Abstract
Aptameric receptors are important biosensor components, yet our ability to identify them depends on the target structures. We analyzed the contributions of individual functional groups on small molecules to binding within 27 target-aptamer pairs, identifying potential hindrances to receptor isolation, for example, negative cooperativity between sterically hindered functional groups. To increase the probability of aptamer isolation for important targets, such as leucine and voriconazole, where multiple previous selection attempts failed, we designed tailored strategies focused on overcoming individual structural barriers to successful selections. This approach enables us to move beyond standardized protocols into functional group-guided searches, relying on sequences common to receptors for targets and their analogs to serve as anchors in regions of vast oligonucleotide spaces wherein useful reagents are likely to be found.
One Sentence Summary:
Concepts from organic and medicinal chemistry guide isolation of aptamers for small molecule targets.
Aptamers are oligonucleotide-based receptors isolated from random libraries through cycles of enrichment based on target affinity coupled to amplifications (1–4). Aptamers can be selected for a variety of small molecules for which antibodies cannot, that is, targets ignored by the immune system even when conjugated to carrier proteins, e.g., neurotransmitters (5) and amino acids (6, 7). Once available, aptamers can be readily engineered into various sensor formats (3,4), including for use as fluorescent (8), electrochemical (9), or electronic biosensors (10).
One of the main obstacles to the broad application of aptamers in biosensing is a lack of aptamers with appropriate affinities for many important low molecular weight targets (3, 4). To exemplify, we were repeatedly unable to isolate DNA aptamers for two clinically important molecules, the amino acid leucine (Leu, 1) and the antifungal agent voriconazole (2) (Fig. 1A). Aptamers to detect blood leucine levels could be used to rapidly clarify false positives during newborn screening for maple syrup urine disease (MSUD) (11, 6). We have sought to expand on our success with vancomycin sensing (12) and isolate receptors that could be used for voriconazole therapeutic monitoring (13). Our attempts were variations of selections based on target-induced stem closure (Fig. 1B) (14, 15). In this approach, oligonucleotide libraries with internal random 36-mer regions are immobilized via 5’-primer regions that hybridize with tethered capture sequences. Potential aptamers hybridized on columns are released by interactions with unmodified targets in solution, which can stabilize stem formation upon displacement (Fig. 1B).
Figure 1. Target functional-group binding free energy analysis for aptamers from stem-loop libraries:
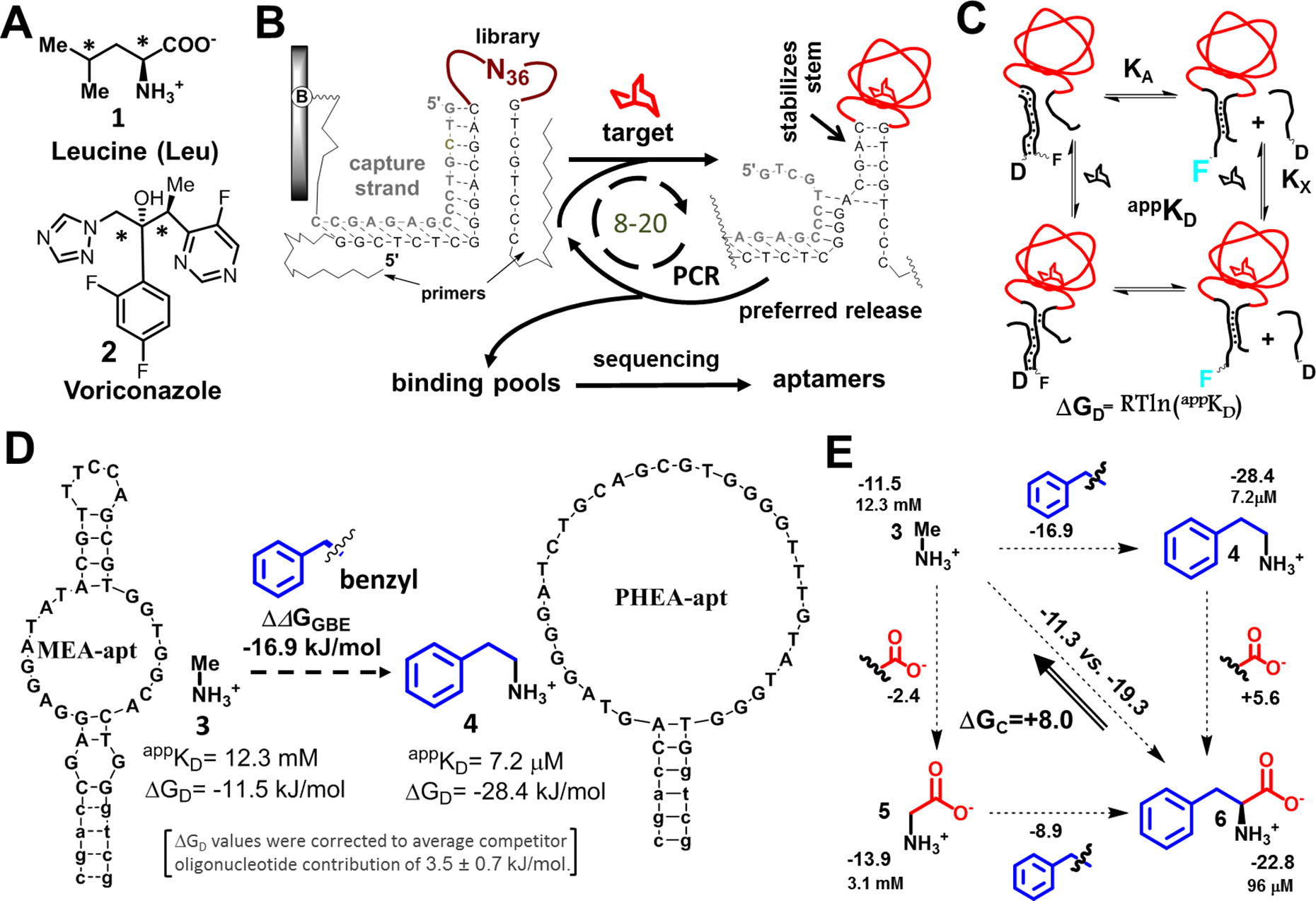
(A) Using standard protocol, we were unable to isolate aptamers for leucine (1) and voriconazole (2), which have congested pairs of carbons (*). (B) Aptamer selections driven by small-molecule-induced stem closures: An oligonucleotide library with a random loop (N36) is hybridized to the complement (capture strand) of a PCR primer. The capture strand is tethered to a column. The column is exposed to target solutions. Sequences that bind the targets and undergo stem stabilization are released, preferentially amplified, and used in the next selection cycle. (C) We measured apparent appKD values for aptamers and from these calculated the free energies of displacement, ΔGD, based on a fluorescence displacement assay associated with the equilibrium between an aptamer (labeled with fluorescein, F) and a complementary oligonucleotide used for capture in selection (labeled with a quencher, dabcyl, D). Target leads to concentration-dependent increases in fluorescence via equilibria shown. KX and KA and are dissociation constants for a target-aptamer complex without competitor and an aptamer-competitor complex without target, respectively. (D) We isolated the contributions of individual functional groups by subtracting individual ΔGD values of aptamer-target pairs, with these values corrected to account for differences in oligonucleotide quenching. Here, the two targets, methylamine (3) and phenylethylamine (4), differ by a benzyl group. The difference in free energy associated with benzyl group addition is ΔΔGGBE (benzyl). Two aptamers used for this calculation are shown. (E) Cooperativity is assessed by double functional group replacement cycles (18). The appKD and ΔGD (normalized to average impact of oligonucleotide on equilibrium, in kJ/mol) values are shown next to the targets, with ΔΔGGBE values shown next to the fragments. A ΔΔGGBE>0 indicates a decrease in affinity upon adding a functional group; the ΔGC value (+8.0 kJ/mol) represents the difference between adding functional groups separately (upper horizontal and left vertical values) vs. at the same time (diagonally), which is interpreted as negative cooperativity when both benzyl group and a carboxylate are together present in a molecule.
Our failure to isolate DNA aptamers for leucine was surprising because RNA aptamers had been previously isolated via leucine-tethered affinity columns (16). Similarly, voriconazole should have been a straightforward target due to its aromatic surfaces and heteroatoms. However, we could neither adapt reported aptamers cross-reactive with the azole class of antifungals (17) as sensor components (8, 12), nor we could isolate new specific aptamers. These two seemingly unrelated targets, with significantly different molecular weights, share proximate pairs of sterically crowded sp3 carbons (Fig. 1A), which inspired us to pursue a broader understanding of the general relationships between target structures and outcomes of highly standardized selections. Our aim was a generalizable approach to aptamer isolation when other, standard methods fail.
Analysis of Free Energies of Oligonucleotide Displacement across Related Targets
We amassed 27 aptamers, 23 of which were newly isolated through this work. The new aptamers emerged directly from selections, without further optimization, identified as the highest affinity receptors targeting amines, amino acids, and their analogs. In the past, while working with individual aptamers, we focused on aptamer dissociation constants obtained by a fluorescence quenching assay that reported fluorescently labeled aptamer competition with a quencher-labeled capture oligonucleotide (Fig. 1C, and Displacement Assay Rationale section in method); the assay could be adapted to a model of allosteric antagonism to account for partial release upon binding (18). To characterize impact of targets on selection outcomes, we instead needed to compare targets in their abilities to outcompete capture oligonucleotides. Thus, here, we focused on appKD (a midpoint response or X50%) of the displacement of oligonucleotide competitor that is used on affinity column during selection, which is related to the free energy of displacement, ΔGD. In contrast to the free energy of binding (ΔGB) obtained, e.g., by isothermal calorimetry, ΔGD governs a comprehensive set of equilibria that impacts the release of aptamers from the column upon target addition. The difference between ΔGD and ΔGB is primarily in the contributions of the capture oligonucleotide present at equilibria.
The targets (Table S3) and their aptamers (Fig. S4–39) were organized in related pairs (Fig. 1D and Fig. S41–45), with each pair differing by the addition of a single functional group or group transformation, e.g., methylamine (3) and phenylethylamine (4) differ by the addition of a seven-carbon benzyl group (Fig. 1D). We defined ΔΔGGBE as the free energy difference related to the equilibria positions impacting the relative outcomes of two selections, attributable to the presence of the additional functional group or transformation. We also assume the portions of ΔGD that govern equilibria unrelated to either target or capture oligonucleotide binding to be similar across all aptamers and that they will largely cancel each other when subtracting two ΔGD values within pairs, which allows us to extract estimates of relative ΔΔGGBE values (Fig. 1D). Related concepts on contributions to the free energy of binding associated with functional groups are often used for ligand optimization in medicinal chemistry (19, 20), where receptors are shared between targets. Two key assumptions, aside from nearly identical selection conditions, were needed to extend the concept of functional-group free-energy contributions to selections:
First, there are ~1021 possible random 36-mers. In selections, we sample only ~1014 of these sequences. Thus, in the absence of extraordinary luck, we do not isolate potentially unique, but only typical receptors (21, 22), which are examples of multiple sequences having similar affinities values broadly distributed over oligonucleotide space. This sparse sampling allows us to treat the properties of the isolated aptamers, represented here by the ‘best’ aptamer from each selection, as characteristic of the highly standardized selection conditions, libraries, and targets. Since selections for new and previously identified aptamers differed mostly in their targets, we attributed large changes in the properties of the aptamers to the impact of structural differences between targets, i.e., to specific functional groups.
Second, functional group contributions to selections can only be based on well-known non-covalent interactions (20, 23). Thus, as a first approximation, within a set of close analogs, we expect to be able to isolate additive effects. When we observe systematic non-additivities in thermodynamic cycles, for example, cooperativity (ΔGC) as estimated through cycles of double replacements of functional groups (24), we can analyze non-additivities to generate hypotheses about barriers to aptamer isolation (Fig. 1E). Reciprocally, if correct in our assumptions, after initial selection failures, we can perform functional group analysis of targets to identify possible structural barriers leading to these failures and design selection protocols to improve our chances of isolating aptamers.
We performed the following three tests with the available aptamers to assess these assumptions. Although each test individually was limited due to small sample sizes, together, they strongly supported our reasoning. First, we analyzed the four highest affinity aptamers for phenylalanine from four separate selections and obtained similar ΔGD values (and estimated ΔGB values), within <3 kJ of each other (Fig. 2B, Table S4). This result is consistent with the affinity of winning aptamers being regularly distributed over oligonucleotide space, and thus, representing a reproducible property of selections. These findings suggest that large differences in target-related ΔGD should reflect differences in functional groups and not different selections.
Fig. 2. Analysis of ΔGD and ΔΔGGBE from a set of 27 aptamers:
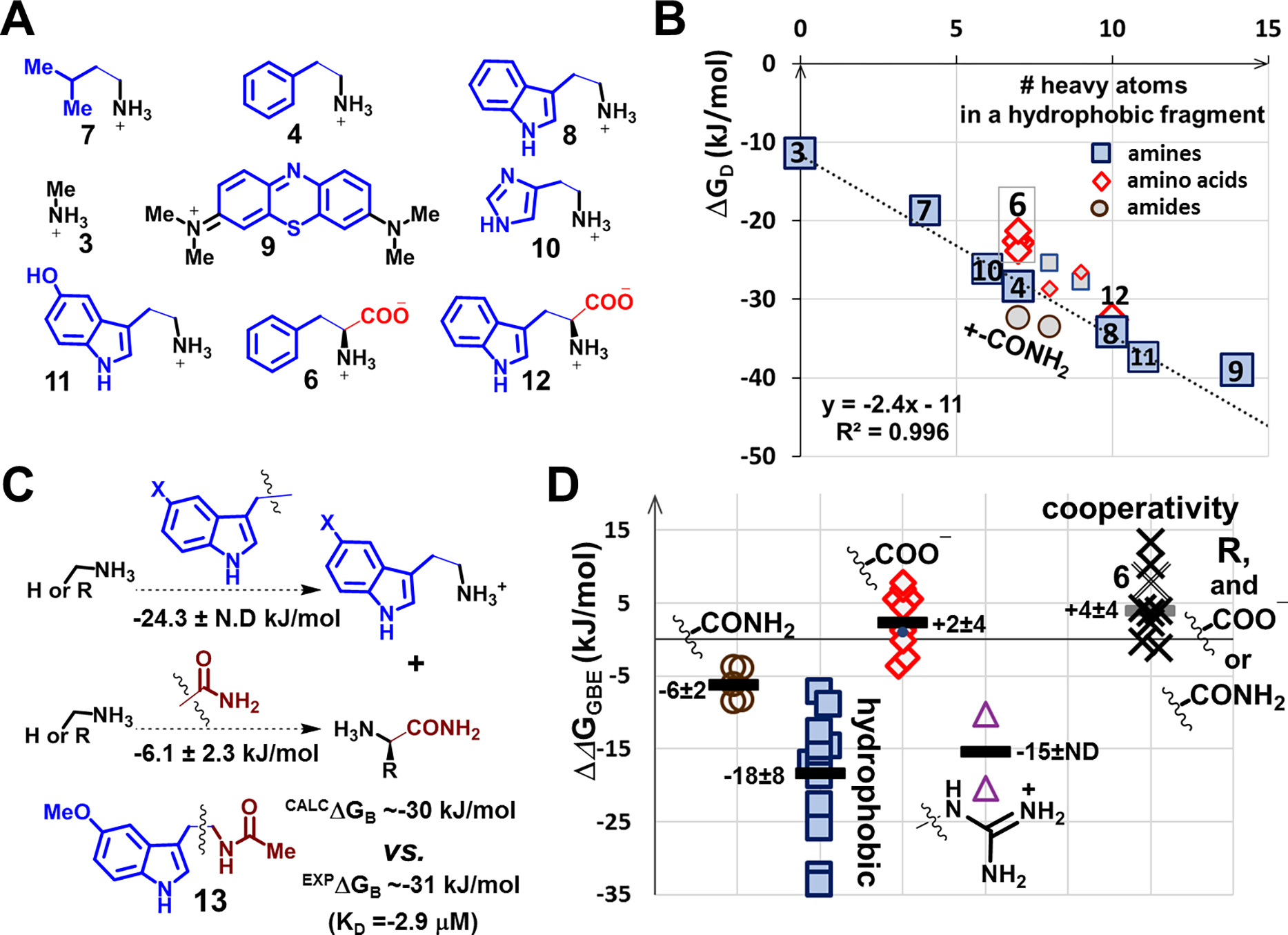
(A) Exemplary targets used to characterize binding optimization to hydrophobic surfaces during selections; hydrophobic/aromatic fragments are shown as brown squares (amines) or green diamonds (amino acids). (B) Regression analysis of target ΔGD vs. # of heavy atoms (other than hydrogen) in aromatic hydrophobic fragments within targets. The regression line including methylamine and two aromatic amines (3, 4, 8) was used to estimate the contributions of the hydrophobic surfaces in two aromatic amino acids (6, 10,) and a non-aromatic hydrophobic amine related to leucine (7). Data for the four aptamers for 6 are shown individually. Methylene blue (9) is the target with the highest affinity for the aptamers isolated directly from N36 libraries. Two amides have ΔGD values above, carboxylates below (diamonds), and histamine (10) and serotonin (11) are on the regression line. Unmarked data points are for tyramine, tyrosine, dopamine and L-DOPA. (C) Additivity of ΔΔGGBE in similar compounds (cf. Fig. S.6): Using the average ΔΔGGBE values of a pair of planar indole-methylene containing molecules and five carboxamides, we estimated ΔGB for the melatonin (13) aptamer. (D) Distributions of ΔΔGGBE contributions of selected functional groups, for carboxylates (open diamonds, we show position of phenylalanine, 6), carboxamides (circles), guanidiniums (triangles), and hydrophobic groups (squares). We show rounded averages (thick lines) and standard deviations in kJ/mol. We also show on the same plot cooperativities (ΔGC) assessed through double functional group replacement cycles (Fig. 1E) for groups added to methylamine together with carboxylates and carboxamides to obtain individual amino acids and their amides (Fig. S41–45). All data points in B-D are results of individual selection experiments, and the uncertainty of this approach can be assessed by four aptamers for phenylalanine, 6, in panel B, which were isolated in four independent selections.
Second, we observed correlations between ΔGD values and the numbers of heavy (non-hydrogen) atoms in the target hydrophobic fragments (Fig. 2A, B). The molecule with the largest hydrophobic surface, methylene blue (9), yielded the highest affinity of all targets. The correlation between methylamine (3) and two planar aromatic amines in our set, 4 and 8, supports an argument that the applied selection pressure directly optimizes affinity in proportion to hydrophobic surfaces, i.e., is based on the functional groups present, and that we can subtract two ΔGD values to isolate the impact of structural changes. We see indications that functional group-based optimization is general, with methylbutylamine (7), histamine (10), and serotonin (11) being very close to the aromatic amine (3, 4, 8) regression line, although caution should be exercised not to overinterpret these results without further structural information (24).
Third, we added average ΔΔGGBE values calculated from two planar indole-containing amines and five primary carboxamides (Fig. S46) to obtain a close match with an experimentally determined ΔGB value for melatonin (13), a planar molecule containing an indole and a secondary carboxamide (Fig. 2C, the addition of ΔΔGGBE values leads to ΔGB). Thus, our protocol simultaneously optimizes the presence of multiple functional groups, and we can use this property to interpret deviations from additivity. Our standard selection protocol (Fig. 1B) depends on target-induced oligonucleotide displacement outcompeting background “noise” in the form of more common ligand-independent oligonucleotide release from the column (20, 25). Combinations of target functional groups that reduce the affinity of common aptamers do so by reducing the target occupancy of an aptamer. This feature decreases the probability of isolating candidate aptamers in the early selection steps, which could be critical for selection success.
We used the aromatic amine (3, 4, 8) regression line (Fig. 2B) to estimate the impact of additional carboxylates on the ΔGD values for the aptamer-target complexes of two related aromatic amino acids, phenylalanine (6) and tryptophan (12), observing that the addition of a carboxyl group is similar to the loss of receptor hydrophobic contacts for between one and two heavy atoms, which is intuitively consistent with the introduction of a polar carboxylate near a primarily hydrophobic pocket. Our analysis of double functional group replacement cycles (Fig. 1E, 2D, S41–45) revealed substantial negative cooperativity while adding negative charge in proximity to mismatched groups, such as hydrophobic residues (in phenylalanine and tryptophan). These structural constellations, then, were identified as likely to reduce the probability of aptamer isolation for leucine.
Functional Group-Guided Selections for Leucine
We extended our analysis to hypothesize that the two out-of-plane carbons and a carboxylate, all in proximity in leucine, act synergistically to minimize contact surfaces and reduce affinities of typical aptamers, thus allowing competing ligand-independent release mechanisms to dominate and suppress the desired outcomes. To overcome this issue, we separated the selection steps for the alkyl (isobutyryl) and α-amino-carboxylate groups (Fig. 3A, B). We first implemented a protocol to identify a sequence iBu.1 certain to contain a binding motif for the isobutyl group. We started with a Cp*Rh(III)-binding aptamer (CpRh1.0), specifically isolated for this purpose, as a temporary placeholder for sequences that interact with the carboxyl and amino groups (26). We inserted a random 22-mer region which will become iBu, into the CpRh1.0, creating a new library (n.b., we can screen complete 22-mer sequence space). From this library, we selected aptamers like CpLeu1.0, which bound leucine in the presence of the Cp*Rh(III) cofactor. Although we could immediately eliminate CpLeu1.0 from further consideration as a Leu sensor, because of its complex mechanism of interactions with leucine, reflected in a sharp threshold behavior of fluorescent sensor (cf. S38), we knew that the inserted sequence, iBu, had to contain binding motifs for the Leu side chain.
Fig.3. Multistep functional group-guided approach to high-affinity aptamers for leucine:
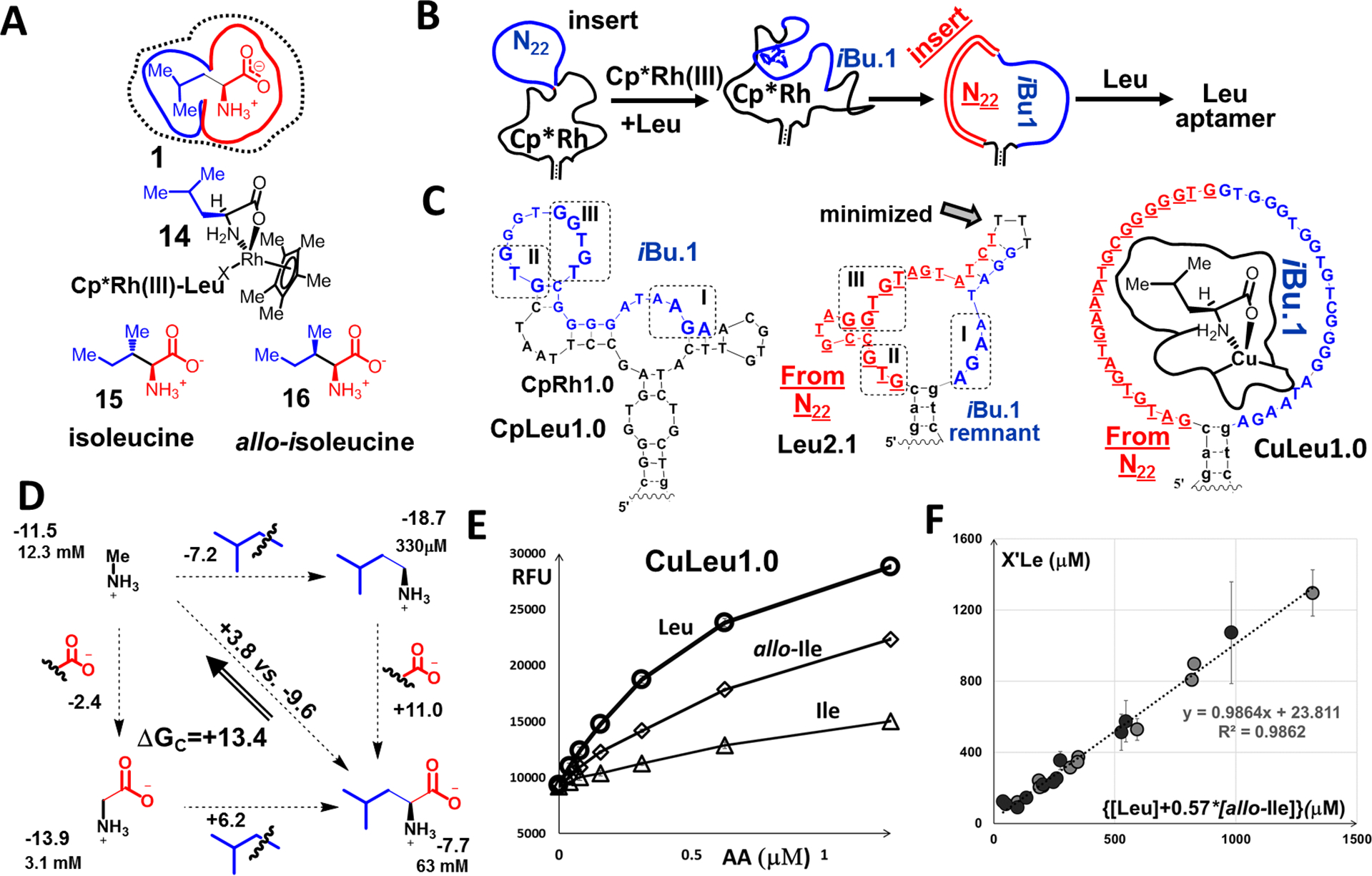
(A) Leucine was broken into two fragments, i.e., isobutyl and 2-aminoethanoate. We designed a selection to isolate aptamers sequentially to recognize one, then both fragments, thereby reducing the target-related barriers in each step and increasing the probability of finding leucine aptamers. Shown are the complex of leucine and Cp*Rh(III), 14, and other amino acids for which we observed challenging aptamer cross-reactivities (15, 16). (B) We started with a Cp*Rh(III) aptamer, performing an N22 random insertion to form a library used to identify the iBu.1 sequence, which recognized the isobutyl group. We used a second N22 library (red) to focus selection pressure on the 2-aminoethoanoate group to arrive at leucine aptamers. (C) Secondary structures of the related CpLeu1.0, Leu2.1 (minimized from Leu2.0, Fig. S.34) and CuLeu1.0. The inserted iBu1 motif is shown in blue in CpLeu1.0 and carried-over sections of the CpRh1.0 aptamer are shown in black. The CpLeu1.0 aptamer binds Leu in the presence of Cp*Rh(III), while Leu2.1 binds leucine on its own. The CuLeu1.0 aptamer binds Leu in the presence of Cu(II) with high affinity (appKD ~170 nM). (D) Double-functional group replacement cycle, methylamine (3) to leucine (1). (E) Fluorescence vs. target concentrations in the presence of 40 μM Cu(II) (displacement assay, cf. Fig. 1C) for CuLeu1.0 and four branched-chain amino acids. (F) Preliminary analytical assessment of CuLeu1.0 in mock human sera samples spiked with branched-chain amino acids to mimic values for patients with maple sugar urine disease (MSUD) (Table S.11). The correlation is between measured values (dilution 1:500, 100 μM Cu(II)) of X’Le (the sensor responsive fraction) vs. added values for{[Leu] + 0.57*[allo-Ile]}. The high correlation indicates that this sensor is suitable component of a minimal cross-reactive array for monitoring in patients, although dilution might have to be adjusted depending on targeted range. Allo-Ile is negligible at birth and during the first several postpartum days (11), thus, we show also correlation in the same mock samples but without allo-Ile (circles). Measurements in E and F are in triplicates with SDs shown.
We then designed a library of 22-mers with iBu.1 positioned next to the stem. From this library, using elutions with leucine without cofactor, we identified Leu2.1 (Fig. 3C). The Leu2.1 aptamer had a KD of almost 10 mM (Fig. S28) and an ~4:1 preference for Leu over Ile (Fig. S49). The negative cooperativity (ΔGC) between the carboxyl and isobutyryl groups was large (>10 kJ/mol), providing an explanation for our initial selection difficulties (Fig. 3D).
We identified homologous regions I-III in CpLeu1.0 and Leu2.1, two of which, II and III, in Leu2.1 originated from the inserted random region outside of iBu.1. The short Leu2.1 aptamer should have been abundant in any initial pool; further, isolation of motifs II and III in control studies of insertion reselection (Fig. S50), indicated that the motif I is not absolutely required in Leu aptamers. However, apparently due to its low affinity, Leu2.1 required the prefixed compatible sequences within I, to increase the probability of isolation via a reduction in the required sequence length in the newly inserted random region.
The Leu2.1 aptamer had a millimolar target affinity insufficient for the intended application of testing newborns with MSUD (11). Further, Leu2.1 preferred phenylalanine over leucine (Fig. S49, which was a dominant problem in our prior selections using Cp*Rh(III) as the cofactor (Fig. S51). Thus, we added an aminophilic cofactor, Cu(II) (27), in the last step of the selection to improve affinity and selectivity. We hypothesized that Cu(II) would serve as a protecting group neutralizing the effects of the carboxylate through the complexation with the 2-amino-ethanoate group. Complexation would allow better access of hydrophobic DNA monomer residues to the leucine side chain, improving affinity and selectivity. Consistent with our hypothesis, we identified aptamer CuLeu1.0 having a 44-mer loop, conserved sections of iBu.1, and high affinity for leucine (KD~170 nM; Fig. 3C).
CuLeu1.0 had selectivity for leucine over isoleucine, valine, and phenylalanine, but we noted strong cross-reactivity with allo-isoleucine (Fig. 3E, Newman projections in S52). The allo-isoleucine metabolite was not used in the aptamer counter-selections because its concentrations are negligible at birth. Currently, newborn screening is performed with mass spectroscopy (11), which integrates isobaric species to provide X’Le values (X’Le = Leu + Ile + allo-Ille + 2OHPro). Thus, our aptamer sensor is a candidate for the development of rapid tests to address false positives in MSUD by showing a lack of steady increase in X’Le values in consecutive measurements, with X’Le defined, e.g., as [Leu] + 0.57*[allo-Ille] (Fig. 3F, S53). After the first few days of life, however, allo-Ile concentrations increase so a monitoring strategy without fully specific aptamers would require a cross-reactive array (26, 28), for which CuLeu1.0 is a suitable component.
The multi-step approach with Cu(II) can be generalized to amino acids that display a side chain away from the Cp*Rh(III) complex, such as Ile (cf. CuIle1.1, Fig. S54–56). This approach would not work for amino acids that carry a chelating group beyond 2-aminoethanoate, e.g., glutamate (Fig. S57). For comparison, we performed a single-step Leu selection with Cu(II) as the cofactor. We isolated receptors with about fivefold lower affinities compared to CuLeu1.0. The two most abundant sequences preferred isoleucine or methionine (Fig. S58–60). These aptamers are candidates for arrays.
A Structure-Guided Approach to Aptamers for Voriconazole
Leucine (1) is closely related to other amino acids in our target set. By contrast, voriconazole (2) is an example of applying a structurally guided approach to unrelated molecules. We initially attributed our voriconazole selection failures to its limited solubility (~200 μM). Nonetheless, selections using a soluble voriconazole phosphate analog also failed. Similar to leucine, we considered that voriconazole has a sterically crowded environment (Fig. 4A) forcing its fragments (structural subunits) into a propeller-like conformation, as revealed in crystal structures (30). This sterically crowded conformation was hypothesized to lead to suboptimal access to hydrophobic surfaces in DNA needed to interact with fragments I-III (Fig. 4A). One possible retrosynthetic disconnection (31) led to a simplified, less congested, and readily synthesized alcohol analog, 2a (Fig. 4A).
Fig. 4. Selection of voriconazole aptamers using an analog:
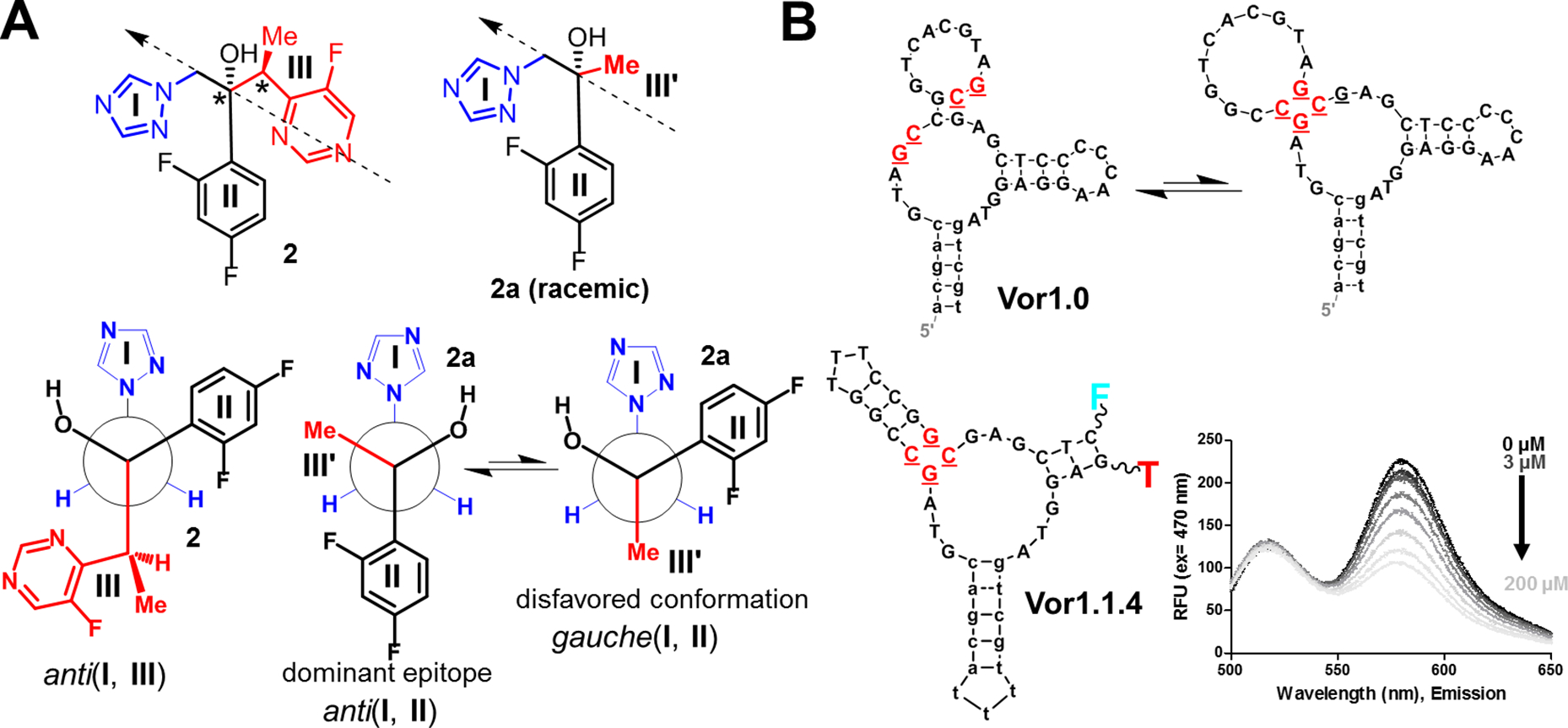
(A) Structure of voriconazole (2) with three fragments (I-III) and its analog 2a, in which fragment III was substituted with a methyl group (III’). The arrows indicate the perspective used to produce the Newman projections below. The anti(I, III) conformation is similar to an observed crystal structure (30). The voriconazole analog 2a simplifies the largest fragment (III) and was designed for reduced complexity and as a more suitable target for selection. Here, the anti (I, II) conformation is likely to be favored and the dominant epitope in selection. (B) The aptamer Vor1.0 was isolated in the selection protocol that used 2 and 2a in parallel. The secondary structure of Vor1.0 is shown as predicted by mFold (top) and as an alternative secondary structure (bottom), which was subsequently confirmed to be the active sensor structure. Structure-switching allows this aptamer to be captured on the column (i.e., the upper structure allows capture) during the initial stages of selection (cf. Fig. S.15). A variant of Vor1.0, Vor1.1.4 (which cannot be captured on the column and, thus, was not isolated during selection) was turned into a quenching-FRET sensor and responded to both 2 and 2.a. Using fluorescence, this sensor detected voriconazole concentrations as low as 3 μM; thus, this oligonucleotide is a candidate for incorporation of electrochemical sensors for in vivo monitoring (12).
Initial attempts starting with 2a at high concentrations, while introducing voriconazole separately in later cycles, failed, yielding exclusively analog-binding aptamers. Further conformational analysis using Newman projections clarified that 2a likely presents a dominant epitope during selection in which fragments I and II are positioned anti. Conversely, in voriconazole, these structural subunits are gauche (Fig. 4A). Inspired by approaches to outflank the immunodominance of epitopes (32), we mixed 2 and 2a at their respective maximal concentrations in the initial selection steps, only gradually phasing out the analog. We hypothesized that this procedure would maximize the probability of release of aptamers that bind similar conformations of the target and its analog, which could be important in the initial rounds of selection. In contrast to previous failures, this change led to two aptamers (Fig. S61–62) responsive to 2 and 2a (Fig. 4B), confirming the advantage of adding the analog.
The mechanisms underlying the improved selection strategy for voriconazole are partially unclear because we cannot exclude the possibility that the analog minimizes target aggregation. Nonetheless, the presence of 2a is certain to improve target-receptor occupancy in the initial cycles, likely buttressing low effective concentrations of monomeric voriconazole in conformations that can elicit aptamers. The isolated aptamers do not bind fluconazole, suggesting they are not class-wide cross-reactive aptamers (28, 29, 17) and that stabilizing interactions occur with group III in 2 (Fig. 4A).
Mutagenesis studies indicated that our lead aptamer, Vor1.0, is a destabilized three-way junction (Fig. 4B), which we engineered into a FRET sensor Vor1.1.4 (Fig. 4B). The latter shows sufficient sensitivity for testing as an electrochemical sensor for in vivo use (12). This specific family of voriconazole-binding three-way junctions, despite being common, are eliminated from direct selections by exceptionally poor interactions with capture oligonucleotides, which was prevented in Vor1.0 by structure switching (Fig. 4B, S61). These observations showcase the complex balance between positive and negative selection pressures in selection protocols. Our procedures, as demonstrated through leucine and voriconazole selections, shift selection balance in our favor by addressing probabilistic barriers assigned to crowded (and other non-optimal) substructures within targets.
Conclusions
In traditional organic synthesis, the functional group abstractions and their reactivities guide us through transformations involving relationships between nuclei and electron clouds (33). Here, in structure-guided aptamer selections, analogous concepts directed random searches through the space of complementary interactions between targets and aptamer receptors. We developed several approaches that can be used in functional-group guided selections. These include insertion reselection, carrying-over and anchoring of partial motifs, the expanded use of metal complexes as “protective” groups matched to targets, placeholders, cross-linkers, and the synthesis of simpler analogs designed to overcome steric or conformational barriers. These approaches can be further studied, optimized, and combined with one another and with traditional protocols (3, 4), organic receptor cofactors (6, 34), and modified bases (35), while considering library designs (25), to enable isolation of high-quality aptamers and engineering of biosensors for previously inaccessible targets.
There are further topics to which our approach, once systematically expanded, is expected to provide new insights: First, there is the question of natural selection of complex functions in the hypothetical, pre-protein, RNA world (36). Behaving as tinkerers (37), we reused simple sequence pieces to find new functions requiring more complex sequences, thus expanding the early work on the use of cofactors in RNA catalysis (38). Second, the approach that applies structural analysis of ligands to find optimal receptors could be inverted, combined with structural methods and insights from a large set of aptamers to improve our ability to design small-molecule drugs that specifically modulate natural nucleic acid targets (39). And third, we provide a substantially expanded set of sequences with confirmed target binding that could be used to improve training sets for computational designs of aptamers (40).
Supplementary Material
Acknowledgments:
We thank Bogdan Solaja, Francine Katz, Henry Hess, Darko Stefanović, Sergei Rudchenko, Alison K. Rinderspacher, Craig Boyle, Virginia Cornish, and John Loeb for their input during the writing of this manuscript. We thank Shixian Deng and Alison Rinderspacher for helping Z. C. with the synthesis of analogs and handling of data. M.N.S. and K.Y. thank the Maple Syrup Urine Disease Family Group for inspiration and guidance, and Kevin Strauss and Karlla Brigatti, Clinic for Special Children, Strasburg, PA, for advice on testing and applications of leucine sensors. K.Y. thanks B.-T. Zhang for introducing her to hypernetwork theory, which led to insertion-reselection designs. M.N.S. dedicates the work to his teachers of organic chemistry, Yoshito Kishi and Bogdan Solaja.
Funding:
The project was supported by the National Institutes of Health (voriconazole, leucine, other small molecules, and general conceptualization of functional-group based approach, GM138843 to M.N.S., neurotransmitters, DA045550 to A.M.A., other small molecules DK126739 to S.M., and EB022015 to M.N.S), the National Science Foundation (aptamers as inputs to molecular computing: CCF1518715 to M.N.S., aptamers for planar compounds to use spiegelmers: 1763632 to M.N.S.), the Defense Threat Reduction Agency (overlapping materials, 16–1-0053 to M.N.S.), the Raymond and Beverly Sackler Center, in Honor of Herbert Prades, at CUIMC (instrumentation), and the Maple Syrup Urine Disease Family Group gift for studies of receptors that bind leucine.
Footnotes
Conflict of interest statement: M.N.S., K.Y., T.S.W., A.M.A., S.T., J.S., and S.M. have (and expect) patents and patent applications regarding aptamers, their uses (Patent Application 20210223240) and sequences (Patent Application 20190136241), including on the use of cofactors in aptamer selection (Patent Number 10155940). M.N.S. and T.P.W. have founders’ shares of a start-up company (Aptatek), are on the scientific board and have expected consulting incomes related to aptamers from companies (M.N.S.: Aptatek and Nutromics, T.S.W.: Aptatek). K.Y. had consulting income from academic institutions and expects one from a company (Nutromics).
Data and materials availability:
All data needed to evaluate the conclusions in the paper are presented in the paper or the supplementary materials, except high-throughput sequencing data for selections, which are deposited in the Sequence Read Archive of the NCBI and can be found using the title of this paper or Accession no. PRJNA947642. No original software was created during these studies.
References and Notes
- 1.Ellington AD, Szostak JW, In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990). [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L, Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990). [DOI] [PubMed] [Google Scholar]
- 3.Ruscito A, DeRosa MC, Small-molecule binding aptamers: Selection strategies, characterization, and applications. Front. Chem. 4, 1–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu H, Alkhamis O, Canoura J, Liu Y, Xiao Y, Advances and challenges in small-molecule DNA aptamer isolation, characterization, and sensor development. Angew. Chem. Int. Ed. 60, 16800–16823 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakatsuko N et al. , Aptamer-field-effect transistors overcome Debye length limitations for small molecule sensing. Science 362, 319–324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang KA et al. , Recognition and sensing of low-epitope targets via ternary complexes with oligonucleotides and synthetic receptors. Nat. Chem. 6, 1003–1008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung KM, et al. , Phenylalanine monitoring via aptamer-field-effect transistor sensors ACS Sensors, 4, 3308–3317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tejavibulya N, et al. , Hydrogel microfilaments toward intradermal health monitoring. iScience, 21, 328–340 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson BS, et al. , Real-time, aptamer-based tracking of circulating therapeutic agents in living animals. Sci. Transl. Med. 5, 213ra165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao C et al. , Implantable aptamer-field-effect transistor neuroprobes for in vivo neurotransmitter monitoring. Science Advances 7:eabj7422 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stroek K, et al. , Evaluation of 11 years of newborn screening for maple syrup urine disease in the Netherlands and a systematic review of the literature: Strategies for optimization. JIMDreports 54, 68–78 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dauphin-Ducharme P et al. , Electrochemical aptamer-based sensors for improved therapeutic drug monitoring and high-precision, feedback-controlled drug delivery. ACS Sens. 4, 2832–2837 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elewa H, El-Mekaty E, El-Bardissy A, Ensom MHH, Wilby KJ, Therapeutic drug monitoring of voriconazole in the management of invasive fungal infections: A critical review, Clin. Pharmacokinet. 54, 1223–1235 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Nutiu R, Li Y, In vitro selection of structure-switching signaling aptamers. Angew. Chem. Int. Ed. 44, 1061–1065 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Rajendran M, Ellington AD, Selection of fluorescent aptamer beacons that light up in the presence of zinc. Anal. Bioanal. Chem. 390, 1067–1075 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Yarus M, Amino acids as RNA ligands: A direct-RNA-template theory for the code’s origin. J. Mol. Evol. 47, 109–117 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Wiedman GR, Zhao Y, Perlin DS, A Novel, Rapid, and low-volume assay for therapeutic drug monitoring of posaconazole and other long-chain azole-class antifungal drugs. mSphere, 3, ee00623–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehlert FJ, Estimation of the affinities of allosteric ligands using radioligand binding and pharmacological null methods.” Mol. Pharm. 33, 187–194 (1987). [PubMed] [Google Scholar]
- 19.Free SM, Wilson JW, A mathematical contribution to structure-activity studies. J. Med. Chem. 7, 395–399 (1964). [DOI] [PubMed] [Google Scholar]
- 20.Bissantz C, Kuhn B, Stahl M, A medicinal chemist’s guide to molecular interactions. J. Med. Chem. 53, 5061–5084 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorsch JR, Szostak JW, Chance and necessity in the selection of nucleic acid catalysts. Acc. Chem. Res. 29, 103–110 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Osborne SE, Ellington AD, Nucleic acid selection and the challenge of combinatorial chemistry. Chem. Rev. 97, 349–370 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Gohlke H, Klebe H, Approaches to the description and prediction of the binding affinity of small-molecule ligands to macromolecular receptors. Angew. Chem. Int. Ed. 41, 2644–2676 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Baum B et al. , Non-additivity of functional group contributions in protein-ligand binding: A comprehensive study by crystallography and isothermal titration calorimetry. J. Mol. Biol. 397, 1042–1054 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Carothers JM, Oestreich SC, Davis JH, Szostak JW, Informational complexity and functional activity of RNA structures. J. Am. Chem. Soc. 126, 5130–5137 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buryak A, Severin K, A chemosensor array for the colorimetric identification of 20 natural amino acids. J. Am. Chem. Soc. 127, 3700–3701 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Liu Z et al. , Tuning the reactivity of nitriles using Cu(II) catalysis – potentially prebiotic activation of nucleotides. Chem. Sci. 35, 7052–7057 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang K-A, Pei R, Stefanovic D, Stojanović M, Optimizing cross-reactivity with evolutionary search for sensors. J. Am. Chem. Soc. 134, 1642–1647, (2012). [DOI] [PubMed] [Google Scholar]
- 29.Yang W, et al. , In vitro isolation of class-specific oligonucleotide-based small-molecule receptors. Nucleic Acids Res. 47, e71 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravikumar K, Sridhar B, Prasad KD, Ak. K. S. Bujanga Rao, Voriconoazole, an antifungal drug. Acta Cryst. E 63, 0565–0567 (2007). [Google Scholar]
- 31.Corey EJ, The Logic of Chemical Synthesis. John Wiley & Sons; (1989). [Google Scholar]
- 32.Angeletti D, Kosic I, Santos JJS, Yewdell JW, Outflanking immunodominance to target subdominant broadly neutralizing epitopes. PNAS, 116, 13474–13479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochiai H, The logical structure of organic chemistry and the empirical adequacy of the classical concept of the molecule. HYLE-Int. J. Phil.Chem. 19, 139–160 (2013). [Google Scholar]
- 34.Manimala JC, Wiskur SL, Ellington AD, Anslyn EV, Tuning the specificity of a synthetic receptor using a selected nucleic acid receptor. J. Am. Chem. Soc. 126, 16515–16519 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Pfeiffer F et al. , Identification and characterization of nucleobase-modified aptamers by click-Selex. Nat. Protoc. 13, 1153–1180 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Saito H, The RNA world “hypothesis”. Nature Rev. Mol. Cell. Bio. 23, 582 (2022). [DOI] [PubMed] [Google Scholar]
- 37.Jacob F, Evolution and tinkering. Science 196, 1161–1166 (1977). [DOI] [PubMed] [Google Scholar]
- 38.Lorsch JR, Szostak JW, In vitro evolution of new ribozymes with polynucleotide kinase activity. Nature, 371, 31–36 (1994). [DOI] [PubMed] [Google Scholar]
- 39.Warner DK, Hajdin CE, Weeks KM, Principles for targeting RNA with drug-like small molecules. Nat. Rev. Drug Discov. 17, 547–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Billings WM, Hedelius B, Millecam T, Wingate D, Corte DD, ProSPr: Democratized implementation of Alphafold protein distance prediction network. bioRxiv doi:. 10.1101/830273 [DOI] [Google Scholar]
- 41.Yang K-A, Pei R, Stojanović MN, In vitro selection and amplification protocols for isolation of aptameric sensors for small molecules. Methods, 106, 58–65 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuker M, Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakatsuka N, Abendroth JM, Yang K-A, Andrews AM, Divalent cation dependence enhances dopamine aptamer biosensing, ACS Appl. Mater. Interfaces, 13, 9425–9435 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Najdi TS, Yang C-R, Shapiro BE, Hatfield GW, Mjolsness ED, Application of a generalized MWC model for the mathematical simulation of metabolic pathways regulated by allosteric enzymes. J. Bioinform. Comp. Bio. 4, 335–355 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Douglas EF, Miller CJ, Sparer G, Shapiro H, Spiegel DA, A Comprehensive mathematical model for three-body binding equlibria. J. Am. Chem. Soc. 135, 6092–6099 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown A, Analysis of cooperativity by isothermal titration calorimetry, Int. J. Mol. Sci. 10, 3457–3477 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are presented in the paper or the supplementary materials, except high-throughput sequencing data for selections, which are deposited in the Sequence Read Archive of the NCBI and can be found using the title of this paper or Accession no. PRJNA947642. No original software was created during these studies.


