Abstract
The present study investigated the prevalence and diagnostic potential of the most commonly reported mutations associated with isoniazid resistance, katG 315Thr, katG 315Asn, inhA −15T, inhA −8A, and the oxyR-ahpC intergenic region, in a population sample of 202 isoniazid-resistant Mycobacterium tuberculosis isolates and 176 randomly selected fully sensitive isolates from England and Wales identified by using a directed oligonucleotide array and limited DNA sequencing. The strains were recovered from patients originating from 29 countries; 41 isolates were multidrug resistant. Mutations affecting katG 315, the inhA promoter, and the oxyR-ahpC intergenic region were found in 62.7, 21.9, and 30% of 169 genotypically distinct isoniazid-resistant isolates, respectively, whereas they were found in 0, 0, and 8% of susceptible strains, respectively. The frequency of mutation at each locus was unrelated to the resistance profile or previous antituberculous drug therapy. The commonest mutation in the oxyR-ahpC intergenic region, ahpC −46A, was present in 23.7% of isoniazid-resistant isolates and 7.5% of susceptible isolates. This proved to be a phylogenetic marker for a subgroup of M. tuberculosis strains originating on the Indian subcontinent, which shared IS6110-based restriction fragment length polymorphism and spoligotype features with the Delhi strain and Central Asian strain CAS1; and this marker is strongly associated with isoniazid resistance and the katG 315Thr mutation. In total, 82.8% of unrelated isoniazid-resistant isolates could be identified by analysis of just two loci: katG 315 and the inhA promoter. Analysis of the oxyR-ahpC intergenic region, although phylogenetically interesting, does not contribute significantly to further identification of isoniazid-resistant isolates.
Tuberculosis (TB) is one of the leading causes of death due to an infectious agent throughout the world, and the increasing rates of drug-resistant TB are of global concern. In England and Wales the incidence of TB has risen from 11.3/100,000 population to 12.7/100,000 population over the last decade (36), although the incidence of drug resistance has remained stable, with isoniazid and rifampin resistance occurring in 6 to 7% and 1% of isolates from new patients, respectively. International and regional studies report an incidence of drug resistance in new cases of TB to any first-line antituberculous drug ranging from 2 to 37% (median, 10.7%) (13), with resistance to isoniazid being the commonest form. Resistance is considerably higher among patients previously treated for TB (median, 23.3%). The present quadruple anti-TB therapy regimens are designed to treat isoniazid-resistant isolates while clinicians await the results of drug susceptibility testing; however, administration of isoniazid is not without risk of side effects. Phenotypic drug susceptibility testing takes between 4 and 6 weeks from the time of receipt of clinical samples. Genotypic analysis, in contrast, can be performed in a matter of hours. Commercial and in-house systems are available for the rapid detection of rifampin-resistant Mycobacterium tuberculosis and take 5 to 8 h from the time of collection of primary sputum samples (9, 52). Unlike rifampin resistance, in which 95% of rifampin-resistant isolates have mutations within an 81-bp region of a single gene encoding the RNA polymerase B subunit, rpoB (43), isoniazid resistance has been associated with mutations in several genes. The most commonly reported mutations occur at three loci: katG, the inhA promoter region, and the oxyR-ahpC intergenic region.
Although previous studies have investigated the role of mutations in katG alone, inhA, or both katG and inhA (2, 11, 18, 24, 28, 29, 30, 31, 38, 44, 46, 48, 50) in isoniazid-resistant M. tuberculosis isolates and the frequency of these mutations in different isolate groups, the value of these studies is often limited. The reasons include the small numbers of isolates sampled from collections (18, 29, 30, 38) or populations (11, 23, 24, 31, 32), the lack of DNA fingerprinting defining the relationship between isolates in population samples (2, 32), a limited number of genotypically distinct isolates (20, 28, 48), restriction of investigation to multidrug resistant (MDR) M. tuberculosis isolates (11, 20, 23, 46), and limited analysis of sensitive isolates. Mutations in the oxyR-ahpC intergenic region have been studied less frequently (19, 20, 27, 32, 34, 35, 41, 42), and in those few studies in which katG 315 and the inhA promoter have also been analyzed, the majority have been performed with isolates from selected populations (20, 32, 34, 42).
The aims of this study were twofold: first, to analyze the prevalence and distributions of the most commonly reported mutations associated with isoniazid resistance (katG 315Thr, katG 315Asn, inhA −15T, inhA −8A, and mutations in the oxyR-ahpC intergenic region) and the katG Arg463Leu polymorphism in a large, diverse, unselected population of both isoniazid-resistant and -susceptible M. tuberculosis isolates and, second, to assess the efficacies of these loci for the genotypic detection of isoniazid resistance by using a low-density directed oligonucleotide array and limited DNA sequencing.
MATERIALS AND METHODS
Bacterial strains.
The 378 M. tuberculosis clinical isolates analyzed were identified in England and Wales between 1 January 1998 and 31 December 1998 and included all viable clinical isolates that were resistant to isoniazid and 176 randomly chosen susceptible isolates sensitive to all first-line antituberculous drugs. Isolates were identified from the National Laboratory Mycobacterium Resistance Network Database and from laboratory records at the National Health Protection Agency Mycobacterium Reference Unit, London, United Kingdom. The first available isolate from each patient was analyzed. Drug susceptibility was determined by the resistance ratio method (8). Isolates were defined as resistant if the resistance ratio was greater than or equal to 4. The strains were characterized by IS6110-based restriction fragment length polymorphism (IS6110 RFLP) analysis and spoligotyping, according to internationally standardized protocols (17, 47). A cluster was defined as two or more strains with indistinguishable IS6110 DNA fingerprints. When the IS6110 copy number was less than five, isolates were considered clustered only if both the IS6110 DNA fingerprints and the spoligotype patterns were identical (17, 47, 49). Epidemiological information was supplemented by the 1998 national tuberculosis survey (36).
Amplification of selected loci.
A multiplex PCR was prepared by using four primer pairs to amplify three gene loci associated with isoniazid resistance and katG 463, as described previously (12, 25). The final PCR mixture volume of 25 μl included 1 μl of template DNA; 2.5 μl of 10× reaction buffer (Qiagen, Crawley, United Kingdom); 400 nM each primers tomkp1 (5′-GGCCCCGAACCCGAGGCTGC-3′), tomkp2 (5′-AACGGGTCCGGGATGGTGCCG-3′), tomkp3 (5′-GCCGACGAGTTCGCCAAGGCC-3′), tomkp4 (5′-ACGACGCCGCCGCCCATGCG-3′), tomap1 (5′-CCGCCGATGAGAGCGGTGAGC-3′), tomap2 (5′-CCACTGCTTTGCCGCCACCGC-3′), tomip1 (5′-CACCCGCAGCCAGGGCCTCG-3′), and tomip2 (5′-CGATCCCCCGGTTTCCTCCGG-3′) (primers tomkp1 and tomkp2 spanned katG codon 315, primers tomkp3 and tomkp4 spanned katG codon 463, primers tomap1 and tomap2 spanned the oxyR-ahpC intergenic region, and primers tomip1 and tomip2 spanned the inhA promoter); 40 μM dGTP, dCTP, and dATP (Bioline, London, United Kingdom); 20 μM dTTP; 20 μM digoxigenin-labeled dUTP (Roche Molecular Biochemicals, Lewes, United Kingdom); and 0.5 U of HotStar Taq DNA polymerase (Qiagen). Thermocycling (9600; Perkin-Elmer, Norwalk, Conn.) consisted of an initial denaturation step of 15 min at 95°C, followed by 30 cycles of 15 s at 95°C, 30 s at 60°C, and 60 s at 72°C, with a final extension for 5 min at 72°C.
Preparation of oligonucleotide array.
The oligonucleotide array technique used has been described previously (1, 7, 25), as has the use of the 11 target oligonucleotides summarized in Table 1 (12, 25). Briefly, the 11 probes (20 mM) and a color development control (digoxigenin-dUTP-labeled oligonucleotide) were spotted in 0.1-μl volumes onto a marked nylon membrane (Osmonics, Minnetonka, Minn.). After the membrane was allowed to dry, the oligonucleotides were cross-linked to the membrane with a UV Stratalinker 1800 cross-linker (Stratagene, La Jolla, Calif.). Unbound oligonucleotide was removed by washing the membrane for 5 min in 0.5× SSC (0.075 M sodium chloride plus 0.0075 M sodium citrate [pH 7.0])-0.1% (wt/vol) sodium dodecyl sulfate. Once the membrane was dry, it was trimmed to produce strips of 5 by 40 mm, which were individually stored in 1-ml plastic tubes at room temperature.
TABLE 1.
Target sequences for isoniazid resistance-directed oligonucleotide PCR and hybridization array
| Locus | Sequence type | Oligonucleotide name (sequence)a |
|---|---|---|
| katG 315 | Wild type | K315WTD20T (5′-CAGCGGCATCGAGGTCGTATTTTTTTTTTTTTTTTTTTTT-3′) |
| katG 315 | Mutant (AAC) | TOMK3159-AE10T (5′-CCAACGGCATCGAGGTCGTATTTTTTTTTT-3′) |
| katG 315 | Mutant (ACC) | TOMK3150-C10T (5′-CGGGATCACCACCGGCATCGTTTTTTTTTT-3′) |
| katG 463 | Arg | TOMK463WB5T (5′-GATCCGGGCATCGGGATTGACTTTTT-3′) |
| katG 463 | Leu | TOMK46399-TG10T (5′-GAGCCAGATCCTGGCATCGGGTTTTTTTTTT-3′) |
| oxyR-ahpC | Wild type | TOMAWT1A (5′-TTTTTTTTTTGCGACATTCCATCGTGCCG-3′) |
| oxyR-ahpC | Wild type | TOMAWT210T (5′-GTGCCGTGAAGTCGCTGTCAGTTTTTTTTTT-3′) |
| oxyR-ahpC | Wild type | TOMAWT320T (5′-TCAGGCAAAGGTGATATATCACACTTTTTTTTTTTTTTTTTTTT-3′) |
| inhA promoter | Wild type | TOMIWT10T (5′-GGCGAGACGATAGGTTGTCGGTTTTTTTTTT-3′) |
| inhA promoter | Mutant (−15T) | TOMIMUT110T (5′-GGCGAGATGATAGGTTGTCGGTTTTTTTTTT-3′) |
| inhA promoter | Mutant (−8A) | TOMIMUT210T (5′-GGCGAGACGATAGGATGTCGGTTTTTTTTTT-3′) |
Boldface indicates the specific nucleotides of interest.
Hybridization and detection.
Hybridization and detection were conducted by using the conditions and buffers described previously (12). Briefly, successful amplification was monitored by electrophoresis of 2.5 μl of the PCR product on a 1.5% (wt/vol) agarose gel visualized under UV light. The product sizes were 250, 212, 238, and 264 bp for the target regions encompassing katG 315, katG 463, the inhA promoter, and the oxyR-ahpC intergenic region, respectively. The remaining 22.5 μl of the PCR products was transferred to a 1-ml plastic tube containing a numbered directed oligonucleotide array and 250 μl of hybridization solution (5× SSC, 0.1% [wt/vol] N-lauroyl sarcosine [Sarkosyl], 0.02% [wt/vol] sodium dodecyl sulfate, 1% [wt/vol] blocking reagent [Roche Molecular Biochemicals]). Following denaturation at 100°C, DNA hybridization was performed for 30 min at 50°C and unbound probe was removed by washing. Hybridization was detected by incubating the membranes for 30 min at room temperature with antidigoxigenin alkaline phosphatase antibody, followed by colorimetric visualization.
Interpretation of the directed PCR-hybridization oligonucleotide array.
Each locus was addressed by two to three directed oligonucleotides (Table 1). katG 315, katG 463, and the inhA promoter were characterized by both wild-type and mutant target oligonucleotide sequences that define the mutations most commonly reported in the literature. The oxyR-ahpC intergenic region was defined by three overlapping wild-type targets. The hybridization stringency was previously optimized to detect a single base pair change within the target sequences (unpublished data). Hybridization to a wild-type target in conjunction with the absence of hybridization to mutant targets at the same locus defines a wild-type strain. Hybridization to a mutant target in conjunction with the absence of hybridization to the wild-type target or an alternative mutant target defines a mutant strain with a characterized mutation (Fig. 1). A strain for which the absence of hybridization to both the wild-type and mutant targets or the unpaired wild-type targets and in which failure of amplification of the locus in the multiplex PCR was ruled out was interpreted as a mutant strain requiring further characterization by DNA sequencing.
FIG. 1.
Hybridization patterns obtained with the directed low-density oligonucleotide PCR and hybridization array. Probe numbers are indicated at the bottom of the array. Probes 1 to 3 target inhA (probe 1, inhA −8A; probe 2, inhA −15T; probe 3, wild-type inhA promoter). Probes 4 to 6 target the oxyR-ahpC intergenic region (probe 4, positions −50 to −27; probe 5, positions −30 to −10; probe 6, positions −12 to +9 with respect to the transcriptional start site). Probes 7 to 8 target the katG 463 polymorphism [probe 7, katG 463Leu(CTG); probe 8, katG 463Arg(CCG)], and probes 9 to 11 target katG 315 [probe 9, katG 315Thr(ACC); probe 10, katG 315Asn(AAC); probe 11, katG 315Ser(AGC)]. Strip inh 11 is the negative control, and strips inh 12 to 18 are examples of hybridization patterns obtained with test isolates.
DNA sequencing.
When the array did not fully characterize a mutation, the individual locus was amplified. The amplification products were purified by precipitation with 20% polyethylene glycol-2.5 M NaCl, and the nucleotide sequence of each DNA strand was determined with internal nested primers and the BigDye Ready Reaction Mix (Applied Biosystems, Foster City, Calif.), according to the instructions of the manufacturer. Unincorporated dye terminators were removed by precipitation with 96% ethanol-0.115 M sodium acetate (pH 4.6). The reaction products were separated and detected with an ABI Prism 3700 or an ABI Prism 377 automated DNA sequencer (Applied Biosystems). In addition, 10 randomly selected isolates representing each polymorphism characterized by the array were sequenced.
RESULTS
Two hundred thirty-two isoniazid-resistant isolates were identified over the 1-year period, of which 202 were viable and available for analysis. One hundred seventy-six fully susceptible isolates were selected at random for comparison. Phenotypically, 107 isolates were isoniazid monoresistant; 46 isolates were resistant to both isoniazid and streptomycin; 47 isolates were MDR M. tuberculosis, i.e., resistant to isoniazid and rifampin (of which 30 were resistant to three or more first-line antituberculous drugs); 1 isolate was resistant to isoniazid and ethambutol; and 1 isolate was resistant to isoniazid, streptomycin, and ethambutol. Information regarding previous tuberculosis history could be determined for 147 of the 202 (72%) patients infected with isoniazid-resistant isolates and 138 of 176 (78.4%) patients infected with fully susceptible isolates. Seventy-five percent (109 of 147) of the patients infected with isoniazid-resistant M. tuberculosis had no history of TB. Of the 38 patients with a history of TB, 20 were known to be infected with MDR M. tuberculosis strains and were already undergoing treatment; however, cultures remained positive. Of the patients infected with fully susceptible M. tuberculosis, 85% (120 of 138) had no history of TB. Although the isolates were identified from patients resident in England and Wales, they were born in 29 different countries. Only 22.8% of the patients with a documented country of birth were born in the United Kingdom.
Hybridization to directed oligonucleotide array.
The oligonucleotide array fully characterized more than 90% of the 378 isolates analyzed. Loci with paired mutant and wild-type target sequences were fully characterized in 92.1 to 99.0% of isolates analyzed (katG 315, 92.1%; inhA promoter, 98.4%; katG 463, 99%). Hybridization to all three target sequences in the oxyR-ahpC intergenic region occurred for 79.1% of the isolates analyzed, consistent with the presence of wild-type DNA sequences. Sequencing was performed for all isolates with uncharacterized loci.
One or more loci of 23 isolates were sequenced to determine the accuracy of the array. Ten isolates, when that number was available, were sequenced for the detection of each locus polymorphism. There was 100% congruency between the sequencing data and the presence or the absence of a mutation, as determined with the directed oligonucleotide array for all but one of the nucleotide sequences; the exception was inhA −8A. The array detected only two isolates that hybridized to this target and that failed to hybridize to the wild-type and the inhA −15T targets. One of the isolates did indeed have the defined base substitution; the other had a inhA T−8C substitution.
Of the isolates not characterized by the array, 30 were uncharacterized at katG 315, 5 were uncharacterized at katG 463, 6 were uncharacterized in the inhA promoter region, and 79 were uncharacterized in the oxyR-ahpC intergenic region. Although 25 of 30 isolates had sequences at the katG 315 locus that should have been characterized by the array (9 wild-type isolates, 12 katG 315Thr isolates, and 4 katG 315Asn isolates), 5 of 30 (16.7%) isolates had other mutations that prevented hybridization. These included one isolate with katG 315Ile (ATC), one isolate with a synonymous nucleotide substitution affecting codon 314 (ACC to ACT [Thr]), one isolate with a double nucleotide substitution in codon 315 (AGC to ACA [Ser to Thr]) previously reported in the strain W of the Beijing family (5), and two isolates with probable whole-gene deletions. The last two isolates were uncharacterized by the array at both katG 315 and katG 463 but were characterized at inhA and oxyR-ahpC. Repeated amplification attempts with a variety of primer pairs including primer sequences 60 to 100 bp external to the katG gene were all unsuccessful, suggesting a probable gene deletion. In the inhA promoter region, one of six (16.7%) of the uncharacterized isolates had another mutation (C→T at position −24) that prevented hybridization to the target, and the remaining five had wild-type sequences.
Sixty-six of 79 (83.5%) isolates uncharacterized by the array had a mutation in the oxyR-ahpC intergenic region. The vast majority (59 of 66 isolates) had a single point mutation (G→A) at position −46 with respect to the ahpC transcriptional start site (33). Two isolates had the point mutation at position −46 plus another mutation (T→A at position −34 or deletion of T at position −34), three isolates had a G→A substitution at position −6, one isolate had a C→T substitution at position −20, and one isolate had a C→T substitution at position −30.
Frequency of mutations within the unselected study population.
The results of sequence characterization at each locus by directed analysis with a low-density array and limited DNA sequencing are shown in Table 2. Of the 202 prevalent isoniazid-resistant isolates from England in 1998, 59.4% (120 of 202) of the isolates had a mutation that produced an amino acid change at katG 315, of which the vast majority (94.2%) possessed the katG AGC315ACC nucleotide substitution. Only five isolates possessed the katG AGC315AAC nucleotide substitution. No nucleotide substitutions were identified at the katG 315 locus in the 176 sensitive isolates. In contrast, the katG Arg463Leu polymorphism was present in both sensitive and resistant isolates in similar proportions: among the isoniazid-resistant isolates, 104 (51.5%) had an arginine and 96 (48%) had a leucine; among the fully susceptible isolates, 106 (60.2%) had an arginine and 70 (39.8%) had a leucine.
TABLE 2.
Frequencies of mutations occurring at three gene loci associated with isoniazid resistance (katG 315, the inhA promoter region, and the oxyR-ahpC intergenic region) in 202 isoniazid-resistant isolates and 176 phenotypically fully susceptible isolates
| Mutation | Sequence
|
No. of isolates
|
||||
|---|---|---|---|---|---|---|
| katG 315 | inhA promoter region | oxyR-ahpC intergenic region | katG 463 | Isoniazid resistant | Isoniazid sensitivea | |
| katG 315 | Ser (AGC) 315 Thr (ACC) | Wild type | Wild type | Leu | 25 | 0 |
| Ser (AGC) 315 Thr (ACC) | Wild type | Wild type | Arg | 51 | 0 | |
| Ser (AGC) 315 Thr (ACC) | Wild type | −6 G→A | Arg | 1 | 0 | |
| Ser (AGC) 315 Thr (ACC) | Wild type | −46 G→A | Leu | 31 | 0 | |
| Ser (AGC) 315 Thr (ACC) | Wild type | −46 G→A and deletion of −34T | Leu | 1 | 0 | |
| Ser (AGC) 315 Asn (AAC) | Wild type | Wild type | Leu | 1 | 0 | |
| Ser (AGC) 315 Asn (AAC) | Wild type | Wild type | Arg | 3 | 0 | |
| Ser (AGC) 315 Asn (AAC) | Wild type | −46 G→A | Leu | 1 | 0 | |
| Ser (AGC) 315 Ile (ATC) | Wild type | Wild type | Arg | 1 | 0 | |
| Ser (AGC) 315 Thr (ACA) | Wild type | Wild type | Arg | 1 | 0 | |
| inhA promoter region | Wild type | −8 T→C | Wild type | Arg | 1 | 0 |
| Wild type | −8 T→A | Wild type | Arg | 0 | (1) | |
| Wild type | −15 C→T | Wild type | Leu | 15 | 0 | |
| Wild type | −15 C→T | Wild type | Arg | 27 | (1) | |
| Wild type | −15 C→T | −6 G→A | Arg | 1 | 0 | |
| Wild type | −15 C→T | −46 G→A | Leu | 7 | 0 | |
| katG 315 and inhA promoter region | Ser (AGC) 315 Thr (ACC) | −15 C→T | Wild type | Leu | 1 | 0 |
| Ser (AGC) 315 Thr (ACC) | −15 C→T | Wild type | Arg | 2 | 0 | |
| Ser (AGC) 315 Thr (ACC) | −24 C→T | −46 G→A | Leu | 1 | 0 | |
| None in katG 315 or inhA promoter region | Wild type | Wild type | −6 G→A | Arg | 1 | 0 |
| Wild type | Wild type | −20 C→T | Arg | 0 | 1 | |
| Wild type | Wild type | −30 C→T | Arg | 1 | 0 | |
| Wild type | Wild type | −46 G→A | Leu | 5 | 13 | |
| Wild type | Wild type | −46 G→A and −34 T→A | Leu | 1 | 0 | |
| Wild type | Wild type | Wild type | Leu | 7 | 56 | |
| Wild type | Wild type | Wild type | Arg | 13 | 104 | |
| Other | ACC (Thr) 314 ACT (Thr) | Wild type | Wild type | Leu | 1 | 0 |
| Probable deletion | Wild type | Wild type | Probable deletion | 1 | 0 | |
| Probable deletion | Wild type | 46 G→A | Probable deletion | 1 | 0 | |
| Total | 202 | 176 | ||||
Parentheses indicate discordant phenotypic and sequence results, i.e., sensitive phenotype but mutations present in inhA.
A mutation in the inhA promoter region was present in 55 of 202 (27.2%) of the isoniazid-resistant isolates, almost all of which (53 of 55) possessed the inhA C−15T nucleotide substitution. However, two isolates reportedly fully susceptible on phenotypic analysis also possessed nucleotide substitutions in the inhA promoter region (one each of inhA T−8A and inhA C−15T).
Fifty-two of 202 isoniazid-resistant isolates possessed nucleotide substitutions in the oxyR-ahpC intergenic region, of which 48 (23.7% overall) possessed a G→A substitution at position −46. This mutation was also found in 13 of 176 (7.5%) fully susceptible isolates. Of the five other mutations identified, four were present exclusively in resistant isolates and one was present in a susceptible isolate (C→T at position −20).
Fully susceptible isolates with mutations in the inhA promoter region or the oxyR-ahpC intergenic region were recorded and blindly submitted for repeat drug susceptibility testing at the HPA Mycobacterium Reference Unit. Both isolates with inhA mutations were reported to have raised resistance ratios. The isolate possessing inhA T−8A had a resistance ratio of 2 (i.e., it was elevated but it did not define the isolate as phenotypically resistant), whereas the isolate possessing the inhA C−15T polymorphism had a resistance ratio of 4 (i.e., it was phenotypically resistant). In contrast, all 13 isolates with mutations in the oxyR-ahpC intergenic region were reported to have a resistance ratio of 1, implying that mutations in the latter region are unrelated to isoniazid resistance
In combination, 85.6% (173 of 202) of isoniazid-resistant isolates from England and Wales had a mutation at either katG 315 or in the inhA promoter region. Four isolates possessed mutations both at katG 315 and in the inhA promoter region. Three were unrelated MDR M. tuberculosis isolates resistant to three or more antituberculous drugs, all of which possessed both katG 315ACC and inhA −15T. The other isolate was isoniazid monoresistant and possessed katG 315ACC and inhA −24T.
Isolates lacking mutations at katG 315 or in the inhA promoter region.
Twenty-nine isoniazid-resistant isolates had a wild-type sequence at both katG 315 and in the inhA promoter region. Eight of the 29 isolates had mutations in the oxyR-ahpC intergenic region (G→A at position −6, C→T at position −30, T→A at position −34, and G→A at position −46). Sequencing of the entire katG gene of these 29 isolates identified other mutations that resulted in amino acid substitutions in katG, excluding the katG 463 polymorphism, in 24.1% (7 of 29) of isolates, as shown in Table 3. Three isolates possessed the katG Ser315Thr mutation, which had not been detected by the array. Two isolates possessed phenotypically silent base substitutions that did not result in amino acid changes. Only two of the isolates that possessed isolated oxyR-ahpC intergenic mutations also possessed mutations in the katG gene; both resulted in a truncated protein product (deletion of A at nucleotide 149 that resulted in a frame shift and Trp107 that resulted in a stop codon). These were ahpC −34A and ahpC −30T, respectively. The ahpC −6A substitution was identified in three isolates and appeared to be unrelated to mutations in katG.
TABLE 3.
Mutations identified by sequencing the entire katG gene of the 29 isoniazid-resistant isolates lacking mutations causing an amino acid substitution at katG 315 or mutations in the inhA promoter region (excluding the katG 463 polymorphism)
| Nucleotide position | Triplet | Codon | Amino acid change | Frequency (no. of isolates) |
|---|---|---|---|---|
| −1 before gene | Insertion of Ca | 1 | ||
| 149 | Deletion of A | 50 | Frame shift | 1 |
| 321 | TGG→TGA | 107 | Trp→stop codon | 1 |
| 571 | TGG→CGG | 191 | Trp→Arg | 1 |
| 701 | GGG→GAG | 234 | Gly→Glu | 1 |
| 723 | CCC→CCGa | 241 | Pro→Pro | 2 |
| 942 | ACC→ACT | 314 | Thr→Thr | 1 |
| 1450 | CGT→AGT | 484 | Arg→Ser | 1 |
| 1469 | CGC→TGC | 490 | Arg→Cys | 1 |
| 2096 | GGG→GAG | 699 | Gly→Gln | 1 |
These mutations occurred together in one isolate.
Influence of strain clustering.
IS6110 RFLP typing was performed with all resistant isolates and 100 sensitive isolates. Secondary typing by spoligotyping was performed with isolates possessing less than five IS6110 copies.
Eighteen clusters involving 51 isolates were identified among the isoniazid-resistant isolates. Two clusters involving seven isolates were identified among the 100 randomly selected fully susceptible isolates typed. The clusters ranged in size from two to seven isolates, with a median cluster size of two. In total, 169 genotypically distinct isoniazid-resistant isolates were present in the population. Of these, 106 (62.7%) possessed a mutation that affected katG 315, 42 (24.9%) possessed a mutation in the inhA promoter region, and 144 (85.8%) possessed either a katG or a inhA mutation. Interestingly, neither the spoligotype nor the IS6110 RFLP type of the isolate with the double nucleotide substitution katG 315ACA was characteristic of the W strain or the Beijing family.
Relationship between genotype and phenotype.
The frequencies of the katG 315ACC mutation and the inhA −15T mutation did not differ significantly between the three major phenotypic groups (isoniazid-monoresistant, streptomycin and isoniazid dual-resistant, and MDR M. tuberculosis isolates) and were unrelated to a history of TB (Table 4).
TABLE 4.
Relationship between mutation frequency, drug resistance phenotype, and patient history of TB for 169 genotypically distinct isoniazid-resistant isolates
| Parameter | Total no. of isolates | No. of isolates with katG 315 mutationa:
|
P for katG 315 | No. of isolates with inhA −15 mutation:
|
P for inhA −15 | ||
|---|---|---|---|---|---|---|---|
| ACC | Other | T | C | ||||
| Resistance profile | |||||||
| Isoniazid monoresistant | 85 | 51 | 33 | 0.82 | 20 | 65 | 0.88 |
| Isoniazid and streptomycin only | 41 | 27 | 14 | 0.42 | 9 | 32 | 0.90 |
| MDR M. tuberculosis | 41 | 21 | 19 | 0.43 | 8 | 33 | 0.79 |
| Other | 2 | 0 | 2 | 1 | 1 | ||
| History of TB | |||||||
| No | 89 | 53 | 36 | 0.33 | 21 | 68 | 0.39 |
| Yes | 26 | 11 | 13 | 4 | 22 | ||
| Unknown | 54 | 35 | 19 | 13 | 41 | ||
| Total | 169 | 99 | 68 | 38 | 131 | ||
For two isolates, katG 315 could not be characterized; the locus could not be amplified due to a probable gene deletion.
ahpC −46A is a polymorphism confined to highly related isolates originating from the Indian subcontinent.
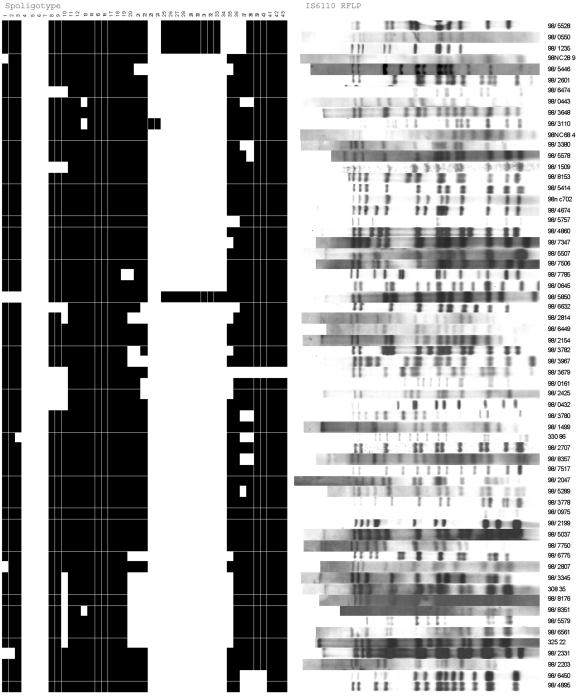
Although 24% of the prevalent isoniazid-resistant isolates from England and Wales possessed an ahpC G−46A nucleotide substitution, this polymorphism was also present in 7.4% of the isoniazid-susceptible isolates. In all cases the ahpC −46A polymorphism occurred in combination with katG 463Leu (CTG), although the latter was also present in isolation. DNA fingerprinting by spoligotyping and IS6110 RFLP analysis confirmed that highly related isolates were characterized by a loss of spacer hybridization at spacers 4 to 7 and 23 to 24 in all isolates, with an additional lack of spacer hybridization at spacers 25 to 34 in the vast majority of isolates and a 12- to 17-band RFLP pattern. No two patterns were identical (Fig. 2). Although 29 countries of birth were represented in the population studied, isolates with ahpC −46A were identified from patients representing only five countries, India, Pakistan, Somalia, the United Kingdom, and Uganda, the vast majority of whom 82.5% originated in India and Pakistan.
FIG. 2.
IS6110 RFLP and spoligotype patterns for isolates possessing the oxyR-ahpC intergenic mutation −46A.
Relationship between isoniazid resistance genotype and M. tuberculosis subdivisions defined by the katG 463 and ahpC −46 polymorphisms.
In view of the significantly higher proportion of genotypically distinct isoniazid-resistant isolates compared with the proportion of sensitive isolates possessing both the katG 463Leu polymorphism (89 of 167 resistant isolates versus 26 of 83 sensitive isolates [P = 0.002]) and the ahpC −46A polymorphisms (48 of 167 resistant isolates versus 7 of 83 sensitive isolates [P = 0.0004]), we looked at the relationship between the isoniazid resistance-conferring mutations katG 315Thr and inhA −15T and each of the polymorphisms. Isoniazid-sensitive and -resistant isolates were analyzed together. We found that both katG 463Leu and ahpC −46A were strongly associated with the katG 315Thr mutation (56 of 113 isoniazid-sensitive isolates versus 43 of 137 isoniazid-resistant isolates for katG 463Leu isolates [P = 0.002] and 34 of 55 isoniazid-sensitive isolates versus 65 of 195 isoniazid-resistant isolates for ahpC −46A isolates [P = 0.0002]) but not with inhA −15T (20 of 113 isoniazid-sensitive isolates versus 18 of 137 isoniazid-resistant isolates for katG 463Leu isolates [P = 0.404] and 8 of 55 isoniazid-sensitive isolates versus 30 of 195 isoniazid-resistant isolates for ahpC −46A isolates [P = 0.95]). When the isolates with katG 463Leu were subdivided according to those with the ahpC −46 polymorphism, only isolates possessing the ahpC −46A polymorphism were significantly associated with isoniazid resistance and katG 315Thr (34 of 58 isolates with ahpC −46A and 20 of 57 isolates without ahpC −46A [P = 0.02]). Among the isoniazid-resistant isolates, only the ahpC −46A polymorphism was significantly associated with katG 315Thr, although the relationship was much less marked (P = 0.048).
DISCUSSION
Isoniazid has been a mainstay of antimicrobial chemotherapy for TB since its discovery more than 50 years ago. The prodrug requires conversion by catalase-peroxidase into its active form, which interferes with mycolic acid biosynthesis. The gene encoding the catalase-peroxidase (katG) gene was identified in 1992 by Zhang et al. (55), who demonstrated that deletion resulted in the loss of catalase-peroxidase activity and resistance to isoniazid. Drug susceptibility could be restored by gene transfer (55), but the loss of catalase-peroxidase has a physiological cost (26), and total or partial gene deletion is rare. More commonly, resistance is associated with a single G-to-C point mutation, revealed by gene sequencing studies, which results in a serine-to-threonine amino acid substitution at codon 315 (33). Mutagenesis experiments confirm that this base substitution confers isoniazid resistance with only a moderate reduction in catalase-peroxidase activity (37, 53). Other resistance-conferring substitutions have been reported at the same site (AGC to AAC [serine to asparagine], AGC to ATC [serine to isoleucine], and AGC to ACA [serine to threonine]) (5), but much less frequently (33).
The target for the active form of isoniazid is not absolutely clear; however, in M. smegmatis, isoniazid resistance results from a single amino acid substitution in inhA, a gene encoding an enoyl acyl carrier protein reductase involved in mycolic acid synthesis (3). This mechanism has rarely been reported in M. tuberculosis, but mutations in the promoter region of inhA have been reported in about 20% of isoniazid-resistant isolates (33) and are thought to result in the upregulation of inhA. Plasmid transformation experiments producing overexpression of inhA confer both isoniazid resistance and ethionamide resistance to M. tuberculosis (22) and M. bovis BCG (3, 22).
The roles of mutations in the oxyR-ahpC locus are less clear. The ahpC gene encodes alkylhydroperoxidase, an enzyme thought to be important in the oxidative stress response (54). Mutations upstream of ahpC have previously been reported in isoniazid-resistant isolates. There is no evidence that overexpression of alkylhydroperoxidase confers isoniazid resistance to susceptible isolates; however, mutations resulting in the upregulation of ahpC have been associated with the absence of catalase-peroxidase activity and are thought to be compensatory mutations important for the survival of the organism (16, 39, 41). It has been argued that mutations in the 60-bp intergenic region upstream of ahpC could be used as a surrogate marker for catalase-negative isoniazid-resistant isolates with missense mutations elsewhere in the 2.7-kb katG gene (35, 42).
In this unselected, population-based study of 202 isoniazid-resistant isolates and 176 sensitive isolates, representing 87% of all isoniazid-resistant M. tuberculosis isolates identified in England and Wales in 1998, the observed frequencies of mutations in katG 315, the inhA promoter region, and the oxyR-ahpC intergenic region were similar to those reported in previous studies: 63% (30, 44, 48, 50), 22% (20, 30, 34), and 30% (41), respectively. However, the range of previously reported mutation frequencies is wide: katG, 39.4% (34, 42) to 91.3% (20, 23, 28); inhA promoter, 4.3% (44, 48) to 34.4% (42); and oxyR-ahpC, 5.7% (20) to 28.5% (41). The prevalence of the katG gene deletion in our M. tuberculosis population was very low. The frequency of katG 315 mutations did not differ significantly between MDR M. tuberculosis isolates and isoniazid-monoresistant isolates, in contrast to previous studies, in which a higher frequency of katG 315 mutations has been reported in MDR M. tuberculosis isolates (2, 20, 23, 28, 32, 46).
It has been suggested that genetic detection of isoniazid resistance should be performed by analysis of katG 315 alone (20); however, this is based on studies from areas with a high prevalence of TB, often with a predominance of Beijing and MDR M. tuberculosis strains and evidence of clonal expansion resulting from high transmission rates. In this study, only 63% of isoniazid-resistant isolates possessed a katG 315 mutation, and mutations in the inhA promoter region were present in a further 21% of the isoniazid-resistant isolates. Combined analysis of just these two loci revealed that 85.6% of the isoniazid-resistant isolates in the population and that 82.8% of the genotypically distinct (unrelated) isolates possessed a nucleotide substitution affecting one or the other locus. The combined prevalence was similar to those in previous studies (56 to 100% of all isolates studied and 56.5 to 100% of genotypically distinct isolates), although the relative contributions of katG 315 and inhA promoter mutations varied significantly between studies (11, 18, 29, 30, 38, 44, 48). We found, in contrast to some previous studies (19), that analysis of the oxyR-ahpC intergenic region contributed little to the molecular detection of isoniazid resistance. Although we identified nucleotide substitutions in the oxyR-ahpC intergenic region in a quarter of the isoniazid-resistant isolates, mutations in this region were also present in 8% of the susceptible isolates. More than 90% of the isolates, including all but one of the susceptible isolates with mutations in the region, possessed a G-to-A substitution at position −46 with respect to the ahpC transcriptional start site. One-fifth of the isoniazid-resistant isolates analyzed lacked resistance-conferring mutations at katG 315 or the inhA promoter; of these, almost a third (8 of 26 isolates) possessed mutations in the oxyR-ahpC intergenic region, but of these only 2 isolates possessed mutations elsewhere in the katG gene. Interestingly, both isolates possessed katG mutations which, on translation, result in premature termination of the catalase-peroxidase protein. One of the associated oxyR-ahpC intergenic mutations (C→T at position −30) is known to upregulate alkylhydroperoxidase (19); the other mutation (ahpC −34A) has not been reported previously. These observations support the compensatory mutation hypothesis; however, not all oxyR-ahpC intergenic mutations are compensatory (19), and other mechanisms are likely to apply. Interestingly, of the two isolates with probable katG gene deletions, only one had an intergenic mutation, and this was ahpC −46A. Five isolates had nonsynonymous nucleotide substitutions elsewhere in katG with no associated oxyR-ahpC intergenic mutation. This is not necessarily surprising. However, these katG mutations are previously unreported, and their effects on catalase-peroxidase activity and isoniazid resistance are unknown.
The ahpC −46A base substitution found in both isoniazid-resistant and -susceptible isolates in our population proved interesting. This base substitution has rarely been reported in the literature but has been found in both susceptible and resistant isolates (41). Immunoblot analysis of ahpC expression in isolates possessing the polymorphism demonstrated a lack of detectable alkylhydroperoxidase production implying that it is not a compensatory mutation for gene upregulation (41). Sixteen percent of all isolates studied in our population possessed the ahpC −46A polymorphism. In all cases, it occurred with the katG 463Leu polymorphism. This was also true of the five isolates that possessed the polymorphism previously reported in the literature (41). The katG Arg463Leu polymorphism is present throughout the M. tuberculosis complex, and in conjunction with the polymorphism gyrA Thr95Ser, it has been used to divide the complex into three clonally derived phylogenetic groups (40). A tight relationship between ahpC −46A and katG 463Leu (CTG) implies that the former is a genetic marker for a clonally related subgroup of M. tuberculosis strains in group 1. DNA fingerprinting analysis confirmed a closely related group of isolates, which were not the result of recent transmission, that possessed a characteristic deletion pattern on spoligotyping and similar IS6110 RFLP patterns. Demographic data revealed that the isolates were obtained predominantly from patients originating from India and Pakistan. The characteristic pattern of spacer deletion obtained on spoligotyping of the strains possessing ahpC −46A closely resembles that described as Central Asian pattern 1 (CAS-1) in the international spoligotyping database (14) and has previously been observed in strains obtained from northern India (4). In addition, the IS6110 RFLP pattern shares the 12.1- and 10.1-kb IS6110-bearing PvuII fragments described as characteristic of the Delhi strain family. Taking the above features together, the ahpC −46A polymorphism defines the CAS-1 and Delhi strain family but may encompass an even larger subgroup of clonally derived strains originating from the Indian subcontinent.
It has been suggested that some strains of M. tuberculosis may have an increased propensity to support genetic mutations. The best example of this is the Beijing strain. Although originally described in isolates originating from Beijing, People's Republic of China (51), the strain has disseminated globally. Studies of Beijing strains in Southeast Asia do not demonstrate any association with drug resistance, whereas those of strains from New York City (6), Estonia (21), Cuba (10), and Russia (12, 45) describe strong associations with drug resistance, most typically MDR M. tuberculosis (15). Only 17 isolates in our study population were Beijing strains, which formed a population too small for subgroup analysis. However, strains possessing the ahpC −46A polymorphism were noted to occur more commonly among isoniazid-resistant isolates than among fully susceptible isolates. Analysis of genetically distinct strains revealed a statistically significant association of the polymorphism with isoniazid resistance and, in particular, with the katG 315Thr resistance-conferring mutation. Although on first analysis it appears that this is also true of all group 1 isolates (of which Beijing strains are also a subgroup), the results are skewed by the ahpC −46A strains. In contrast, MDR M. tuberculosis strains were not associated with either of the two common isoniazid resistance-conferring mutations. The strong association between ahpC −46A and katG 315Thr may reflect an increased ability of strains to support the katG 315Thr mutation or may purely reflect the widespread use of isoniazid monotherapy in India and Pakistan following successful trials in the early 1950s and the acquisition of a resistance-conferring mutation that does not significantly compromise viability or virulence.
The population of M. tuberculosis isolates from England and Wales has provided a unique insight into the genotypic analysis of isoniazid resistance conferred by changes at three commonly used loci. Although the oxyR-ahpC intergenic region does not contribute to the genotypic detection of isoniazid resistance, a single nucleotide polymorphism within the region can define a previously geographically restricted group of strains. By analyzing just two loci, katG codon 315 and the inhA promoter region, by a multiplex PCR and directed low-density oligonucleotide array analysis with limited DNA sequencing when necessary, 82.8% of genotypically distinct isoniazid-resistant isolates were identified. PCR and directed oligonucleotide array analysis provides a highly cost-effective approach for the rapid genotypic detection of isoniazid resistance.
Acknowledgments
We thank all the research and reference unit staff at the HPA Mycobacterium Reference Unit for assistance with IS6110 RFLP analysis and spoligotyping and Jo Herbert, HPA Colindale, for help with demographic analysis.
L.V.B. was a British Lung Foundation research fellow. This work was funded by the British Lung Foundation.
REFERENCES
- 1.Anthony, R. M., T. J. Brown, and G. L. French. 2000. Rapid diagnosis of bacteremia by universal amplification of 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J. Clin. Microbiol. 38:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakonyte, D., A. Baranauskaite, J. Cicenaite, A. Sosnovskaja, and P. Stakenas. 2003. Molecular characterization of isoniazid-resistant Mycobacterium tuberculosis clinical isolates in Lithuania. Antimicrob. Agents Chemother. 47:2009-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee, A., E. Dubnau, A. Quemard, V. Balasubramanian, K. S. Um, T. Wilson, D. Collins, G. de Lisle, and W. R. Jacobs, Jr. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227-230. [DOI] [PubMed] [Google Scholar]
- 4.Bhanu, N., D. van Soolingen, J. van Embden, L. Dar, R. Pandey, and P. Seth. 2002. Predominace of a novel Mycobacterium tuberculosis genotype in the Delhi region of India. Tuberculosis (Edinburgh) 82:105. [DOI] [PubMed] [Google Scholar]
- 5.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-52. [DOI] [PubMed] [Google Scholar]
- 6.Bifani, P. J., B. B. Plikaytis, V. Kapur, K. Stockbauer, X. Pan, M. L. Lutfey, S. L. Moghazeh, W. Eisner, T. M. Daniel, M. H. Kaplan, J. T. Crawford, J. M. Musser, and B. N. Kreiswirth. 1996. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA 275:452-457. [PubMed] [Google Scholar]
- 7.Brown, T. J., and R. M. Anthony. 2000. The addition of low numbers of 3′ thymine bases can be used to improve the hybridization signal of oligonucleotides for use within arrays on nylon supports. J. Microbiol. Methods 42:203-207. [DOI] [PubMed] [Google Scholar]
- 8.Collins, C., J. Grange, and M. D. Yates. 1997. Tuberculosis, bacteriology, organisation and practice, 2nd ed. Butterworth-Heinemann, Oxford, United Kingdom.
- 9.de Beenhouwer, H., Z. Lhiang, G. Jannes, W. Mijs, L. Machtelinckx, R. Rossau, H. Traore, and F. Portaels. 1995. Rapid detection of rifampicin resistance in sputum and biopsy specimens from tuberculosis patients by PCR and line probe assay. Tuber. Lung Dis. 76:425-430. [DOI] [PubMed] [Google Scholar]
- 10.Diaz, R., K. Kremer, P. E. de Haas, R. I. Gomez, A. Marrero, J. A. Valdivia, J. D. Van Embden, and D. van Soolingen. 1998. Molecular epidemiology of tuberculosis in Cuba outside of Havana, July 1994-June 1995: utility of spoligotyping versus IS6110 restriction fragment length polymorphism. Int. J. Tuberc. Lung Dis. 2:743-750. [PubMed] [Google Scholar]
- 11.Dobner, P., S. Rusch-Gerdes, G. Bretzel, K. Feldmann, M. Rifai, T. Loscher, and H. Rinder. 1997. Usefulness of Mycobacterium tuberculosis genomic mutations in the genes katG and inhA for the prediction of isoniazid resistance. Int. J. Tuberc. Lung Dis. 1:365-369. [PubMed] [Google Scholar]
- 12.Drobniewski, F., Y. Balabanova, M. Ruddy, L. Weldon, K. Jeltkova, T. Brown, N. Malomanova, E. Elizarova, A. Melentyey, E. Mutovkin, S. Zhakharova, and I. Fedorin. 2002. Rifampin- and multidrug-resistant tuberculosis in Russian civilians and prison inmates: dominance of the Beijing strain family. Emerg. Infect. Dis. 8:1320-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espinal, M. A., A. Laszlo, L. Simonsen, F. Boulahbal, S. J. Kim, A. Reniero, S. E. Hoffner, H. L. Rieder, N. Binkin, C. Dye, R. Williams, M. C. Raviglione, et al. 2001. Global trends in resistance to antituberculosis drugs. N. Engl. J. Med. 344:1294-1303. [DOI] [PubMed] [Google Scholar]
- 14.Filliol, I., J. R. Driscoll, D. van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valetudie, D. D. Anh, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. H. Haas, H. Heersma, G. Kallenius, E. Kassa-Kelembho, T. Koivula, H. M. Ly, A. Makristathis, C. Mammina, G. Martin, P. Mostrom, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. De Waard, C. Sola, and N. Rastogi. 2002. Global distribution of Mycobacterium tuberculosis spoligotypes. Emerg. Infect. Dis. 8:1347-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heym, B., E. Stavropoulos, N. Honore, P. Domenech, B. Saint-Joanis, T. M. Wilson, D. M. Collins, M. J. Colston, and S. T. Cole. 1997. Effects of overexpression of the alkyl hydroperoxide reductase ahpC on the virulence and isoniazid resistance of Mycobacterium tuberculosis. Infect. Immun. 65:1395-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapur, V., L. L. Li, M. R. Hamrick, B. B. Plikaytis, T. M. Shinnick, A. Telenti, W. R. Jacobs, Jr., A. Banerjee, S. Cole, and K. Y. Yuen. 1995. Rapid Mycobacterium species assignment and unambiguous identification of mutations associated with antimicrobial resistance in Mycobacterium tuberculosis by automated DNA sequencing. Arch. Pathol. Lab. Med. 119:131-138. [PubMed] [Google Scholar]
- 19.Kelley, C. L., D. A. Rouse, and S. L. Morris. 1997. Analysis of ahpC gene mutations in isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 41:2057-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiepiela, P., K. S. Bishop, A. N. Smith, L. Roux, and D. F. York. 2000. Genomic mutations in the katG, inhA and ahpC genes are useful for the prediction of isoniazid resistance in Mycobacterium tuberculosis isolates from Kwazulu Natal, South Africa. Tuber. Lung Dis. 80:47-56. [DOI] [PubMed] [Google Scholar]
- 21.Kruuner, A., S. E. Hoffner, H. Sillastu, M. Danilovits, K. Levina, S. B. Svenson, S. Ghebremichael, T. Koivula, and G. Kallenius. 2001. Spread of drug-resistant pulmonary tuberculosis in Estonia. J. Clin. Microbiol. 39:3339-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen, M. H., C. Vilcheze, L. Kremer, G. S. Besra, L. Parsons, M. Salfinger, L. Heifets, M. H. Hazbon, D. Alland, J. C. Sacchettini, and W. R. Jacobs, Jr. 2002. Overexpression of inhA, but not kasA, confers resistance to isoniazid and ethionamide in Mycobacterium smegmatis, M. bovis BCG and M. tuberculosis. Mol. Microbiol. 46:453-466. [DOI] [PubMed] [Google Scholar]
- 23.Marttila, H. J., H. Soini, E. Eerola, E. Vyshnevskaya, B. I. Vyshnevskiy, T. F. Otten, A. V. Vasilyef, and M. K. Viljanen. 1998. A Ser315Thr substitution in katG is predominant in genetically heterogeneous multidrug-resistant Mycobacterium tuberculosis isolates originating from the St. Petersburg area in Russia. Antimicrob. Agents Chemother. 42:2443-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marttila, H. J., H. Soini, P. Huovinen, and M. K. Viljanen. 1996. katG mutations in isoniazid-resistant Mycobacterium tuberculosis isolates recovered from Finnish patients. Antimicrob. Agents Chemother. 40:2187-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melzer, M., T. J. Brown, G. L. French, A. Dickens, T. D. McHugh, L. R. Bagg, R. A. Storring, and S. Lacey. 2002. Molecular analysis of drug resistant TB. Thorax 57:562-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Middlebrook, G., and M. Cohn. 1953. Some observations on the pathogenicity of isoniazid resistant variants of tubercle bacilli. Science 118:297-299. [DOI] [PubMed] [Google Scholar]
- 27.Mokrousov, I., N. V. Bhanu, P. N. Suffys, G. V. Kadival, S. F. Yap, S. N. Cho, A. M. Jordaan, O. Narvskaya, U. B. Singh, H. M. Gomes, H. Lee, S. P. Kulkarni, K. C. Lim, B. K. Khan, D. van Soolingen, T. C. Victor, and L. M. Schouls. 2004. Multicenter evaluation of reverse line blot assay for detection of drug resistance in Mycobacterium tuberculosis clinical isolates. J. Microbiol. Methods 57:323-335. [DOI] [PubMed] [Google Scholar]
- 28.Mokrousov, I., O. Narvskaya, T. Otten, E. Limeschenko, L. Steklova, and B. Vyshnevskiy. 2002. High prevalence of katG Ser315Thr substitution among isoniazid-resistant Mycobacterium tuberculosis clinical isolates from northwestern Russia, 1996 to 2001. Antimicrob. Agents Chemother. 46:1417-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris, S., G. H. Bai, P. Suffys, L. Portillo-Gomez, M. Fairchok, and D. Rouse. 1995. Molecular mechanisms of multiple drug resistance in clinical isolates of Mycobacterium tuberculosis. J. Infect. Dis. 171:954-960. [DOI] [PubMed] [Google Scholar]
- 30.Musser, J. M., V. Kapur, D. L. Williams, B. N. Kreiswirth, D. van Soolingen, and J. D. Van Embden. 1996. Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: restricted array of mutations associated with drug resistance. J. Infect. Dis. 173:196-202. [DOI] [PubMed] [Google Scholar]
- 31.Nachamkin, I., C. Kang, and M. P. Weinstein. 1997. Detection of resistance to isoniazid, rifampin, and streptomycin in clinical isolates of Mycobacterium tuberculosis by molecular methods. Clin. Infect. Dis. 24:894-900. [DOI] [PubMed] [Google Scholar]
- 32.Piatek, A. S., A. Telenti, M. R. Murray, H. el Hajj, W. R. Jacobs, Jr., F. R. Kramer, and D. Alland. 2000. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob. Agents Chemother. 44:103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 34.Ramaswamy, S. V., R. Reich, S. J. Dou, L. Jasperse, X. Pan, A. Wanger, T. Quitugua, and E. A. Graviss. 2003. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rinder, H., A. Thomschke, S. Rusch-Gerdes, G. Bretzel, K. Feldmann, M. Rifai, and T. Loscher. 1998. Significance of ahpC promoter mutations for the prediction of isoniazid resistance in Mycobacterium tuberculosis. Eur. J. Clin. Microbiol. Infect. Dis. 17:508-511. [DOI] [PubMed] [Google Scholar]
- 36.Rose, A. M., J. M. Watson, C. Graham, A. J. Nunn, F. Drobniewski, L. P. Ormerod, J. H. Darbyshire, and J. Leese. 2001. Tuberculosis at the end of the 20th century in England and Wales: results of a national survey in 1998. Thorax 56:173-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rouse, D. A., J. A. DeVito, Z. Li, H. Byer, and S. L. Morris. 1996. Site-directed mutagenesis of the katG gene of Mycobacterium tuberculosis: effects on catalase-peroxidase activities and isoniazid resistance. Mol. Microbiol. 22:583-592. [DOI] [PubMed] [Google Scholar]
- 38.Rouse, D. A., Z. Li, G. H. Bai, and S. L. Morris. 1995. Characterization of the katG and inhA genes of isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 39:2472-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherman, D. R., K. Mdluli, M. J. Hickey, T. M. Arain, S. L. Morris, C. E. Barry III, and C. K. Stover. 1996. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science 272:1641-1643. [DOI] [PubMed] [Google Scholar]
- 40.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sreevatsan, S., X. Pan, Y. Zhang, V. Deretic, and J. M. Musser. 1997. Analysis of the oxyR-ahpC region in isoniazid-resistant and -susceptible Mycobacterium tuberculosis complex organisms recovered from diseased humans and animals in diverse localities. Antimicrob. Agents Chemother. 41:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Telenti, A., N. Honore, C. Bernasconi, J. March, A. Ortega, B. Heym, H. E. Takiff, and S. T. Cole. 1997. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J. Clin. Microbiol. 35:719-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 44.Torres, M. J., A. Criado, N. Gonzalez, J. C. Palomares, and J. Aznar. 2002. Rifampin and isoniazid resistance associated mutations in Mycobacterium tuberculosis clinical isolates in Seville, Spain. Int. J. Tuberc. Lung Dis. 6:160-163. [PubMed] [Google Scholar]
- 45.Toungoussova, O. S., P. Sandven, A. O. Mariandyshev, N. I. Nizovtseva, G. Bjune, and D. A. Caugant. 2002. Spread of drug-resistant Mycobacterium tuberculosis strains of the Beijing genotype in the Archangel Oblast, Russia. J. Clin. Microbiol. 40:1930-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tracevska, T., I. Jansone, L. Broka, O. Marga, and V. Baumanis. 2002. Mutations in the rpoB and katG genes leading to drug resistance in Mycobacterium tuberculosis in Latvia. J. Clin. Microbiol. 40:3789-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Rie, A., R. Warren, I. Mshanga, A. M. Jordaan, G. D. van der Spuy, M. Richardson, J. Simpson, R. P. Gie, D. A. Enarson, N. Beyers, P. D. van Helden, and T. C. Victor. 2001. Analysis for a limited number of gene codons can predict drug resistance of Mycobacterium tuberculosis in a high-incidence community. J. Clin. Microbiol. 39:636-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Soolingen, D., M. W. Borgdorff, P. E. de Haas, M. M. Sebek, J. Veen, M. Dessens, K. Kremer, and J. D. Van Embden. 1999. Molecular epidemiology of tuberculosis in The Netherlands: a nationwide study from 1993 through 1997. J. Infect. Dis. 180:726-736. [DOI] [PubMed] [Google Scholar]
- 50.van Soolingen, D., P. E. de Haas, H. R. van Doorn, E. Kuijper, H. Rinder, and M. W. Borgdorff. 2000. Mutations at amino acid position 315 of the katG gene are associated with high-level resistance to isoniazid, other drug resistance, and successful transmission of Mycobacterium tuberculosis in The Netherlands. J. Infect. Dis. 182:1788-1790. [DOI] [PubMed] [Google Scholar]
- 51.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. Van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watterson, S. A., S. M. Wilson, M. D. Yates, and F. A. Drobniewski. 1998. Comparison of three molecular assays for rapid detection of rifampin resistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 36:1969-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wengenack, N. L., J. R. Uhl, A. L. St. Amand, A. J. Tomlinson, L. M. Benson, S. Naylor, B. C. Kline, F. R. Cockerill III, and F. Rusnak. 1997. Recombinant Mycobacterium tuberculosis katG(S315T) is a competent catalase-peroxidase with reduced activity toward isoniazid. J. Infect. Dis. 176:722-727. [DOI] [PubMed] [Google Scholar]
- 54.Wilson, T. M., and D. M. Collins. 1996. ahpC, a gene involved in isoniazid resistance of the Mycobacterium tuberculosis complex. Mol. Microbiol. 19:1025-1034. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, Y., B. Heym, B. Allen, D. Young, and S. Cole. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591-593. [DOI] [PubMed] [Google Scholar]