Abstract
Hydrogels are attractive materials for tissue engineering, but efforts to date have shown limited ability to produce the microstructural features necessary to promote cellular self-organization into hierarchical 3D organ models. Here, we develop a hydrogel ink containing pre-fabricated gelatin fibers to print 3D organ-level scaffolds that recapitulate the intra- and inter-cellular organization of the heart. The addition of pre-fabricated gelatin fibers to hydrogels enables tailoring ink rheology, allowing for a controlled sol-gel transition to achieve precise printing of free-standing 3D structures without additional supporting materials. Shear-induced alignment of fibers during ink extrusion provides microscale geometric cues that promote self-organization of cultured human cardiomyocytes into anisotropic muscular tissues in vitro. The resulting 3D printed ventricle in vitro model exhibited biomimetic anisotropic electrophysiological and contractile properties.
The intra- and inter-cellular organization within cardiac tissues helps facilitate coordinated electromechanical coupling and efficient muscle contraction in the heart. To achieve these organized structures, extracellular matrix (ECM) proteins act as topological and biochemical cues for the cells, which are crucial for tissue development1, 2. This is particularly important in the heart, where the hierarchical organization of cardiac tissues3 is needed to regulate the spatiotemporal dynamics of excitation-contraction coupling, resulting in the cardiac cycle4, 5. However, recreating the structure-function relationships of the heart in vitro represents a significant challenge, requiring control over tissue self-organization both at the cellular scale and at the macroscopic organ level.
In an effort to replicate cardiac cellular organization, researchers have engineered biopolymers and hydrogels through microcontact printing6–10, photolithography11, directional freeze drying12, fiber spinning13–15, and microphysiological chip fabrication4, 16–19. These methods are however limited in their ability to produce tissues on complex geometries that fully replicate cardiac function. Specifically, to build organ models that mimic the 3D geometry requires additive manufacturing strategies15, 20. Particularly, 3D printing has been increasingly used for engineering tissue models for enabling facile design and editing of 3D structure prior to printing, and fabrication of complex 3D structures with flexible formulation of biomaterial inks21–23. To this end, considerable efforts were made in developing hydrogel-based inks or printing techniques to enhance 3D stability and shape fidelity, such as printing multimaterial inks with combined nozzle system24, 25, photo-crosslinkable hydrogels17, 26, 27, adding viscosifiers28–30, or printing in sacrificial supporting materials31–33. However, these efforts provide limited control over tissue self-organization and alignment. As a result, bridging the length scales from the micro- and nanometer scale to the centimeter scale to induce both intra- and intercellular organization within engineered heart models remains a major challenge in 3D printing functional organs and tissues32, 34, 35.
We reasoned that printing 3D scaffolds containing pre-fabricated microscale fibers would provide self-supportive scaffolds for cardiomyocytes to self-organize to form a cardiac chamber. Inspired by the ECM networks of the heart, we designed our ink by infusing gelatin fibers into a gelatin and alginate (Gel-Alg) hydrogel matrix (Fig. 1a). In contrast to cellulose nanofiber and carbon nanotubes, previously used in inks 36–38, fibronectin-coated gelatin fibers contain arginine–glycine–aspartic (RGD) acid peptide binding domains that promote matrix-cell adhesion through integrins39. Furthermore, incorporating fibers into the hydrogel modifies the ink rheology, allowing for creating accurate and complex 3D scaffolds without any supporting structures or materials. We hypothesized that incorporating both topological and chemical cues in the form of aligned gelatin microfibers, 3D ventricle scaffolds printed in this manner would promote the self-organization of cardiomyocytes into anisotropic muscle tissues (Fig. 1b).
Fig. 1. Development of gelatin fiber infused gel (FIG) inks for free-standing 3D printed tissue scaffolds with cellular alignment cues.
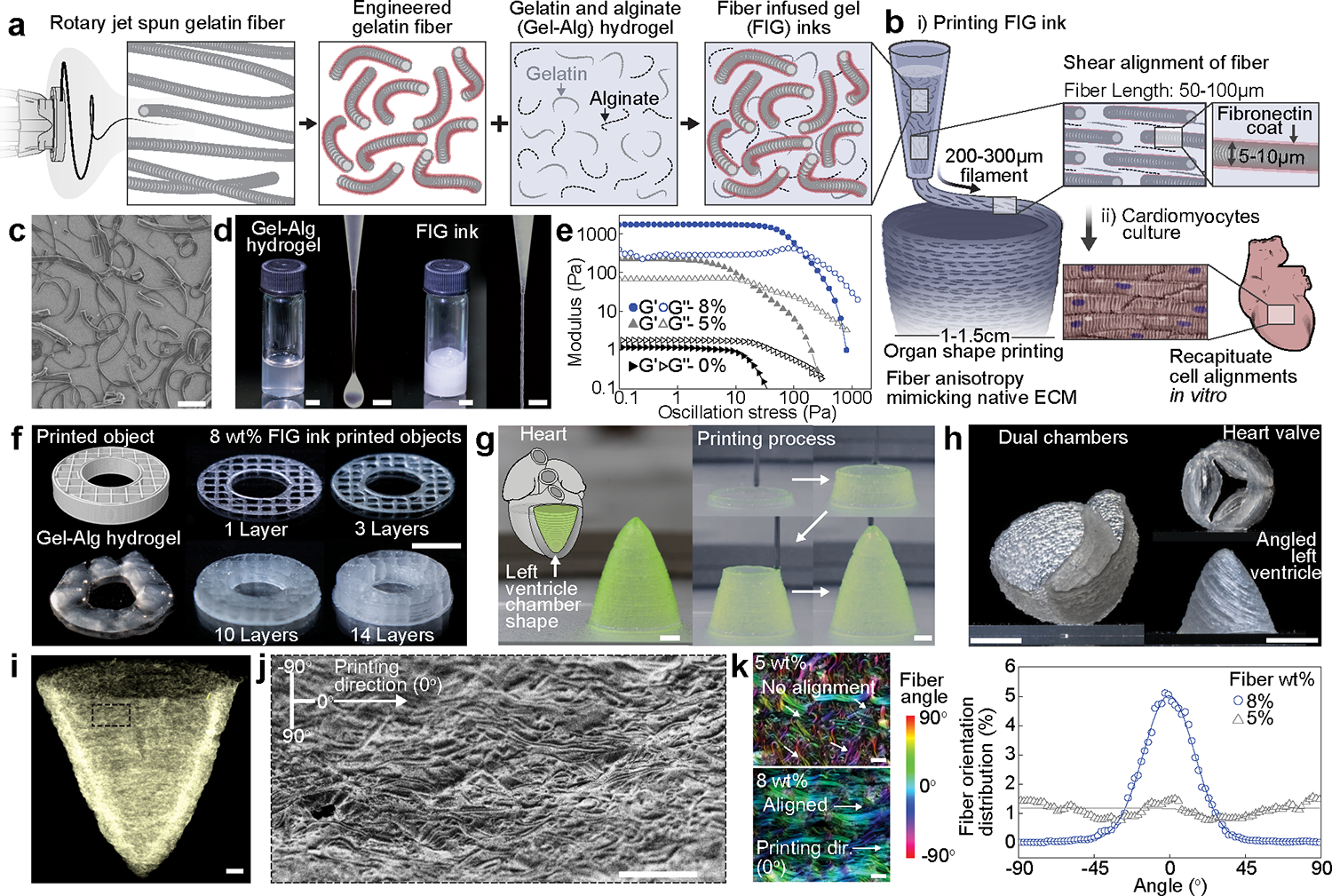
a-b, Schematic illustration of FIG ink components. As fragmented gelatin fibers are combined with Gel-Alg hydrogels (Red, Fibronectin (FN)) (a), the ink viscosity increases and displays a solid-like behavior, which allows for 3D printing of ventricle scaffolds with hierarchical structure (b). Fiber alignment occurred under shear stress (τ) during 3D printing leads to native ECM anisotropic structural features in 3D scaffolds, promoting tissue alignment and organization to recapitulate in vivo heart muscle (b). c, A scanning electron microscopy image showing truncated gelatin fibers. Scale bar, 100 μm. d, Comparison of Gel-Alg ink (0 wt% fiber, left panel) or FIG ink (8 wt% fiber, right panel). The Gel-Alg hydrogel has low viscosity liquid-like behavior (left). FIG inks behave solid-like at rest and extrude in a continuous stream (right). Scale bar, 2mm. e, Oscillation stress sweep test to measure storage (Gʹ) and loss (Gʺ) modulus, showing concentration- and strain-dependent shear thinning behavior and sol-gel transition f, 3D donut shape with rectilinear infill pattern with increasing number of stacking layers, showing high shape retention of using FIG inks. g, A cone-shaped model of the self-supporting inverted left ventricle printed in circumferential direction. Scale bars, 2 mm. h, Self-supportive dual ventricle chambers and a heart valve printed in circumferential direction and an angled left ventricle printed in diagonal (30°) titled direction. Scale bars, 5 mm. i, A micro-computed tomography image of the 3D printed ventricle scaffold after critical point drying, showing fiber structure in the 3D printed geometry. Scale bars, 1 mm. j, A scanning electron microscopy image of the 3D printed ventricle scaffold showing fiber alignments in printing direction. Scale bar, 200 μm. k, Analysis of fiber alignment from confocal images of the 3D printed ventricle FIG scaffolds with 5 and 8 wt% fibers and corresponding fiber orientation angular distribution graph. 0° indicates printing direction. Scale bars, 100 μm.
Results
Printed 3D hydrogel scaffolds provide microscale anisotropy
We fabricated fiber infused gel (FIG) inks for printing 3D scaffolds consisting of gelatin fibers (Fig. 1c) and Gel-Alg hydrogel (Fig. 1d and Supplementary Fig. 1a). Gelatin fibers produced by rotary jet spinning (Supplementary Fig. 1b) 14, 15 were truncated by ultrasonication to a length of 85.9 ± 2.53 μm with a diameter of 4.20 ± 0.23 μm (mean ± SEM), reducing variance in length and enabling passage through the printing nozzle with a diameter of 200 μm (Supplementary Fig. 1c–d). Afterwards, we chemically crosslinked the fibers with N-(3-Dimethylaminopropyl)-Nʹ-ethylcarbodiimidehydrochlorideN-hydroxysuccinimide (EDC/NHS) and fibronectin (Supplementary Fig. 1e). Unlike Gel-Alg hydrogels where 3D printing is limited due to their fluidity, incorporating 8 wt% pre-fabricated gelatin fibers into Gel-Alg hydrogels rendered the hydrogels into a predominantly elastic gel (8 wt% fiber, 2.4 wt% gelatin, 2.4% alginate in PBS, Fig. 1d–f, Supplementary Fig. 2a–d). During printing, shear stresses in the nozzle was sufficient to turn the elastic FIG ink into a liquid, and its elastic stability was restored upon extrusion (Supplementary Fig. 2e). These nonlinear rheological properties allow for continuous extrusion of FIG inks with minimal post-extrusion swelling and spreading (Fig. 1d and e and Supplementary Fig. 3) and enables printing of layered structures for increasingly complex geometries without sacrificial layers/bath as supporting structures (Fig. 1f). This allowed us to print self-supportive hollow 3D structures such as a cardiac ventricle with a broad range of design parameters, reproducibility, and accuracy (Fig. 1g, Supplementary Fig. 4a–b, and Supplementary Video. 1). More complex structures, such as heart valves, dual chambered hearts, and left ventricles with varying angular orientations, could also be printed using FIG inks (Fig. 1h and Supplementary Fig. 4c–d). The high elastic modulus of gel-like FIG inks provided sufficient gel stability to print self-supportive walls with thicknesses ranging between 200 μm and 350 μm without the need for scaffolds or sacrificial baths (Supplementary Fig. 2a–g, and Rheology of FIG inks in Supplementary information).
Inducing cardiac muscle cells to recapitulate structure-function relationships of heart muscle requires structural anisotropy and biochemical guidance cues with micron scale precision. To achieve this, we exploited the shear thinning property of FIG inks (Supplementary Fig. 2h–i), which is not only essential for printing, but also for inducing fiber alignment along the direction of printing40. This allowed for the formation of aligned fiber embedded scaffolds, which can be used to direct cardiomyocyte alignment. Following this process, shear induced fiber alignment on the micron scale was achieved across the entire length scale of the printed structure (Fig. 1i–j). Alignment of the incorporated fibers was well-maintained post extrusion due to the gel stability of FIG inks (Fig. 1k and Supplementary Fig. 5), which was further enhanced by lowering the printing bed temperature to 6°C compared to printing at room temperature (Supplementary Fig. 2f). By ionic crosslinking of alginate (1 wt% calcium chloride) and enzymatic crosslinking of gelatin (~8 wt% microbial transglutaminase), the printed scaffolds and their microscale architecture remained intact in water (Supplementary Fig. 4b). The resulting printed scaffolds had a resolution closer to the fiber diameter (~5 μm) which acted as geometric cues for cell alignment despite the printing resolution being limited by the nozzle diameter (200–300 μm).
Fiber infused gel scaffolds potentiate cellular alignment
To evaluate the ability of FIG printed scaffolds to potentiate intra- and inter-cellular organization of cardiomyocytes, myofibrillar organization, and alignment, we first seeded neonatal rat ventricular cardiomyocytes (NRVMs) on printed 2D FIG scaffolds (8 wt% fiber, 2.4 wt% gelatin, and 2.4 wt% alginate, Supplementary Fig. 6a). This resulted in the formation of highly aligned cardiac tissues (Fig. 2a–b, Supplementary Fig. 6b), which retained comparable cell viabilities relative to pleomorphic culture conditions (Supplementary Fig. 7). We then measured the degree of structural tissue anisotropy by computing the orientational order parameter (OOP),41 which quantifies sarcomeric α-actinin and cytoskeletal F-actin alignment, representative of intra- and inter-cellular organization (Fig. 2c and Supplementary Fig. 6c). In physiologically healthy cardiac tissues, cytoskeletal structures including sarcomeres42, 43 and actin filaments44, 45, are highly aligned with OOP values close to unity. Here, both sarcomeric and F-actin OOP values were significantly higher for tissues on FIG scaffolds (0.578±0.060 for α-actinin, 0.657±0.021 for F-actin, mean ± SEM, n = 5) in comparison to control Gel-Alg scaffolds that contain a similar chemical composition to FIG scaffolds without microscale topologies (4 wt% gelatin and 4 wt% alginate, 0.245±0.033 for α-actinin, 0.314±0.052 for F-actin, mean ± SEM, n = 5). Consequently, the pre-stress generated by intra- and inter-cellular cytoskeletal structures, determined nuclear elongation and orientation46, 47. Nuclei were elongated on FIG scaffolds with a mean eccentricity ratio (length of major axis/length of minor axis) of 2.37± 0.17 compared to nuclei on control Gel-Alg scaffolds that remained relatively round (eccentricity ratio = 1.40±0.02, mean ± SEM, Fig. 2d and e). Nuclear orientation of NRVMs on FIG scaffolds also aligned along the printing direction (0°) in contrast to those on control Gel-Alg scaffolds that showed no specific orientation (Fig. 2d and f, and Supplementary Fig. 6d).
Fig. 2. Anisotropic intra- and inter-cellular organization of cardiac tissues cultured on printed FIG scaffolds.
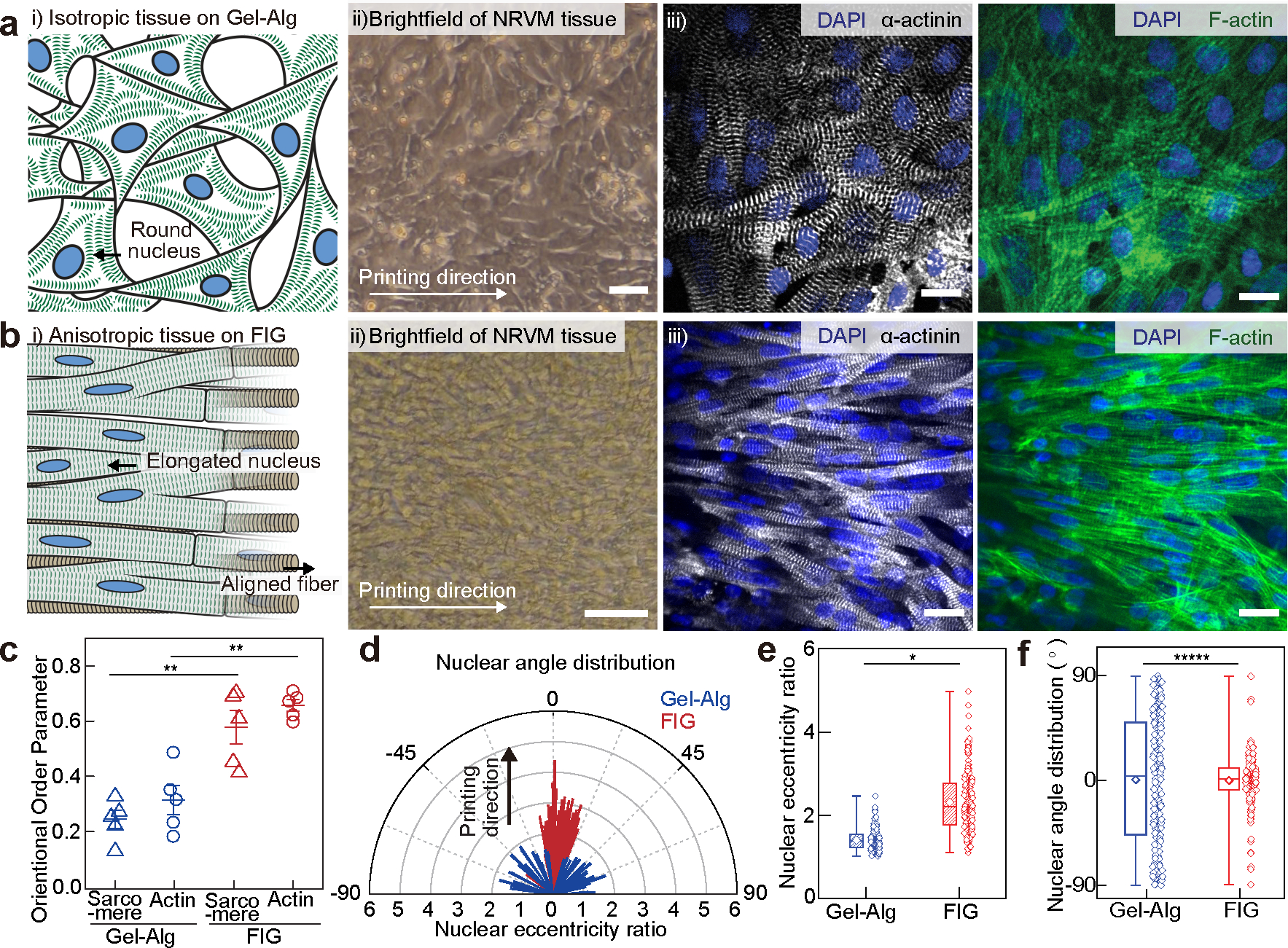
a-b, i) Schematic illustrations showing neonatal rat ventricular myocyte (NRVM) tissue formation on the 2D printed Gel-Alg hydrogel (a) and FIG (b) scaffolds, ii) brightfield images of NRVM cultured on each scaffold, and iii) representative immunostained images of nuclei (blue), α-actinin (grey), and F-actin (green). Scale bars, 20 μm. c, Normalized sarcomeric α-actinin and F-actin alignment with their alignment quantified on a scale of 0 (random) to 1 (aligned) using an orientation order parameter. Statistical analysis was performed using a two-tailed student’s t-test with unequal variance, **P=0.000252 and 0.00149 for sarcomere and F-actin, respectively. n=5 tissues per scaffold condition. Data are presented as mean values +/− SEM. d, Representative distribution of nuclear shape (line length: eccentricity ratio) and orientation (line angle) with printing direction at 0°. e-f, Nuclear eccentricity ratio (e) and angle (f) from −90° to 90° (n = 203 and 183 nuclei from 3 tissues on Gel-Alg and FIG scaffolds, respectively). Statistical analysis was performed using a two-tailed student’s t-test with unequal variance, *P=0.0295 (e) and a two-sample Kolmogorov-Smirnov test, *****P=3.46e-10. (f). n =203 and 183 nuclei from 3 independent tissues on Gel-Alg and FIG scaffolds, respectively. Box plot: center diamond, box limits, and whiskers indicates mean, the first and third quartiles, and max-min, respectively.
Cardiac tissues on FIG recapitulate functional anisotropy
The goal of engineering cardiomyocyte alignment and organization is to better replicate the spatiotemporal excitation and contraction dynamics of the heart in vitro. To test the ability of FIGs to induce cardiomyocytes to self-organize along the fiber structures and form a functional syncytium of the heart5, 48, we measured the functional consequence of cell-cell junction architecture (Supplementary Fig. 6e) and their electrophysiological properties. Cardiac impulses propagate in aligned cardiac tissues approximately two times faster in the longitudinal () than the transverse direction () due to rapid conduction along the longitudinal axis and intercellular conduction at gap junctions49. We used optical mapping to measure calcium (Ca2+) transient propagation as an indicator of cardiac impulse conduction along the longitudinal and transverse axes relative to the printing direction. We assessed anisotropic propagation by calculating the longitudinal to transverse propagation speed ratio (, Fig. 3a–b). Cardiac tissues on FIG scaffolds using NRVMs and human induced pluripotent stem cell derived cardiomyocytes (hiPSC-CM, Supplementary Fig. 8, Supplementary Video. 2) exhibited of 1.67 ± 0.085 and 1.62 ± 0.18, respectively (mean ± SEM, Fig. 3b, Supplementary Fig. 6f–g and 8, and Supplementary Video. 3–4). In contrast, NRVM tissues on control scaffolds without fibers exhibited a propagation speed ratio close to unity (0.96 ± 0.067 at Day 5, mean ± SEM) indicating their isotropy. Cardiac tissues on FIG scaffolds also propagated Ca2+ waves over a 6 cm serpentine pattern, demonstrating robust electrical coupling along the direction of printing (Supplementary Fig. 9 and Supplementary Video. 5).
Fig. 3. Dynamics of electromechanical coupling of multi-directional anisotropic cardiac tissues.
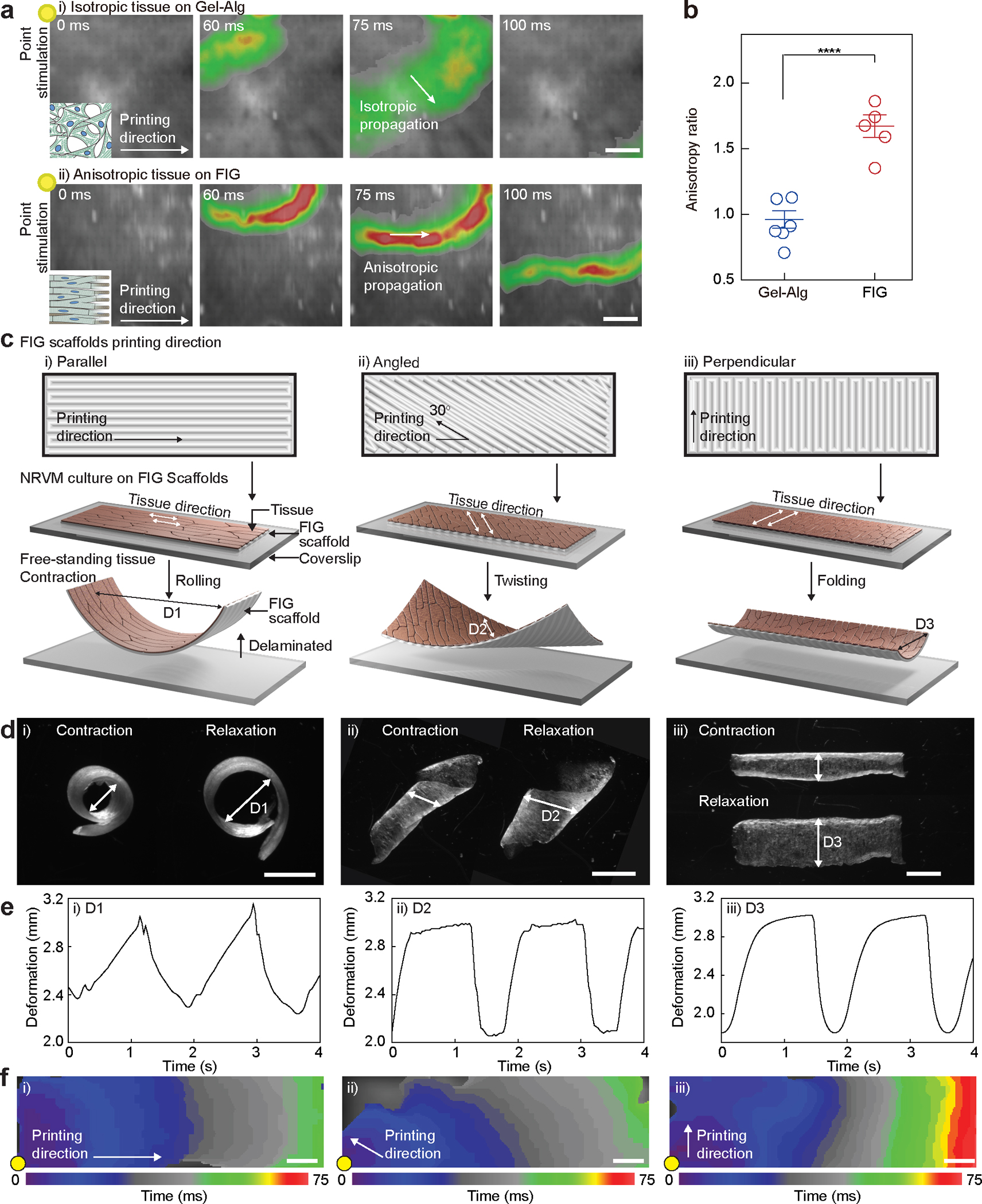
a, Representative Ca2+ transient propagation images from Ca2+ optical mapping of NRVM tissues cultured on Gel-Alg scaffolds (i) and FIG scaffolds (ii), resulting in isotropic (i) and anisotropic (ii) Ca2+ propagation, respectively, under 1 Hz point electrical stimulation at the top-left corner (yellow dot). Scale bar, 2 mm. b, The anisotropy ratio () for NRVM tissues on Gel-Alg scaffolds and FIG scaffolds, quantifying anisotropic Ca2+ propagation. Statistical analysis was performed using a two-tailed student’s t-test with unequal variance. ****P = 0.000174. n=6,5 tissues per scaffold condition. Data are presented as mean values +/− SEM. c,d, Schematic illustration (c) and microscope images (d) of NRVM tissue cultured on rectangular shaped FIG scaffolds printed in parallel (i), angled (ii), and perpendicular (iii) direction to the long side of scaffold geometry. Delaminated free-standing tissue layers from the coverslip showed different contractile motions, rolling (i), twisting (ii), and bending (iii). Scale bar, 2mm. e, Diameter (arrow in (d)) of the deformed shape continuously changes in time under 0.5 Hz field electrical stimulation. f, Isochrone mapping images demonstrating Ca2+ propagation of NRVM tissues on parallel (i), angled (ii), and perpendicular (iii) patterned FIG scaffolds. Point electrical stimulation at 1 Hz (yellow dot) initiates Ca2+ propagation, showing fast propagation in parallel and angled patterns but slow propagation in the perpendicular pattern throughout the scaffold geometry. Scale bar, 1 mm
Further, to assess the effect of fiber anisotropy on the contractile patterns in electrically coupled cardiac tissues, we cultured NRVM on the rectangular scaffolds printed in parallel, angled, and perpendicular directions (10 mm x 3 mm, Supplementary Fig. 10a–c and Supplementary Video 6). Each FIG orientation resulted in distinct contractile patterns such as rolling, twisting, and folding, respectively (Fig. 3c–e). Preferential electromechanical coupling of cardiomyocytes along the printing direction enabled cyclic deformations in response to electrical stimulation (Fig. 3e–f). Alternating alignment directions in one scaffold also showed that cardiomyocyte organization follows the local fiber direction, resulting in corresponding local tissue contractile patterns (Supplementary Fig. 10b). This enables control of local dynamic contractile motions similar to those seen in laminar muscles around the ventricle50.
As FIG scaffolds can be printed in a highly scalable and reproducible manner, they can also easily be adapted to rapidly fabricate previously published standardized metrics of quantifying cellular contractile force via muscular thin films (MTFs) (Supplementary Fig. 10d)9. The average contractile force of Day 7 NRVM tissues on FIG MTFs was 1661.48 ± 209.46 Pa, comparable to NRVMs on polydimethylsiloxane MTFs (1801.66 ± 428.94 Pa, mean ± SEM, Supplementary Fig. 10e–f), and they responded to increasing pacing frequencies up to 3 Hz (Supplementary Fig. 10g). In addition, FIG MTFs enabled us to assess the contractility of hiPSC-CM tissue. We found the contractile stress of hiPSC-CM tissues to be 455.13 ± 130.33 Pa (mean ± SEM) at day 14 (Supplementary Fig. 10d–f), which was significantly lower than the more mature NRVM tissues.
Engineered 3D ventricle models exhibit cyclic contractility
Given the ability to efficiently control tissue alignment and thus, electromechanical coupling, we questioned whether we could build 3D ventricle shaped cardiac tissues in vitro, where the 3D geometry would induce synchronized contractile pumping motions as seen in human hearts. We cultured NRVMs and hiPSC-CMs on a 3D printed ventricle scaffold (8 wt% fiber, 2.4 wt% gelatin, and 2.4 wt% alginate) with circumferential fiber alignment, forming laminar anisotropic cardiac tissues in a 3D ventricular shape (Fig. 4a–b, Supplementary Fig. 11, and Supplementary Video. 7–8). The in vitro ventricle models were maintained for 14 days in culture with a spontaneous beat rate of ~0.71 beats per second. This suggested that the scaffolds were durable over ~800,000 mechanical load cycles without visible degradation. Following point electrical stimulation of NRVMs cultured ventricle models, we observed that Ca2+ transient propagations were faster in the transverse (direction of printing) compared to the longitudinal direction (Supplementary Fig. 12a–b, and Supplementary Video. 9). This confirms that impulse propagation in the 3D printed ventricle scaffolds also occurred along tissue alignment, which is determined by the printing direction. Similarly, in hiPSC-CM cultured ventricle models, directional Ca2+ transient propagations indicated the confluency and synchronization of cardiomyocytes across the 3D printed ventricle scaffolds (Fig. 4a, Supplementary Fig. 12c, and Supplementary Video. 10). This led to synchronized and coordinated contractions of hiPSC-CM cultured ventricle models, which were demonstrated by deformation mapping analysis measuring displacement of the in vitro ventricle models (Fig. 4c–d and Supplementary Video. 11). To assess the contractile performance of hiPSC-CM ventricle models, we measured its fluid-dynamic output at the basal opening (shown in Fig. 4c, grey box) using particle imaging velocimetry (PIV). Fluorescent microparticles flow corresponded with the fluid movement out of and into the in vitro ventricle models (Fig. 4e, Supplementary Fig. 12d, and Supplementary Video. 12). The direction and velocity of particle flow allowed us to estimate the mass flux of fluid movement during contraction and relaxation (Fig. 4f). We calculated our hiPSC-CM based ventricular volumetric change between peak contraction and peak relaxation to be 5.94 ± 1.66 % (n = 4, mean ± SEM, Fig. 4g), which is 2~5 and 8~20 times higher than values previously reported (ejection fraction) for NRVM and hiPSC-CM in vitro ventricle models, respectively13, 15, 32.
Fig. 4. Structural, electrophysiological, and contractile properties of human stem cell-based tissue-engineered 3D ventricle models.
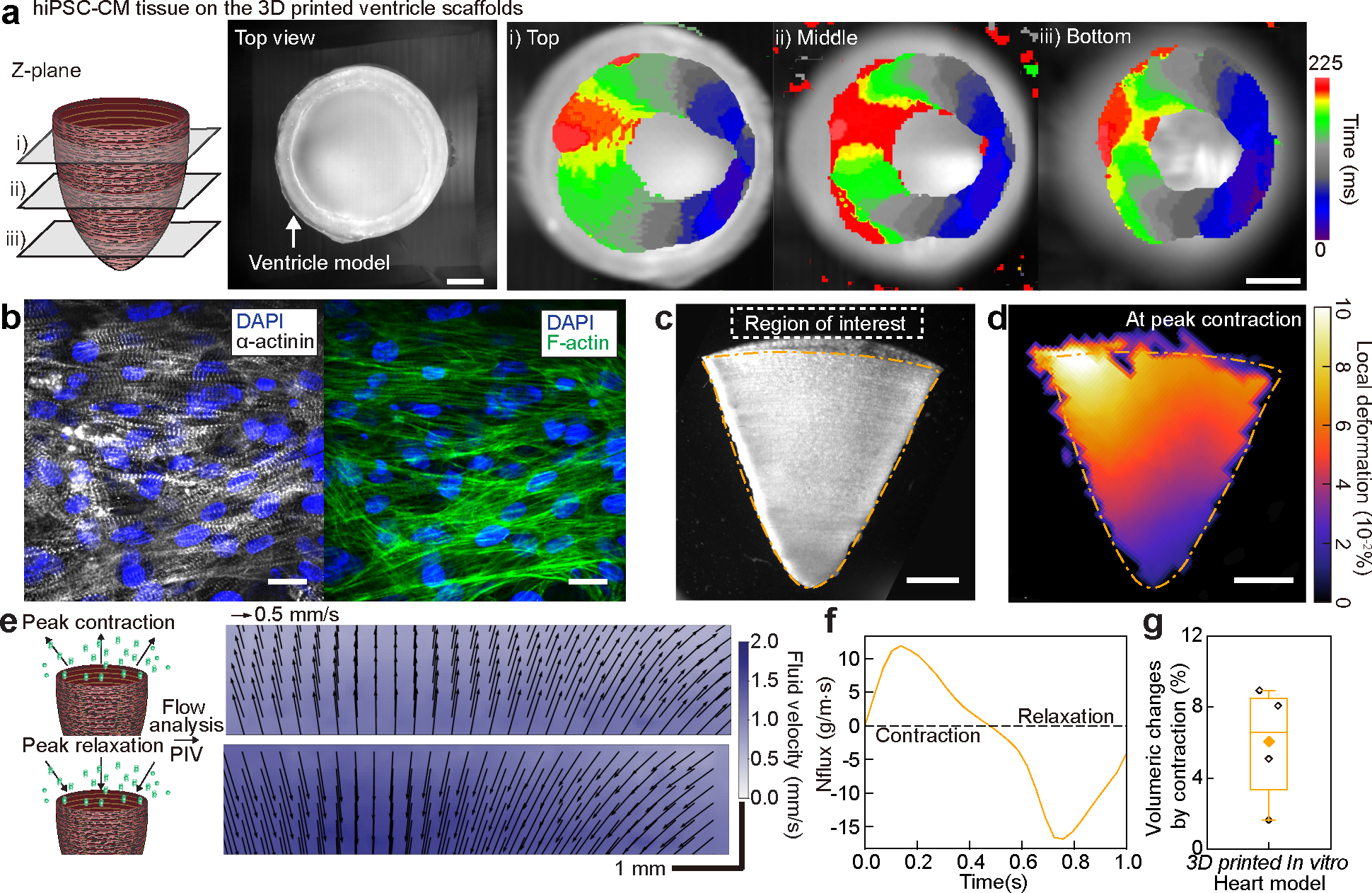
a, Spontaneous Ca2+ transient propagation on a human induced pluripotent stem cell-derived cardiomyocyte (hiPSC-CM) 3D printed ventricle model (top view), showing anisotropic Ca2+ propagation along the printing direction. Scale bar, 2 mm. b, Immunostained images of hiPSC-CM tissues cultured in 3D printed model ventricle scaffolds. Scale bar, 20 μm. c, A brightfield image of the tissue-engineered 3D ventricle model after hiPSC-CMs were cultured for 14 days. Scale bar, 2 mm. d, Representative deformation map of the 3D hiPSC-CM ventricle model at peak contraction. Scale bar, 2 mm. e, Fluid velocity maps at peak contraction (top) and peak relaxation (bottom) analyzed by tracking fluorescent bead displacement in a region of interest under 1 Hz electrical field stimulation. Scale bar, 1 mm. f, Instantaneous mass flux in a region of interest for a representative ventricle during a complete cycle of contraction and relaxation. g, Volumeric changes by contraction of hiPSC-CM 3D ventricle models. n = 4 ventricle models. Box plot: center diamond, box limits, and whiskers indicates mean, the first and third quartiles, and max-min, respectively.
Outlook
We have demonstrated the ability to 3D print tissue-engineered ventricle models with hydrogel-based FIG ink, which simultaneously recapitulates the heart’s microstructural ECM architecture and macrostructural organ-level geometry. Here, infused gelatin fibers acted as a rheological modifier in our hydrogel inks, allowing for the printing of complex 3D objects without the use of sacrificial baths. In addition, these fibers also provided biochemical and microstructural cues, which promoted cell adhesion and self-organization into a functional syncytium. Resulting in vitro ventricle models showed cyclic contractile motions of the heart, however, to enhance cardiac performance, multilayered tissue ventricle models are required in the future. To that end, FIG inks and its biochemical and structural markers can be further investigated to promote cell infiltration, and vary local muscle direction, fabricate multilayered tissues, and integrate multiple cell types, for example, to achieve vascularization. This work suggests that 3D FIG ink printing will play an important role in promoting standardized and accessible bioprinting processes to recapitulate native organ geometries with microstructural precision.
Method
Preparation of gelatin fiber filler
Gelatin fibers were produced by the focused rotary jet spinning method as previously described in more detail 15. Gelatin (type A, gel strength ~300 g Bloom, Sigma Aldrich) was dissolved in 1,1,1,3,3,3-hexafuoro-2-propanol (HFIP; Oakwood Chemical 003409) at 5 w/v%. The gelatin/HFIP solution was then injected into a customized spinneret at 1.8 mL/min using an automated syringe pump (Harvard Apparatus, part No. 703007). The spinneret was rotated at 10,000 rpm, extruding the gelatin solution out of three 400 μm holes into long fiber streams which were collected on a rotating collector (Heidolph Hei-Torque Core) at 300 rpm (Supplementary Fig. 1a, i). Distance from the spinneret to the collector was 15–20 cm, allowing enough time between the spinneret and collection for HFIP evaporation and fiber formation. Fibers were pre-cut to about 1 mm using an array of razor blades and dispersed into anhydrous ethanol at 0.004 g/ml. The pre-cut fiber solution (40 mL total) was placed in an ice bath and underwent ultrasonication (FB-505, Fisher Scientific, USA) to fragment further the gelatin fibers (Supplementary Fig. 1a, ii). The length of the gelatin fibers can be controlled by power amplitude and duration of sonication. The 13 mg/ml of gelatin fibers were then crosslinked with a 5:2 molar ratio of N,N-(3-dimethylaminopropyl)-N’-ethyl-carbodiimide hydrochloride (EDC, Sigma Aldrich) and N-hydroxysuccinimide (NHS, Sigma Aldrich) in anhydrous ethanol for 3 hours while stirring at 260 rpm (Supplementary Fig. 1a, iii). After the gelatin fibers were washed with water and separated by centrifuge, the fibers were immersed in a solution containing fibronectin protein (50 μg/ml fibronectin, Corning), 2 μg/ml EDC (Thermofisher Scientific, USA), and 5.5 μg/ml N-hydroxysulfosuccinimide (sulfo-NHS, Thermofisher Scientific, USA) diluted in PBS and incubated overnight at 4°C (Supplementary Fig. 1a, iv). Fluorescent fibronectin (Green Fluorescent, HiLyte 488, Cytoskeleton, Inc.) was used to confirm fibronectin coating on the fibers (Supplementary Fig. 1e). The fibers were then washed and centrifuged with DI water and 2x concentrated PBS (2x PBS) (Supplementary Fig. 1a, v–vi). 2x PBS was made of 16 mM NaH2PO4, 272mM NaCl, 4mM KH2PO4, and 5.2mM KCl in DI water. A schematic illustration of the procedure to prepare fiber fragmentation materials is described in Supplementary Fig. 1a. Fiber length distribution was measured from bright-field images taken by 10x EVOS FL microscope and diameter distribution was measured from confocal microscope (Olympus ix83, USA) images that were captured by LUCPlanFLNPh 20× objective.
Fabrication of FIG inks and crosslinking process.
FIG inks were prepared by mechanically mixing 5–10 wt% pre-fabricated fibers into a 65 °C prewarmed solution consisting of 2.4 wt% of gelatin (type A, gel strength ~175 g Bloom, Sigma Aldrich) and 2.4 wt% of sodium alginate (Sigma Aldrich) in the 2x PBS (Supplementary Fig. 1a, vii). After mixing thoroughly, the contents were transferred to a 3 ml syringe and centrifuged at 450 g for 2 mins to remove air bubbles before printing (Supplementary Fig. 1a, viii). FIG inks were printed using a 3D bio-printer (Bio X™, Cellink, USA). 2D FIG scaffolds were prepared ~100 μm thickness. After printing, FIG scaffolds were treated with a 1 wt% calcium chloride (Sigma, USA) solution for 5 minutes to crosslink alginate and a 2–8 wt% microbial transglutaminase (Activa TI Transglutaminase, Ajinomoto Co., Japan) solution for 1 hour at room temperature to crosslink the printed FIG scaffolds. The printed scaffold was then treated with 0.1M ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA, Sigma Aldrich, USA) solution for 15 mins at room temperature. The Gel-Alg hydrogel inks with 4 wt% gelatin/4 wt% alginate in the 2x PBS were used for printing the 3D printed control models in Fig. 1–3 and Gel-Alg hydrogel inks with both 4 wt% gelatin/4 wt% alginate and 2.4 wt% gelatin/2.4 wt% alginate in the 2x PBS were used to compare line printability and cell alignment in Supplementary Fig. 3 and 6a, respectively. All post-procedures after printing were the same as FIG printed scaffolds.
Rheological test of FIG inks
The rheological properties were measured using a Discovery Hybrid 3 Rheometer (TA instrument, USA). 0% fiber condition used as a control ink (without fiber) for rheology in Fig. 1e and Supplementary Fig. 2 was prepared with 2.4 wt% of gelatin and 2.4 wt% alginate in the 2x PBS. A 60 mm cone geometry for 0% Gel-Alg inks and a flat plane geometry for FIG inks were used. The plane geometry experimental setups used a 500 μm gap height. Viscosity was measured by a shear rate sweep from 0.1 to 1000 1/s (Supplementary Fig. 2h). An oscillation amplitude sweep was performed by a shear stress sweep from 0.1 to 1000 Pa at a frequency of 1 Hz (Supplementary Fig. 2b), and the yield stress was evaluated and determined at the cross over point of the storage and loss moduli (Supplementary Fig. 2d). The yield stress for the 0% fiber hydrogel inks could not be quantified using the oscillation amplitude sweep tests as the value ranges were too low. Therefore, the yield stress for the 0% fiber hydrogel inks was calculated using the stress-shear rate graphs (Supplementary Fig. 2c) which showed a plateau region as the shear rate decreased. The cyclic strain test was measured with oscillatory strain change between 1% and 100% every 1 minute at a frequency of 1 Hz and stress of 0.1 Pa (Supplementary Fig. 2e). All measurements were performed at 25°C. The temperature sweep test was performed from 10°C to 40°C with 2°C steps at a frequency of 1 Hz and stress of 0.1Pa (Supplementary Fig. 2f).
3D printing
G-code file, the computational language used to guide 3D printer nozzle pathways, was prepared by slicer software (slic3r), 3D bio-printer (Bio X™, Cellink, USA), or hand-script. Gel-Alg hydrogel and FIG scaffolds/patterns were printed onto petri-dishes or gelatin layer coated coverslips by a direct-ink-writing method. The gelatin layer coated coverslip was used to enhance adhesion between printed scaffolds and coverslip. It was fabricated by printing 14 mm by 14 mm mesh square onto coverslips, ultraviolet-ozone (UVO) treated for 5 mins, using 4 wt% gelatin (type A, gel strength ~175 g Bloom, Sigma Aldrich) solution. The printing patterns were designed by hand-script G-code, MATLAB (MathWorks) script generated G-code, or 3D bio-printer (Bio X™, Cellink, USA). The printing line distance for 2D scaffolds was set to 0.3 mm. STL files of 3D free-standing objects were processed and printed by the 3D bio-printer with a layer height of 0.2 mm for 23G straight nozzles (Inner diameter, I.D. 0.337mm) and 0.15 mm for 27G tapered nozzles (I.D. 0.21mm). The G-code for the 3D cone shape ventricle chamber was prepared by 3D coordination of cone shape derived from parabola equations (Supplementary Fig. 4a) with a layer height of 0.2 mm using MATLAB. The G-codes were visualized with software (Repetier-Host). The printing condition for the scaffolds was set at a range of 50–100 kPa of printing pressure and 5–20 mm/s of printing speed with 23G straight or 27G tapered nozzles. 3D free-standing objects were printed with 70–120 kPa of pressure and 5–10 mm/s of printing speed using 23G straight nozzles and 27G tapered nozzles at 6°C of printing bed temperature.
Scaffold structural and biochemical imaging analysis
Gelatin fiber structure in the printed 3D FIG ventricle scaffold was captured by a spinning disk confocal microscope (Olympus ix83, USA) with UPLSAPO 10x objective after staining the gelatin fibers using 1μl/ml of NHS-Fluorescein (Thermofisher Scientific, USA). Fiber alignments were analyzed by ImageJ Plugin (Orientationl J) with 2° bin size and colorized the images based on fiber angle 51. The colorized images were then processed using a custom MATLAB script to output an orientation order parameter value, falling on a normalized scale of 0 (perfectly random) to 1 (perfectly aligned), based on the distribution of reported angles from the OrientationJ processed image41. Scanning Electron Microscopy images (FESEM Ultra Plus, SmartSEM, Zeiss, Germany) and Micro-CT (Xradia Versa 620, Zeiss, Germany) images of 3D printed scaffold were taken after dehydration using a critical-point dryer (931 GL 2.5, Tousimis, USA). Micro-CT images of 3D structures were imaged by Versa 620 X-ray Microscope (Carl Zeiss, Inc., Pleasanton, California, USA) with a tube voltage of 50 kV and current of 90 μA microfocus X-ray source using 4x objective with wide-field mode at Harvard University’s Center for Nanoscale Systems (CNS). 4501 projection images were captured per sample on a 16-bit, 2048 × 2048 4X objective detector with achievable voxel resolutions of 3.19 μm. 3D data visualization was performed using Dragonfly Pro software.
NRVM and iPSC-CM cell culture
Neonatal rat ventricular myocytes (NRVMs):
The animal protocol numbered 24–01-2 is approved by the Harvard Animal Care and Use Committee and is based on previously published methods9. Ventricular tissue was removed from two-day-old Sprague-Dawley rats. The tissue was minced by scissors, rinsed in Hank’s Balanced Salt Solution, and digested in 1 mg/ml trypsin in HBSS solution at 4°C for 14 hours overnight. Next day, the tissue was washed in customized media (details below) at 37°C for 4 minutes. The tissue was then further dissociated using 1 mg/ml collagenase in HBSS solution at 37°C for 2 minutes. Cells dissociated from the tissue were collected in a pre-chilled tube containing fresh HBSS without collagenase to quench the digestion. The collagenase step was repeated a total of four times until most of the tissue was digested. The dissociated cells were centrifuged at 250 g for 8 minutes, resuspended in chilled HBSS and filtered through a 40 μm strainer to remove any remaining bulk tissue. The cells were then centrifuged again at 250 g for 8 minutes and plated in a T175 flask for 2 hours and 15 minutes to remove the fast-adhering fibroblasts. The non-adhering cells were collected as the primary NRVMs and plated on tissue-engineered scaffolds. Culture media for NRVM were Medium 199 supplemented with 0.01M HEPES Buffer Solution, 1% MEM non-essential amino acids, 20 mM Glucose, 2mM L-Glutamine, 1.5μM of Vitamin B12, 50 U/ml Penicillin. 10 % heat-inactivated fetal bovine serum (FBS) were supplemented to the media for the first two days and 2% FBS supplement were used starting the 3rd day and changed every other day onwards.
Human induced pluripotent stem cell cardiomyocytes (hiPSC-CMs) culture:
hiPSCs (WTC-11, Coriell Institute, GM25256) were seeded onto Geltrex (LDEV-free matrix, Thermofisher Scientific)-coated 6 well plates and maintained in Essential 8 (E8) medium. At 60~70% confluency, hiPSCs were passaged using Versene (Life Technologies, USA) into 6 well plates for maintenance and 12 well plates for hiPSC cardiomyocyte (hiPSC-CMs) differentiation. 5 μM Y-27632 (R&D) added E8 medium was used on the first day of passage. The differentiation process for hiPSC-CMs was conducted for 15 days following published protocols52. Differentiation started (Day 0) at 60~70% confluence using 6 μM CHIR909921 (STEMCELL Technologies) with a differentiation medium consisting of RPMI-1640 (Life Technologies) and a B-27 minus insulin supplement (RPMI/B27(−)) after short rinse with PBS. On Day 2, the medium was replaced with the differentiation medium for 24 hours and changed to the differentiation medium with 5 μM IWR-1 (STEMCELL Technologies) (Day 3). After 48 hours of incubation and a quick PBS rinse, the differentiation medium was replaced 48 hours. On Day 9, cardiomyocytes selection was conducted using 0.4 mM Lactate (# L7022, Sigma Aldrich)) added the differentiation medium for 48 hours and 0.4 mM Lactate added RPMI-1680 for the next 48 hours. After cardiomyocyte selection, cells were cultured in the differentiation medium until hiPSC-CMs isolation on Day 15–17. After visual confirmation of cardiomyocytes contractility (Supplementary Video. S2), hiPSC-CMs were isolated after incubating in Accutase (STEMCELL Technologies) for 30 mins. Isolated hiPSC-CMs were seeded using STEMdiff™ Cardiomyocyte Support Medium (STEMCELL Technologies). The culture medium was changed to RPMI/B27(−) every other day.
Cell culture on the printed FIG scaffolds
Before cell seeding, printed scaffolds were sterilized with a 70 % ethanol wash for 5 min and UVO exposure for 4 min. Subsequently, scaffolds were incubated with 50 μg/ml solution of fibronectin (BD Biosciences) in phosphate-buffered saline (PBS; Gibco, Thermo Fisher Scientific, USA) at 37 °C for 1 h. After the fibronectin solution was washed with PBS, the 2D scaffolds were seeded with 2M NRVM and hiPSC-CM at 12 well plates. For 3D printed ventricle chamber seeding, we inserted a printed ventricle scaffold into a PDMS mold made by casting the same 3D printed ventricle plastic model (VEROCLEAR, Object30, Stratasys, USA). Considering the cells are less likely to be evenly distributed in the 3D structure, NRVM and hiPSC-CMs were seeded for 3D printed ventricle models with 5M and 8M and turned the ventricles upside down after 1 hr and 0.5 hr later, respectively.
Tissue immunostaining and structural analysis
For Live/dead cell staining, we cultured NRVM on the fibronectin-coated glass coverslips and the FIG scaffolds with 0.5 M/cm2 for 96 hr. Live/dead cell stained images were captured by a confocal microscope (Olympus ix83, USA) with LUCPlanFLNPh 20× objective after incubating cells for 5 minutes at 1 μg/ml of Hoechst 33342 in media for live cell staining and ReadyProbes™ Cell Viability Imaging Kit, Green (1~2 drops in the 1ml of media, ThermoFisher) for dead cells staining. Tissue structure analysis was conducted using confocal microscope immunostaining images of NRVM tissue cultured on the scaffold for 12 days and ventricle for 7–12 days. Washed samples were fixed with 4 v/v% paraformaldehyde in PBS for 15min and permeabilized with 0.1 v/v% Triton-X 100 solution in PBS for 15 min at room temperature. The samples were then incubated in 5 w/v% bovine serum albumin (BSA) solution in PBS for 30 mins at room temperature. Samples were then incubated overnight with 1:100 diluted monoclonal sarcomeric α-actinin (clone EA-53; abcam) or 1:100 Anti-Connexin-43 antibody produced in rabbit (Sigma-Aldrich, C-6219) primary antibody in 1 wt% of BSA solution at 4°C. After being washed three times in 0.5 wt% of BSA solution for 5 mins, the samples were counterstained with 1:200 diluted Alexa Fluor 546-conjugated anti-mouse secondary antibody (Invitrogen, A-11003), 1:1000 diluted Alexa Fluor 633-conjugated Phalloidin (Life Technologies, A22284), and 1:500 diluted DAPI (Invitrogen) in 1 wt% of BSA solution for 2 hrs at room temperature. After the samples were washed three times with 0.5 wt% of BSA solution, they were mounted on the glass to observe immunostaining images using a spinning disk confocal microscope (Olympus ix83, USA). Images were captured by LUCPlanFLNPh 20× and 40× objectives on a Hamamatsu Orca Flash 4.0 C11440 at 16-bit depth. Background subtraction and z-stack max projection were applied for displayed confocal microscope images in figures using Image J software. We used 40x confocal microscope images with 1024 × 1024 pixels to quantify nucleus alignment, eccentricity ratio, and sarcomere and F-actin alignments after rotating those images to position the print orientation horizontally. We analyzed the angle and length of nuclei in the major and minor axis using Image J plug-in (Analyze particle). We subtracted 90 degrees from the angle results to ensure that the print direction was oriented towards 0 degrees. The eccentricity ratio was calculated by dividing the length of the major axis by the length of the minor axis. Sarcomere and F-actin images were processed with image J plugin (OrientationJ) to get color-coded maps based on cytoskeletal fiber orientation. Based on the distribution of angular orientations, we calculated OOP values that range from zero (random organization) to one (perfect alignment) using custom-made MATLAB (Mathwork) code, as previously published41.
Optical mapping experiments of cardiac tissues
After observing spontaneous beating of NRVM tissue at Day 3 and iPSC-CM tissue at Day 5, we observed Ca2+ activities of cardiac tissues on the 3D printed scaffolds using optical mapping system at Day 5 and Day 7, respectively. For hiPSC-CM tissues on the 3D printed ventricle scaffolds, we cultured for 14 days before conducting optical mapping experiment to ensure the structural and functional coupling of the cells in tissues due to higher number of cells in 3D structures. The optical mapping system includes a modified tandem-lens macroscope (Scimedia, Costa Mesa, CA) equipped with a high-speed camera (MiCAM Ultima, Scimedia, Costa Mesa, CA), a plan APO 0.63× or 1x objective, a collimator (Lumencor, Beaverton, OR), a 200mW Mercury lamp (X-Cite exacte, Lumen Dynamics, Canada), a high-spatial resolution sCMOS camera (pco.edge, PCO AG), and 880 nm darkfield LED light. For Ca2+ imaging, we used an excitation filter with 580/14 nm, a dichroic mirror with 593 nm cut-off, and an emission filter with 641/75 nm (Semrock, Rochester, NY). For dark-field imaging, we added a dichroic mirror with 685 nm cut-off and long pass emission filter with 664 nm cut-off filter (Semrock, Rochester, NY). Cultured tissues were incubated with 2 μM X-Rhod-1 (Invitrogen) for 60 min at 37 °C, rinsed, and incubated in dye-free media for an additional 15 min at 37 °C before recording. The recording was conducted in the Tyrode’s solution (1.8 mM CaCl2, 5 mM glucose, 5 mM HEPES, 1 mM MgCl2, 5.4 mM KCl, 135 mM NaCl, and 0.33 mM NaH2PO4 in deionized water, pH 7.4, at 37°C) for NRVM tissue and fresh RPMI/B27(−) for iPSC-CM tissue. Point stimulation was applied to in vitro cardiac tissues using two platinum electrodes (Sigma-Aldrich) with 1 mm spacing with 10V amplitude and 10 ms duration. The platinum electrodes were located at 1.0 mm from the corner of the tissue samples (IonOptix myopacer). For each recording, Ca2+ and dark field images were acquired with frame rates of 400 frames per second (FPS) for 10 s. The recording of the top view was performed after the 3D printed in vitro ventricles moved to the PDMS mold without external stimulation. Post-processing of data was conducted with custom software written in MATLAB (MathWorks) and MiCAM imaging software (MiCAM Ultima, Scimedia, Costa Mesa, CA). A spatial filter with 3 × 3 pixels was applied to improve the signal-noise ratio. Activation time of each pixel was calculated at the average maximum upstroke slope of multiple pulses of X-Rhod-1 signals over a 10-second recording window. Velocity was derived from the difference in activation time that occurred between 1–5 mm.
Muscular thin film (MTF) analysis and contractile force measurement
FIG MTF chips were manufactured with cantilever designs53. G-code was made by hand script to print FIG ink in the direction of cantilevers and the printing model is shown in Supplementary Fig. 10d, left. MTFs were printed on the coverslip. A cantilever of MTFs dimension was a width of 1 mm and a length of 2.5–3 mm. MTF experiment was performed after 5 days of NRVMs culture in a 3.5 mm petri dish. Video micrographs were recorded on a Zeiss Discovery V12 stereomicroscope with 30 frames per second for 5–10 secs. IonOptix myopacer was used to apply 1 Hz field electrical stimulation with two parallel platinum electrode wires placed ~3 cm apart. Contractile stress was calculated as previously reported53. MTF cantilever images were threshold to binary images. The radius (r) of curvature of each cantilever was calculated using x-projection (blue line in Supplementary Fig. 10d) and original length (white box in Supplementary Fig. 8c). Briefly, numerically approximate was determined by following equations:
where is the original cantilever length, and x is the projected length of each cantilever extracted from the cantilever movie using NIH’s Image . The contractile stress was calculated by applying modified Stoney’s equation.
where is the elastic modulus, and are thickness of the FIG scaffolds and tissue, respectively. The average FIG scaffold thickness and tissue thickness for FIG MTFs were 70 μm, and 10 μm, respectively, and the average PDMS thickness and tissue thickness for PDMS MTFs were 20 μm and 5 μm, respectively. The elastic modulus of FIG was 51 kPa measured by compressing in fiber direction at 1 Hz oscillation using a dynamic mechanical analyzer (DMA, DMA1-STAR, Mettler Toledo, USA). Twitch stress was derived by subtracting peak diastolic stress (minimum stress of contractile stress) from the contractile stresses.
Particle Imaging Velocimetry (PIV) analysis
iPSC-CMs were cultured for 15 days in printed 3D FIG ventricle chamber scaffolds. The in vitro model of the heart chamber moved to the 35mm petri-dish filled with RPMI/B27(−) with 0.1 μg/ml of fluorescence beads (MG – Green Fluorescent Microspheres, Cospheric, Santa Barbara, CA, USA). 1 Hz Field electrical stimulation (IonOptix myopacer) was applied with 10–20V using two parallel platinum electrodes positioned ~30 cm apart. Bead movements were recorded using a stereoscopic microscope (Zeiss Discovery.V12 stereomicroscope) with a HBO 100 fluorescent light source at a 14x magnification with frame rates of 30 FPS. Bright-field images were recorded with Basler electric ACA2500–14UC USB 3.0 camera at an 8.5 x magnification and 30 FPS with and without 1 Hz field electrical stimulation (spontaneous contraction). Fluorescent image sequences were prepared for analysis using a rolling ball background subtraction (NIH ImageJ) and were analyzed by custom software based on the open-source package OpenPIV to reconstruct velocity maps 54. This resulted in velocity-field profiles as a function of time. The setime-dependent velocity profiles were then phase averaged over the entire cycle to smooth out local fluctuations in performance. As quantitative metrics of cardiac performance, we examined the total mass flux (cardiac output) and ejection fraction resulting from the 3D printed ventricles as previously reported 15. Briefly, here we summed the velocity profiles over the basal opening and normalized this value by the fluid density to obtain a 2D instantaneous mass flux with units of g/m. Further integrating this 2D value over the basal opening resulted in a 3D estimate of the total mass flux occurring during systole. Finally, knowing this mass flux value, the density of the fluid, and the initial starting volume of the ventricle, we could estimate the fraction of fluid ejected during contraction. Based on printing dimensions, the width of 10 mm, the height of 10 mm, and the depth of 11 mm were used to estimate the starting volume, assuming the shape of a half-oblate ellipsoid.
Deformation map analysis of in vitro 3D ventricle models
Deformation mapping was performed as previously reported15. Here in vitro models of the heart chamber were soaked in an RPMI/B27(−) media with 0.1 μg/ml of fluorescence beads for 20–30 minutes to allow the fluorescence beads to adhere to the printed ventricle scaffolds. After the printed in vitro ventricle moved to a fresh RPMI/B27(−), beads on the in vitro ventricle were recorded with the same system for PIV data recording. The open-source package OpenPIV54 was used to measure displacements in corresponding images by digital image cross-correlation to obtain deformation maps. One frame at the peak diastole was selected as a given reference image, and changes in deformation were measured relative to this frame using OpenPIV. Deformation maps were then normalized by the total ventricle height to produce strain values. To remove null frames and artifacts, images were then masked using a thresholding process (huang thresholding) with outliers filtered using a rolling kernel density estimate (kernel of size 3×3). Here the standard deviation in strain for each kernel was measured, and values that locally exceeded a certain threshold (2σ) were replaced with mean kernel values.
Statistics and Reproducibility
Statistical analysis was conducted to compare cell viability, tissue alignment, nucleus aspect ratio, nucleus angle distribution, tissue contractility, Ca2+ conduction anisotropy ratio in the longitudinal direction versus the transverse direction between tissues on the printed Gel-Alg hydrogel and the FIG scaffolds. All error bars are given as the standard error of the mean (SEM) unless otherwise noted. Statistical analyses were performed with a two-tailed unpaired Student’s t-test, assuming unequal variance, a one-way analysis of variance (ANOVA) with Tukey post-hoc test within the groups, and a two-sample Kolmogorov-Smirnov test. and was conducted unless otherwise noted. P values < 0.05 were considered statistically significant. Samples sizes are given for each experimental condition in the figure legends. For representative data and images given in the figures, at least three or more independent experiments were conducted, showing similar results.
Supplementary Material
Acknowledgments
The authors would like to thank Michael Rosnach for photography and illustrations and Dr. A.G. Kleber, M.D. for discussions regarding cardiac physiology. This work was sponsored by the John A. Paulson School of Engineering and Applied Sciences at Harvard University, the National Science Foundation through the Harvard University Materials Research Science and Engineering Center (DMR-1420570, DMR-2011754 to K.K.P), the National Institutes of Health and National Center for Advancing Translational Sciences (UH3HL141798 to W.H.P and K.K.P, UG3TR003279 to W.H.P and K.K.P). This work was also performed in part at the Harvard University Center for Nanoscale Systems (CNS); a member of the National Nanotechnology Coordinated Infrastructure Network (NNCI), which is supported by the National Science Foundation under NSF award no. ECCS-2025158. Micro-CT imaging reported in this publication was supported by Harvard University, CNS under National Institutes of Health award number S10OD023519. H.A.M.A. would like to thank the American Chemical Society for support through the Irving S. Sigal Postdoctoral Fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing interests
Harvard University filed for intellectual property relevant to this manuscript, listing L.A.M. and K.K.P. as inventors. The remaining authors declare no competing interests.
Code availability
G-code files used in this study are provided in the Supplementary Code. Contractility analysis of in vitro ventricle models was analyzed by custom made code that are published in Science 377, 180–255 (2022) and available at https://doi.org/10.5281/zenodo.6547775.
Data availability
All data generated or analysed during this study are included in this published article and available at a Figshare repository55.
References
- 1.Frantz C, Stewart KM, Weaver VM The extracellular matrix at a glance. J. Cell Sci. 123, 4195–4200 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blazeski A, Lowenthal J, Zhu R, Ewoldt J, Boheler KR, Tung L Functional properties of engineered heart slices incorporating human induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Reports 12, 982–995 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeGrice I, Pope A, Smaill B The architecture of the heart: Myocyte organization and the cardiac extracellular matrix. In: Villarreal FJ (ed). Interstitial fibrosis in heart failure. Springer New York: New York, NY, 2005, pp 3–21. [Google Scholar]
- 4.Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, et al. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556, 239–243 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleber AG, Rudy Y Basic mechanisms of cardiac impulse propagation and associated arrhythmias. Physiol. Rev. 84, 431–488 (2004) [DOI] [PubMed] [Google Scholar]
- 6.Park SJ, Zhang D, Qi Y, Li Y, Lee KY, Bezzerides VJ, et al. Insights into the pathogenesis of catecholaminergic polymorphic ventricular tachycardia from engineered human heart tissue. Circulation 140, 390–404 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, et al. Modeling the mitochondrial cardiomyopathy of barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 20, 616–623 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCain ML, Agarwal A, Nesmith HW, Nesmith AP, Parker KK Micromolded gelatin hydrogels for extended culture of engineered cardiac tissues. Biomaterials 35, 5462–5471 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feinberg AW, Feigel A, Shevkoplyas SS, Sheehy S, Whitesides GM, Parker KK Muscular thin films for building actuators and powering devices. Science 317, 1366–1370 (2007) [DOI] [PubMed] [Google Scholar]
- 10.Li RA, Keung W, Cashman TJ, Backeris PC, Johnson BV, Bardot ES, et al. Bioengineering an electro-mechanically functional miniature ventricular heart chamber from human pluripotent stem cells. Biomaterials 163, 116–127 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karp JM, Yeo Y, Geng W, Cannizarro C, Yan K, Kohane DS, et al. A photolithographic method to create cellular micropatterns. Biomaterials 27, 4755–4764 (2006) [DOI] [PubMed] [Google Scholar]
- 12.Wu X, Liu Y, Li X, Wen P, Zhang Y, Long Y, et al. Preparation of aligned porous gelatin scaffolds by unidirectional freeze-drying method. Acta Biomater. 6, 1167–1177 (2010) [DOI] [PubMed] [Google Scholar]
- 13.MacQueen LA, Sheehy SP, Chantre CO, Zimmerman JF, Pasqualini FS, Liu X, et al. A tissue-engineered scale model of the heart ventricle. Nat. Biomed. Eng. 2, 930–941 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capulli AK, Emmert MY, Pasqualini FS, Kehl D, Caliskan E, Lind JU, et al. Jetvalve: Rapid manufacturing of biohybrid scaffolds for biomimetic heart valve replacement. Biomaterials 133, 229–241 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang H, Liu Q, Zimmerman JF, Lee KY, Jin Q, Peters MM, et al. Recreating the heart’s helical structure-function relationship with focused rotary jet spinning. Science 377, 180–185 (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Lee SJ, Cheng HJ, Yoo JJ, Atala A 3d bioprinted functional and contractile cardiac tissue constructs. Acta Biomater. 70, 48–56 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue K, Trujillo-de Santiago G, Alvarez MM, Tamayol A, Annabi N, Khademhosseini A Synthesis, properties, and biomedical applications of gelatin methacryloyl (gelma) hydrogels. Biomaterials 73, 254–271 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lind JU, Busbee TA, Valentine AD, Pasqualini FS, Yuan H, Yadid M, et al. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat. Mater. 16, 303–308 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, Rafatian N, Feric NT, Cox BJ, Aschar-Sobbi R, Wang EY, et al. A platform for generation of chamber-specific cardiac tissues and disease modeling. Cell 176, 913–927 e918 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capulli AK, MacQueen LA, Sheehy SP, Parker KK Fibrous scaffolds for building hearts and heart parts. Advanced Drug Delivery Reviews 96, 83–102 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi HG, Jeong YH, Kim Y, Choi YJ, Moon HE, Park SH, et al. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat. Biomed. Eng. 3, 509–519 (2019) [DOI] [PubMed] [Google Scholar]
- 22.Daly AC, Davidson MD, Burdick JA 3d bioprinting of high cell-density heterogeneous tissue models through spheroid fusion within self-healing hydrogels. Nat. Commun. 12, 753 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brassard JA, Nikolaev M, Hubscher T, Hofer M, Lutolf MP Recapitulating macro-scale tissue self-organization through organoid bioprinting. Nat. Mater. 20, 22–29 (2021) [DOI] [PubMed] [Google Scholar]
- 24.Jia W, Gungor-Ozkerim PS, Zhang YS, Yue K, Zhu K, Liu W, et al. Direct 3d bioprinting of perfusable vascular constructs using a blend bioink. Biomaterials 106, 58–68 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashammakhi N, Ahadian S, Xu C, Montazerian H, Ko H, Nasiri R, et al. Bioinks and bioprinting technologies to make heterogeneous and biomimetic tissue constructs. Mater Today Bio 1, 100008 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouyang L, Armstrong JPK, Lin Y, Wojciechowski JP, Lee-Reeves C, Hachim D, et al. Expanding and optimizing 3d bioprinting capabilities using complementary network bioinks. Sci Adv 6, (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi JR, Yong KW, Choi JY, Cowie AC Recent advances in photo-crosslinkable hydrogels for biomedical applications. BioTechniques 66, 40–53 (2019) [DOI] [PubMed] [Google Scholar]
- 28.Markstedt K, Mantas A, Tournier I, Martinez Avila H, Hagg D, Gatenholm P 3d bioprinting human chondrocytes with nanocellulose-alginate bioink for cartilage tissue engineering applications. Biomacromolecules 16, 1489–1496 (2015) [DOI] [PubMed] [Google Scholar]
- 29.Nadernezhad A, Caliskan OS, Topuz F, Afghah F, Erman B, Koc B Nanocomposite bioinks based on agarose and 2d nanosilicates with tunable flow properties and bioactivity for 3d bioprinting. ACS Appl. Bio Mater. 2, 796–806 (2019) [DOI] [PubMed] [Google Scholar]
- 30.Schwab A, Levato R, D’Este M, Piluso S, Eglin D, Malda J Printability and shape fidelity of bioinks in 3d bioprinting. Chem. Rev. 120, 11028–11055 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noor N, Shapira A, Edri R, Gal I, Wertheim L, Dvir T 3d printing of personalized thick and perfusable cardiac patches and hearts. Adv. Sci. 6, 1900344 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kupfer ME, Lin WH, Ravikumar V, Qiu K, Wang L, Gao L, et al. In situ expansion, differentiation, and electromechanical coupling of human cardiac muscle in a 3d bioprinted, chambered organoid. Circulation Research 127, 207–224 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee A, Hudson AR, Shiwarski DJ, Tashman JW, Hinton TJ, Yerneni S, et al. 3d bioprinting of collagen to rebuild components of the human heart. Science 365, 482–487 (2019) [DOI] [PubMed] [Google Scholar]
- 34.Campostrini G, Windt LM, van Meer BJ, Bellin M, Mummery CL Cardiac tissues from stem cells: New routes to maturation and cardiac regeneration. Circ Res 128, 775–801 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahrens JH, Uzel SGM, Skylar-Scott M, Mata MM, Lu A, Kroll KT, et al. Programming cellular alignment in engineered cardiac tissue via bioprinting anisotropic organ building blocks. Adv. Mater. 34, e2200217 (2022) [DOI] [PubMed] [Google Scholar]
- 36.Sydney Gladman A, Matsumoto EA, Nuzzo RG, Mahadevan L, Lewis JA Biomimetic 4d printing. Nat. Mater. 15, 413–418 (2016) [DOI] [PubMed] [Google Scholar]
- 37.Siqueira G, Kokkinis D, Libanori R, Hausmann MK, Gladman AS, Neels A, et al. Cellulose nanocrystal inks for 3d printing of textured cellular architectures. Adv. Funct. Mater. 27, (2017) [Google Scholar]
- 38.Jinkins KR, Chan J, Jacobberger RM, Berson A, Arnold MS Substrate-wide confined shear alignment of carbon nanotubes for thin film transistors. Advanced Electronic Materials 5, (2018) [Google Scholar]
- 39.Caliari SR, Burdick JA A practical guide to hydrogels for cell culture. Nat. Methods 13, 405–414 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hausmann MK, Ruhs PA, Siqueira G, Lauger J, Libanori R, Zimmermann T, et al. Dynamics of cellulose nanocrystal alignment during 3d printing. ACS Nano 12, 6926–6937 (2018) [DOI] [PubMed] [Google Scholar]
- 41.Pasqualini FS, Sheehy SP, Agarwal A, Aratyn-Schaus Y, Parker KK Structural phenotyping of stem cell-derived cardiomyocytes. Stem Cell Reports 4, 340–347 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bray MA, Sheehy SP, Parker KK Sarcomere alignment is regulated by myocyte shape. Cell Motil Cytoskeleton 65, 641–651 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuo PL, Lee H, Bray MA, Geisse NA, Huang YT, Adams WJ, et al. Myocyte shape regulates lateral registry of sarcomeres and contractility. The American Journal of Pathology 181, 2030–2037 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geisse NA, Sheehy SP, Parker KK Control of myocyte remodeling in vitro with engineered substrates. In Vitro Cellular & Developmental Biology - Animal 2009 45:7 45, 343–350 (2009) [DOI] [PubMed] [Google Scholar]
- 45.Grosberg A, Kuo P-L, Guo C-L, Geisse NA, Bray M-A, Adams WJ, et al. Self-organization of muscle cell structure and function. PLOS Computational Biology 7, e1001088 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bray MA, Adams WJ, Geisse NA, Feinberg AW, Sheehy SP, Parker KK Nuclear morphology and deformation in engineered cardiac myocytes and tissues. Biomaterials 31, 5143–5150 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee H, Adams WJ, Alford PW, McCain ML, Feinberg AW, Sheehy SP, et al. Cytoskeletal prestress regulates nuclear shape and stiffness in cardiac myocytes. Experimental Biology and Medicine 240, 1543–1554 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carmeliet E Conduction in cardiac tissue. Historical reflections. Physiol Rep 7, e13860 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kleber AG, Jin Q Coupling between cardiac cells-an important determinant of electrical impulse propagation and arrhythmogenesis. Biophys Rev (Melville) 2, 031301 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buckberg G, Hoffman JI, Mahajan A, Saleh S, Coghlan C Cardiac mechanics revisited: The relationship of cardiac architecture to ventricular function. Circulation 118, 2571–2587 (2008) [DOI] [PubMed] [Google Scholar]
Methods-only references
- 51.Rezakhaniha R, Agianniotis A, Schrauwen JT, Griffa A, Sage D, Bouten CV, et al. Experimental investigation of collagen waviness and orientation in the arterial adventitia using confocal laser scanning microscopy. Biomech. Model. Mechanobiol. 11, 461–473 (2012) [DOI] [PubMed] [Google Scholar]
- 52.Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating wnt/beta-catenin signaling under fully defined conditions. Nat. Protoc. 8, 162–175 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Al Tanoury Z, Zimmerman JF, Rao J, Sieiro D, McNamara HM, Cherrier T, et al. Prednisolone rescues duchenne muscular dystrophy phenotypes in human pluripotent stem cell-derived skeletal muscle in vitro. Proc. Natl. Acad. Sci. U. S. A. 118, (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alex Liberzon DL, Aubert Mathias, Bachant Pete, jakirkham, ranleu, tomerast, Käufer Theo, Borg Joe, Dallas Cameron, & Vodenicharski Boyko. Openpiv/openpiv-python: Fixed windows conda-forge failure with encoding. 10.5281/zenodo.3566451 (2019) [DOI] [Google Scholar]
- 55.Choi S, Lee KY, Kim SL, Macqueen LA, Chang H, Zimmerman JF, Jin Q, Peters MM, Ardoña HAM, Liu X, Heiler A-C, Richardson C, Gabardi R, Pu WT, Bausch AR, and Parker KK Fiber Infused Gel Scaffolds Guide Cardiomyocyte Alignment in 3D Printed Ventricles, Figshare, 10.6084/m9.figshare.22787714, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and available at a Figshare repository55.


