Abstract
A new natural TEM derivative with extended-spectrum β-lactamase activity, TEM-134, was identified in a ceftazidime-resistant clinical isolate of Citrobacter koseri. Compared to TEM-1, TEM-134 contains the following mutations: Q39K, E104K, R164H, and G238S. The blaTEM-134 gene was not transferable by conjugation and, apparently, was chromosomally encoded. Expression studies with Escherichia coli revealed efficient cefotaximase and ceftazidimase activity for TEM-134.
Class A β-lactamases are the most widespread plasmid-encoded β-lactamases among gram-negative pathogens (3). The extensive use of expanded-spectrum β-lactams in clinical practice has selected for a plethora of natural derivatives of the older plasmid-encoded β-lactamases, namely TEM-1/2 and SHV-1, with an extended-spectrum activity toward the oxyimino-cephalosporins and monobactams (2). These extended-spectrum β-lactamases (ESBLs) are now widespread in members of the family Enterobacteriaceae circulating in hospital settings on a worldwide scale (2, 11). Overall, the TEM- and SHV-type derivatives are the most prevalent acquired ESBLs among Enterobacteriaceae, although other plasmid-mediated ESBLs, such as those of the CTX-M type, may prevail in some areas (1, 2, 13).
The original TEM-1 and TEM-2 enzymes are those from which the largest number of variants with ESBL activity have been generated (K. Bush and G. A. Jacoby, 2004 [http://www.lahey.org/studies/webt.htm]), suggesting a greater evolutionary plasticity for these enzymes.
In 2003, an Italian nationwide survey on ESBL production in Enterobacteriaceae was undertaken to compare the epidemiological situation to that observed in a previous survey (12, 16). In this work we describe the identification of a new natural TEM-type derivative with ESBL activity, named TEM-134, from a Citrobacter koseri clinical isolate collected during the 2003 survey.
C. koseri VA-535/03 was isolated from the urine of a patient admitted at the emergency room of the Varese University Hospital (northern Italy) in July 2003. The isolate was identified by using the Phoenix System (Becton Dickinson, Milan, Italy). Susceptibility testing was carried out with E-test (AB Biodisk, Solna, Sweden). The isolate was resistant to penicillins, narrow-spectrum cephalosporins, ceftazidime, and fluoroquinolones while retaining susceptibility to penicillins plus β-lactamase inhibitor combinations, imipenem, cefepime, cefotaxime, and aztreonam (although MICs of cefotaxime and aztreonam were >1 μg/ml) (Table 1 and data not shown). The ESBL phenotypic confirmatory test recommended by the National Committee for Clinical Laboratory Standards for Klebsiella spp. and Escherichia coli (8), based on cefotaxime-clavulanate and ceftadizime-clavulanate disks, was positive (>5-mm increase of the inhibitory zone diameter in the presence of the inhibitor) with either β-lactam. A double-disk test, carried out as described previously (16), revealed synergy between clavulanate and cefotaxime, ceftazidime, ceftriaxone, or aztreonam. Analytical isoelectric focusing of a crude extract of VA-535/03, carried out as described previously (9), revealed the presence of two β-lactamase bands of approximately pI 5.2 and 7.8, respectively (data not shown).
TABLE 1.
Antimicrobial susceptibilities of C. koseri VA535/03 and of E. coli HB101(pMGP-134), carrying the blaTEM-134 gene cloned into the plasmid vector pBC-SK
| Agenta | MIC (μg/ml) of agent for strain:
|
||
|---|---|---|---|
| VA-535/03 | HB101(pMGP-134)b | HB101(pBC-SK)d | |
| AMX | >128 | >64 | 4 |
| AMC | 8/4 | 64/32 | 4/2 |
| PIP | 64 | 32 | 0.5 |
| TZPe | 2/4 | 2/4 | 0.5/4 |
| CFZ | 128 | >64 | 2 |
| CTX | 4 | 32 | ≤0.125 |
| CAZ | 64 | >64 | ≤0.125 |
| ATM | 8 | 16 | ≤0.125 |
| IPM | 0.125 | 0.25 | ≤0.125 |
| FEP | 1 | 0.25 | ≤0.125 |
| CAR | NDc | >128 | 8 |
| TIC | ND | >256 | 4 |
AMX, amoxicillin; AMC, amoxicillin-clavulanic acid; PIP, piperacillin; TZP, piperacillin-tazobactam; CFZ, cefazolin; CTX, cefotaxime; CAZ, ceftazidime; ATM, aztreonam; IPM, imipenem; FEP, cefepime; CAR, carbenicillin; TIC, ticarcillin.
In pMGP-134, the cloned blaTEM-134 gene was oriented to be expressed under the control of the Plac promoter flanking the pBC-SK polylinker.
ND, not determined.
Shown for comparison.
Tazobactam at fixed concentration of 4 μg/ml.
DNA probes specific for blaTEM and blaSHV alleles, made of PCR products containing the entire blaTEM-1 or blaSHV-1 genes and labeled with 32P by the random priming technique, were used for hybridization experiments. In a colony blot experiment, carried out as described previously (16), the isolate yielded a positive hybridization signal with the blaTEM-1 probe but not with the blaSHV-1 probe. Amplification of the blaTEM allele by PCR was carried out from a single bacterial colony, using primers TEM/F (5′-GGGGGATCCATAAAATTCTTGAAGAC) and TEM/R (5′-GGGGGATCCTTACCAATGCTTAATCA), as described previously (12), using the Hot Start DNA polymerase (Eppendorf, Hamburg, Germany). Direct sequencing of the resulting 0.9-kb amplicon on both strands was carried out as described previously (12). The nucleotide sequence revealed a blaTEM allele encoding a variant that, compared to TEM-1, carried the following amino acid substitutions: Q39K, E104K, R164H, and G238S. This combination of point mutations was original, and the new variant was assigned the name TEM-134 (http://www.lahey.org/studies/webt.htm).
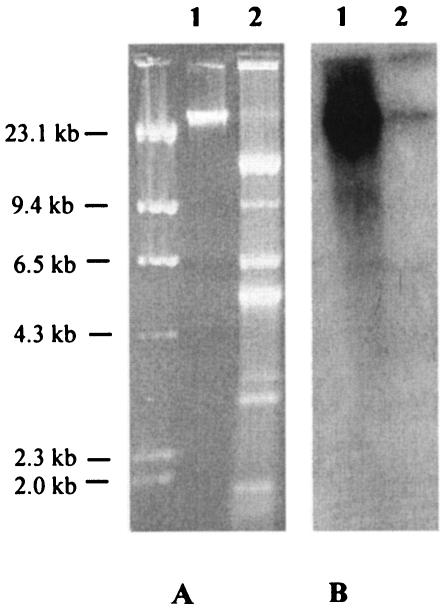
Plasmid DNA was extracted from C. koseri VA-535/03 by the alkaline lysis method (14). Chromosomal DNA was extracted as described previously (14). The plasmid DNA preparation revealed the presence of plasmid DNA (Fig. 1). Conjugation experiments were carried out in solid medium as described previously (10), using Escherichia coli MKD-135 (argH rpoB18 rpoB19 recA rpsL) as a recipient, and cefotaxime (2 μg/ml) or ampicillin (200 μg/ml) plus rifampin (300 μg/ml) for the selection of transconjugants. No conjugational transfer of cefotaxime resistance was observed in repeated experiments (under the adopted experimental conditions, the detection sensitivity of the assay was ≥5 × 10−7 transconjugants per recipient). Electroporation of the plasmid DNA preparation from VA-535/03 into the E. coli strain DH5α (Life Technologies, Milan, Italy), was carried out using a Bio-Rad Gene Pulser apparatus (Bio-Rad Laboratories, Richmond, Calif.) with approximately 500 ng of the plasmid DNA preparation and under the conditions recommended by the manufacturer. No transformants were obtained on medium containing either cefotaxime (2 μg/ml) or ampicillin (200 μg/ml).
FIG. 1.
(A) Agarose gel electrophoresis of a chromosomal DNA preparation (lane 1) and of a plasmid DNA preparation (lane 2) from the C. koseri isolate VA-535/03. (B) Results of a Southern blot analysis of the gel shown in section A, using the blaTEM probe. DNA size standards are shown on the left side.
In a Southern blot experiment, carried out directly on a dried gel as described previously (17), the blaTEM-1 probe yielded a strong hybridization signal in correspondence with the band of chromosomal DNA, while no hybridization signal was observed with bands of plasmid DNA (Fig. 1).
The blaTEM-134 gene, amplified by PCR as described above, was cloned in the BamHI site of the plasmid vector pBC-SK (Stratagene, Inc., La Jolla, Calif.) to obtain the recombinant plasmid pMGP-134. E. coli HB101(pMGP-134) was resistant or intermediate to penicillins, narrow- and expanded-spectrum cephalosporins (except cefepime), and aztreonam; tazobactam was able to restore susceptibility to piperacillin (Table 1). These results confirmed the ESBL activity of TEM-134 and indicated that the new enzyme exhibits notable activity against both ceftazidime and cefotaxime.
TEM-134 is a new natural TEM-type ESBL derivative carrying a unique combination of amino acid substitutions, Q39K, E104K, R164H, and G238S, which exhibits efficient activity against both ceftazidime and cefotaxime in agreement with the presence of the R164H and G238S mutations (4, 7). TEM-134 could be a derivative of either TEM-3 or TEM-16, by acquisition of the R164H or the G238S mutation, respectively. Since TEM-3 essentially behaves as a cefotaximase (15) while TEM-16 behaves as a ceftazidimase (5), in either case the evolution toward TEM-134 would result in a further broadening of the substrate specificity and could reflect an extensive usage of different types of extended-spectrum cephalosporins. Interestingly, TEM-134 is one of the very few examples of TEM-type ESBLs carrying mutations at both positions 164 and 238. The two other variants are TEM-8, found in Klebsiella pneumoniae (5), which is identical to TEM-134 except for the presence of a serine instead of a histidine residue at position 164, and TEM-107, found in K. pneumoniae, which compared to TEM-134 lacks the Q39K mutation but incorporates the M182T global suppressor (6). It would be interesting to understand why the combination of the R164H and G238S mutations has emerged so rarely in natural TEM-type variants.
Nucleotide sequence accession number.
The nucleotide sequence data for TEM-134 will appear in the GenBank database under accession number AY574271.
Acknowledgments
This work was supported by a research grant from Wyeth Pharmaceuticals (Italy).
REFERENCES
- 1.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, P. A. 2001. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantu, C., and T. Palzkill. 1998. The role of residue 238 of TEM-1 β-lactamase in the hydrolysis of extended-spectrum antibiotics. J. Biol. Chem. 273:26603-26609. [DOI] [PubMed] [Google Scholar]
- 5.Chanal, C. M., D. L. Sirot, A. Petit, R. Labia, A. Morand, J. L. Sirot, and R. A. Cluzel. 1989. Multiplicity of TEM-derived β-lactamases from Klebsiella pneumoniae strains isolated at the same hospital and relationships between the responsible plasmids. Antimicrob. Agents Chemother. 33:1915-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang, W., and T. Palzkill. 1997. A natural polymorphism in β-lactamase is a global suppressor. Proc. Natl. Acad. Sci. USA 94:8801-8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knox, J. R. 1995. Extended-spectrum and inhibitor-resistant TEM-type β-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob. Agents Chemother. 39:2593-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial susceptibility testing; 13th informational supplement. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 9.Oliva, B., M. C. Marinucci, G. Amicosante, A. Oratore, and P. M. Bennet. 1987. Citrobacter diversus ULA27 produces two forms of a chromosomal β-lactamase. J. Antimicrob. Chemother. 20:23-35. [DOI] [PubMed] [Google Scholar]
- 10.Pagani, L., E. Mantengoli, R. Migliavacca, E. Nucleo, S. Pollini, M. Spalla, R. Daturi, E. Romero, and G. M. Rossolini. 2004. Multifocal detection of multidrug-resistant Pseudomonas aeruginosa producing the PER-1 extended-spectrum β-lactamase in northern Italy. J. Clin. Microbiol. 42:2523-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patterson, D. L., K. M. Hujer, A. M. Hujer, B. Yeiser, L. B. Rice, and R. A. Bonomo. 2003. Extended-spectrum β-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV- and CTX-M-type beta-lactamases. Antimicrob. Agents Chemother. 47:3554-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perilli, M., E. Dell'Amico, B. Segatore, M. R. De Massis, C. Bianchi, F. Luzzaro, G. M. Rossolini, A. Toniolo, G. Nicoletti, and G. Amicosante. 2002. Molecular characterization of extended-spectrum β-lactamases produced by nosocomial isolates of Enterobacteriaceae from an Italian nationwide survey. J. Clin. Microbiol. 40:611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinteros, M., M. Radice, N. Gardella, M. M. Rodriguez, N Costa, D. Korbenfeld, E. Couto, G. Gutkind, and the Microbiology Study Group. 2003. Extended-spectrum β-lactamases in Enterobacteriaceae in Buenos Aires, Argentina, Public Hospitals. Antimicrob. Agents Chemother. 47:2864-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Sougakoff, W., S. Goussard, and P. Courvalin. 1988. The TEM-3 β-lactamase, which hydrolyzes broad-spectrum cephalosporins, is derived from the TEM-2 penicillinase by two amino acid substitutions. FEMS Microbiol. Lett. 56:343-348. [Google Scholar]
- 16.Spanu, T., F. Luzzaro, M. Perilli, G. Amicosante, A. Toniolo, G. Fadda, and The Italian ESβL Study Group. 2002. Occurrence of extended-spectrum β-lactamases in members of the family Enterobacteriaceae in Italy: implications for resistance to β-lactams and other antimicrobial drugs. Antimicrob. Agents Chemother. 46:196-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsao, S. G., C. F. Brunk, and R. E. Pearlman. 1983. Hybridization of nucleic acids directly in agarose gels. Anal. Biochem. 131:365-372. [DOI] [PubMed] [Google Scholar]