Abstract
MurG and MraY, essential enzymes involved in the synthesis of bacterial peptidoglycan, are difficult to assay because the substrates are lipidic and hard to prepare in large quantities. Based on the use of Escherichia coli membranes lacking PBP1b, we report a high-throughput method to measure the activity of MurG and, optionally, MraY as well. In these membranes, incubation with the two peptidoglycan sugar precursors results in accumulation of lipid II rather than the peptidoglycan produced by wild-type membranes. MurG was assayed by addition of UDP-[3H]N-acetylglucosamine to membranes in which lipid I was preformed by incubation with UDP-N-acetyl-muramylpentapeptide, and the product was captured by wheat germ agglutinin scintillation proximity assay beads. In a modification of the assay, the activity of MraY was coupled to that of MurG by addition of both sugar precursors together in a single step. This allows simultaneous detection of inhibitors of either enzyme. Both assays could be performed using wild-type membranes by addition of the transglycosylase inhibitor moenomycin. Nisin and vancomycin inhibited the MurG reaction; the MraY-MurG assay was inhibited by tunicamycin as well. Inhibitors of other enzymes of peptidoglycan synthesis—penicillin G, moenomycin, and bacitracin—had no effect. Surprisingly, however, the β-lactam cephalosporin C inhibited both the MurG and MraY-MurG assays, indicating a secondary mechanism by which this drug inhibits bacterial growth. In addition, it inhibited NADH dehydrogenase in membranes, a hitherto-unreported activity. These assays can be used to screen for novel antibacterial agents.
Cell wall-related targets are attractive for the discovery of novel antibacterial drugs. Since peptidoglycan is unique to the bacterial cell, has no mammalian counterpart, and is present in most bacterial cell walls, agents inhibiting its synthesis have the potential to become broad-spectrum antibiotics and are of special interest.
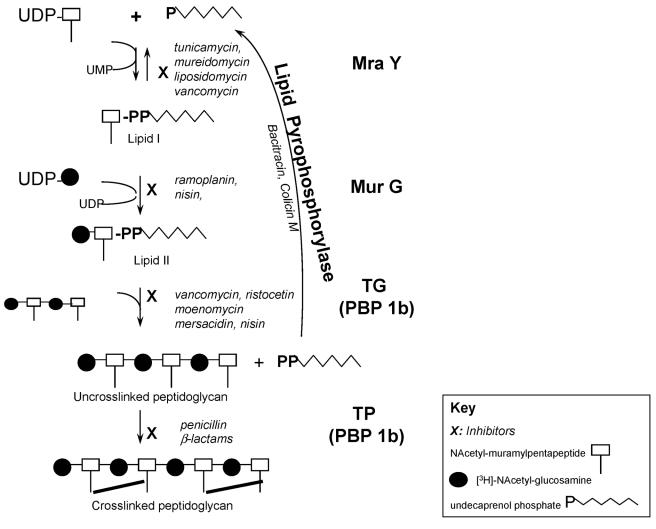
MraY is the first enzyme in the membrane stage of peptidoglycan synthesis. It catalyzes the transfer of muramylpentapeptide from UDP-muramylpentapeptide [UDP-MurNAc(pp)] to the lipid carrier undecaprenol phosphate to form lipid I. MurG catalyzes the transfer of the N-acetylglucosamine (GlcNAc) residue from UDP-GlcNAc to undecaprenyl-pyrophosphoryl-MurNAc(pp) (lipid I) to yield lipid II, i.e., GlcNAc-MurNAc(pp)-pyrophosphoryl-undecaprenol (Fig. 1).
FIG. 1.
Schematic of membrane steps of peptidoglycan synthesis. Cross-linked peptidoglycan is formed as a result of the catalytic activities of five enzymes (shown in boldface to the right of the schematic). Inhibitors of each enzyme are shown in italics. TG, transglycosylase; TP, transpeptidase.
MurG is associated with the cytoplasmic surface of the membrane. It can be eluted from the membrane and purified (10), and its structure with one substrate (UDP-GlcNAc) bound has been elucidated (17, 21). In addition, inhibitors that compete with UDP-GlcNAc have been reported (20). This offers the possibility of structure-based drug design, making it more attractive than the other membrane-bound enzymes of peptidoglycan synthesis for a drug discovery program.
However, MurG is difficult to screen in high-throughput format, since the substrate is lipidic and difficult to isolate in large quantities. Traditionally, the enzyme has been assayed in bacterial membranes, where the lipid substrate was made in situ by the action of MraY in membranes preincubated with UDP-MurNAc(pp). In a second step, MurG was assayed by the addition of radiolabeled UDP-GlcNAc, and radiolabeled lipid II was separated from the substrate by extraction with butanol or by paper chromatography (24). The purified enzyme can also be assayed in solution using artificial, synthetic substrates. These soluble substrates have shorter lipid chains than the natural substrate (1, 23, 26), and this method is good for kinetic characterization of the enzyme (8, 18), but scaling up the synthesis to allow high-throughput screening is difficult.
Here, we report a high-throughput method to select inhibitors of MurG and MraY. The assay was performed with Escherichia coli membranes using radiolabeled UDP-GlcNAc, and the product, lipid II, was measured using scintillation proximity assay beads (9). A problem encountered when assaying MraY and MurG in membranes is that lipid II is converted to peptidoglycan by the transglycosylase present in the same membranes. If activity of MurG is to be monitored, this conversion of lipid II must be stopped by inactivation of the transglycosylase. This was accomplished either by using membranes from a strain of E. coli lacking PBP1b or by using moenomycin, a transglycosylase inhibitor, in combination with wild-type membranes, since PBP1b contributes the major transglycosylase activity under these conditions (6, 14).
(Part of this work was presented at the 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, Ill., 14 to 17 September 2003 [25]).
MATERIALS AND METHODS
Materials.
Wheat germ agglutinin-coated scintillation proximity assay (WGA-SPA) beads (catalog no. RPNQ0001; PVT beads) were from Amersham International plc. (United Kingdom). UDP-[3H]GlcNAc was from Dupont, NEN Research Products (Boston, Mass.). Flavomycin (moenomycin) was a gift from Hoechst (Mumbai, India). Antibiotic medium 3 was from Difco Laboratories (Detroit, Mich.). Chromatography materials were from Bio-Rad Laboratories (Richmond, Calif.) or from Whatman (Clifton, N.J.). All other chemicals were from Sigma-Aldrich Corp. (St. Louis, Mo.). Optiphase scintillation fluid was from Wallac (Turku, Finland). E. coli AMA1004 lacking PBP1b (AMA1004 ponB::Spcr) was generated in house as described earlier (29) and was grown in Luria-Bertani broth containing 50 μg of spectinomycin/ml. UDP-MurNAc(pp) was purified from Bacillus cereus 6A1 as described earlier (6, 14).
Enzyme preparation.
Membranes were prepared from E. coli AMA1004 (wild type) or AMA1004 ponB::Spcr as described earlier (6, 14). Briefly, the cells (in 50 mM Tris-HCl, pH 7.5, 0.1 mM MgCl2) were lysed in a French pressure cell. After clarification by low-speed centrifugation, the membranes were pelleted at 150,000 × g and washed once. The membrane preparation was stored in aliquots at −70°C. Protein was estimated using the Coomassie blue dye-binding reagent from Pierce Chemical Co. (Rockford, Ill.). The quality of each membrane batch was monitored as described previously (6) or by measuring the lipid II synthesized by different quantities of protein in the MraY-MurG assay. The radioactivity incorporated in the blank reaction (see below) was also taken into consideration for quality assurance of membrane preparations. Little variation between batches was observed.
MurG assay.
The MurG assay was performed in flexible 96-well plates (catalog no. 1450-401) from Wallac. In the first step, the MurG substrate was made in situ by incubating membranes of E. coli AMA1004 ponB::Spcr (4 μg of protein) for ∼40 min at 37°C with 75 μM UDP-MurNAc(pp) in 50 mM HEPES-ammonia, pH 7.5-10 mM MgCl2 in a final volume of 15 μl. In the second step, the MurG enzyme in the same mixture was assayed by the addition of UDP-[3H]GlcNAc (to 2.5 μM concentration; 0.7 μCi), and dimethyl sulfoxide (DMSO) (to 8% concentration) in HEPES-ammonia, pH 7.5, making up the volume to 25 μl. Following incubation for 5 min at room temperature, the enzyme reaction was stopped by the addition of 5 μl of 90 mM EDTA, UDP-GlcNAc to 200 μM, and 500 μg of WGA-SPA beads in 50 mM HEPES-ammonia (pH 7.5), making up the volume to 200 μl. Reactions were carried out in duplicate. Radioactivity was measured directly in a Microbeta Trilux (Wallac) 3 to 16 h after addition of the beads. Since it is difficult to determine counting efficiency for the SPA, the results are represented as unadjusted counts per minute.
A reaction without the first sugar nucleotide [UDP-MurNAc(pp)] was run in parallel. This was treated as a blank, and the counts per minute obtained in this reaction were subtracted from that of reactions containing both sugar precursors (complete, or 100%, reaction) as a measure of peptidoglycan synthesis. After the blank was subtracted from each well, percent inhibition was calculated as follows: 100 − (counts per minute of well containing inhibitor × 100/counts per minute of control well without inhibitor).
MraY-MurG assay.
The MraY-MurG assay reaction was performed, in a similar manner by incubating E. coli AMA1004 ponB::Spcr membranes (4 μg of protein) for ∼5 min at 37°C with 15 μM UDP-MurNAc(pp)-2.5 μM UDP-[3H]GlcNAc (0.7 μCi)-8% DMSO in a buffer of 50 mM HEPES-ammonia (pH 7.5)-10 mM MgCl2 in a final volume of 25 μl. The enzyme reaction was stopped by the addition of 5 μl of 90 mM EDTA, UDP-GlcNAc to 200 μM, and 500 μg of WGA-SPA beads as for the MurG assay. Reactions were performed in duplicate.
Peptidoglycan synthesis assay: paper chromatography and butanol extraction.
For analysis of the radioactive products formed under different assay conditions (Table 1), parallel sets of reactions were set up, and these were analyzed in four different ways. Membranes (4 μg of protein) were incubated at 37°C for 90 min with 15 μM UDP-MurNAc(pp)-2.5 μM UDP-[3H]GlcNAc (1 μCi)-8% DMSO in 50 mM HEPES-ammonia (pH 7.5)-10 mM MgCl2 in a final volume of 25 μl. All reactions were performed in 96-well plates.
TABLE 1.
Lipid II accumulates in a PBP1b mutant or in wild-type membranes in the presence of moenomycin
| Membrane source | Avg cpma
|
||||
|---|---|---|---|---|---|
| Paper chromatographyc
|
Butanol extract | SPA | SPA + Sarkosyl | ||
| Peptidoglycan | Lipid II | ||||
| E. coli AMA 1004 | |||||
| Activityb | 27,945 ± 2,101 | 1,433 ± 27 | 2,457 ± 131 | 23,072 ± 105 | 23,771 ± 139 |
| Blank | 836 ± 142 | 1,497 ± 202 | 8,768 ± 219 | 9,931 ± 153 | 6,042 ± 878 |
| E. coli AMA 1004 + 0.3 μM moenomycin | |||||
| Activity | 1,597 ± 137 | 7,436 ± 74 | 22,188 ± 1,299 | 18,840 ± 45 | −132 |
| Blank | 301 ± 12 | 2,021 ± 96 | 10,898 ± 1,317 | 9,377 ± 332 | 5,707 ± 233 |
| E. coli AMA 1004 ponB::Spcr | |||||
| Activity | 2,097 ± 117 | 10,723 ± 1,930 | 28,549 ± 815 | 20,979 ± 3 | 438 ± 468 |
| Blank | 188 ± 40 | 1,849 ± 279 | 11,814 ± 448 | 9,576 ± 86 | 5,428 ± 33 |
Shown are the average cpms for the respective conditions and standard deviation. Membranes were incubated with UDP-MurNAc(pp) and UDP-[3H] GlcNAc, as described in the text, in four parallel sets of reactions, which were analyzed by paper chromatography, butanol extraction, capture by WGA-SPA beads, or capture by WGA-SPA beads in the presence of Sarkosyl. For the first two sets of analysis, only a fraction of the total reaction was counted, as described in Materials and Methods.
Activity represents the cpm values of the complete reaction after the respective blank has been subtracted; the blank values [enzyme reactions where UDP-MurNAc(pp) was omitted] for each are indicated.
For the paper chromatography analysis, the counts at the origin represent peptidoglycan, whereas those with a retardation factor of ≈0.9 represent the radioactivity in lipid II.
For paper chromatography analysis, the reaction was stopped by the addition of 5 μl of 90 mM EDTA. Twenty-four microliters of the reaction material was spotted on Whatman 3M chromatography paper and separated using isobutyric acid-1 M ammonia (5:3 [vol/vol]). The chromatogram was cut into 1-cm-wide pieces and counted in 3 ml of Optiphase scintillation fluid (Wallac) in a Microbeta Trilux scintillation counter (Perkin-Elmer-Wallac).
For butanol extraction of radiolabeled lipid II, the reaction was stopped by the addition of 200 μl of butanol-6 M pyridinium acetate (2:1 [vol/vol]; pH 4.1) (3), and the lipids were extracted by vortexing the mixture. The butanol extract (100 μl) was transferred to another Eppendorf tube and washed three times (with 100 μl of butanol-saturated water each time), and 50 μl of the butanol extract was counted in 3 ml of scintillation fluid.
For SPA analysis, the reaction was stopped with 5 μl of 90 mM EDTA, and then 500 μg of SPA beads was added in a volume of 170 μl of 50 mM HEPES-ammonia buffer (pH 7.5) without detergent or with N-lauryl sarcosine (Sarkosyl) so that the final concentration (in 200 μl) was 0.2%.
NADH dehydrogenase assay.
NADH dehydrogenase, part of the respiratory chain, was assayed in the same membranes by incubating 0.5 or 2 μg of membrane protein in 100 μl of 50 mM HEPES-ammonia buffer (pH 7.5)-10 mM MgCl2-0.3 mM NADH-8% DMSO in a 96-well microtiter plate. The oxidation of NADH was monitored at 340 nm in a Spectramax 250 microtiter plate reader (Molecular Devices).
RESULTS
Lipid II accumulates in a PBP1b mutant.
In wild-type membranes incubated with UDP-MurNAc(pp) and UDP-[3H]GlcNAc, the major product formed was peptidoglycan, as shown by paper chromatography; negligible quantities of lipid II were formed (5% of the quantity of peptidoglycan) (Table 1). In contrast, in wild-type membranes incubated with moenomycin or in the PBP1b mutant membranes, the major product (>80%) was lipid II (Table 1), and very little peptidoglycan was formed.
The same conclusion can be drawn from the results of the lipid extraction, in which insignificant quantities of radioactive lipid were formed in wild-type membranes but significant quantities were formed in the presence of moenomycin or in the PBP1b mutant. We conclude that incubation of the E. coli PBP1b mutant membranes with the peptidoglycan sugar precursors results in synthesis of lipid II and that such a system can be used to monitor the MurG reaction; the same can be achieved by using wild-type membranes in the presence of moenomycin.
Capture of the reaction products by WGA-SPA beads in the presence of detergent reflects the quantity of cross-linked peptidoglycan synthesized (6) and indicates the formation of peptidoglycan in the wild-type membranes but not in the same membranes treated with moenomycin or in the PBP1b mutant membranes (Table 1). However, in the absence of detergent, the WGA-SPA beads captured the lipid II formed in membranes of the PBP1b mutant and in wild-type membranes in the presence of moenomycin. This suggested that the WGA-SPA beads could be used to develop a high-throughput assay for MurG and that it could replace analysis of lipid II using butanol extraction.
MurG assay.
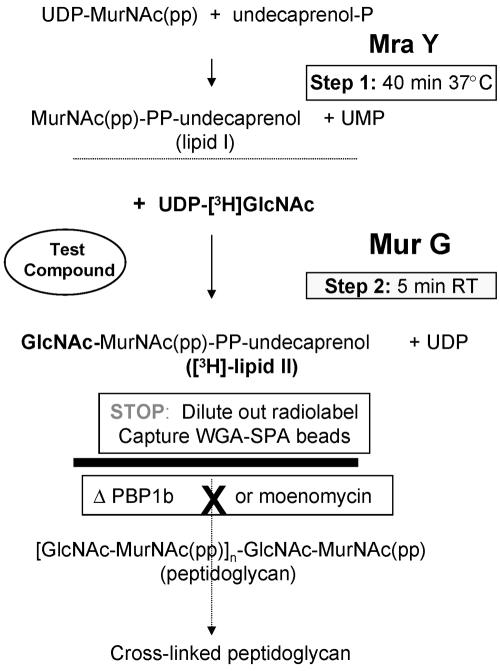
The lipid I substrate for MurG is difficult to isolate in large quantities, but it is easily made in situ, in the same membranes, by incubating them with UDP-MurNAc(pp) (Fig. 2, step1). In the second step, the MurG enzyme can be assayed by the addition of radiolabeled UDP-GlcNAc, as described earlier (24).
FIG. 2.
Schematic of MurG assay In the first step, by preincubating the membranes with UDP-MurNAc(pp), the MurG substrate, lipid I, is made by MraY present in the membranes. In the second step, the MurG reaction is initiated by providing its second substrate, UDP-[3H]GlcNAc. In a PBP1b mutant, or in the presence of moenomycin in wild-type membranes, lipid II is not converted to peptidoglycan and can be captured by WGA-SPA beads; under these conditions, the pathway of reactions stops at lipid II (solid line).
The traditional butanol extraction MurG assay was set up in membranes that had been treated with lysozyme (24). However, capture of lipid II by the addition of WGA-SPA beads to these membranes was not efficient. Hence, the MurG assay was developed using E. coli membranes prepared as described in Materials and Methods. Since the use of wild-type membranes requires the addition of moenomycin, a compound that was not commercially available, we focused on the assay using the PBP1b mutant membranes. A schematic of how this was accomplished is shown in Fig. 2.
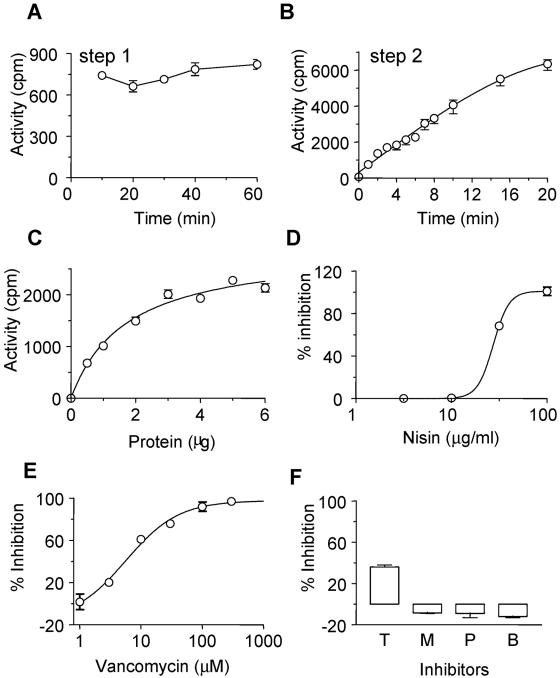
Using the PBP1b mutant membranes, the synthesis of lipid I at 37°C was studied. The incubation time for step 1 was varied, and the effect of this was monitored in terms of MurG activity in the second step, keeping the time for the MurG reaction (step 2) constant at 5 min (Fig. 3A). Synthesis of lipid I saturated within ∼10 min. However, for convenience while setting up a large number of reactions, the incubation time for this step was fixed at 40 min for routine assays.
FIG. 3.
Optimization of the MurG assay using AMA1004 ponB::Spcr membranes. (A) Time dependence. To determine the time course for lipid I formation, membranes (4 μg) were incubated at 37°C for different periods with UDP-MurNAc(pp). Following this, UDP-[3H]GlcNAc was added, and the MurG reaction mixture was incubated for 5 min at room temperature. (B) Time course. The MurG reaction was initiated by addition of UDP-[3H]GlcNAc, and the reaction was stopped at different times at room temperature; step1 was for 40 min at 37°C. (C) Protein dependence. The quantity of protein was varied under the standard conditions described for the assay. (D) Effect of nisin on MurG. (E) Effect of vancomycin on MurG. (F) Effects of 10 μg of tunicamycin (T)/ml, 1 μM moenomycin (M), 100 μM penicillin G (P), and 10 μM bacitracin (B), inhibitors of MraY, transglycosylase, transpeptidase, and lipid pyrophosphorylase, respectively. All values represent activity, i.e., after subtracting the blank values from those of the complete reactions. The error bars indicate standard errors of the means (SEM).
Subsequently, the first step was performed for 40 min at 37°C and the second step (Fig. 2) was performed for various times at room temperature (Fig. 3B). Based on the results, an incubation of 5 min at room temperature was chosen for the MurG assay. The quantity of membrane protein required to give maximum activity under the above-mentioned conditions was found to be ∼4 μg (Fig. 3C), and this was used in routine assays.
The MurG reaction was not completely stopped by the addition of 5 mM EDTA, in contrast to the peptidoglycan synthesis pathway of reactions (6). Hence, reactions were stopped by the addition of EDTA, along with an excess (∼100-fold) of cold UDP-GlcNAc; diluting out the specific activity of the radiolabel would essentially prevent further monitoring of the MurG reaction.
Using these reaction conditions, known inhibitors of MurG, as well as those of other enzymes in peptidoglycan synthesis, were tested to check the specificity of the reaction being measured. The inhibitors were added at the start of step 2, just before initiation of the MurG assay with UDP-[3H]GlcNAc. As expected, only inhibitors of MurG (nisin and vancomycin) showed inhibition, with 50% inhibitory concentrations (IC50s) of 34 ± 5 and 10 ± 4 μg/ml, respectively (Fig. 3D and E).
Bacitracin, moenomycin, and penicillin G, inhibitors of the lipids pyrophosphorylase, transglycosylase, and transpeptidase, respectively, had no effect (Fig. 3F); these were tested at concentrations of ∼10 times their IC50s in the peptidoglycan synthesis assay (6). Tunicamycin, an inhibitor of MraY, inhibited the MurG assay (∼40%) at 10 μg/ml. However, similar degrees of inhibition were observed at all concentrations of tunicamycin (0.03 to 30 μg/ml), i.e., no dose response was observed (data not shown). If, however, tunicamycin was also added to the blank reaction and each blank was subtracted from the corresponding complete reaction, no inhibition was observed. Tunicamycin inhibits transfer, by the wecA gene product, of UDP-GlcNAc to the lipid carrier in E. coli membranes (22). This reaction contributes to the background activity in the MurG assay, and hence, a blank lacking UDP-MurNAc(pp) is used. Thus, the inhibition of MurG by tunicamycin is an artifact of the effect of tunicamycin on the blank reaction and does not reflect its effect on MurG. We concluded that this assay specifically measures MurG activity.
MraY-MurG coupled assay.
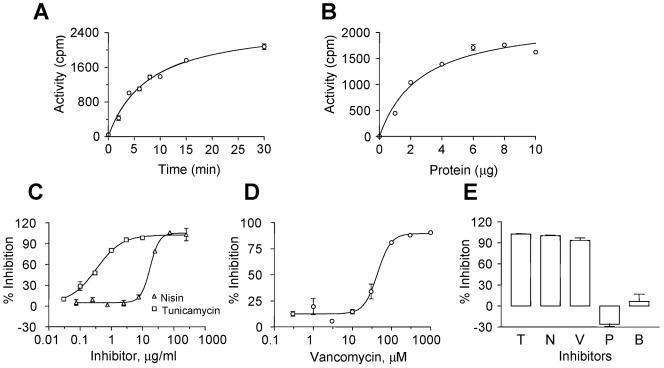
If desired, it is possible to measure MraY together with the MurG enzyme by simultaneously incubating both sugar precursors with membranes of E. coli AMA1004 ponB::Spcr. This assay was performed at 37°C, since the background reaction [in which UDP-MurNAc(pp) was omitted] was higher if the reaction was performed at room temperature. The effect of varying protein, using a 5-min reaction time (Fig. 4B), as well as varying the time of reactions containing 4 μg of protein (Fig. 4A), led to the use of 4 μg of protein and 5 min for screening of inhibitors. Unlike the MurG assay, the MraY-MurG coupled assay could be stopped by EDTA alone. The assay is more convenient to perform than the MurG assay, since both sugar precursors were added in a single step.
FIG. 4.
Optimization of the MraY-MurG coupled assay. (A to D) Membranes of E. coli AMA1004 ponB::Spcr were incubated with UDP-MurNAc(pp) and UDP-[3H]GlcNAc at 37°C. (A) Time dependence; 4 μg of membrane protein was used, and the reaction was terminated at different times. (B) Protein dependence; various quantities of membrane protein were used, and the reaction was terminated after 5 min. (C) Effects of tunicamycin (IC50, ∼0.3 μg/ml) and nisin (IC50, ∼16 μg/ml). (D) Effect of vancomycin (IC50, ∼9 μM). In panels C and D, the assay was performed under the standard conditions described in Materials and Methods. (E) MraY-MurG assay in wild-type membranes in the presence of 0.3 μM moenomycin. Shown are the effects of tunicamycin (T; 4 μg/ml), nisin (N; 312 μg/ml), vancomycin (V; 300 μM), penicillin G (P; 200 μM), and bacitracin (B; 4 U/ml). The error bars indicate SEM.
As expected, nisin and vancomycin also inhibited the MraY-MurG assay, with IC50s of ∼16 and ∼9 μM, respectively. In addition, tunicamycin inhibited the MraY-MurG assay, with an IC50 of ∼0.3 μg/ml (Fig. 4C and D).
Cephalosporin C.
While developing a test to differentiate β-lactam inhibition of peptidoglycan synthesis from that of other inhibitors in E. coli membranes (7), we noticed that cephalosporin C and cefsulodin behaved unlike the other β-lactams and more like inhibitors of MraY and MurG, i.e., they inhibited reactions captured by type A WGA-SPA beads (which most β-lactams did not). We interpreted this to mean that these two compounds inhibited MraY, MurG, or lipid pyrophosphorylase and prompted us to test these two compounds in the MraY-MurG assay.
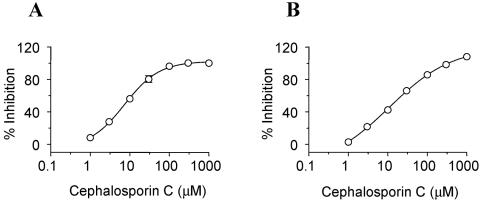
Cephalosporin C inhibited the MraY-MurG assay, with an IC50 of ∼6 ± 3 μM (Fig. 5A). To check which of the two enzymes was inhibited, we tested it in the specific MurG assay. Cephalosporin C inhibited the MurG assay, with an IC50 of 16 ± 5 μM (Fig. 5B). Cefsulodin, too, inhibited both the MraY-MurG and MurG assays, with IC50s of 11 ± 5 and ∼6 ± 3 μM, respectively, while it had no effect on the MraY assay (27). Since this β-lactam, in particular, is unstable in solution (12, 15), variable results may be obtained; DMSO solutions of cefsulodin were less potent than aqueous solutions. Hence, it is likely that inhibition was due to a breakdown product of the drug.
FIG. 5.
Effect of cephalosporin C. (A) MraY-MurG assay (IC50, ∼8 μM). (B) MurG assay (IC50, ∼14 μM). The assays were performed under standard conditions. The error bars indicate SEM.
The β-lactams are accepted as inhibitors of transpeptidase that act by covalently binding to the penicillin binding proteins, so it was surprising that cephalosporin C inhibited MurG. The major transglycosylase-transpeptidase activity measured in vitro under the conditions we used is that of PBP1b (6, 7). However, PBP1b is absent from the ponB::Spcr membranes used for the MurG assay, so the inhibition must be via another interaction.
Specificity of inhibition.
SPAs are subject to color artifacts, and colored compounds may appear as false positives in a screen. With such compounds, addition of a compound after the enzyme reaction will generate IC50 curves similar to those in which the compound is added before the start of the enzyme reaction; however, color quench programs associated with the counting instrument take care of this artifact. Another caution is that membranes are used as a source of the enzymes, so membrane-perturbing agents can interfere with the assay.
We developed a means to distinguish such inhibitors from genuine enzyme inhibitors. The principle is to test the effect of the compounds on an unrelated enzyme present in the same membrane preparation; compounds that inhibit due to membrane perturbation should show the same rank order of potency in the two unrelated enzyme assays.
Accordingly, we tested the effects of inhibitors on NADH dehydrogenase in the same membrane preparation. As expected, Triton X-100 inhibited the enzyme, with an IC50 of ∼0.005%, a value 10 times lower than its IC50 in the peptidoglycan synthesis assay (6). However, specific inhibitors of the enzymes of peptidoglycan synthesis, e.g., vancomycin and moenomycin, did not inhibit NADH dehydrogenase. Tunicamycin inhibited ∼50%, but at a concentration ∼100 times its IC50 for MraY.
Cephalosporin C also inhibited NADH dehydrogenase, with an IC50 of ∼8 μM; data from wild-type E. coli membranes are shown in Table 2, but similar results were obtained with membranes from AMA1004 ponB::Spcr. This is very similar to the concentration at which it inhibited MurG (IC50, ∼16 μM). Cefsulodin had no effect on NADH dehydrogenase. Thus, cephalosporin C appears to be a somewhat nonspecific inhibitor, a surprising observation for a β-lactam. The inhibition of NADH dehydrogenase and MurG could be due to membrane perturbation or a nonspecific interaction with both proteins. The interaction with MurG can be tested in the assay with the pure enzyme (18). It will be interesting to see if this compound inhibits all enzymes involved in peptidoglycan synthesis or any other membrane enzymes.
TABLE 2.
Effects of compounds on NADH dehydrogenasea
| Test compound | Concn | % Inhibition |
|---|---|---|
| Triton X-100 | 0.005% | 50 |
| Vancomycin | 100 μM | −1 |
| Moenomycin | 1 μM | 5 |
| Nisin | 100 μg/ml | 19 |
| Tunicamycin | 30 μg/ml | 47 |
| Penicillin | 100 μM | 13 |
| Cefsulodin | 100 μM | 13 |
| Cephalosporin C | 100 μM | 89 |
| Cephalosporin C | 30 μM | 76 |
Compounds were tested on E. coli AMA1004 membranes under the conditions described in the text. For the experiment with cefsulodin and cephalosporin C, 0.5 μg of membrane protein was used; for all others, the quantity of protein used was 2 μg.
MurG or MraY-MurG assays in wild-type membranes.
Both the MurG and MraY-MurG assays can be performed in wild-type membranes by using 0.3 μM moenomycin so that the transglycosylase is inhibited and further processing of the MurG product, lipid II, to peptidoglycan is prevented. All other reaction conditions, as well as the capture by WGA-SPA beads, were the same. Similar data were obtained (Fig. 4E). This method is advantageous if the assays need to be performed in a different bacterial strain or genetic background, if a mutant deficient in PBP1b is not available, or if the assay is to be set up in another bacterial species in which the genetic manipulation is not easy or the gene for the major transglycosylase activity has not been identified.
DISCUSSION
Many high-throughput assays that measure MurG together with MraY or as part of a cascade of other enzymes have been described, but these assays do not distinguish inhibitors of MurG from those of other enzymes (2, 6, 28) or require low-throughput methods, like thin-layer chromatography or paper chromatography, to do so (13). Here, we have described a high-throughput method to assay the activity of MurG specifically; in a modification, the activity of MraY can be coupled to that of MurG to allow selection of inhibitors of either enzyme simultaneously. To save on the cost of high-throughput screening, the MraY-MurG assay could be used for a primary screen, and inhibitors of MraY or MurG can be subsequently deconvoluted using the individual MurG or MraY assay (27). The MraY-MurG assay is easier to perform, since it has fewer steps, but both assays have a short incubation time, which is feasible, albeit not optimal, for a high-throughput format. Despite this, we were able to screen several 96-well plates per day; the assay is quite reproducible, giving a z′ value (31) between 0.6 and 0.8. We were also able to pick up an inhibitor (from our compound collection) with an IC50 of ∼20 μM that is competitive with UDP-GlcNAc; this compound did not inhibit NADH dehydrogenase.
Both the MurG and MraY-MurG assays have the advantages that they are easy to set up and do not require chemical synthesis of the lipid I substrate (1, 23, 26) nor its modification, e.g., labeling of lipid I with biotin to enable capture of lipid II (4). In addition, in the assays we describe, the MurG and MraY enzymes are associated with the membrane and are in the ratio present in native membranes, which is ideal for screening for inhibitors, since it most closely reflects the natural situation. However, in this assay system, the concentration of the lipid substrate cannot be controlled and mechanistic studies are not possible.
Much progress has been made with kinetic studies using purified MurG and synthetic analogs of lipid I and assays of MurG in solution phase using these artificial substrates (1, 8, 11, 23). These are artificial systems in which the natural substrate is not as effective an acceptor as the analogs. Unfortunately, unusual assay conditions may be required (e.g., 33% DMSO) (1), and the assays are not suitable for high-throughput screening. In addition, inhibitors of the enzyme in solution may not inhibit the membrane-associated enzyme (26), so assays that more closely resemble the native situation are more likely to pick up enzyme inhibitors that will have an MIC.
Recently, two elegant assays to measure the MraY or coupled MraY-MurG activities have been described. One uses wild-type membranes supplemented with decaprenol phosphate and phosphatidylglycerol, and the lipid product is captured using hydrophobic HP20ss beads (22). However, the beads have to be washed using a filter plate, which would lower the throughput. A second assay uses membranes overexpressing MraY together with purified recombinant MurG, undecaprenol phosphate, phosphatidyl glycerol, and the sugar precursors (30). Radiolabeled lipid II was monitored either by filtering the reaction mixture through an Immobilon-P membrane plate or by performing the reaction in the presence of WGA-coated SPA beads. This assay is similar to the one we describe here, except that the enzyme preparation is more tedious and it requires lipid supplements. However, specific measurement of MurG activity was not described for either of the above-mentioned assays.
A major problem with setting up a specific MurG assay is to prevent further conversion of the MurG product, lipid II, to peptidoglycan. Traditionally, using membranes isolated from spheroplasts prevented this (24). We have circumvented this problem by using moenomycin or membranes lacking PBP1b. In the other high-throughput assays reported, this is presumably prevented by the presence of detergent and the higher quantities of MraY and MurG relative to the quantity of PBPs (4, 22, 30).
We have shown inhibition of MurG by vancomycin. Given its mode of action—binding to the terminal region of the stem peptide—it would be expected to inhibit MurG, but it is surprising that there are no reports showing this. One reason could be the relatively large quantity of enzyme (and hence preformed peptidoglycan) that is used in those assays. In addition, the IC50 of vancomycin is influenced by the quantity of UDP-MurNAc(pp) used in the assay, since it binds to vancomycin and neutralizes its inhibitory effect.
We report novel inhibitory activities of cephalosporin C. The inhibition of MurG and NADH dehydrogenase could be a secondary mechanism by which this compound inhibits bacterial growth. Although many inhibitors in the peptidoglycan synthesis pathway act on more than one enzyme (e.g., vancomycin inhibits MraY [27], MurG [25], and the transglycosylase and the transpeptidase [16]), the β-lactams are thought of as specific inhibitors of the transpeptidase activity of the PBPs. There is, however, one report that imipenem, besides inhibiting the transpeptidase activities of many PBPs, also inhibited the transglycosylase activity of PBP1a, although there, too, the mechanism was not understood (19). Since both the transpeptidase and transglycosylase activities are on the same polypeptide, inhibition of the transglycosylase by a transpeptidase inhibitor can be interpreted as steric hindrance; it is more difficult to explain the inhibition of MurG or that of NADH dehydrogenase.
There are two approaches to finding new antibacterial drugs. One is to screen compounds for their MICs for the target bacterium, and the second is to find inhibitors of an essential enzyme and improve the potencies of such inhibitors to obtain antibacterial activity. In the first approach, one starts with compounds that have an MIC, improvement of the MIC is largely empirical, and it is often difficult to prove the molecular target of such compounds, especially when there are multiple targets in the cell. In the second approach, the crystal structure of the target protein and cocrystals with the inhibitor can be used to improve the potency of inhibition of the target protein, but achieving an MIC is a major challenge. We have described assays for MurG and MraY-MurG that can be used in the second approach. Although both enzymes are essential, inhibition of either by a compound, even if very potent, does not guarantee that it will have an MIC; even if it does, the MIC may not be due to inhibition of MurG-MraY. Vancomycin and cephalosporin C inhibit other enzymes in the cell, e.g., the transglycosylase and transpeptidase activities of the PBPs, and it is generally believed that their antibacterial activities are due to inhibition of these two enzymes, respectively. Our assay reveals that these compounds inhibit MraY-MurG in addition to the transglycosylase and transpeptidase, which earlier assay methods did not reveal. This inhibition of MraY-MurG is insufficient to claim that it is the mechanism by which these compounds kill bacteria, and we have no evidence to indicate this. In fact, vancomycin has no MIC for wild-type E. coli, but it does for an envA mutant of E. coli (data not shown). Studies of a different nature are required to prove that the MIC is due to inhibition of a particular molecular target, e.g., the mapping of mutations that cause resistance to the compound or an increase in the MIC for a strain overexpressing the specific protein target.
In summary, we have developed high-throughput assays for the MurG or coupled MraY-MurG enzymes that can be used to discover novel antibacterial agents.
Acknowledgments
The first hint that cefsulodin and cephalosporin C inhibited MraY, MurG, or lipid pyrophosphorylase came from experiments done by R. K. Shandil and B. Chandrakala. We thank Noel de Souza, formerly of Hoechst, India, for the gift of flavomycin. We thank R. K. Roy and Anand Kumar for critical comments on the manuscript.
REFERENCES
- 1.Auger, G., J. van Heijenoort, D. Mengin-Lecreulx, and D. Blanot. 2003. A MurG assay which utilizes a synthetic analog of lipid I. FEMS Microbiol. Lett. 219:115-119. [DOI] [PubMed] [Google Scholar]
- 2.Barbosa, M. D. F. S., H. O. Ross, M. C. Hillman, R. P. Meade, M. G. Kurilla, and D. L. Pompliano. 2002. A multitarget assay for inhibitors of membrane-associated steps of peptidoglycan biosynthesis. Anal. Biochem. 306:17-22. [DOI] [PubMed] [Google Scholar]
- 3.Brandish, P. E., M. K. Burnham, J. T. Lonsdale, R. Southgate, M. Inukai, and T. D. H. Bugg. 1996. Slow binding inhibition of phospho-N-acetylmuramyl-pentapeptide-translocase (Escherichia coli) by mureidomycin A. J. Biol. Chem. 271:7609-7614. [DOI] [PubMed] [Google Scholar]
- 4.Branstrom, A. A., S. Midha, C. B. Longley, K. Han, E. R. Baizman, and H. R. Axelrod. 2000. Assay for identification of inhibitors for bacterial MraY translocase or MurG transferase. Anal. Biochem. 280:315-319. [DOI] [PubMed] [Google Scholar]
- 5.Bupp, K., and J. van Heijenoort. 1993. The final step of peptidoglycan subunit assembly in Escherichia coli occurs in the cytoplasm. J. Bacteriol. 175:1841-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandrakala, B., B. C. Elias, U. Mehra, N. S. Umapathy, P. Dwarakanath, T. S. Balganesh, and S. M. de Sousa. 2001. Novel scintillation proximity assay for measuring membrane-associated steps of peptidoglycan biosynthesis in Escherichia coli. Antimicrob. Agents Chemother. 45:768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandrakala, B., R. K. Shandil, U. Mehra, S. Ravishankar, P. Kaur, V. Usha, B. Joe, and S. M. de Sousa. 2004. High-throughput screen for inhibitors of transglycosylase and/or transpeptidase activities of Escherichia coli penicillin binding protein 1b. Antimicrob. Agents Chemother. 48:30-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, L., H. Men, S. Ha, X. Y. Ye, L. Brunner, Y. Hu, and S. Walker. 2002. Intrinsic lipid preferences and kinetic mechanism of Escherichia coli MurG. Biochemistry 41:6824-6833. [DOI] [PubMed] [Google Scholar]
- 9.Cook, N. D. 1996. Scintillation proximity assay: a versatile high throughput screening technology. Drug Discov. Today 1:87-294. [Google Scholar]
- 10.Crouvoisier, M., D. Mengin-Lecreulx, and J. van Heijenoort. 1999. UDP-N-acetylglucosamine:N-acetylmuramoyl-(pentapeptide) pyrophosphoryl undecaprenol N-acetylglucosamine transferase from Escherichia coli: overproduction, solubilization, and purification. FEBS Lett. 449:289-292. [DOI] [PubMed] [Google Scholar]
- 11.Cudic, P., D. C. Behanna, M. K. Yu, R. G. Kruger, L. M. Szewczuk, and D. G. McCafferty. 2001. Synthesis of P1-citronellyl-P2-a-D-pyranosyl pyrophosphates as potential substrates for the E. coli undecaprenyl-pyrophosphoryl-N-acetylglucosaminyl transferase MurG. Bioorg. Med. Chem. Lett. 11:3107-3110. [DOI] [PubMed] [Google Scholar]
- 12.Das Gupta, V., and K. R. Stewart. 1981. Stability of cefsulodin in aqueous buffered solutions and some intravenous admixtures. J. Clin. Hosp. Pharm. 9:21-27. [DOI] [PubMed] [Google Scholar]
- 13.deCenzo, M., M. Kuranda, S. Cohen, J. Babiak, Z.-D. Jiang, D. Sun, M. Hickey, P. Sanc, P. A. Bradford, P. Youngman, S. Projan, and D. M. Rothstein. 2002. Identification of compounds that inhibit the late stages of peptidoglycan synthesis in bacteria. J. Antibiot. (Tokyo) 55:288-295. [DOI] [PubMed] [Google Scholar]
- 14.den Blaauwen, T., M. Aarsman, and N. Nanninga. 1990. Interaction of monoclonal antibodies with the enzymatic domains of penicillin-binding protein 1b of Escherichia coli. J. Bacteriol. 172:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita, T., and A. Koshiro. 1984. Kinetics and mechanism of the degradation and epimerisation of sodium cefsulodin in aqueous solution. Chem. Pharm. Bull. 32:3651-3661. [DOI] [PubMed] [Google Scholar]
- 16.Ge, M., Z. Chen, H. R. Onishi, J. Kohler, L. L. Silver, R. Kerns, S. Fukuzawa, S. Thompson, and D. Kahne. 1999. Vancomycin derivatives that inhibit peptidoglycan synthesis without binding D-ala-D-ala. Science 284:507-511. [DOI] [PubMed] [Google Scholar]
- 17.Ha, S., D. Walker, Y. Shi, and S. Walker. 2000. The 1.9 Å crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis. Protein Sci. 9:1045-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ha, S., E. Chang, M.-C. Lo, H. Men, P. Park, M. Ge, and S. Walker. 1999. The kinetic characterization of Escherichia coli MurG using synthetic substrate analogues. J. Am. Chem. Soc. 121:8415-8426. [Google Scholar]
- 19.Hashizume, T., F. Ishino, J. Nakagawa, S. Tamaki, and M. Matsuhashi. 1984. Studies on the mechanism of action of imipenem (N-formimidoylthienamycin) in vitro: binding to the penicillin-binding proteins (PBPs) in Escherichia coli and Pseudomonas aeruginosa, and inhibition of enzyme activities due to the PBPs in E. coli. J. Antibiot. (Tokyo) 37:394-400. [DOI] [PubMed] [Google Scholar]
- 20.Helm, J. S., Y. Hu, L. Chen, B. Gross, and S. Walker. 2003. Identification of active-site inhibitors of MurG using a generalizable, high-throughput glycosyltransferase screen. J. Am. Chem. Soc. 125:11168-11169. [DOI] [PubMed] [Google Scholar]
- 21.Hu, Y., L. Chen, S. Ha, B. Gross, B. Falcone, D. Walker, M. Mokhtarzadeh, and S. Walker. 2003. Crystal structure of the MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases. Proc. Natl. Acad. Sci. USA 100:845-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyland, S. A., and M. S. Anderson. 2003. A high-throughput solid-phase extraction assay capable of measuring diverse polyprenyl phosphate:sugar-1-phosphate transferases as exemplified by the WecA, MraY, and MurG proteins. Anal. Biochem. 317:156-165. [DOI] [PubMed] [Google Scholar]
- 23.Men, H., P. Park, M. Ge, and S. Walker. 1998. Substrate synthesis and activity assay for MurG. J. Am. Chem. Soc. 120:2484-2485. [Google Scholar]
- 24.Mengin-Lecreulx, D., L. Texier, M. Rousseau, and J. van Heijenoort. 1991. The murG gene of Escherichia coli codes for the UDP-N-acetylglucosamine:N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. J. Bacteriol. 173:4625-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravishankar, S., V. P. Kumar, B. Chandrakala, S. M. Solapure, and S. M. de Sousa. 2003. Scintillation proximity assay for inhibitors of Escherichia coli MurG and, optionally, the MraY as well. 43rd Intersci. Conf. Antimicrob. Agents Chemother., poster F-1454. [DOI] [PMC free article] [PubMed]
- 26.Silva, D. J., C. L. Bowe, A. A. Branstrom, E. R. Baizman, and M. J. Sofia. 2000. Synthesis and biological evaluation of analogues of bacterial lipid I. Bioorg. Med. Chem. Lett. 10:2811-2813. [DOI] [PubMed] [Google Scholar]
- 27.Solapure, S. M., C. N. Gayathri, K. Das, B. Chandrakala, and S. M. de Sousa. 2003. High-throughput screen for inhibitors of E. coli MraY. 43rd Intersci. Conf. Antimicrob. Agents Chemother., poster F-1455.
- 28.Sun, D., S. Cohen, N. Mani, C. Murphy, and D. M. Rothstein. 2002. A pathway-specific cell based screening system to detect bacterial cell wall inhibitors. J. Antibiot. 55:279-287. [DOI] [PubMed] [Google Scholar]
- 29.Yousif, S. Y., J. K. Broome-Smith, and B. G. Spratt. 1985. Lysis of Escherichia coli by beta-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J. Gen. Microbiol. 131:2839-2845. [DOI] [PubMed] [Google Scholar]
- 30.Zawadzke, L. E., P. Wu, L. Cook, L. Fan, M. Casperson, M. Kishnani, D. Calambur, S. J. Hofstead, and R. Padmanabha. 2003. Targeting the MraY and MurG bacterial enzymes for antimicrobial therapeutic intervention. Anal. Biochem. 314:243-252. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, J.-H., T. D. Y. Chung, and K. R. Oldenburg. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67-73. [DOI] [PubMed] [Google Scholar]