Abstract
Parasitic weeds such as broomrapes (Phelipanche ramosa and Orobanche cumana) cause severe damage to crops and their development must be controlled. Given that phloroglucinol compounds (PGCs) produced by environmental Pseudomonas could be toxic towards certain plants, we assessed the potential herbicidal effect of the bacterial model Pseudomonas ogarae F113, a PGCs‐producing bacterium, on parasitic weed. By combining the use of a mutagenesis approach and of pure PGCs, we evaluated the in vitro effect of PGC‐produced by P. ogarae F113 on broomrape germination and assessed the protective activity of a PGC‐producing bacteria on oilseed rape (Brassica napus) against P. ramosa in non‐sterile soils. We showed that the inhibition of the germination depends on the PGCs molecular structure and their concentrations as well as the broomrape species and pathovars. This inhibition caused by the PGCs is irreversible, causing a brown coloration of the broomrape seeds. The inoculation of PGCs‐producing bacteria limited the broomrape infection of P. ramosa, without affecting the host growth. Moreover, elemental profiling analysis of oilseed rape revealed that neither F113 nor applied PGCs affected the nutrition capacity of the oilseed rape host. Our study expands the knowledge on plant‐beneficial Pseudomonas as weed biocontrol agents and opens new avenues for the development of natural bioherbicides to enhance crop yield.
The present work expands our knowledge about the role that DAPG‐producing Pseudomonas play as natural bioherbicides to ward off parasitic plant infections, which are responsible for important yield losses in crops.
INTRODUCTION
Broomrapes are parasitic plants causing significant damage to different crops in agroecosystems (Parker, 2012). They belong to the Orobanche and Phelipanche genera from the Oroabanchaceae family (Bennett & Mathews, 2006; Joel, 2009). These parasitic plants are obligate root holoparasites, entirely dependent on their host plant to survive as they are not capable of photosynthesis (Westwood, 2013). Indeed, these parasitic plants obtain all the resources they need while maintaining their host alive for accomplishing their entire life cycle (Cartry et al., 2021; Mutuku et al., 2021). The seed germination and haustorium formation are specifically induced by allelochemical signals such as strigolactones released from the host roots (Aliche et al., 2020). The activity spectrum of these parasitic plants can be either specific (e.g. Orobanche cumana, which can only parasitize sunflower) or generalist (e.g. Phelipanche ramosa able to parasitize tomato (Solanum lycopersicum), potato (Solanum tuberosum), hemp (Cannabis sativa), tobacco (Nicotiana tabacum) and oilseed rape (Brassica napus)) (Cartry et al., 2021; Parker, 2012). Broomrapes produce a large number of small seeds (less than 3 mm) that can survive in soil for several decades. This constitute the main problem to constrain their deleterious impact on crops (Haring & Flessner, 2018). The survival of the seeds depends on various abiotic factors (pH, humidity, climate) (Rubiales et al., 2003) and biotic factors (host plants, soil and rhizosphere microbiota; (Huet et al., 2020; Kawa et al., 2022; Martinez et al., 2023; Mutuku et al., 2021). Different agricultural strategies attempt to regulate broomrape populations in agroecosystems, such as crop rotation, triggering the suicidal germination of the plant parasitic seeds or the use of resistant host plant varieties or chemical herbicides (Cartry et al., 2021). However, biological control solutions are emerging to limit broomrape infestation, including the use of microorganisms (Cartry et al., 2021; Masteling et al., 2019). Indeed, several microorganisms inhibit the germination of different broomrape species, including, among others, Fusarium oxysporum (Hasannejad et al., 2006), Azospirillum brasilense (Dadon et al., 2004) or Pseudomonas fluorescens (Balthazar et al., 2021). Although the use of microorganisms represents a promising alternative to ward off parasitic plants, their mode of action as well as the identification of the metabolites responsible for their inhibition effect remains often uncharted.
Several environmental Pseudomonas are well known as plant‐colonizing bacteria (Silby et al., 2011). These Pseudomonas usually display a large arsenal of secondary metabolites encoded within their genomes that act on plant health and development (Haas & Défago, 2005; Loper et al., 2012). Among these metabolites, 2,4‐diacetylphloroglucinol (DAPG) has been studied notably for its role in plant protection. DAPG and its biosynthetic intermediates, phloroglucinol (PG) and monoacetylphloroglucinol (MAPG) are the main phloroglucinol compounds (PGCs) produced by Pseudomonas belonging to the P. protegens and P. corrugata subgroups (Almario et al., 2017). The production of DAPG relies on the presence of the phl gene cluster composed out of nine genes (Achkar et al., 2005; Biessy & Filion, 2021). The initiation of the synthesis of PG from malonyl‐CoA is mediated by phlD encoding a polyketide synthase, while phlABC encode for enzymes implicated into the transformation of PG to MAPG and subsequently to DAPG. The transformation of MAPG to DAPG is reversible through a hydrolase encoded by phlG. The remaining genes, phlF/phlH and phlE, are involved in the regulation as well as the secretion of these PGCs, respectively. The production of PGCs by Pseudomonas is influenced by environmental factors including (i) carbon sources (Shanahan et al., 1992), (ii) specific metabolites found in the root exudates such as flavonoids, apigenin and phloretin (Yu et al., 2020) or (iii) iron availability and interaction with other microorganisms (Laveilhé et al., 2022).
In addition to having been studied for its role in plant pathogen suppression, DAPG acts as a signal molecule affecting gene expression of plant‐beneficial traits in other microorganisms. Indeed, DAPG was described as an inducer of the production of PGCs and a repressor of the production of pyoluteorin in other Pseudomonas (Brodhagen et al., 2004; Maurhofer et al., 2004). Moreover, DAPG produced by Pseudomonas also activates the expression of genes involved in the production of auxins by Azospirillum baldaniorum Sp245, another plant‐beneficial microorganism (Combes‐Meynet et al., 2011). Since PGC‐producing Pseudomonas are residing in the vicinity of or on plant roots, PGCs produced diffuse and also interact directly with plant root cells. Thus, it was shown that DAPG elicited the plant‐induced systemic resistance (ISR), protecting partially the plant leaves from the oomycete Peronospora parasitica (Bakker et al., 2007; Iavicoli et al., 2003). On the root part, the addition of DAPG triggered a massive increase in the efflux of amino acids by plant root cells (Phillips et al., 2004). It was also demonstrated that DAPG modulates auxin‐dependent plant signalling pathway leading to significant modifications of plant root development (Brazelton et al., 2008; Vacheron et al., 2018; Weller et al., 2012). Moreover, following an exposition to DAPG, the germination as well as the development of different crop plants could be impacted (Brazelton et al., 2008; Keel, 1992; Khan et al., 2022). Nevertheless, this herbicidal effect remains variable according to the plant species and was observed following the exposure to high concentrations that do not reflect those produced in vivo.
In this study, we aimed to investigate the impact of PGCs on the germination of the two main parasitic plants, Phelipanche and Orobanche. To evaluate the herbicidal effects of these PGCs, we have investigated the inhibitory effect of PGCs‐producing strains and pure molecules in different in vitro and in planta experimental systems. First, the impact of the PGCs‐producing strain Pseudomonas ogarae F113 and its mutants, a PGC‐deficient and PGC overproducers, have been studied on broomrape germination by applying culture supernatants. Then, the role of PGCs on the germination of four different broomrapes was assessed at different concentrations under in vitro experiment. Finally, we evaluated the ability of PGCs‐producing bacteria and DAPG application to protect oilseed rape against broomrape in greenhouse, in non‐sterile soil.
EXPERIMENTAL PROCEDURES
Bacterial strains and media
We used the plant‐beneficial model strain Pseudomonas ogarae F113 (Garrido‐Sanz et al., 2021; Shanahan et al., 1992) and several of its mutants (Vacheron et al., 2018). The bacterial strains used in this study as well as their characteristics are listed in Table S1. The different bacterial strains were incubated at 28°C in King's B (King et al., 1954) medium or in a modified AB medium (ABm) supplemented with gentamycin (15 μg/mL when necessary) to maintain plasmid pBBR1‐MCS5‐phlD. ABm was composed of salts [MgSO4 (1.2 mM), CaCl2 (70 μM), NH4Cl (18 mM), KCl (2 mM), FeSO4 (9 μM)], a phosphate buffer diluted 10‐fold containing K2HPO4 (1.725 mM) and NaH2PO4 (960 μM) and fructose (20 mM) as carbon source.
Plant material
Seeds of Phelipanche ramosa were collected in France as described in Huet et al., 2020 on winter oilseed rape (Phelipanche ramosa pv. oilseed rape), tobacco (Phelipanche ramosa pv. tobacco) and hemp (Phelipanche ramosa pv. hemp). Seeds of Orobanche cumana that parasites sunflower were provided by Terres Inovia in 2016 (Huet et al., 2020). Seeds of Brassica napus cultivar AMAZZONITE (broomrape‐sensitive) were provided by the breeder companies RAGT 2n (France).
Chemicals
The germination of broomrape seeds was triggered using the synthetic strigolactone analogue GR24 (Chiralix, Nijmegen, NL). It was first suspended in acetone (4.79 mg/mL), then diluted at 10 μM with a phosphate buffer (1 mM sodium‐potassium phosphate buffer at pH 7.5).
Phloroglucinol (PG, Sigma‐Aldrich), mono‐acetyl‐phloroglucinol (MAPG, Cayman Chemical), 2,4‐diacetylphloroglucinol (DAPG, ChemCruz) and tri‐acetyl‐phloroglucinol (TAPG, Santa Cruz Biotechnology) were suspended in methanol (20 mM). These solutions were diluted with 1 mM phosphate buffer to obtain different stock solutions at different concentrations (66.60, 33.30, 16.65, 8.33, 4.16 μM). As methanol might have an effect on the germination of broomrapes, the final concentration was adjusted to 0.33% in all these stock solutions to prevent an effect of the dilution.
Quantification of phloroglucinol compounds produced in bacterial supernatants
The quantification of PGCs was conducted on 1.5 mL of bacterial culture supernatants for each condition. First, supernatants were lyophilized (Martin Christ Alpha 1–4 LSC, Osterode, Germany) prior solid/liquid extraction with methanol. Samples were sonicated 20 min, then centrifuged for 20 min at 15,000 g and the supernatants recovered. The extraction protocol was repeated, leading to a total extracted volume of 3 mL per sample. The organic phase (methanol) was dried using a SpeedVac (Centrivap Cold Trap Concentrator; LABCONCO Co., MO, USA). Dried extracts were suspended in 200 μL of methanol and centrifuged for 5 min at 12,000 g to pellet the remaining solid phase.
Two hundred microlitres of the supernatant were then transferred into vials and were proceeded for ultra‐high pressure liquid chromatography coupled with UV (UHPLC‐UV) analysis, as previous described. (Rieusset et al., 2020). Chromatograms were analysed with MassHunter Qualitative Analysis B.07.00 software (Agilent Technologies) and the quantification of DAPG and other PGCs was done according to a standard curve with commercial PGCs.
Inhibition of broomrape germination in vitro
Broomrape seeds were surface‐disinfected with minor modifications to the method as previously described (Pouvreau et al., 2013). Briefly, broomrape seeds were soaked 5 min in a bleach solution (9.6% active chlorine) and then washed 5 times with sterile water. After washing, 1 mM phosphate buffer supplemented with plant agar 0.1% and PPM 0.2% (Plant Preservative Mixture; Plant Cell Technology) was added to obtain a density of approximatively 2000 seeds/mL. These solutions containing the seeds were conditioned in sealed tubes for 10 days at 21°C in the dark in a cooled incubator (LMS, model 120, Kent, UK). The supernatant of conditioned seeds was removed and replaced by fresh phosphate buffer supplemented with plant agar 0.1% and PPM 0.2%. Fifteen microlitres of this seed suspension were distributed in a 96‐well plate (Cellstar; Greiner Bio‐One, France), corresponding to approximatively 30 seeds per well. Then, 10 μL of GR24 solution were then added in each well (final concentration of 1 μM). Seventy‐five microlitres of either bacterial supernatants (3‐fold diluted), or cocktails or individual PGCs were added to obtain a final volume of 100 μL per well. For the addition of PGCs, the 75 μL were taken from the different stock solutions described above to obtain final concentrations of 50, 25, 12.5, 6.25 and 3.125 μM. Negative controls were realized using 75 μL of fresh ABm medium fructose 20 mM for supernatant (3‐fold diluted) or phosphate buffer with 0.33% of methanol. After 10 days of incubation at 21°C in the dark, the percentage of broomrape germinated seeds was counted under a binocular (Leica, Switzerland) using the software Zen 2.3.
Greenhouse experiments
Brassica napus plants were grown on a soil mix containing 1/3 of a natural loamy soil collected at the experimental farm in La Côte‐St‐André (France; 16.2% clay, 43.9% silt and 39.9% sand, pH 7.0, in water; 2.1% organic matter (El Zemrany et al., 2006)), 1/3 of vermiculite and 1/3 of TS3 peat‐based substrate (Klasmann‐Deilmann GmbH, Geeste, Germany). The humidity of the soil mix was maintained at 70% of field capacity. Each pot was filled with 1 L of free‐broomrape soil mix and then further filled with another litre of soil mix contaminated with non‐disinfected seeds of Phelipanche ramosa pv. oilseed rape leading to a final density of 3.9 mg of seeds per pot corresponding to approximatively 300 seeds per litre of soil. Seeds of Brassica napus cultivar AMAZZONITE were sown in pot after being pre‐germinated 24 h in the dark at 21°C in Petri dish containing water‐soaked Whatman paper.
The protective effect of the inoculation of F113 was compared to its mutant impaired in the production of PGCs (ΔphlD). The different bacterial strains were cultured 24 h in King's B medium at 28°C. The bacteria were centrifuged at 4500 rpm during 10 min and washed with a MgSO4 10 mM solution before being adjusted to a bacterial concentration of 2 × 106 CFU/mL. Five millilitres of these bacterial suspensions were sprayed at the base of the plant stem. Five millilitres of a MgSO4 10 mM solution were applied as non‐inoculated control. In parallel, the impact of DAPG application was also assessed (more details in supporting information, Figures S5 and S6).
Twenty pots per conditions were used for the inoculation of bacteria. The experiment was conducted under controlled conditions with a 16 h light and 8 h dark photoperiod, at 25°C with 50%–70% relative humidity in a greenhouse for 50 days. Treatments were applied twice during the experiment. The first bacterial application was performed when B. napus had between 2 to 4 leaves. The second application was done at 6 to 8 leaves. After 50 days, the root systems were sampled and the adhering soil was very thoroughly removed by hands. Broomrapes attached to each root system were all collected, classified according to their development stage (using the infectivity scale described in Figure S4) and counted. The shoot and root dry biomasses of B. napus were also measured at the end of the experiment after 3 days at 70°C in an oven.
Elemental profiling analysis of B. napus
After 50 days, dry samples from shoot of rapeseed from greenhouse experiment were ground into fine powder using a metal ball in a 50 mL plastic tube. For the elemental profiling analysis, 5 independent samples of 4 plants for strains‐inoculation and of 2 plants for DAPG treatment were collected. The concentrations of 17 elements (Na, Mo, Cd, Sb, Be, B, Mg, P, S, Ca, Mn, Fe, Co, Ni, Cu, Zn and K) were measured by High Resolution Inductively Coupled Plasma Mass Spectrometry (HR ICP‐MS, Thermo Scientific, Element 2TM, Bremen, Germany) as previously described (Lurthy et al., 2020).
Data processing and statistical analysis
Data were analysed using R studio (v.4.2.1) and considered significantly different when p‐value <0.05. The data were assessed for Normal distribution and variance homogeneity using Shapiro–Wilk tests and Bartlett tests, respectively. When these parameters were respected, we performed ANOVA coupled with either HSD‐Tukey test (more than 3 conditions to compare) or LSD‐Fisher test (3 conditions to compare). Otherwise, Kruskal‐Wallis tests applying Bonferroni correction were used to detect differences between conditions. A two‐way ANOVA was performed to assess the effect of the different variable tested, such as the effect of the pathovar and / or the PGCs applied on broomrape seed germination.
RESULTS
The DAPG produced by Pseudomonas contributes to the inhibition of the germination of Phelipanche ramosa in vitro
We tested the ability of Pseudomonas ogarae F113, a PGC‐producing Pseudomonas, to inhibit the germination of four different broomrapes selected according to their host specificity, their parasitism cycle and their associated microbiota (Huet et al., 2020): P. ramosa pv. oilseed rape, O. cumana sunflower, P. ramosa pv. tobacco and P. ramosa pv. hemp.
We first determined the concentration of four different PGCs (PG, MAPG, DAPG and TAPG, Figure 1C) by UHPLC‐UV in the supernatant of P. ogarae F113 wild type (F113) as well as in the supernatants of a mutant impaired in the production of DAPG (ΔphlD), a complemented mutant (ΔphlD Comp.) and a strain engineered to overexpress the gene phlD (Over phlD) involved in the production of PG, the precursor of the DAPG (Figure 1A). Three out of the four different PGCs measured (PG, MAPG and DAPG) were detected in all supernatants, except in the supernatant of ΔphlD (Figure 1B). The concentration of DAPG in the supernatant of the complemented and the overproducing strains were in the same order of magnitude than F113. However, ΔphlD Comp. and Over phlD accumulated between 10 to 20 times more PG and MAPG in their supernatants than F113. Finally, TAPG was not detected in any of the bacterial supernatants (Figure 1B).
FIGURE 1.
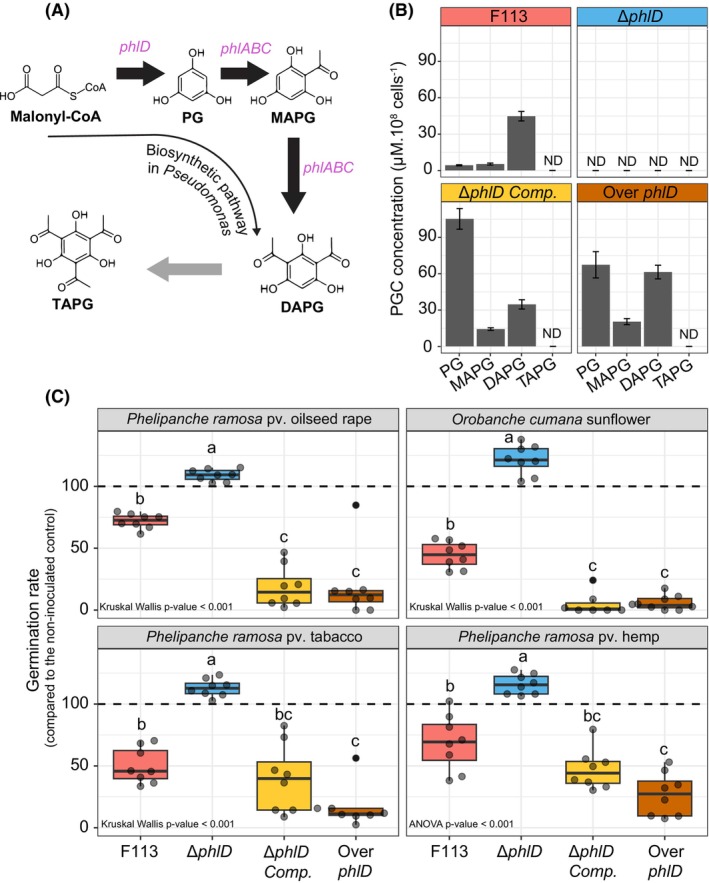
Impact of bacterial supernatants of PGC‐producing Pseudomonas on the germination rate of different broomrapes. (A) Biosynthetic pathway of the DAPG in Pseudomonas cells (according to Biessy & Filion, 2021). The genes involved in the different transformation steps are written in italic. (B) Quantification of Phloroglucinol (PG), monoacetylphloroglucinol (MAPG), diacetylphloroglucinol (DAPG) and triacetylphloroglucinol (TAPG) in the bacterial supernatants of Pseudomonas ogarae F113 and its mutant derivatives: ΔphlD (mutant impaired in the production of DAPG); ΔphlD Comp. (complemented mutant strain overproducing PG) and Over phlD (DAPG, MAPG and PG overproducing strain). Error bars correspond to the standard deviation; ND: Not detected. (C) Impact of these supernatants on the germination capacity of different P. ramosa pathovars and O. cumana in vitro. The supernatants as well as the control condition were supplemented with 1 μM of the germination stimulant (GR24). Results are expressed as percentage of germination of the non‐inoculated ABm medium control. Statistical differences were assessed by ANOVA and Kruskal‐Wallis test using a Bonferroni correction and are indicated with letters. The horizontal lines indicate the interquartile range with the centre representing the median.
Then, we tested the capacity of the different bacterial supernatants to inhibit the germination of the different broomrapes (Figure 1C). We observed that the supernatant of F113 reduced the germination rate of all broomrapes tested (from 28% to 55%). However, the supernatant of ΔphlD did not inhibit the germination rate and appears, on the contrary, to slightly promote it compared to the control (>100%) (Figure 1C). The highest inhibition of the germination was observed with the supernatants of the complemented and the overproducing strains (Figure 1C). Furthermore, the sensitivity to the bacterial supernatants is different according to the broomrape tested (e.g. P. ramosa pv. tobacco being more inhibited by the supernatant of F113 than P. ramosa pv. oil seed rape) (Figure S1).
Each PGC contributes differentially to the inhibition of broomrape germination
To assess the contribution of the PGCs detected in bacterial supernatants on broomrape germination, we composed three different cocktails made of commercially available PGCs, mimicking the proportions detected in the supernatants of the different F113 derivatives (Figure 2A). The application of these cocktails on seeds of P. ramosa pv. oilseed rape allowed us to determine whether the effect observed with the complex supernatants was mainly due to the presence of these PGCs and not to other compounds produced by the bacteria.
FIGURE 2.
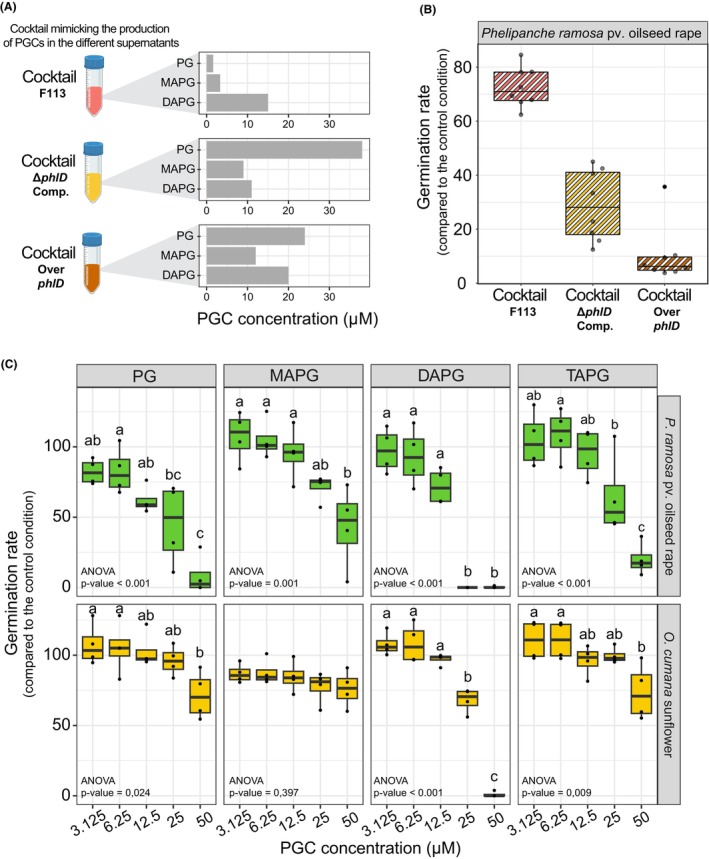
Individual contribution of the different PGCs to the inhibition of the germination of P. ramosa pv. oilseed rape and O. cumana sunflower. (A) Quantification of the pure PGCs added into the different cocktails mimicking the concentration detected in the bacterial supernatants. PG: Phloroglucinol; MAPG: monoacetylphloroglucinol; DAPG: Diacetylphloroglucinol; TAPG: Triacetylphloroglucinol. (B) Effect of the PGCs cocktails on the germination rate of P. ramosa pv oilseed rape. (C) Impact of PG, MAPG, DAPG and TAPG on the germination capacity of different P. ramosa pv. oilseed rape and O. cumana sunflower in vitro. In all the conditions, broomrape seed germination was induced by adding the germination stimulant GR24 (1 μM). Results are expressed as a percentage of germination of the non‐inoculated methanol control. Statistical differences were assessed by ANOVA and Kruskal‐Wallis test using a Bonferroni correction and are indicated with letters. The horizontal lines indicate the interquartile range with the centre representing the median. This experiment was repeated four times independently.
The same levels of inhibition were observed with the different molecular cocktails as in the experiment with bacterial supernatants. Indeed, the cocktails mimicking the concentration of PGCs in the supernatants of complemented and the overproducing strains most inhibited the germination of P. ramosa pv. oilseed rape (Figure 2B). Thus, as observed with complex supernatants, the inhibition of P. ramosa germination is dependent on the composition of the PGCs and their relative concentrations (Figure 2A,B).
Furthermore, we wanted to determine the individual contribution of these PGCs to the inhibition of broomrape germination by testing five different concentrations of each PGC (Figure 2C). First, we performed this inhibition assays on the four previously used broomrape species. However, the presence of 0.33% of methanol in the PGC solutions inhibited seed germination of P. ramosa pv. tobacco and P. ramosa pv. hemp (Data not shown). Contrariwise to what was observed with the bacterial supernatants (Figure S1), the germination rate of P. ramosa pv. oilseed rape was more affected following the exposure to PGCs than O. cumana sunflower (two‐way ANOVA showed a broomrape effect, p‐value <0.001) (Figure 2C). Indeed, a reduction of more than 50% of O. cumana germination was observed only where the seeds were exposed to 50 μM of DAPG. Remarkably, MAPG did not affect the germination rate of O. cumana. On the contrary, the germination rate of P. ramosa pv. oilseed rape started to be affected by the addition of PGCs at 12.5 μM. DAPG was the most effective PGC to disable broomrape germination with 100% of inhibition at 25 μM and 50 μM for P. ramosa pv. oilseed rape and 100% inhibition at 50 μM for O. cumana sunflower. Indeed, the estimated median lethal concentration (LC50) is the lowest for the DAPG for both broomrape species tested (20.0 μM for P. ramosa pv. oilseed rape and 29.1 μM for O. cumana (Figure S2). At the end of the experiment, we recovered the seeds of P. ramosa treated with 50 μM of the different PGCs and washed those 3 times with phosphate buffer to remove PGC traces. Then we added again the GR24 germination stimulant. We observed that the inhibition by the PGC is irreversible since none of the seed treated with PGC germinated. Moreover, for all PGCs except for DAPG, between 25 and 50 μM, the inhibition effects were associated with a brown coloration of the seeds or the radicles (Figure S3).
The inoculation of PGC‐producing Pseudomonas reduced the infection of P. ramosa on Brassica napus
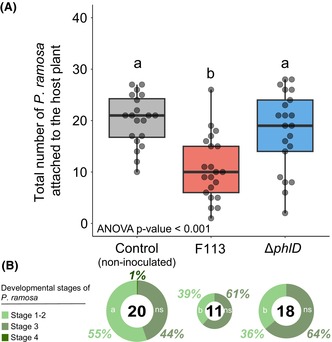
As the DAPG showed the best inhibition of broomrape germination in vitro, we compared the ability of F113 and its ΔphlD mutant to reduce the infection of Brassica napus by P. ramosa pv. oilseed rape in greenhouse conditions. We evaluated the developmental stage of P. ramosa using the development scale available in the Figure S4 and based on Martinez et al. (2023). We observed a significant reduction (47%) in the number of P. ramosa attached to the root of oilseed rape when F113 was inoculated (Figure 3A). This effect was not detected when the ΔphlD mutant was inoculated. Interestingly, at the end of the experiment, we observed a significant reduction in the proportion of early‐stage infections (stage 1 and 2; qualitative infection scale available in Figure S4) in the condition inoculated with F113 or ΔphlD compared to the control (Figure 3B). The proportion of broomrapes in a more advanced infection stage (i.e. particularly from stage 3, buds) did not change with bacterial inoculation. However, the DAPG‐producing F113 strain tended to decrease the average number of broomrapes in an advanced stage as it reduced significantly the total number of broomrapes per root system. We also measured the effect of the bacterial inoculation on the physiology of Brassica napus (Figure 4A–D ). We observed that the biomass of the root system was significantly lower than the control condition when F113 was inoculated (Figure 4B). We also looked at the nutrition capacity of B. napus using an elemental profiling approach (i.e. ionomic) to investigate potential switch of elements according to the treatments we applied. For all the tested conditions, we did not observe a main significant switch of the ion profile inside the shoots of B. napus (Figure 4C,D; Table S2). However, the proportions of certain ions changed according to the treatment applied. The inoculation of F113 led to a significant increase in sodium and a decrease in manganese quantity, whereas the mutant ΔphlD was associated with a decrease in potassium (P) (Figure 4C).
FIGURE 3.
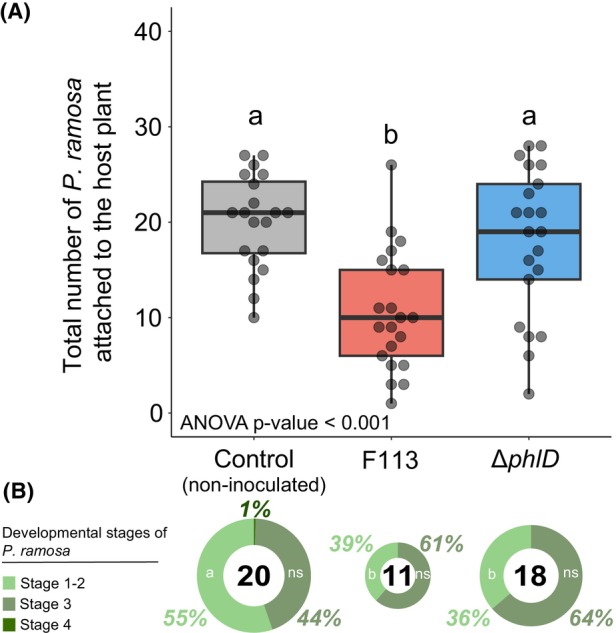
Impact of the inoculation of PGC‐producing Pseudomonas on the infection level by P. ramosa pv. oilseed rape on Brassica napus in greenhouse conditions. (A) Evaluation of the number of attached P. ramosa on the root system of Brassica napus after 50 days in the greenhouse. Different treatments were applied as follows: bacterial inoculants (F113 and ΔphlD impaired in the production of DAPG, approximately five millilitres per pot of solutions at 2 × 106 bacteria/mL) and the control condition which corresponds to the application of 5 mL with MgSO4 10 mM. This experiment was performed in a mixture containing natural soil artificially infested with approximatively 300 P. ramosa seeds per litre of soil. (B) Proportion of broomrapes attached to the root of B. napus according to their developmental stage. The developmental stage of P. ramosa was estimated according to the developmental scale available in Figure S4. The number in the centre of pie chart represents the mean of attached P. ramosa on B. napus roots, and the size of pie charts is proportional to this number. For A and B, Statistical differences are indicated with letters (ANOVA and Fisher's LSD tests, p < 0.05). The horizontal lines indicate the interquartile range with the centre representing the median.
FIGURE 4.
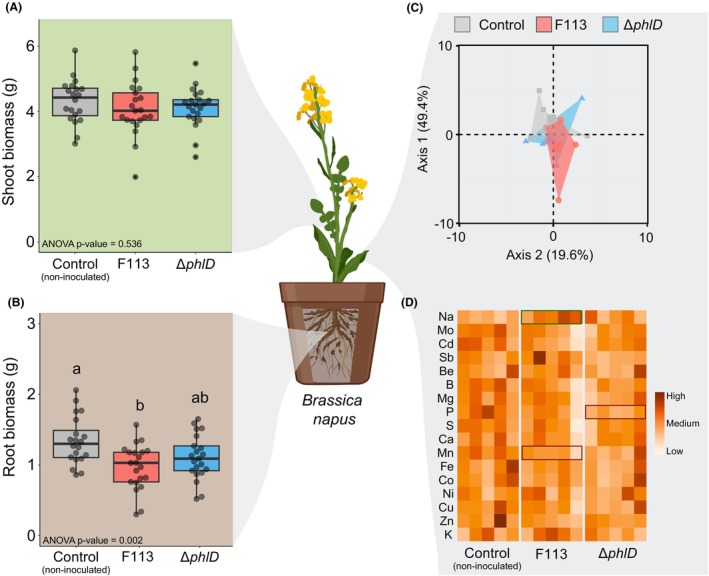
Effect of the inoculation of bacteria on the shoot (A) and root (B) biomasses and, on the ion profile (C,D) of the shoot of Brassica napus. (A,B) We measured for all the tested conditions (presented in Figure 3) the shoot and root biomasses of Brassica napus, 50 days after sowing. The control condition of the bacterial inoculant experiment corresponds to the application of 5 mL with MgSO4 10 mM. The brown background of the boxplots corresponds to the root biomass data while the green background is associated with shoot biomass data. Statistical differences are indicated with letters (ANOVA and Fisher's LSD tests, p < 0.05). The horizontal lines indicate the interquartile range with the centre representing the median. (C) Principal component analysis of the element composition of the shoot of Brassica napus according to the different treatments applied. (D) Heatmap showing the elemental profile of the different conditions. Significant differences between treatments and control were obtained with Student's t‐test. When Student's t‐test assumptions were not met, a Welch t‐test was performed. Red and green boxes correspond to significant negative and positive impact on ionome, respectively. The detailed data are available in Table S2.
On the contrary, the application of DAPG at 50 or 250 μM (v = 5 mL) did not affect the infection of P. ramosa (Figure S5A,B), nor rapeseed biomasses (Figure S7A). Regarding elemental profiling analysis, the addition of DAPG did not switch as well and was associated with a significant increase of antimony (Sb), beryllium (Be) and a reduction of nickel (Ni) in rapeseed shoots (Figure S7B, C; Table S3).
DISCUSSION
PGC‐producing Pseudomonas are well described for their capacity to protect different crop plants from the infection by plant pathogens. Their biocontrol activity has been shown to be dependent on the production of antimicrobial compounds, including DAPG. In this study, we assessed whether the biocontrol activity of such bacterial strains can be extended to the protection of crops towards parasitic plants. We focused on the model environmental Pseudomonas, P. ogarae F113, which is known to display different plant‐beneficial properties including the production of DAPG (Redondo‐Nieto et al., 2012; Vacheron et al., 2018).
Except for TAPG whose bacterial production remains to be demonstrated, F113 is able to synthesize, in different amounts, a cocktail of PGCs composed of PG, MAPG and DAPG (Figure 1B). It is worth noting that the production and the proportion of PGCs are variable among Pseudomonas strains (Duffy & Défago, 1999) and depend on many environmental factors such as carbon sources (Shanahan et al., 1992), bacterial lifestyle (e.g. planktonic or biofilm) (Rieusset et al., 2020) or composition of plant exudates (de Werra et al., 2011; Rieusset et al., 2021). The introduction of the phlD gene on a low‐copy plasmid in the phlD mutant (ΔphlD Comp.) and in the wild‐type F113 strain (Over phlD) modified the production of PGCs, these strains accumulating more PG and MAPG in their supernatants than F113. The production of different PGCs (as in the Over phlD F113 derivative) leads to a stronger inhibitory effect on the germination of the different broomrapes tested (Figure 1C). We also confirmed these results using a cocktail of commercial PGCs in the same proportions (Figure 2A,B) and highlighted that each of the PGCs displayed a different inhibition capacity towards the germination of broomrapes (Figure 2C). Indeed, we showed that the DAPG provided the highest inhibition results. Islam and von Tiedemann (2011) tested the effect of DAPG and its derivatives on the zoosporogenesis and the motility of zoospores from Plasmopara viticola and Aphanomyces cochlioides. They also observed that DAPG displayed the highest inhibitory effect compared to its derivatives. Similar results were observed on the mycelial growth of the plant pathogen Pythium ultimum (de Souza et al., 2003). Altogether, these results highlight the importance to consider the amount of the different precursors of DAPG produced by PGCs‐producing Pseudomonas strains as they could also act as active compounds.
Although DAPG had been well described for its antimicrobial activity towards different kind of plant pathogens (e.g. fungi, oomycetes) (Biessy & Filion, 2021), its toxic effect on plants was also investigated on crop plants and appeared to be associated with its concentration and the plant species (Keel, 1992). A recent study analysed the impact of DAPG at 50 μg mL−1 (equivalent to 240 μM) on the germination rate of 69 wheat cultivars and showed strong differences according to cultivars (Yang et al., 2021). This is in line with our results where the inhibition of germination was dependent on the concentration of DAPG and also different according to the broomrapes species and pathovar. Chae et al. (2020) identified several genes implicated in the sensitivity of Arabidopsis thaliana to exposure to DAPG (Chae et al., 2020). These genes are involved in different metabolic pathways such as the tryptophan and monocarboxylic acid metabolisms and iron management (Chae et al., 2020). The development of molecular tools to modify broomrape genomes would provide new insights on the molecular determinants responsible for their resistance and sensitivity towards PGCs.
The mode of action of DAPG and its derivatives at the cellular level remains elusive. In our study, we observed a brown coloration of the broomrape radicles treated with PGCs (Figure S3). This coloration had previously been observed on tomato seedlings following the addition of 50 μM of DAPG (Brazelton et al., 2008). At cellular levels, the sensitivity to DAPG was correlated with disruption of F‐actin cytoskeleton in A. cochlioides hyphae (Islam & Fukushi, 2010) and with alterations of major physiological functions in yeast including the regulation of cellular responses to reactive oxygen stress and cell homeostasis (Kwak et al., 2011). Thus, the brown coloration of root tissues could be attributed to cell wall disorganization (Islam & Fukushi, 2010), impairment of mitochondrial functions (Kwak et al., 2011; Troppens et al., 2013) and induction of oxidative burst (Kwak et al., 2011). Furthermore, in our experiment, the damage caused by the DAPG on broomrape seeds was irreversible since none of the broomrape seeds treated with DAPG were able to germinate after several washing steps and the addition of the GR24 germination inductor.
To determine if PGCs‐producing Pseudomonas strains are good candidates to prevent the infection of broomrapes, we performed a greenhouse experiment with a natural soil, which was infected with broomrape seeds. The density of broomrape seeds used in this experiment was in the same range of population levels than those used in literature (Bernhard et al., 1998; Chen et al., 2016), leading to a number of attached broomrapes to Brassica napus root system close to what we observed in fields. We observed that the wild‐type F113 was able to significantly reduce the number of broomrapes bound to B. napus roots compared to the mutant impaired in DAPG production (Figure 3). Interestingly, the number of broomrapes associated with new host infection (stage 1 and 2) in the condition where the bacteria were inoculated was significantly lower than in the control condition. Thus, the reduction of the infection is delayed and did not start immediately after the inoculation of the bacterial strains. This lag phase could be interpreted as the time needed for the bacterial inoculant to establish itself within the soil and/or root microbiome, and/or to produce DAPG in sufficient amounts. We observed that the ΔphlD mutant was also able to reduce the proportion of broomrapes in an early infection stage. This could be due to the fact bacteria can exert, as well, an indirect positive activity on oilseed rape leading to a decreased sensitivity to broomrape infection, independently of the production of DAPG, as cited previously by Cartry et al. (2021). Moreover, several reports showed that DAPG act as a signalling molecule inducing the plant systemic resistance (Weller et al., 2012) or the expression of its own biosynthetic genes (Maurhofer et al., 2004). Thus, the enrichment of the B. napus rhizo‐microbiota with PGC‐producing pseudomonads could stimulate other PGC‐producing strains already present in the rhizosphere, leading to an increase of PGC concentrations in soil, thereby leading to detrimental effects on broomrape germination.
In addition, the inoculation of F113 leads to a decrease of the root system biomass of B. napus (Figure 4). It has been shown that the inoculation of PGC‐producing Pseudomonas was linked to a decrease of the root length in Arabidopsis thaliana and other crop plants (Brazelton et al., 2008; De Leij et al., 2002; Vacheron et al., 2018). Thus, PGCs‐producing strains could limit the germination of broomrapes by two modes of action. The first one can be direct via the production of DAPG that inhibit the parasite seed germination whereas the second affects the root length of the host plant that in fine decreases the probability of contact between the host plant roots and broomrape seeds within the rhizosphere soil.
Conversely, the application of DAPG did not limit the infection of P. ramosa on B. napus roots (Figure S5). The DAPG applied may have not only specifically targeted the broomrape seeds but also interacted with soil particles or the resident soil/root microbiomes as well as the oilseed plant. Thus, the amount of DAPG targeting the broomrape seeds might be insufficient to deliver a significant reduction of P. ramosa infection. Moreover, the soil chemical properties could impact the efficiency of DAPG inhibition. Indeed, Kwak and collaborators documented that DAPG has a half‐life of less than 6 h in soil (Kwak et al., 2012). Increasing the DAPG concentration over 250 μM might induce negative effects on the environment as any phytochemical products. Moreover, its field application would be too expensive. So, the application of pure DAPG has an herbicide against broomrape is neither realistic nor desirable. Conversely, we showed that a DAPG‐producing strain was able to significantly reduce broomrapes' infectivity under greenhouse conditions, suggesting that inoculation of PGC‐producing strains represents a better biocontrol solution against broomrapes. Except using genetically modified organisms, such as our strain Over phlD, which is banned in Europe, the application of PGC‐overproducing strains may improve considerably biocontrol effect.
Elemental analyses highlighted no unwanted changes on plant host nutrition induced by bacterial inoculant (Figure 4), or by pure DAPG amendment (Figure S7). It's an important concern in field where oilseed rape nutrition must be unchanged to achieve high yield. Shoot accumulation of sodium (p‐value <0.05 for F113 treatment and <0.1 for DAPG 50 μM treatment) appears to be linked to less parasitism. Sodium accumulation is generally considered as toxic for plants depending on plant species (Kronzucker et al., 2013). In our study, the accumulation of sodium does not affect the plant host but is correlated to a reduction of P. ramosa infection. Bacterial inoculation with Pseudomonas or Azospirillum was rather associated with a decrease of sodium in rapeseed leaf when exposed to harmful effects of salt stress (Farhangi‐Abriz et al., 2020). Thus, improvement of sodium uptake in the host by DAPG‐producing bacteria could be another factor associated with broomrape protection. As the nutrient flux from the host plant to the parasite is driven by an osmotic pressure differential between them (Shen et al., 2006), we hypothesize that the increase of sodium content in the host may reduce the level of nutrient uptake by the parasite and may impact its growth. Thus, sodium accumulation could be an interesting factor to study on oilseed rape cultivars in response to biocontrol agents. Nonetheless, the signals emitted by the broomrape and the B. napus roots during the haustoriogenesis that may attract biocontrol agents or participate to the regulation of their activities, are not yet precisely known, and their identification deserved particular attention.
Here, we showed that the F113 DAPG‐producing strain combined different direct and indirect effects to protect oilseed rape against broomrape. DAPG and its derivatives could be interesting bioherbicides produced by Pseudomonas for preventing parasitic plant infestation as we did not observe any toxicity towards the host plant. The soil and root microbiome compositions were previously associated with natural suppressiveness towards parasitic weeds. Indeed, several studies claimed that the suppressiveness of Orobanche sp. as well as Striga hermonthica is associated with the presence of specific bacterial taxa, including Pseudomonas (Kawa et al., 2022; Zermane et al., 2007). Since, PGC‐producing Pseudomonas can be followed in soil via qPCR approaches (Almario et al., 2013), determining the correlation between the community of PGC‐producing Pseudomonas and the level of parasitic plant infection would bring new insights on the ecological role of these bacteria in suppressive soils.
Finally, this study reinforces the interest of using microorganisms as natural solutions to regulate populations of pests and plant pathogens (Vurro, 2023). Indeed, this study is the first to discover and demonstrate the inhibitory effect of DAPG on parasitic plant germination, under in vitro conditions expanding the repertoire of plant‐beneficial properties of environmental pseudomonads. Furthermore, we evidenced that PGCs produced by P. ogarae are the main compounds that irreversibly inhibit the germination of broomrape species tested. Without the use of adjuvants able to enhance their biocontrol activity (Campos et al., 2023) and taking into account that PGC production could be modulated by B. napus and other microorganisms in the plant rhizosphere (Laveilhé et al., 2022), we obtained promising results during the experiments carried out in natural soil in greenhouse condition. Indeed, the inoculation of P. ogarae halved the infection of P. ramosa on its host plant, Brassica napus. The present work significantly expands our knowledge about the role that these plant‐beneficial Pseudomonas play in the environment. It provides new alternative direction for the development of natural bioherbicides to ward off parasitic plant infections, which are responsible for important yield losses in crops.
AUTHOR CONTRIBUTIONS
Tristan Lurthy: Conceptualization (lead); data curation (lead); investigation (lead); methodology (lead); writing – original draft (lead); writing – review and editing (lead). Ségolène Perot: Investigation (equal). Florence Gerin‐Eveillard: Investigation (equal). Marjolaine Rey: Conceptualization (equal); investigation (equal). Florence Wisniewski‐Dyé: Conceptualization (supporting); investigation (supporting); writing – review and editing (lead). Jordan Vacheron: Conceptualization (lead); data curation (lead); investigation (lead); writing – review and editing (lead). Claire Prigent‐Combaret: Conceptualization (lead); data curation (lead); funding acquisition (lead); investigation (lead); project administration (lead); writing – review and editing (lead).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Supporting information
supporting information
ACKNOWLEDGMENTS
We thank the research team US2B of Nantes and more specifically Jean‐Bernard Pouvreau for providing the broomrape seeds. We thank the company RAGT for providing seeds of the oilseed rape cultivars. We thank members of the Rhizo team of the Microbial Ecology unit at University Lyon 1 for their help in enumeration of broomrapes bound to B. napus roots in greenhouse. We are most grateful to PLATIN’ (Plateau d'Isotopie de Normandie) core facility for all element and isotope analysis used in this study. The platform ‘Serre’ of FR BioEEnViS (University Lyon 1) was used to carry out this work. This research and TL were supported by a grant from the French national research agency ‘Ecophyto Maturation’ (ANR‐19‐ECOM‐0002 WeedsBiocontrol) and by a maturation grant from the Pulsalys Technology Transfer Acceleration Company. JV was supported by the Swiss National Centre of Competence in Research (NCCR) Microbiomes (no. 51NF40_180575).
Lurthy, T. , Perot, S. , Gerin‐Eveillard, F. , Rey, M. , Wisniewski‐Dyé, F. , Vacheron, J. et al. (2023) Inhibition of broomrape germination by 2,4‐diacetylphloroglucinol produced by environmental Pseudomonas . Microbial Biotechnology, 16, 2313–2325. Available from: 10.1111/1751-7915.14336
Jordan Vacheron and Claire Prigent‐Combaret co‐senior authors.
Contributor Information
Tristan Lurthy, Email: tristan.lurthy@univ-lyon1.fr.
Jordan Vacheron, Email: jordan.vacheron@unil.ch.
Claire Prigent‐Combaret, Email: claire.prigent-combaret@univ-lyon1.fr.
REFERENCES
- Achkar, J. , Xian, M. , Zhao, H. & Frost, J.W. (2005) Biosynthesis of phloroglucinol. Journal of the American Chemical Society, 127, 5332–5333. [DOI] [PubMed] [Google Scholar]
- Aliche, E.B. , Screpanti, C. , De Mesmaeker, A. , Munnik, T. & Bouwmeester, H.J. (2020) Science and application of strigolactones. The New Phytologist, 227, 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almario, J. , Bruto, M. , Vacheron, J. , Prigent‐Combaret, C. , Moënne‐Loccoz, Y. & Muller, D. (2017) Distribution of 2,4‐diacetylphloroglucinol biosynthetic genes among the Pseudomonas spp. reveals unexpected polyphyletism. Frontiers in Microbiology, 8, 1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almario, J. , Moënne‐Loccoz, Y. & Muller, D. (2013) Monitoring of the relation between 2,4‐diacetylphloroglucinol‐producing Pseudomonas and Thielaviopsis basicola populations by real‐time PCR in tobacco black root‐rot suppressive and conducive soils. Soil Biology and Biochemistry, 57, 144–155. [Google Scholar]
- Bakker, P.A.H.M. , Pieterse, C.M.J. & van Loon, L.C. (2007) Induced systemic resistance by fluorescent Pseudomonas spp. Phytopathology, 97, 239–243. [DOI] [PubMed] [Google Scholar]
- Balthazar, C. , Joly, D.L. & Filion, M. (2021) Exploiting beneficial Pseudomonasspp. for cannabis production. Frontiers in Microbiology, 12, 833172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, J.R. & Mathews, S. (2006) Phylogeny of the parasitic plant family Orobanchaceae inferred from phytochrome A. American Journal of Botany, 93, 1039–1051. [DOI] [PubMed] [Google Scholar]
- Bernhard, R.H. , Jensen, J.E. & Andreasen, C. (1998) Prediction of yield loss caused by Orobanche spp. in carrot and pea crops based on the soil seedbank. Weed Research (Oxford), 38, 191–197. [Google Scholar]
- Biessy, A. & Filion, M. (2021) Phloroglucinol derivatives in plant‐beneficial Pseudomonas spp.: biosynthesis, regulation, and functions. Metabolites, 11, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazelton, J.N. , Pfeufer, E.E. , Sweat, T.A. , Gardener, B.B.M. & Coenen, C. (2008) 2,4‐Diacetylphloroglucinol alters plant root development. Molecular Plant‐Microbe Interactions, 21, 1349–1358. [DOI] [PubMed] [Google Scholar]
- Brodhagen, M. , Henkels, M.D. & Loper, J.E. (2004) Positive autoregulation and signaling properties of pyoluteorin, an antibiotic produced by the biological control organism Pseudomonas fluorescens Pf‐5. Applied and Environmental Microbiology, 70, 1758–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos, E.V.R. , Ratko, J. , Bidyarani, N. , Takeshita, V. & Fraceto, L.F. (2023) Nature‐based herbicides and micro−/nanotechnology fostering sustainable agriculture. ACS Sustainable Chemistry & Engineering,11, 9900–9917. [Google Scholar]
- Cartry, D. , Steinberg, C. & Gibot‐Leclerc, S. (2021) Main drivers of broomrape regulation. A review. Agronomy for Sustainable Development, 41, 17. [Google Scholar]
- Chae, D.‐H. , Kim, D.‐R. , Cho, G. , Moon, S. & Kwak, Y.‐S. (2020) Genome‐wide investigation of 2,4‐diacetylphloroglucinol protection genes in Arabidopsis thaliana . Molecular Plant‐Microbe Interactions, 33, 1072–1079. [DOI] [PubMed] [Google Scholar]
- Chen, J. , Xue, Q.H. , McErlean, C.S.P. , Zhi, J.H. , Ma, Y.Q. , Jia, X.T. et al. (2016) Biocontrol potential of the antagonistic microorganism Streptomyces enissocaesilis against Orobanche cumana . BioControl, 61, 781–791. [Google Scholar]
- Combes‐Meynet, E. , Pothier, J.F. , Moënne‐Loccoz, Y. & Prigent‐Combaret, C. (2011) The Pseudomonas secondary metabolite 2,4‐diacetylphloroglucinol is a signal inducing rhizoplane expression of Azospirillum genes involved in plant‐growth promotion. Molecular Plant‐Microbe Interactions, 24, 271–284. [DOI] [PubMed] [Google Scholar]
- Dadon, T. , Nun, N.B. & Mayer, A.M. (2004) A factor from Azospirillum brasilense inhibits germination and radicle growth of Orobanche aegyptiaca . Israel Journal of Plant Sciences, 52, 83–86. [Google Scholar]
- De Leij, F.A. , Dixon‐Hardy, J.E. & Lynch, J.M. (2002) Effect of 2,4‐diacetylphloroglucinol‐producing and non‐producing strains of Pseudomonas fluorescens on root development of pea seedlings in three different soil types and its effect on nodulation by Rhizobium . Biology and Fertility of Soils, 35, 114–121. [Google Scholar]
- de Souza, J.T. , Arnould, C. , Deulvot, C. , Lemanceau, P. , Gianinazzi‐Pearson, V. & Raaijmakers, J.M. (2003) Effect of 2,4‐diacetylphloroglucinol on pythium: cellular responses and variation in sensitivity among propagules and species. Phytopathology, 93, 966–975. [DOI] [PubMed] [Google Scholar]
- de Werra, P. , Huser, A. , Tabacchi, R. , Keel, C. & Maurhofer, M. (2011) Plant‐ and microbe‐derived compounds affect the expression of genes encoding antifungal compounds in a pseudomonad with biocontrol activity. Applied and Environmental Microbiology, 77, 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy, B.K. & Défago, G. (1999) Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Applied and Environmental Microbiology, 65, 2429–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Zemrany, H. , Cortet, J. , Peter Lutz, M. , Chabert, A. , Baudoin, E. , Haurat, J. et al. (2006) Field survival of the phytostimulator Azospirillum lipoferum CRT1 and functional impact on maize crop, biodegradation of crop residues, and soil faunal indicators in a context of decreasing nitrogen fertilisation. Soil Biology and Biochemistry, 38, 1712–1726. [Google Scholar]
- Farhangi‐Abriz, S. , Tavasolee, A. , Ghassemi‐Golezani, K. , Torabian, S. , Monirifar, H. & Rahmani, H.A. (2020) Growth‐promoting bacteria and natural regulators mitigate salt toxicity and improve rapeseed plant performance. Protoplasma, 257, 1035–1047. [DOI] [PubMed] [Google Scholar]
- Garrido‐Sanz, D. , Redondo‐Nieto, M. , Martin, M. & Rivilla, R. (2021) Comparative genomics of the Pseudomonas corrugata subgroup reveals high species diversity and allows the description of Pseudomonas ogarae sp. nov. Microbial Genomics, 7, 000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, D. & Défago, G. (2005) Biological control of soil‐borne pathogens by fluorescent pseudomonads. Nature Reviews. Microbiology, 3, 307–319. [DOI] [PubMed] [Google Scholar]
- Haring, S.C. & Flessner, M.L. (2018) Improving soil seed bank management. Pest Management Science, 74, 2412–2418. [DOI] [PubMed] [Google Scholar]
- Hasannejad, S. , Zad, S.J. , Alizade, H.M. & Rahymian, H. (2006) The effects of Fusarium oxysporum on broomrape (Orobanche egyptiaca) seed germination. Communications in Agricultural and Applied Biological Sciences, 71, 1295–1299. [PubMed] [Google Scholar]
- Huet, S. , Pouvreau, J.‐B. , Delage, E. , Delgrange, S. , Marais, C. , Bahut, M. et al. (2020) Populations of the parasitic plant Phelipanche ramosa influence their seed microbiota. Frontiers in Plant Science, 11, 1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iavicoli, A. , Boutet, E. , Buchala, A. & Métraux, J.‐P. (2003) Induced systemic resistance in Arabidopsis thaliana in response to root inoculation with Pseudomonas fluorescens CHA0. Molecular Plant‐Microbe Interactions, 16, 851–858. [DOI] [PubMed] [Google Scholar]
- Islam, M.T. & Fukushi, Y. (2010) Growth inhibition and excessive branching in Aphanomyces cochlioides induced by 2,4‐diacetylphloroglucinol is linked to disruption of filamentous Actin cytoskeleton in the hyphae. World Journal of Microbiology and Biotechnology, 26, 1163–1170. [DOI] [PubMed] [Google Scholar]
- Islam, M.T. & von Tiedemann, A. (2011) 2,4‐Diacetylphloroglucinol suppresses zoosporogenesis and impairs motility of Peronosporomycete zoospores. World Journal of Microbiology and Biotechnology, 27, 2071–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel, D.M. (2009) The new nomenclature of Orobanche and Phelipanche . Weed Research, 49, 6–7. [Google Scholar]
- Kawa, D. , Thiombiano, B. , Shimels, M. , Taylor, T. , Walmsley, A. , Vahldick, H.E. et al. (2022) The soil microbiome reduces Striga infection of sorghum by modulation of host‐derived signaling molecules and root development. bioRxiv. 10.1101/2022.11.06.515382 [DOI]
- Keel, C. (1992) Suppression of root diseases by Pseudomonas fluorescens CHA0: importance of the bacterial secondary metabolite 2,4‐diacetylphloroglucinol. Molecular Plant‐Microbe Interactions, 5, 4. [Google Scholar]
- Khan, F. , Tabassum, N. , Bamunuarachchi, N.I. & Kim, Y.‐M. (2022) Phloroglucinol and its derivatives: antimicrobial properties toward microbial pathogens. Journal of Agricultural and Food Chemistry, 70, 4817–4838. [DOI] [PubMed] [Google Scholar]
- King, E.O. , Ward, M.K. & Raney, D.E. (1954) Two simple media for the demonstration of pyocyanin and fluorescin. The Journal of Laboratory and Clinical Medicine, 44, 301–307. [PubMed] [Google Scholar]
- Kronzucker, H.J. , Coskun, D. , Schulze, L.M. , Wong, J.R. & Britto, D.T. (2013) Sodium as nutrient and toxicant. Plant and Soil, 369, 1–23. [Google Scholar]
- Kwak, Y.‐S. , Bonsall, R.F. , Okubara, P.A. , Paulitz, T.C. , Thomashow, L.S. & Weller, D.M. (2012) Factors impacting the activity of 2,4‐diacetylphloroglucinol‐producing Pseudomonas fluorescens against take‐all of wheat. Soil Biology and Biochemistry, 54, 48–56. [Google Scholar]
- Kwak, Y.‐S. , Han, S. , Thomashow, L.S. , Rice, J.T. , Paulitz, T.C. , Kim, D. et al. (2011) Saccharomyces cerevisiae genome‐wide mutant screen for sensitivity to 2,4‐diacetylphloroglucinol, an antibiotic produced by Pseudomonas fluorescens . Applied and Environmental Microbiology, 77, 1770–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laveilhé, A. , Fochesato, S. , Lalaouna, D. , Heulin, T. & Achouak, W. (2022) Phytobeneficial traits of rhizobacteria under the control of multiple molecular dialogues. Microbial Biotechnology, 15, 2083–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper, J.E. , Hassan, K.A. , Mavrodi, D.V. , Davis, E.W. , Lim, C.K. , Shaffer, B.T. et al. (2012) Comparative genomics of plant‐associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genetics, 8, e1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurthy, T. , Cantat, C. , Jeudy, C. , Declerck, P. , Gallardo, K. , Barraud, C. et al. (2020) Impact of bacterial siderophores on iron status and ionome in pea. Frontiers in Plant Science, 11, 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, L. , Pouvreau, J.‐B. , Montiel, G. , Jestin, C. , Delavault, P. , Simier, P. et al. (2023) Soil microbiota promotes early developmental stages of Phelipanche ramosa L. Pomel during plant parasitism on Brassica napus L. Plant and Soil, 483, 667–691. [Google Scholar]
- Masteling, R. , Lombard, L. , de Boer, W. , Raaijmakers, J.M. & Dini‐Andreote, F. (2019) Harnessing the microbiome to control plant parasitic weeds. Current Opinion in Microbiology, 49, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurhofer, M. , Baehler, E. , Notz, R. , Martinez, V. & Keel, C. (2004) Cross talk between 2,4‐diacetylphloroglucinol‐producing biocontrol pseudomonads on wheat roots. Applied and Environmental Microbiology, 70, 1990–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutuku, J.M. , Cui, S. , Yoshida, S. & Shirasu, K. (2021) Orobanchaceae parasite–host interactions. New Phytolog, 230, 46–59. [DOI] [PubMed] [Google Scholar]
- Parker, C. (2012) Parasitic weeds: a world challenge. Weed Science, 60, 269–276. [Google Scholar]
- Phillips, D.A. , Fox, T.C. , King, M.D. , Bhuvaneswari, T.V. & Teuber, L.R. (2004) Microbial products trigger amino acid exudation from plant roots. Plant Physiology, 136, 2887–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouvreau, J.‐B. , Gaudin, Z. , Auger, B. , Lechat, M.‐M. , Gauthier, M. , Delavault, P. et al. (2013) A high‐throughput seed germination assay for root parasitic plants. Plant Methods, 9, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo‐Nieto, M. , Barret, M. , Morrisey, J.P. , Germaine, K. , Martínez‐Granero, F. , Barahona, E. et al. (2012) Genome sequence of the biocontrol strain Pseudomonas fluorescens F113. Journal of Bacteriology, 194, 1273–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieusset, L. , Rey, M. , Gerin, F. , Wisniewski‐Dyé, F. , Prigent‐Combaret, C. & Comte, G. (2021) A cross‐metabolomic approach shows that wheat interferes with fluorescent Pseudomonas physiology through its root metabolites. Metabolites, 11, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieusset, L. , Rey, M. , Muller, D. , Vacheron, J. , Gerin, F. , Dubost, A. et al. (2020) Secondary metabolites from plant‐associated Pseudomonas are overproduced in biofilm. Microbial Biotechnology, 13, 1562–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubiales, D. , Alcántara, C. , Pérez‐de‐Luque, A. , Gil, J. & Sillero, J. (2003) Infection of chickpea (Cicer arietinum) by crenate broomrape (Orobanche crenata) as influenced by sowing date and weather conditions. Agronomy, 23, 359–362. [Google Scholar]
- Shanahan, P. , O'sullivan, D.J. , Simpson, P. , Glennon, J.D. & O'gara, F. (1992) Isolation of 2,4‐diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Applied and Environmental Microbiology, 58, 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, H. , Ye, W. , Hong, L. , Huang, H. , Wang, Z. , Deng, X. et al. (2006) Progress in parasitic plant biology: host selection and nutrient transfer. Plant Biology (Stuttgart, Germany), 8, 175–185. [DOI] [PubMed] [Google Scholar]
- Silby, M.W. , Winstanley, C. , Godfrey, S.A.C. , Levy, S.B. & Jackson, R.W. (2011) Pseudomonas genomes: diverse and adaptable. FEMS Microbiology Reviews, 35, 652–680. [DOI] [PubMed] [Google Scholar]
- Troppens, D.M. , Dmitriev, R.I. , Papkovsky, D.B. , O'Gara, F. & Morrissey, J.P. (2013) Genome‐wide investigation of cellular targets and mode of action of the antifungal bacterial metabolite 2,4‐diacetylphloroglucinol in Saccharomyces cerevisiae . FEMS Yeast Research, 13, 322–334. [DOI] [PubMed] [Google Scholar]
- Vacheron, J. , Desbrosses, G. , Renoud, S. , Padilla, R. , Walker, V. , Muller, D. et al. (2018) Differential contribution of plant‐beneficial functions from Pseudomonas kilonensis F113 to root system architecture alterations in Arabidopsis thaliana and Zea mays . Molecular Plant‐Microbe Interactions, 31, 212–223. [DOI] [PubMed] [Google Scholar]
- Vurro, M. (2023) Are root parasitic broomrapes still a good target for bioherbicide control? Pest Management Science. Available from: 10.1002/ps.7360 [DOI] [PubMed] [Google Scholar]
- Weller, D.M. , Mavrodi, D.V. , van Pelt, J.A. , Pieterse, C.M.J. , van Loon, L.C. & Bakker, P.A.H.M. (2012) Induced systemic resistance in Arabidopsis thaliana against Pseudomonas syringae pv. tomato by 2,4‐diacetylphloroglucinol‐producing Pseudomonas fluorescens . Phytopathology, 102, 403–412. [DOI] [PubMed] [Google Scholar]
- Westwood, J.H. (2013) The physiology of the established parasite–host association. In: Joel, D.M. , Gressel, J. & Musselman, L.J. (Eds.) Parasitic Orobanchaceae: parasitic mechanisms and control strategies. Berlin, Heidelberg: Springer, pp. 87–114. [Google Scholar]
- Yang, M. , Thomashow, L.S. & Weller, D.M. (2021) Evaluation of the phytotoxicity of 2,4‐diacetylphloroglucinol and Pseudomonas brassicacearum Q8r1‐96 on different wheat cultivars. Phytopathology, 111, 1935–1941. [DOI] [PubMed] [Google Scholar]
- Yu, X.‐Q. , Yan, X. , Zhang, M.‐Y. , Zhang, L.‐Q. & He, Y.‐X. (2020) Flavonoids repress the production of antifungal 2,4‐DAPG but potentially facilitate root colonization of the rhizobacterium Pseudomonas fluorescens . Environmental Microbiology, 22, 5073–5089. [DOI] [PubMed] [Google Scholar]
- Zermane, N. , Souissi, T. , Kroschel, J. & Sikora, R. (2007) Biocontrol of broomrape (Orobanche crenata Forsk. and Orobanche foetida Poir.) by Pseudomonas fluorescens isolate Bf7‐9 from the faba bean rhizosphere. Biocontrol Science and Technology, 17, 483–497. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supporting information


