Abstract
Bacteriophage‐derived endolysins are a novel class of antimicrobials known to rapidly kill bacteria, including antibiotic‐resistant strains. We here engineered endolysins against the bovine mastitis pathogens Streptococcus uberis, Streptococcus agalactiae and Streptococcus dysgalactiae, also targeting intracellular survival and biofilm formation. For this purpose, high‐throughput DNA assembly was used to create a library with >80,000 theoretical endolysin variants for screening of their bacteriolytic activity against Gram‐positive isolates from (sub)clinically affected cows. This lytic activity was evaluated by turbidity reduction and time‐kill assays in phosphate‐buffered saline and pasteurized whole cow's milk to allow a rank up of the most potent leading candidates. A top candidate was selected with a 4.0 log killing efficacy against S. uberis, also showing similar activity against S. agalactiae and S. dysgalactiae. This top candidate eradicated S. uberis biofilm and showed intracellular activity in two bovine mammary epithelial cell lines as was confirmed by confocal microscopy. A potentiating effect on cloxacillin, a beta‐lactam penicillin used to intramammarily treat bovine Gram‐positive mastitis, was observed for this top candidate endolysin in raw cow's milk from (sub)clinically infected udders. Our in vitro results indicate that engineered endolysins may have a future role as add‐on in the treatment of bovine streptococcal mastitis.
Bacteriophage‐derived endolysins are a novel class of antimicrobials known to rapidly kill bacteria, including antibiotic‐resistant strains. We here engineered endolysins against the bovine mastitis pathogens Streptococcus uberis, Streptococcus agalactiae and Streptococcus dysgalactiae, also targeting intracellular survival and biofilm formation. For this purpose, high‐throughput DNA assembly was used to create a library with > 80,000 theoretical endolysin variants for screening of their bacteriolytic activity against Gram‐positive isolates from (sub)clinically affected cows.
INTRODUCTION
Bovine mastitis, an inflammation of the cow's udder, remains the most important disease impacting the dairy industry mainly due to decreased milk production, treatment costs and premature culling of diseased animals (Geary et al., 2012; Hogeveen et al., 2011; Rollin et al., 2015). Udder inflammation is usually the result of an intramammary infection, with the most frequently isolated Gram‐positive bacteria being Streptococcus uberis, Streptococcus dysgalactiae, Staphylococcus aureus and to a lesser extent Streptococcus agalactiae (de Jong et al., 2018; Persson et al., 2011; Saini et al., 2012; Verbeke et al., 2014). Despite progress in the prevention of bovine mastitis by applying novel housing techniques, additional hygiene measures, vaccination and the use of probiotic agents (Collado et al., 2018; De Vliegher et al., 2018; Gomes & Henriques, 2016; Pereira et al., 2011; Rainard & Foucras, 2018; Verbeke et al., 2014), mastitis still frequently occurs in the dairy industry. Farmers not only treat diseased animals with antibiotics, but also often intramammarily administer antibiotics as a preventive strategy known as dry cow treatment (Gomes & Henriques, 2016). This (ab)use of antibiotics has several disadvantages. The milk of treated animals needs to be discarded, which results in food waste, an additional loss of income and an exposure of the environment to the antibiotics used (Oliver et al., 2011). Bovine streptococci are able to form biofilms and penetrate the mammary epithelial cells, causing the udder infection to persist during the next lactation even after antibiotic treatment (Bardiau et al., 2016; Niemi et al., 2021; Schönborn et al., 2017; Tamilselvam et al., 2006; Varhimo et al., 2011). Antibiotics that are considered critical by the World Health Organization (WHO) such as cephalosporines belonging to the third and fourth generation (e.g. cefaperazone, cefquinome) are still registered for bovine mastitis, while the use of antimicrobials in animal agriculture should be diminished as much as possible to counteract resistance development from a one health approach (Garcia et al., 2019; Krömker & Leimbach, 2017; World Health Organization, 2017). Emphasizing the urgent need for novel antimicrobials, the WHO announced that resistance to current antimicrobials is a major public health issue since increasing resistance levels are reported (World Health Organization, 2021b). Moreover, the WHO also warned that the number of antimicrobials under development are insufficient to tackle the challenge of increasing emergence and spread of antimicrobial resistance (World Health Organization, 2021a).
Over the past two decades, endolysins derived from bacteriophages have gained increasing attention due to their rapid activity, high specificity for the target bacteria and most of all their efficacy against antibiotic‐resistant bacterial strains (Fischetti, 2010; Haddad Kashani et al., 2018; Linden et al., 2021; Nelson et al., 2012). Indeed, endolysins are peptidoglycan hydrolases and degrade the Gram‐positive cell wall resulting in osmotic lysis of the bacterial cell (Nelson et al., 2012). This method of action circumvents the intrinsic and acquired resistance mechanisms of antibiotic‐resistant strains. More specifically, endolysins are also active against dormant bacteria in biofilms and can act on intracellular pathogens (Becker et al., 2016; Gutiérrez et al., 2014, 2015; Nelson et al., 2012; Röhrig et al., 2020). This explains why they are one of the major novel antimicrobials under development, with SAL‐200, CF‐301 (a.k.a. PlySs2), P128 and Staphefekt being staphylococcal endolysins currently being evaluated in diverse human clinical trials (World Health Organization, 2021a). Also within the field of Gram‐positive bovine mastitis, a few promising studies about the application of endolysins exist (Scholte et al., 2018; Vander Elst et al., 2020; Vander Elst & Meyer, 2018). We previously evaluated the wild‐type endolysins PlySs2 and PlySs9 against S. uberis, and other endolysins have been reported with in vitro and in vivo activity against bovine mastitis streptococci (PlyC, Ply700, λSA2 and B30) (Celia et al., 2008; Schmelcher et al., 2015; Scholte et al., 2018), staphylococci (trx‐SA1, phi11, phiH5, Lys109 and LysRODI) (Donovan et al., 2006; Fan et al., 2016; Gutiérrez et al., 2020; Obeso et al., 2008; Son et al., 2021) or both (PlySs2, ClyR) (Huang et al., 2015; Vander Elst et al., 2020; Yang et al., 2015). In addition, recent advances in the field of protein engineering have been applied to enhance and/or extend the activity of wild‐type endolysins. Indeed, endolysins targeting Gram‐positive bacteria have a modular design consisting of so‐called enzymatically active domains (EADs) and cell wall‐binding domains (CBDs). This allows the creation of engineered endolysins with improved activities compared to their wild types (Gerstmans et al., 2018; Nelson et al., 2012). Recently, we developed the high‐throughput DNA assembly technique VersaTile to rapidly assemble such engineered endolysins (Duyvejonck et al., 2021; Gerstmans et al., 2020; Gutiérrez et al., 2021). First, a repository of tiles corresponding to the individual wild‐type endolysin EADs and CBDs is made, which is subsequently combinatorially assembled. In addition, other protein engineering approaches can be applied to improve the potency of wild‐type endolysins, such as prolongation of their serum half‐life or enhancement of their intracellular delivery (Röhrig et al., 2020; Seijsing et al., 2018).
Here, we describe the development of an optimized engineered endolysin with activity in raw cow's milk against the bovine streptococcal mastitis pathogens S. uberis, S. agalactiae and S. dysgalactiae by engineering wild‐type endolysins with known activity against these targeted pathogens. We therefore use the VersaTile technique to create large combinatorial libraries and follow an elaborated hit‐to‐lead screening approach against these streptococcal bovine mastitis pathogens as well as Sta. aureus. The endolysins were combined with cell‐penetrating peptides (CPPs) to enhance their intracellular killing capacity.
RESULTS
EAD, CBD and CPP sourcing for VersaTile assembly
At the start of the study, a collection of endolysin building blocks (CBDs, EADs) and CPPs was made to create a combinatorial endolysin library with the VersaTile technique. First, a selection of endolysins with known activity against S. uberis, S. agalactiae, S. dysgalactiae and Sta. aureus was made (Appendix S1). This included 11 EADs and 11 CBDs from PlySs2, PlySs9, PlyC, Ply700, λSA2, LysRODI, LysC1C, LysA72 and PlyGRCS (Celia et al., 2008; Gutiérrez et al., 2021; Linden et al., 2015; Schmelcher et al., 2015; Scholte et al., 2018; Vander Elst et al., 2020). Subsequently, this selection was supplemented with bio‐informatically discovered EADs and CBDs in intact prophage elements present in both S. uberis and S. suis genomes derived from the NCBI Genbank. The S. suis genomes were also included because we and others previously reported endolysins in S. suis prophages with activity against various streptococci and Sta. aureus, including bovine mastitis‐derived isolates (Gilmer et al., 2013; Vander Elst et al., 2020). A multiple sequence alignment was performed and the protein sequences of the endolysin subdomains that shared <85.0% and ≤87.5% amino acid sequence similarity for S. suis and S. uberis prophage endolysins, respectively, were selected for tile construction (Appendix S2). These delineated domains or tiles consistently included the C‐terminal linkers. More specifically, the amidase_5, glucosaminidase and CW_7 of S. suis HA0609, amidase_5 and CW_7 of S. suis 1081, CW_7 of S. suis SH0104 and SH3_4 of S. uberis NCTC3858 and NCTC4674 were found eligible. To also improve intracellular penetration, five CPPs (i.e. oligoarginine, TAT, Penetratin, Pep‐1 and H2) were selected (Appendix S3).
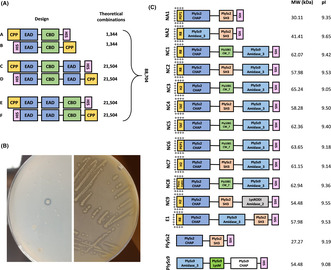
These selected EADs, CBDs and CPPs were used to create engineered endolysins by VersaTile assembly, combining six designs in a four‐way assembly system to construct a total of 88,704 theoretical variants (Figure 1A). More specifically, three paired designs A/B, C/D and E/F were created with each library consisting of the same backbone, but mirroring the end positions of the CPP and polyhistidine tag (His6‐tag). Combinations without a CPP were also screened. This resulted in 1344 (= 6 × 16 × 14) theoretical combinations for both libraries A/B, and 21,504 (= 6 × 16 × 16 × 14) for each of the libraries C/D and E/F (Appendix S4).
FIGURE 1.
VersaTile assembly, screening and hit‐to‐lead selection of candidate engineered endolysins. (A) Graphic display of six designs to screen for bispecific engineered endolysins, with the number of theoretical combinations per design. CPP, EAD, CBD and HIS: cell penetrating peptide, enzymatically active domain, cell wall‐binding domain and polyhistidine tag. Combinations without a CPP were also screened. (B) Transformed Escherichia coli BL21 (DE3) pLysS with one colony expressing an engineered endolysin active against autoclaved Staphylococcus aureus N305 (left). This colony was subsequently picked and streaked out on the same agar containing autoclaved Streptococcus uberis 0140J (right). This engineered endolysin was regarded a hit since clearing zones (i.e. halos) were observed. (C) Graphic display of the 12 engineered endolysins withheld after screening the VersaTile assembled libraries with lytic activity against S. uberis 0140J and Sta. aureus N305 peptidoglycan, with their molecular weight (MW) and isoelectric point (pI). PlySs2 and PlySs9 were reassembled through VersaTile and included as positive controls.
High‐throughput screening and hit‐to‐lead selection of VersaTile assembled endolysins
Following the construction of the six libraries by VersaTile assembly, these variants were first screened with a halo‐based assay on agar containing autoclaved Sta. aureus N305 (Figure 1B). The initial screening against autoclaved Sta. aureus cells was included for the possibility to select for variants with bispecific activity against both bovine mastitis streptococci as well as Sta. aureus. Subsequently, colonies showing a halo were picked to screen on agar containing autoclaved S. uberis 0140J. If clearing zones were again visible, these Escherichia coli clones were grown for long‐term storage and plasmids were Sanger sequenced. Eighteen engineered endolysins (i.e. ‘hits’) were identified: 10 hits in design A, 7 hits in design C and 1 hit in design E (Table 1). No active engineered endolysins were found in designs B, D and F. Thus, more hits occurred in designs A (p < 0.001) and C (p = 0.021), but not in B, D, E and F (p > 0.016) (Table 1). Within these hits, having no CPP at position 1 was significantly (p < 0.016) favoured over having a CPP fused to the construct (Table 2). Regarding the EADs, PlySs2 CHAP and PlySs9 amidase_3 were significantly (both p < 0.006) overrepresented at position 2, LysRODI CHAP was significantly (p < 0.006) overrepresented at position 4 and PlySs2 SH3_5 and Ply1081 CW_7 were significantly (p < 0.007) overrepresented at position 3 (Table 2). To narrow down the designs and building blocks, a second screening was performed by only including the overrepresented designs A and C, hereafter referred to as narrow‐down designs A (NA) and C (NC). Only the significantly overrepresented tiles in positions 2 and 3 were included as candidate building blocks (i.e. PlySs2 CHAP and PlySs9 amidase_3 in position 2 and PlySs2 SH3_5 and Ply1081 CW_7 in position 3 respectively). To create engineered endolysins with improved cell penetrating capacities, all CPPs were included and the blank in position 1 was excluded. In position 4 (i.e. only for NC), all building blocks were included due to the limited number of seven hits found in the first screening. By narrowing down the designs to NA and NC with the selected five CPPs in position 1, two EADs in position 2, two CBDs in position 3 and four EADs in position 4, a library with 100 [= (5 × 2 × 2) + (5 × 2 × 2 × 4)] theoretical combinations remained (Appendix S4). Similar to the first screening, a halo‐based plating method followed by Sanger sequencing was used to screen NA and NC in a saturating way for active engineered endolysins (Table 1). This narrow down to NA and NC caused an increased hit rate corresponding to 11‐ and 53‐fold enrichments respectively.
TABLE 1.
The number of active engineered endolysins or ‘hits’ retrieved from each screened VersaTile assembled library.
| Screening | Design | Approximately screened | Bispecific hits | Binomial t test | |||
|---|---|---|---|---|---|---|---|
| Numeric | % a | Numeric | % | p | Significance | ||
| 1 | A | 3900 | 29.17 | 10 | 0.26 | <0.001 | b |
| B | 3900 | 29.17 | 0 | 0 | Not determined | ||
| C | 5120 | 23.81 | 7 | 0.17 | 0.021 | ns | |
| D | 6780 | 31.53 | 0 | 0 | Not determined | ||
| E | 16,220 | 75.43 | 1 | 0.01 | 0.341 | ns | |
| F | 4250 | 19.76 | 0 | 0 | Not determined | ||
| Total | 40,170 | 45.29 | 18 | 0.04 | Not determined | ||
| 2 | NA | 145 | 725.00 | 4 | 2.76 | Not determined | |
| NC | 275 | 343.15 | 25 | 9.09 | Not determined | ||
| Total | 420 | 420.00 | 29 | 6.90 | Not determined | ||
Note: A binomial t test with α = 0.016 was performed in the first screening to identify libraries that statistically contained more active engineered endolysins.
Abbreviation: ns, non‐significance.
Assuming that every possible engineered endolysin occurred only once in the performed screening.
Significantly different distribution compared to a uniform distribution (p < 0.016).
TABLE 2.
Overrepresentation of endolysin subdomains or ‘tiles’ present in the retrieved active engineered endolysins or ‘hits’ in position 1 for designs A, C and E, positions 2 and 3 for designs A and C and position 4 for design C.
| Position | Tile | Times observed | Binomial t test | |||
|---|---|---|---|---|---|---|
| Numeric | % | p | Significance | |||
| 1 | CPP | No CPP | 16 | 88.89 | <0.001 | a |
| 1 | CPP | R8 (oligoarginine) | 2 | 11.11 | 0.755 | ns |
| 2 | EAD | PlySs2 CHAP | 8 | 47.10 | <0.001 | a |
| 2 | EAD | PlySs9 amidase_3 | 7 | 41.20 | <0.001 | a |
| 2 | EAD | LysRODI CHAP | 1 | 5.85 | 1.000 | ns |
| 2 | EAD | LysRODI amidase_2 | 1 | 5.85 | 1.000 | ns |
| 3 | CBD | PlySs2 SH3_5 | 10 | 58.80 | <0.001 | a |
| 3 | CBD | Ply1081 CW_7 | 5 | 29.40 | 0.005 | a |
| 3 | CBD | PlySH0104 CW_7 | 1 | 5.90 | 1.000 | ns |
| 3 | CBD | PlyλSA2 CW_7 | 1 | 5.90 | 1.000 | ns |
| 4 | EAD | LysRODI CHAP | 4 | 57.10 | <0.001 | a |
| 4 | EAD | LysRODI amidase_2 | 1 | 14.30 | 0.364 | ns |
| 4 | EAD | PlySs9 amidase_3 | 1 | 14.30 | 0.364 | ns |
| 4 | EAD | PlyC CHAP | 1 | 14.30 | 0.364 | ns |
Note: A binomial t test with α = 0.016 for every CPP, α = 0.006 for every EAD and α = 0.007 for every CBD was performed to identify the tiles significantly overrepresented at their respective position.
Abbreviation: ns, non‐significance.
Significantly different distribution compared to a uniform distribution (CPP: p < 0.016, EAD: p < 0.006, CBD: p < 0.007).
Eventually, 12 engineered endolysins were discovered with lytic activity against autoclaved Sta. aureus N305 and S. uberis 0140J in the halo‐based assay (Figure 1C). Two and nine hits belonged to designs NA and NC respectively. The hit found in library E during the first screening was also included. All hits carried the PlySs2 CHAP domains in position 2, except for hit NA2 that contained the PlySs9 amidase_3. Similarly, all hits in design NC carried the PlySs9 amidase_3 in position 4, except for hit NC9 that had the LysRODI amidase_2 instead. The hits in design NC always combined a CHAP domain with an amidase. Another important observation was that PlySs2 CHAP + Ply1081 CW_7 + PlySs9 amidase_3 occurred in combination with all five CPPs. The selected CBDs in position 3 were more or less equally present in the nine hits retrieved from design NC, whereas only PlySs2 SH3_5 was observed in both hits of design NA. Hit E1 consisted of the same building blocks as hit NC2, but in a different order. The MW and pI of the 12 hits ranged from 30.11 to 65.24 kDa and 9.05 to 9.65 respectively.
Ranking of candidate engineered endolysins
The 12 selected engineered endolysins were first expressed and semi‐purified (Appendix S5). NC4 could not be included for further evaluation as it disintegrated and precipitated during the preceding dialysis step required to remove the elution buffer's imidazole from the purified proteins. PlySs2 and PlySs9 were reassembled through VersaTile and included as positive controls (Figure 1C). It is important to note that VersaTile assembly adds one alanine and one glycine residue at each junction between the domains, meaning these PlySs2 and PlySs9 controls were not fully identical to their wild‐type sequences. Stationary phase Gram‐positive bovine mastitis reference strains Sta. aureus N305 and S. uberis 0140J, as well as a randomly chosen clinical isolate of Sta. aureus, S. uberis, S. agalactiae and S. dysgalactiae were comparatively challenged with a final concentration of approximately 0.25 μM of each engineered endolysin and the PlySs2 and PlySs9 controls in a 2‐h time‐kill assay (TKA) in phosphate‐buffered saline (PBS) versus pasteurized whole cow's milk (Appendix S6). The latter endolysin concentration was chosen to ensure stringency of the assay in order to differentiate between less and better performing variants, also allowing their ranking (Figure 2A). Regarding both Sta. aureus isolates challenged in PBS, all Δlog10 were <0.50. However, a Δlog10 of 1.58 ± 0.07 and 5.14 ± 0.49 was demonstrated for NC2 and PlySs2 against Sta. aureus N305 in pasteurized whole milk. NC2 also had a 1.27 ± 0.18 Δlog10 against the clinical Sta. aureus isolate, again observed in milk. After challenging S. uberis 0140J in PBS (Appendix S6A), Δlog10 of 0.67 ± 0.03 and 0.62 ± 0.38 for NC3 and PlySs2 were observed respectively. Furthermore, Δlog10 of 1.68 ± 0.07 was observed for both NC5 and NC8 against the clinical S. uberis isolate, attaining the limit of detection (LOD). For S. agalactiae, Δlog10 of >3.0 was measured after challenge with NC3, NC5, NC6 and NC8, whereas all hits derived from design C as well as the PlySs9 positive control caused Δlog10 of ≥2.0 for S. dysgalactiae, also frequently reaching the LOD. Proceeding with the bacterial killing in pasteurized whole milk (Appendix S6B), PlySs2 caused a 1.29 ± 0.14 and 0.49 ± 0.23 Δlog10 against S. uberis 0140J and the clinical S. uberis strain respectively. While no remarkable activity was present for S. agalactiae, Δlog10 of 3.00 ± 0.30 and 2.71 ± 0.41 was observed against S. dysgalactiae for NC5 and PlySs2 respectively.
FIGURE 2.
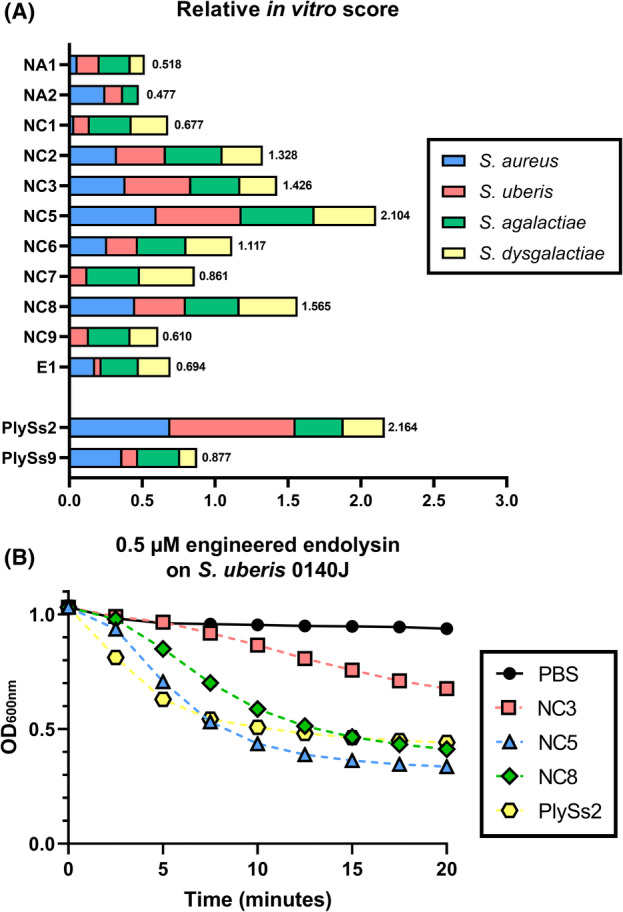
Ranking of candidate engineered endolysins. (A) Relative in vitro scoring system of the engineered endolysins in PBS and pasteurized whole milk against the bovine mastitis pathogens Streptococcus uberis, S. agalactiae, S. dysgalactiae and Sta. aureus. NC4 could not be included due to its insufficient stability in PBS. PlySs2 and PlySs9 were included as positive controls. A cumulative score of 3.0 corresponds to a theoretical candidate with the maximum score in each of the individual conditions performed. (B) Comparative turbidity reduction assay performed with 0.5 μM of the engineered endolysins NC3 (pink square), NC5 (blue triangle) and NC8 (green diamond) and the PlySs2 positive control (yellow hexagon) against stationary phase S. uberis 0140J. Bacteria in PBS (black circle) served as negative control.
Subsequently, the Δlog10 values under both the PBS and pasteurized whole milk conditions were converted to a relative in vitro scoring system, which allowed a ranking of the leading candidates within these 12 engineered endolysins (Figure 2A and Appendix S7A–D). In a next narrow‐down step, it was decided to continue with the four overall best‐performing engineered candidates, which were NC2, NC3, NC5 and NC8. When NC3, NC5 and NC8 (which all share the same backbone, differing only in their CPP) were comparatively incubated at a concentration of 0.5 μM with S. uberis 0140J in a turbidity reduction assay (TRA), NC5 lysed the bacterial cells fastest (Figure 2B). Therefore, NC5 was compared with NC2 (the only of the four best‐performing engineered endolysins which has a different CBD) to select one of both engineered endolysins as final candidate.
Final selection of one top candidate engineered endolysin
The lytic and antibacterial activities of the two remaining candidates NC2 and NC5 were extensively evaluated and compared by TRAs and TKAs in order to select a lead candidate (Table 3, Figure 3A–C). An initial test panel comprised the reference strain S. uberis 0140J and nine additional isolates mainly belonging to the global clonal complex (GCC) sequence types (STs) ‐5 and ‐143, which are typically associated with (sub)clinical bovine mastitis (Davies et al., 2016; Käppeli et al., 2019; Tomita et al., 2008). The sensitivity was strain dependent, but lytic activity was demonstrated for all S. uberis isolates challenged by NC2 and NC5. Overall, no significant (p > 0.05) pairwise differences in Δlog10 by TKA against S. uberis were seen for NC2 compared to NC5. However, NC5 lysed all these S. uberis isolates significantly (p < 0.01) faster (shown for S. uberis 0140J in Figure 3A). Therefore, we proceeded with NC5 as the lead candidate. Bacterial killings up to 4.05 ± 0.07 and 3.02 ± 0.37 Δlog10 were observed for 0.3 μM NC5 for a GCC ST‐5 and ‐143 S. uberis isolate respectively. Three additional isolates of S. agalactiae and S. dysgalactiae, other relevant bovine mastitis species, were similarly challenged with 0.3 μM NC5 to analyse the spectrum of NC5. NC5 showed consistent lytic activity on all isolates with bacterial killings up to 1.50 ± 0.02 and 1.77 ± 0.43 Δlog10 respectively (Table 3, Figure 3B,C). Since we included Sta. aureus to potentially generate engineered endolysins with possible bispecific activity, we also tested the activity of NC5 against reference strain Sta. aureus N305 and nine additional isolates of one cap 5+ agr I and eight cap 8+ agr I, II or III isolates. Such isolates are typically associated with (sub)clinical bovine mastitis (Bardiau et al., 2014, 2016; Rossi et al., 2021). The lytic activity as evaluated by TRA was generally poor or absent [≤0.01 (ΔOD600/min)/μM] against these Sta. aureus strains and no killing was observed (Appendix S8). Also, when a threefold increased concentration of NC5 (0.9 μM) was used against two relevant Sta. aureus isolates belonging to cap 5+ agr I and cap 8+ agr II (Appendix S9), no significant (p > 0.05) activity could be observed against these Sta. aureus isolates. Thus, the selected top candidate NC5 is a potent endolysin against S. uberis, S. agalactiae and S. dysgalactiae, but not against Sta. aureus. For completion, protein integrity of the selected top candidate NC5 was checked by ELISAs in addition to SDS‐PAGE, which confirmed the presence of the TAT peptide on NC5 (Appendix S10A–D).
TABLE 3.
Determination of the Δlog10 reductions (CFU/mL) and lysis rate [(ΔOD600/min)/μM] of engineered endolysins NC2 and NC5 for Gram‐positive mastitis isolates of (sub)clinically infected dairy cows.
| Gram‐positive bovine mastitis isolate | MALDI‐TOF score (confidence level) | Genomic characteristics | NC2 | NC5 | ||
|---|---|---|---|---|---|---|
| ΔLog10 (CFU/mL) | (ΔOD600/min)/μM | ΔLog10 (CFU/mL) | (ΔOD600/min)/μM | |||
| S. uberis 0140J | Not determined | ST 1, GCC 5 | 0.90 ± 0.03 | 0.05 ± 0.00 | 1.16 ± 0.06 | 0.14 ± 0.00 |
| Clinical S. uberis 1 | 2.44 (High) | Unknown ST and GCC | 0.00 ± 0.48 | 0.03 ± 0.00 | 0.03 ± 0.26 | 0.06 ± 0.00 |
| Clinical S. uberis 2 | 2.35 (High) | Unknown ST, GCC 5 a | 4.16 ± 0.15 | 0.08 ± 0.01 | 4.05 ± 0.07 | 0.11 ± 0.00 |
| Clinical S. uberis 3 | 2.31 (High) | Unknown ST, GCC 5 a | 1.66 ± 0.17 | 0.22 ± 0.02 | 1.82 ± 0.18 | 0.44 ± 0.01 |
| Clinical S. uberis 4 | 2.43 (High) | ST 815, GCC 143 a | 0.01 ± 0.21 | 0.04 ± 0.00 | 0.00 ± 0.22 | 0.05 ± 0.00 |
| Clinical S. uberis 5 | 2.33 (High) | ST 944, GCC 5 | 0.86 ± 0.11 | 0.12 ± 0.01 | 0.71 ± 0.13 | 0.21 ± 0.00 |
| Subclinical S. uberis 1 | 2.42 (High) | ST 808, GCC 143 | 2.84 ± 0.12 | 0.18 ± 0.01 | 3.02 ± 0.37 | 0.43 ± 0.01 |
| Subclinical S. uberis 2 | 2.47 (High) | Unknown ST and GCC | 1.02 ± 0.20 | 0.06 ± 0.01 | 1.13 ± 0.12 | 0.11 ± 0.00 |
| Subclinical S. uberis 3 | 2.28 (High) | Unknown ST, GCC 5 a | 1.31 ± 0.25 | 0.18 ± 0.01 | 1.24 ± 0.10 | 0.39 ± 0.01 |
| Subclinical S. uberis 4 | 2.34 (High) | Unknown ST and GCC | 2.84 ± 0.37 | 0.15 ± 0.01 | 2.48 ± 0.34 | 0.28 ± 0.00 |
| S. agalactiae 1 | 2.45 (High) | Not determined | 1.49 ± 0.12 | 0.15 ± 0.01 | ||
| S. agalactiae 2 | 2.30 (High) | 1.50 ± 0.02 | 0.08 ± 0.00 | |||
| S. agalactiae 3 | 2.36 (High) | 0.00 ± 0.20 | 0.12 ± 0.00 | |||
| S. dysgalactiae 1 | 2.05 (High) | 1.77 ± 0.43 | 0.25 ± 0.01 | |||
| S. dysgalactiae 2 | 2.36 (High) | 0.15 ± 0.07 | 0.30 ± 0.00 | |||
| S. dysgalactiae 3 | 2.24 (High) | 1.11 ± 0.17 | 0.25 ± 0.01 | |||
Note: Bacteria were challenged during 1 h with 0.3 μM of either NC2 or NC5. MALDI‐TOF MS was performed to confirm the bacterial species of the isolates. A score to each isolate is assigned indicating a non‐ (<1.70) to low (≥1.70) or high (≥2.00) confidence level. Whole genome sequencing was performed to assign a known sequencing type (ST) or global clonal complex (GCC) to the Streptococcus uberis isolates. These sequencing data confirmed the bacterial species in addition to MALDI‐TOF MS for S. uberis.
indicates the GCC was estimated based on sequence alignment against known GCCs from the PubMLST database.
FIGURE 3.
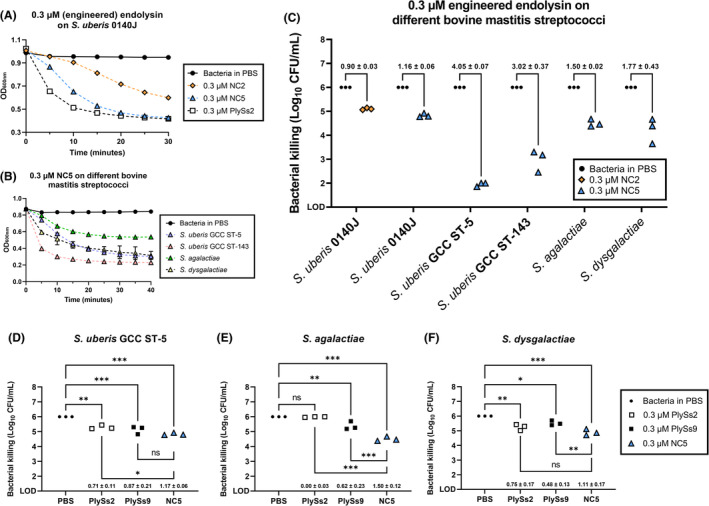
Bactericidal activity of candidate engineered endolysins against multiple bovine mastitis‐derived streptococcal isolates. (A) Turbidity reduction assay with 0.3 μM NC2 (orange diamond), NC5 (blue triangle) and PlySs2 (open square) on stationary phase Streptococcus uberis 0140J, the same bacteria in PBS served as the negative control (black circle). (B) Turbidity reduction assay with 0.3 μM NC5 on bovine mastitis‐isolated stationary phase S. uberis belonging to the global clonal complex (GCC ST‐) 5 and 143, S. agalactiae and S. dysgalactiae (purple, red, yellow and green triangles respectively). S. uberis GCC ST‐5 in PBS served as a representative negative control for all strains tested (black circle). (C) Time‐kill assay with 0.3 μM NC2 on S. uberis 0140J (orange diamond) and NC5 (blue triangles) on S. uberis 0140J and bovine mastitis‐isolated stationary phase S. uberis belonging to the global clonal complex (GCC ST‐) 5 and 143, as well as S. agalactiae and S. dysgalactiae. Time‐kill assays with 0.3 μM PlySs2 (open square), PlySs9 (filled square) and NC5 (blue triangle) on bovine mastitis‐isolated stationary phase (D) S. uberis GCC ST‐5, (E) S. agalactiae and (F) S. dysgalactiae. Bacteria in PBS (standardized to 106 CFU/mL) served as negative controls (black circle). The difference in log10 (CFU/mL) is represented as mean ± standard deviation. LOD: limit of detection; *, ** and *** indicate p < 0.05, p < 0.01 and p < 0.001, whereas ns indicates non‐significance corresponding to a p > 0.05.
The top candidate engineered endolysin equals or outperforms the VersaTile reassembled PlySs2 and PlySs9 endolysins against S. uberis, S. agalactiae and S. dysgalactiae in PBS
The bactericidal activity of NC5 was compared in PBS with that of PlySs2 and PlySs9, which were reassembled by VersaTile, against a randomly selected mastitis‐derived S. uberis GCC ST‐5, S. agalactiae and S. dysgalactiae at a differentiating dose of 0.3 μM (Figure 3D–F). NC5 consistently showed the highest bacterial killing, with significant (p < 0.001) Δlog10 of 1.17 ± 0.06, 1.50 ± 0.12 and 1.11 ± 0.17 against S. uberis GCC ST‐5, S. agalactiae and S. dysgalactiae respectively. PlySs2 and PlySs9 also caused significant Δlog10 of 0.71 ± 0.11 (p < 0.01) and 0.87 ± 0.21 (p < 0.001) against S. uberis GCC ST‐5, of which the PlySs2 reduction was significantly (p < 0.05) lower than that of NC5 (Figure 3D). No significant (p > 0.05) log10 reduction was caused by PlySs2 against S. agalactiae in comparison with the PBS negative control, to which NC5 performed significantly (p < 0.001) better (Figure 3E). PlySs9 caused a significant (p < 0.01) Δlog10 of the selected S. agalactiae strain of 0.62 ± 0.23, which differed significantly (p < 0.001) from NC5. Regarding S. dysgalactiae, PlySs2 and PlySs9 caused significant Δlog10 of 0.75 ± 0.17 (p < 0.01) and 0.48 ± 0.13 (p < 0.05), which did not differ significantly (p > 0.05) and differed significantly (p < 0.01) from NC5 respectively (Figure 3F). Overall, NC5 killed S. uberis GCC ST‐5 (p < 0.01) and S. agalactiae (p < 0.001) significantly better than PlySs2, and S. dysgalactiae (p < 0.01) significantly better than PlySs9, and did not show inferior bactericidal activity in PBS compared to these VersaTile reassembled PlySs2 and PlySs9 endolysins against these randomly selected streptococcal mastitis‐derived strains.
The top candidate engineered endolysin kills S. uberis in the bovine mammary epithelium
To assess whether NC5 enters bovine mammary epithelial cells (boMECs), lysate from MAC‐T cells incubated with 2.5 μM NC5 was analysed by western blotting after washing away the extracellular protein and compared to lysate of untreated cells (Appendix S11A,B). A band at the expected molecular weight of NC5 was detected, which was absent in the lysate of untreated cells. To further support that NC5 was present intracellularly, MAC‐T cells that were likewise treated with 2.5 μM NC5 were visualized by confocal microscopy (Figure 4A), which clearly revealed the intracellular presence of NC5 in the MAC‐T cells. To assure that the observed fluorescent signal was specific for endolysin NC5, autofluorescence of the MAC‐T was determined in addition to subjecting endolysin‐untreated MAC‐T to the same staining protocol. Neither autofluorescence nor aspecific binding of the primary and fluorescent secondary antibodies were detected (Appendix S12A–I). Taken together, these data show that NC5 is able to enter MAC‐T cells.
FIGURE 4.
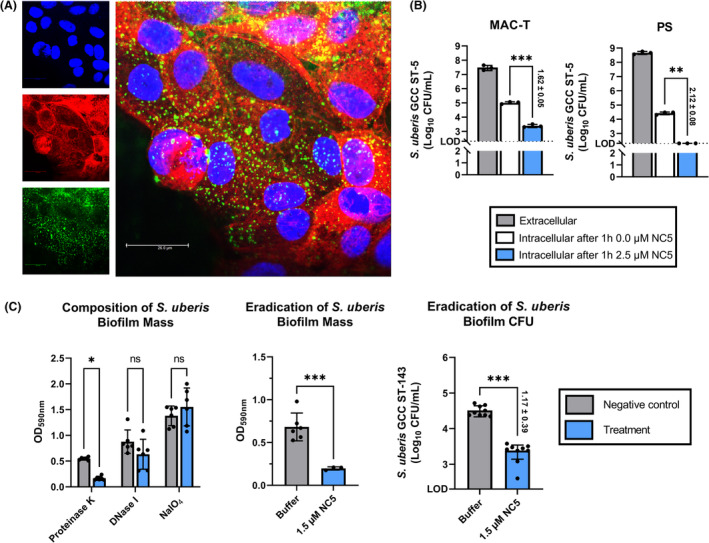
Intracellular presence and biofilm eradicating activity of the top candidate engineered endolysin. (A) Confocal microscopy‐based optical section through the centre of MAC‐T bovine mammary epithelial cells treated with 2.5 μM NC5. Nuclei were stained using Hoechst 33342 (blue), actin cytoskeleton using phalloidin‐iFluor 594 (red) and NC5 using Alexa Fluor 488‐labelled antibody for the His6 tag (green). The intracellular presence of NC5 in MAC‐T is evident from the overlay of the different fluorescence channels. (B) Intracellular killing of Streptococcus uberis GCC ST‐5 observed in the bovine mammary epithelial cell lines MAC‐T and PS after incubating infected MAC‐T and PS cells with 2.5 μM NC5 at 37°C during 1 h. MAC‐T and PS were infected at a multiplicity of infection of approximately 250 and 500 respectively. Culture medium without NC5 served as negative control. (C) Determination of the biofilm composition of S. uberis GCC ST‐143 by proteinase K, DNase I and NaIO4 treatment, as well as eradication of both the biofilm biomass and the number of CFU after incubation with 1.5 μM NC5 during 2 h 30 min at 37°C on a shaker. The difference in log10 (CFU/mL) is represented as mean ± standard deviation. LOD: Limit of detection (200 CFU/mL); *, ** and *** indicate p < 0.05, p < 0.01 and p < 0.001, whereas ns indicates non‐significance corresponding to a p > 0.05.
Subsequently, MAC‐T and PS bovine mammary epithelial cell lines were challenged with a S. uberis GCC ST‐5 isolate sensitive to NC5 and with predetermined intracellular invasion (i.e. clinical S. uberis 5 in Table 3). After 3 h co‐incubation of S. uberis with both boMEC, 7.46 ± 0.14 and 8.66 ± 0.08 log10 were extracellularly retrieved from MAC‐T and PS cells, respectively, corresponding to a multiplicity of infection (MOI) of approximately 250 and 500 _(Figure 4B). After gentamicin treatment to kill these extracellular S. uberis, infected MAC‐T and PS were incubated during 1 h with 2.5 μM NC5 in culture medium. The same medium without the addition of NC5 served as negative control. From this negative control, 5.00 ± 0.06 and 4.42 ± 0.08 log10 were respectively retrieved from the infected MAC‐T and PS cells (Figure 4B). In comparison, both boMECs showed a reduction of the intracellular number of S. uberis fraction after NC5 treatment. More specifically, a significant (p < 0.001) Δlog10 of 1.62 ± 0.05 was observed for the infected MAC‐T, whereas this was at least (p < 0.01) 2.12 ± 0.08 Δlog10 for the infected PS as the remaining S. uberis numbers all were below or equal to the LOD of 200 colony‐forming units (CFU)/mL (Figure 4B). Overall these data indicate that NC5 was able to eradicate S. uberis intracellularly in boMEC.
The top candidate engineered endolysin eradicates S. uberis biofilm
Streptococcus uberis GCC ST‐143, which was found sensitive to NC5 (i.e. subclinical S. uberis 1 in Table 3), was the only isolate in the S. uberis panel that reproducibly formed a biofilm in vitro. This biofilm was mainly composed of protein, as proteinase K treatment significantly (p < 0.05) eradicated its mass (Figure 4C). Upon exposing this biofilm to a concentration of 1.5 μM NC5 during 2 h 30 min, a significant (p < 0.001) reduction of the biofilm mass of ±70% was observed, corresponding to a significant (p < 0.001) 1.17 ± 0.39 Δlog10 reduction (Figure 4C).
The top candidate engineered endolysin potentiates cloxacillin against S. uberis in raw mastitic cow's milk
Raw mastitic cow's milk from four cows with a confirmed S. uberis infection was challenged during 8 h with either 0.5 μM NC5, 50 μg/mL cloxacillin, a combination of both (i.e. the combination therapy) or PBS as a negative control (Figure 5). Cloxacillin is a penicillin antibiotic frequently used to treat streptococcal mastitis in dairy cows (de Jong et al., 2018). Treatment with 0.5 μM NC5 alone caused a reduction in all mastitic milk samples after 8 h ranging from 0.31 ± 0.12 to 2.61 ± 0.35 Δlog10, which were only significant (p < 0.05) compared to the PBS control in two out of four samples. These log10 reductions caused by 0.5 μM NC5 were inferior compared to those caused by cloxacillin, except for cow 2 at ≤6 h. The Δlog10 caused by cloxacillin alone ranged from 2.20 ± 0.14 to 4.55 ± 0.15 at 8 h. In contrast, the cloxacillin and combination therapy caused significant (p < 0.05) reductions in all mastitic milk samples at 8 h, ranging from 2.20 ± 0.14 to 4.83 ± 0.43 Δlog10, and attained the LOD in two out of four samples. A key finding was that all mastitic milk samples showed a faster decrease of the S. uberis load if cloxacillin treatment was combined with 0.5 μM NC5 (Figure 5). Indeed, the amount of surviving S. uberis in the combination therapy was significantly (p < 0.05) decreased compared to cloxacillin alone during at least 2 h in the mastitic milk samples in three out of four cows with Δlog10 in the range of 2.20 ± 0.17 to 5.78 ± 0.67. In one cow, these Δlog10 were non‐significantly (p > 0.05) decreased. For completion, the killing rate was analysed for two raw mastitic cow's milk samples using a 30‐min time interval during 2 h, confirming the highest Δlog10 in the combination therapy group vs. the negative control after 2 h with 0.80 ± 0.09 and 0.89 ± 0.13 (Appendix S13).
FIGURE 5.
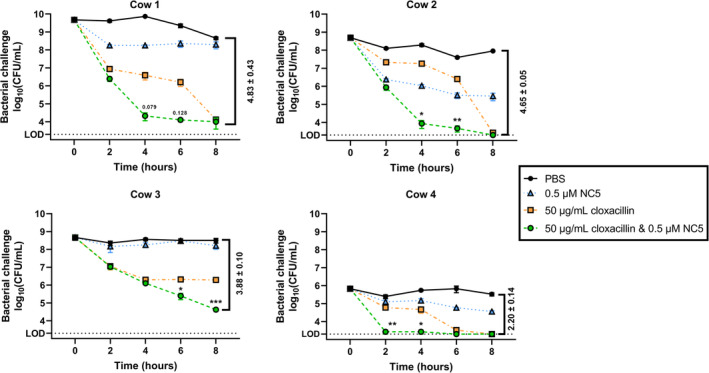
Activity of the top candidate engineered endolysin in Streptococcus uberis mastitic raw cow's milk. Raw mastitic cow's milk from four S. uberis infected animals was incubated during 8 h with either 0.5 μM NC5 (blue triangle), 50 μg/mL cloxacillin (orange square), a combination of both (green circle) or PBS (black circle) as a negative control. The number of surviving bacteria was determined respecting a 2‐h interval. Difference in log10 (CFU/mL) between the combination treatment and PBS negative control are indicated as mean ± standard deviation; LOD: limit of detection (2000 CFU/mL); p values between the cloxacillin and combination treatment group are represented as numbers or as *, ** and *** indicating p < 0.05, p < 0.01 and p < 0.001 respectively.
DISCUSSION
Bovine Gram‐positive mastitis, frequently caused by S. uberis, S. dysgalactiae, Sta. aureus and to a lesser extent S. agalactiae, is a high‐impact disease for the dairy industry's profitability (Geary et al., 2012; Hogeveen et al., 2011; Rollin et al., 2015). Given the rising dairy consumer's awareness on the use of antibiotics in dairy agriculture, the overuse of antibiotics both preventively for dry cow treatment and therapeutically for mastitis are increasingly questioned. Proof of the latter are new governmental policies imposing restrictions on the abuse of antibiotics in the dairy sector (e.g. European Green Deal, WHO and a new EU Regulation on Veterinary Medicines 2019/06), indicating the need to develop alternatives that can phase out especially the critical antibiotics from animal production. Our group proposed bacteriophage‐derived endolysins as replacement or add‐on strategies and also previously characterized the S. suis wild‐type endolysins PlySs2 and PlySs9 with potent lytic activity against S. uberis (Vander Elst et al., 2020; Vander Elst & Meyer, 2018). In addition, we reported that improved engineered endolysins can be conveniently engineered through domain swapping by the VersaTile DNA assembly technique (Duyvejonck et al., 2021; Gerstmans et al., 2020). Therefore, we here aimed at creating engineered endolysins by VersaTile with enhanced (i) activity in raw cow's milk, (ii) intracellular penetration and killing activity and (iii) biofilm eradicating properties against bovine Gram‐positive mastitis‐causing streptococci as well as Sta. aureus.
Although the assembly, screening and hit‐to‐lead selection of endolysins with activity against either streptococci or staphylococci has been reported (Keller et al., 2022; Röhrig et al., 2020; Son et al., 2021; Verbree et al., 2018; Yang et al., 2015), the maximal number of endolysins screened was limited to <500 (Röhrig et al., 2020). We here at first performed the high‐throughput assembly of >88,704 theoretical endolysin variants against bovine mastitis S. uberis and Sta. aureus. From these libraries, 40,170 constructs were screened. Throughout this random screening certain variants stochastically occurred more than once. This approach overcame a major limitation in previous reports that required protein expression of the assembled endolysins in liquid media, limiting the throughput of screening assays to the capacity of multiwell plates (Duyvejonck et al., 2021; Keller et al., 2022; Röhrig et al., 2020). Instead, we employed a new method to extent this screening to tens of thousands of endolysins simultaneously, by inducing protein expression in single colonies harbouring the assembled endolysins on agar plates containing autoclaved targeted cells. Following identification of the active colonies and sequencing of their plasmids harbouring these endolysin genes, design rules complemented with statistics were applied to identify optimal designs and the significantly overrepresented tiles at certain positions in these designs. A stepwise narrow‐down selection or ‘funnel’ of the significantly overrepresented domains within such a design was then applied to hit‐to‐lead select the top candidates for further downstream assays. This approach was successfully applied, as the hit rate increased in the range of 10‐ to 50‐fold by implementing design rules in two of the narrow‐down libraries. We here identified the optimal design of streptococcal endolysins to be (CPP‐)EAD‐CBD or (CPP‐)EAD‐CBD‐EAD, with a pI in the range 9.05–9.65. These designs and their in silico predicted pI follow that of most wild‐type streptococcal endolysins as was previously identified for PlySs2 and PlySs9 by our group and others (Linden et al., 2021; Vander Elst et al., 2020). It should be noted that the terminal EAD in the (CPP‐)EAD‐CBD‐EAD design contains one LysM repeat in case of the PlySs9 amidase_3 domain, which was initially missed but became apparent with the availability of ColabFold in the course of our work (Mirdita et al., 2022).
The creation of engineered endolysins in combination with CPPs to obtain intracellular activity has also been reported (Becker et al., 2016; Keller et al., 2022; Röhrig et al., 2020). Although the presence of a CPP is not always necessary to translocate endolysins intracellularly, increased cellular uptake upon fusion of a CPP has been demonstrated by different groups (Becker et al., 2016; Keller et al., 2022; Röhrig et al., 2020). These latter studies evaluated fusion of the CPPs TAT, R8 and penetratin to engineered endolysins. Remarkably, they all independently reported a decrease of the lytic activity upon fusion of a CPP. This study confirms this observation, as our screening data showed that the designs EAD‐CBD or EAD‐CBD‐EAD were significantly overrepresented in comparison with CPP‐EAD‐CBD or CPP‐EAD‐CBD‐EAD respectively. However, our data also revealed that small and cationic CPPs can favour intracellular killing and retain most of the endolysin's lytic activity. This key finding corroborates a previous report from our group (Rodríguez‐Rubio et al., 2016). While the effect of the absence versus presence of a CPP was not further investigated in the current study, the TAT peptide of HIV‐1 in lead candidate NC5 has been identified as one of the most promising CPPs to obtain intracellular killing, while at the same time minimizing loss in lytic activity (Becker et al., 2016; Keller et al., 2022; Röhrig et al., 2020). Additionally, we identified an optimal position of this TAT at the N‐terminus, while the cited studies all rationally fused this specific CPP C‐terminally. For completion, it can be remarked that the PlySs2 SH3_5 and PlyCb domains included in this study have also been attributed certain CPP functionalities (Shen et al., 2016; Yang et al., 2021).
The comparative evaluation of the antibacterial activity of our selected engineered endolysins was assessed in PBS, ultra‐high temperature (UHT)‐pasteurized whole milk and raw mastitic milk. While PBS does not allow growth of the incubated pathogens, this occurs in milk resulting in a major difference between incubation conditions (Schmelcher et al., 2015). This has important consequences when interpreting our results. First, the Δlog10 reductions in CFU/mL observed in PBS versus pasteurized and raw milk are not expected to correlate (Röhrig et al., 2020). Even screening of engineered endolysins in pasteurized milk cannot guarantee an effective bacterial killing in raw cow's milk (Verbree et al., 2018). Second, endolysins are enzymes with a limited lifetime and turnover, which was for example only 16 min for the pneumococcal endolysin Cpl‐1 in cerebrospinal fluid (Grandgirard et al., 2008). More specifically, they are expected to be sensitive to proteases produced and released during the innate immune response, also present in raw mastitic cow's milk (Meyer‐Hoffert & Wiedow, 2011). Third, the activity of endolysins in PBS can either be reduced or improved in pasteurized or raw milk. Reduced activity of the engineered endolysins in pasteurized and mastitic milk compared to PBS was frequently observed in our study, similar to previous reports (Celia et al., 2008; Verbree et al., 2018; Yang et al., 2015). An exception has been reported for PlySs2. This wild‐type endolysin has been characterized with the unique property to bind (lact)albumin, which also and simultaneously occurs for certain mastitis pathogens such as Sta. aureus, resulting in increased killing by co‐localization of PlySs2 and Sta. aureus on (lact)albumin (Indiani et al., 2019). Our results showed a similar observation, with increased bacterial killing in pasteurized milk versus PBS for PlySs2 as well as NC2. Of note, NC2 contains the full PlySs2 endolysin. Proceeding with the activity in raw mastitic milk, an inconsistent killing of S. uberis by our lead candidate NC5 as stand‐alone therapy was observed in different field samples. This variable activity can most likely be attributed to differences in the raw milk sample composition, pH and/or to S. uberis strain differences, also taking into account different genetic backgrounds of the mastitic cows. Although NC5 did not show convincing potent activity in the mastitic milk samples as stand‐alone therapy, it significantly potentiated cloxacillin treatment as a combination therapy by eliminating S. uberis faster than cloxacillin treatment alone. Indeed, to circumvent the limitations of endolysins as stand‐alone antimicrobials, we suggest to exploit additive or synergistic effects by combining multiple endolysins targeting different peptidoglycan types or bonds, or in combination with antibiotics or bacteriocins (e.g. lysostaphin) (Daniel et al., 2010; Schmelcher et al., 2012, 2015). We deliberately used cloxacillin since it is a non‐critical, narrow‐spectrum penicillin derivative frequently used to combat streptococcal infections in the udder (de Jong et al., 2018). Combination therapy to pursue synergy with antibiotics has also been proposed for the staphylococcal endolysin trxSA‐1 after evaluation of trxSA‐1 as stand‐alone treatment in Sta. aureus‐infected udder quarters (Fan et al., 2016). Future studies will now investigate if this cloxacillin potentiating effect of NC5 can be retained in vivo, as it is known that these additive or synergistic effects are sometimes lost in the mammary gland (Schmelcher et al., 2015).
In our screening approach, we had initially foreseen the option to obtain bispecific endolysins also targeting Sta. aureus in addition to S. uberis, S. agalactiae and S. dysgalactiae. While the initial hits scored positive in the halo‐based assay against autoclaved Sta. aureus cells, purified proteins from hits such as NC5 eventually did not consistently kill all Sta. aureus isolates with the doses used, in contrast to S. uberis. This discrepancy can be explained by the high sensitivity of the halo‐based assay because of the prolonged incubation (18 h) and the use of dead cells. The concept of creating an engineered endolysin against several Gram‐positive strains has also been pursued previously (Yang et al., 2015). The latter report showed that the engineered endolysin ClyR, consisting of the C‐terminal CHAP of PlyC and the N‐terminal SH3_5 of PlySs2, displayed activity against both streptococcal and staphylococcal strains, including S. uberis, S. agalactiae, S. dysgalactiae and Sta. aureus. Although both domains were included in our extensive screening, ClyR was not retrieved. This might be due to several differences between ClyR and our study such as screening conditions and differences in delineation of the subdomains (Röhrig et al., 2020; Vander Elst et al., 2020). Notably, follow‐up studies with ClyR only focused on streptococcal strains (Xu et al., 2018; Yang et al., 2016, 2021; Zhu et al., 2021).
CONCLUSION
We here report for the first time the assembly, screening, selection and identification of engineered endolysins against bovine Gram‐positive mastitis employing VersaTile, followed by a new approach to high‐throughput screen engineered endolysins on individual colony level. The identified hits showed potent antibacterial activity against S. uberis, S. agalactiae and S. dysgalactiae and harboured the designs CPP‐EAD‐CBD or CPP‐EAD‐CBD‐EAD. The lead candidate NC5 was able to lyse S. uberis in bovine mammary epithelial cells, eradicate S. uberis biofilm and potentiate cloxacillin treatment in raw mastitic cow's milk. We will now proceed with in vivo studies to evaluate the potential of NC5 in a murine model for bovine streptococcal mastitis as an add‐on to the currently used intramammary antibiotics.
MATERIALS AND METHODS
Bioinformatic analyses
Streptococcus uberis or S. suis genomes (latter inspired by Vander Elst et al., 2020) were derived from NCBI GenBank and screened for intact prophages by Phaster (Arndt et al., 2016; Clark et al., 2016). Subsequently, genes in the discovered prophage genomes were annotated to identify endolysins (Brettin et al., 2015). Multiple sequence alignments were consistently performed by Clustal Omega using the MAFFT algorithm in combination with a Pearson/FASTA output (Madeira et al., 2019). The percentages given in this work were always derived from the percentage identity matrix of the aligned amino acid sequences. Endolysin subdomains were delineated for tile assembly by combining InterPro and Phyre2 analyses (Blum et al., 2021; Kelley et al., 2015). Linkers were identified as proline‐ and lysin‐rich regions, and the domains were consistently delineated by including their C‐terminal linker.
Cloning of endolysin genes and VersaTile assembly of chimers
The 14 selected wild‐type endolysins were codon optimized for expression in E. coli and synthesized (Twist Bioscience, USA) with a 5′ and 3′ BsaI restriction site. Subsequently, these synthesized gene fragments were introduced into pVTD3 (as destination vector) by Type IIs cloning (Duyvejonck et al., 2021; Gerstmans et al., 2020). The endolysin subdomains were thereafter subcloned starting from the full‐length endolysins in the entry vector (pVTEIII), yielding tiles (Gerstmans et al., 2020). Briefly, tiles were made in the entry vector pVTEIII by VersaTile cloning with the restriction enzyme SapI. Subsequently, chimers were assembled with BsaI in a cyclic restriction ligation reaction. A four‐way system was chosen with pVTD1, pVTD2 or pVTD3 as destination vectors depending on the number and order of tiles to be assembled and the desired position of the His6‐tag (Gerstmans et al., 2020). All these destination vectors contain a T7 promotor.
Bacterial strains and culture
Bovine mastitis isolates S. uberis 0140J (ATCC BAA‐854) and Sta. aureus Newbould 305 (N305, ATCC 29740) were used as reference strains. All other S. uberis, S. dysgalactiae, S. agalactiae and Sta. aureus strains included are recent (<5 years) isolates from Milk Control Center Flanders from (sub)clinical bovine mastitis cases originating from various dairy farms located in Flanders (Belgium). These isolates were complementary verified by MALDI‐TOF MS (Vander Elst et al., 2020). Streptococci and Sta. aureus were grown at 37°C in brain heart infusion (BHI) or tryptic soy broth (Oxoid, Belgium) respectively. Plating was conducted on BHI with the addition of 15.0 g/L agar (BHI agar) (Chem Lab, Belgium) for streptococci or tryptic soy agar (TSA) (Oxoid, Belgium) for Sta. aureus. E. coli harbouring the cloned constructs were grown in lysogeny broth (LB) (for 1.0 L: 10.0 g tryptone, 10.0 g NaCl and 5.0 g yeast extract) or terrific broth (TB) (for 1.0 L: 12.0 g tryptone, 24.0 g yeast extract, 4.0 mL 100% glycerol, 0.72 M K2HPO4 and 0.17 M KH2PO4) containing either 100.0 μg/mL ampicillin or 50.0 μg/mL kanamycin (Carl Roth, Germany). Selective plates consisted of the same reagents, to which 15.0 g/L agar (VWR, Belgium) was added.
Whole genome sequencing, sequence typing and determination of the agr and capsular (sero)types of bovine mastitis isolates
DNA was extracted from 18 h grown bacterial cultures according to the manufacturer's protocol using the DNeasy Ultraclean Microbial kit (Qiagen, USA). Subsequently, the genomic DNA was sequenced using a Illumina MiniSeq platform. A library was prepared using the Nextera Flex DNA Library Kit (Illumina, USA) for each sample, tagged with a unique adapter sequence. The quality of each library preparation was controlled using an Agilent Bioanalyzer 2100. Genome assembly was performed using the Galaxy platform (SPAdes assembly algorithm: version 3.12.0). Quality of the reads was verified using FASTQC (version 1.1.5). Regarding S. uberis, multilocus sequence typing was performed as described previously (Zouharova et al., 2022). The sequencing data in FASTA format were submitted to the PubMLST database (https://pubmlst.org/organisms/streptococcus‐uberis) (accessed on 5 January 2022) to confirm the bacterial species and identify allelic matches. Each S. uberis isolate was defined by an allelic profile, which corresponds to the allele numbers at the seven loci in the order arcC, ddl, gki, recP, tdk, tpi and yqiL. According to the combination of alleles, the sequence type (ST) and global clonal complex (GCC) were determined. If an unknown ST emerged, the GCC was estimated based on multiple sequence alignment against a set of S. uberis genomes with a known GCC derived from the PubMLST database. This was also done if the allelic profile yielded a ST that was not yet assigned to a GCC in the PubMLST database. Regarding Sta. aureus, the agr and capsular (sero)type were determined inspired by Bardiau et al. (2016) and Rossi et al. (2021). Multiple sequence alignment using BLASTn was performed with each Sta. aureus genome against the agr types I, II, III and IV genes, as well as the genes encoding capsular serotype 5 or 8.
Halo‐based high‐throughput screening of active fusion proteins
Chemocompetent E. coli TOP10, BL21 (DE3) or BL21 (DE3) pLysS (NEB, USA) were transformed with 5.0 ng/μL assembly mixture via heat shock during 45 s at 42°C after a 30‐min incubation on melting ice. The transformed cells were then incubated for 1 h at 37°C in super optimal broth (for 1.0 L: 0.5% yeast extract, 2.0% tryptone, 10.0 mM NaCl, 2.5 mM KCl, 10.0 mM MgCl2, 10 mM MgSO4 and 20 mM d‐glucose) after a 5‐min recovery on melting ice. E. coli cells were subsequently plated. For rationally assembled constructs (i.e. PlySs2 and PlySs9 as positive controls), one colony from the 18‐h incubated plate was inoculated and grown in LB with the addition of a selective antibiotic and again incubated for 18 h. For randomly assembled VersaTile constructs (e.g. NC5), transformed E. coli BL21 (DE3) pLysS were plated on LB agar that contained 50.0 g/mL kanamycin and 5% sucrose (Carl Roth, Germany) as selection markers, 1 mM isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) (Carl Roth, Germany) as inducing agent for protein expression and 2.0 or 1.0% autoclaved, washed S. uberis 0140J or Sta. aureus N305 cells respectively. These plates were then incubated at 37°C for 48 h followed by incubation at 22°C for at least another 24 h. Colonies harbouring an active engineered endolysin will be featured by a halo due to release of the recombinant protein from disintegrating cells within the colony and diffusion into the agar where the embedded target cells are lysed. Colonies displaying a halo against Sta. aureus N305 were picked and streaked out on the same agar, but containing S. uberis 0140J peptidoglycan instead. Constructs that yielded halos against both bacterial species were along with the rationally assembled constructs inoculated and grown in LB with the addition of 50.0 μg/mL kanamycin. Long‐term storage was performed at −80°C by adding 20% glycerol in a cryovial. Pure plasmid was obtained from an 18‐h culture using the GeneJET plasmid miniprep kit (Thermo Fisher Scientific, USA) and sent to LGC Genomics (Berlin, Germany) for Sanger sequencing. All DNA manipulations, DNA engineering and DNA alignments were executed via Benchling (Biology Software, USA).
Protein expression and purification of active fusion proteins
A 5.0‐mL 18‐h culture of the transformed E. coli BL21 (DE3) was poured into a 2.0‐L baffled Erlenmeyer flask filled with 0.5 L LB (or alternatively TB) and the addition of 50.0 μg/mL kanamycin. When the culture obtained an optical density measured at 600 nm (OD600) of 0.6–0.8, protein expression was induced by addition of 1 mM IPTG. After expression (18 h at 16°C), E. coli were pelleted, supernatant was decanted and the pellet was stored at −80°C until further processing. After thawing on melting ice, the pellet was resuspended in PBS containing 10 mM imidazole (Carl Roth, Germany), 1 mM phenylmethylsulphonyl fluoride (Carl Roth, Germany) and 1 mM DNase I (NEB, USA). The E. coli suspension was then sonicated on melting ice and centrifuged (20,000 g, 4°C, 20 min). Subsequently, the supernatant was poured over a His GraviTrap column (Sigma‐Aldrich, USA) for Ni‐NTA chromatography. The column was washed with 10 mM imidazole in PBS (pH 7.4), 1 M NaCl (Carl Roth, Germany) in PBS (pH 7.4) and 20 mM 2‐(N‐morpholino) ethane sulphonic acid (Carl Roth, Germany) in PBS (pH 6.0). Finally, the target protein was eluted from the column with increasing concentrations of imidazole (20, 50, 100, 250 and 500 mM) in PBS, each time with the addition of 0.5 M NaCl and 10% glycerol (pH 7.4). The target protein in the eluted fractions was detected by SDS‐PAGE on a 12% polyacrylamide gel (Bio‐Rad Laboratories, USA), stained with Coomassie brilliant blue (Carl Roth, Germany) and decolorized in distilled water. Buffer exchange and concentrating the protein were executed by using Pierce™ Protein Concentrators PES with a molecular weight cut‐off (MWCO) of 10 kDa (Thermo Fisher Scientific, USA). Finally, protein was filter sterilized (PVDF membrane, 0.45 μm) by means of a syringe filter and the protein concentration was estimated by the Bradford protein assay (Bio‐Rad, USA).
Evaluation of the enzymatic and antibacterial activity of active fusion proteins in PBS and pasteurized whole cow's milk
For the time‐kill assay (TKA), an 18‐h bacterial culture was pelleted (4000 g, 10 min), washed with PBS and diluted 1:10 for S. uberis, S. agalactiae and S. dysgalactiae or 1:100 for Sta. aureus in either PBS or UHT‐pasteurized whole cow's milk. For both conditions, 100 μL was combined with an equal volume of either the engineered endolysin dissolved in PBS or PBS (as negative control) and incubated in a covered 96‐well microtiter plate at 37°C for 2 h. The endolysin concentration was low (i.e. in the range of 0.25–0.30 μM) to differentiate between less and better performing variants, allowing their ranking. After 1:10 serial dilution in PBS, 5 μL of each fraction was spotted on either TSA or BHI agar and incubated 18 h at 37°C. The following day, CFU were counted to calculate reductions in CFU/mL compared to the negative control. These reductions are presented in this work as Briggs logarithms (i.e. log10). The relative scores were calculated by dividing the observed logarithmic reduction for each engineered endolysin by the maximal logarithmic reduction observed for that bacterial strain under each specific condition, in analogy with Röhrig et al. (2020). Subsequently, these values were divided by 4 to arbitrarily set the maximal relative score of the best‐performing engineered endolysin within a certain condition and strain to 0.25. When accumulating these individual scores over the different strains and both conditions tested (i.e. 12 = 6 bacterial strains × 2 conditions), the endolysin that overall performs superior is expected to yield the maximal score of 3.0 (= 12 × 0.25). The turbidity reduction assay (TRA) was performed as described previously (Vander Elst et al., 2020). In short, a 18‐h bacterial culture was pelleted (4000 g, 10 min), washed with PBS, resuspended in PBS and diluted in a 1:1 ratio with PBS to an OD600 of approximately 1.0. Subsequently, 100 μL of the resuspended bacterial culture was combined with an equal volume of 0.6 μM endolysin in PBS (to obtain a final concentration of 0.3 μM) or PBS as negative control. Next, the OD600 was kinetically measured during 1 h every 15 s at 37°C using a plate reader (Infinite 200 PRO Tecan, Switzerland), shaking the 96‐well plate between each measurement. From these measurements, the enzymatic activity was calculated as (ΔOD600/min)/μM, which has been standardized for endolysins as described previously (Briers et al., 2007).
Cell culture conditions of bovine mammary epithelial cells
The bovine MAC‐T cell line (established 1991) (Huynh et al., 1991) in passage 10–15 was grown in culture flasks at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) with 10% foetal bovine serum (FBS), 0.5% insulin–transferrin–selenium and 100 U/mL penicillin–streptomycin mix (all Gibco, USA). The bovine PS cell line (established 2015) (Roussel et al., 2015) in passage 10–15 was grown in culture flasks at 37°C and 5% CO2 in advanced DMEM Ham's F‐12 with 20 mM HEPES, 2 mM glutamine, 100 U/mL penicillin–streptomycin mix (all Gibco, USA), 1 μg/mL hydrocortisone 21‐hemisuccinate, 5 ng/mL epidermal growth factor (both Sigma‐Aldrich, USA), 10 ng/mL insulin‐like growth factor‐I and 5 ng/mL fibroblast growth factor (both Peprotech, France). Cell cultures were routinely checked for bacterial and mycoplasma contamination, by plating cell culture media on Columbia blood agar plates (VWR, Belgium) and by means of a PlasmoTest (InvivoGen, USA) respectively. After washing with Dulbecco's PBS (dPBS), cells were harvested using 0.25% trypsin–ethylenediaminetetraacetic acid (Sigma‐Aldrich, USA), which was inactivated once the cells were detached from the culture flask by adding an equal volume of 20% FBS in dPBS. Cells were centrifuged (200 g for 5 min), resuspended in cell culture medium, counted by means of a Bürker chamber and seeded (ratio 1:3 and 1:5 for MAC‐T and PS respectively).
Intracellular detection of the top candidate engineered endolysin by immunofluorescence analyses
Bovine MAC‐T cells in cell culture medium were seeded in a glass four‐chamber slide (Novolab, Belgium) at 50,000 cells/chamber. BoMECs were grown to 90%–100% confluency and cell culture medium was replaced by pre‐warmed NC5 in DMEM (buffer exchange performed at 4–8°C by means of a Pierce™ Protein Concentrator PES 10 kDa MWCO), followed by incubation during 1 h at 37°C and 5.0% CO2. DMEM alone served as the negative control. Following incubation, the supernatant was aspirated and cells were washed 5× with pre‐warmed dPBS. Subsequently, MAC‐T cells were fixed and permeabilized on chamber slides by means of an inside stain kit (Miltenyi Biotec, the Netherlands). Primary antibody, being rabbit monoclonal IgG anti‐hexa‐histidine (Abcam, the Netherlands) was administered to the fixed cells at 1:100 in inside perm solution (IPS) and allowed to bind at 22°C during 1 h. Next, cells were washed 3× with IPS and treated at 22°C for 30 min with IPS containing 2 μg/mL Hoechst 33342 (Thermo Fisher Scientific, USA), 1:1000 Phalloidin‐iFluor 594 and 1:200 AlexaFluor 488‐labelled goat monoclonal IgG anti‐rabbit (both Abcam, the Netherlands), all diluted in IPS. Finally, cells were washed 3× with IPS and mounted on a cover slide using anti‐fading glycerol mounting medium with DABCO (Abcam, the Netherlands). Cells were visualized using the Leica DMI8 and Leica TCS SP2 microscopes (Leica Microsystems GmbH, Germany).
Intracellular antibacterial activity of the top candidate engineered endolysin
Bovine MAC‐T and PS cells were seeded in a 24‐well plate at 50,000 cells/well in their culture media and supplemented with 25 μg/mL calf skin collagen type I (Sigma‐Aldrich, USA). The latter was solubilized in 0.1 M acetic acid. When the BoMECs reached 90%–100% confluency, culture media were aspirated and BoMECs were washed 3× with dPBS to remove residual penicillin–streptomycin mix. An 18‐h culture of a clinical S. uberis isolate with predetermined intracellular invasion activity in BoMECs (i.e. clinical S. uberis 5 in Table 3) was pelleted (4000 g, 10 min), washed with dPBS and resuspended in an equal volume of pre‐warmed cell culture media without the addition of penicillin–streptomycin mix. Subsequently, 0.5 mL thereof was incubated per well during 3 h at 37°C to allow intracellular invasion. Culture media containing S. uberis were thereafter aspirated from the BoMECs, 1:10 serially diluted and plated as described earlier to determine the number of extracellular bacteria. Cells were again washed 3× with dPBS and 0.5 mL of pre‐warmed cell culture media was added with 200 μg/mL gentamicin (Sigma‐Aldrich, USA) and incubated during 3 h at 37°C to kill remaining extracellular and adherent S. uberis. Next, the gentamicin‐containing cell culture media were aspirated from the BoMECs and plated on BHI agar to evaluate if the extracellular S. uberis were indeed successfully killed. Thereafter, BoMECs were again 3× washed with dPBS and a final concentration of 2.5 μM of pre‐warmed NC5 in 0.5 mL cell culture media without penicillin–streptomycin mix was added to each well during 1 h to eradicate the intracellular bacteria. Buffer exchange was performed as described earlier. Subsequently, cells were 3× washed with dPBS and detached from the wells by trypsinization as described earlier. Their viability was evaluated by trypan blue staining (Sigma‐Aldrich, USA). Cells were pelleted and resuspended in dPBS containing 0.1% Triton X‐100 to lyse the BoMECs and release intracellular S. uberis. Subsequently, this lysate was 1:10 serially diluted in dPBS, plated on BHI agar and incubated during 18 h at 37°C to determine the amount of intracellular bacteria the next day. Additional controls were included to evaluate robustness of the assay and consisted of MAC‐T which were not challenged with S. uberis, as well as S. uberis in the absence of MAC‐T.
Streptococcus uberis biofilm growth, characterization and eradication
Biofilms were grown by transferring mid‐log phase (OD600 = 0.6) S. uberis (i.e. subclinical S. uberis 1; Table 3) grown in BHI containing 0.25% α‐d‐glucose (Thermo Fisher Scientific, USA) to a Nunclon delta‐treated, U‐shaped bottom 96‐well microplate (Thermo Fisher Scientific, USA). This plate was incubated for 21–24 h in a box containing a wet tissue to prevent drying out. Subsequently, media and planktonic cells were removed and 1.5 μM of endolysin was allowed to eradicate the biofilms during 2 h and 30 min at 37°C on a shaker at 120 rotations per minute (rpm). Biofilm characterization was performed by proteinase K (100 μg/mL in 100 mM NaCl and 20 mM tris; pH 7.5) (Thermo Fisher Scientific, USA), DNase I (100 μg/mL in 150 mM NaCl, 1 mM CaCl2 and 20 mM Tris; pH 7.5) (Sigma‐Aldrich, USA) or NaIO4 (10 mM in 50 mM sodium acetate buffer; pH 4.5) (Sigma‐Aldrich, USA) treatment for 1 h on a shaker at 120 rpm and 37°C. Biomass was fixated with 100% ethanol (Thermo Fisher Scientific, USA) and stained with 0.1% crystal violet (Sigma‐Aldrich, USA), 5% methanol and isopropanol (Thermo Fisher Scientific, USA) in PBS. Triple washing with PBS by inverting the plate was executed before solubilizing the stained biomass in 30% acetic acid (Chem‐Lab, Belgium). OD590 nm was measured using a CLARIOstar Plus plate reader (BMG Labtech, the Netherlands). Logarithmic reductions were determined by scraping biomass from the wells with a sterile tooth picker followed by resuspension in PBS, which was subsequently serially diluted and spotted on BHI agar. CFU/mL were counted after 18 h incubation at 37°C.
Antibacterial activity in raw cow's milk
Raw cow's milk samples from infected dairy cows that yielded a monoculture for S. uberis were supplied by the Milk Control Center Flanders. Upon arrival after transport at 4°C, the milk was immediately 1:10 serially diluted in PBS and plated on BHI agar as described earlier. After incubation during 18 h, this allowed (i) to critically evaluate the uniformity of colony morphology for S. uberis, (ii) to confirm the isolates to be S. uberis by picking one of these uniform colonies for MALDI‐TOF MS analysis and (iii) to determine the S. uberis load in the milk samples. Only if samples had >105 log10 CFU/mL S. uberis and no other microorganisms were observed, they were included in this assay. If needed, the raw milk was incubated during 8 h at 37°C to artificially increase S. uberis CFUs. Next, triplicates of 160 μL mastitic milk were supplemented in a 96‐well microtiter plate with 40 μL of either dPBS, 2.5 μM NC5 in dPBS, 250 μg/mL cloxacillin sodium in dPBS (Sigma‐Aldrich, USA) or a combination of the two latter (i.e. the combination therapy). This resulted in final concentrations of 0.5 μM and 50 μg/mL of NC5 and cloxacillin sodium in 4:5 diluted mastitic milk respectively. This mixture was incubated at 37°C and at every 2 h time interval, 20 μL was transferred from each well to 180 μL dPBS, serially diluted in a 1:10 ratio and plated on BHI agar as described earlier. Plates were incubated during 18 h at 37°C and CFU/mL were calculated the next day. Undiluted mastitic milk was always pipetted with ClipTip pipette tips (Westburg, the Netherlands) due to high viscosity.
Statistics
Data were analysed using GraphPad Prism (version 9.5.1) to calculate p values and determine statistically significant differences (p < 0.05). The data obtained from the high‐throughput screening and hit‐to‐lead selection by the halo‐based assay were analysed as a binomial distributed dataset. The occurrences of observed tiles at their predetermined position in each design were compared to an expected distribution, to assess if they differed significantly or not. A Bonferroni correction for multiple comparisons was included. Other data were analysed as normally distributed datasets, which was confirmed by an Anderson–Darling test or by quantile–quantile plotting the residuals. If needed, data underwent a log10 transformation, leading to a normal distribution of the data with equal variances. Two groups were compared with unpaired, two‐tailed t tests, and multiple groups with analysis of the variance (ANOVA) and a Bonferroni post hoc test.
AUTHOR CONTRIBUTIONS
Niels Vander Elst: Conceptualization (lead); data curation (lead); formal analysis (lead); funding acquisition (lead); investigation (lead); methodology (lead); project administration (lead); software (lead); validation (lead); visualization (lead); writing – original draft (lead); writing – review and editing (lead). Joni Bert: Data curation (supporting). Herman Favoreel: Conceptualization (supporting); data curation (supporting); investigation (supporting); methodology (supporting); resources (supporting); software (supporting); writing – review and editing (supporting). Rob Lavigne: Conceptualization (equal); formal analysis (equal); funding acquisition (lead); investigation (equal); methodology (equal); project administration (equal); resources (equal); software (equal); supervision (lead); validation (equal); visualization (equal); writing – original draft (equal); writing – review and editing (equal). Evelyne Meyer: Conceptualization (equal); formal analysis (equal); funding acquisition (lead); investigation (equal); methodology (equal); project administration (equal); resources (equal); software (equal); supervision (lead); validation (equal); visualization (equal); writing – original draft (equal); writing – review and editing (equal). Yves Briers: Conceptualization (lead); data curation (lead); formal analysis (lead); funding acquisition (lead); investigation (lead); methodology (lead); project administration (lead); resources (lead); software (lead); supervision (lead); validation (lead); visualization (lead); writing – original draft (lead); writing – review and editing (lead).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
PATENT
The engineered endolysins of this work are the subject of an issued and pending patent, submitted at the European Patent Office with application number EP 23164960.9.
Supporting information
Data S1.
ACKNOWLEDGMENTS
We acknowledge Milk Control Center Flanders for providing the (sub)clinical bovine mastitis‐derived strains and the group of Sarne De Vliegher (UGent) for verifying the identity of these strains by MALDI‐TOF. We thank Els Van Coillie and Pierre Germon for providing the MAC‐T and PS cell lines respectively. We also acknowledge the technical assistance of Kristel Demeyere, Alison Kerremans and Cliff Van Waesberghe, as well as the scientific advice of Diana Gutiérrez Fernández, Roberto Vazquez Fernandez and Vincent De Maesschalck. We thank Célia Pas for processing and assembling the sequenced bacterial genomes. This research was funded by the Research Foundation of Flanders (FWO Vlaanderen) grant number 1.S.236.20N for NV. The MALDI‐TOF MS was financed by the Research Foundation of Flanders (FWO Vlaanderen) as Hercules project (G0H2516N, AUGE/15/05).
Vander Elst, N. , Bert, J. , Favoreel, H. , Lavigne, R. , Meyer, E. & Briers, Y. (2023) Development of engineered endolysins with in vitro intracellular activity against streptococcal bovine mastitis‐causing pathogens. Microbial Biotechnology, 16, 2367–2386. Available from: 10.1111/1751-7915.14339
Rob Lavigne and Evelyne Meyer contributed equally to this work.
DATA AVAILABILITY STATEMENT
All data generated or analysed during this study are included in this published article, or can be requested upon reasonable request.
REFERENCES
- Arndt, D. , Grant, J.R. , Marcu, A. , Sajed, T. , Pon, A. , Liang, Y. et al. (2016) PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Research, 44(W1), W16–W21. Available from: 10.1093/nar/gkw387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardiau, M. , Caplin, J. , Detilleux, J. , Graber, H. , Moroni, P. , Taminiau, B. et al. (2016) Existence of two groups of Staphylococcus aureus strains isolated from bovine mastitis based on biofilm formation, intracellular survival, capsular profile and agr‐typing. Veterinary Microbiology, 185, 1–6. Available from: 10.1016/j.vetmic.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Bardiau, M. , Detilleux, J. , Farnir, F. , Mainil, J.G. & Ote, I. (2014) Associations between properties linked with persistence in a collection of Staphylococcus aureus isolates from bovine mastitis. Veterinary Microbiology, 169(1–2), 74–79. Available from: 10.1016/j.vetmic.2013.12.010 [DOI] [PubMed] [Google Scholar]
- Becker, S.C. , Roach, D.R. , Chauhan, V.S. , Shen, Y. , Foster‐Frey, J. , Powell, A.M. et al. (2016) Triple‐acting lytic enzyme treatment of drug‐resistant and intracellular Staphylococcus aureus . Scientific Reports, 6, 1–10. Available from: 10.1038/srep25063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, M. , Chang, H.Y. , Chuguransky, S. , Grego, T. , Kandasaamy, S. , Mitchell, A. et al. (2021) The InterPro protein families and domains database: 20 years on. Nucleic Acids Research, 49(D1), D344–D354. Available from: 10.1093/nar/gkaa977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettin, T. , Davis, J.J. , Disz, T. , Edwards, R.A. , Gerdes, S. , Olsen, G.J. et al. (2015) RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Scientific Reports, 5, 8365. Available from: 10.1038/srep08365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briers, Y. , Lavigne, R. , Volckaert, G. & Hertveldt, K. (2007) A standardized approach for accurate quantification of murein hydrolase activity in high‐throughput assays. Journal of Biochemical and Biophysical Methods, 70(3), 531–533. Available from: 10.1016/j.jbbm.2006.10.009 [DOI] [PubMed] [Google Scholar]
- Celia, L.K. , Nelson, D. & Kerr, D.E. (2008) Characterization of a bacteriophage lysin (Ply700) from Streptococcus uberis . Veterinary Microbiology, 130(1–2), 107–117. Available from: 10.1016/j.vetmic.2007.12.004 [DOI] [PubMed] [Google Scholar]
- Clark, K. , Karsch‐Mizrachi, I. , Lipman, D.J. , Ostell, J. & Sayers, E.W. (2016) GenBank. Nucleic Acids Research, 44(D1), D67–D72. Available from: 10.1093/nar/gkv1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado, R. , Montbrau, C. , Sitjà, M. & Prenafeta, A. (2018) Study of the efficacy of a Streptococcus uberis mastitis vaccine against an experimental intramammary infection with a heterologous strain in dairy cows. Journal of Dairy Science, 101(11), 10290–10302. Available from: 10.3168/jds.2018-14840 [DOI] [PubMed] [Google Scholar]
- Daniel, A. , Euler, C. , Collin, M. , Chahales, P. , Gorelick, K.J. & Fischetti, V.A. (2010) Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin‐resistant Staphylococcus aureus . Antimicrobial Agents and Chemotherapy, 54(4), 1603–1612. Available from: 10.1128/AAC.01625-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, P.L. , Leigh, J.A. , Bradley, A.J. , Archer, S.C. , Emes, R.D. & Green, M.J. (2016) Molecular epidemiology of Streptococcus uberis clinical mastitis in dairy herds: strain heterogeneity and transmission. Journal of Clinical Microbiology, 54(1), 68–74. Available from: 10.1128/JCM.01583-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, A. , Garch, F.E. , Simjee, S. , Moyaert, H. , Rose, M. , Youala, M. et al. (2018) Monitoring of antimicrobial susceptibility of udder pathogens recovered from cases of clinical mastitis in dairy cows across Europe: VetPath results. Veterinary Microbiology, 213, 73–81. Available from: 10.1016/j.vetmic.2017.11.021 [DOI] [PubMed] [Google Scholar]
- De Vliegher, S. , Ohnstad, I. & Piepers, S. (2018) Management and prevention of mastitis: a multifactorial approach with a focus on milking, bedding and data‐management. Journal of Integrative Agriculture, 17(6), 1214–1233. Available from: 10.1016/S2095-3119(17)61893-8 [DOI] [Google Scholar]
- Donovan, D.M. , Lardeo, M. & Foster‐Frey, J. (2006) Lysis of staphylococcal mastitis pathogens by bacteriophage phi11 endolysin. FEMS Microbiology Letters, 265(1), 133–139. Available from: 10.1111/j.1574-6968.2006.00483.x [DOI] [PubMed] [Google Scholar]
- Duyvejonck, L. , Gerstmans, H. , Stock, M. , Grimon, D. , Lavigne, R. & Briers, Y. (2021) Rapid and high‐throughput evaluation of diverse configurations of engineered lysins using the versatile technique. Antibiotics, 10(3), 293. Available from: 10.3390/antibiotics10030293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J. , Zeng, Z. , Mai, K. , Yang, Y. , Feng, J. , Bai, Y. et al. (2016) Preliminary treatment of bovine mastitis caused by Staphylococcus aureus, with trx‐SA1, recombinant endolysin of S. Aureus bacteriophage IME‐SA1. Veterinary Microbiology, 191, 65–71. Available from: 10.1016/j.vetmic.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Fischetti, V.A. (2010) Bacteriophage endolysins: a novel anti‐infective to control gram‐positive pathogens. International Journal of Medical Microbiology, 300(6), 357–362. Available from: 10.1016/j.ijmm.2010.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, S.N. , Osburn, B.I. & Cullor, J.S. (2019) A one health perspective on dairy production and dairy food safety. One Health, 7, 100086. Available from: 10.1016/j.onehlt.2019.100086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary, U. , Lopez‐Villalobos, N. , Begley, N. , McCoy, F. , O'Brien, B. , O'Grady, L. et al. (2012) Estimating the effect of mastitis on the profitability of Irish dairy farms. Journal of Dairy Science, 95(7), 3662–3673. Available from: 10.3168/jds.2011-4863 [DOI] [PubMed] [Google Scholar]
- Gerstmans, H. , Criel, B. & Briers, Y. (2018) Synthetic biology of modular endolysins. Biotechnology Advances, 36(3), 624–640. Available from: 10.1016/j.biotechadv.2017.12.009 [DOI] [PubMed] [Google Scholar]
- Gerstmans, H. , Grimon, D. , Gutiérrez, D. , Lood, C. , Rodríguez, A. , van Noort, V. et al. (2020) A VersaTile‐driven platform for rapid hit‐to‐lead development of engineered lysins. Science Advances, 6(23), 1–12. Available from: 10.1126/sciadv.aaz1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmer, D.B. , Schmitz, J.E. , Euler, C.W. & Fischetti, V.A. (2013) Novel bacteriophage lysin with broad lytic activity protects against mixed infection by Streptococcus pyogenes and methicillin‐resistant Staphylococcus aureus . Antimicrobial Agents and Chemotherapy, 57(6), 2743–2750. Available from: 10.1128/AAC.02526-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, F. & Henriques, M. (2016) Control of bovine mastitis: old and recent therapeutic approaches. Current Microbiology, 72, 377–382. Available from: 10.1007/s00284-015-0958-8 [DOI] [PubMed] [Google Scholar]
- Grandgirard, D. , Loeffler, J.M. , Fischetti, V.A. & Leib, S.L. (2008) Phage lytic enzyme Cpl‐1 for antibacterial therapy in experimental pneumococcal meningitis. The Journal of Infectious Diseases, 197(11), 1519–1522. Available from: 10.1086/587942 [DOI] [PubMed] [Google Scholar]
- Gutiérrez, D. , Garrido, V. , Fernández, L. , Portilla, S. , Rodríguez, A. , Grilló, M.J. et al. (2020) Phage lytic protein LysRODI prevents staphylococcal mastitis in mice. Frontiers in Microbiology, 11, 7. Available from: 10.3389/fmicb.2020.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez, D. , Rodríguez‐Rubio, L. , Ruas‐Madiedo, P. , Fernández, L. , Campelo, A.B. , Briers, Y. et al. (2021) Design and selection of engineered lytic proteins with Staphylococcus aureus decolonizing activity. Frontiers in Microbiology, 12, 1–16. Available from: 10.3389/fmicb.2021.723834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez, D. , Ruas‐Madiedo, P. , Martínez, B. , Rodríguez, A. & García, P. (2014) Effective removal of staphylococcal biofilms by the endolysin LysH5. PLoS ONE, 9(9), e107307. Available from: 10.1371/journal.pone.0107307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez, D. , Vandenheuvel, D. , Martínez, B. , Rodríguez, A. , Lavigne, R. & García, P. (2015) Two phages, phiIPLA‐RODI and phiIPLA‐C1C, lyse mono‐and dual‐species staphylococcal biofilms. Applied and Environmental Microbiology, 81(10), 3336–3348. Available from: 10.1128/AEM.03560-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad Kashani, H. , Schmelcher, M. , Sabzalipoor, H. , Seyed Hosseini, E. & Moniri, R. (2018) Recombinant endolysins as potential therapeutics against antibiotic‐resistant Staphylococcus aureus: current status of research and novel delivery strategies. Clinical Microbiology Reviews, 31(1), e00071‐17. Available from: 10.1128/CMR.00071-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogeveen, H. , Huijps, K. & Lam, T.J.G.M. (2011) Economic aspects of mastitis: new developments. New Zealand Veterinary Journal, 59(1), 16–23. Available from: 10.1080/00480169.2011.547165 [DOI] [PubMed] [Google Scholar]
- Huang, Y. , Yang, H. , Yu, J. & Wei, H. (2015) Molecular dissection of phage lysin PlySs2: integrity of the catalytic and cell wall binding domains is essential for its broad lytic activity. Virologica Sinica, 30(1), 45–51. Available from: 10.1007/s12250-014-3535-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh, H.T. , Robitaille, G. & Turner, J.D. (1991) Establishment of bovine mammary epithelial cells (MAC‐T): an in vitro model for bovine lactation. Experimental Cell Research, 197(2), 191–199. Available from: 10.1016/0014-4827(91)90422-q [DOI] [PubMed] [Google Scholar]
- Indiani, C. , Sauve, K. , Raz, A. , Abdelhady, W. , Xiong, Y.Q. , Cassino, C. et al. (2019) The Antistaphylococcal lysin, CF‐301, activates key host factors in human blood to potentiate methicillin‐resistant Staphylococcus aureus bacteriolysis. Antimicrobial Agents and Chemotherapy, 63(4), e02291‐18. Available from: 10.1128/AAC.02291-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käppeli, N. , Morach, M. , Zurfluh, K. , Corti, S. , Nüesch‐Inderbinen, M. & Stephan, R. (2019) Sequence types and antimicrobial resistance profiles of Streptococcus uberis isolated from bovine mastitis. Frontiers in Veterinary Science, 6, 234. Available from: 10.3389/fvets.2019.00234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, A.P. , Ly, S. , Daetwyler, S. , Eichenseher, F. , Loessner, M.J. & Schmelcher, M. (2022) Chimeric peptidoglycan hydrolases kill staphylococcal mastitis isolates in raw milk and within bovine mammary gland epithelial cells. Viruses, 14(12), 2801. Available from: 10.3390/v14122801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, L.A. , Mezulis, S. , Yates, C.M. , Wass, M.N. & Sternberg, M.J.E. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols, 10(6), 845–858. Available from: 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krömker, V. & Leimbach, S. (2017) Mastitis treatment—reduction in antibiotic usage in dairy cows. Reproduction in Domestic Animals, 52, 21–29. Available from: 10.1111/rda.13032 [DOI] [PubMed] [Google Scholar]
- Linden, S.B. , Alreja, A.B. & Nelson, D.C. (2021) Application of bacteriophage‐derived endolysins to combat streptococcal disease: current state and perspectives. Current Opinion in Biotechnology, 68, 213–220. Available from: 10.1016/j.copbio.2021.01.012 [DOI] [PubMed] [Google Scholar]
- Linden, S.B. , Zhang, H. , Heselpoth, R.D. , Shen, Y. , Schmelcher, M. , Eichenseher, F. et al. (2015) Biochemical and biophysical characterization of PlyGRCS, a bacteriophage endolysin active against methicillin‐resistant Staphylococcus aureus . Applied Microbiology and Biotechnology, 99(2), 741–752. Available from: 10.1007/s00253-014-5930-1 [DOI] [PubMed] [Google Scholar]
- Madeira, F. , Park, Y. , Lee, J. , Buso, N. , Gur, T. , Madhusoodanan, N. et al. (2019) The EMBL‐EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Research, 47(W1), W636–W641. Available from: 10.1093/nar/gkz268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer‐Hoffert, U. & Wiedow, O. (2011) Neutrophil serine proteases: mediators of innate immune responses. Current Opinion in Hematology, 18(1), 19–24. Available from: 10.1097/MOH.0b013e32834115d1 [DOI] [PubMed] [Google Scholar]
- Mirdita, M. , Schütze, K. , Moriwaki, Y. , Heo, L. , Ovchinnikov, S. & Steinegger, M. (2022) ColabFold: making protein folding accessible to all. Nature Methods, 19(6), 679–682. Available from: 10.1038/s41592-022-01488-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.C. , Schmelcher, M. , Rodriguez‐Rubio, L. , Klumpp, J. , Pritchard, D.G. , Dong, S. et al. (2012) Endolysins as antimicrobials. Advances in Virus Research, 83, 299–365. Available from: 10.1016/B978-0-12-394438-2.00007-4 [DOI] [PubMed] [Google Scholar]
- Niemi, R.E. , Hovinen, M. , Vilar, M.J. , Simojoki, H. & Rajala‐Schultz, P.J. (2021) Dry cow therapy and early lactation udder health problems—associations and risk factors. Preventive Veterinary Medicine, 188, 105268. Available from: 10.1016/j.prevetmed.2021.105268 [DOI] [PubMed] [Google Scholar]
- Obeso, J.M. , Martínez, B. , Rodríguez, A. & García, P. (2008) Lytic activity of the recombinant staphylococcal bacteriophage PhiH5 endolysin active against Staphylococcus aureus in milk. International Journal of Food Microbiology, 128(2), 212–218. Available from: 10.1016/j.ijfoodmicro.2008.08.010 [DOI] [PubMed] [Google Scholar]
- Oliver, S.P. , Murinda, S.E. & Jayarao, B.M. (2011) Impact of antibiotic use in adult dairy cows on antimicrobial resistance of veterinary and human pathogens: a comprehensive review. Foodborne Pathogens and Disease, 8(3), 337–355. Available from: 10.1089/fpd.2010.0730 [DOI] [PubMed] [Google Scholar]
- Pereira, U.P. , Oliveira, D.G.S. , Mesquita, L.R. , Costa, G.M. & Pereira, L.J. (2011) Efficacy of Staphylococcus aureus vaccines for bovine mastitis: a systematic review. Veterinary Microbiology, 148(2–4), 117–124. Available from: 10.1016/j.vetmic.2010.10.003 [DOI] [PubMed] [Google Scholar]
- Persson, Y. , Nyman, A.K.J. & Grönlund‐Andersson, U. (2011) Etiology and antimicrobial susceptibility of udder pathogens from cases of subclinical mastitis in dairy cows in Sweden. Acta Veterinaria Scandinavica, 53(1), 1–8. Available from: 10.1186/1751-0147-53-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainard, P. & Foucras, G. (2018) A critical appraisal of probiotics for mastitis control. Frontiers in Veterinary Science, 5, 1–13. Available from: 10.3389/fvets.2018.00251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Rubio, L. , Chang, W.L. , Gutiérrez, D. , Lavigne, R. , Martínez, B. , Rodríguez, A. et al. (2016) “Artilysation” of endolysin λSa2lys strongly improves its enzymatic and antibacterial activity against streptococci. Scientific Reports, 6, 1–11. Available from: 10.1038/srep35382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhrig, C. , Huemer, M. , Lorgé, D. , Luterbacher, S. , Phothaworn, P. , Schefer, C. et al. (2020) Targeting hidden pathogens: cell‐penetrating enzybiotics eradicate intracellular drug‐resistant Staphylococcus aureus . MBio, 11(2), e00209‐20. Available from: 10.1128/mBio.00209-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollin, E. , Dhuyvetter, K.C. & Overton, M.W. (2015) The cost of clinical mastitis in the first 30 days of lactation: an economic modeling tool. Preventive Veterinary Medicine, 122(3), 257–264. Available from: 10.1016/j.prevetmed.2015.11.006 [DOI] [PubMed] [Google Scholar]
- Rossi, B.F. , Bonsaglia, E.C.R. , Pantoja, J.C.F. , Santos, M.V. , Gonçalves, J.L. , Fernandes Júnior, A. et al. (2021) Short communication: association between the accessory gene regulator (agr) group and the severity of bovine mastitis caused by Staphylococcus aureus . Journal of Dairy Science, 104(3), 3564–3568. Available from: 10.3168/jds.2020-19275 [DOI] [PubMed] [Google Scholar]
- Roussel, P. , Cunha, P. , Porcherie, A. , Petzl, W. , Gilbert, F.B. , Riollet, C. et al. (2015) Investigating the contribution of IL‐17A and IL‐17F to the host response during Escherichia coli mastitis. Veterinary Research, 46(1), 56. Available from: 10.1186/s13567-015-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini, V. , McClure, J.T. , Léger, D. , Keefe, G.P. , Scholl, D.T. , Morck, D.W. et al. (2012) Antimicrobial resistance profiles of common mastitis pathogens on Canadian dairy farms. Journal of Dairy Science, 95(8), 4319–4332. Available from: 10.3168/jds.2012-5373 [DOI] [PubMed] [Google Scholar]
- Schmelcher, M. , Powell, A.M. , Becker, S.C. , Camp, M.J. & Donovan, D.M. (2012) Chimeric phage lysins act synergistically with lysostaphin to kill mastitis‐causing Staphylococcus aureus in murine mammary glands. Applied and Environmental Microbiology, 78(7), 2297–2305. Available from: 10.1128/AEM.07050-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelcher, M. , Powell, A.M. , Camp, M.J. , Pohl, C.S. & Donovan, D.M. (2015) Synergistic streptococcal phage λSA2 and B30 endolysins kill streptococci in cow milk and in a mouse model of mastitis. Applied Microbiology and Biotechnology, 99(20), 8475–8486. Available from: 10.1007/s00253-015-6579-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholte, C.M. , Nelson, D.C. , Garcia, M. , Linden, S.B. , Elsasser, T.H. , Kahl, S. et al. (2018) Short communication: recombinant bacteriophage endolysin PlyC is nontoxic and does not alter blood neutrophil oxidative response in lactating dairy cows. Journal of Dairy Science, 101(7), 6419–6423. Available from: 10.3168/jds.2017-13908 [DOI] [PubMed] [Google Scholar]
- Schönborn, S. , Wente, N. , Paduch, J.H. & Krömker, V. (2017) In vitro ability of mastitis causing pathogens to form biofilms. Journal of Dairy Research, 84(2), 198–201. Available from: 10.1017/S0022029917000218 [DOI] [PubMed] [Google Scholar]
- Seijsing, J. , Sobieraj, A.M. , Keller, N. , Shen, Y. , Zinkernagel, A.S. , Loessner, M.J. et al. (2018) Improved biodistribution and extended serum half‐life of a bacteriophage endolysin by albumin binding domain fusion. Frontiers in Microbiology, 9, 1–9. Available from: 10.3389/fmicb.2018.02927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y. , Barros, M. , Vennemann, T. , Gallagher, D.T. , Yin, Y. , Linden, S.B. et al. (2016) A bacteriophage endolysin that eliminates intracellular streptococci. eLife, 5, 1–26. Available from: 10.7554/eLife.13152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son, B. , Kong, M. , Lee, Y. & Ryu, S. (2021) Development of a novel chimeric endolysin, Lys109 with enhanced lytic activity against Staphylococcus aureus . Frontiers in Microbiology, 11, 1–12. Available from: 10.3389/fmicb.2020.615887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamilselvam, B. , Almeida, R.A. , Dunlap, J.R. & Oliver, S.P. (2006) Streptococcus uberis internalizes and persists in bovine mammary epithelial cells. Microbial Pathogenesis, 40(6), 279–285. Available from: 10.1016/j.micpath.2006.02.006 [DOI] [PubMed] [Google Scholar]
- Tomita, T. , Meehan, B. , Wongkattiya, N. , Malmo, J. , Pullinger, G. , Leigh, J. et al. (2008) Identification of Streptococcus uberis multilocus sequence types highly associated with mastitis. Applied and Environmental Microbiology, 74(1), 114–124. Available from: 10.1128/AEM.01373-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Elst, N. , Linden, S.B. , Lavigne, R. , Meyer, E. , Briers, Y. & Nelson, D.C. (2020) Characterization of the bacteriophage‐derived endolysins plyss2 and plyss9 with in vitro lytic activity against bovine mastitis Streptococcus uberis . Antibiotics, 9(9), 1–14. Available from: 10.3390/antibiotics9090621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Elst, N. & Meyer, E. (2018) Potential therapeutic application of bacteriophages and phage‐derived endolysins as alternative treatment of bovine mastitis. Vlaams Diergeneeskundig Tijdschrift, 87, 181–187. [Google Scholar]
- Varhimo, E. , Varmanen, P. , Fallarero, A. , Skogman, M. , Pyörälä, S. , Iivanainen, A. et al. (2011) Alpha‐ and β‐casein components of host milk induce biofilm formation in the mastitis bacterium Streptococcus uberis . Veterinary Microbiology, 149(3–4), 381–389. Available from: 10.1016/j.vetmic.2010.11.010 [DOI] [PubMed] [Google Scholar]
- Verbeke, J. , Piepers, S. , Supré, K. & de Vliegher, S. (2014) Pathogen‐specific incidence rate of clinical mastitis in Flemish dairy herds, severity, and association with herd hygiene. Journal of Dairy Science, 97(11), 6926–6934. Available from: 10.3168/jds.2014-8173 [DOI] [PubMed] [Google Scholar]
- Verbree, C.T. , Dätwyler, S.M. , Meile, S. , Eichenseher, F. , Donovan, D.M. , Loessner, M.J. et al. (2018) Corrected and republished from: identification of peptidoglycan hydrolase constructs with synergistic staphylolytic activity in cow's milk. Applied and Environmental Microbiology, 84(1), e02134‐17. Available from: 10.1128/AEM.02134-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . (2017) WHO list of critically important antimicrobials for human medicine (WHO CIA list) 6th rev. Geneva, Switzerland: World Health Organization. 6th revision. Available at: http://who.int/foodsafety/publications/antimicrobials‐fifth/en/ [Google Scholar]
- World Health Organization . (2021a) 2020 Antibacterial agents in clinical and preclinical development: an overview and analysis .
- World Health Organization . (2021b) Global antimicrobial resistance and use surveillance system (GLASS) report 2021. Geneva, Switzerland: World Health Organization. Available at: http://www.who.int/glass/resources/publications/early‐implementation‐report‐2020/en/ [Google Scholar]
- Xu, J. , Yang, H. , Bi, Y. , Li, W. , Wei, H. & Li, Y. (2018) Activity of the chimeric lysin ClyR against common gram‐positive oral microbes and its anticaries efficacy in rat models. Viruses, 10(7), 380. Available from: 10.3390/v10070380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Bi, Y. , Shang, X. , Wang, M. , Linden, S.B. , Li, Y. et al. (2016) Antibiofilm activities of a novel chimeolysin against Streptococcus mutans under physiological and cariogenic conditions. Antimicrobial Agents and Chemotherapy, 60(12), 7436–7443. Available from: 10.1128/AAC.01872-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Linden, S.B. , Wang, J. , Yu, J. , Nelson, D.C. & Wei, H. (2015) A chimeolysin with extended‐spectrum streptococcal host range found by an induced lysis‐based rapid screening method. Scientific Reports, 5, 17257. Available from: 10.1038/srep17257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Xu, J. , Gong, Y. , Tang, Y. , Li, W. , Zheng, Z. et al. (2021) Internal cell‐penetrating peptide‐mediated internalization enables a chimeric lysin to target intracellular pathogens. International Journal of Pharmaceutics, 599, 120449. Available from: 10.1016/j.ijpharm.2021.120449 [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Yan, J. , Mujtaba, B.M. , Li, Y. , Wei, H. & Huang, S. (2021) The dual anti‐caries effect of carboxymethyl chitosan nanogel loaded with chimeric lysin ClyR and amorphous calcium phosphate. European Journal of Oral Sciences, 129(3), e12784. Available from: 10.1111/eos.12784 [DOI] [PubMed] [Google Scholar]
- Zouharova, M. , Nedbalcova, K. , Kralova, N. , Slama, P. , Matiaskova, K. & Matiasovic, J. (2022) Multilocus sequence genotype heterogeneity in Streptococcus uberis isolated from bovine mastitis in The Czech Republic. Animals, 12, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1.
Data Availability Statement
All data generated or analysed during this study are included in this published article, or can be requested upon reasonable request.


