated, for 1 h prior to the addition of a biotinylated trans-lactam probe, compound 6. Samples were analyzed by Western blotting using an HRP-neutravidin probe and were visualized by using SuperSignal reagent. Wt, wild-type vaccinia virus-infected cell extract; NS3-5, rVV NS3-5-infected cell extract.
DISCUSSION
Vaccinia virus has been extensively used as a vector for the cytoplasmic expression of proteins in mammalian cells. Infection with a recombinant vaccinia virus results in rapid inhibition of host cell nucleic acid and protein synthesis, and the majority of proteins observed after metabolic labeling are derived from the recombinant vaccinia virus. By using BT7H cells, which constitutively express T7 RNA polymerase, the expression of the recombinant proteins, in this case HCV nonstructural proteins from rVV NS3-5, is even more pronounced. Bands corresponding to all the nonstructural proteins of HCV can be observed in cells infected with rVV NS3-5, as has been reported by others (7). The identity of these proteins was verified by immunoprecipitation with specific antisera. The addition of a trans-lactam, an inhibitor of the HCV NS3/4A protease, to the system resulted in the disappearance of mature, fully processed proteins and the appearance of uncleaved or partially cleaved precursor NS3-5 polyprotein. These high-molecular-weight proteins were not evident when the same compounds were added to wild-type vaccinia virus-infected cells. They were verified as being HCV derived by immunoprecipitation with anti-NS5 antibodies, indicating that these proteins contained NS5 sequences. The largest precursor protein represents the uncleaved NS3-5 polyprotein, estimated to be ∼220 kDa, while the other major precursor protein probably represents NS4A-NS4B-NS5A-NS5B, since it reacts with antisera against all these proteins (data not shown) and the first cleavage event occurs in cis between NS3 and NS4A. The trans-lactams inhibit NS3/4A protease by acylating the active-site serine (residue 1165) irreversibly (N. M. Gray, unpublished data). In this acylated form, the protease is probably unstable and most likely turned over more rapidly by the cell. This is illustrated by the loss of the NS3 band in metabolic labeling experiments. There does not appear to be a stoichiometric relationship between the loss of NS3 and the appearance of high-molecular-weight proteins, because the small amount of high-molecular-weight protein does not appear to account for the large decrease in the amount of mature proteins. It is speculated that this is probably due to increased turnover of modified NS3 by the cell, since an NS3-5 polyprotein with a nonfunctional NS3/4A protease is also found to be present at a very low level (unpublished data).
It has been shown that the half-life of NS3 differs greatly depending on whether NS3 is expressed alone or in conjunction with NS4A. In a tetracycline-inducible cell system, where a continuous human cell line expressed NS3 either alone or with NS4A, but only in the presence of tetracycline, the protein half-life of NS3 was 3 h when NS3 was expressed alone but increased to 26 h when NS3 was coexpressed with NS4A. This suggests that NS4A increases the stability of NS3 in this cellular context (35). In addition, it has been suggested that, in a transient protein expression system in COS-1 cells (33), NS3 does not exist stably in the cell in the absence of NS4A. Most of the NS3 processed from a construct consisting of NS2, NS3, NS4B, NS5A, and NS5B, but with NS4A deleted, was localized in the cytosol and was degraded promptly. In our system, NS3 is expressed in the context of the replication complex, which consists of NS3, NS4A, NS4B, NS5A, and NS5B proteins. If the trans-lactam compound, by acylating the active site, prevents NS3 from associating with NS4A and the replication complex and causes the subsequent failure in processing of the remaining NS proteins, this could result in rapid turnover of the acylated NS3, and hence in the decrease in the amount of NS3 in the system.
At the time of these studies, this class of inhibitor was the only in-house series capable of specific intracellular activity against NS3/4A, and it was possible that the compounds might exhibit nonspecific or cytotoxic effects such as down-regulation of transcription and/or translation. There was no effect on transcription with either the active enantiomer or the inactive compound at concentrations at which levels of NS3, and the other NS proteins, had diminished significantly with the active enantiomer relative to levels of the untreated control (Fig. 3 and 4). This demonstrates two things; first, it is unlikely that these compounds are exerting their effect through a toxic effect on transcription; second, the inhibitory effect is specifically associated with the active enantiomer. To investigate the effects on translation, another rVV expressing the HCV structural proteins under the control of the same promoter and with the same IRES background was tested with the compound. No effect was found on expression of the structural proteins at concentrations of the compound which markedly depleted the amount of NS3, implying that interference with translation was unlikely. There was also no effect on expression of wild-type vaccinia virus proteins. To further confirm the lack of cytotoxicity of these compounds, data were obtained from a Vero cell assay where the cells were in the exponential phase of growth throughout. The results verified that the potencies of the compounds in the gel-based assay, and later in the ELISA, were not related to gross cytotoxicity (Table 1).
In the system used in this study, the NS3-5 polyprotein is expressed continuously, but the assay is performed by pulse-labeling the cells at 19 to 21 h postinfection and detects only newly synthesized proteins. It was necessary to establish whether large amounts of NS3 protein present in the cells prior to labeling would affect the assay. Therefore, an assay was carried out in which the compound was added to the sample only during the 1-h methionine starvation and/or the 2-h labeling stage. Western blot analysis was performed on parallel samples to visualize the total amount of NS3 produced. While overnight incubation with 10 μM compound 1b was sufficient to deplete most, if not all, of the NS3, interestingly, when the compound was added only from 19 to 21 h postinfection, inhibition of newly synthesized protein was still possible within 2 h, despite the high levels of preexisting NS3 (Fig. 5). This gave us confidence that even in a demanding system, where NS3 is expressed to levels much higher than those thought to exist in infected hepatocytes, NS3/4A protease activity could still be inhibited.
The observed decrease in levels of mature NS proteins enables end points to be determined for active trans-lactam compounds. We arbitrarily chose to define the end point as the lowest concentration at which NS4B was no longer visible and there was a concurrent appearance of the high-molecular-weight precursors. NS4B was selected because it was very easy to visualize after metabolic labeling. It was more difficult to evaluate the decreases in the levels of the other NS proteins, NS3, NS5A, and NS5B, because they were located in an area of the gel that contained other proteins, although careful examination of the gels gave the same results. Determination of the end point was utilized to rank compounds according to their potencies in this system. In the example shown in Fig. 6, compound 1a had an end point of 3 μM while that of compound 3 was 0.3 μM. The biochemical potencies of these compounds against a purified protease domain with exogenous NS4A were 300 and 1,529 M−1 s−1, respectively, as measured by the kinetic parameter kobs/I (1). This parameter was found to be a robust kinetic measure of potency and enabled structure-activity relationships to be determined. In general, the larger the kobs/I value, the greater the potency of the compound. From previous studies, it was evident that prerequisites for potency in the cell-based assays were good cellular penetration and a kobs/I value greater than 150 M−1 s−1 (2).
In a separate investigation, a biotinylated trans-lactam was generated and used as a probe to detect active NS3 in the rVV NS3-5 system. Lysates from cells infected with rVV NS3-5 were preincubated for 1 h with an unlabeled trans-lactam, compound 1q, before probing with a biotinylated trans-lactam probe, compound 6. Biotinylated NS3 could then be visualized by probing with HRP-neutravidin (see Materials and Methods). IC50 could be obtained by using a range of concentrations of unlabeled trans-lactam (Fig. 8). The concept behind this assay is that preincubation of NS3 with an unlabeled trans-lactam for a specific time allows active NS3 to be covalently modified at a rate dependent on the kobs/I and concentration of the compound. The subsequent addition of the biotinylated trans-lactam is designed to covalently modify any remaining active NS3. The 45-min reaction with the biotinylated trans-lactam, compound 6, will have gone to completion (kobs/I, 2,001 M−1 s−1), allowing a relative degree of potency to be assessed. It should be noted that, although compound 1q has very good biochemical potency in vitro, it has poor cell penetration and so is inactive in both the gel-based assay and the ELISA (see Table 1). The assay described above avoids the issue of cell permeability by using cell extracts, enabling IC50 to be determined.
Although the gel-based assay is initially very useful, its limitations are obvious. It uses radioisotopes, is very labor-intensive, and has low throughput, and the results are subjective and only semiquantitative. For these reasons, we developed the assay into an ELISA format, which immediately increased the throughput, was nonisotopic, and resulted in quantitative results in the form of IC50. This type of assay was made possible by the observation that in the presence of NS3/4A inhibitors, not only was processing disrupted, but the absolute levels of NS proteins decreased. A panel of commercially available monoclonal antibodies directed against HCV NS proteins was evaluated. Antibodies raised against NS4, NS5, and the C100 antigen were found most suitable in that there was a particularly good signal/noise ratio between rVV NS3-5-infected cells and the background of uninfected cells (Fig. 7A). It was thought prudent to use an antibody raised against a protein other than NS3 for the evaluation of NS3/4A protease inhibitors; consequently, the anti-C100 and anti-NS5A antibodies were selected.
A panel of trans-lactams that had been ranked on the basis of potency in the gel-based assay was tested in the ELISA using the anti-C100 and anti-NS5A antibodies. Both assays ranked the compounds in essentially the same order (Table 1), confirming the correlation between inhibition of cleavage by NS3 and levels of total protein. Thus, the ELISA could be used to quantify the NS3 inhibition visualized qualitatively in the gel-based assay. One technical difference between the vaccinia virus gel-based assay and the ELISA was that the compound was fed twice in the gel-based assay, once during the methionine starvation stage and again during the labeling stage, whereas there was only a single 18-h incubation with the compound in the ELISA. However, since it was established by Western blotting that the total level of the NS3 protein was reduced after an 18-h incubation with the compound (see Fig. 5), this difference was not considered important.
With the advent of an HCV replicon system, consisting of Huh-7 cells stably transfected with an HCV minigenome expressing NS3-5 (23), the vaccinia virus ELISA was modified to use these cells. The replicon clone 5-15 (23) was selected because it produced the best signal:noise ratio with our antibodies. The replicon ELISA has several advantages over the vaccinia virus ELISA. Huh-7 cells are human hepatocarcinoma derived, whereas the BT7H cells used in the vaccinia virus ELISA are monkey kidney cells. Vaccinia virus infection results in the death of the cell, whereas the replicon does not kill the Huh-7 host cell but behaves more like a persistent infection. Perhaps the greatest advantage is that the replicon system is not only useful for assessment of protease inhibitors but also can be used to screen potential inhibitors of other viral targets, including polymerase and helicase, and also possibly replication complex inhibitors.
The two ELISA systems (vaccinia virus and replicon) were compared by using a series of trans-lactams and produced IC50 which ranked similarly (Table 2). These generally correlated well with the vaccinia virus gel-based assay, which gives a direct visual measure of inhibition of NS3/4A protease activity. This is encouraging, in view of the fact that the systems are quite different in a number of aspects. In the replicon ELISA, the compound is in contact with the cells for 3 days, whereas in the vaccinia virus ELISA this was only an overnight incubation. Different HCV sequences are utilized in the rVV NS3-5 assays and the replicon ELISA; however, they both belong to the 1b genotype. We do not yet have any data using any other genotypes (recently, trans-lactams have been shown to be active against a genotype 1a replicon [M. Gartland and E. M. Amphlett, personal communication]).
In conclusion, we have established a number of assays for evaluation of HCV NS3/4A protease inhibitors. The gel-based assay allows visualization of the inhibition of proteolysis of the NS3-5 polyprotein by NS3/4A, and the rVV NS3-5 ELISA exploits the phenomenon that absolute levels of mature NS proteins decrease in the presence of NS3/4A inhibitors. The replicon ELISA exhibits similar ranking of compounds but in a context more relevant physiologically than vaccinia virus-infected cells. These systems have proved to be invaluable in evaluating potential protease inhibitors, specifically the trans-lactams, which have in turn been useful for validating our assays.
Acknowledgments
We thank David Andrews, Martin Slater, Martin Johnson, Gail Mills, and Paul Jones for synthesis of compounds; Michael Barnes for synthesis of the biotinylated trans-lactam probe; Liz Amphlett for the Vero cell cytotoxicity data; Ralf Bartenschlager for provision of replicon cell lines; and Bernie Moss for the pTM3 vaccinia virus transfer vector.
REFERENCES
- 1.Andrews, D. M., S. J. Carey, H. Chaignot, B. A. Coomber, N. M. Gray, S. L. Hind, P. S. Jones, G. Mills, J. E. Robinson, and M. J. Slater. 2002. Pyrrolidine-5,5-trans-lactams. 1. Synthesis and incorporation into inhibitors of hepatitis C virus NS3/4A protease. Org. Lett. 4:4475-4478. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, D. M., H. M. Chaignot, B. A. Coomber, A. C. Good, S. L. Hind, M. R. Johnson, P. S. Jones, G. Mills, J. E. Robinson, T. Skarzynski, M. J. Slater, and D. O. Somers. 2002. Pyrrolidine-5,5-trans-lactams. 2. The use of X-ray crystal structure data in the optimisation of P3 and P4 substituents. Org. Lett. 4:4479-4482. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, D. M., P. S. Jones, G. Mills, S. L. Hind, M. J. Slater, N. Trivedi, and K. J. Wareing. 2003. Design and synthesis of spiro-cyclopentenyl and spiro-[1,3]-dithiolanyl substituted pyrrolidine-5,5-trans-lactams as inhibitors of hepatitis C virus NS3/4A protease. Bioorg. Med. Chem. Lett. 13:1657-1660. [DOI] [PubMed] [Google Scholar]
- 4.Andrews, D. M., H. M. Chaignot, B. A. Coomber, M. D. Dowle, S. L. Hind, M. R. Johnson, P. S. Jones, G. Mills, A. Patikis, T. J. Pateman, J. E. Robinson, M. J. Slater, and N. Trivedi. 2003. The design of potent, non-peptidic inhibitors of hepatitis C protease. Eur. J. Med. Chem. 38:339-343. [DOI] [PubMed] [Google Scholar]
- 5.Andrews, D. M., M. C. Barnes, M. D. Dowle, S. L. Hind, M. R. Johnson, P. S. Jones, G. Mills, A. Patikis, T. J. Pateman, T. J. Redfern, J. E. Robinson, M. J. Slater, and N. Trivedi. 2003. Pyrrolidine-5,5-trans-lactams. 5. Pharmacokinetic optimisation of inhibitors of hepatitis C virus NS3/4A protease. Org. Lett. 5:4631-4634. [DOI] [PubMed] [Google Scholar]
- 6.Bartenschlager, R. 2002. Hepatitis C virus replicons: potential role for drug development. Nat. Rev. Drug Discov. 1:911-916. [DOI] [PubMed] [Google Scholar]
- 7.Bartenschlager, R., L. Ahlborn-Laake, K. Yasargil, J. Mous, and H. Jacobsen. 1995. Substrate determinants for cleavage in cis and in trans by the hepatitis C virus NS3 proteinase. J. Virol. 69:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beard, M. R., G. Abell, M. Honda, A. Carroll, M. Gartland, B. Clarke, K. Suzuki, R. Lanford, D. V. Sangar, and S. M. Lemon. 1999. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology 30:316-324. [DOI] [PubMed] [Google Scholar]
- 9.Boulant, S., M. Becchi, F. Penin, and J.-P. Lavergne. 2003. Unusual multiple recoding events leading to alternative forms of hepatitis C virus core protein from genotype 1b. J. Biol. Chem. 278:45785-45792. [DOI] [PubMed] [Google Scholar]
- 10.Cho, Y. G., H. S. Moon, and Y. C. Sung. 1997. Construction of hepatitis C-SIN virus recombinants with replicative dependency on hepatitis C virus serine protease activity. J. Virol. Methods 65:201-207. [DOI] [PubMed] [Google Scholar]
- 11.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 12.De Francesco, R., and C. Steinkühler. 2000. Structure and function of the hepatitis C virus NS3-NS4A serine protease. Curr. Top. Microbiol. Immunol. 242:149-169. [DOI] [PubMed] [Google Scholar]
- 13.De Francesco, R., P. Neddermann, L. Tomei, C. Steinkühler, P. Gallinari, and A. Folgori. 2000. Biochemical and immunologic properties of the non-structural proteins of the hepatitis C virus: implications for the development of antiviral agents and vaccines. Semin. Liver Dis. 20:69-83. [DOI] [PubMed] [Google Scholar]
- 14.De Francesco, R., L. Tomei, S. Altamura, V. Summa, and G. Migliaccio. 2003. Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antivir. Res. 58:1-16. [DOI] [PubMed] [Google Scholar]
- 15.Falkner, F. O., and B. Moss. 1988. Escherichia coli gpt gene provides dominant selection for vaccinia virus open reading frame expression vectors. J. Virol. 62:1849-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuda, K., D. Vishnuvardhan, S. Sekiya, J. Hwang, N. Kakiuchi, K. Taira, K. Shimotohno, P. K. R. Kumar, and S. Nishikawa. 2000. Isolation and characterisation of RNA aptamers specific for the hepatitis C virus nonstructural protein 3 protease. Eur. J. Biochem. 267:3685-3694. [DOI] [PubMed] [Google Scholar]
- 17.Hahm, B., S. H. Back, T. G. Lee, E. Wimmer, and S. K. Jang. 1996. Generation of a novel poliovirus with a requirement of hepatitis C virus protease activity. Virology 226:318-326. [DOI] [PubMed] [Google Scholar]
- 18.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong, A. D., J. L. Kim, G. Rao, D. Lipovsek, and S. A. Raybuck. 1998. Hepatitis C virus NS3/4A protease. Antivir. Res. 40:1-18. [DOI] [PubMed] [Google Scholar]
- 20.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bös, D. R. Cameron, M. Cartier, M. G. Cordingley, A.-M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagacé, S. R. LaPlante, H. Narjes, M-A. Poupart, J. Rancourt, R. E. Sentjens, R. St. George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C.-L. Yong, and M. Linàs-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 21.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41-52. [DOI] [PubMed] [Google Scholar]
- 22.Lee, J. C., Y. F. Shih, S. P. Hsu, T. Y. Chang, L. H. Chen, and J. T. A. Hsu. 2003. Development of a cell-based assay for monitoring specific hepatitis C virus NS3/4A protease activity in mammalian cells. Anal. Biochem. 316:152-170. [DOI] [PubMed] [Google Scholar]
- 23.Lohmann, V., F. Korner, J.-O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 24.Macdonald, S. J., M. D. Dowle, L. A. Harrison, P. Shah, M. R. Johnson, G. G. Inglis, G. D. Clarke, R. A. Smith, D. Humphreys, C. R. Molloy, A. Amour, M. Dixon, G. Murkitt, R. E. Goodward, T. Padfield, T. Skarzynski, O. M. Singh, K. A. Kumar, G. Fleetwood, S. T. Hodgson, G. W. Hardy, and H. Finch. 2001. The discovery of a potent, intracellular, orally bioavailable, long duration inhibitor of human neutrophil elastase—GW311616A a development candidate. Bioorg. Med. Chem. Lett. 11:895-898. [DOI] [PubMed] [Google Scholar]
- 25.McPhee, F., K. S. Yeung, A. C. Good, and N. A. Meanwell. 2003. Hepatitis C virus NS3 serine protease as a drug discovery target. Drug Future 28:465-488. [Google Scholar]
- 26.Moradpour, D., V. Brass, R. Gosert, B. Wölk, and H. E. Blum. 2002. Hepatitis C: molecular virology and antiviral targets. Trends Mol. Med. 8:476-482. [DOI] [PubMed] [Google Scholar]
- 27.Moss, B., O. Elroy-Stein, T. Mizukami, W. A. Alexander, and T. R. Fuerst. 1990. New mammalian expression vectors. Nature 348:91-92. [DOI] [PubMed] [Google Scholar]
- 28.Narjes, F., U. Koch, and C. Steinkühler. 2003. Recent developments in the discovery of hepatitis C virus serine protease inhibitors—towards a new class of antiviral agents? Expert Opin. Investig. Drugs 12:153-163. [DOI] [PubMed] [Google Scholar]
- 29.Pacini, L., L. Bartholomew, A. Vitelli, and G. Migliaccio. 2004. Reporter substrates for assessing the activity of the hepatitis C virus NS3-4A serine protease in living cells. Anal. Biochem. 331:46-59. [DOI] [PubMed] [Google Scholar]
- 30.Rice, C. M. 1996. Flaviviridae: the viruses and their replication, p. 931-959. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 31.Slater, M. J., D. M. Andrews, G. Baker, S. S. Bethell, S. J. Carey, H. M. Chaignot, B. Clarke, B. Coomber, M. Ellis, A. Good, N. M. Gray, G. Hardy, P. S. Jones, G. Mills, and J. E. Robinson. 2002. Design and synthesis of ethyl pyrrolidine-5,5-trans-lactams as inhibitors of hepatitis C virus NS3/4A protease. Bioorg. Med. Chem. Lett. 12:3359-3362. [DOI] [PubMed] [Google Scholar]
- 32.Slater, M. J., E. M. Amphlett, D. M. Andrews, P. Bamborough, S. J. Carey, M. R. Johnson, P. S. Jones, G. Mills, N. R. Parry, D. O. Somers, A. J. Stewart, and T. Skarzynski. 2003. Pyrrolidine-5,5-trans-lactams. 4. Incorporation of a P3/P4 urea leads to potent intracellular inhibitors of hepatitis C virus NS3/4A protease. Org. Lett. 5:4627-4630. [DOI] [PubMed] [Google Scholar]
- 33.Tanji, Y., M. Hijikata, S. Satoh, T. Kaneko, and K. Shimotohno. 1995. Hepatitis C virus-encoded non-structural protein NS4A has versatile functions in viral protein processing. J. Virol. 69:1575-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomei, L., C. Failla, E. Santolini, R. DeFrancesco, and N. La Monica. 1993. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J. Virol. 67:4017-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wölk, B., D. Sansonno, H.-G. Kräusslich, F. Dammacco, C. M. Rice, H. E. Blum, and D. Moradpour. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]