Abstract
Pseudomonas sp. strain ADP metabolizes atrazine to cyanuric acid via three plasmid-encoded enzymes, AtzA, AtzB, and AtzC. The first enzyme, AtzA, catalyzes the hydrolytic dechlorination of atrazine, yielding hydroxyatrazine. The second enzyme, AtzB, catalyzes hydroxyatrazine deamidation, yielding N-isopropylammelide. In this study, the third gene in the atrazine catabolic pathway, atzC, was cloned from a Pseudomonas sp. strain ADP cosmid library as a 25-kb EcoRI DNA fragment in Escherichia coli. The atzC gene was further delimited by functional analysis following transposon Tn5 mutagenesis and subcloned as a 2.0-kb EcoRI-AvaI fragment. An E. coli strain containing this DNA fragment expressed N-isopropylammelide isopropylamino hydrolase activity, metabolizing N-isopropylammelide stoichiometrically to cyanuric acid and N-isopropylamine. The 2.0-kb DNA fragment was sequenced and found to contain a single open reading frame of 1,209 nucleotides, encoding a protein of 403 amino acids. AtzC showed modest sequence identity of 29 and 25%, respectively, to cytosine deaminase and dihydroorotase, both members of an amidohydrolase protein superfamily. The sequence of AtzC was compared to that of E. coli cytosine deaminase in the regions containing the five ligands to the catalytically important metal for the protein. Pairwise comparison of the 35 amino acids showed 61% sequence identity and 85% sequence similarity. AtzC is thus assigned to the amidohydrolase protein family that includes cytosine deaminase, urease, adenine deaminase, and phosphotriester hydrolase. Similar sequence comparisons of the most highly conserved regions indicated that the AtzA and AtzB proteins also belong to the same amidohydrolase family. Overall, the data suggest that AtzA, AtzB, and AtzC diverged from a common ancestor and, by random events, have been reconstituted onto an atrazine catabolic plasmid.
Despite several years of research, little information concerning the genes involved in the metabolism of atrazine and other s-triazine compounds is available. An inducible set of genes that encode the enzymes for melamine (1,3,5-triazine-2,4,6-triamine) metabolism was isolated from Pseudomonas sp. strain NRRL B-12227 (15, 16). While NRRL B-12227 did not degrade atrazine, it metabolized melamine via six enzymatic steps to liberate six ammonia molecules. Three of the genes involved in the melamine degradation pathway, trzB, trzC, and trzD, have been cloned. Similar degradative genes have been isolated from Pseudomonas sp. strain NRRL B-12228 and Klebsiella pneumoniae 99 (15, 16). More recently, it has been shown that the genes encoding ammelide aminohydrolase (trzC) and cyanuric acid amidohydrolase (trzD) (from strain NRRLB 12227) are located on a large IncI plasmid in K. pneumonia 99 (21).
Genes encoding atrazine degradation activity from Rhodococcus sp. strains have been reported (27–29). In Rhodococcus sp. strain TE1, N dealkylation of atrazine is mediated by a single gene, atrA (33). Rhodococcus corallinus NRRL B-15544R has the ability to dechlorinate the s-triazines desethylsimazine and desethylatrazine (26). This strain, however, does not metabolize atrazine or simazine. The gene responsible for the dechlorination-deamination has been sequenced and is termed trzA (35). A Rhodococcus cytochrome P-450 multicomponent monooxygenase system, encoded by the thcBCD genes (34), catalyzes the N dealkylation of atrazine to desethylsimazine and desethylatrazine (27, 28). A recombinant Rhodococcus strain containing atrA and trzA catalyzes multiple steps in atrazine metabolism but not the complete mineralization of atrazine (35).
We recently used functional and transposon Tn5 mutagenesis approaches to isolate and characterize gene regions encoding atrazine catabolism by Pseudomonas sp. strain ADP (2, 10–12). Pseudomonas sp. strain ADP (23) uses atrazine as a sole source of nitrogen for growth and transforms the ring and side-chain atoms to carbon dioxide. The first gene in the degradation pathway, atzA, catalyzes atrazine dechlorination to hydroxyatrazine (10), directly forming a nonphytotoxic metabolite (2). The atzA gene was localized to a 21.5-kb EcoRI genomic DNA fragment, designated pMD1, and shown to encode atrazine degradation activity in Escherichia coli DH5α. Atrazine degradation was demonstrated by a zone-clearing assay on agar medium containing crystalline atrazine (10). AtzA, a polypeptide of 473 amino acids, was purified to homogeneity by a rapid purification procedure (11) and found to be a homotetramer with a native molecular mass of about 245 kDa.
The second step in the atrazine catabolic pathway is encoded by atzB (2). Transposon Tn5 mutagenesis localized atzA and atzB to the same (21.5-kb) genomic DNA fragment, pMD1, as atzA. The atzB gene encodes a 481-amino-acid polypeptide that transforms hydroxyatrazine to N-isopropylammelide [2,4-dihydroxy-6-(isopropylamino)-s-triazine] by the hydrolytic removal of the N-ethyl group. The atzA and atzB genes are separated by approximately 8.7 kb in Pseudomonas sp. strain ADP (2, 12).
Transposons have been reported to be a significant factor affecting the evolution of novel degradative pathways (4). While there have been many reports of transposable elements that carry antibiotic resistance determinants, a smaller number have described catabolic transposons that specify metabolic pathways for the degradation of organic compounds (36). We recently reported that the Pseudomonas sp. strain ADP atzA gene was flanked by DNA showing greater than 95% sequence identity to insertion sequence IS1071 from Alcaligenes sp. strain BR60 and that the atzA and atzB genes are located on a 96-kb self-transmissible plasmid, pADP-1 (12). Moreover, six atrazine-degrading microorganisms which were recently isolated from geographically separated sites exposed to atrazine contain homologous atrazine degradation genes (12). Taken together, these results indicate that atrazine catabolism via hydroxyatrazine is widespread and suggests a potential molecular mechanism for the global dispersion of the atzA and atzB genes.
Protein sequence analyses have indicated that AtzA and AtzB are 41 and 25% homologous, respectively, to TrzA, a protein that catalyzes hydrolytic deamination of the s-triazine substrates melamine and 2-chloro-4,6-diamino-s-triazine, but not atrazine (2, 26). TrzA has very recently been identified as a member of a broad class of bacterial amidohydrolases that includes dihydroorotase, cytosine deaminase, urease, and adenine deaminase (18). AtzB is also an amidohydrolase, but its evolutionary relationship with other proteins has not been described.
In this study, we used N-isopropylammelide to screen a Pseudomonas sp. strain ADP gene library for the gene encoding the degradation of the AtzB product. This gene, atzC, is shown here to encode an enzyme catalyzing the hydrolytic deamidation of N-isopropylammelide to cyanuric acid and isopropylamine. Moreover, the gene sequence reveals that AtzC is a new member of the broad family of amidohydrolases that includes TrzA. Most surprisingly, AtzA, AtzB, and AtzC contain significant sequence identity near or in the regions of the amidohydrolase consensus metal-binding amino acids. The data suggests that AtzA, AtzB, and AtzC have all diverged from a common ancestor and have now been assembled on a catabolic plasmid for the purpose of metabolizing the herbicide atrazine.
MATERIALS AND METHODS
Chemicals.
Authentic samples of atrazine (2-chloro-4-isopropylamino-6-isopropylamino-1,3,5-s-triazine) and N-isopropylammelide (2,4-dihydroxy-6-isopropylamino-s-triazine) were obtained from Novartis Crop Protection, Greensboro, N.C. Cyanuric acid (2,4,6-trihydroxy-1,3,5-s-triazine) and biuret (imidocarbonic diamide) were obtained from Aldrich Chemical Co., Milwaukee, Wis.
Bacteria and growth conditions.
Atrazine-degrading Pseudomonas sp. strain ADP was previously described (23) and was grown at 37°C in minimal salt medium (10) or Luria-Bertani (LB) medium (32). E. coli DH5α (32) was used for all molecular manipulations. E. coli(pLTD4) was grown in LB medium supplemented with tetracycline (30 μg/ml), and E. coli containing plasmid pTD2 or pTD2.5 was grown in LB medium containing ampicillin (25 μg/ml).
Library screening.
A Pseudomonas sp. strain ADP DNA library containing 2,000 clones was obtained as described previously (10). E. coli colonies were grouped into 20 sets of 100 clones each. Each set of 100 clones was grown as a mixture in a single test tube with 5 ml of LB medium containing 50 μg of N-isopropylammelide per ml. After 18 h of incubation at 37°C, the mixed clones were screened by high-performance liquid chromatography (HPLC) analysis for the disappearance of N-isopropylammelide (see below). Positive mixtures were further analyzed by dividing the 100 original clones into 10 subgroups and repeating the HPLC screening procedure. Individual clones with N-isopropylammelide-metabolizing ability were subsequently identified. One clone, pLTD4, was chosen for further analysis.
Plasmids and molecular manipulations.
Subcloning and plasmid purification were performed as described previously (32). Plasmid pLTD4 contains a 25-kb EcoRI genomic DNA fragment from Pseudomonas sp. strain ADP cloned in cosmid pLAFR3 (10). A 2.7-kb EcoRI fragment from plasmid pLTD4 was subcloned into the EcoRI site of pUC18, generating plasmid pTD2. Plasmid pTD2 was further subcloned as a 2.0-kb EcoRI-AvaI fragment in pUC18, producing plasmid pTD2.5.
DNA sequencing.
The DNA sequence of plasmid pTD2.5 was obtained by use of custom synthesized primers (Gibco BRL, Gaithersberg, Md.). The DNA sequence was generated by fluorescence sequencing with the Applied Biosystems (Foster City, Calif.) Prism DyeDeoxy Terminator cycle sequencing kit. Sequencing reaction mixtures were prepared with a TempCycler II thermal cycler (Coy Laboratory Products, Inc., Ann Arbor, Mich.), purified through Centri-Sep spin columns (Princeton Separations, Inc., Adelphia, N.J.), and analyzed on an Applied Biosystems model 373 DNA sequencer. DNA sequence data were compiled with the GeneWorks 2.45 software package (IntelliGenetics, Inc., Mountain View, Calif.).
Sequence analyses.
DNA and protein sequence analyses were done with GCG sequence analysis software (version 8.1-Unix; Genetics Computer Group, Madison, Wis.). Searches of protein and nucleic acid sequence data banks were performed at the National Center for Biotechnology Information by use of the BLAST and Blitz network services. The codon preference and third-position GC bias of possible coding regions were compared to a codon usage table for Pseudomonas sp. genes (PSE.COD) by use of the GCG program CODONPREFERENCE. Consensus patterns were found by use of the ProfileScan subroutine of the GCG program, and amino acid sequences were aligned manually.
Metabolic studies with resting cells.
E. coli DH5α cells containing pLTD4, pTD2, or pDT2.5 were grown overnight in LB medium with the appropriate antibiotics and harvested by centrifugation at 14,000 × g for 10 min. Cells were washed three times in 50 mM sodium phosphate buffer (pH 7.2) and resuspended in the same buffer to an A600 of 1.0. N-Isopropylammelide or isopropylamine was added to a final concentration of 50 μg/ml or 1 mM, and cell suspensions were incubated at 37°C for 12 h.
Analysis of N-isopropylammelide and metabolites.
N-Isopropylammelide and cyanuric acid were determined by HPLC analysis. Samples of resting cell suspensions were prepared for HPLC analysis by centrifugation at 14,000 × g for 10 min. Supernatant fractions were added to an equal volume of methanol and centrifuged at 14,000 × g for 10 min to remove salts and other insoluble materials. HPLC analysis was performed as described previously (14) with a Hewlett-Packard HP 1090 liquid chromatography system equipped with a photodiode array detector and interfaced to an HP 79994A Chemstation. N-Isopropylammelide and cyanuric acid were resolved by use of an analytical Lichrosorb RP-18 reverse-phase HPLC column (Alltech Associates, Deerfield, Ill.; 5-μm spherical packing; 250 by 4.6 μm). The isocratic mobile phase was 0.1 M potassium phosphate buffer (pH 7) at a flow rate of 1.0 ml min−1. For all compounds, spectral data for the column eluent were acquired at between 200 and 400 nm (12-nm bandwidth per channel) with a sampling frequency of 640 ms. Spectra were referenced against a signal at 550 nm and compared to those obtained with authentic samples of N-isopropylammelide and cyanuric acid. Concentrations of N-isopropylammelide and cyanuric acid were quantified by integrating peak areas at 220 nm. Under these conditions, N-isopropylammelide and cyanuric acid eluted from the column at about 20 and 4 min, respectively. Isopropylamine in supernatants from resting cell suspensions was determined with 2,4-dinitrofluorobenzene as described by McIntire and coworkers (25) and modified by Dubin (13).
Mass spectrometry.
The products of N-isopropylammelide degradation by E. coli DH5α(pLTD4) and E. coli DH5α(pTD2) were identified by mass spectrometry analysis. A 100-ml culture of E. coli DH5α(pLTD4) or DH5α(pTD2) was grown in LB medium containing tetracycline (30 μg/ml) or ampicillin (25 μg/ml), respectively, for 24 h at 37°C. Resting cells were prepared as described above and resuspended in 1 ml of 50 mM potassium phosphate buffer (pH 7.0) amended with 50 μg of N-isopropylammelide per ml. Cells were incubated, with shaking, at 37°C for 12 h. Cells were removed by centrifugation, and the supernatant was subjected to HPLC analysis as described above. Samples were collected from the targeted peak and subjected to mass spectrometry analysis. Direct-insertion mass spectrometry was performed by use of a glycerol matrix with a Kratos (Ramsey, N.J.) mass spectrometer operated in the fast atom bombardment mode with xenon.
Tn5 mutagenesis.
Tn5 mutagenesis of plasmid pTD2.5 was done with λ::Tn5 (λ 467 b221 rex::Tn5 cI857 Oam29 Pam80) as described by de Bruijn and Lupski (8). Tn5 insertions in cloned insert DNA were identified and mapped by restriction enzyme analysis and by Southern hybridization. To determine whether Tn5 insertions in pTD2.5 affected N-isopropylammelide degradation activity, E. coli cells containing mutated plasmids were grown in LB medium containing ampicillin (30 μg/ml), and resting cells were cultured with 50 μg of N-isopropylammelide per ml. N-Isopropylammelide in the culture medium was determined by HPLC analysis as described above.
Protein purification and electrophoresis.
E. coli DH5α and E. coli DH5α(pLTD4) were grown overnight in LB medium containing ampicillin (25 μg/ml), when required. Cultures were centrifuged at 14,000 × g for 10 min at 4°C and washed in 0.85% NaCl, and cell pellets were resuspended in 25 mM morpholinepropanesulfonic acid (MOPS) buffer (pH 6.9) on ice. Cold cell suspensions were broken by three consecutive freeze-thaw cycles, followed by sonication with a Biosonik sonicator (Bronwill Scientific, Rochester, N.Y.). Sonication was carried out three times at 80% probe intensity with intermittent cooling on ice. The broken cell suspensions were centrifuged at 17,000 × g for 90 min at 4°C to obtain crude cell extracts. The cell extracts from E. coli DH5α and E. coli DH5α(pTD2.5) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) with 7.5% acrylamide and a MiniProtean II gel apparatus (Bio-Rad Laboratories, Hercules, Calif.) as described previously (11, 22). The AtzC protein was electroblotted onto a polyvinylidene difluoride membrane and processed for N-terminal sequencing as described previously (24).
Amino acid analysis.
The N-terminal amino acid sequence of gel-purified AtzC was determined with an Applied Biosystems 477A protein sequencer at the Microchemical Facility, Human Genetics Institute, University of Minnesota.
Nucleotide sequence accession number.
The DNA sequence of atzC and the protein sequence of AtzC have been submitted to the GenBank Nucleotide Sequence Database under accession no. AF017572.
RESULTS
Cloning of genes involved in the degradation of N-isopropylammelide.
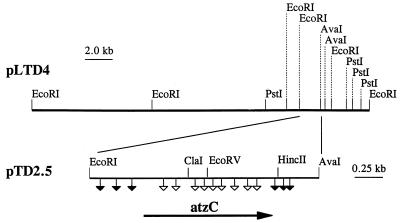
A library of Pseudomonas sp. strain ADP genomic DNA (10) in E. coli DH5α was screened for the ability to degrade N-isopropylammelide, the product of atrazine transformation by the sequential action of AtzA and AtzB. HPLC analysis identified four clones that degraded 50 μg of N-isopropylammelide per ml within 12 h. One cosmid clone, pLTD4, was selected for further study. pLTD4 contained a 25-kb DNA insert (Fig. 1) cloned in pLAFR3.
FIG. 1.
Restriction map of the 25-kb Pseudomonas sp. strain ADP genomic DNA fragment cloned in plasmid pLTD4 and containing the atzC gene. Symbols: ▿, mutations which disrupted the ability of cells to convert N-isopropylammelide to cyanuric acid and isopropylamine; ▾, mutations not affecting the ability of cells to degrade N-isopropylammelide. A 2.0-kb EcoRI-AvaI fragment was subcloned into pUC18 to produce plasmid pTD2.5. The direction of transcription is indicated by the arrow.
In order to localize the gene region involved in N-isopropylammelide metabolism, restriction fragments were subcloned into pUC18. A plasmid containing a 2.7-kb EcoRI insert, pTD2, was obtained, and the insert was further subcloned as a 2.0-kb EcoRI-AvaI fragment in pUC18 to yield pTD2.5. Clones containing either plasmid metabolized N-isopropylammelide, as determined by HPLC analysis.
Enzymatic activity in resting cell suspensions.
E. coli clones containing pTD2 or pTD2.5 were evaluated for their ability to degrade N-isopropylammelide. With the HPLC conditions used in this study, N-isopropylammelide, which eluted at 20 min, was observed to decrease over time, with a concomitant increase in a peak eluting at 4 min. Since authentic cyanuric acid also eluted at 4 min, our results suggested that the cloned DNA region encoded the ability to degrade N-isopropylammelide to cyanuric acid, the third step in the Pseudomonas sp. strain ADP atrazine degradation pathway (Fig. 2). The same apparent product was also detected in culture supernatants of Pseudomonas sp. strain ADP growing with N-isopropylammelide (data not shown).
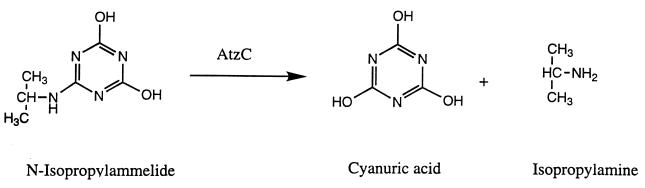
FIG. 2.
Third enzymatic step in atrazine degradation by Pseudomonas sp. strain ADP. The AtzC enzyme, N-isopropylammelide isopropylaminohydrolase, transforms N-isopropylammelide to cyanuric acid and isopropylamine via a hydrolytic deamination reaction.
The unknown metabolite was purified by HPLC and subjected to mass spectrometry. Fast atom bombardment-mass spectrometry indicated that the metabolite had a molecular weight of 129 with prominent ion fragments having molecular weights of 86 and 70. Authentic cyanuric acid gave the same mass spectrum (data not shown). Taken together, these data showed that the N-isopropyl group of N-isopropylammelide had been replaced by a hydroxyl group, producing cyanuric acid. Based on this metabolite, the other product of this reaction likely was isopropylamine. Isopropylamine was detected (data not shown) in resting cell culture supernatants of E. coli(pLTD4) and E. coli(pTD2.5) by a colorimetric assay with 2,4-dinitrofluorobenzene.
The reaction stoichiometry was investigated with both Pseudomonas sp. strain ADP and E. coli clones (Table 1) to confirm that N-isopropylammelide was transformed to cyanuric acid and isopropylamine in a single physiologically relevant reaction catalyzed by AtzC. Within 12 h, Pseudomonas sp. strain ADP completely degraded 1 mM N-isopropylammelide to 0.7 mM cyanuric acid and 1.2 mM isopropylamine. The concentration of cyanuric acid was less than 1 mM because it was further degraded by Pseudomonas sp. strain ADP. The catabolism of exogenously added isopropylamine by Pseudomonas sp. strain ADP was very slow. Resting cells of the bacterium required 6 days to metabolize 1 mM isopropylamine (data not shown). E. coli(pLTD4), however, completely degraded 1 mM N-isopropylammelide to 1.0 mM cyanuric acid and 1.1 mM isopropylamine within 12 h and failed to further metabolize cyanuric acid or isopropylamine. E. coli(pTD2) gave the same stoichiometry but metabolized N-isopropylammelide more slowly. It transformed 0.5 mM N-isopropylammelide to 0.5 mM cyanuric acid and 0.6 mM isopropylamine within the 12-h incubation period.
TABLE 1.
Expression of AtzC in Pseudomonas sp. strain ADP and E. coli clones
| Strain | Concna (mM) of:
|
||
|---|---|---|---|
| N-Isopropylammelideb | Cyanuric acid | Isopropylamine | |
| Pseudomonas sp. strain ADP | NDc | 0.67 ± 0.03 | 1.2 ± 0.10 |
| E. coli DH5α | 0.96 ± 0.07 | ND | ND |
| E. coli(pLTD4) | ND | 0.98 ± 0.06 | 1.1 ± 0.23 |
| E. coli(pTD2) | 0.48 ± 0.09 | 0.53 ± 0.10 | 0.56 ± 0.10 |
Values are means of triplicate samples ± standard error of means.
Resting cells were incubated for 12 h with N-isopropylammelide at a calculated initial concentration of 1.0 mM.
ND, not detected. Heat-treated resting cells of Pseudomonas sp. strain ADP had 1.07 ± 0.04 mM N-isopropylammelide after 12 h of incubation.
Tn5 mutagenesis analysis.
Random Tn5 mutagenesis was used to more precisely delineate the region of pTD2.5 encoding the enzyme(s) metabolizing N-isopropylammelide. Tn5 insertion sites in 32 mutated plasmids were determined by restriction enzyme analysis; the locations of the 15 unique Tn5 insertions in pTD2.5 are shown in Fig. 1. Nine transposon insertions abolished N-isopropylammelide degradation activity. The Tn5 insertions in all other regions of pTD2.5 did not affect N-isopropylammelide degradation. Results of this mutagenesis study indicated that the region essential for N-isopropylammelide degradation was limited to the central 1.2-kb region of pTD2.5.
DNA sequence analysis.
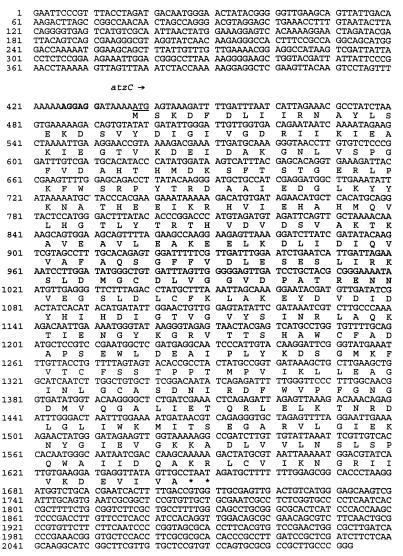
The 2.0-kb EcoRI-AvaI fragment cloned in pTD2.5 was sequenced in both directions. The DNA sequence is shown in Fig. 3. The sequenced region contained one large open reading frame (ORF) beginning at base 438. Results of codon usage and preference analyses and the Tn5 data indicated that this ORF comprised the N-isopropylammelide degradation gene. This gene, designated atzC, consisted of 1,209 nucleotides predicted to encode a protein of 403 amino acids, with a calculated molecular weight of 44,938 and a pI of 5.3. A putative ribosome binding site (AGGAGG) was identified 11 nucleotides upstream of the translation initiation codon (ATG). Two consecutive stop codons (TAA TAG) were located at the end of the coding region, beginning at nucleotide 1647.
FIG. 3.
Sequence of the atzC gene. The DNA sequence of atzC was determined with pTD2.5. The DNA sequence of the 2,093-nucleotide region was determined on both strands. The atzC ORF is indicated by the arrow. The start codon of the atzC ORF is underlined, a possible ribosome binding site upstream of the start site is in boldface, and stop codons are represented by asterisks. The translation of the atzC ORF is shown below the first nucleic acid of each codon.
Expression of the recombinant protein.
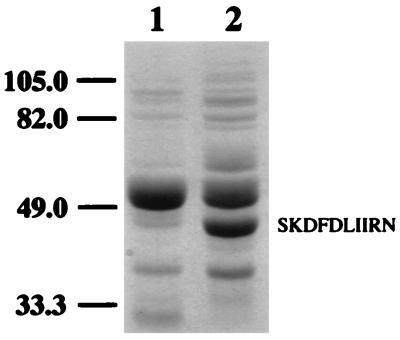
Protein extracts from E. coli DH5α and E. coli DH5α(pLTD4) were separated by denaturing PAGE. Figure 4 shows that relative to the E. coli control, E. coli(pLTD4) produced a unique, intensely stained protein band with a molecular weight of approximately 44,000. This size is in close agreement with the calculated molecular weight of AtzC (44,938), based on translation of the 403-amino-acid ORF. The putative AtzC protein was electroblotted onto polyvinylidene difluoride membranes and subjected to N-terminal sequence analysis. The first 10 amino acids detected (SKDFDLIIRN) were identical to those predicted by translation of the atzC ORF, with the exception of the posttranslational removal of the N-terminal methionine (Fig. 4).
FIG. 4.
Denaturing PAGE of total cell protein from E. coli DH5α (lane 1) and E. coli DH5α(pLTD4) (lane 2). Molecular mass markers (in kilodaltons) are on the left. The first 10 amino acids detected by N-terminal sequence analysis of the approximately 44,000-molecular-weight protein encoded by atzC are shown on the right.
Protein sequence analysis.
The amino acid sequence derived from translation of the atzC ORF was compared with those of other proteins in the SwissProt and PIR databases and those derived from translation of genes in the GenBank and EMBL databases. AtzC had less than 30% overall sequence identity with any known proteins. The highest identities were observed with cytosine deaminase from E. coli (29%) and dihydroorotase from Bacillus subtilis (25%). Moreover, AtzC had about 20% identity with the first two enzymes in the atrazine degradation pathway, AtzA and AtzB.
A search for conserved sequence motifs, however, was more informative. AtzC matches the first 5 residues of the N-terminal dihydroorotase signature pattern (Prosite entry PS00482), simplified here as D-X-H-X-H, and has significant similarity (79%) to dihydroorotase (PyrC) from B. subtilis in 13 residues adjacent to the N-terminal side of this pattern. Moreover, multisequence alignment of this 18-residue motif region indicated that AtzC clearly fits in with dihydroorotase and other metal-binding hydrolases acting largely on nitrogenous heterocyclic ring substrates (Fig. 5A). E. coli AdeC, which encodes adenine deaminase, and Dan, an N-acyl-d-glutamate amidohydrolase from Alcaligenes xylosoxidans, also match AtzC in 10 of 18 amino acids. The conserved H-X-H region is proposed to provide ligands to a zinc atom bound by dihydroorotase (3), suggesting a need for functional conservation of this region of the protein. In all cases, the conserved motif is found in a region near the N terminus of each protein.
FIG. 5.
Alignment of N-terminal (A) and C-terminal (B) deduced amino acid sequences of Pseudomonas sp. strain ADP AtzA (atrazine chlorohydrolase; U55933), AtzB (hydroxyatrazine ethylaminohydrolase; U66917), and AtzC (N-isopropylammelide isopropylaminohydrolase; AF017572); B. subtilis PyrC (dihydroorotase; M59757); E. coli AdeC (adenine deaminase; L10328); and A. xylosoxidans Dan (N-acyl-d-glutamate amidohydolase; D45919). The amino acids were numbered according to the N terminus of each protein as reported in GenBank, and boxes indicate identical amino acids.
The alignment in Fig. 5A also reveals a heretofore-unrecognized homology among AtzC, AtzB, and AtzA, the first three proteins in the atrazine degradation pathway of Pseudomonas sp. strain ADP. All three proteins show significant identity or similarity in amino acids upstream of the H-X-H motif, and all contain the conserved H-X-H motif in similar regions. While AtzA shares 10 of 18 amino acids with AtzC in the motif region, AtzB shares 5 of 18 amino acids with AtzC and 8 of 18 amino acids with AtzA. Moreover, AtzC also shares conserved amino acids with AtzA (13 of 20) and AtzB (8 of 20) in the C termini of the proteins (Fig. 5B). While the sequence relatedness of AtzB was the least, AtzB showed its greatest overall sequence identity with proteins in the amidohydrolase protein superfamily: s-triazine hydrolase from Rhodococcus corallinus (25% identity) and dihydroorotase from Lactobacillus leichmannii (22% identity).
Origins of AtzC.
It is also instructive to compare the atzA, -B and -C gene sequences and the respective DNA sequences flanking each of their coding regions. First, atzA and -B have mol% G+C contents of 58.3 and 64.1, respectively, within the range of moles percent G+C contents found in total Pseudomonas sp. DNA (58 to 70%). In contrast, the atzC gene has a 39.5 mol% G+C content, well outside the range for total Pseudomonas sp. DNA.
Previous evidence indicated that atrazine catabolism by Pseudomonas sp. strain ADP may have had recent evolutionary origins (12). Part of the evidence was the non-operon-like structure of the atzA, -B and -C genes and the discovery of insertion element sequences flanking the atzA gene (12). In this study, DNA sequence analysis revealed the presence of insertion sequence-like elements downstream of atzC between nucleotides 1836 and 2035 (Fig. 3). The IS-like sequence showed the greatest sequence identity with portions of IS1051 (73%) from Xanthomonas campestris pv. dieffenbachiae (1), IS52 (69%) from Pseudomonas savastanoi subsp. savastanoi (37), ISXW5 and ISXW4 (73%) from Xanthomonas campestris pv. campestris (18a; GenBank), and tnpA (68%) from plasmid pEST1226 (30). In total, the data suggest that atzC is derived from an ancient family of amidohydrolases and has been brought together with two other long-divergent members of that family by recent evolutionary events.
DISCUSSION
Studies on the biochemistry and genetics of microbiologically mediated atrazine degradation are proving useful for our understanding of how bacteria evolve catabolic pathways in response to new chemical inputs into the environment. For the last 40 years, more than 1 billion pounds of atrazine has been applied to soils. This application has provided selection pressure for the evolution of new pathways of microbial atrazine metabolism.
In this paper, we describe the cloning, sequencing, and analysis of the third gene in the atrazine degradation pathway of Pseudomonas sp. strain ADP. This gene, atzC, encodes an enzyme, N-isopropylammelide isopropylaminohydrolase, which transforms N-isopropylammelide to cyanuric acid and isopropylamine (Fig. 2). The identities of these products were unequivocally established by HPLC, mass spectrometry, and chemical analysis.
The cloning, expression in E. coli, and elucidation of the reaction products of atzC revealed the overall metabolic logic of atrazine catabolism by Pseudomonas sp. strain ADP. Metabolism proceeds by a series of hydrolases that are unique in each reaction and that catalyze dechlorination and the sequential hydrolytic removal of N-ethylamine and N-isopropylamine. The resultant product of the three enzyme-catalyzed reactions is cyanuric acid, a cyclic polyamide in its preferred tautomeric form which is known to undergo hydrolytic ring opening (15), a reaction similar to that catalyzed by dihydroorotase. Cyanuric acid, biuret, and urea are known to undergo hydrolytic C—N bond cleavage reactions, and the occurrence of these reactions in Pseudomonas sp. strain ADP could explain previous observations that all three ring carbon atoms from atrazine are liberated as CO2 (23). Moreover, cyanuric acid and related compounds are readily catabolized by many soil bacteria (5, 6, 17, 20) and by Pseudomonas sp. strain ADP to carbon dioxide and ammonia (23), providing evolutionary pressure for the atzA, -B, and -C genes to allow bacterial growth on atrazine.
While the first two genes encoding enzymes for atrazine catabolism in Pseudomonas sp. strain ADP, atzA and atzB, have been localized to the same 21.5-kb genomic DNA fragment, cloned in plasmid pMD1 (2, 10, 11), the exact location of the atzC gene is currently not known. Preliminary data indicate that atzC is located at least 25 kb from atzA and atzB, since pLTD4 did not hybridize to atzA or atzB or overlap with pMD1. However, our recent data indicate that all three genes are located on a 96-kb self-transmissible plasmid, pADP-1, in Pseudomonas sp. strain ADP (12).
The most surprising finding in this report is the placement of AtzA, AtzB, and AtzC in an ancient class of amidohydrolases that are distributed throughout three kingdoms of life: Eubacteria, Archaea, and Eukaryota. AtzA, AtzB, and AtzC are each comprised of polypeptides of similar sizes. Moreover, they all catalyze hydrolytic reactions with nitrogen heterocyclic ring substrates, as does dihydroorotase (PyrC) and adenine deaminase (AdeC). However, the sequence identity in pairwise comparisons among AtzA, AtzB, and AtzC was only on the order of 20%, so the idea for even an ancient common evolutionary origin was initially missed. Examination of the sequences in the region of the conserved motif for the dihydroorotase (PyrC) protein family, as defined in the ProfileScan database, indicated that the AtzA, AtzB, and AtzC protein sequences were reasonably well conserved in the critical N-terminal H-X-H region proposed to contain metal-coordinating histidine residues in the members of this family of proteins (Fig. 5). This proposition is strengthened by the recent analysis of the amidohydrolase protein family, which includes dihydroorotase, adenine deaminase, ureases, cytosine deaminases, and s-triazine hydrolase (18). This study established an amidohydrolase superfamily of proteins containing over 70 members, some of which show sequence identities in pairwise comparisons that are lower than those observed in pairwise comparisons among AtzA, AtzB, and AtzC. However, analysis of the sequences in the context of the known structures of adenine deaminase, ureases, and phosphotriester hydrolase revealed that sequence conservation is highest in the region of the metal-coordinating amino acids. Moreover, based on comparisons to the crystal structure and enzyme architecture of ureases and adenine deaminase (18, 19), it is proposed that these proteins all share a common fold.
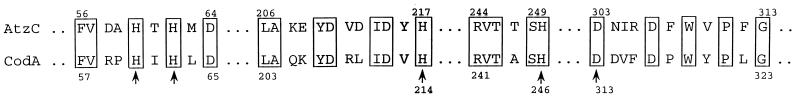
One of the members of the amidohydrolase protein superfamily is cytosine deaminase (CodA) from E. coli (7), and its pairwise alignment with AtzC reveals how a selective alignment in the regions of proposed metal ligands can show a striking homology between proteins that might otherwise be missed by a comparison of the entire protein sequences. Figure 6 shows a comparison of the AtzC sequence and the sequence of cytosine deaminase (CodA) in the regions of the five amino acids that coordinate the transition metal implicated in its catalytic activity. While the overall sequence identity is 29%, the identity in the regions shown in Fig. 6 is 61% and the similarity is 85%. This high sequence relatedness argues that these regions are critical in the functioning of AtzC and hence have been conserved despite extensive evolutionary divergence. The high degree of identity, the observation that the common residues appear in nearly identical regions, and the fact that the similarities are spread across five regions which are most conserved across the entire amidohydrolase superfamily strongly argue for divergent, rather than convergent, evolution.
FIG. 6.
Pairwise alignment of amino acid sequences from AtzC and E. coli cytosine deaminase (CodA; P25524) in the regions of CodA indicated to be involved in active-site metal coordination. The five metal-liganding amino acids of CodA are indicated with vertical arrows. Amino acid numbers were determined by counting from the N terminus of each protein.
A key mechanistic feature of the amidohydrolase superfamily is that many of the enzymes contain mononuclear or binuclear metal centers that are essential for catalytic activity. Cytosine deaminase, for example, can bind zinc, manganese, iron, or cobalt and is maximally activated by iron (31). The role of metals in the catalytic mechanisms of AtzA, AtzB, or AtzC is largely uninvestigated. Previously, it was reported that AtzA purified from a recombinant E. coli strain contained only substoichiometric quantities of transition metals. However, more recent experiments indicated that atrazine hydrolysis by AtzA is strongly activated by iron(II) and cobalt(II) salts (9). These data suggest a functional significance for the H-X-H motif found in that protein (Fig. 5). The conservation of all cytosine deaminase metal-binding amino acids in AtzC suggests that the latter protein might use a metal ion catalytically. Further studies on the purification and characterization of AtzC will resolve this question.
ACKNOWLEDGMENTS
This work was supported in part by a grant from Novartis Crop Protection, Inc., and by grant 94-34339-1122 from the United States-Israel Binational Agricultural Research and Development (BARD) Fund.
We thank Janis Mcfarland and Steven Dumford of Novartis for providing s-triazine compounds, William Koskinen, Tom Krick, and Eric Eccleston for experimental assistance, and Lynda Ellis for helpful critical comments about the manuscript.
Footnotes
Manuscript 971250037 in the University of Minnesota Agricultural Experiment Station series.
REFERENCES
- 1.Berthier Y, Thierry D, Lemattre M, Guesdon J L. Isolation of an insertion sequence (IS1051) from Xanthomonas campestris pv. dieffenbachiae with potential use for strain identification and characterization. Appl Environ Microbiol. 1994;60:377–384. doi: 10.1128/aem.60.1.377-384.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boundy-Mills K L, de Souza M L, Mandelbaum R T, Wackett L P, Sadowsky M J. The atzB gene of Pseudomonas sp. strain ADP encodes the second enzyme of a novel atrazine degradation pathway. Appl Environ Microbiol. 1997;63:916–923. doi: 10.1128/aem.63.3.916-923.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown D C, Collins K D. Dihydroorotase from Escherichia coli: substitution of Co(II) for the active site Zn(II) J Biol Chem. 1991;266:1597–1604. [PubMed] [Google Scholar]
- 4.Chakrabarty A M. Microbial degradation of toxic chemicals: evolutionary insights and practical considerations. ASM News. 1996;62:130–137. [Google Scholar]
- 5.Cook A M, Bellstein P, Grossenbacher H, Hutter R. Ring cleavage and degradative pathway of cyanuric acid in bacteria. Biochem J. 1985;231:25–30. doi: 10.1042/bj2310025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook A M. Biodegradation of s-triazine xenobiotics. FEMS Microbiol Rev. 1987;46:93–116. [Google Scholar]
- 7.Danielsen S, Kilstrup M, Barilla K, Jochimsen B, Neuhard J. Characterization of the Escherichia coli codBA operon encoding cytosine permease and cytosine deaminase. Mol Microbiol. 1992;6:1335–1344. doi: 10.1111/j.1365-2958.1992.tb00854.x. [DOI] [PubMed] [Google Scholar]
- 8.de Bruijn F J, Lupski J R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids. Gene. 1984;27:131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]
- 9.de Souza, M. L. Unpublished data.
- 10.de Souza M L, Wackett L P, Boundy-Mills K L, Mandelbaum R T, Sadowsky M J. Cloning, characterization, and expression of a gene region from Pseudomonas sp. strain ADP involved in the dechlorination of atrazine. Appl Environ Microbiol. 1995;61:3373–3378. doi: 10.1128/aem.61.9.3373-3378.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Souza M L, Sadowsky M J, Wackett L P. Atrazine chlorohydrolase from Pseudomonas sp. strain ADP: gene sequence, enzyme purification, and protein characterization. J Bacteriol. 1996;178:4894–4900. doi: 10.1128/jb.178.16.4894-4900.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Souza M L, Wackett L P, Sadowsky M J. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. 1997. Highly homologous atrazine degradation genes widespread in recently isolated atrazine degrading organisms, abstr. Q-405; p. 522. [Google Scholar]
- 13.Dubin D T. The assay and characterization of amines by means of 2,4-dinitrofluorobenzene. J Biol Chem. 1960;235:783–786. [PubMed] [Google Scholar]
- 14.Eady R R, Jarman T R, Large P J. Microbial oxidation of amines. Biochem J. 1971;125:449–459. doi: 10.1042/bj1250449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eaton R W, Karns J S. Cloning and comparison of the DNA encoding ammelide aminohydrolase and cyanuric acid amidohydrolase from three s-triazine-degrading bacterial strains. J Bacteriol. 1991;173:1363–1366. doi: 10.1128/jb.173.3.1363-1366.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eaton R W, Karns J S. Cloning and analysis of s-triazine catabolic genes from Pseudomonas sp. strain NRRLB-12227. J Bacteriol. 1991;173:1215–1222. doi: 10.1128/jb.173.3.1215-1222.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson L E, Lee K H. Degradation of atrazine and related s-triazines. Crit Rev Environ Control. 1989;19:1–14. [Google Scholar]
- 18.Holm L, Sander C. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins. 1997;28:72–82. [PubMed] [Google Scholar]
- 18a.Hsieh, Y., and J. Chen. Unpublished data.
- 19.Jabri E, Carr M B, Hausinger R P, Karplus P A. The crystal structure of urease from Klebsiella aerogenes. Science. 1995;268:998–1004. [PubMed] [Google Scholar]
- 20.Jutzi K, Cook A M, Hutter R. The degradative pathway of the s-triazine melamine. Biochem J. 1982;208:679–684. doi: 10.1042/bj2080679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karns J S, Eaton R W. Genes encoding s-triazine degradation are plasmid-borne in Klebsiella pneumoniae strain 99. J Agric Food Chem. 1997;45:1017–1022. [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Mandelbaum R T, Allan D L, Wackett L P. Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine. Appl Environ Microbiol. 1995;61:1451–1457. doi: 10.1128/aem.61.4.1451-1457.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 25.McIntire F C, Clements L M, Sproull M. 1-Fluoro-2,4-dinitrobenzene as a quantitative reagent for primary and secondary amines. Anal Chem. 1953;25:1757–1758. [Google Scholar]
- 26.Mulbry W W. Purification and characterization of an inducible S-triazine hydrolase from Rhodococcus corallinus NRRL B-15444R. Appl Environ Microbiol. 1994;60:613–618. doi: 10.1128/aem.60.2.613-618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy I, Compernolle F, Ghys K, Vanderleyden J, de Mot R. A single cytochrome P-450 system is involved in degradation of the herbicides EPTC (S-ethyldipropylthiocarbamate) and atrazine by Rhodococcus sp. strain NI86/21. Appl Environ Microbiol. 1995;61:2056–2060. doi: 10.1128/aem.61.5.2056-2060.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagy I, Schoofs G, Compernolle F, Proost P, Vanderleyden J, Demot R. Degradation of the thiocarbamate herbicide EPTC (S-ethyl dipropylcarbamothioate) and biosafening by Rhodococcus sp. strain NI86/21 involve an inducible cytochrome P-450 system and aldehyde dehydrogenase. J Bacteriol. 1995;177:676–687. doi: 10.1128/jb.177.3.676-687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagy I, Verheijen S, De Schrijver A, Van Damme J, Proost P, Schoofs G, Vanderleyden J, De Mot R. Characterization of the Rhodococcus sp. NI86/21 gene encoding alcohol: N,N′-dimethyl-4-nitrosoaniline oxidoreductase inducible by atrazine and thiocarbamate herbicides. Arch Microbiol. 1995;163:439–446. doi: 10.1007/BF00272133. [DOI] [PubMed] [Google Scholar]
- 30.Nurk A, Kasak L, Kivisaar M. Sequence of the gene (pheA) encoding phenol monooxygenase from Pseudomonas sp. EST1001: expression in Escherichia coli and Pseudomonas putida. Gene. 1991;102:13–18. doi: 10.1016/0378-1119(91)90531-f. [DOI] [PubMed] [Google Scholar]
- 31.Porter D J, Austin E A. Cytosine deaminase: the role of divalent metal ions in catalysis. J Biol Chem. 1993;268:24005–24011. [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Shao Z Q, Behki R. Cloning of the genes for degradation of the herbicides EPTC (S-ethyldipropylthiocarbamate) and atrazine from Rhodococcus sp. strain TE1. Appl Environ Microbiol. 1995;61:2061–2065. doi: 10.1128/aem.61.5.2061-2065.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao Z Q, Behki R. Characterization of the expression of the thcB gene, coding for a pesticide-degrading cytochrome P-450 in Rhodococcus strains. Appl Environ Microbiol. 1996;62:403–407. doi: 10.1128/aem.62.2.403-407.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shao Z Q, Sefens W, Mulbry W, Behki R M. Cloning and expression of the s-triazine hydrolase gene (trzA) from Rhodococcus corallinus and development of Rhodococcus recombinant strains capable of dealkylating and dechlorinating the herbicide atrazine. J Bacteriol. 1995;177:5748–5755. doi: 10.1128/jb.177.20.5748-5755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wyndham R C, Cashore A E, Nakatsu C H, Peel M C. Catabolic transposons. Biodegradation. 1994;5:323–342. doi: 10.1007/BF00696468. [DOI] [PubMed] [Google Scholar]
- 37.Yamada T, Lee P, Kosuge T. Insertion sequence of Pseudomonas savastanoi: nucleotide sequence and homology with Agrobacterium tumefaciens transfer DNA. Proc Natl Acad Sci USA. 1986;83:8263–8267. doi: 10.1073/pnas.83.21.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]