Abstract
The extent and efficacy of DNA end resection at DNA double-strand breaks (DSB) determine the repair pathway choice. Here we describe how the 53BP1-associated protein DYNLL1 works in tandem with the Shieldin complex to protect DNA ends. DYNLL1 is recruited to DSBs by 53BP1, where it limits end resection by binding and disrupting the MRE11 dimer. The Shieldin complex is recruited to a fraction of 53BP1-positive DSBs hours after DYNLL1, predominantly in G1 cells. Shieldin localization to DSBs depends on MRE11 activity and is regulated by the interaction of DYNLL1 with MRE11. BRCA1-deficient cells rendered resistant to PARP inhibitors by the loss of Shieldin proteins can be resensitized by the constitutive association of DYNLL1 with MRE11. These results define the temporal and functional dynamics of the 53BP1-centric DNA end resection factors in cells.
Resection of a broken DNA end plays a determinant role in the choice of DNA DSB repair pathways. Homologous recombination-mediated DSB repair occurs in the S phase and requires extended DNA end resection promoted by the tumor suppressor BRCA11. Minimal end resection facilitates nonhomologous end joining (NHEJ), an efficient pathway that remains active throughout the cell cycle2,3. Extensive work in the past two decades has defined the end resection machinery, specifically the large number of factors that process DNA ends, such as the exo- and endonucleases and the helicases that function therein4. In the last decade, 53BP1 has emerged as a focal antiresection factor that counters BRCA1 to favor NHEJ5–10. More recently, factors that functionally and biochemically associate with 53BP1 to restrict end resection have been discovered in genetic and biochemical screens11. These include the Shieldin complex12–14, composed of REV7, a known 53BP1-pathway component, and three uncharacterized proteins: C20orf196 (also known as SHLD1), FAM35A (SHLD2) and CTC-534A2.2 (SHLD3). Shieldin localizes to DSB sites in a 53BP1-dependent manner and directly binds to single-stranded DNA via the OB-fold domains of the SHLD2 subunit to protect DNA ends from hyper-resection12. Next, the CST complex (CTC1–STN1–TEN1) interacts with Shieldin components and localizes with Polα to sites of DNA damage in a 53BP1- and Shieldin-dependent manner to fill in resected DSBs and counteract end resection7,10. Depleting components of either the Shieldin or CST complexes promote resection and homologous recombination, thereby suppressing the sensitivity of BRCA1-null cells to PARP inhibitors (PARPi)7,10. The spatiotemporal dynamics of how these repair proteins function together to inhibit end resection remains to be determined.
We discovered that the cytoplasmic motor protein, DYNLL1, was an antiresection factor recruited to DSBs and directly bound the nuclease MRE11 to impede DNA end resection in vitro and in cells15. Very similar to the Shieldin and CST complex, loss of DYNLL1 conferred resistance to PARPi sensitivity in BRCA1-mutant cell lines. Several key questions emerged from this work: (1) how and when is DYNLL1 recruited to DSBs? (2) How does DYNLL1 impede the activity of MRE11? (3) What is the relative order of recruitment of DYNLL1 and the Shieldin complex? (4) Why does the cell use multiple independent complexes for its antiresection activity at DNA ends? Here we address these questions and define the dynamics of these end resecting factors and how they regulate DSB repair. We observed that DYNLL1 recruitment to DSBs depended solely on 53BP1 and no other known 53BP1-associated antiresection proteins. This is distinct from the Shieldin and CST complexes, which require REV7 and RIF1 proteins7,12. DYNLL1 localization at DNA lesions occurred concurrently with 53BP1 to block the initiation of DNA end resection. Biochemical mapping of the interaction domain of MRE11 with DYNLL1 combined with in vitro studies revealed that DYNLL1 interferes with the dimerization of MRE11 and potentially removes it from chromatin. The Shieldin complex localized to a subset of 53BP1-positive DSBs primarily in the G1 phase, hours after DNA damage. MRE11 and CtIP were necessary for Shieldin localization. In accordance with these findings, the loss of DYNLL1 promoted Shieldin recruitment to DSBs. Conversely, constitutive interaction of DYNLL1 with MRE11 impaired Shieldin recruitment to DSBs. Finally, PARPi resistance induced in BRCA1-mutant cells by depletion of Shieldin complex proteins was ‘rescued’ by the expression of a phosphomimetic DYNLL1 mutant that constitutively binds MRE11. Together, our results establish DYNLL1 as a partner of 53BP1 in the antiresection machinery that functions upstream of other 53BP1 interacting proteins to prevent DNA end resection.
RESULTS
DYNLL1 recruitment to DSBs is dependent on 53BP1
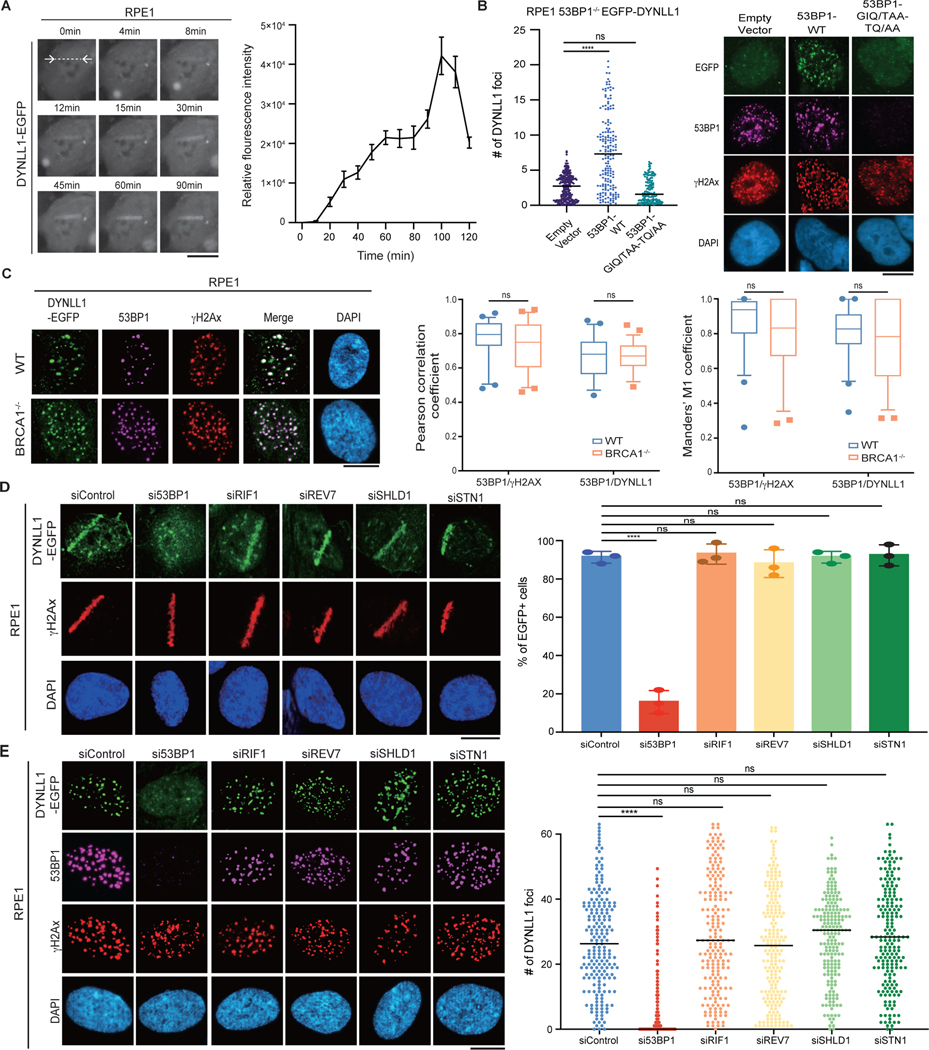
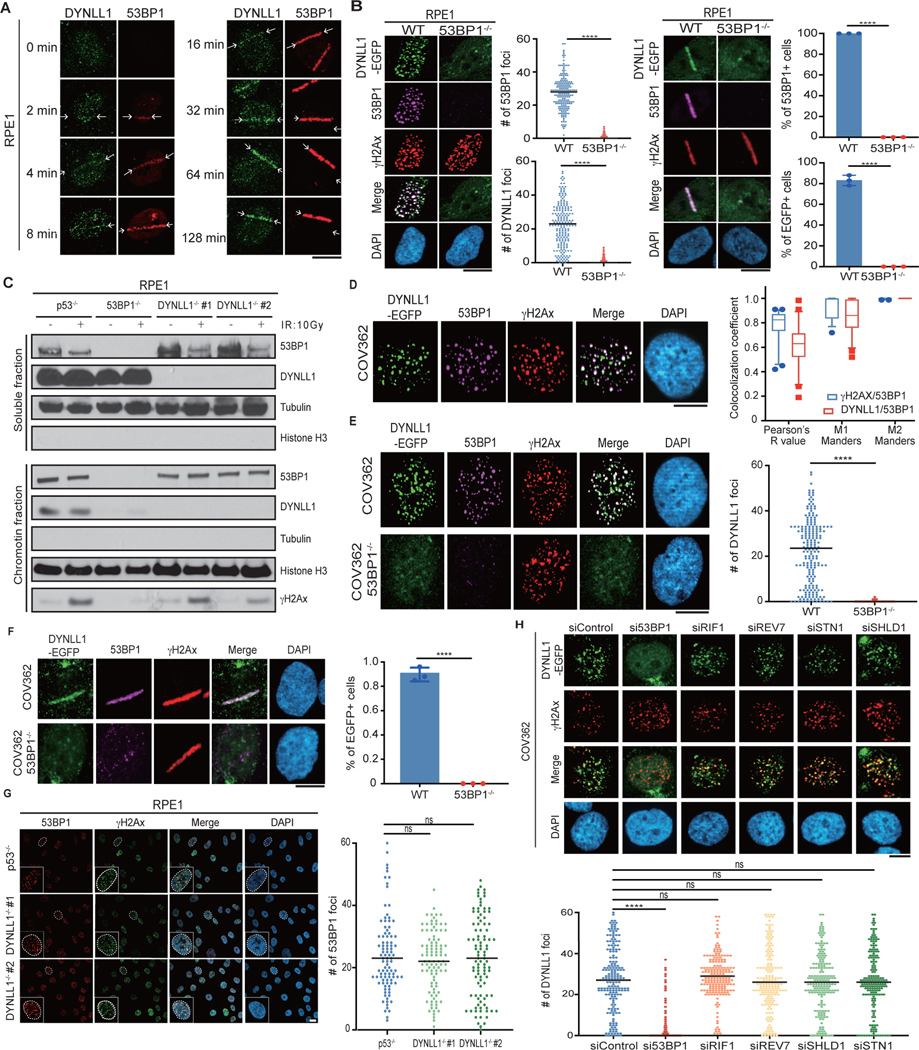
We observed that DYNLL1 is recruited to DSBs15. Using enhanced GFP (EGFP)-tagged DYNLL1 and laser microirradiation to induce DSBs, we determined that DYNLL1 localized to DNA lesions rapidly within minutes (Fig. 1a), with kinetics comparable to 53BP1 (Extended Data Fig. 1a). This is consistent with observations that DYNLL1 directly interacts with 53BP1 near the oligomerization domain (1,113–1,177 amino acids (aa))16–18. Therefore, the constitutive interaction with 53BP1 allows DYNLL1 to be recruited with 53BP1 to DSBs. Loss of 53BP1 strongly impaired recruitment of DYNLL1 to laser micro-irradiated or irradiation-induced DSBs (Extended Data Fig. 1b). Consistent with previous data, loss of 53BP1 also decreased DYNLL1 chromatin binding17 both in the presence and absence of damage (Extended Data Fig. 1c). Alanine substitution of the three anchor residues (GIQ and TQT) in both the DYNLL1-binding motifs of 53BP1 blocks oligomerization domain-independent 53BP1 localization18, thereby inhibiting DYNLL1 localization to chromatin17 and DYNLL1 recruitment to DSBs (Fig. 1b). This 53BP1 mutant localizes to DSBs and forms foci but does not recruit DYNLL1 (Fig. 1b). It is noteworthy that BRCA1 deficiency had no effect on DYNLL1 foci (Fig. 1c and Extended Data Fig. 1d–f). We also confirmed previous findings that the loss of DYNLL1 alone did not notably affect the recruitment of 53BP1 to DSBs15,18 (Extended Data Fig. 1g). The other 53BP1 interactors, RIF1, PTIP, and REV7, associate with 53BP1 after DNA damage-induced ATM-mediated phosphorylation of the N terminus11. Along with 53BP1, RIF1 and REV7 are necessary for recruitment of Shieldin complex to DSBs7,10,12,19. The Shieldin complex, specifically SHLD1, is required for the CST complex to be localized at DSBs7,10. Therefore, we asked whether any of these proteins play a role in the recruitment or retention of DYNLL1 to DSBs. While recruitment of DYNLL1 to DSBs was dependent on 53BP1, depletion of 53BP1-associated factors, RIF1, REV7, or components of the Shieldin and CST complexes did not affect DYNLL1 localization to DSBs (Fig. 1d,e and Extended Data Fig. 1h). From these results, we infer that recruitment of DYNLL1 to DSBs is solely dependent on 53BP1 and not the associated proteins.
Fig. 1 |. DYNLL1 recruitment to DSBs is dependent on 53BP1 but independent of other 53BP1-associated factors.
a, Representative live cell images of RPE1 cells expressing DYNLL1-EGFP after laser microirradiation. b, RPE1 53BP1−/− cells coexpressing EGFP-DYNLL1 and 53BP1–DYNLL1-binding mutants were subjected to 2 Gy of irradiation and 2 h later were processed for immunofluorescence. c, Representative immunofluorescent images of wild-type (WT) RPE1 or BRCA1−/− cells 2 h after exposure to 2 Gy of irradiation. Box plots show the mean, upper and lower quartiles (boxes) and range (whiskers). d,e, Representative images of RPE1 cells coexpressing EGFP-tagged DYNLL1 and short interfering RNA (siRNA) constructs and subjected to laser microirradiation (d) or 2 Gy of irradiation (e) and fixed 1 h later. siControl denotes siRNA against non-targeting control. In a–e, n = 3 biologically independent experiments, counting at least 100 cells per experiment. Error bars represent mean ± s.e.m. P values were determined by Mann–Whitney test (b, c, e) or two-sided unpaired t-tests (d). ****P < 0.0001.
NS, not significant (P < 0.05). Black lines in dot plots represent the median. DAPI, 4,6-diamidino-2-phenylindole. Scale bars, 20 μm.
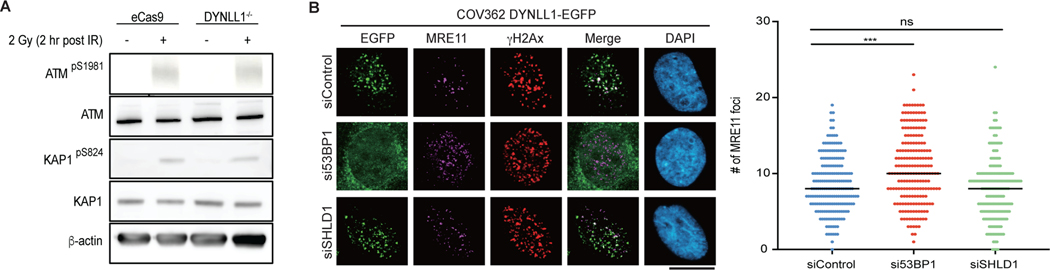
DYNLL1 regulates MRE11 activity independent of 53BP1
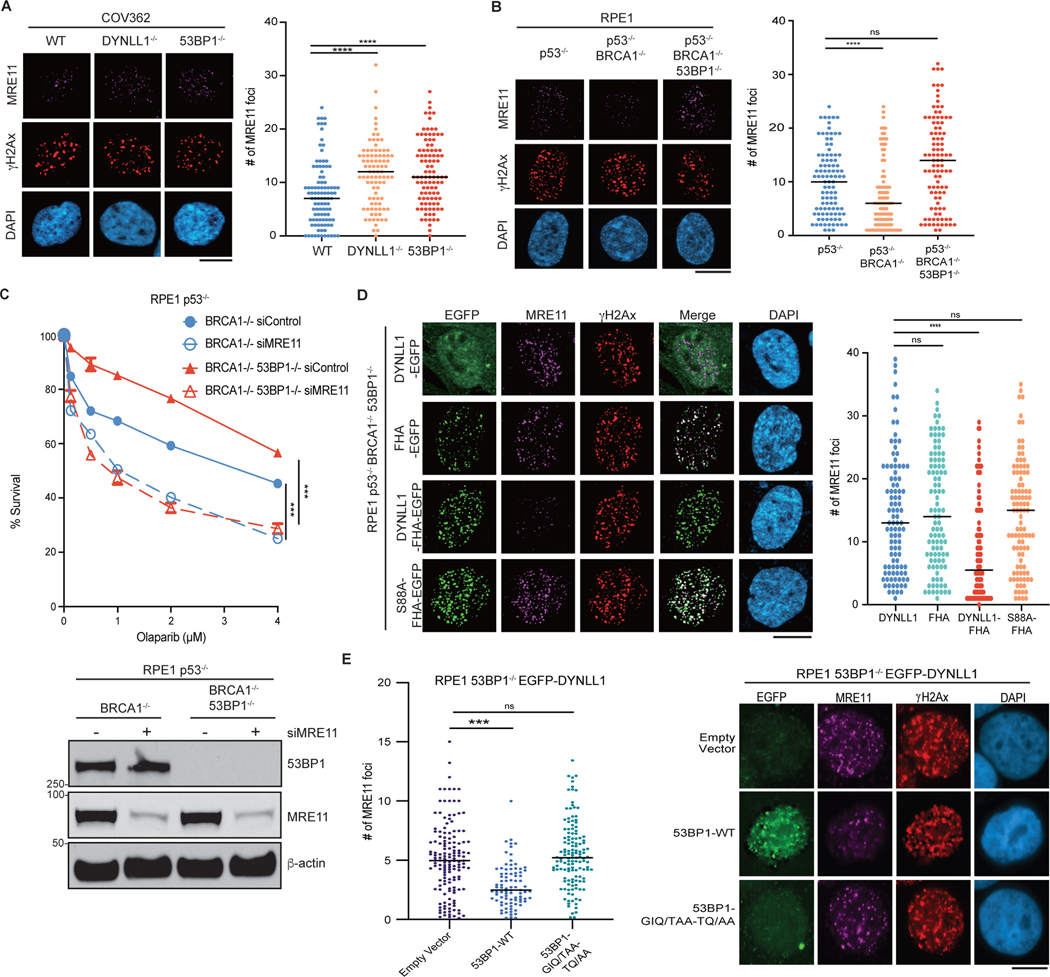
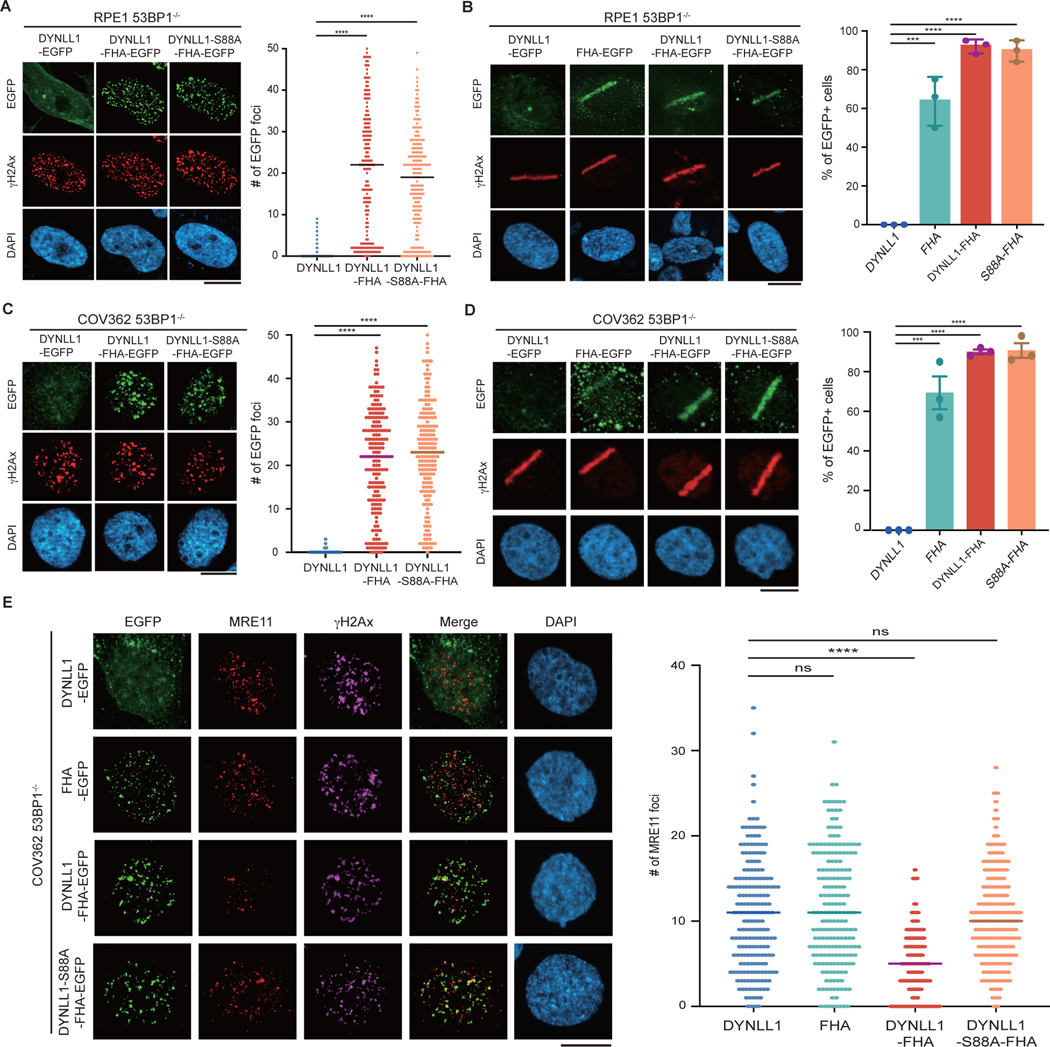
The factor 53BP1 influences DSB repair pathway choice and PARPi sensitivity by regulating DNA end resection at DSBs. In response to DNA damage, DYNLL1 loss increased MRE11 foci formation in BRCA1-deficient cells15 (Fig. 2a). Consistent with its effect on DYNLL1 foci formation, loss of 53BP1 also increased MRE11 foci formation in BRCA1-deficient and/or mutant cells (Fig. 2a). BRCA1 loss diminished MRE11 foci, and this was ‘rescued’ by the codeletion of 53BP1 (Fig. 2b). Next we depleted MRE11 to investigate its importance in the BRCA1–53BP1–DYNLL1 regulatory loop in the context of PARPi treatment. PARPi sensitivity in BRCA1−/− (or BRCA1 mutant) 53BP1−/− (53BP1 is also known as TP53BP1) cells was rescued on depletion of MRE11 (Fig. 2c). This suggested that MRE11 functioned downstream of 53BP1 in regulating DNA end resection. The depletion of MRE11 further sensitized BRCA1−/− cells to PARPi (Fig. 2c). The implication was that the effect of MRE11 on PARPi sensitivity is not limited to BRCA1 function. Therefore, we asked whether 53BP1 is required for the role of DYNLL1 in impeding MRE11 foci formation. To address this question, we fused the FHA domain from RNF8 to the C terminus of DYNLL112. This strategy has been used in multiple studies to direct proteins to DSBs circumventing other regulatory modules10,12. DYNLL1-EGFP–FHA localized to DNA lesions generated by either irradiation or laser microirradiation in BRCA proficient and BRCA1-mutant cell lines in 53BP1 depleted cells (Extended Data Fig. 2a–d). Compared to the expression of untethered DYNLL1-EGFP, expression of DYNLL1-EGFP–FHA substantially diminished MRE11 foci after irradiation-induced DNA damage in BRCA1-deficient cells depleted of 53BP1 or expressing the 53BP1–DYNLL1-binding mutant (Fig. 2d,e and Extended Data Fig. 2e). Additionally, expression of the FHA-tagged MRE11 binding mutant, DYNLL1-S88A, failed to suppress MRE11 foci formation after DNA damage (Fig. 2d and Extended Data Fig. 2e). Together, our results indicate that 53BP1 brings DYNLL1 to DSBs to interact with and block MRE11-dependent end resection. However, force tethering DYNLL1 to DSBs ‘rescues’ the DYNLL1–MRE11 interaction, thereby impeding MRE11 foci formation in the absence of 53BP1. Although this signaling cascade centers around 53BP1, it is distinct from the Shieldin–CST complexes.
Fig. 2: DYNLL1 regulates MRE11 activity independent of 53BP1.
a,b, COV362 cells depleted of DYNLL1 or 53BP1 (a) and RPE1 cells depleted of p53, BRCA1 and/or 53BP1 (b) using CRISPR–Cas9 were exposed to 2 Gy of irradiation for 2 h. Cells were fixed and immunostained using antibodies against MRE11 and γH2AX. c, RPE1 p53−/− BRCA1−/− and p53−/− BRCA1−/− 53BP1−/− cells were depleted of MRE11 using siRNA (siMRE11), and treated with indicated concentrations of olaparib for 6 days. Survival was determined via a cell viability assay (top), and a representative western blot shows successful knockdown of MRE11 using siRNA (bottom). d, Immunofluorescence of RPE1 p53−/− BRCA1−/− 53BP1−/− cells expressing EGFP-tagged DYNLL1 constructs exposed to 2 Gy of irradiation for 2 h, and stained using antibodies against MRE11, GFP (DYNLL1) and γH2AX.
e, Immunofluorescence of RPE1 53BP1−/− cells coexpressing EGFP-DYNLL1 and 53BP1–DYNLL1-binding mutants exposed to 2 Gy of irradiation for 2 h, and stained using antibodies against MRE11, 53BP1 and γH2AX. In a–e, n = 3 biologically independent experiments, counting at least 100 cells per experiment. P values determined by two-sided unpaired t-tests (a,b,d,e) or nonregression curve analysis (c). ***P < 0.001, ****P < 0.0001. Black lines in dot plots represent medians. Scale bars, 20 μm.
Functional effect of DYNLL1 at DSBs in 53BP1-deficient cells
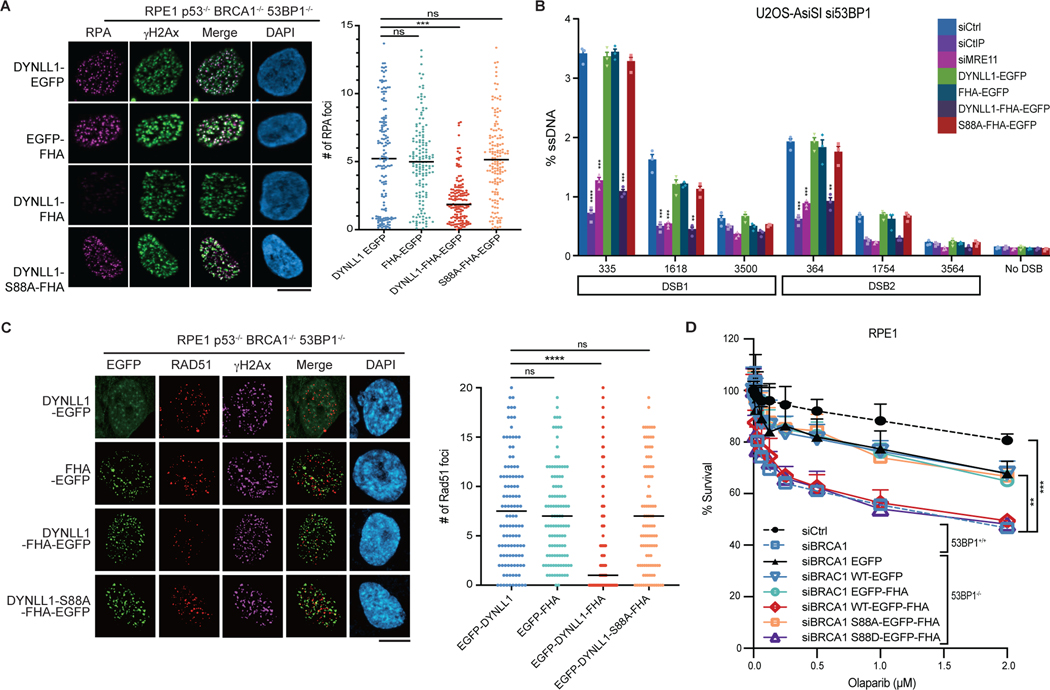
Replication protein A (RPA) directly participates in DSB repair by stimulating 5′–3′ end resection by the BLM helicase and DNA2 endonuclease20,21. To directly test the effect of DYNLL1 on the homologous recombination pathway independently of 53BP1, we evaluated RPA foci formation upon expression of DYNLL1–FHA. Expression of FHA–DYNLL1 in BRCA1−/− 53BP1−/− cells reduced the number of RPA foci (Fig. 3a). To directly evaluate the effect of DYNLL1 on end resection, we introduced the AsiSI endonuclease fused to estrogen receptor (ER-AsiSI)22 in BRCA1- and 53BP1-depleted cells and used a quantitative PCR-based method23,24 to measure single-stranded DNA (ssDNA). Upon expression of DYNLL1–FHA, we observed a decrease in ssDNA generated from two specific DSBs (Fig. 3b). Downstream of the DNA end-resecting step is the loading of RAD51 onto ssDNA, ultimately leading to successful homologous recombination-mediated DSB repair. BRCA1 loss impairs RAD51 loading, and concurrent loss of 53BP1 restores RAD51 foci6. Therefore, we evaluated RAD51 foci formation upon expression of DYNLL1–FHA. Expression of FHA–DYNLL1 in BRCA1−/− 53BP1−/− cells reduced the number of RAD51 foci (Fig. 3c and Extended Data Fig. 3a–c) and resensitized these cells to PARPi (Fig. 3d and Extended Data Fig. 3d) when compared to cells expressing untethered EGFP-DYNLL1. Together, these observations suggested that 53BP1 recruits DYNLL1 to DSBs to block MRE11 activity, and this step is sufficient to block homologous recombination-mediated DSB repair and sensitize BRCA1-mutant cells to PARPi.
Fig. 3: Functional effect of DYNLL1 at DSBs in 53BP1-deficient cells.
a, Immunofluorescence of RPE1 p53−/− BRCA1−/− 53BP1−/− cells expressing EGFP-tagged DYNLL1 or FHA–DYNNL1 constructs exposed to 2 Gy of irradiation for 4 h, and stained with, antibodies against GFP (DYNLL1), RPA32 and γH2AX. b, U2OS AsiSI cells cotransfected with siRNA against 53BP1 and EGFP-tagged DYNLL1 or FHA–DYNLL1 constructs were treated with 300 nM 4-OHT for 4 h, and ssDNA formation was determined at various sites downstream of the break via PCR quantification. siCtrl denotes non-targeting control siRNA; siCtIP denotes siRNA against CtIP (also known as RBBP8). c, Immunofluorescence of RPE1 p53−/− BRCA1−/− 53BP1−/− cells expressing EGFP-tagged DYNLL1 or FHA–DYNNL1 constructs exposed to 2 Gy of irradiation for 4 h. Cells were stained with antibodies against GFP (DYNLL1), RAD51 and γH2AX. d, RPE1 p53−/− 53BP1−/− cells were cotransfected with siRNA against BRCA1 (siBRCA1) and EGFP-tagged DYNLL1 or FHA–DYNLL1 constructs, and treated with indicated concentrations of olaparib for 6 days. Survival was determined via a cell viability assay. In a–d, n = 3 biologically independent experiments, counting at least 100 cells per experiment. Error bars represent mean ± s.e.m. P values were calculated using two-sided unpaired t-tests (a,c) or nonregression curve analysis (d). **P < 0.01, ***P < 0.001, ****P < 0.0001. Black lines in dot plots represent medians. Scale bars, 20 μm.
DYNLL1 disrupts MRE11 dimerization and its retention on DNA
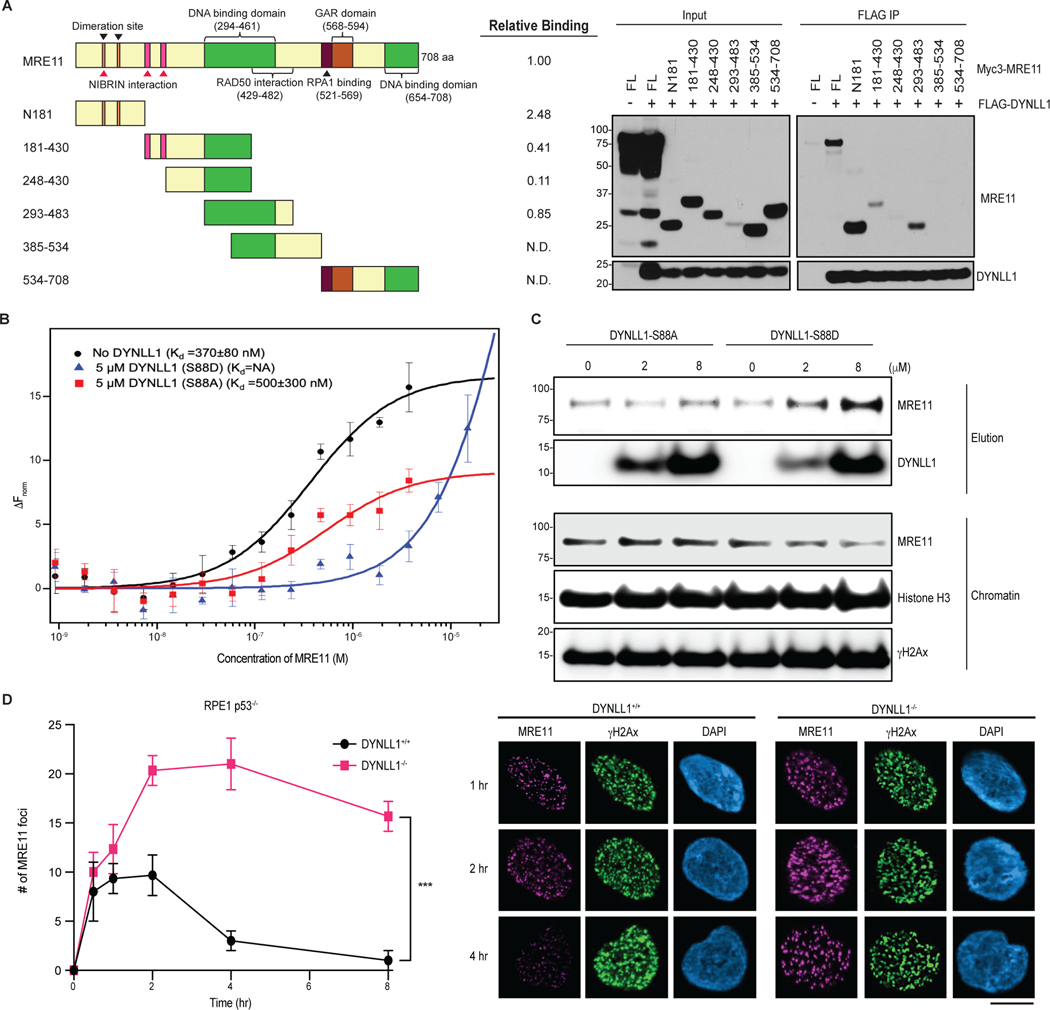
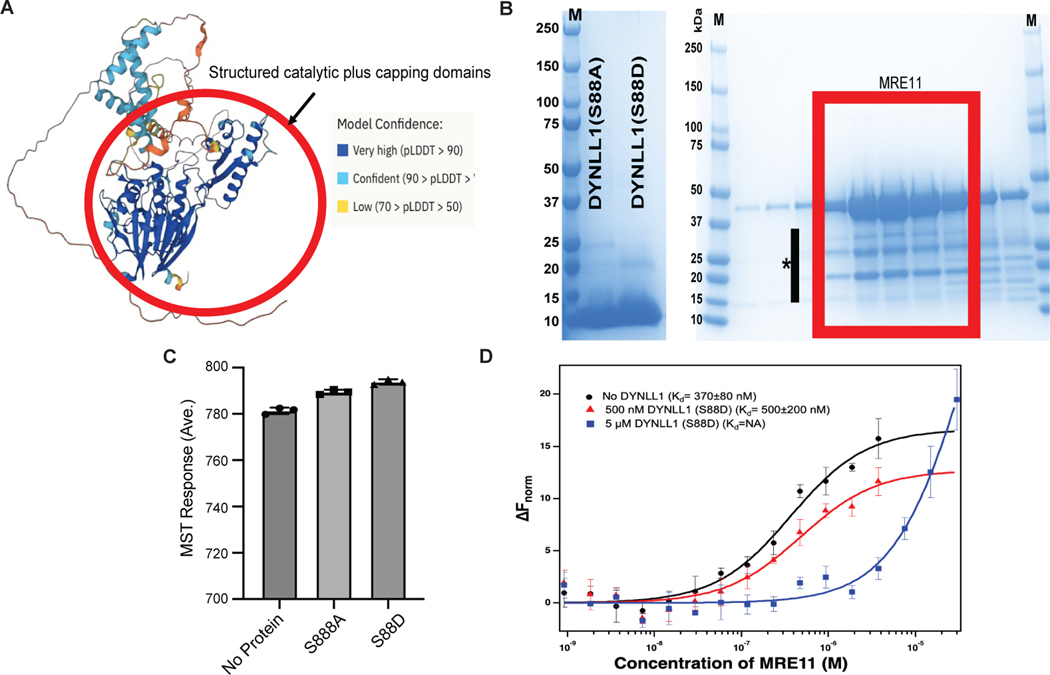
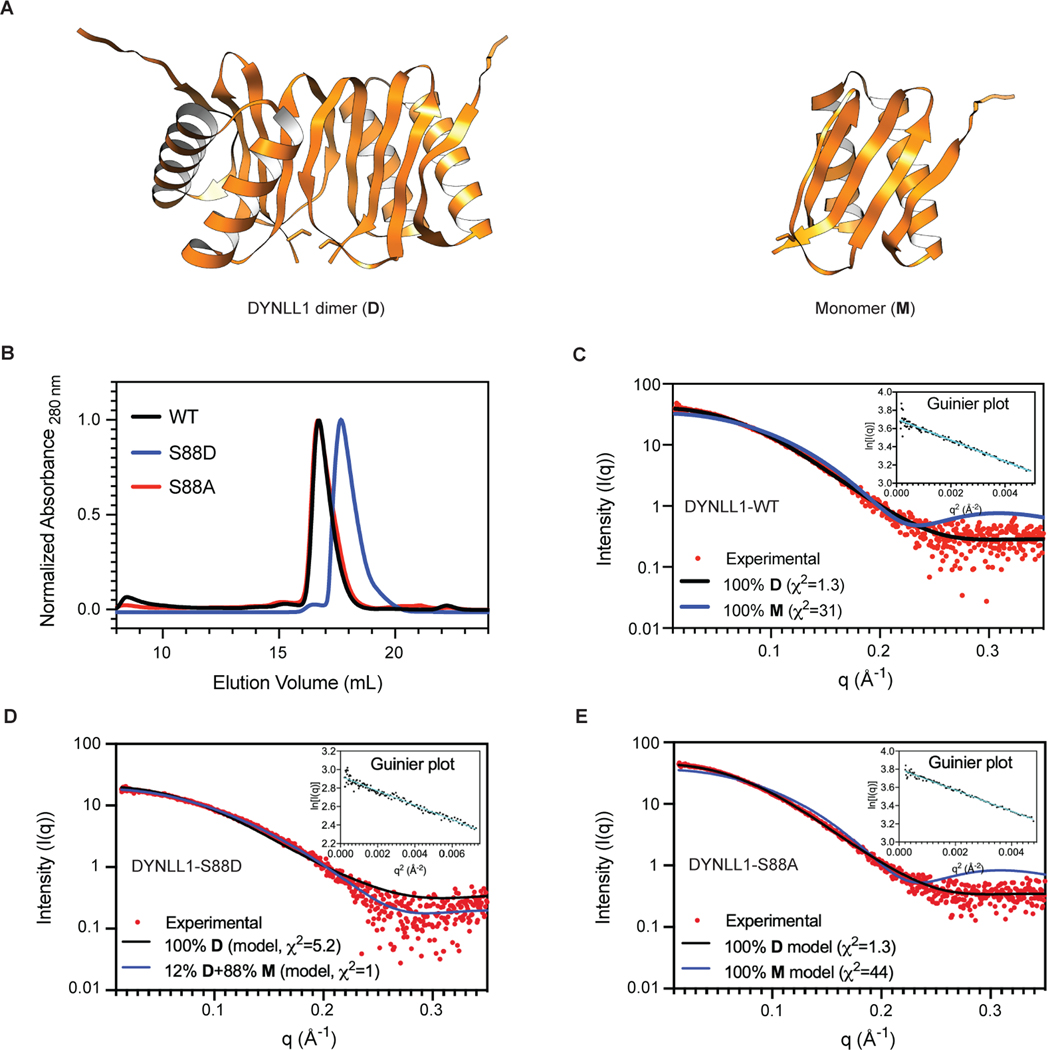
How does DYNLL1 inhibit MRE11 function? To address this question, we generated MRE11 truncation mutants and evaluated their interaction with DYNLL1 via immunoprecipitation (Fig. 4a). We have previously shown that DYNLL1 directly binds to MRE11 in vitro15. Two distinct fragments of MRE11 can bind DYNLL1 in cells. One is the N-terminal fragment (residues 1–181), whereas the other fragment (residues 293–483) encompasses a DNA-binding domain of MRE11. The N-terminal fragment of MRE11 resides in the structured-nuclease domain, which includes its dimerization domain, while the rest of MRE11 is largely disordered (AlphaFold Model-Extended Data Fig. 4a). MRE11 is functionally conserved across species25 and crystal structures of the nuclease domain of MRE11 protein from different species have confirmed that dimerization of MRE11 is evolutionarily conserved26–29. Since one of the DYNLL1-binding sites resides in the N-terminus domain of MRE11, we hypothesized that DYNLL1 affects MRE11 dimerization.
Fig. 4: DYNLL1 disrupts MRE11 dimerization to impair its retention on chromatin.
a, Left: schematic of MRE11 truncation mutants. Right: relative binding of MRE11 mutants and DYNLL1. Immunoprecipitation (IP) from cells coexpressing Flag–DYNLL1 and Myc3–MRE11 truncation mutants. ND, not detected. b, Change in the normalized fluorescence (ΔFnorm) as a result of thermophoresis in the MST experiment plotted as a function of the concentration of unlabeled MRE11. The resulting curves represent MRE11 dimerization in the absence of any DYNLL1 (black circles), or in the presence of 5 μM DYNLL1-S88D (blue triangles) or 5 μM DYNLL1-S88A (red squares). The dissociation constant (Kd) values are measured by fitting the curves with the Kd model in the analysis software. Data are mean and s.d. of three independent measurements. c, Western blot analysis of purified DYNLL1-S88D and DYNLL1-S88A proteins (Extended Data Fig. 4b) incubated with pre-extracted chromatin from HEK293T cells after 5 Gy of irradiation. Incubation of recombinant DYNLL1-S88D but not DYNLL1-S88A protein resulted in increased MRE11 elution from damaged chromatin. d, Immunofluorescence of DYNLL1+/+ and DYNLL1−/− cells exposed to 2 Gy of irradiation and fixed at various time points after irradiation. a–d, n = 3 biologically independent experiments, counting at least 100 cells per experiment. Error bars represent s.d. (b) or mean ± s.e.m. (d). P values determined by two-sided unpaired t-tests. ***P < 0.001. Scale bar, 20 μm.
To assess the effect of DYNLL1 on the dimerization of MRE11 we used microscale thermophoresis (MST)30. First, we used MST to determine the dimerization Kd of the nuclease domain of MRE11 by titrating unlabeled MRE11 into Alexa-647-labeled MRE11. In near-physiological buffer condition MRE11 forms a dimer with a dimerization Kd of 370 ± 80 nM. Next, we repeated MRE11-dimerization MST experiments in the presence of fixed concentrations of either DYNLL1-S88D or DYNLL1-S88A mutant in the MST buffer. In the absence of unlabeled MRE11, we see the effect of DYNLL1 binding to labeled MRE11 in the case of DYNLL1-S88D and to a lesser extent with DYNLL1-S88A (Extended Data Fig. 4c). Consistent with this, we observed that MRE11 can still dimerize in the presence of 5 μM DYNLL1-S88A in the MST buffer whereas MRE11 dimerization was almost completely abolished at the same concentration of DYNLL1-S88D (Fig. 4b). This is consistent with our observation that DYNLL1-S88A mutant does not bind MRE11 in cells and S88D binds more efficiently than wild-type DYNLL1. We also tested the tenfold lower concentration of DYNLL1-S88D in the MRE11-dimerization experiment and observed that 500 nM of DYNLL1-S88D is not effective in inhibiting MRE11 dimerization (Extended Data Fig. 4d). Size-exclusion chromatography (SEC) and SEC-coupled to small-angle X-ray scattering (SAXS) data showed that both wild-type protein and S88A form dimers in solution, whereas S88D mostly exists as monomer (Extended Data Fig. 5a–e). Therefore, we used DYNLL1-S88A as a proxy for wild-type protein in these experiments. Together, we concluded that phosphorylated DYNLL1 binds to MRE11 to disrupt its dimerization, which is critical for its function including localization to chromatin.
As a member of the MRN complex, MRE11 is one of the earliest responders to DSBs and is necessary for ATM activation and the expansion of γH2AX foci across megabases surrounding the break site3,31,32. The 53BP1-dependent recruitment of DYNLL1 to chromatin and/or DSBs is unlikely to precede MRE11 localization to the DNA repair foci. Therefore, we speculated that DYNLL1 is not preventing the recruitment of MRE11 to DSBs and not interfering with ATM activation. DYNLL1 is more likely to destabilize and remove MRE11 from chromatin by interfering with its dimerization. First, we ascertained that DYNLL1 loss does not affect ATM activation (Extended Data Fig. 6a) but leads to increased MRE11 in chromatin15. To directly test the effect of DYNLL1 on chromatinized MRE11, we isolated chromatin from human embryonic kidney 293T (HEK293T) cells after irradiation (5 Gy) and added recombinant DYNLL1-S88A or DYNLL1-S88D and evaluated MRE11 levels. We observed that DYNLL1-S88A had no effect, but the addition of DYNLL1-S88D induced a dose-dependent release of MRE11 from chromatin and an increase in the soluble fraction (Fig. 4c). To further validate that DYNLL1 does not inhibit MRE11 recruitment to DSBs, but evicts MRE11 from damaged chromatin, we evaluated MRE11 foci formation at irradiation-induced DSBs in wild-type and DYNLL1−/− cells. In wild-type cells, we observe rapid foci formation of MRE11 within 30 min and a reduction after 2 h, whereas in cells depleted of DYNLL1, we observe an increase in the number of MRE11 foci and also prolonged retention of MRE11 (Fig. 4d). Together with our in vitro data, we infer that DYNLL1 interferes with MRE11 dimerization and promotes its release from damaged chromatin.
Functional comparison of DYNLL1 and the Shieldin complex
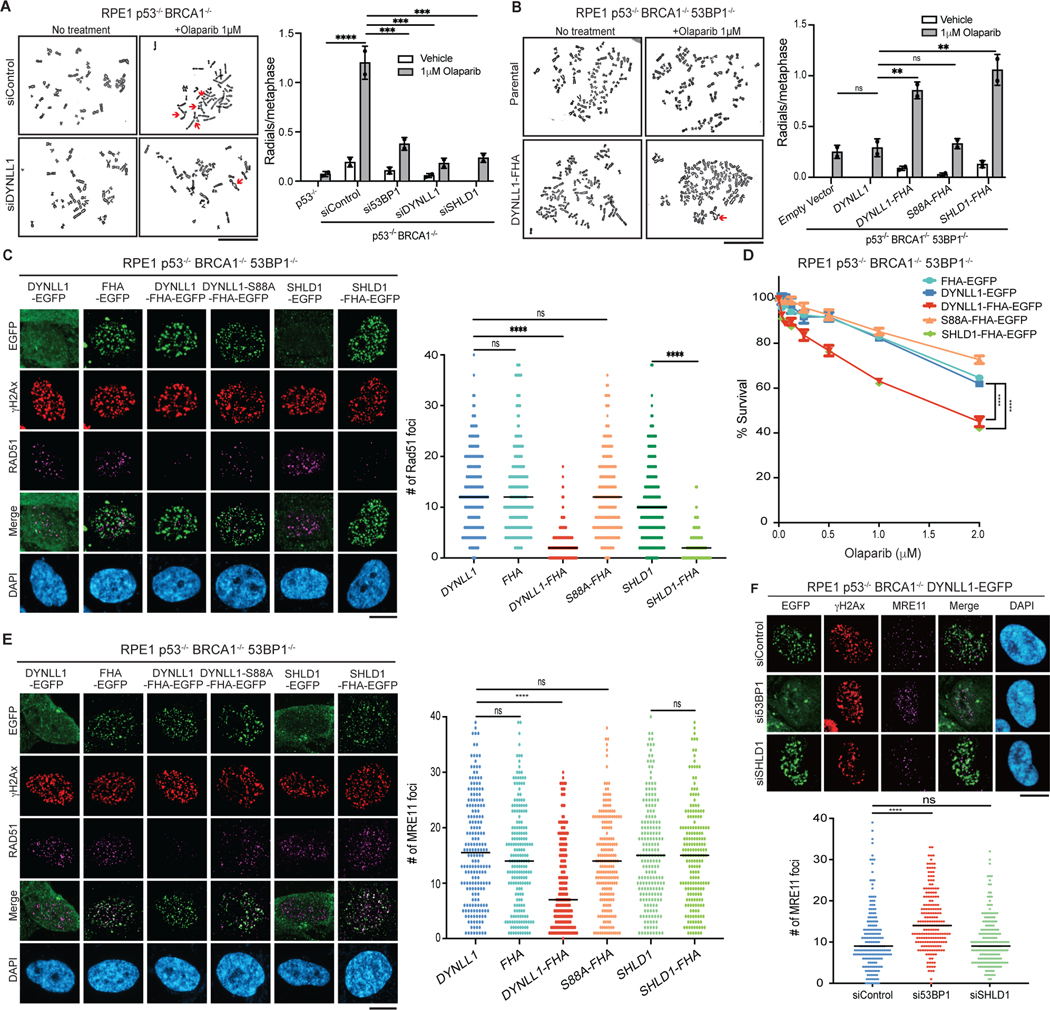
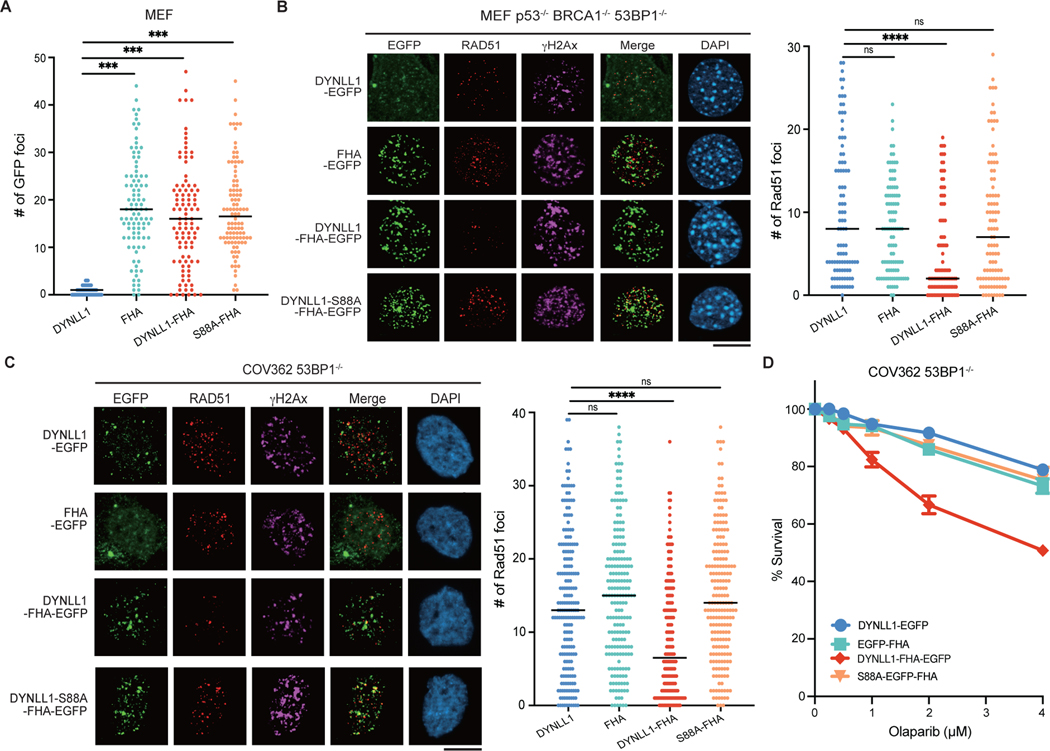
A key question is why cells require multiple and redundant methods to inhibit end resection downstream of 53BP1. We first questioned whether loss of either DYNLL1 or components of the Shieldin complex had similar functional consequences. Chromosomal aberrations, specifically radials, are a readout of the PARPi response33,34. We compared the effect of DYNLL1 and SHLD1 on radials in the context of PARPi exposure. Depletion of 53BP1, DYNLL1 or SHLD1 in BRCA1-deficient cells had a comparable effect and substantially decreased the number of radials per cell (Fig. 5a). Conversely, expression of DYNLL1–FHA and SHLD1–FHA in BRCA1−/− 53BP1−/− cells increased the number of radials per cell (Fig. 5b), diminished the number of RAD51 foci (Fig. 5c) and resensitized cells to PARPi (Fig. 5d). However, a notable difference was that the expression of DYNLL1–FHA reduced MRE11 foci, but SHLD1– FHA had no effect (Fig. 5e). Consistent with these results, depletion of SHLD1 also had no effect on MRE11 foci (Fig. 5f and Extended Data Fig. 6b). These results indicated that the Shieldin complex functions downstream of MRE11 and independently of RAD51 recruitment to impede DNA end resection.
Fig. 5: Functional comparison of DYNLL1 and the Shieldin complex.
a, RPE1 p53−/− BRCA1−/− cells depleted of 53BP1, DYNLL1 or SHLD1 were treated with olaparib. Representative images of metaphase spreads (left) and quantification of the number of redials per cell (right) are shown. b, RPE1 p53−/− BRCA1−/− 53BP1−/− cells expressing DYNLL1–FHA or SHLD1–FHA constructs were treated with olaparib. Representative images of metaphase spreads (left) and quantification of the number of radials per cell (right) are shown. c, Immunofluorescence of cells expressing EGFP-tagged DYNLL1, FHA–DYNLL1 or SHLD1–FHA constructs exposed to 2 Gy of irradiation for 4 h, and stained using antibodies against RAD51, GFP (DYNLL1) and γH2AX. d, RPE1 p53−/− BRCA1−/− 53BP1−/− cells expressing EGFP-tagged DYNLL1, FHA–DYNLL1 or SHLD1–FHA constructs were treated with indicated concentrations of olaparib for 6 days, and cell survival was determined via a cell viability assay. e, Immunofluorescence of cells expressing EGFP-tagged DYNLL1, FHA–DYNLL1, or SHLD1–FHA constructs exposed to 2 Gy of irradiation for 2 h, and stained using antibodies against MRE11, GFP (DYNLL1) and γH2AX. f, Immunofluorescence of RPE1 p53−/− BRCA1−/− 53BP1−/− cells coexpressing EGFP- tagged DYNLL1 and indicated siRNAs exposed to 2 Gy of irradiation for 2 h, and stained using antibodies against GFP, γH2AX and MRE11. n = 2 (a,b) or n = 3 (c–f) biologically independent experiments, counting at least 100 cells per experiment. Error bars represent mean ± s.e.m. P values were determined by two-sided unpaired t-tests (a–c,e,f) or nonregression curve analysis (d). **P < 0.01, ***P < 0.001, ****P < 0.0001. Black lines in dot plots represent medians. Scale bars, 20 μm.
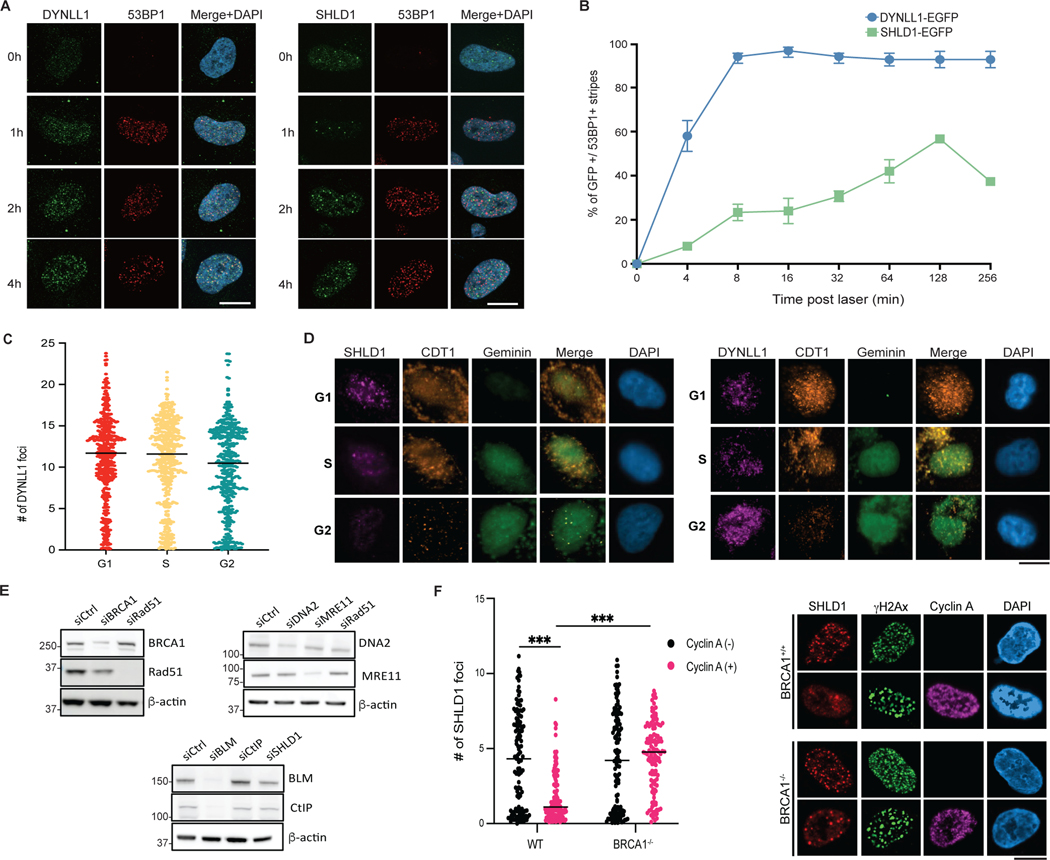
Kinetics and dependencies of Shieldin recruitment to DSBs
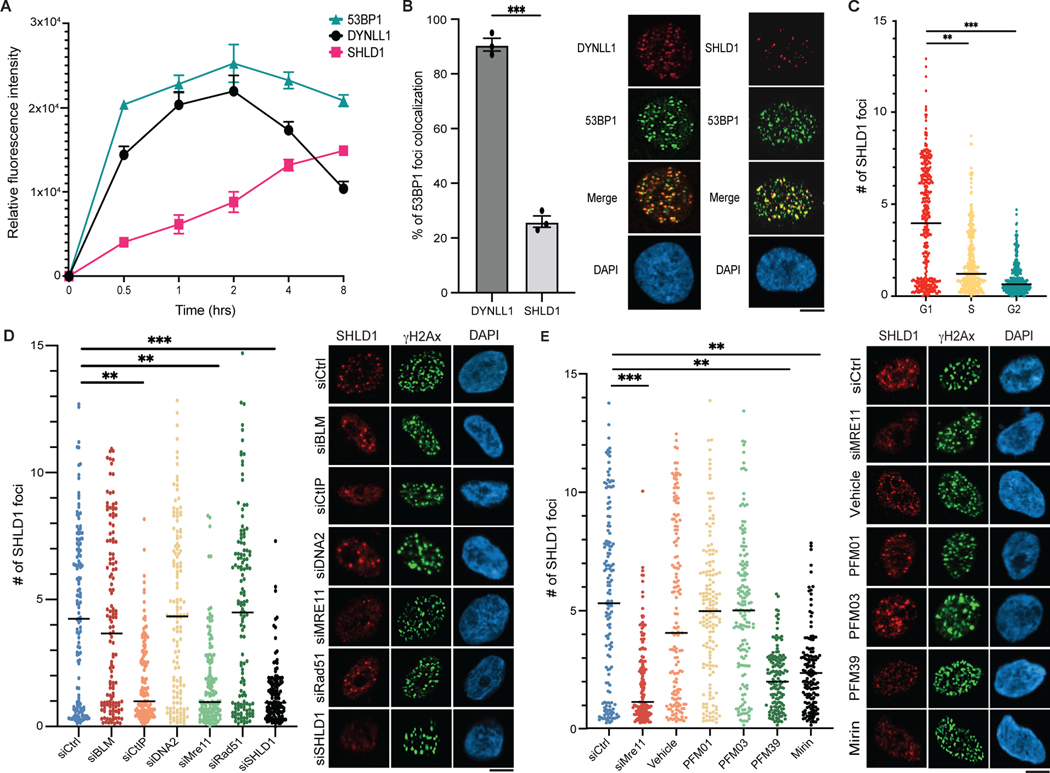
Next, to better understand the dynamics of end resection regulation, we evaluated the kinetics of recruitment of the Shieldin complex relative to 53BP1 and DYNLL1 using laser microirradiation and irradiation. SHLD1 (representative of the Shieldin complex) was recruited to DNA lesions marked by 53BP1 more than 1 h or more after DYNLL1 (Fig. 6a and Extended Data Fig. 7a,b). When evaluating the kinetics of Shieldin localization to DSBs, we observed that about 20% of 53BP1 foci were occupied by SHLD1 foci, compared to DYNLL1, which occupied about 90% of 53BP1 foci (Fig. 6b), suggesting that Shieldin is recruited to a subset of DSBs.
Fig. 6: Kinetics and dependencies of Shieldin complex recruitment to DSBs.
a, RPE1 cells were pretreated with EdU, exposed to 10 Gy of irradiation, and fixed at indicated time points. Cells were processed for immunofluorescence using antibodies against 53BP1, DYNLL1, SHLD1 and γH2AX. Relative fluorescence intensity is normalized to γH2AX. EdU-negative cells were quantified. b, Immuofluorescence of RPE1 cells pretreated with EdU, exposed to 10 Gy of irradiation for 4 h, and stained using antibodies against DYNLL1 and 53BP1, or SHLD1 and 53BP1. Colocalization of DYNLL1 or SHLD1 foci with 53BP1 foci in EdU-negative cells is quantified. c, RPE1 cells were transduced with lentivirus composed of the Fucci system reporter assay. Cells were exposed to 10 Gy of irradiation, fixed 6 h later and processed using antibodies against geminin, CDT1 and SHLD1. d,e, Cells were transfected with the indicated siRNA (d) or treated with MRE11 endo- and exonuclease inhibitors (e). Cells were pretreated with EdU for 30 min, then exposed to 10 Gy of irradiation and fixed 6 h later to be processed for immunofluorescence. EdU-negative cells were quantified. a–e, n = 3 biologically independent experiments, counting at least 100 cells per experiment. Error bars represent mean ± s.e.m. P values were determined using two-sided unpaired t-tests. **P < 0.01, ***P < 0.001, ****P < 0.0001. Black lines in dot plots represent medians. Scale bars, 20 μm.
Next, we aimed to determine the cell cycle dynamics of DYNLL1 and Shieldin recruitment to DSBs. We used the Fucci cell cycle sensor system, a two-color indicator that uses red and green fluorescent proteins (GFP) to follow cell division35, and determined that most SHLD1 foci were formed within the G1 phase, whereas DYNLL1 recruitment to DSBs was not dependent on cell cycle phase18 (Fig. 6c and Extended Data Fig. 7c,d). Given that the Shieldin complex binds to ssDNA via the OB-fold (oligonucleotide/oligosaccharide-binding fold) domains of the SHLD2 component12, we hypothesized that Shieldin is recruited to ssDNA downstream of end resection in G1. End resection in G1 uses specific factors that include MRE11 and CtIP36. Depletion of either MRE11 or CtIP decreased SHLD1 foci formation in G1 phase cells (Fig. 6d and Extended Data Fig. 7e). There was no change in SHLD1 foci formation on knockdown of G2-dependent end resection factors, BLM and DNA2 (Fig. 6d and Extended Data Fig. 7e). MRE11 endonuclease activity is dispensable for end resection in G1, whereas its exonuclease activity is necessary to promote limited resection at slowly repairing DSBs in G136. Upon inhibition of exonuclease activity, a decrease in SHLD1 foci is observed, whereas cells treated with inhibitors of MRE11 endonuclease activity did not display any effect on SHLD1 localization to DSBs (Fig. 6e). Together, we identify that Shieldin is recruited to a small subset of minimally resected ssDNA in G1 phase cells.
However, Shieldin foci formation in G1 is counterintuitive, considering that the Shieldin complex influences PARPi sensitivity in BRCA1-mutant tumors. Therefore, we hypothesized that BRCA1 regulates Shieldin localization in the context of the cell cycle, thereby influencing PARPi sensitivity. In support of this notion, SHLD1 foci formation is known to be increased in BRCA1-mutant cells13. In addition, we observed an increase in SHLD1 foci formation in cyclin A+ (indicative of S phase) BRCA1-depleted cells in response to DNA damage (Extended Data Fig. 7f), suggesting a BRCA1-mediated regulation of the Shieldin complex in S phase.
Loss of DYNLL1 increases Shieldin at DSBs
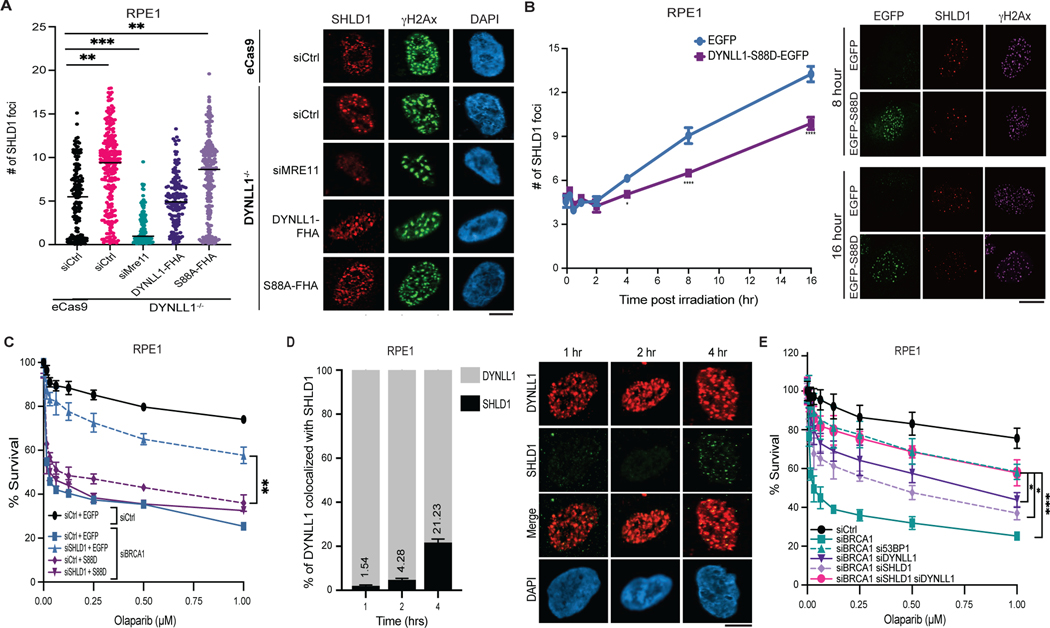
Thus far, we have shown that DYNLL1 functions to remove MRE11 from chromatin. In addition, we have shown that MRE11-dependent minimal end resection is necessary for the recruitment of Shieldin to DSBs. Therefore, we sought to determine whether DYNLL1 affects Shieldin recruitment to DSBs. In cells depleted of DYNLL1, we observe an increase of SHDL1 foci at DSBs, and this was negated by expressing DYNLL1–FHA and not by DYNLL1-S88A–FHA (Fig. 7a). Furthermore, expression of DYNLL1-S88D decreased SHLD1 foci formation (Fig. 7b) and rescued PARPi sensitivity in BRCA1 and SHLD1 depleted cells (Fig. 7c). This supports a potential model in which phosphorylated DYNLL1 negatively regulates Shieldin localization due to impaired MRE11 activity. Therefore, DSBs that are occupied by the Shieldin complex are ones in which DYNLL1 is likely to be unphosphorylated and has failed to suppress MRE11 activity. DYNLL1 and SHLD1 colocalize at DSBs at late time points postirradiation (Fig. 7d). This suggests that in a subset of DSBs, 53BP1 bound DYNLL1 remains unphosphorylated allowing MRE11 activity. This in turn leads to ssDNA promoting the recruitment of the Shieldin complex. Therefore, both DYNLL1 and the Shieldin complex are required for restricting DNA end resection. Indeed, codepletion of DYNLL1 and SHLD1 increased resistance of RPE1 p53−/− BRCA1−/− cells to PARPi compared to single knockdown alone (Fig. 7e). These results indicate that the Shieldin complex serves as a backup for preventing resection in specific scenarios where DYNLL1 has failed to impede MRE11.
Fig. 7: DYNLL1 is required for Shieldin loading to DSBs.
a, Wild-type or DYNLL1−/− RPE1 cells expressing DYNLL1–FHA constructs were pretreated with EdU for 30 min, exposed to 10 Gy of irradiation, and fixed 6 h later for immunofluorescence. EdU-negative cells were quantified. b, RPE1 cells expressing EGFP-tagged wild-type DYNLL1 or its phosphomimetic, S88D, were exposed to 10 Gy of irradiation and fixed at the indicated time points. Cells were stained using antibodies against GFP (DYNLL1), γH2AX and SHLD1. c, BRCA1-depleted RPE1 cells coexpressing DYNLL1 constructs and siRNA against SHLD1 (siSHLD1) were treated with indicated concentrations of olaparib. Cell viability assays were performed 6 days after treatment. d, RPE1 cells expressing
EGFP-DYNLL1 were pretreated with EdU then exposed to 10 Gy of irradiation for 4 h. Cells were fixed and processed using antibodies against GFP (DYNLL1), SHLD1 and 53BP1. Colocalization between DYNLL1 and SHLD1 is quantified in EdU-negative cells. e, RPE1 cells depleted of BRCA1, 53BP1, DYNLL1 and/or SHLD1 were treated with indicated concentrations of olaparib. Cell viability assays were performed 6 days after treatment. For all experiments, n = 3 biologically independent experiments, counting at least 100 cells per experiment. Error bars represent mean ± s.e.m. P values determined using two-sided unpaired t-tests (a,b,d) or nonregression curve analysis (c,e). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Black lines in dot plots represent medians. Scale bars 20 μm.
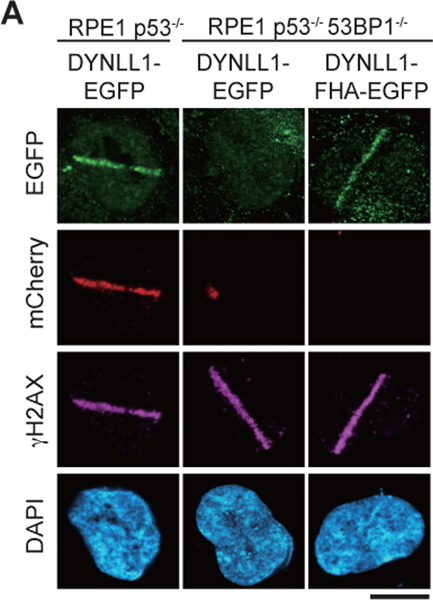
The 53BP1-deficient cells have no Shieldin or DYNLL1 at DSBs. Notably, expressing DYNLL1-EGFP or DYNLL1–FHA in these cells cannot rescue Shieldin loss (Extended Data Fig. 8a), suggesting that DYNLL1 is not sufficient for the initial localization of Shieldin to DSBs.
Discussion
DNA end resection is a critical step that entails the transient loss of genetic material. In normal cells, as the resected ssDNA forms a RAD51 nucleofilament and finds the homologous template, the genetic material is restored with de novo DNA synthesis. However, the balance of DNA end resection is of paramount importance as excess resection or insufficient resection will impede homologous recombination-mediated DSB repair. This equilibrium is often lost in cancer cells as the end resection machinery is harnessed to cause mutagenesis leading to unrestricted proliferation or, in other scenarios, inducing resistance to DNA-damaging cancer therapy. Here we focused on understanding the interplay of the anti-end resection factors centered around the master regulator 53BP1. Our results have defined how distinct 53BP1 interacting proteins work in synchronized chronology to regulate DNA end resection.
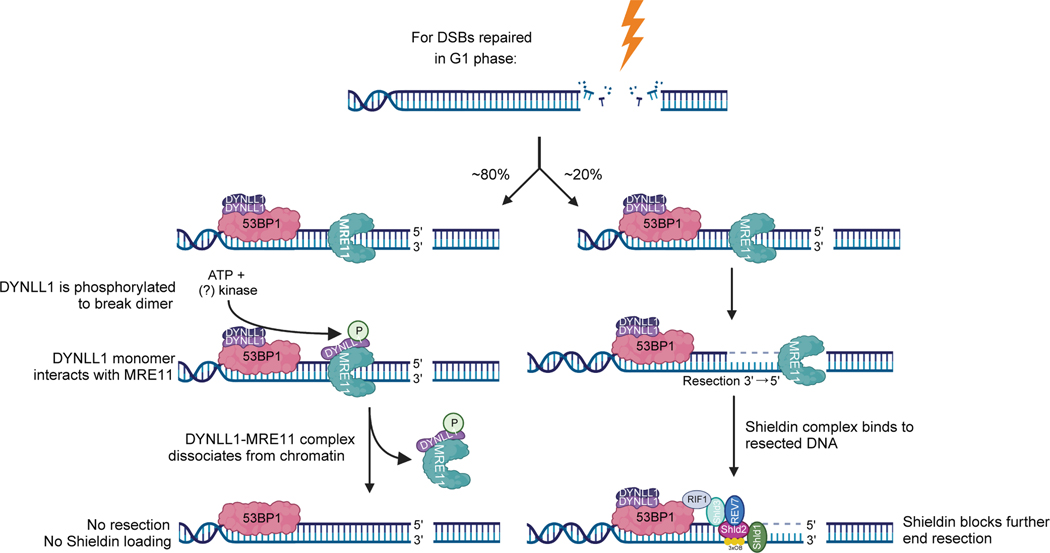
DYNLL1 is constitutively associated with 53BP1 and is recruited to DSBs within minutes of damage, and this is dependent on 53BP1 and no other known associated proteins. MRE11 is one of the earliest factors recruited to DSBs, with multiple proteins involved in this process31,37. DYNLL1 directly binds MRE11 to remove it from DNA lesions15. This step is critical for impeding the initiation of DNA end resection and potentially promoting NHEJ. In scenarios where MRE11 has been recruited and end resection has occurred, the Shieldin complex is recruited. Therefore, Shieldin complex assembly and recruitment follows DYNLL1 with detectable levels of Shieldin proteins visible at DSBs only hours after DNA damage. However, the Shieldin complex recruitment to DSBs does not require DYNLL1 activity but depends on DYNLL1 being inactive. This is supported by our observations that loss of DYNLL1 increases SHLD1 foci formation, and both DYNLL1 and SHLD1 colocalize at a subset of 53BP1-occupied DSBs indicating that DYNLL1 is bound to 53BP1 and cannot suppress MRE11 activity, thereby permitting ssDNA formation and Shieldin recruitment. Phosphorylated DYNLL1 blocks Shieldin localization as it impairs the initiation of DNA end resection. This end resection occurs in a relatively small subset of DSBs (roughly 20%), predominantly in the G1 phase, and is dependent on MRE11 and CtIP. Therefore, Shieldin localizes downstream of MRE11 and upstream of RAD51.
By contrast, DYNLL1 and 53BP1 have roles that are upstream of MRE11 and RAD51. In support of this model (Fig. 8), we observed that loss of DYNLL1 and 53BP1 enhanced MRE11 and RAD51 foci and forced localization of DYNLL1 at DSBs via the FHA-chimeric protein diminished both MRE11 and RAD51 foci. Tethering SHLD1 blocked only the RAD51 foci and not the MRE11 foci. Although BRCA1 regulates MRE11 function, the effect of MRE11 loss on homologous recombination and PARPi sensitivity is beyond BRCA138. This is evident from our observation that MRE11 depletion further sensitizes BRCA1-mutant and/or deficient cells to PARPi. This further highlights the importance of DYNLL1 mediated regulation of MRE11 activity.
Fig 8: Phosphorylated DYNLL1 negatively regulates end resection and Shieldin complex recruitment at DSBs.
The model shows that in about 80% of DSB repair that does not require end resection, phosphorylated DYNLL1-S88 binds to and removes MRE11 from chromatin to inhibit resection. However, in a subset of DSBs, 53BP1 bound DYNLL1 remains unphosphorylated allowing MRE11 activity. MRE11 exonuclease activity creates ssDNA permitting Shieldin complex binding and inhibition of long-range end resection.
Several questions remain unanswered from these studies. How does the hand-off of DYNLL1 from 53BP1 to MRE11 occur at DSBs? What are the factors involved in this regulatory step? DYNLL1 and Shieldin/ CST recruitment to DNA lesions occur independently of one another, although both are reliant on 53BP1. However, improper oligomerization of 53BP1 caused by DYNLL1 loss17,18 may also facilitate MRE11 short-range resection. Therefore, DYNLL1 may regulate MRE11 activity and Shieldin binding to ssDNA at multiple levels. The recruitment of the Shieldin complex to damage sites depends on the ability of SHLD2 to bind resected ssDNA7,12,39. However, we need to reconcile how the Shieldin complex inhibits end resection, albeit requiring end resection for its binding to damaged DNA. One possible explanation is that the Shieldin complex inhibits further MRE11-dependent end resection in G1 as part of a salvage pathway to redirect the DSB to NHEJ after fill-in synthesis by the CST complex (model in Fig. 8). In that scenario, we hypothesize that the Shieldin complex is recruited to cleave the ssDNA overhang created by MRE11 end resection using ASTE140, and Polα-primase is then recruited for fill-in synthesis and end-joining repair10,40. Another key issue is how the Shieldin complex primarily functioning in G1 influences PARPi sensitivity in BRCA1-mutant tumors. PARPi has been proposed to induce DNA lesions during DNA replication, which are then reliant on the BRCA-centric homologous recombination pathway. It is feasible that BRCA1 regulates Shieldin proteins and the function of this complex is altered in the context of BRCA1 deficiency. In support of this notion, there was increased SHLD1 foci formation in BRCA1-mutant cells13 in the S phase (Extended Data Fig. 7f). Furthermore, PARPi sensitivity has now been closely tied to replication fork stability and ssDNA gap formation41. Loss of REV742 and the CST complex43 have been shown to destabilize the fork, which should cause PARPi sensitivity. This is in contrast to PARPi resistance in BRCA1-mutant cells. Together, they indicate that the Shieldin and CST complex may have a differential function in the absence of BRCA1. Future studies will address these questions and provide further insight into the complex mechanism of DNA end resection and PARPi sensitivity.
Methods
Cell lines and transfection
The 293T and COV362 cells were cultured in high glucose DMEM (catalog no. 11995065, Gibco) with 10% (v/v) heat-inactivated fetal calf serum (catalog no. 10–437-028, Gibco) and 1% (v/v) penicillin-streptomycin (catalog no. 15140–122, Gibco). RPE1 cells were cultured in DMEM/F12 (catalog no. 10565018, Gibco) with 10% heat-inactivated fetal calf serum and 1% penicillin-streptomycin. U2OS cells were cultured in McCoy’s 5A media with 10% (v/v) heat-inactivated fetal calf serum (catalog no. 10–437-028, Gibco) and 1% (v/v) penicillin-streptomycin (catalog no. 15140–122, Gibco). Cells were maintained in 20% O2, 5% CO2 and 37 °C. Mouse embryonic fibroblasts (MEFs) were cultured in high glucose and phenol red-free DMEM (catalog no. 21063045, Gibco) with 10% heat-inactivated fetal calf serum. MEF cells were maintained in hypoxic conditions (3% oxygen). All cell lines tested negative for mycoplasma.
All plasmids were transfected using Lipofectamine 2000 (catalog no. 11668019, Invitrogen), and siRNA were transfected by Lipofectamine RNAiMax (catalog no. 13778150, Invitrogen) according to the manufacturer’s protocol.
Retrovirus and lentivirus production and transduction HEK293T cells were transfected with various lentiviral or retroviral constructs and viral packaging plasmids. The medium was changed 24 h after transfection. The virus was collected at 48 and 72 h after transfection. All viruses were filtered using a 0.45 μm SFCA membrane (catalog no. 723–2545, Thermo Scientific) and used immediately or aliquoted and stored at −80 °C.
Virus medium mixed with fresh medium (1:1) was added to 10 cm or six-well plates with 10 μg ml−1 polybrene (catalog no. TR-1003, Sigma Aldrich). 24 h after infection, the medium was discarded and changed to a fresh medium. Cells expressing GFP or mCherry were selected for by fluorescence-activated cell sorting.
Western blots showing the generation of cell lines are displayed in Supplemental Fig.1.
Gene knockout and cell line generation
Two CRISPR guide RNAs were selected from the GeCKO library44. Single-guide RNAs (sgRNAs) targeting TP53, BRCA1, TP53BP1 and DYNLL1 were cloned in the pLentiGuide-puro vector (catalog no. 52963, Addgene). 24 h after viral transduction, cells were selected with puromycin. Three days later, cells were serially diluted, followed by single clonal selection. All knockout cell lines were verified by western blotting.
Sequences of sgRNA can be found in Supplementary Table 1. Western blots showing the generation of cell lines are displayed in Supplementary Fig.1.
MEFs
MEFs were generated from E13.5 embryos grown in DMEM supplemented with 15% heat-inactivated fetal bovine serum and 1% penicillin-streptomycin. Primary MEFs between passages 2–4 were transiently transfected with avector encoding SV40T-antigen (pCMV-SV40T) to establish immortalized MEF cell lines. SV40-immortalized MEFs were routinely cultured in DMEM supplemented with 10% FBS.
Plasmids, antibodies and reagents
Antibodies and siRNA sequences used in this study can be found in Supplementary Table 1. The siRNAs were used at a final concentration of 30 pmol. Olaparib (AZD2281) was purchased from Selleckchem (catalog no. S1060). The concentration and duration of treatment are indicated in the corresponding figure legends.
CellTiter-Glo cell viability assay
Around 300–500 cells per well were plated in a 96-well plate. After 8 h, cells were treated with increasing concentrations of Olaparib (as indicated in corresponding figures) and maintained in the drug for 6 days. The CellTiter-Glo luminescent cell viability assay kit (catalog no. G9242, Promega) was used to measure cell viability after treatment. The plates were scanned with a luminescence microplate reader. The surviving fraction of drug-treated cells was normalized to values from the dimethylsulfoxide-treated control. Untreated and treated conditions were performed in technical triplicates, and each experiment was repeated at least three times. Survival and statistics were determined by GraphPad Prism software.
Cell lysis and immunoblotting
For whole-cell protein extraction, cells were gathered using scrapers (catalog no. CLS3008, Sigma) and first lysed in buffer containing 500 mM NaCl, 20 mM HCl (pH 7.5), 0.5% NP-40, 5 mM EDTA, 5% glycerol
and a protease and phosphatase cocktail inhibitor (Roche). Lysates were rotated at 4 °C for 20 min. An equal volume of lysis buffer (0 mM NaCl, 20 mM HCl (pH 7.5), 0.5% NP-40, 5 mM EDTA, 5% glycerol with protease and phosphatase cocktail inhibitor (Roche)) was added, then rotated at 4 °C for 20 min and spun down at 15,000 rpm for 10 min at 4 °C. The supernatant was moved to a fresh Eppendorf tube. Protein concentration was measured using the Bradford assay (catalog no. 23225, ThermoFisher Scientific). Next, 50 μg of lysate was mixed with SDS loading buffer and loaded into 4–12% precast gels (catalog no. NP0335, Life Technologies). For BRCA1 western blotting, 100 μg of the lysate was loaded into 3–8% precast gels (catalog no. EA0378, Invitrogen).
For cellular fractionation protein extraction, cells were gathered and lysed in a buffer containing 150 mM NaCl, 20 mM HCl (pH 7.5), 0.5% NP-40, 5 mM EDTA, 5% glycerol and a protease and phosphatase cocktail inhibitor (Roche). Lysates were vortexed to homogenize and incubated on ice for 10 min. Lysates were spun down at 2,000 rpm for 4 min. The supernatant was transferred to an Eppendorf tube as the soluble fraction. The pellet was gently washed twice in CE buffer (10 mM HEPES, 60 mM KCl, 1 mM EDTA, 1 mM dithiothreitol and 1 mM phenylmethyl sulfonyl fluoride, adjusted to pH 7.6). The pellet was resuspended in a buffer containing 500 mM NaCl, 20 mM HCl (pH 7.5), 0.5% NP-40, 5 mM EDTA, 5% glycerol and a protease and phosphatase cocktail inhibitor (Roche). The pellet was vortexed to homogenize and incubated on ice for 10 min. Lysates were spun at 15,000 rpm for 10 min. The supernatant was transferred to an Eppendorf tube as the chromatin fraction. For western blotting, 50 μg of the soluble fraction or 30 μg of the chromatin fraction was mixed with SDS loading buffer and loaded into a precast gel.
Immunoprecipitation
Immunoprecipitation was carried out by incubating 1 mg of protein lysate with Anti-Flag Affinity beads (catalog no. A2220, Sigma) and rotating overnight at 4 °C. The beads were washed and eluted in the SDS loading buffer the following day.
Immunofluorescence assays
Cells were plated on glass coverslips in 12-well plates. The following day, cells were irradiated at indicated doses and fixed or gathered at indicated time points. Cells were pre-extracted with 0.5% Triton X-100 in CSK buffer (20 mM HEPES pH 7.6, 100 mM NaCl, 300 mM sucrose and 3 mM MgCl2, with a phosphatase inhibitor) for 5 min on ice, followed by fixation with 4% paraformaldehyde in CSK buffer for 30 min at room temperature. The wells were washed three times in PBS containing 3% BSA. Blocking buffer (1% BSA, 10% donkey serum (catalog no. ab7475, Abcam), 0.1% Triton X-100 in PBS) was added to the plates for 1 h at room temperature. Cells were incubated with primary antibody overnight at 4 °C. Cells were then washed in PBS three times and then incubated with appropriate secondary antibodies for 1 h at room temperature. Coverslips were rinsed with PBS three times and then mounted using Prolong Gold mounting reagent with 4,6-diamidino-2-phenylindole (catalog no. p36931, Invitrogen). Images were acquired with an Olympus BX41 microscope equipped with a digital camera at ×63 magnification.
For experiments requiring EdU (5-ethynyl-2-deoxyuridine) labeling, cells were treated with 10 μM EdU for 30 min. After 30 min, EdU was removed by rinsing and replacing with fresh media. Cells were then irradiated and fixed at indicated time points, followed by pre-extraction with 0.5% Triton X-100 in CSK buffer (20 mM HEPES pH 7.6, 100 mM NaCl, 300 mM sucrose and 3 mM MgCl2, with a phosphatase inhibitor) for 5 min on ice, followed by fixation with 4% paraformaldehyde in CSK buffer for 30 min at room temperature. The wells were washed three times in PBS containing 3% BSA. EdU was stained using Click-iT technology (catalog no. C10640, Life Technologies; following the manufacturer’s protocol). Cells were then rinsed in 3% BSA followed by blocking and antibody incubation as described above.
Metaphase spreads and chromosomal aberrations
Approximately 1 × 106 RPE1 p53−/− BRCA1−/− 53BP1−/− cells were seeded in 10 cm plates. After 48 h, cells were treated with 2 μM olaparib for 24 h. Cells were then treated with colcemid (0.1 μg ml−1) for 2 h and collected using a 0.075 M KCl hypotonic solution and fixed with 3:1 methanol:acetic acid. Slides were stained with Wright’s stain, and 50 metaphase spreads were scored for aberrations. The relative number of chromosomal breaks and radials was calculated relative to control cells or empty vector control (indicated in the figure legends).
Laser microirradiation
Approximately 0.5 × 106–1 × 106 cells were plated in a 35 mm μ-Dish with a glass bottom (catalog no. 81158, Ibidi). On the day of the experiment, the medium was replaced with 1 ml of fresh medium. A Zeiss PALM microdissection microscope equipped with a 360 nm ultraviolet laser at the 38% energy dose was used to create DSBs. All images were analyzed by Fiji (v.2.1.0/1.53c).
ER-AsiSI resection assay
The percentage of resection adjacent to a specific DSB (DSB1 or DSB2) was measured as previously described23. The primer pairs for DSB1 and DSB2 are across BsrG1 and BamHI restriction sites. ER-AsiSI U2OS cells were treated with 4-OHT for 4 h to allow for the nuclear translocation of AsiSI and induction of DSBs. Cells were collected, and genomic DNA was digested with BsrGI or BamHI enzymes or mock digested overnight at 37 °C. Digested samples were used as a template for quantitative PCR. Primers used are listed in Supplementary Table 1 (ref. 23). For each sample, a ΔCt was calculated by subtraction the Ct value of the mock-digested sample from the Ct value of the digested sample.
Protein expression and purification
N-terminal 6XHis-tagged MRE11 structured-domain (1–411 aa) construct was a gift from J. Newman. This construct was expressed and purified as described previously45 with minor modifications. Briefly, the protein was purified using TALON resin, and His-tag was subsequently cleaved with tobacco etch virus (TEV) protease during dialysis (in the buffer: 25 mM Tris (pH 8), 300 mM NaCl, 2.5% glycerol, 5 mM B-ME) in the cold room and the tag-cleaved protein was recovered by passing through the TALON resin again. Protein fractions were concentrated and loaded onto a pre-equilibrated Superdex 200 Increase 10/300 gel-filtration column for further polishing using SEC buffer (20 mM HEPES (pH 7.5), 200 mM NaCl, 1 mM TCEP (pH 7.5)). Wild-type and S88A or S88D DYNLL1 His-tagged recombinant proteins were expressed in chemical competent Rosetta cells. DYNLL1 proteins were purified using a protocol similar to MRE11 using TALON resin. After His-tag cleavage with TEV protease, untagged proteins were concentrated in the dialysis buffer and flash-frozen in liquid nitrogen. To determine the oligomeric state, wild-type and mutant DYNLL1 were further verified by running on a pre-equilibrated Superdex 200 Increase 10/300 gel-filtration column.
As expected, DYNLL1-S88D preferentially forms a monomer, whereas the wild-type and S88A mutant DYNLL1 form a dimer. Protein quality was further verified by running a protein gel and Coomassie Brilliant Blue G-250 staining.
SEC–SAXS
All SEC–SAXS experiments were performed at the SIBYLS beamline 12.3.1 (Advanced Light Source, Berkeley) using their mailinSAXS program46,47. DYNLL1-WT, S88D and S88A at 10 mg ml−1 were run on a pre-equilibrated Shodex KW802.5 column at a flow rate of 0.65 ml min−1 using the running buffer (25 mM Tris (pH 8.0), 150 mM NaCl, 1 mM TCEP). The SAXS data were collected with X-ray wavelength set to λ = 1.24 Å with the sample-to-detector distance of 2,075 mm resulting in scattering vectors (q) ranging from 0.01 to 0.46 Å−1. The scattering vector is defined as q = 4πsinθ/λ, where 2θ is the scattering angle. The scattering data were collected in 2 s exposures continuously over 25 min of elution using a Pilatus 3X 2M detector (DECTRIS). Normalization and integration of each image were processed as previously described48. SCÅTTER (https://bl1231.als.lbl.gov/scatter/) program was used for buffer subtraction, data merging and Guinier analyses.
AlphaFold2 and AlphaFold2-Multimer
AlphaFold2 and the AlphaFold2-Multimer (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb#scrollTo=G4yBrceuFbf3) were used to generate the full-length DYNLL1 monomer and dimer structural models, respectively49. The full-length sequence of DYNLL1 included an additional three residues at the N terminus as Ser-Asn-Ala residues were left behind after the cleaving of the His-tag. Default settings in the AlphaFold-Coalb were used to predict the models of DYNLL1 monomer (using alphafold2_ptm as model_type) and dimer (using alphafold2_multimer_v3 as model_type). The AlphaFold model for the full-length MRE11 was downloaded from the AlphaFold Protein Structure Database using UniProt accession number P49959. Experimental SAXS curves were fit to DYNLL1 model-derived SAXS curves using either FoXS or MultiFoXS web-based server (https://modbase.compbio.ucsf.edu/foxs/)46,50. The goodness of the fit was determined by comparing the χ2 values.
MST to measure the MRE11 dimerization
The N-terminus nuclease domain of MRE11 (1–411 aa) dimerization Kd was measured by titrating unlabeled MRE11 into labeled MRE11. MST experiments were performed on a Monolith NT.115Pico system (NanoTemper). Purified MRE11 (1–411 aa) was fluorescently labeled with an amine-reactive AlexaFluor-647 dye. All MST experiments were performed in the following buffer: 20 mM HEPES (pH 7.5), 150 mM NaCl, 1.5% glycerol, 0.1 mM EDTA, 1 mM MnCl2, 1 mM TCEP and 0.05% T-20. Both ligand (unlabeled MRE11) and target (labeled MRE11) dilutions were prepared in the MST buffer. Equal volumes of 10 nM AlexaFluor-647-labeled MRE11 and serially 2×-diluted unlabeled MRE11 were mixed to get a fixed concentration of labeled MRE11 (5 nM) and variable concentration of unlabeled MRE11 (final concentrations ranging from 1.8 nM to 30 μM). The mixture was then incubated for 10 min at room temperature before loading into regular Monolith NT.115 capillaries for the MST measurements. After validating the assay condition, the following instrument settings were used for the binding affinity experiments: a 6% excitation power in the Pico-RED channel and high MST power with other default settings, including experiment temperature set to 25 °C. Each binding experiment was performed in three independent runs, and the resulting MST data loaded to MO.Affinity Analysis Software (NanoTemper) to estimate the Kd for MRE11 dimerization. To study the effect of DYNLL1 mutants (S88D and S88A) on MRE11 dimerization, MST experiments were performed in the MST buffer with either 5 μM DYNLL1-S88D or 5 μM DYNLL1-S88A. MST data are presented as a change in normalized fluorescence (due to thermophoresis) as a function of unlabeled MRE11 concentration.
The MST data were fit to the Kd model from the MO.Affinity Analysis software. Both raw and fit data were exported and plotted in IgorPro9 software (WaveMetrics) for presentation.
MRE11 elution from chromatin
HEK293T cells were treated with 5 Gy of irradiation and pre-extracted with 5% NP-40 lysis buffer with proteinase and phosphatase inhibitors. Insoluble chromatin was washed with 1× TBS + 0.05% BSA and incubated with indicated recombinant DYNLL1 protein at 37 °C for 30 min. Elution was collected, and insoluble chromatin was washed with 1× TBS, whereas the chromatin binding protein was extracted with 5% NP-40 lysis buffer + 1% SDS + 0.1 mM EDTA and boiled at 95 °C for 10 min. Elution and chromatin-bound fractions were subject to western blot analysis.
Quantification and statistical analysis
Immunofluorescence foci were counted by CellProfiler (v.4.2.0). Colocalization analyses were done in ImageJ (v.1.53a). All statistical analyses were performed by Prism v.8 and 9.5 (GraphPad). All experiments were done in triplicate. P values for the percentage ‘laser positive’ analysis were calculated using unpaired t-tests. P values for foci formation and colocalization were calculated using the Mann–Whitney test. P value measurements for cell survival curves were assessed by nonregression curve analysis in GraphPad. Error bars represent mean ± s.e.m. as described in the figure legends. In all cases, NS, not significant (P ≥ 0.05): *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data
Extended Data Fig. 1: 53BP1 is necessary for chromatin localization of DYNLL1. Related to Fig. 1.
(a) Representative immunofluorescence images of RPE1 cells subjected to laser microirradiation. Cells were fixed at indicated time points post laser microirradiation and processed for immunofluorescence with DYNLL1 and 53BP1 antibodies. (b) Representative images of RPE1 wild-type or 53BP1−/− cells 2 h after exposure to 2 Gy irradiation or laser microirradiation. Cells were fixed and processed for immunofluorescence using antibodies against 53BP1, GFP (DYNLL1), and γH2AX. (c) RPE1 cells depleted of p53, 53BP1, or DYNLL1 using CRISPR/Cas9 were exposed to 10 Gy irradiation. Protein was collected after 3 h. Localization of DYNLL1 to chromatin was evaluated by subcellular fractionation followed by immunoblotting for DYNLL1. (d–f) Representative images of COV362 cells (D) and COV362 wild-type or 53BP1−/− cells (e, f) exposed to 2 Gy irradiation (d, e) or laser microirradiation (F). 2 h post-recovery cells were fixed and processed for immunofluorescence using antibodies against 53BP1, GFP (DYNLL1), and γH2AX. Box plots show mean and center, quartiles (boxes), and range (whiskers)(d). (g) Representative immunofluorescence images of RPE1 cells depleted of DYNLL1 using CRISPR/Cas9 and exposed to 2 Gy irradiation. 1h post-irradiation, cells were fixed and processed for immunofluorescence using antibodies against 53BP1 and γH2AX. (a-g) n = 3 biologically independent experiments, counting ≥ 100 cells per experiment. Error bars represent the mean±s.e.m. P-values for foci quantification and “laser positive” cell analysis were calculated using two-sided unpaired t-tests. P values are indicated by nonsignificant (P >0.05), *(P < 0.05), **(P < 0.01), ***(P < 0.001), ****(P<0.0001). Black line in dot plots represent median. Scale bar = 20μm.
Extended Data Fig. 2: Force tethering DYNLL1 to chromatin inhibits MRE11 foci formation. Related to Fig. 2.
(a, b) RPE1 53BP1−/− cells were transfected with EGFP-tagged DYNLL1 or DYNLL1-FHA constructs. Cells were subjected to 2 Gy irradiation (A) or laser microirradiation (B). 2 h later cells were fixed and processed for immunofluorescence using antibodies against GFP (DYNLL1) and γH2AX. (c, d) COV362 53BP1−/− cells were transfected with a EGFP-tagged DYNLL1 and DYNLL1-FHA constructs. Cells were subjected to 2 Gy irradiation (c) or laser microirradiation (d). 2 h later cells were fixed and processed for immunofluorescence using antibodies against GFP (DYNLL1) and γH2AX. (e) COV362 53BP1−/− cells were transfected with EGFP-tagged DYNLL1 and DYNLL1- FHA constructs. Cells were exposed to 2 Gy irradiation. 2 h post-irradiation cells were fixed and processed for immunofluorescence using antibodies against MRE11, GFP (DYNLL1) and γH2AX. (A-E) n = 3 biologically independent experiments, counting ≥ 100 cells per experiment. Error bars represent the mean±s.e.m. P-values for foci quantification and “laser positive” cell analysis were calculated using two-sided unpaired t-tests. P values are indicated by nonsignificant (P >0.05), *(P < 0.05), **(P < 0.01), ***(P < 0.001), ****(P<0.0001). Black line in dot plots represent median. Scale bar = 20μm.
Extended Data Fig. 3: DYNLL1 chromatin binding suppresses 53BP1 loss- induced restoration of HR in BRCA1 deficient cells. Related to Fig. 3.
(a) MEFs expressing EGFP-tagged DYNLL1, and EGFP-tagged DYNLL1-FHA domains constructs were exposed to 2 Gy irradiation. 2 h after irradiation cells were fixed and processed for immunofluorescence using a GFP (DYNLL1) antibody. (b, c) MEF p53−/− BRCA1−/− 53BP1−/− cells (b) and COV362 53BP1−/− cells (c) were transfected with EGFP-tagged DYNLL, or EGFP-tagged DYNNL1-FHA constructs. Cells were exposed to 2 Gy irradiation. 2 h later, cells were fixed and processed for immunofluorescence using antibodies against GFP (DYNLL1), RAD51, and γH2AX. (d) COV362 53BP1−/− cells were transfected EGFP-tagged DYNLL, or EGFP-tagged DYNNL1-FHA constructs. Cells were treated with indicated concentrations of Olaparib for 6 days. Percent survival was determined via a cell viability assay. (a-d) n = 3 biologically independent experiments, counting ≥ 100 cells per experiment. Error bars represent the mean±s.e.m. P-values for foci quantification were calculated using two-sided unpaired t-tests. P-value measurements for cell survival curves were assessed by non-regression curve analysis. P values are indicated by nonsignificant (P >0.05), *(P < 0.05), **(P < 0.01), ***(P < 0.001),****(P<0.0001). Black line in dot plots represent median. Scale bar = 20μm.
Extended Data Fig. 4: DYNLL1 interferes with MRE11 dimerization. Related to Fig. 4.
(a) Predicted structure of full-length MRE11 created by AlphaFold Monomer V2 for Uniprot Accession number P49959. The structured catalytic domain of MRE11 is highlighted with a red circle. The model is color coded in terms of confidence in prediction and respective color schemes for the confidence is given in the figure. In general, disordered regions have less confidence in model prediction, thus indicating the unstructured regions of MRE11 beyond capping domain. (b) Coomassie-stained protein gels indicating the quality of the recombinant protein used in the current study. Left: DYNLL1 mutants after cleaving the His-tag with TEV protease. Right: MRE11 catalytic domain after the gel-filtration purification step. The red rectangle indicates the fractions that are combined. M indicates the protein standards and * indicates the MRE11 degradation bands. (c) Average MST response (n = 3) measured from labeled MRE11 in the MST buffer or MST buffer with 5 μM DYNLL1-S88A or DYNLL1-S88D mutant. (d) Change in the normalized fluorescence (ΔFnorm) as result of thermophoresis in the MST experiment plotted as a function of concentration of unlabeled MRE11. The resulting curves represents MRE11 dimerization in the absence of any DYNLL1 (black circles), in the presence of 500 nM DYNLL1-S88D (red triangles) or in the presence of 5 μM DYNLL1-S88D (blue squares). The Kd values are measured by fitting the curves with Kd model in the analysis software. (B-D) The data points represent average of three independent measurements and error bars represents standard deviation.
Extended Data Fig. 5: Solution structures of DYNLL1-WT and mutants.
(a) AlphaFold2 predicted models of DYNLL1 dimer (left) and monomer (right). (b) Size exclusion chromatography elution profiles for DYNLL1-WT, S88A and S88D mutants. For clarity, initial 8 mL (pre-void volume with no peaks) were omitted from the chromatograms. (c–e) FoXS fitting of experimental X-ray scattering data (red dots) to theoretical SAXS profiles (solid lines) derived from structural models of DYNLL1 monomer (M) and dimer (D) or mixture of monomer and dimer state. The goodness of the fit is evaluated by χ2. Guinier plots from the measured scattering intensity (I(q)) as a function of scattering vector (q) in the low q region shown as insets for WT and mutant proteins.
Extended Data Fig. 6: Depletion of the Shieldin complex does not affect MRE11 recruitment. Related to Fig. 5.
(a) Protein expression from lysates collected from RPE1 wild-type or DYNLL1−/− cells 2 h after 2 Gy irradiation. (b) COV362 cells overexpressing EGFP-DYNLL1 and transfected with siRNA targeting either 53BP1, or SHLD1 were exposed to 2 Gy irradiation. Cells were fixed 2 h post exposure and processed for immunofluorescence using antibodies against GFP (DYNLL1), γH2AX, and MRE11. (a, b) n = 3 biologically independent experiments, counting ≥ 100 cells per experiment. P-values for foci quantification were calculated using two-sided unpaired t-tests. P values are indicated by nonsignificant (P >0.05), *(P < 0.05), **(P < 0.01), ***(P < 0.001), ****(P<0.0001). Black line in dot plots represent median. Scale bar = 20μm.
Extended Data Fig. 7: Shieldin is recruited to DSBs later than DYNLL1 and in G1 phase only. Related to Fig. 6.
(a) Representative images from Fig. 6a. (b) Quantification of GFP positive stripes for cells expressing DYNLL1-EGFP or SHLD1-EGFP after exposure to laser microirradiation and fixed at indicated time points. (c) Quantification of number of DYNLL1 foci for RPE1 cells transduced with lentivirus comprised of the Fucci system reporter assay. Cells were exposed to 10 Gy irradiation, fixed 6 h later, and processed using antibodies against Geminin, Cdt1, and DYNLL1. (d) Representative images for Fig. 6c and Extended Data Fig. 6c. (e) Representative western blots showing knockdown of indicated proteins for Fig. 6d. (f) RPE1 BRCA1−/− were subjected to 10 Gy IR and 4 h later were fixed and processed for immunofluorescence using antibodies against SHLD1, Cyclin A, and γH2AX. (a-f) n = 3 biologically independent experiments, counting ≥ 100 cells per experiment. Error bars represent the mean±s.e.m. P-values for foci quantification and ‘laser positive’ cell analysis was calculated using two-sided unpaired t-tests. P values are indicated by nonsignificant (P >0.05), *(P < 0.05), **(P < 0.01), ***(P < 0.001), ****(P<0.0001). Black line in dot plots represent median. Scale bar = 20μm.
Extended Data Fig. 8: Shieldin functions downstream of DYNLL1, but is not dependent on DYNLL1 for its localization to chromatin. Related to Fig. 7.
(a) RPE1 p53−/− and RPE1 p53−/− 53BP1−/− cells were transfected with EGFP-tagged DYNLL1 or EGFP-tagged DYNLL1-FHA. Cells were then transfected with mCherry-SHLD1 and subjected to laser microirradiation. 2 h after laser microirradiation cells were fixed and processed for immunofluorescence using antibodies against GFP (DYNLL1), mCherry (SHLD1), and γH2AX. n = 3 biologically independent experiments. Scale bar = 20μm.
Supplementary Material
Acknowledgements
D.C. is supported by grants R01 CA208244 and R01 CA264900, Gray Foundation Team Science Award, DOD Ovarian Cancer Award W81XWH-15-0564/OC140632, Tina’s Wish Foundation, Detect Me If You Can, V Foundation Award and the Claudia Adams Barr Program in Innovative Basic Cancer Research. M.L.S. is supported by National Institutes of Health (NIH) grant F32 GM149115. J.A.T. and A.S. were partly supported by NIH grants P01 CA092548 and R35 CA220430, plus Cancer Prevention Research Institute of Texas grant RP180813 and an endowed Robert A. Welch Chemistry Chair supported by the Welch Foundation. B.T. was supported by the Polish National Agency for Academic Exchange (grant PPN/WAL/2019/1/00018) and by the Foundation for Polish Science (START Program). We thank M. Hammel and K. H. Burnett for their help in SEC-SAXS data collection at the SIBYLS beamline. The SEC-SAXS data collection at SIBYLS is supported in part by the NIH-NIGMS grant P30 GM124169-01 (ALS-ENABLE). We thank the Center for Macromolecular Interactions at Harvard Medical School for access to their Monolith NT.115Pico (NanoTemper) system. We also thank J. Newman for providing the MRE11 expression construct to produce the recombinant protein. The model in Fig. 8 was created with BioRender.com.
Competing interests
A.D.D. reports consulting for AstraZeneca, Bayer AG, Blacksmith/ Lightstone Ventures, Bristol Myers Squibb, Cyteir Therapeutics, EMD Serono, Impact Therapeutics, PrimeFour Therapeutics, Pfizer, Tango Therapeutics and Zentalis Pharmaceuticals/Zeno Management; is an Advisory Board member for Cyteir and Impact Therapeutics; a stockholder in Cedilla Therapeutics, Cyteir, Impact Therapeutics and PrimeFour Therapeutics, and reports receiving commercial research grants from Bristol Myers Squibb, EMD Serono, Moderna and Tango Therapeutics. The remaining authors declare no competing interests.
Data availability
Numerical source data are provided with this paper. A description of the AlphaFold modeling can be found in the Methods. All other data reported in this paper will be shared by the lead contact upon request.
References
- 1.Zhao W, Wiese C, Kwon Y, Hromas R. & Sung P. The BRCA Tumor Suppressor Network in Chromosome Damage Repair by Homologous Recombination. Annu Rev Biochem 88, 221–245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao B, Rothenberg E, Ramsden DA & Lieber MR The molecular basis and disease relevance of non-homologous DNA end joining. Nat Rev Mol Cell Biol 21, 765–781 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hustedt N. & Durocher D. The control of DNA repair by the cell cycle. Nat Cell Biol 19, 1–9 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Gnugge R. & Symington LS DNA end resection during homologous recombination. Curr Opin Genet Dev 71, 99–105 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunting SF et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–54 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouwman P. et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 17, 688–95 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirman Z. et al. 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polalpha-dependent fill-in. Nature 560, 112–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zimmermann M. & de Lange T. 53BP1: pro choice in DNA repair. Trends Cell Biol 24, 108–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ochs F. et al. 53BP1 fosters fidelity of homology-directed DNA repair. Nat Struct Mol Biol 23, 714–21 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Mirman Z, Sasi NK, King A, Chapman JR & de Lange T. 53BP1-shieldin-dependent DSB processing in BRCA1-deficient cells requires CST-Polalpha-primase fill-in synthesis. Nat Cell Biol 24, 51–61 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mirman Z. & de Lange T. 53BP1: a DSB escort. Genes Dev 34, 7–23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noordermeer SM et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature 560, 117–121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dev H. et al. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat Cell Biol 20, 954–965 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta R. et al. DNA Repair Network Analysis Reveals Shieldin as a Key Regulator of NHEJ and PARP Inhibitor Sensitivity. Cell 173, 972–988 e23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He YJ et al. DYNLL1 binds to MRE11 to limit DNA end resection in BRCA1-deficient cells. Nature 563, 522–526 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lo KW et al. The 8-kDa dynein light chain binds to p53-binding protein 1 and mediates DNA damage-induced p53 nuclear accumulation. J Biol Chem 280, 8172–9 (2005). [DOI] [PubMed] [Google Scholar]
- 17.West KL et al. LC8/DYNLL1 is a 53BP1 effector and regulates checkpoint activation. Nucleic Acids Res 47, 6236–6249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker JR et al. The ASCIZ-DYNLL1 axis promotes 53BP1-dependent non-homologous end joining and PARP inhibitor sensitivity. Nat Commun 9, 5406 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noordermeer SM & van Attikum H. PARP Inhibitor Resistance: A Tug-of-War in BRCA-Mutated Cells. Trends Cell Biol 29, 820–834 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Chen H, Lisby M. & Symington LS RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol Cell 50, 589–600 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cejka P. DNA End Resection: Nucleases Team Up with the Right Partners to Initiate Homologous Recombination. J Biol Chem 290, 22931–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iacovoni JS et al. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J 29, 1446–57 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Caron P, Legube G. & Paull TT Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res 42, e19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drane P. et al. TIRR regulates 53BP1 by masking its histone methyl-lysine binding function. Nature 543, 211–216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Syed A. & Tainer JA The MRE11-RAD50-NBS1 Complex Conducts the Orchestration of Damage Signaling and Outcomes to Stress in DNA Replication and Repair. Annu Rev Biochem 87, 263–294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hopfner KP et al. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105, 473–85 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Lammens K. et al. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell 145, 54–66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiller CB et al. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat Struct Mol Biol 19, 693–700 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park YB, Chae J, Kim YC & Cho Y. Crystal structure of human Mre11: understanding tumorigenic mutations. Structure 19, 1591–602 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P. & Duhr S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol 9, 342–53 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paull TT 20 Years of Mre11 Biology: No End in Sight. Mol Cell 71, 419–427 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Paull TT & Lee JH The Mre11/Rad50/Nbs1 complex and its role as a DNA double-strand break sensor for ATM. Cell Cycle 4, 737–40 (2005). [DOI] [PubMed] [Google Scholar]
- 33.de Lange T. Shelterin-Mediated Telomere Protection. Annu Rev Genet 52, 223–247 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Maciejowski J. & de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol 18, 175–186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakaue-Sawano A. et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–98 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Biehs R. et al. DNA Double-Strand Break Resection Occurs during Non-homologous End Joining in G1 but Is Distinct from Resection during Homologous Recombination. Mol Cell 65, 671–684 e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye Z. et al. GRB2 enforces homology-directed repair initiation by MRE11. Sci Adv 7(2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stracker TH & Petrini JH The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol 12, 90–103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Setiaputra D. & Durocher D. Shieldin - the protector of DNA ends. EMBO Rep 20(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao F. et al. ASTE1 promotes shieldin-complex-mediated DNA repair by attenuating end resection. Nat Cell Biol 23, 894–904 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Cantor SB Revisiting the BRCA-pathway through the lens of replication gap suppression: “Gaps determine therapy response in BRCA mutant cancer”. DNA Repair (Amst) 107, 103209 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paniagua I. et al. MAD2L2 promotes replication fork protection and recovery in a shieldin-independent and REV3L-dependent manner. Nat Commun 13, 5167 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lyu X. et al. Human CST complex protects stalled replication forks by directly blocking MRE11 degradation of nascent-strand DNA. EMBO J 40, e103654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanjana NE, Shalem O. & Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenberg DJ, Syed A, Tainer JA & Hura GL Monitoring Nuclease Activity by X-Ray Scattering Interferometry Using Gold Nanoparticle-Conjugated DNA. Methods Mol Biol 2444, 183–205 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneidman-Duhovny D, Hammel M, Tainer JA & Sali A. FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res 44, W424–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenberg DJ, Hura GL & Hammel M. Size exclusion chromatography coupled small angle X-ray scattering with tandem multiangle light scattering at the SIBYLS beamline. Methods Enzymol 677, 191–219 (2022). [DOI] [PubMed] [Google Scholar]
- 48.Hura GL et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat Methods 6, 606–12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mirdita M. et al. ColabFold: making protein folding accessible to all. Nat Methods 19, 679–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneidman-Duhovny D, Hammel M, Tainer JA & Sali A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys J 105, 962–74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Numerical source data are provided with this paper. A description of the AlphaFold modeling can be found in the Methods. All other data reported in this paper will be shared by the lead contact upon request.