Abstract
The HIV-1 epidemic in the US has historically been dominated by subtype B. HIV subtype diversity has not been extensively examined in most US cities to determine whether non-B variants have become established, as has been observed in many other global regions. We describe the diversity of non-B variants and present evidence of local transmission of non-B HIV in San Francisco. Viral sequences collected from patients between 2000 and 2016 were matched to the San Francisco HIV/AIDS case registry. HIV subtype was determined using COMET. Phylogenies were reconstructed using the pol region of subtypes A, C, D, G, CRF01_AE, CRF02_AG, and CRF07_BC, with reference sequences from the LANL HIV database. Associations of non-B subtypes and circulating recombinant forms (CRFs) with patient characteristics were assessed using multivariable logistic regression. Out of 11,381 sequences, 10,669 were from 7,235 registry cases, of which 141 (2%) had non-B subtypes and CRFs and 72 (1%) had unique recombinant forms. CRF01_AE (0.8%) and subtype C (0.5%) were the most prevalent non-B forms. The frequency of non-B subtypes and CRFs increased in San Francisco during years 2000-2016. Out of 146 transmission events involving non-B study sequences, 18% indicated local transmission within the study population and 74% appeared to be inward migration of the virus. Compared to 7,016 cases with only subtype B, 141 cases with non-B sequences were more likely to be of non-US country of birth (aOR=11.02; p<0.001), of Asian/Pacific-Islander race/ethnicity (aOR=3.17; p<0.001), and diagnosed after 2009 (aOR=4.81; p<0.001). Results suggest that most non-B infections were likely acquired outside the US and that local transmission of non-B forms has occurred but so far has not produced extensive transmission networks. Thus, non-B variants were not widely established in San Francisco, an observation that differs from cities worldwide with more diverse epidemics.
Keywords: HIV, phylogeny, non-B subtype, circulating recombinant forms, epidemiology, San Francisco
1. Introduction
Since the introduction of HIV-1 to the US in the late 1960s or early 1970s, subtype B has been the dominant variant of HIV in North America (Gilbert et al., 2007). In many global regions where subtype B predominates, minority subtypes and circulating recombinant forms (CRFs) are common and comprise an important portion of the epidemic (Aggarwal et al., 2006; Dilernia et al., 2007; Giuliani et al., 2009; González-Alba et al., 2011; Pérez-Alvarez et al., 2003; Sallam et al., 2017; Semaille et al., 2007; UK Collaborative Group on HIV Drug Resistance, 2014; Von Wyl et al., 2011). Non-B variants are often associated with transmission through heterosexual sex and injection drug use, and their expansion can indicate a shift in transmission mode (Beloukas et al., 2016; Chibo and Birch, 2012; Delgado et al., 2019; Kuiken et al., 2000).
In the US, non-B HIV is most documented in immigrant communities (Achkar et al., 2004; Lin et al., 2006; Sides et al., 2005). Broader surveillance and survey data indicate the presence of non-B subtypes and CRFs, with subtype C being most common, followed by subtype A and CRF02_AG (Dennis et al., 2017; Germer et al., 2015; Oster et al., 2017; Pyne et al., 2013; Sey et al., 2014; Wheeler et al., 2010). Studies in the Northeastern US report 1–13% prevalence of non-B forms among HIV cases and suggest that prevalence may vary across locations and may be increasing (Achkar et al., 2004; Carr et al., 2010; Chan et al., 2014; Dennis et al., 2017; Lin et al., 2006; Oster et al., 2017; Pérez-Losada et al., 2017; Pyne et al., 2013; Sides et al., 2005; Weidle et al., 2000). Local onward transmission in the US has been demonstrated in a few local jurisdictions and multi-state studies using phylogenetic methods (Chan et al., 2014; Dennis et al., 2017; Sey et al., 2014; Wertheim et al., 2016).
San Francisco is a historical epicenter of HIV, particularly among men who have sex with men (MSM) (San Francisco Department of Public Health, 2018). Given the predominance of subtype B in North America and the association between subtype B and MSM seen in much of the world, local transmission networks of non-B variants in San Francisco would be surprising. San Francisco, however, is also an immigration gateway and potentially a link between the US and the genetically diverse global HIV epidemic (U.S. Census Bureau). If non-B variants are present in San Francisco, it is important to know whether they are transmitted locally and among the same risk groups as the majority of the epidemic. To date, San Francisco’s HIV epidemic has not been described in terms of viral diversity, local transmission of non-B subtypes, or how transmission risk groups may differ between B and non-B subtypes. Genetic data necessary to conduct these analyses are now available because HIV drug-resistance testing has been common since the late 2000s, and viral genotypes have been reportable to the San Francisco Department of Public Health (SFDPH) since 2014 (56% complete for diagnoses during 2014–2016) (San Francisco Department of Public Health, 2018; Truong et al., 2015; Truong, O’Keefe et al., 2019; Truong, Pipkin et al., 2019).
We examine the most complete set of HIV genetic sequences available for San Francisco, in combination with demographic and risk data from the SFDPH HIV/AIDS case registry. We characterize the frequency and diversity of non-B variants. We employ phylogenetic methods to examine whether non-B subtypes and CRFs are locally established and to gauge the extent of local transmission, and we examine the characteristics of patients with non-B subtypes or CRFs. Knowledge of non-B HIV diversity, transmission patterns, and epidemiology in US cities like San Francisco may point to ways in which the HIV epidemic is changing and may illuminate connections between Western US cities and the global epidemic.
2. Materials and Methods
2.1. Viral sequences
HIV-1 full protease and partial reverse transcriptase sequences (bases 2253–3554 on HXB2, GenBank: K03455) from drug-resistance testing were obtained from two sources. The ARI-UCSF Laboratory of Clinical Virology (LCV) provided sequences from public and community-based clinics in San Francisco, generated using TRUGENE (TRUGENE HIV-1 Genotyping Assay, Siemens, Malvern, PA) for samples dated January 2000–June 2015 and ViroSeq (ViroSeq HIV-1 Genotyping System v2.0, Abbott Molecular, Inc., Des Plains, IL) for samples dated July 2015–December 2016 (Truong et al., 2015). Specimens with >99% sequence similarity to previous specimens in the same laboratory were investigated. Sequences were matched deterministically by patient identifying information to the SFDPH HIV/AIDS case registry to obtain demographic and risk characteristics: sex at birth, race/ethnicity, country of birth, HIV transmission risk category, age at HIV diagnosis, and year of HIV diagnosis (first diagnosis in the US). Additional sequences dated 2014–2016, ordered by private San Francisco health providers or by external providers for San Francisco residents, were provided with demographic and risk data by SFDPH Molecular HIV Surveillance (MHS).
Reference HIV-1 sequences were downloaded from Los Alamos National Laboratories. For seven non-B subtypes and CRFs, all sequences complete for bases 2253–3554 and with a known geographic origin were downloaded: 3,703 subtype A (including 3,632 of subtype A1); 9,096 of C; 1,315 of D; 801 of G; 3,315 of CRF01_AE; 2,151 of CRF02_AG; and 285 of CRF07_BC (Los Alamos National Laboratory). We also downloaded 51 sequences from the Subtype Reference Alignment (SRA) 2010 update, representing all HIV-1 group M subtypes, CRF01_AE, and groups N and O (Leitner et al., 2005).
2.2. Subtyping and phylogenetics
Study sequences were subtyped using COMET stand-alone version 1.1.4, provided by the authors (Struck et al., 2014). Sequences of subtypes and CRFs found in more than three individuals were aligned by subtype using ClustalW within BioEdit v7.2.5 followed by manual adjustment (Hall, 1999; Thompson et al., 1994). Alignments included all subtype- and CRF-specific reference sequences and all SRA references sequences. Sixty-seven codons where substitutions associated with drug resistance are located were excluded (Wensing et al., 2015). A phylogeny was reconstructed from each non-B subtype- and CRF-specific alignment using FastTree 2.1.7 with the generalized-time-reversible nucleotide substitution model and a single substitution rate per site (GTR+CAT) and rooted by groups N and O (Price et al., 2010). Furthermore, all LCV sequences and SRA reference sequences were aligned together and included in a multi-subtype phylogeny which was used to assign the final subtype for six sequences for which COMET had indicated two possible subtypes. One sequence was excluded from analyses because the observed phylogenetic position contradicted COMET.
Extent of local transmission was evaluated based on the distribution of study sequences among reference sequences in the non-B subtype-and CRF-specific phylogenies. A “close” relationship between sequences was defined as a separation by one to two nodes (branch points) on the phylogeny. A “distant” relationship was defined as a separation by more than three nodes. Nodes with low Shimodaira-Hasegawa support (SH <0.7) were considered unresolved, and the relationships between surrounding sequences were assigned a maximum and a minimum number of nodes of separation to account for uncertainty. The maximum was used for identifying close relationships, and the minimum for identifying distant relationships. Multiple closely related sequences from the same individual were treated as resulting from a single transmission. Distantly related sequences from the same individual were considered separate transmissions.
We counted the number of local transmissions on each phylogeny as follows. A study sequence (or set of sequences from a single individual) indicated local transmission if it had at least one close relationship to another study sequence or to a branch that contained only study sequences, without double counting any relationship. We did not speculate on directionality or whether transmission was direct or through some intermediate host.
Similarly, on each phylogeny we counted the number of inward migrations of the virus. A study sequence (or set of sequences from a single individual) was identified as resulting from inward migration if it was distantly related to all other study sequences and to branches that contained only study sequences. Where a group of study sequences from n individuals occurred surrounded by reference sequences, we counted one inward migration plus n-1 local transmissions among the individuals, without speculating which individual migrated.
Sequences that met neither requirement were not classified as local transmission or inward migration, e.g., sequences separated by three nodes or poorly supported phylogenetic relationships.
2.3. Epidemiology
Demographic and risk characteristics of registry cases with any sequence of non-B subtype or CRF were compared to characteristics of cases with only subtype B sequences. Cases missing year of diagnosis were excluded. Associations were assessed using the Pearson Chi square test and multivariable logistic regression. Multivariable models were run with only main terms and with two-way interactions (sex at birth x year of HIV diagnosis, country of birth x year of diagnosis, age at diagnosis x year of diagnosis, sex at birth x country of birth). Interactions with p-values ≥0.10 were eliminated stepwise. Multivariable analyses were performed with SAS version 9.3 proc logistic and included Firth’s bias correction (Heinze, 2006).
We examined how the distribution and overall frequency of non-B subtypes and CRFs changed over year of diagnosis among registry cases. Confidence intervals were calculated using the Wilson score method with continuity correction (Brown et al., 2001). Exact Cochran-Armitage test for trend over year of diagnosis was performed in SAS proc freq.
To assess possible selection bias in cases for whom sequences were available, we compared our sample to all HIV cases on record in the city of San Francisco between 2000 and 2016.
2.4. Study approval
The study received approval from the Institutional Review Board at the University of California, San Francisco.
3. Results
3.1. Sample
The study analyzed a total of 11,381 sequences and 7,235 registry cases with demographic and risk data. LCV contributed 8,459 sequences (2000–2016), 7,746 of which were from 5,394 registry cases. MHS contributed another 2,922 sequences (2014–2016) from 1,841 additional registry cases. The majority of cases (70%) were diagnosed between 1995 and 2012, on average 283 cases per year 1995–2012. Cases with sequences represented 21% of all cases who received care in San Francisco during 2000–2016.
3.2. Prevalence and frequencies
Among registry cases, 7,036 (97%) had a subtype B sequence, 141 (2%) had a non-B sequence or a CRF, including 54 cases (0.8%) with CRF01_AE and 33 (0.5%) with subtype C (Table 1). Seventy-two cases (1%) contributed unique recombinant forms (URFs) of 36 different compositions, most of which involved subtype B, including 10 cases (0.1%) with B-C recombinants and nine (0.1%) with A1-B recombinants (Suppl. table 1). Twelve registry cases had both a subtype B sequence and a sequence of another form. No case had sequences of multiple non-B subtypes or CRFs. Two cases had a non-B subtype or CRF plus a URF. Three cases had URFs of different compositions.
Table 1:
The subtypes and circulating recombinant forms (CRFs) (n=15) observed among sequences collected in San Francisco from 2000 to 2016, and the number of San Francisco HIV/AIDS registry cases that contributed sequences of each subtype or CRF.
| Observed Subtype/CRF | Number (%) of Registry Cases |
|---|---|
| B | 7,036 (97.2) |
| CRF01_AE | 54 (0.8) |
| C | 33 (0.5) |
| CRF02_AG | 17 (0.2) |
| CRF07_BC | 10 (0.1) |
| A1 | 9 (0.1) |
| G | 5 (0.1) |
| D | 4 (0.1) |
| CRF12_BF | <4 (<0.1) |
| CRF06_cpx | <4 (<0.1) |
| CRF17_BF | <4 (<0.1) |
| CRF18_cpx | <4 (<0.1) |
| CRF20_BG | <4 (<0.1) |
| CRF44_BF | <4 (<0.1) |
| F1 | no matched cases |
3.3. Phylogenetics
The seven non-B subtypes and CRF-specific phylogenies included study sequences from 144 individuals. Geographic structure by collection location was apparent among reference sequences. However, most study sequences were interspersed among reference sequences. The most notable exceptions were two groups of closely related subtype C sequences from four individuals (Fig. 1a–b), two groups of closely related CRF01_AE sequences from four individuals (Fig. 1c–d) and one group of closely related CRF07_BC sequences from five individuals (Fig. 1e).
Figure 1:
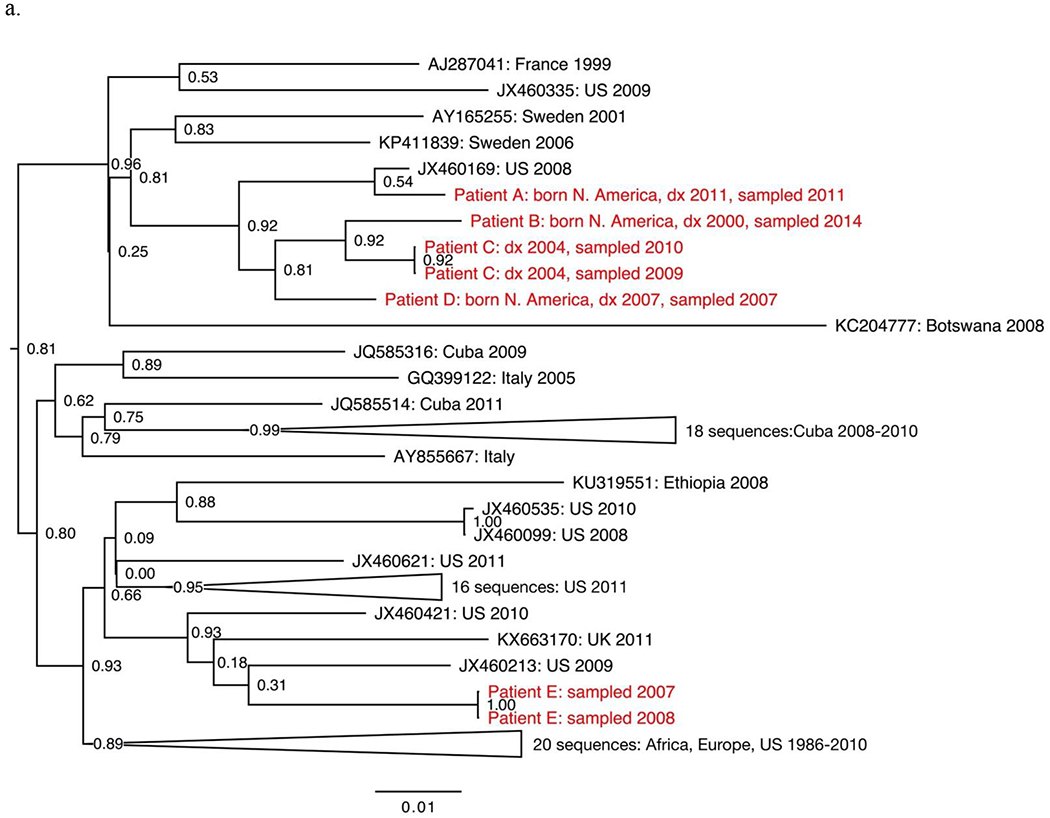
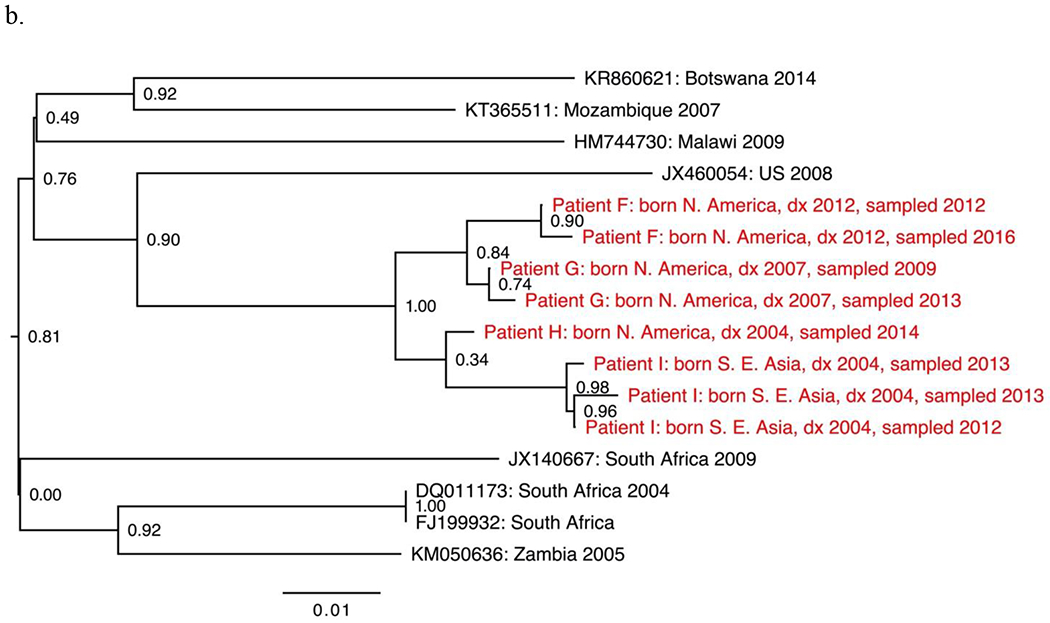
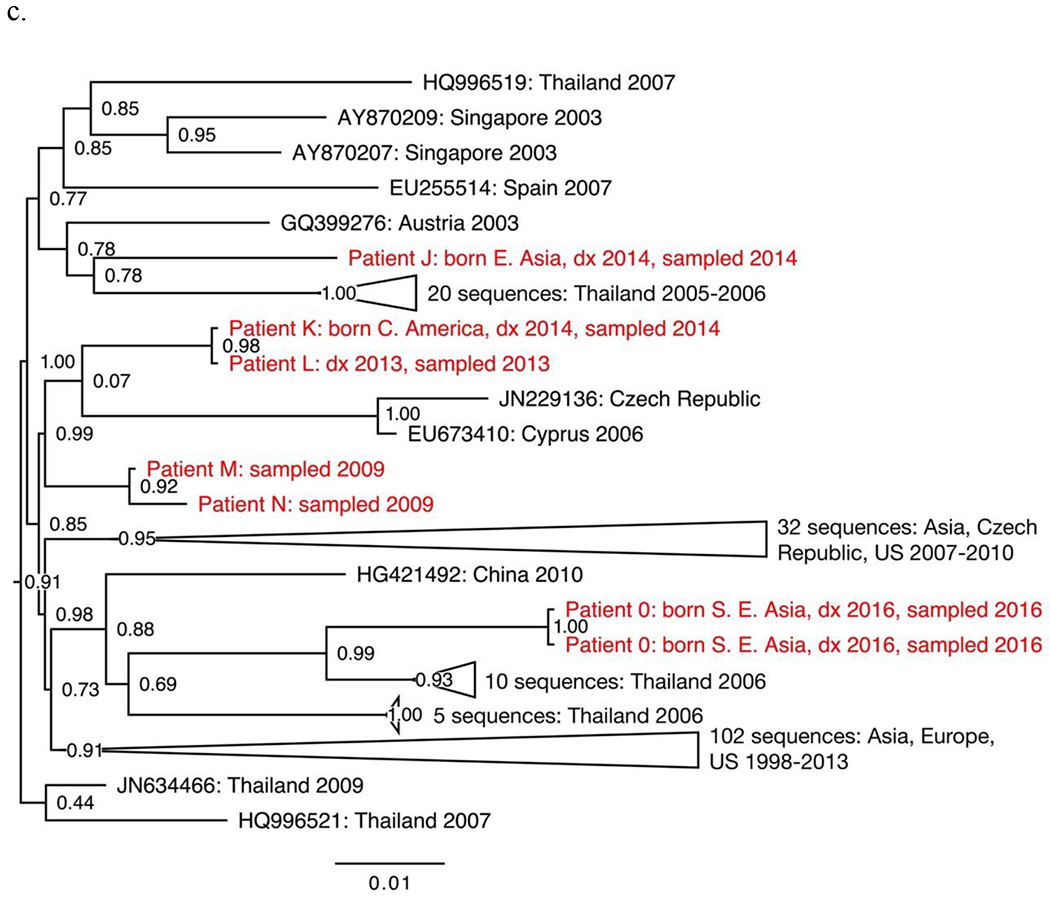
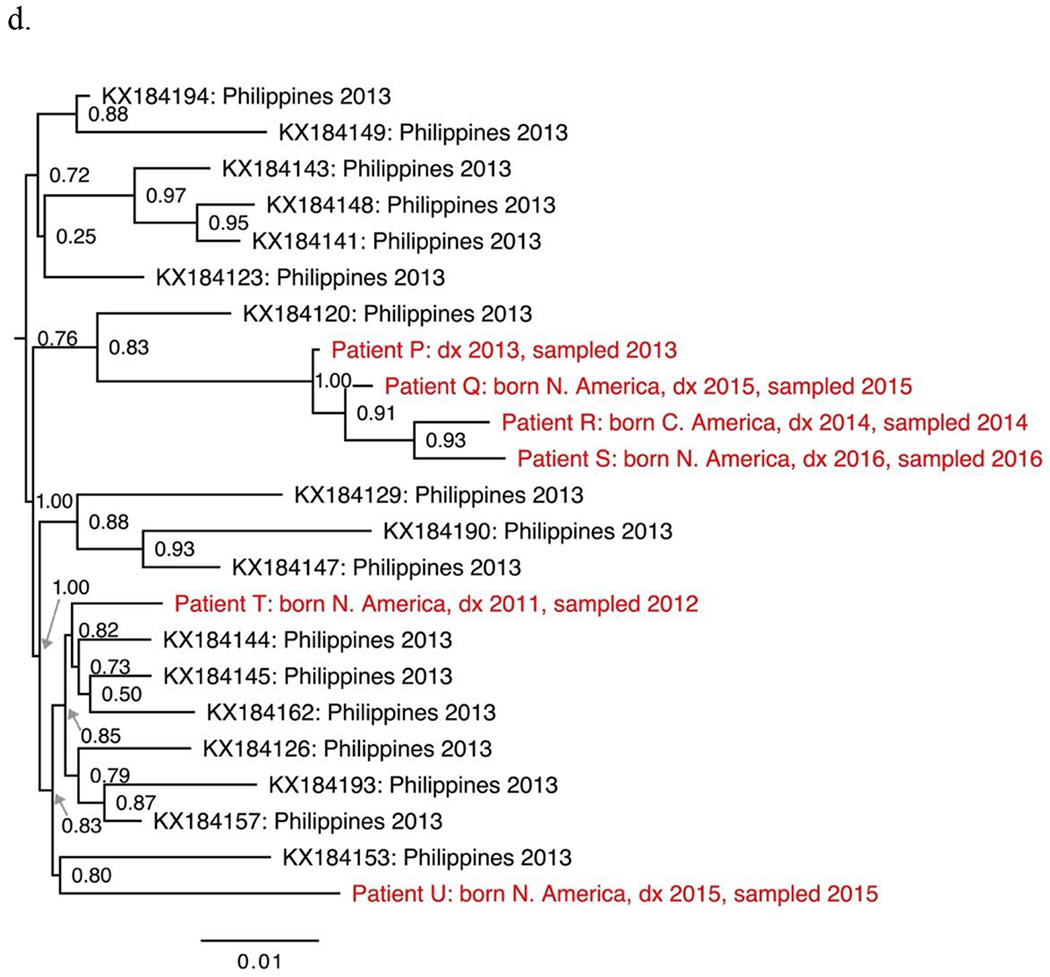
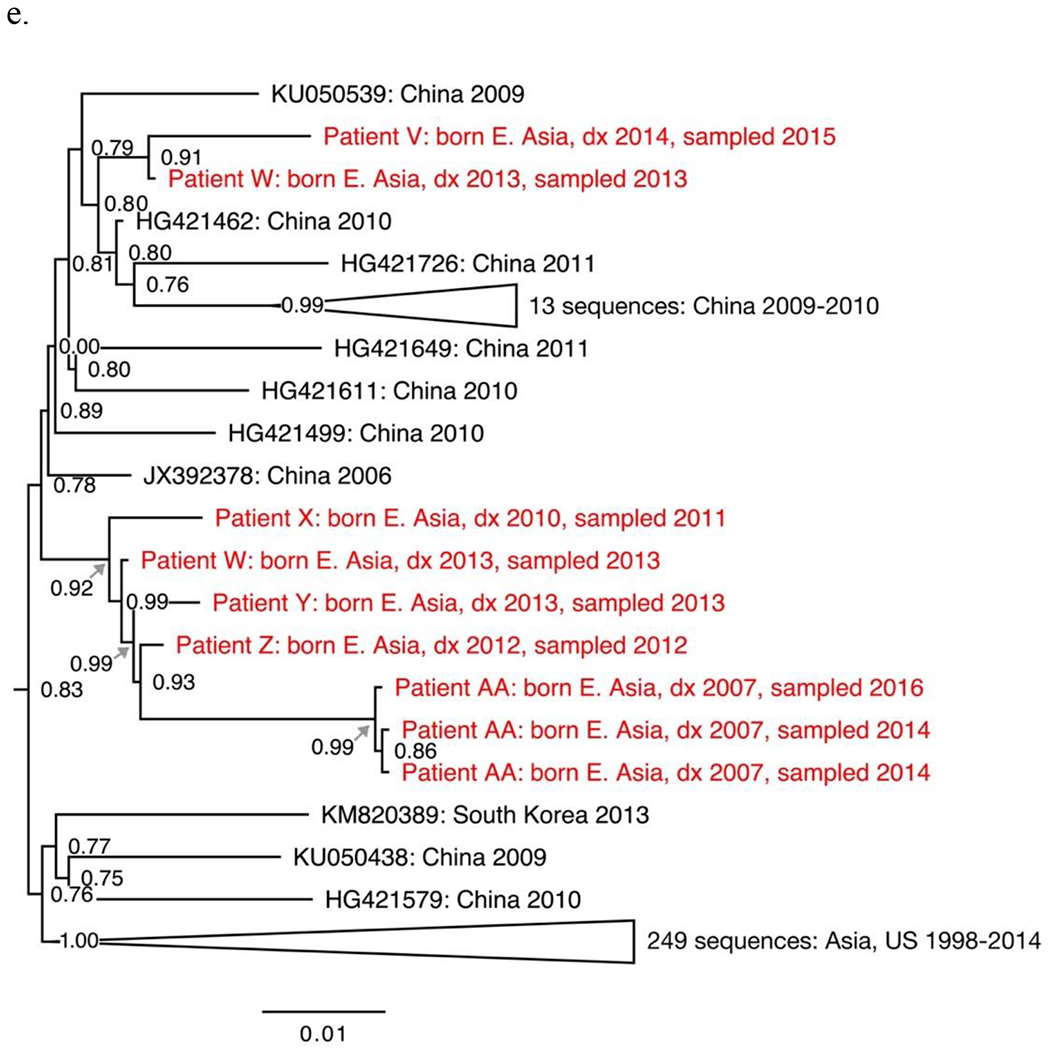
Sections from the phytogenies of HIV sequences of subtype C (a-b), CRF01_AE (c-d) and CRF07_BC (e) sampled in San Francisco during 2000-2016. Relationships between sequences from patients A-D, F-I, and P-S each suggest three local transmissions. Sequences from patients K-N suggest two or three local transmissions. (Two were counted.) Relationships between sequences from patients W-Z and AA suggest four local transmissions. The pair of sequences from patients V and W (top of e) were counted as one local transmission. The solitary position (among reference sequences) of sequences from patients E, J, O, T and U indicate migration from outside the study population.
Phylogenies depicted 146 transmissions, including two transmissions to each of two individuals. We classified 27 transmissions (18%) as likely local transmissions, of which 11 were CRF01_AE (Table 2). Another 108 transmissions (74%) appeared to represent inward migrations of the virus, including 39 of CRF01_AE and 30 of subtype C. The other 11 transmissions (7%) were not classified. Local transmission was indicated in all non-B subtypes and CRFs except D (four inward migrations) and A1 (ten inward migrations).
Table 2:
Number of non-B HIV-1 transmissions by subtype/circulating recombinant form (CRF), classified as local transmissions or inward migrations (or left unclassified) based on phylogenies of viral sequences collected in San Francisco from 2000 to 2016.
| Subtype/CRF | ||||||||
|---|---|---|---|---|---|---|---|---|
| Transmission Type | A1 | C | D | G | CRF01_AE | CRF02_AG | CRF07_BC | Total |
| Local transmission | 0 | 7 | 0 | 1 | 11 | 3 | 5 | 27 |
| Inward migration | 10 | 30 | 4 | 4 | 39 | 15 | 6 | 108 |
| Unclassified transmission | 0 | 3 | 0 | 0 | 7 | 1 | 0 | 11 |
| Total | 10 | 40 | 4 | 5 | 57 | 19 | 11 | 146 |
3.4. Epidemiology
In analyzing patient characteristics, we excluded eight cases (all subtype B) with unknown year of HIV diagnosis. The demographic and risk characteristics of 7,157 cases are presented in Table 3. Among cases with non-B subtypes or CRFs, 24.1% were female, and 58.2% were born outside the US. Asian/Pacific-Islander race/ethnicity (aOR=3.17; 95% CI: 1.84–5.46), black race/ethnicity (aOR=1.84; 95% CI: 1.09–3.12), non-US place of birth (aOR=11.02; 95% CI: 6.64–18.28), and diagnosis after 2009 (aOR=4.81; 95% CI: 3.25–7.11) were associated with having non-B subtype or CRF (Table 4). MSM (aOR=0.25; 95% CI: 0.15–0.44) and people who inject drugs (PWID) (aOR=0.35; 95% CI: 0.20–0.60) were less likely than cases with other transmission risks to have a non-B subtype or CRF. In the interactions model, non-US country of birth was a strong predictor of non-B HIV in both sexes but especially among females (aOR=2.84; 95% CI: 1.00–8.08); diagnosis after 2009 was a strong predictor of non-B HIV, particularly in cases diagnosed at age 50 or older (aOR=12.38; 95% CI: 0.71–217.52).
Table 3:
Demographic and risk characteristics of San Francisco HIV/AIDS registry cases who contributed viral sequences from 2000 to 2016, by subtype/circulating recombinant form (CRF).
| Subtype/CRF | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristic | Only B n (%) |
A1 n (%) |
C n (%) |
D n (%) |
G n (%) |
CRF01_AE n (%) |
CRF02_AG n (%) |
CRF07_BC n (%) |
Other CRF n (%) |
Total non-B n (%) |
Total n (%) |
| Sex at Birth | |||||||||||
| Female | 634 (9.0) | 4 (44.4) | 14 (42.4) | 2 (50.0) | 3 (60.0) | 2 (3.7) | 5 (29.4) | 3 (30.0) | 1 (11.1) | 34 (24.1) | 668 (9.3) |
| Male | 6,382 (91.0) | 5 (55.6) | 19 (57.6) | 2 (50.0) | 2 (40.0) | 52 (96.3) | 12 (70.6) | 7 (70.0) | 8 (88.9) | 107 (75.9) | 6,489 (90.7) |
| Race/Ethnicity | |||||||||||
| Asian/Pacific Islander | 337 (4.8) | 4 (44.4) | 9 (27.3) | 2 (50.0) | 0 (0.0) | 33 (61.1) | 3 (17.6) | 10 (100.0) | 1 (11.1) | 62 (44.0) | 399 (5.6) |
| Black, not Hispanic | 1,432 (20.4) | 1 (11.1) | 14 (42.4) | 1 (25.0) | 4 (80.0) | 2 (3.7) | 9 (52.9) | 0 (0.0) | 1 (11.1) | 32 (22.7) | 1,464 (20.5) |
| Hispanic | 1,593 (22.7) | 1 (11.1) | 2 (6.1) | 0 (0.0) | 0 (0.0) | 5 (9.3) | 0 (0.0) | 0 (0.0) | 1 (11.1) | 9 (6.4) | 1,602 (22.4) |
| White, not Hispanic | 3,333 (47.5) | 3 (33.3) | 7 (21.2) | 0 (0.0) | 1 (20.0) | 12 (22.2) | 4 (23.5) | 0 (0.0) | 5 (55.6) | 32 (22.7) | 3,365 (47.0) |
| Other/Unknown | 321 (4.6) | 0 (0.0) | 1 (3.0) | 1 (25.0) | 0 (0.0) | 2 (3.7) | 1 (5.9) | 0 (0.0) | 1 (11.1) | 6 (4.3) | 327 (4.6) |
| Country of Birth | |||||||||||
| USA | 5,446 (77.6) | 3 (33.3) | 7 (21.2) | 1 (25.0) | 4 (80.0) | 14 (25.9) | 8 (47.1) | 1 (10.0) | 3 (33.3) | 41 (29.1) | 5,487 (76.7) |
| Non-USA | 1,032 (14.7) | 6 (66.7) | 22 (66.7) | 3 (75.0) | 1 (20.0) | 31 (57.4) | 6 (35.3) | 9 (90.0) | 4 (44.4) | 82 (58.2) | 1,114 (15.6) |
| Unknown | 538 (7.7) | 0 (0.0) | 4 (12.1) | 0 (0.0) | 0 (0.0) | 9 (16.7) | 3 (17.6) | 0 (0.0) | 2 (22.2) | 18 (12.8) | 556 (7.8) |
| Transmission Risk | |||||||||||
| Heterosexual sex | 342 (4.9) | 5 (55.6) | 13 (39.4) | 1 (25.0) | 2 (40.0) | 7 (13.0) | 4 (23.5) | 5 (50.0) | 1 (11.1) | 38 (27.0) | 380 (5.3) |
| Injection drug use | 813 (11.6) | 0 (0.0) | 3 (9.1) | 0 (0.0) | 2 (40.0) | 2 (3.7) | 3 (17.6) | 1 (10.0) | 0 (0.0) | 11 (7.8) | 824 (11.5) |
| Male sex with men | 4,058 (57.8) | 4 (44.4) | 9 (27.3) | 1 (25.0) | 1 (20.0) | 42 (77.8) | 8 (47.1) | 3 (30.0) | 8 (88.9) | 76 (53.9) | 4,134 (57.8) |
| Both injection drug use & male sex with men | 1,690 (24.1) | 0 (0.0) | 2 (6.1) | 1 (25.0) | 0 (0.0) | 2 (3.7) | 1 (5.9) | 0 (0.0) | 0 (0.0) | 6 (4.3) | 1,696 (23.7) |
| Other/unknown | 113 (1.6) | 0 (0.0) | 6 (18.2) | 1 (25.0) | 0 (0.0) | 1 (1.9) | 1 (5.9) | 1 (10.0) | 0 (0.0) | 10 (7.1) | 123 (1.7) |
| Age at HIV Diagnosis | |||||||||||
| Under 25 years old | 1,010 (14.4) | 4 (44.4) | 2 (6.1) | 2 (50.0) | 0 (0.0) | 11 (20.4) | 4 (23.5) | 1 (10.0) | 2 (22.2) | 26 (18.4) | 1,036 (14.5) |
| 25-29 years old | 1,316 (18.8) | 2 (22.2) | 6 (18.2) | 0 (0.0) | 1 (20.0) | 12 (22.2) | 4 (23.5) | 0 (0.0) | 1 (11.1) | 26 (18.4) | 1,342 (18.8) |
| 30-39 years old | 2,731 (38.9) | 0 (0.0) | 8 (24.2) | 2 (50.0) | 0 (0.0) | 12 (22.2) | 4 (23.5) | 4 (40.0) | 5 (55.6) | 35 (24.8) | 2,766 (38.6) |
| 40-49 years old | 1,488 (21.2) | 2 (22.2) | 12 (36.4) | 0 (0.0) | 4 (80.0) | 14 (25.9) | 4 (23.5) | 3 (30.0) | 0 (0.0) | 39 (27.7) | 1,527 (21.3) |
| 50 years old or older | 469 (6.7) | 1 (11.1) | 4 (12.1) | 0 (0.0) | 0 (0.0) | 5 (9.3) | 1 (5.9) | 2 (20.0) | 1 (11.1) | 14 (9.9) | 483 (6.7) |
| Unknown | 2 (0.0) | 0 (0.0) | 1 (3.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.7) | 3 (0.0) |
| Year of HIV Diagnosis | |||||||||||
| Before 1990 | 447 (6.4) | 0 (0.0) | 1 (3.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.7) | 448 (6.3) |
| 1990-1999 | 2,192 (31.2) | 0 (0.0) | 2 (6.1) | 0 (0.0) | 2 (40.0) | 2 (3.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 6 (4.3) | 2,198 (30.7) |
| 2000-2009 | 2,813 (40.1) | 4 (44.4) | 13 (39.4) | 2 (50.0) | 2 (40.0) | 11 (20.4) | 6 (35.3) | 2 (20.0) | 1 (11.1) | 41 (29.1) | 2,854 (39.9) |
| 2010-2016 | 1,564 (22.3) | 5 (55.6) | 17 (51.5) | 2 (50.0) | 1 (20.0) | 41 (75.9) | 11 (64.7) | 8 (80.0) | 8 (88.9) | 93 (66.0) | 1,657 (23.2) |
| TOTAL | 7,016 (100.0) | 9 (100.0) | 33 (100.0) | 4 (100.0) | 5 (100.0) | 54 (100.0) | 17 (100.0) | 10 (100.0) | 9 (100.0) | 141 (100.0) | 7,157 (100.0) |
Table 4:
Comparison of demographic and risk characteristics of San Francisco HIV/AIDS registry cases with sequences of non-B subtypes or circulating recombinant forms (CRFs) to characteristics of cases with only subtype B sequences from 2000 to 2016.
| Characteristic | Unadjusted OR (95% CI) | p-value | Adjusted OR (95% CI) | p-value |
|---|---|---|---|---|
| Sex at Birth | ||||
| Male | 1·00 | 1·00 | ||
| Female | 3·20 (2·16, 4·75) | < 0·01 | 1·40 (0·76, 2·57) | 0·28 |
| Race/Ethnicity | ||||
| White, not Hispanic | 1·00 | 1·00 | ||
| Asian/Pacific Islander | 19·16 (12·33, 29·79) | < 0·01 | 3·17 (1·84, 5·46) | < 0·01 |
| Black, not Hispanic | 2·33 (1·42, 3·81) | < 0·01 | 1·84 (1·09, 3·12) | < 0·02 |
| Hispanic | 0·59 (0·28, 1·24) | 0·16 | 0·10 (0·05, 0·23) | < 0·01 |
| Other/Unknown | 1·95 (0·8, 4·69) | n/aa | 1·03 (0·41, 2·58) | 0·94 |
| Country of Birth | ||||
| USA | 1·00 | 1·00 | ||
| Non-USA | 10·55 (7·21, 15·45) | < 0·01 | 11·02 (6·64, 18·28) | < 0·01 |
| Unknown | 4·44 (2·54, 7·79) | < 0·01 | 2·67 (1·49, 4·80) | < 0·01 |
| Sex-Related Transmission | ||||
| Not male-male sex | 1·00 | 1·00 | ||
| Male-male sex | 0·31 (0·22, 0·43) | < 0·01 | 0·25 (0·15, 0·44) | < 0·01 |
| Injection-related Transmission | ||||
| Not injection drug use | 1·00 | 1·00 | ||
| Injection drug use | 0·25 (0·15, 0·41) | < 0·01 | 0·35 (0·20, 0·60) | < 0·01 |
| Age at HIV Diagnosis | ||||
| 50 years old or older | 1·00 | 1·00 | ||
| Less than 50 years old | 0·64 (0·37, 1·13) | 0·12 | 1·10 (0·60, 2·04) | 0·75 |
| Year of HIV Diagnosis | ||||
| Before 2010 | 1·00 | 1·00 | ||
| 2010 or later | 6·75 (4·75, 9·61) | < 0·01 | 4·81 (3·25, 7·11) | < 0·01 |
Chi-square test was not performed for the Other/Unknown category due to a small expected value
Across years of diagnosis, frequency of non-B subtypes and CRFs increased from 0.2% (95% CI: <0.1%–1.4%) before 1990 to 8.2% (95% CI: 6.0%–11.0%) in 2014–2016 (p<0.001), increasing most substantially starting in 2007 (Fig. 2). The rise in frequency of non-B subtypes and CRFs between 2005–2007 and 2011–2013 corresponds to a more than quadrupling of the number of diagnoses of cases with non-B subtypes and CRFs. The relative frequencies of the different non-B subtypes and CRFs shifted across year of diagnosis, with CRF01_AE becoming the most common non-B subtype or CRF in the mid-2000s.
Figure 2:
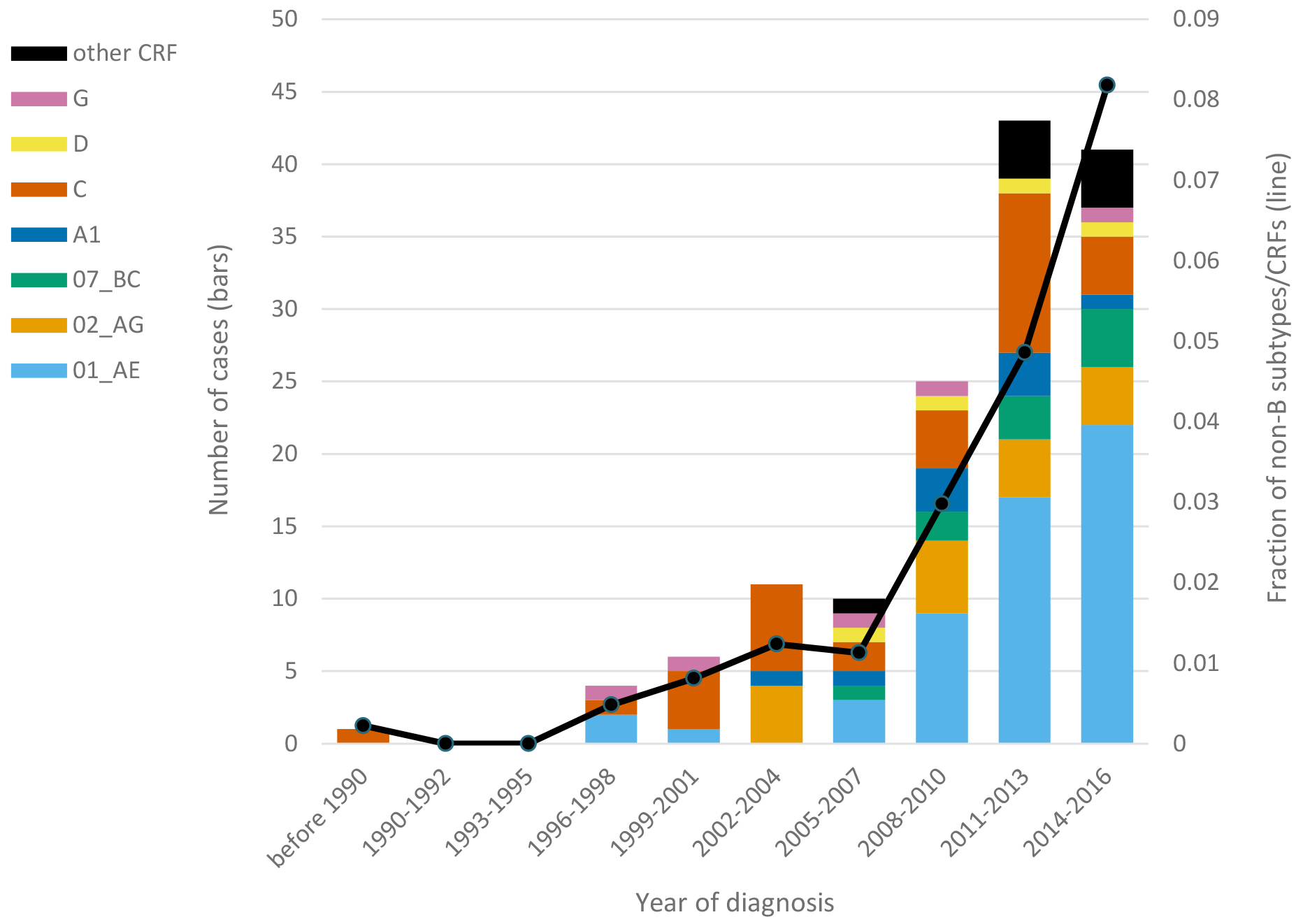
Number (bars) of cases with HIV-1 subtypes and circulating recombinant forms (CRFs) and overall frequency (line) of non-B subtypes and CRFs over year of diagnosis (pre-1990 to 2016) among San Francisco HIV/AIDS registry cases. Data for 2015–2016 were subject to reporting delays.
Compared to all HIV cases on record in the city between 2000 and 2016, our sample had a similar proportion of cases with non-US birthplace but had a higher fraction of cases with black and Hispanic race/ethnicity, PWID and MSM-PWID, cases diagnosed before age 30, and cases diagnosed in 2000 or later (data not shown).
4. Discussion
This study is one of the first detailed examinations of non-B HIV across a US city. Non-B subtypes, CRFs, and URFs were present in 3% of HIV cases in San Francisco. We found evidence of some local transmission of non-B subtypes and CRFs, but no indication of a well-established local non-B epidemic. Non-B subtypes and CRFs were most prevalent among people born outside the US and of Asian/Pacific Islander race/ethnicity, which suggests that the majority of non-B HIV may have come to San Francisco from outside the US. The frequency of non-B subtypes and CRFs has been rising in San Francisco and is probably higher than 6% among recently diagnosed cases.
The overall frequency of non-B variants observed was similar to a previous estimate for California during 2004–2011 but higher than an older estimate for Northern California from years 1997–2000 (Gonzales et al., 2001; Pyne et al., 2013;). The observed frequency of CRF01_AE (38%) among non-B subtypes and CRFs was greater than the 5%-29% reported by other US-based studies (Chan et al., 2014; Dennis et al., 2017; Germer et al., 2015; Oster et al., 2017; Pyne et al., 2013; Sey et al., 2014; Wheeler et al., 2010). CRF01_AE is common in South-East Asia, the origin of about half of San Francisco’s large immigrant population (Angelis et al., 2015; U.S. Census Bureau). The second most common non-B form in San Francisco was subtype C, the most prevalent subtype globally and the most commonly observed non-B subtype in other large US studies (Dennis et al., 2017; Germer et al., 2015; Hamelaar et al., 2011; Oster et al., 2017; Pyne et al., 2013; Wheeler et al., 2010). Subtype C has been associated with African immigrant communities in the US (Chan et al., 2014; Lin et al., 2006; Sides et al., 2005).
About one-third of non-B infections were classified as URFs, higher than the 1%–24% found by other large studies, although methods of identifying recombinants varied (Dennis et al., 2017; Germer et al., 2015; Oster et al., 2017; Pyne et al., 2013). URFs are by definition not known to be widespread and may be very transient (Bbosa et al. 2019). Our analyses focused on non-B subtypes and CRFs, because they are known to be circulating in other geographic regions and are relevant to the question of local transmission versus inward migration of the virus. Future examination of the complex histories of the URFs, would add a further dimension to the local evolutionary dynamics of HIV in San Francisco.
We used phylogenetics to reconstruct relationships between non-B HIV genetic sequences and a large international set of reference sequences in order to examine whether non-B subtypes and CRFs formed local transmission networks within our San Francisco population. Rather than taking a distance-based or cluster-based approach (Hughes et al., 2009; Von Wyl et al., 2011), we presented counts of close topological relationships among San Francisco sequences under the rationale that these are most likely explained by local transmission. Counts of local transmission and inward migration of the virus were intended as a rough gauge of the importance of these two processes, rather than a precise estimate of their occurrence, which may remain elusive until sampling is very complete. We chose a conservative criterion for local transmission, separation by only one or two nodes, to identify probable local transmission. While no standard exists for what branch support is reliable, we considered nodes with SH support values of <0.70 to be unreliable based on our experience comparing likelihood-based phylogenies of small portions of the genetic dataset. Relying only on support values at or above 0.9, still yields 20 (14%) local transmissions.
The lack of extensive local transmission networks of non-B sequences and the predominance of inward migration over local transmission suggest that non-B subtypes and CRFs were largely present in the population due to migration of an infected individual or may have resulted from transmission to or from a person in transit. This contrasts with extensive local transmission, illustrated by a cluster of 29 CRF02_AG, found previously in a sample from several US states (Wertheim et al., 2016). An optimistic inference would be that treatment and prevention programs have succeeded in limiting onward transmission. Alternatively, patients with non-B HIV may be geographically transient or not highly connected to sexual and needle-sharing networks.
Patient demographic and risk characteristics, especially a strong association with non-US birth, suggest that non-B subtypes and CRFs migrated to San Francisco from abroad. The association of non-B subtypes and CRFs with Asian/Pacific Islander and black race/ethnicity is also what one would expect from inward global migration of our most prevalent non-B variants, CRF01_AE and subtype C (Hamelaar et al., 2011). We found non-B subtypes and CRFs in all three major transmission categories (male-male sex, injection drug use, and heterosexual sex). The association of non-B subtypes and CRFs with heterosexual sex, especially compared to prevalent patterns in San Francisco, points to possible links to non-US epidemics dominated by non-B subtypes and more heterosexual transmission (San Francisco Department of Public Health, 2018).
Like other large US studies, ours found increasing frequency of non-B subtypes and CRFs over the last two decades (Dennis et al., 2017; Oster et al., 2017; Pyne et al., 2013). In San Francisco, the trend began before 2010, the year when the federal ban on entry and immigration of persons with HIV/AIDS was lifted (Winston et al., 2011). Increasing frequency was not due to a shrinking sample, as the number of cases with available sequences declined substantially only after diagnosis year 2013, although the total number of HIV diagnoses in San Francisco has been falling since 2008 (San Francisco Department of Public Health, 2018). Furthermore, the rise in frequency was not diminished in 2014–2016 by the inclusion of sequences from private facilities, where we might expect fewer immigrants and racial minorities to receive care.
Our study used available HIV sequences from local public and private healthcare facilities. LCV provided a complete or nearly complete history of resistance testing at public facilities, and sequences were available from more than half of cases diagnosed in 2014–2016. Nevertheless, study sequences represented a minority of the entire local epidemic, as resistance testing was less common before the late 2000s and sequences from private facilities before 2014 were unavailable (San Francisco Department of Public Health, 2018). Sampling bias towards black race/ethnicity and recent diagnosis, both positively associated with non-B subtypes and CRFs, may have caused over-estimation of the frequency of non-B variants. Phylogenies likely did not capture the full membership of local transmission networks. Where transmission partners were missing, we may have underestimated the prevalence of local transmission. Despite this limitation, our results indicate that local transmission of non-B subtypes and CRFs has occurred.
Data security and privacy requirements of SFDPH prohibited web-based sequence analysis. We were able to subtype the sequences using the stand-alone version of COMET. Although no single subtyping program is perfect, COMET has reasonable sensitivity and specificity relative to other programs and also detects likely recombinant forms.
Due to the rarity of non-B sequences, we included in the non-B phylogenies non-B sequences that did not match cases in the SFDPH HIV/AIDS registry, although we had little information about the individuals who contributed these sequences. Most were likely non-residents who received some care in San Francisco public facilities. Some were possibly residents not yet in the registry, while others may have been in the registry under other names, which could result in overestimation of local transmission.
Our analysis aggregated all non-B subtypes and CRFs together. Since the non-B subtypes and CRFs are as different from each other as they are from subtype B, this aggregation was not ideal but necessary due to the small number of registry cases per subtype/CRF. We observed signs of potential differences in characteristics of patients with different subtypes and CRFs. Given a broader set of US data, stratifying the analysis by subtype/CRF might produce important insights.
Our understanding of HIV in San Francisco would be incomplete without an appreciation of the diversity of HIV and the epidemiology of its less-common varieties. Our study investigated the diversity, transmission patterns, and epidemiology of non-B HIV. Results indicate that non-B subtypes and CRFs have been transmitted locally but are not widely established in the San Francisco area. The small and growing presence of non-B subtypes and CRFs is a sign of dynamic connection with HIV in other regions of the world, particularly Asia. San Francisco, however, remains dominated by subtype B, and thus differs from cities worldwide that have already seen a more diverse epidemic. This study provides a first step toward understanding how the HIV landscape in San Francisco fits into the global HIV epidemic.
Supplementary Material
Highlights.
Three percent of HIV infections in San Francisco were non-subtype B variants.
Variants included subtypes A, C, D and G and CRF01_AE, CRF02_AG and CRF07_BC.
Prevalence of non-B subtypes and CRFs increased to 6% of new diagnoses in 2015–2016.
Phylogenetics indicated that about 18% of non-B infections were transmitted locally.
Patient characteristics suggested most non-B infections were acquired outside the US.
Funding Support
This work was supported by National Institutes of Health Grant number R01 MH096642 (PI: H.M. Truong). The funding source were not involved in the conduct of the research and the submission of the article for publication.
Declarations of interest:
Dr. Truong reports receiving grant funding from the US National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Credit Author Statement
K.J.O.: Conceptualization, Methodology, Data curation, Investigation, Formal analysis, Writing – Original draft
S.P.: Data Curation, Investigation, Writing – Review & editing
R.F.: Formal analysis, Writing – Review & editing
S.S.: Resources, Writing – Review & editing
T.L.: Resources. W.F.: Writing – Review & editing
W.M.: Writing – Review & editing
R.M.G.: Writing – Review & editing
H.M.T.: Conceptualization, Funding acquisition, Supervision, Writing – Review & editing
References
- Achkar JM, Burda ST, Konings FA, Urbanski MM, Williams CA, Seifen D, et al. Infection with HIV type 1 group M non-B subtypes in individuals living in New York City. J Acquir Immune Defic Syndr 2004; 36(3):835–44. [DOI] [PubMed] [Google Scholar]
- Aggarwal I, Smith M, Tatt ID, Murad S, Osner N, Geretti AM, et al. Evidence for onward transmission of HIV-1 non-B subtype strains in the United Kingdom. J Acquir Immune Defic Syndr 2006; 41(2):201–209. [DOI] [PubMed] [Google Scholar]
- Angelis K, Albert J, Mamais I, Magiorkinis G, Hatzakis A, Hamouda O, et al. Global dispersal pattern of HIV type 1 subtype CRF01_AE: a genetic trace of human mobility related to heterosexual sexual activities centralized in Southeast Asia. J Infect Dis 2015; 211(11):1735–1744. [DOI] [PubMed] [Google Scholar]
- Bbosa N, Kaleebu P, Ssemwanga D. HIV subtype diversity worldwide. Curr Opin HIV AIDS 2019; 14(3):153–160 [DOI] [PubMed] [Google Scholar]
- Beloukas A, Psarris A, Giannelou P, Kostaki E, Hatzakis A, Parakevis D. Molecular epidemiology of HIV-1 infection in Europe: an overview. Infect Genet Evol 2016; 46:180–189. [DOI] [PubMed] [Google Scholar]
- Brown LD, Cai TT, DasGupta A. Interval Estimation for a Binomial Proportion. Statistical Science 2001; 16(2):101–133. [Google Scholar]
- Carr JK, Osinusi A, Flynn CP, Gilliam BL, Maheshwari V, Zhao RY. Two independent epidemics of HIV in Maryland. J Acquir Immune Defic Syndr 2010; 54(3):297–303. [DOI] [PubMed] [Google Scholar]
- Chan PA, Reitsma MB, DeLong A, Boucek B, Nunn A, Salemi M, et al. Phylogenetic and geospatial evaluation of HIV-1 subtype diversity at the largest HIV center in Rhode Island. Infect Genet Evol 2014; 28:358–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibo D, Birch C. Increasing diversity of human immunodeficiency virus type 1 subtypes circulating in Australia. AIDS Res Human Retroviruses 2012; 28:578–583. [DOI] [PubMed] [Google Scholar]
- Delgado E, Benito S, Montero V, Cuevas MT, Fernández-García A, Sánchez-Martínez M, et al. Diverse large HIV-1 non-subtype B clusters are spreading among men who have sex with men in Spain. Front Microbiol 2019; 10:655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis AM, Hué S, Learner E, Sebastian J, Miller WC, Eron JJ. Rising prevalence of non-B HIV-1 subtypes in North Carolina and evidence for local onward transmission. Virus Evol 2017; 3(1):vex013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilernia DA, Gomez AM, Lourtau L, Marone R, Losso MH, Salomón H, et al. HIV type 1 genetic diversity surveillance among newly diagnosed individuals from 2003 to 2005 in Buenos Aires, Argentina. AIDS Res Hum Retroviruses 2007; 23(10):1201–7. [DOI] [PubMed] [Google Scholar]
- Germer JJ, Wu P, Soderberg JD, Mandrekar JN, Yao JD. HIV-1 subtype diversity among clinical specimens submitted for routine antiviral drug resistance testing in the United States. Diagn Microbiol Infect Dis 2015; 83:257–260. [DOI] [PubMed] [Google Scholar]
- Gilbert MT, Rambaut A, Wlasiuk G, Spira TJ, Pitchenik AE, Worobey M. The emergence of HIV/AIDS in the Americas and beyond. Proc Natl Acad Sci 2007; 104:18566–18570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani M, Montieri S, Palamara G, Latini A, Alteri C, Pemo CF, et al. Non-B HIV type 1 subtypes among men who have sex with men in Rome, Italy. AIDS Res Hum Retroviruses 2009; 25(2):157–64. [DOI] [PubMed] [Google Scholar]
- Gonzales MJ, Machekano RN, Shafer RW. Human immunodeficiency virus type 1 reverse-transcriptase and protease subtypes: classification, amino acid mutation patterns, and prevalence in a northern California clinic-based population. J Infect Dis 2001; 184:998–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Alba JM, Holguín A, Garcia R, García-Bujalance S, Alonso R, Suárez A, et al. Molecular surveillance of HIV-1 in Madrid, Spain: a phylogeographic analysis. J Virol 2011; 85(20):10755–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999; 41:95–98. [Google Scholar]
- Heinze G A comparative investigation of methods for logistic regression with separated or nearly separated data. Stat Med 2006; 25:4216–4226. [DOI] [PubMed] [Google Scholar]
- Hemelaar J, Gouws E, Ghys PD, Osmanov S, WHO-UNAIDS Network for HIV Isolation and Characterisation. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 2011; 25:679–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GJ, Feamhill E, Dunn D, Lycett SJ, Rambaut A, Leigh Brown AJ, et al. Molecular phylodynamics of the heterosexual HIV epidemic in the United Kingdom. PLoS Pathog 2009; 5(9):e1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken C, Thakallapalli R, Esklid A, de Ronde A. Genetic analysis reveals epidemiological patterns in the spread of human immunodeficiency virus. Am J Epidemiol 2000; 152:814–822. [DOI] [PubMed] [Google Scholar]
- Leitner T, Korber B, Daniels M, Calef C, Foley B. HIV-1 subtype and circulating recombinant form (CRF) reference sequences, 2005. HIV Sequence Compendium 2005. Los Alamos (NM): Theoretical Biology and Biophysics, Los Alamos National Laboratory. Available from: https://www.hiv.lanl.gov/content/sequence/HIV/REVIEWS/RefSeqs2005/RefSeqs05.html. [Google Scholar]
- Lin HH, Gaschen BK, Collie M, El-Fishaway M, Chen Z, Korber BT, et al. Genetic characterization of diverse HIV-1 strains in an immigrant population living in New York City. J Acquir Immune Defic Syndr 2006; 41(4):399–404. [DOI] [PubMed] [Google Scholar]
- [dataset] Los Alamos National Laboratory. HIV sequence database and Subtype Reference Alignment 2010. Downloaded 12/19/2016 and 10/20/2016, respectively. Available from: https://www.hiv.lanl.gov/.
- Oster AM, Switzer WM, Hernandez AL, Saduvala N, Wertheim JO, Nwangwu-Ike N, et al. Increasing HIV-1 subtype diversity in seven states, United States, 2006-2013. Ann Epidemiol 2017; 27(4):244–251. [DOI] [PubMed] [Google Scholar]
- Pérez-Alvarez L, Carmona R, Muñoz M, Delgado E, Thomson MM, Contreras G, et al. High incidence of non-B and recombinant HIV-1 strains in newly diagnosed patients in Galicia, Spain: study of genotypic resistance. Antivir Ther 2003; 8(4):355–60. [PubMed] [Google Scholar]
- Pérez-Losada M, Castel AD, Lewis B, Kharfen M, Cartwright CP, Huang B, et al. Characterization of HIV diversity, phylodynamics and drug resistance in Washington, DC. PLoS One 2017; 12(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 2010; 5(3):e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne MT, Hackett J Jr, Holzmayer V, Hillyard DR. Large-scale analysis of the prevalence and geographic distribution of HIV-1 non-B variants in the United States. J Clin Microbiol 2013; 51(8):2662–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallam M, Esbjörnsson J, Baldvinsdóttir G, Indriðason H, Björnsdóttir TB, Widell A, et al. Molecular epidemiology of HIV-1 in Iceland: Early introductions, transmission dynamics and recent outbreaks among injection drug users. Infect Genet Evol 2017; 49:157–163. [DOI] [PubMed] [Google Scholar]
- San Francisco Department of Public Health. HIV Epidemiology Annual Report 2017. 2018. Available from: https://www.sfdph.org/dph/files/reports/RptsHIVAIDS/AnnualReport2017-Green-20180904-Web.pdf.
- Semaille C, Barin F, Cazein F, Pillonel J, Lot F, Brand D, et al. Monitoring the dynamics of the HIV epidemic using assays for recent infection and serotyping among new HIV diagnoses: experience after 2 years in France. J Infect Dis 2007; 196:377–383. [DOI] [PubMed] [Google Scholar]
- Sey K, Ma Y, Lan YC, Song N, Hu YW, Ou Y, et al. Prevalence and circulation patterns of variant, atypical and resistant HIV in Los Angeles County (2007–2009). J Med Virol 2014; 86:1639–1647. [DOI] [PubMed] [Google Scholar]
- Sides TL, Akinsete O, Henry K, Wotton JT, Carr PW, Bartkus J. HIV-1 subtype diversity in Minnesota. J Infect Dis 2005; 192(1):37–45. [DOI] [PubMed] [Google Scholar]
- Struck D, Lawyer G, Ternes AM, Schmit JC, Bercoff DP. COMET: adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res 2014; 42(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 1994; 22(22):4673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong HM, O’Keefe KJ, Pipkin S, Liegler T, Scheer S, Wilson E et al. How are transgender women acquiring HIV? Insights from phylogenetic transmission clusters in San Francisco. AIDS 2019; 33(13):2073–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong HM, Pipkin S, Grant RM, Liegler T, O’Keefe KJ, Scheer S. Increased uptake of early initiation of antiretroviral therapy and baseline drug resistance testing in San Francisco between 2001 and 2015. PLoS One 2019; 14(3):e0213167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong HM, Pipkin S, O’Keefe KJ, Louie B, Liegler T, McFarland W et al. Recent infection, sexually transmitted infections, and transmission clusters frequently observed among persons newly diagnosed with HIV in San Francisco. J Acquir Immune Defic Syndr 2015; 69(5):606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UK Collaborative Group on HIV Drug Resistance. The increasing genetic diversity of HIV-1 in the UK, 2002-2010. AIDS 2014; 28(5):773–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [dataset]U.S. Census Bureau. American Community Survey, tables B05006 2007–2015. Accessed 5/10/2019. Available from: https://factfinder.census.gov.
- Von Wyl V, Kouyos RD, Yerly S, Böni J, Shah C, Bürgisser P, et al. The role of migration and domestic transmission in the spread of HIV-1 non-B subtypes in Switzerland. J Infect Dis 2011; 204(7):1095–103. [DOI] [PubMed] [Google Scholar]
- Weidle PJ, Ganea CE, Irwin KL, Pieniazek D, McGowan JP, Olivo N, et al. Presence of human immunodeficiency virus (HIV) type 1, group M, non-B subtypes, Bronx, New York: a sentinel site for monitoring HIV genetic diversity in the United States. J Infect Dis 2000; 181(2):470–5. [DOI] [PubMed] [Google Scholar]
- Wensing AM, Calvez V, Günthard HF, Johnson VA, Paredes R, Pillay D, et al. 2015 update of the drug resistance mutations in HIV-1. Top Antivir Med 2015; 23(4):132–141. [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Oster AM, Hernandez AL, Saduvala N, Ocfemia MCB, Hall HI. The International Dimension of the U.S. HIV Transmission Network and Onward Transmission of HIV Recently Imported into the United States. AIDS Res Hum Retroviruses 2016; 32(10–11):1046–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler WH, Ziebell RA, Zabina H, Pieniazek D, Prejean J, Bodnar UR, et al. Prevalence of transmitted drug resistance associated mutations and HIV-1 subtypes in new HIV-1 diagnoses, U.S.-2006. AIDS 2010; 24(8):1203–12. [DOI] [PubMed] [Google Scholar]
- Winston SE, Beckwith CG. The impact of removing the immigration ban on HIV-infected persons. AIDS Patient Care STDS 2011; 25(12):709–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


