Some integral membrane proteins can reach the plasma membrane in a Golgi-bypassing manner, a process also known as type IV UcPS. Here, Li et al. report that RAB-8– and RAB-11–positive endosomes are utilized as intermediate carriers in the unconventional apical protein transport.
Abstract
Multiple physiology-pertinent transmembrane proteins reach the cell surface via the Golgi-bypassing unconventional protein secretion (UcPS) pathway. By employing C. elegans–polarized intestine epithelia, we recently have revealed that the small GTPase RAB-8/Rab8 serves as an important player in the process. Nonetheless, its function and the relevant UcPS itinerary remain poorly understood. Here, we show that deregulated RAB-8 activity resulted in impaired apical UcPS, which increased sensitivity to infection and environmental stress. We also identified the SNARE VTI-1/Vti1a/b as a new RAB-8–interacting factor involved in the apical UcPS. Besides, RAB-11/Rab11 was capable of recruiting RABI-8/Rabin8 to reduce the guanine nucleotide exchange activity of SMGL-1/GEF toward RAB-8, indicating the necessity of a finely tuned RAB-8/RAB-11 network. Populations of RAB-8– and RAB-11–positive endosomal structures containing the apical UcPS cargo moved toward the apical side. In the absence of RAB-11 or its effectors, the cargo was retained in RAB-8– and RAB-11–positive endosomes, respectively, suggesting that these endosomes are utilized as intermediate carriers for the UcPS.
Introduction
Unconventional protein secretion (UcPS) refers to ER-Golgi-independent synthetic pathway (Zhang and Schekman, 2013; Malhotra, 2013; Rabouille, 2017; Filaquier et al., 2022; Zheng and Ge, 2022). As the discovery of cargo proteins continues to grow, it is believed that UcPS is conserved across species and involved in various physiological conditions. The malfunction of UcPS is associated with the development of several diseases, neurodegeneration, and cancer (Ejlerskov et al., 2013; Lock et al., 2014; Son et al., 2016; Kim et al., 2018). The UcPS cargoes include both soluble and membrane proteins, which are delivered outside of the cell or to the plasma membrane (PM). Soluble cargoes lack signal peptides and thus do not enter the ER. On the other hand, membrane cargoes with signal peptides, after being synthesized in the ER, are transported via a route bypassing the Golgi.
The release of leaderless proteins, such as prototypical cytokine IL-1β, utilizes multiple unconventional secretory routes. In addition to the release through gasdermin pores during pyroptosis, cytosolic IL-1β can be translocated into the lumen of the ER-Golgi intermediate compartment (Zhang et al., 2015, 2020; Broz et al., 2020). Furthermore, autophagosomes and endolysosomes are reported to mediate the unconventional secretion of soluble leadless proteins (Duran et al., 2010; Dupont et al., 2011; Zhang et al., 2015; Ponpuak et al., 2015). For unconventional secretion of membrane cargoes, Golgi reassembly stacking protein (GRASP) has been identified as one of the prime regulators of the Golgi-bypassing UcPS (Rabouille, 2017). The GRASP-dependent UcPS facilitates the transport of mutant cystic fibrosis transmembrane conductance regulator (ΔF508-CFTR) and α integrin to the plasma membrane. However, GRASP does not regulate the UcPS of other membrane cargoes such as cilia peripherin and mutant pendrin (Schotman et al., 2008; Gee et al., 2011; Tian et al., 2014; Jung et al., 2016). By making the effort to establish the Caenorhabditis elegans intestine as an in vivo multicellular model, our recent work has revealed the involvement of RAB-8 in the Golgi-bypassing unconventional apical protein transport (Wang et al., 2022). However, the nature of post-ER intermediate carriers traversed by membrane cargoes en route to the PM and the regulatory mechanism remains to be clarified.
Endosomes constitute highly dynamic vesicular/tubular compartments, where small GTPase Rabs play a pivotal role in orchestrating the specificity of membrane trafficking (Stenmark, 2009). The specificity of Rabs generally arises from the selective recruitment and activation on specific membranes. Guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) mediate the switch between an active GTP-bound state and an inactive GDP-bound state (Cherfils and Zeghouf, 2013; Lamber et al., 2019). Polarized cells such as epithelia have distinct apical and basolateral membrane domains with unique protein and lipid composition. This necessitates a complicated endosomal architecture and an orchestrated polarized exocytosis regulated by diverse Rabs (Apodaca et al., 2012; Perez Bay et al., 2016). Experiments conducted with polarized Madin–Darby canine kidney (MDCK) cells and other epithelial cells have demonstrated that newly synthesized membrane proteins take an indirect route to the PM. Apical and basolateral proteins transit through Rab8-positive common recycling endosomes (CREs) and Rab11-positive apical recycling endosomes (AREs) following the exit from Golgi (Ang et al., 2004; Cresawn et al., 2007; Cramm-Behrens et al., 2008; Weisz and Rodriguez-Boulan, 2009; Thuenauer et al., 2014). Rab8 and Rab11a are similarly required for the correct intestinal apical protein localization in mouse (Sobajima et al., 2014; Sato et al., 2007). Notably, during primary ciliogenesis, endosomal trafficking is controlled by a Rab11–Rabin8–Rab8 cascade that also directs exocytic carrier vesicles toward the apical surface for de novo lumen generation (Knödler et al., 2010; Bryant et al., 2010; Westlake et al., 2011; Gálvez-Santisteban et al., 2012; Donovan and Bretscher, 2012). However, the role of Rab11-positive endosomes during the Golgi-bypassing UcPS and how Rab8 and Rab11 cooperate are yet to be elucidated.
By attempting to establish the soil nematode C. elegans as a genetically tractable model, we have recently studied the transport of a Golgi-bypassing secretory cargo, PGP-1/ATP-binding cassette (ABC) transporter (Wang et al., 2022). PGP-1 is localized to the apical surface of intestinal epithelia, providing a protective mechanism against environmental stressors, such as natural toxins, heavy metals, and pathogens (Broeks et al., 1995, 1996; Mahajan-Miklos et al., 1999). Our findings revealed the importance of RAB-8 activity during the unconventional apical transport of PGP-1 and identified SMGL-1 as a RAB-8 GEF (Wang et al., 2022). Here, we further show that hyperactive RAB-8 also impairs the Golgi-bypassing transport of membrane cargoes and the associated resistance to environmental stressors. Recruited by RAB-11, RABI-8 deactivated RAB-8 on endosomes. RAB-11 deficiency caused the retention of PGP-1 on RAB-8–positive endosomes, while knockdown of RAB-11 effectors retained PGP-1 on RAB-11–positive endosomes. Altogether, our work indicates the significance of the fine-tuned RAB-8 activity and the involvement of RAB-8/RAB-11–positive endosomes as intermediate carriers during Golgi-bypassing unconventional apical protein transport.
Results
Loss of RABI-8 impairs the Golgi-bypassing unconventional transport of apical membrane proteins
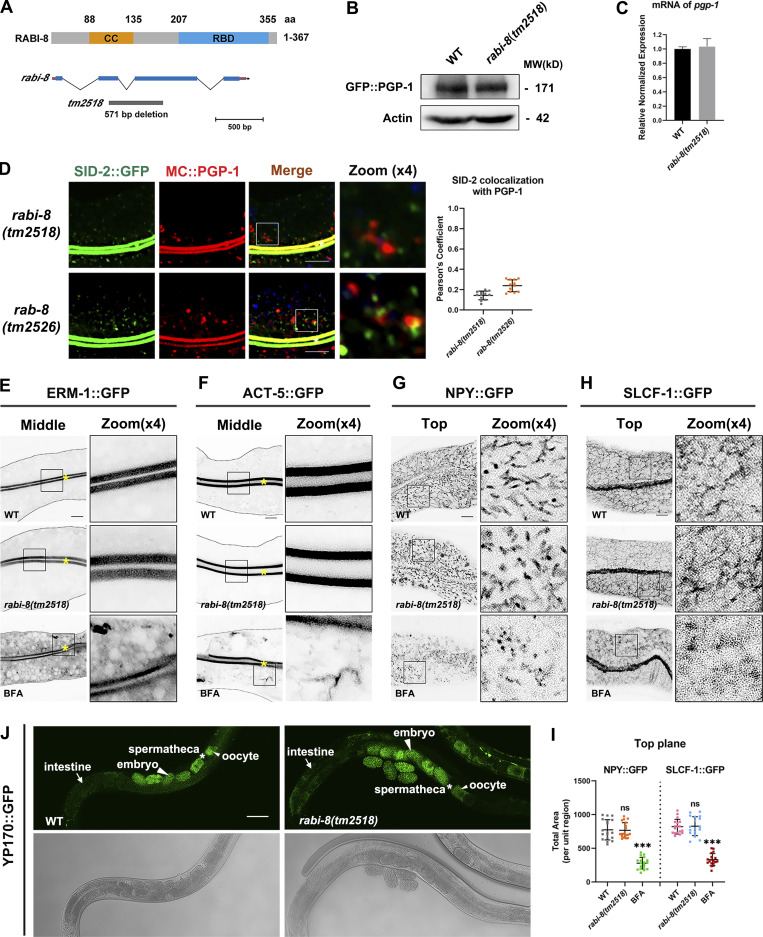
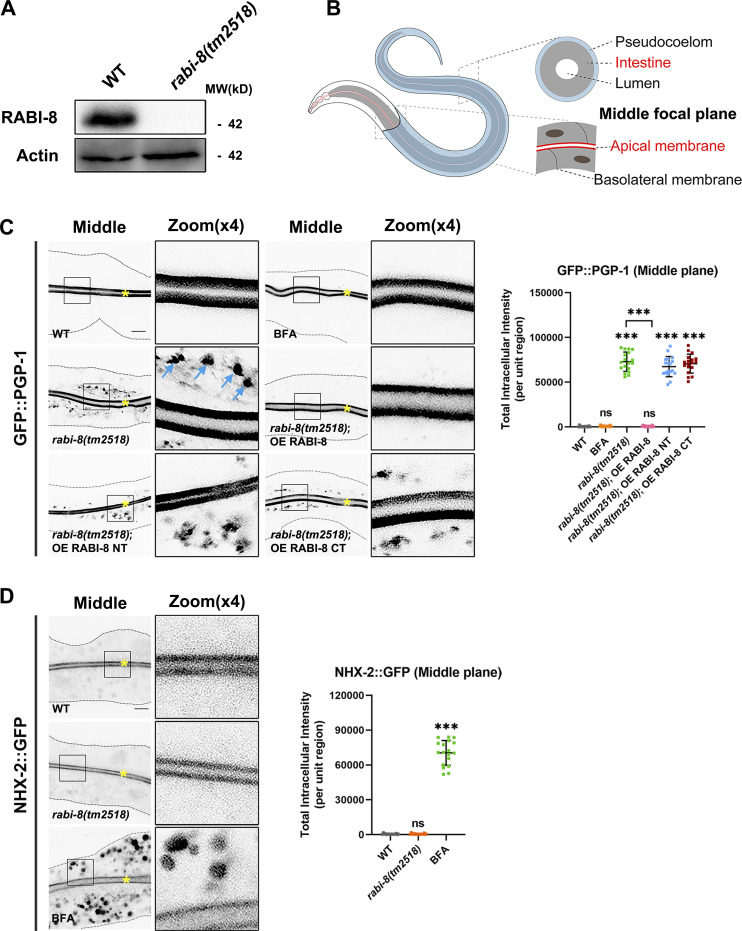
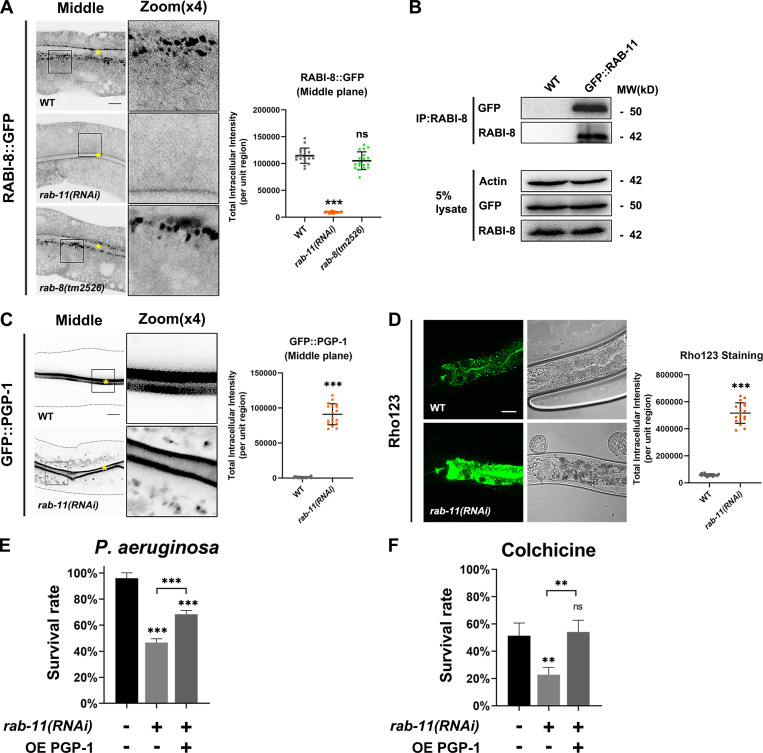
Loss of RABI-8 leads to increased RAB-8 activity, as indicated by increased fluorescence intensity and membrane-to-cytosol fraction of GFP-tagged RAB-8 (Wang et al., 2022). To gain a better understanding of the role of RABI-8 in Golgi-bypassing UcPS, we set out to analyze the localization of two established membrane cargoes, PGP-1 (ABC transporter) and SID-2 (double-stranded RNA [dsRNA] receptor) by confocal examination (Wang et al., 2022). The tm2518 allele of rabi-8 contains a 571-bp deletion that removes the majority of the second and third exons, generating a premature stop codon (Fig. S1 A). A polyclonal antibody against the recombinant RABI-8 recognized a single band of ∼42 kD that was absent in rabi-8(tm2518) mutants (Fig. 1 A). Thus, tm2518 is supposed to be null allele of rabi-8 and was used in our assays. Compared with the normal apical localization of GFP::PGP-1 and SID-2::GFP of wild-type animals from L4 larvae stage onward, RABI-8 mutation caused the abnormal intracellular accumulation of GFP::PGP-1 and SID-2::GFP near the apical area (Fig. 1 C and Fig. S1 D), suggestive of apical transport defects. Of note, we actually would not expect a complete block, as exocytosis can occur via various pathways, whereas aberrant intracellular cargo accumulation can still be utilized as a prominent indicator of UcPS insufficiency (Wang et al., 2022). Western blot and RT-PCR analysis revealed comparable levels of PGP-1 expression following RABI-8 depletion (Fig. S1, B and C), indicating that the observed defects cannot be attributed to expression level discrepancies. Importantly, the trafficking defects of PGP-1 in rabi-8(tm2518) mutants were rescued by RABI-8 overexpression (Fig. 1 C), thus demonstrating the absence of RABI-8 is responsible for the phenotypes. RABI-8 features two significant domains (Fig. S1 A): the predicted coiled-coil domain (88–135 aa) and the Rab-binding domain (RBD; 207–355 aa). Overexpression of either the N-terminus (NT; 1–200 aa) or C-terminus (CT; 200–367 aa) of RABI-8 failed to rescue the PGP-1 defects of rabi-8 mutants, suggesting that the full-length of RABI-8 is essential for the unconventional apical protein transport (Fig. 1 C).
Figure S1.
Loss of RABI-8 does not affect the exocytosis of other membrane and soluble cargoes. (A) The schematic domain structure of RABI-8 and rabi-8 locus. The coiled-coil (CC) domain is depicted in orange. The RBD is depicted in blue. The rabi-8 gene structure is shown with filled boxes representing exons and thin lines indicating introns. The arrow delineates the direction of transcription. The gray bars below the transcript show the size and position of the deleted regions in tm2518. (B) A presentative western blot showing expressional levels of GFP::PGP-1 in wild-type and rabi-8(tm2518) mutant animals. (C) RT-PCR showing mRNA expressional levels of PGP-1 in wild-type animals and rabi-8(tm2518) mutants. Each is the average of three replicates. (D) Confocal images and quantification showing the localization pattern of MC::PGP-1 and SID-2::GFP in rabi-8(tm2518) and rab-8(tm2526) mutants. Pearson’s correlation coefficients for GFP and MC signals were calculated (n = 12 animals). The signals from the apical membrane were avoided by manual ROI selection. White asterisks indicate intestinal lumen. Scale bars: 10 μm. (E and F) Compared to wild-type animals, the localization of ERM-1::GFP (E), and ACT-5::GFP (F) remained unchanged in rabi-8(tm2518). ERM-1::GFP (E) and ACT-5::GFP (F) both aberrantly accumulated in the cytoplasm of wild-type animals upon BFA treatment. Scale bars: 10 μm. Yellow asterisks indicate intestinal lumen. Dashed lines indicate the outline of the intestine. (G–I) The basolateral membrane protein NPY::GFP (G) and SLCF-1::GFP (H) exhibited similar tubular localization in wild-type and rabi-8(tm2518) animals. BFA treatment disrupted the normal localization. (I) Quantification of the total area of NPY::GFP in G and SLCF-1::GFP in H. Data are shown as mean ± SD (n = 18 each, six animals of each genotype sampled in three different unit regions of each intestine defined by a 100 × 100 [pixel2] box positioned at random). Statistical significance was determined using a one-way ANOVA followed by a post-hoc test (Dunn’s Multiple Comparison Test) for multiple comparisons. ***P < 0.001; ns, no significance. Data distribution was assumed to be normal but this was not formally tested. About one cell length of the intestine is shown in each panel. Scale bars: 10 μm. (J) Compared to wild-type animals, the distribution pattern of YP170::GFP remained unchanged in rabi-8(tm2518). Spermathecas are indicated by stars. Embryos and oocytes are indicated by big and small arrowheads, respectively. Intestines are indicated by arrows. Scale bars: 100 μm. Source data are available for this figure: SourceData FS1.
Figure 1.
Loss of RABI-8 impairs the delivery of apical Golgi-bypassing membrane cargoes. (A) A representative immunoblot of RABI-8 in lysates from wild-type and rabi-8(tm2518) mutant animals. Compared with wild-type, levels of RABI-8 are dramatically decreased in rabi-8(tm2518) mutants. (B) A schematic diagram of C. elegans intestine, showing the apical and basolateral membranes. The imaging plane where the apical membrane and intestinal lumen can be observed is defined as the middle focal plane of the confocal microscopy. (C) Confocal images of intestinal cells expressing GFP-tagged PGP-1. GFP::PGP-1 is predominantly localized at the apical membrane of intestinal epithelia in L4 larvae and young adults of wild-type animals. BFA treatment did not alter the localization. In contrast, GFP::PGP-1 displayed intracellular accumulation indicated by blue arrows in rabi-8(tm2518) mutants. Overexpression of full-length RABI-8 but not the NT or CT rescued the localization defects of PGP-1 in rabi-8(tm2518) mutants. (D) Confocal images of intestinal cells expressing GFP-tagged NHX-2. In contrast to the predominantly apical-localized NHX-2::GFP in L4 larvae or young adults of wild-type animals, NHX-2::GFP aberrantly accumulated in the cytoplasm of wild-type animals upon BFA treatment. NHX-2::GFP remained unchanged in rabi-8(tm2518) mutants. For quantification, the signals from the apical membrane were avoided by manual ROI selection. Data are shown as mean ± SD (n = 18 each, six animals of each genotype sampled in three different unit regions of each intestine defined by a 100 × 100 [pixel2] box positioned at random). Statistical significance was determined using a one-way ANOVA followed by a post-hoc test (Dunn’s Multiple Comparison Test) for multiple comparisons. ***P < 0.001; ns, no significance. Data distribution was assumed to be normal but this was not formally tested. About one cell length of the intestine is shown in each panel. Scale bars: 10 μm. Yellow asterisks indicate intestinal lumen. Dashed lines indicate the outline of the intestine. Source data are available for this figure: SourceData F1.
In contrast to PGP-1 and SID-2, RABI-8 mutation did not alter the distribution of other tested apical membrane proteins such as NHX-2::GFP, ERM-1::GFP, or ACT-5::GFP (Fig. 1 D and Fig. S1, E and F), which are all sensitive to brefeldin A (BFA) treatment and have been shown to travel along the traditional secretion pathway (Wang et al., 2022). The intracellular PGP-1 and SID-2 signals were largely separate in rabi-8(tm2518) and rab-8(tm2526) mutant animals (Fig. S1 D), which is consistent with the previous data that SID-2 can be transported through the conventional secretory pathway as well (Wang et al., 2022). The C. elegans intestine is a polarized single-layer epithelium, with the apical membrane facing the lumen and the basolateral membrane contacting the body cavity (Fig. 1 B). We further tested the effects of RABI-8 mutation on the steady-state localization of basolateral cargo proteins NPY::GFP (neuropeptide Y) and SLCF-1::GFP (monocarboxylate transporter; Wang et al., 2022). Both cargoes remained intact in rabi-8(tm2518) mutants (Fig. S1, G–I). Yolk protein YP170 is synthesized in the intestine and secreted into the body cavity from which it is endocytosed by oocytes (Balklava et al., 2007). Likewise, RABI-8 mutation did not affect the distribution of YP170::GFP (Fig. S1 J). Collectively, RABI-8 appears to selectively regulate the apical transport of PGP-1 and SID-2. These findings argue that RABI-8 participates in the apical Golgi-bypassing protein transport in the intestine.
Tight regulation of RAB-8 activity is required for the unconventional apical protein transport
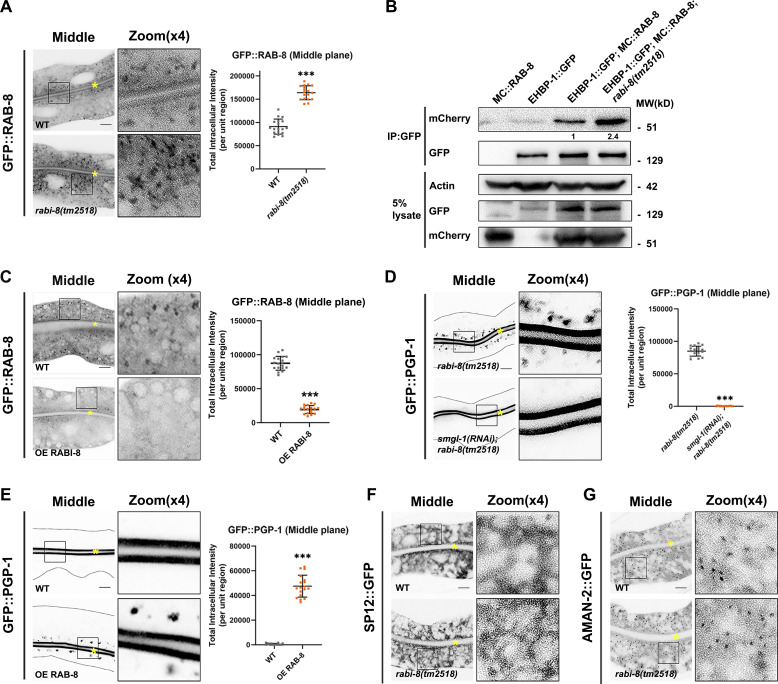
The PGP-1 transport defect prompted us to study the significance of RAB-8 activity regulation during the unconventional apical protein transport. To first confirm the level of active RAB-8 in rabi-8 animals, in addition to the fluorescence data and membrane fraction analysis (Fig. 2 A; Wang et al., 2022), we analyzed the amount of EHBP-1, a known RAB-8 effector protein, to be pulled down in the co-immunoprecipitation (Co-IP) experiments, which reflect the abundance of GTP-bound active RAB-8 (Shi et al., 2010). More than twofold EHBP-1 was pulled down from lysates of rabi-8 mutants in comparison with wild-type (Fig. 2 B), which is indicative of increased RAB-8 activity. Consistently, overexpression of RABI-8 caused a significant decrease in the number of punctate GFP::RAB-8, suggesting the redistribution of RAB-8 from endosomal compartments to the cytosol (Fig. 2 C). These results are not consistent with the role of RABI-8 as an RAB-8 GEF and rather support the possibility that RABI-8 functions to dampen the activity of RAB-8.
Figure 2.
Animals lacking RABI-8 exhibit increased RAB-8 activity. (A) Confocal images of intestinal cells expressing GFP-tagged RAB-8. Compared with wild-type animals, the intensity of GFP::RAB-8–labeled puncta was increased in rabi-8(tm2518) mutants. (B) mCherry (MC)::RAB-8 coimmunoprecipitated by EHBP-1::GFP were increased in rabi-8 mutant animals. Levels of MC::RAB-8 are normalized to the corresponding GFP (set to 1 in wild-type animals). (C) The fluorescence intensity and the number of GFP::RAB-8 puncta were diminished in RABI-8–overexpressing animals. (D) Confocal images of intestinal cells expressing GFP-tagged PGP-1. Knockdown of SMGL-1 significantly reduced the cytosol accumulation of GFP::PGP-1 in rabi-8(tm2518) mutants. (E) Compared with wild-type animals, GFP::PGP-1 aberrantly accumulated in the cytoplasm of animals overexpressing RAB-8. For quantification, the signals from the apical membrane were avoided by manual ROI selection. Data are shown as mean ± SD (n = 18 each, six animals of each genotype sampled in three different unit regions of each intestine defined by a 100 × 100 [pixel2] box positioned at random). Statistical significance was determined using a two-tailed, unpaired Student’s t test. ***P < 0.001. Data distribution was assumed to be normal but this was not formally tested. (F and G) The localization of fluorescence-tagged ER maker SP12 (F) and Golgi marker AMAN-2 (G) was unaltered in rabi-8(tm2518) mutants. About one cell length of the intestine is shown in each panel. Scale bars: 10 μm. Yellow asterisks indicate intestinal lumen. Dashed lines indicate the outline of the intestine. Source data are available for this figure: SourceData F2.
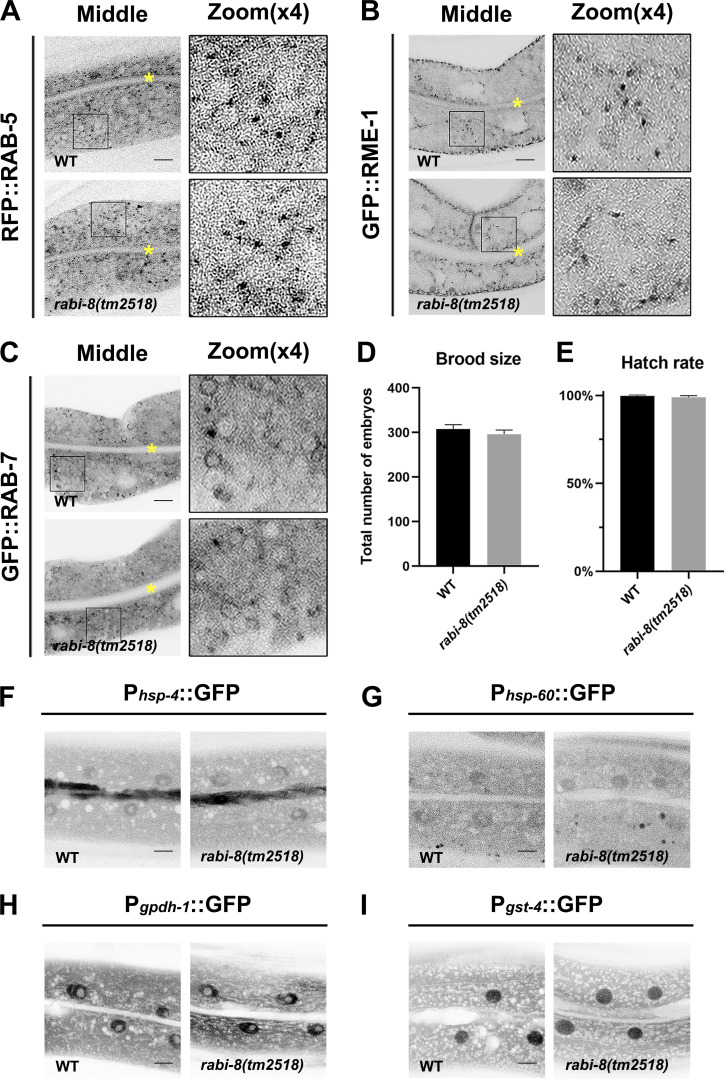
We next sought to determine whether the defect in PGP-1 secretion of rabi-8 mutants is the consequence of overactive RAB-8. If this is the case, downregulation of RAB-8 activity would be expected to at least partially restore the PGP-1 defects. Indeed, RNAi-mediated knockdown of smgl-1, the previously identified RAB-8–activating protein (Wang et al., 2022), restored the PGP-1 accumulation in rabi-8 mutants (Fig. 2 D). Loss of RABI-8 had no discernible effects on the localization of various markers on the secretory and endocytic pathways, such as SP12-labeled ER, AMAN-2–labled Golgi, RAB-5–labeled early/sorting endosome, RME-1–labeled basolateral recycling endosome, and RAB-7–labeled late endosome (Fig. 2, F and G; and Fig. S2, A–C), indicating the specific role of RABI-8 in the RAB-8 modulation during unconventional apical protein transport. Notably, the postulation that overactive RAB-8 could also be detrimental to the unconventional apical protein transport was further supported by the observation of aberrant intracellular PGP-1 accumulation upon RAB-8 overexpression (Fig. 2 E), similar to RAB-8–deficient animals (Wang et al., 2022).
Figure S2.
Some endolysosomal markers, the brood size, hatch rate, and several stress reporters are not affected in rabi-8 mutant animals. (A–C) The localization of fluorescence-tagged early endosome marker RAB-5 (A), basolateral recycling endosome marker RME-1 (B), and late endosome marker RAB-7 (C) were unaltered in rabi-8(tm2518) mutants. About one cell length of the intestine is shown in each panel. Scale bars: 10 μm. Yellow asterisks indicate intestinal lumen. (D and E) Mutation in rabi-8 had no effect on the brood size (D) or hatch rate (E), compared with wild-type animals. Three independent experiments were performed and data are shown as mean ± SD. (F–I) Confocal images of animals expressing Phsp-4::GFP (ER stress reporter) (F), Phsp-60::GFP (mitochondrial stress reporter) (G), Pgpdh-1::GFP (osmolyte accumulation) (H), and Pgst-4::GFP (detoxification) (I) in wild-type and rabi-8(tm2518) mutant animals. All the reporters displayed the similar expression and distribution upon rabi-8 depletion. Scale bars: 10 μm.
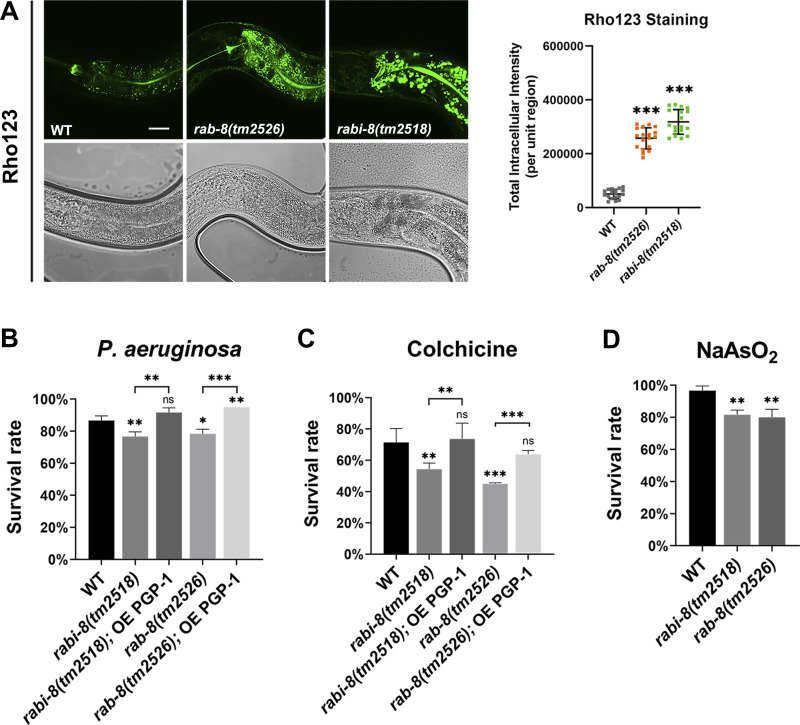
To further evaluate the effects of hypo- or hyper-RAB-8 activity, we conducted PGP functional assays, as ABC transporters such as P-glycoproteins (PGP) and multidrug-resistant proteins (MDRs) are associated with drug efflux functions. We tested the accumulation of the known mammalian fluorescent PGP substrate rhodamine123 (rho123; Kerboeuf and Guégnard, 2011; Ménez et al., 2019). Accumulation of this xenobiotic was five times higher in rab-8 mutants than that of wild-type animals (Fig. 3 A), implying that RAB-8 functionality is required for PGP-mediated transport of xenobiotics. In addition, supporting the postulation that hyperactive RAB-8 is also detrimental to PGP-1 secretion, similar dramatic accumulation of rho123 was observed in rabi-8 mutants (Fig. 3 A). Taken together, these results suggest that the tight regulation of RAB-8 activity is essential for Golgi-bypassing unconventional transport of PGP-1.
Figure 3.
Dysregulation of RAB-8 leads to reduced resistance to multiple environmental stressors. (A) Confocal images of wild-type, rab-8(tm2526), and rabi-8(tm2518) animals after a 48-h incubation with fluorescent dye rho123. Compared with wild-type animals, the fluorescence intensity of rho123 increased sixfold in rabi-8(tm2518) and fivefold in rab-8(tm2526), respectively. Data are shown as mean ± SD (n = 18 each, six animals of each genotype sampled in three different unit regions of each intestine defined by a 100 × 100 [pixel2] box positioned at random). Statistical significance was determined using a one-way ANOVA followed by a post-hoc test (Dunn’s Multiple Comparison Test) for multiple comparisons. ***P < 0.001. Data distribution was assumed to be normal but this was not formally tested. Scale bars: 20 μm. (B) Compared with wild-type animals, the survival rate of rab-8(tm2526) and rabi-8(tm2518) mutants was reduced after 24 h of P. aeruginosa exposure. PGP-1 overexpression (OE) improved the survival rate of both mutants. (C) Compared with wild-type animals, the survival rate of rab-8(tm2526) and rabi-8(tm2518) mutants was reduced upon colchicine (2 mM) treatment. PGP-1 overexpression improved the survival rate of both mutants. (D) Mutations in rab-8 and rabi-8 reduced the resistance to arsenite (2 mM). Data are shown as mean ± SD. Statistical significance was determined using a one-way ANOVA followed by a post-hoc test (Dunn’s Multiple Comparison Test) for multiple comparisons. *P < 0.05; **P < 0.01; ***P < 0.001; ns, no significance. Data distribution was assumed to be normal but this was not formally tested.
PGP-1 overexpression restores the decrease in resistance to environmental stressors caused by dysregulation of RAB-8 activity
Our research thus far has established the necessity of accurate regulation of RAB-8 activity for the apical exocytosis of PGP-1. We next addressed the potential physiological effects of the dysregulation of RAB-8 activity. In C. elegans, members of PGP/MDR family proteins have been associated with resistance to environmental stressors, such as various drugs, pathogens, and heavy metals (Broeks et al., 1995, 1996; Mahajan-Miklos et al., 1999). Accordingly, we scored the survival rate following Pseudomonas aeruginosa infection as well as colchicine and arsenite treatment. Under standard culture conditions, no alterations in either the brood size or hatch rate of the rabi-8 mutant animals were observed (Fig. S2, D and E). The mutants developed and behaved in a manner similar to wild-type animals. However, rabi-8 mutants exhibited a considerable decrease in their survival rate when exposed to all the tested environmental stressors (Fig. 3, B–D). To ascertain whether rabi-8 mutation induces stress responses under steady-state condition, we observed the fluorescent pattern of several stress reporters, including Phsp-4::GFP (ER stress), Phsp-60::GFP (mitochondrial stress), Pgpdh-1::GFP (osmolyte accumulation), and Pgst-4::GFP (detoxification). No significant differences were found between wild-type and rabi-8 mutant animals (Fig. S2, F–I). Thus, the sensitivity phenotype of rabi-8 is not caused by general development defects or nonspecific sickness. In clinical oncology, a high level of PGP contributes to multidrug resistance in cancer cells. Consistently, the crystal structure of PGP-1 has been determined and its drug-modulated ATPase function characterized in vitro supports the role in multidrug export (Jin et al., 2012). Hence, we speculated that the reduced tolerance to environmental stressors could be attributed, to some extent, to the excessive RAB-8 activity and the resultant disruption of PGP-1 transport. To validate this speculation, we overexpressed PGP-1 in rabi-8 mutants and evaluated the survival rate. It was remarkable that an overabundance of PGP-1 significantly ameliorated the reduced resistance of rabi-8 mutants to P. aeruginosa infection as well as colchicine treatment (Fig. 3, B and C).
We further assessed the impact of a decrease in RAB-8 activity on stressor resistance. The survival rate of smgl-1 knockdown animals and rab-8(tm2526) deletion mutants was drastically diminished following P. aeruginosa, arsenite, and colchicine treatment (Fig. 3, B–D; Wang et al., 2022). Similarly, we observed a significant enhancement of resistance to P. aeruginosa and colchicine in animals that overexpressed PGP-1 (Fig. 3, B and C). Together, these findings provide additional evidence for the functional link between PGPs and stressor resistance and highlight the significance of the fine-tuned RAB-8 activity in the PGP-1–mediated resistance to environmental stressors.
RABI-8 attenuates the SMGL-1’s GEF activity toward RAB-8 by affecting its oligomerization state
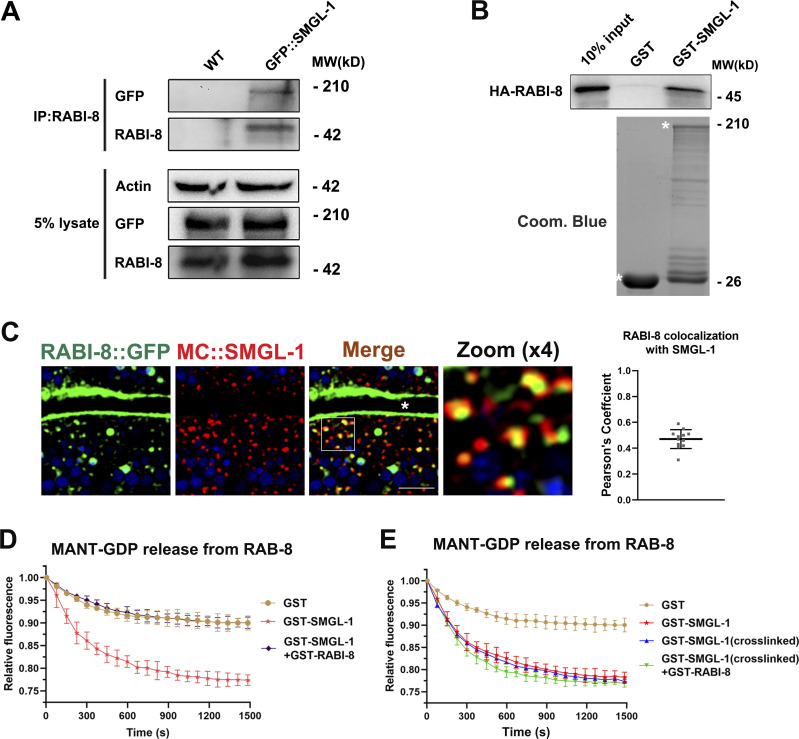
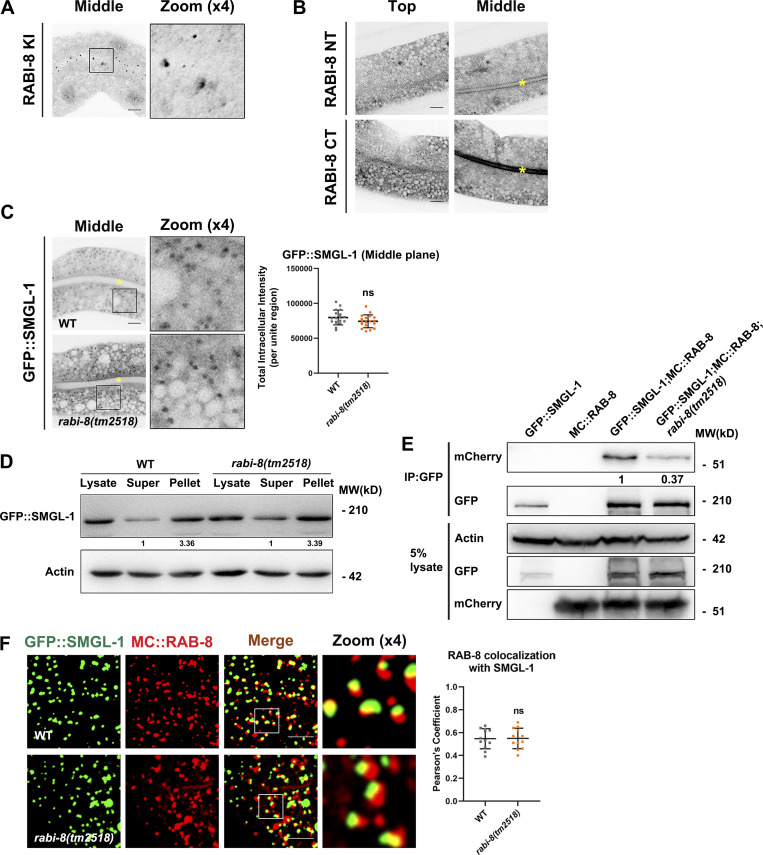
To gain insight into the mechanism by which RABI-8 regulates the activity of RAB-8, we set out to examine the potential interaction between RABI-8 and SMGL-1, the latter has been shown to activate RAB-8 in intact living worms (Wang et al., 2022). Using animals expressing GFP fusion of SMGL-1, Co-IP assays indicated that endogenous RABI-8 coprecipitated with SMGL-1 (Fig. 4 A). In vitro GST pull-down assays further revealed the direct interaction between RABI-8 and SMGL-1 (Fig. 4 B). Moreover, we compared the subcellular localization of RABI-8 and SMGL-1 using a GFP-tagged reporter of RABI-8 and CRISPR/Cas9-mediated RABI-8 knock-in line due to the poor performance of the RABI-8 antibody in immunostaining. RABI-8 was detected as punctate signal that was prominent in the apical cortical areas (Fig. 4 C and Fig. S3 A), while some additional puncta were also seen in the deep regions of the cytoplasm (Fig. 4 C). It is noteworthy that RABI-8::GFP showed considerable overlap with MC::SMGL-1 puncta, particularly at places near the apical end (Fig. 4 C). In addition, the fluorescence signal of RABI-8 NT (1–200 aa) containing the coiled-coil domain was mainly diffused in the cytoplasm, whereas the CT (200–367 aa) containing the RBD domain displayed a similar localization to that of the wild-type RABI-8 (Fig. S3 B), indicating the importance of RBD domain in the subcellular localization. These data provide the collective evidence that RABI-8 could physically interact with SMGL-1.
Figure 4.
RABI-8 interacts with SMGL-1 and reduces its GEF activity toward RAB-8. (A) RABI-8 precipitated GFP::SMGL-1 in a representative Co-IP assay. (B) A representative immunoblot showing the GST-tagged SMGL-1 was pulled down by in vitro translated HA-RABI-8. (C) Confocal images and quantification showing colocalization between RABI-8::GFP and MC::SMGL-1 in the intestinal cells. Broad-spectrum intestinal autofluorescent lysosome-like organelles can be seen in blue. Pearson’s correlation coefficients for GFP and mCherry signals were calculated (n = 12 animals). The signals from the apical membrane were avoided by manual ROI selection. Scale bars: 10 μm. White asterisk indicates the intestinal lumen. (D) In vitro GEF assay. MANT-GDP release from RAB-8 was measured by adding GST only, GST-SMGL-1, and GST-SMGL-1 and GST-RABI-8. GST-SMGL-1 promoted the release of MANT-GDP from RAB-8, while the presence of RABI-8 inhibited the promotion. Data are shown as mean ± SD from three independent replicates. (E) In vitro GEF assay. MANT-GDP release from RAB-8 was facilitated by adding GST only, GST-SMGL-1, GST-SMGL-1 (crosslinked) and GST-SMGL-1 (crosslinked), and GST-RABI-8. Crosslinking assay was performed using DSG. Data are shown as mean ± SD from three independent replicates. Source data are available for this figure: SourceData F4.
Figure S3.
RABI-8 does not affect the normal localization of SMGL-1 or the association of SMGL-1 with RAB-8. (A) Knock-in (KI) FLAG-RABI-8 via the CRISPR/Cas9 system to add an epitope tag (2xFLAG) to the CT of endogenous RABI-8. (B) Confocal images of intestinal cells expressing GFP-tagged NT and CT of RABI-8. (C) Confocal images of intestinal cells expressing GFP-tagged SMGL-1. Mutation in rabi-8 did not change the localization of GFP::SMGL-1. Data are shown as mean ± SD (n = 18 each, six animals of each genotype sampled in three different unit regions of each intestine defined by a 100 × 100 [pixel2] box positioned at random). Statistical significance was determined using a two-tailed, unpaired Student’s t test. ns, no significance. Data distribution was assumed to be normal but this was not formally tested. Yellow asterisks indicate intestinal lumen. (D) The membrane-to-cytosol ratio of GFP::SMGL-1 did not change notably in rabi-8(tm2518) mutants. Actin served as the loading control. Levels of SMGL-1 were normalized to the corresponding controls and set to one in the supernatant fraction. (E) GFP::SMGL-1 precipitated less MC::RAB-8 in rabi-8(tm2518) mutants in a representative Co-IP assay. (F) Confocal images and quantification showing partial colocalization between GFP::SMGL-1 and MC::RAB-8 in wild-type and rabi-8(tm2518) mutant animals. Pearson’s correlation coefficients for GFP and mCherry signals were calculated (n = 12 animals). Data are shown as mean ± SD (n = 12, 12 animals of each genotype of each intestine defined by a 100 × 100 [pixel2] box positioned at random). Statistical significance was determined using a two-tailed, unpaired Student’s t test. ns, no significance. Scale bars: 10 μm. Source data are available for this figure: SourceData FS3.
We next assessed whether this physical interaction is functionally related to the increased RAB-8 activity of rabi-8 mutants. SMGL-1 possesses the GEF activity toward RAB-8 both in vitro and in vivo (Wang et al., 2022); we therefore tested whether RABI-8 could affect the GEF activity of SMGL-1. GEF promotes the exchange from GDP to GTP and thus in the in vitro GEF assays, and the fluorescent intensity of N-Methylanthraniloyl (MANT)-GDP was facilitated to be released from RAB-8 in the presence of the GEF protein SMGL-1 (Fig. 4 D; Wang et al., 2022). However, the introduction of RABI-8 to the reaction system caused a considerable decrease in the amount of MANT-GDP released, bringing the fluorescent intensity of MANT-GDP close to the control level (Fig. 4 D). These results indicate that RABI-8 attenuates the GEF activity of SMGL-1 in vitro, which is consistent with the augmented RAB-8 activity observed when RABI-8 is depleted in vivo.
We further delved into the mechanism by which RABI-8 diminished the GEF activity of SMGL-1 toward RAB-8. To this end, we examined whether RABI-8 could affect the normal localization of SMGL-1 or the association of SMGL-1 with RAB-8. Fluorescence data and the membrane fraction analysis both revealed no significant alterations in the localization of SMGL-1 of rabi-8 mutants (Fig. S3, C and D). No more but even less RAB-8 was pulled down by SMGL-1 in rabi-8 mutants in the Co-IP assays (Fig. S3 E). In addition, the fluorescent signal overlap between RAB-8 and SMGL-1 remained unchanged after RABI-8 depletion (Fig. S3 F). Together, these data suggest that RABI-8 does not affect the normal localization of SMGL-1 or reduce the association of SMGL-1 with RAB-8.
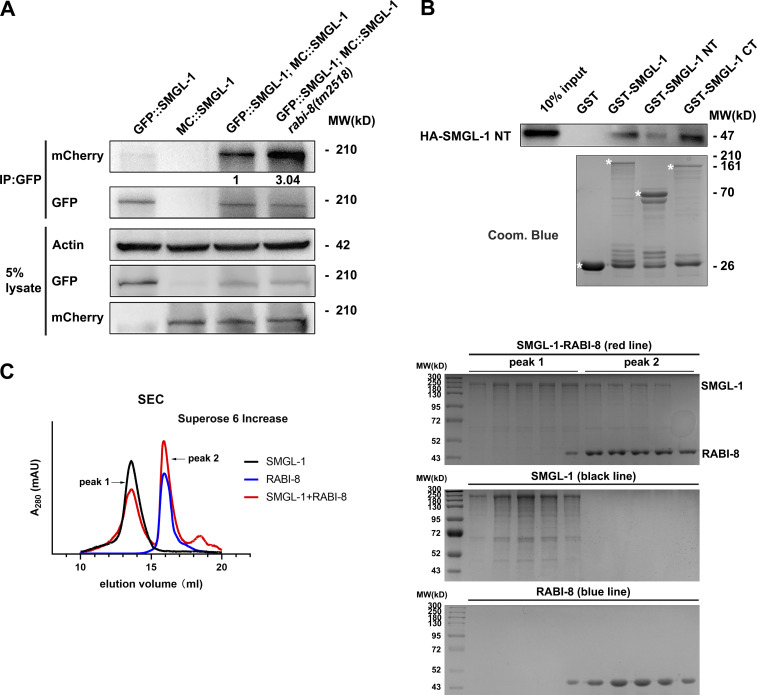
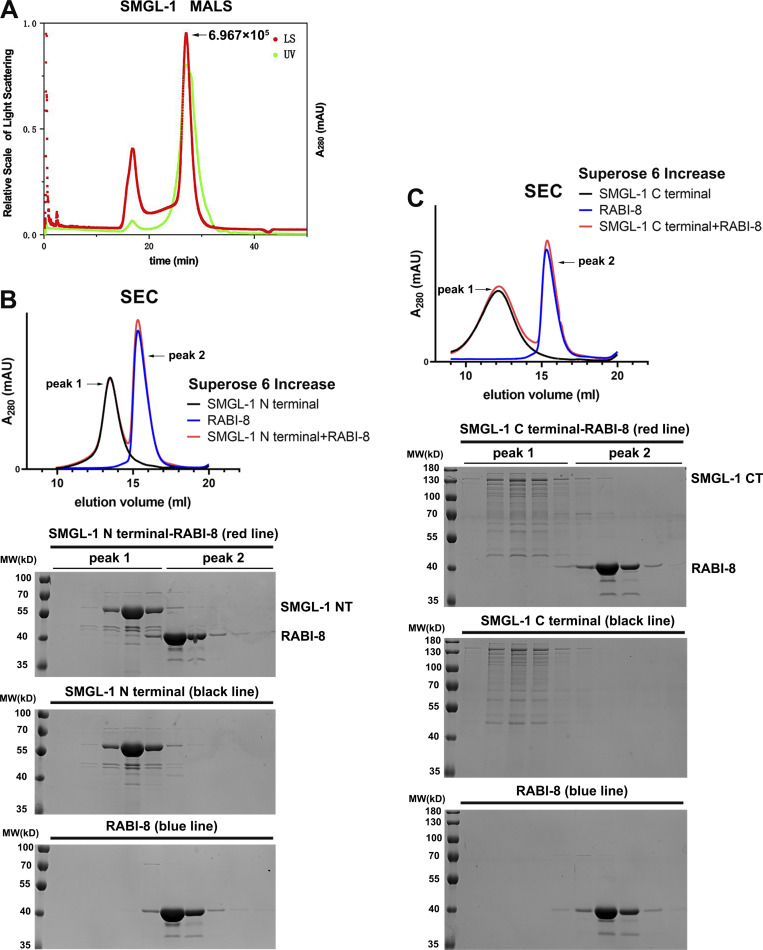
Multiple studies have revealed that certain GEF proteins function in the form of dimerization or oligomerization (Bos et al., 2007; Müller and Goody, 2018); therefore, we sought to determine whether SMGL-1 is capable of self-association and the effect of RABI-8 on its oligomerization state, especially taking into account the N-terminal WD40 repeats on SMGL-1. Using transgenic lines that co-expressed the mCherry and GFP-fusion SMGL-1, we affirmed the self-association features of SMGL-1 in the Co-IP assays (Fig. 5 A). Further pull-down assays revealed that the NT (1–399 aa) directly interacted with itself, the α-helices–containing CT (393–1,615 aa), and the full-length SMGL-1 (Fig. 5 B). Based on these results, by employing size-exclusion chromatography (SEC) and multiangle light scattering, we observed that the full-length SMGL-1 was capable of forming a stable homotetramer (Fig. 5 C and Fig. S4 A). Of note, in the presence of RABI-8, the height of the tetrameric SMGL-1 peak in the SEC was decreased and some SMGL-1 was eluted together with RABI-8, likely in the form of a heterologous protein complex (Fig. 5 C). This data is consistent with the interaction between RABI-8 and SMGL-1 and also suggest that RABI-8 can affect the oligomerization state of SMGL-1. To confirm this, we again performed the Co-IP assays to compare the amount of SMGL-1 to be precipitated in wild-type animals with that in animals lacking RABI-8. Data showed that in rabi-8(tm2518) mutants, threefold MC::SMGL-1 was precipitated by GFP::SMGL-1, which supports the in vitro result (Fig. 5 A). Oligomerization was also observed with the SMGL-1 NT and CT regions, yet RABI-8 appeared to have no discernible effect on their oligomerization state (Fig. S4, B and C). Together, our data suggest that RABI-8 interferes with the oligomeric state of SMGL-1, and this disturbance involves the full-length SMGL-1.
Figure 5.
RABI-8 affects the oligomerization state of SMGL-1. (A) GFP::SMGL-1 precipitated MC::SMGL-1 in a representative Co-IP assay. Loss of RABI-8 increased the amount of precipitated MC::SMGL-1. Levels of MC::SMGL-1 are normalized to the corresponding GFP (set to 1 in wild-type animals). (B) The NT of SMGL-1 was pulled down by the full-length, NT, and CT of SMGL-1 in a representative protein pull-down assay. (C) Superose 6 increase SEC elution profile of SMGL-1 (black), RABI-8 (blue), and a mixture of SMGL-1 and RABI-8 (red). Right, from top to bottom: SDS-PAGE of the peak fractions from the SMGL-1–RABI-8, SMGL-1, and RABI-8 elution. Source data are available for this figure: SourceData F5.
Figure S4.
RABI-8 does not alter the oligomerization state of SMGL-1 NT or CT. (A) A static light scattering assay indicated that SMGL-1 forms oligomers. The x-axis represents the elution time. The parameters monitored by LS (laser-light scattering) and UV (ultraviolet light) detectors were plotted as curves. The molecular weight was calculated according to LS measurements. Peak: 6.967 × 105. MALS, multiangle light scattering. (B) Superose 6 increase SEC elution profile of SMGL-1 NT (black), RABI-8 (blue), and a mixture of SMGL-1 NT and RABI-8 (red). Below, from top to bottom: SDS-PAGE of the peak fractions from the SMGL-1 NT-RABI-8, SMGL-1 NT, and RABI-8 elution. (C) Superose 6 increase SEC elution profile of SMGL-1 CT (black), RABI-8 (blue), and a mixture of SMGL-1 CT and RABI-8 (red). Below, from top to bottom: SDS-PAGE of the peak fractions from the SMGL-1 CT-RABI-8, SMGL-1 CT, and RABI-8 elution.
To ascertain whether the oligomerization disturbance is responsible for the inhibition of SMGL-1’s GEF activity in vitro, we applied crosslinking reagent disuccinimidyl glutarate (DSG) to retain SMGL-1 in its tetrameric state. Under this condition, crosslinked SMGL-1 was still capable of promoting the release of MANT-GDP from the RAB-8; however, this GEF activity toward RAB-8 could no longer be repressed by RABI-8 (Fig. 4 E). These results collectively suggested that RABI-8 attenuates the GEF activity of SMGL-1 by interfering with its oligomerization state.
RAB-11 plays a role in the Golgi-bypassing unconventional apical protein transport
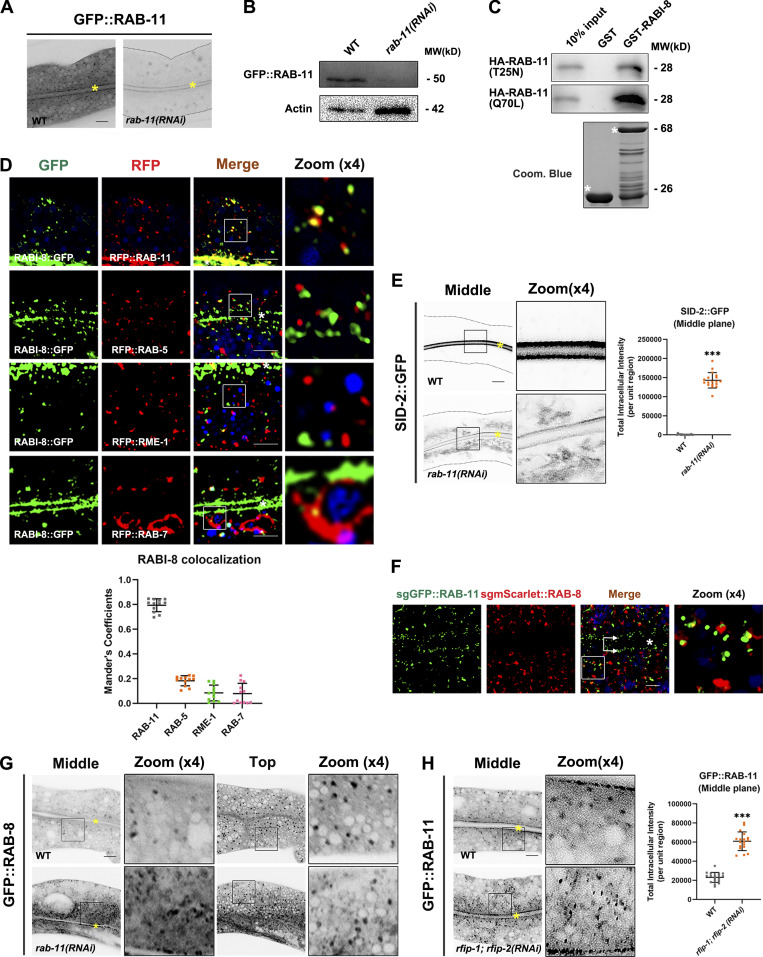
As a key regulator in protein secretion, Rab11 was reported to facilitate the membrane recruitment of Rabin8 during primary ciliogenesis (Knödler et al., 2010; Westlake et al., 2011; Feng et al., 2012). We thus tested the effect of RAB-11 deficiency on the subcellular localization of RABI-8. Due to the lethality of rab-11 null alleles, we used RNAi-mediated rab-11 knockdown animals. Significant reduction in GFP::RAB-11 levels was achieved by feeding L1 worms with rab-11 RNAi, as assessed by fluorescence confocal and Western blot, confirming the knockdown efficiency (Fig. S5, A and B). We observed a significant decrease in the clustered RABI-8 puncta in the apical cytoplasm of rab-11 knockdown animals (Fig. 6 A), consolidating the role of Rab11 in the recruitment of Rabin8 to the vesicular membrane. In contrast, the localization of RABI-8 was not affected in rab-8 mutants (Fig. 6 A). RABI-8 and RAB-11 displayed direct binding in GST pull-down assays (Fig. S5 C). The interaction was further confirmed by Co-IP of endogenous RABI-8 with GFP::RAB-11 (Fig. 6 B). Moreover, RABI-8 tended to bind to the constitutively active RAB-11 (Q70L; Fig. S5 C), which suggests that RABI-8 is likely a direct downstream effector of RAB-11. In agreement with biochemical results, a substantial colocalization between RABI-8 and RAB-11 (79.03 ± 1.6% of all RABI-8 colocalized with RAB-11) was observed in the subapical region (Fig. S5 D). As controls, RABI-8 did not colocalize with the early/sorting endosome marker RAB-5, basolateral recycling endosome marker RME-1, or late endosome marker RAB-7 (Fig. S5 D).
Figure S5.
The subcellular localization of RABI-8, RAB-8, and RAB-11. (A) Confocal images of intestinal cells expressing GFP-tagged RAB-11. RNAi-mediated knockdown of RAB-11 dramatically reduced the fluorescence intensity of GFP::RAB-11. Image brightness and contrast were adjusted identically for both images to reveal the contour of the intestinal cells in rab-11(RNAi) animals. Yellow asterisks indicate intestinal lumen. (B) A representative western blot showing expressional levels of GFP::RAB-11 in wild-type and rab-11(RNAi) animals. (C) GST-RABI-8 pulled down more HA-RAB-11(Q70L) than HA-RAB-11(T25N) in a representative pull-down assay. (D) Confocal images and quantification showing colocalization between RABI-8::GFP and organelle markers in the intestinal cells. RABI-8::GFP puncta overlapped with the subpopulation of RFP:RAB-11. RABI-8::GFP puncta were separated from RAB-5–labeled early endosomes, RME-1–labeled basolateral recycling endosomes and RAB-7–labeled late endosomes. In each set of images, broad-spectrum intestinal autofluorescent lysosome-like organelles can be seen in blue. Manders’ correlation coefficients for GFP and RFP signals were calculated (n = 12 animals). The signals from the apical membrane were avoided by manual ROI selection. Scale bars: 10 μm. White asterisks indicate intestinal lumen. (E) Confocal images of intestinal cells expressing GFP-tagged SID-2. Compared with wild-type animals, SID-2::GFP accumulated in intracellular structures in rab-11(RNAi) knockdown animals. For quantification, the signals from the apical membrane were avoided by manual ROI selection. Data are shown as mean ± SD (n = 18 each, six animals of each genotype sampled in three different unit regions of each intestine defined by a 100 × 100 [pixel2] box positioned at random). Statistical significance was determined using a two-tailed, unpaired Student’s t test. ***P < 0.001. Data distribution was assumed to be normal but this was not formally tested. Scale bars: 10 μm. Yellow asterisks indicate intestinal lumen. Dashed lines indicate the outline of the intestine. (F) Single-copy GFP::RAB-11; mScarlet::RAB-8 animals were imaged by structured illumination microscopy. Double arrows indicate the apical enriched RAB-11 sub-population. Scale bars: 5 μm. White asterisk indicates intestinal lumen. (G) Confocal images of intestinal cells expressing GFP-tagged RAB-8 in wild-type and rab-11(RNAi) animals. (H) Compared with wild-type animals, the GFP::RAB-11 was diffusive and formed smaller puncta in rfip-1; rfip-2 (RNAi) knockdown animals. Data are shown as mean ± SD (n = 18 each, six animals of each genotype sampled in three different unit regions of each intestine defined by a 100 × 100 [pixel2] box positioned at random). Statistical significance was determined using a two-tailed, unpaired Student’s t test. ***P < 0.001. Data distribution was assumed to be normal but this was not formally tested. Scale bars: 10 μm. Yellow asterisks indicate intestinal lumen. Source data are available for this figure: SourceData FS5.
Figure 6.
RAB-11 recruits RABI-8 to vesicular endosomal compartments and regulates unconventional apical protein transport. (A) Confocal images of intestinal cells expressing GFP-tagged RABI-8. Compared with wild-type animals, rab-11 knockdown significantly reduced the number and intensity of RABI-8::GFP puncta, while mutation in rab-8 had no obvious effect. Data are shown as mean ± SD (n = 18 each, six animals of each genotype sampled in three different unit regions of each intestine defined by a 100 × 100 [pixel2] box positioned at random). Statistical significance was determined using a one-way ANOVA followed by a post-hoc test (Dunn’s Multiple Comparison Test) for multiple comparisons. ***P < 0.001; ns, no significance. Data distribution was assumed to be normal but this was not formally tested. Scale bars: 10 μm. (B) RABI-8 precipitated GFP::RAB-11 in a representative Co-IP assay. (C) Confocal images of intestinal cells expressing GFP-tagged PGP-1. Compared with wild-type animals, GFP::PGP-1 accumulated in intracellular structures in rab-11(RNAi) knockdown animals. For quantification, the signals from the apical membrane were avoided by manual ROI selection. Data are shown as mean ± SD (n = 18 each, six animals of each genotype sampled in three different unit regions of each intestine defined by a 100 × 100 [pixel2] box positioned at random). Statistical significance was determined using a two-tailed, unpaired Student’s t test. ***P < 0.001. Data distribution was assumed to be normal but this was not formally tested. About one cell length of the intestine is shown in each panel. Scale bars: 10 μm. Yellow asterisks indicate intestinal lumen. Dashed lines indicate the outline of the intestine. (D) Confocal images of wild-type and rab-11(RNAi) knockdown animals after a 48-h incubation with fluorescent dye rho123. Accumulation of rho123 was about 10-fold higher in rab-11(RNAi) than in wild-type animals. Data are shown as mean ± SD (n = 18 each, six animals of each genotype sampled in three different unit regions of each intestine defined by a 100 × 100 [pixel2] box positioned at random). Statistical significance was determined using a two-tailed, unpaired Student’s t test. ***P < 0.001. Data distribution was assumed to be normal but this was not formally tested. Scale bars: 20 μm. (E) Compared with wild-type animals, rab-11 knockdown reduced the survival rate after 24 h of P. aeruginosa exposure. PGP-1 overexpression (OE) improved the survival rate of rab-11(RNAi) animals. (F) Compared with wild-type animals, rab-11 knockdown reduced the survival rate upon colchicine (2 mM) treatment. PGP-1 overexpression improved the survival rate of rab-11(RNAi) animals. Statistical significance was determined using a one-way ANOVA followed by a post-hoc test (Dunn’s Multiple Comparison Test) for multiple comparisons. **P < 0.01; ***P < 0.001; ns, no significance. Data distribution was assumed to be normal but this was not formally tested. Source data are available for this figure: SourceData F6.
To date, whether RAB-11 plays a role in the UcPS pathway remains unknown. We thus assessed the localization of PGP-1 and SID-2 in rab-11(RNAi) knockdown animals. Fluorescence data showed that PGP-1 and SID-2 both aberrantly accumulated in the cytosol of rab-11 knockdown cells (Fig. 6 C and Fig. S5 E). Consistently, PGP substrate rho123 showed dramatic intestinal accumulation (Fig. 6 D). In addition, rab-11 knockdown caused a dramatic decrease in the survival rates when exposed to P. aeruginosa and colchicine, whereas the expression of PGP-1 was able to mitigate this hypersensitivity (Fig. 6, E and F). Taken together, these findings demonstrate that RAB-11 plays an important role in the Golgi-bypassing unconventional apical protein transport.
PGP-1 transits RAB-8– and RAB-11–positive endosomes en route to the apical surface
Our data thus far have suggested that RAB-8 and RAB-11 both regulate the Golgi-bypassing unconventional apical protein transport. To explore their functional relationship in the process, we began by taking a close examination of RAB-8 and RAB-11 subcellular localization. Although the well-established transgenic lines of RAB-8 and RAB-11 used in our assays were originally generated by microparticle bombard methods to produce the low expression level (Chen et al., 2006; Schweinsberg and Grant, 2013), we additionally generated single-copy transgenic reporters of RAB-8 and RAB-11 by CRISPR/Cas9 (Fig. S5 F). Using structured illumination microscopy (SIM; Fig. S5 F), we examined the subcellular localization of RAB-8 and RAB-11, and data suggest the presence of two subpopulations of RAB-11. In line with the prior study in C. elegans (Winter et al., 2012), one subset of RAB-11 aligns along the apical membrane, with another scattered through the cytosol and concentrated subapically (Fig. S5 F). Nevertheless, fluorescence-labeled RAB-8 appears to be more evenly distributed throughout the cytosol, with less residing at regions beneath the apical surface (Fig. S5 F). Furthermore, there was partial overlapping between the apical punctate structures of RAB-8 and RAB-11 (Fig. S5 F). This localization pattern of RAB-11 and RAB-8 is similar to that observed in the mouse intestine (Sobajima et al., 2014), suggesting that RAB-11/Rab11 and RAB-8/Rab8 have overlapping yet distinct functions in the apical exocytotic process.
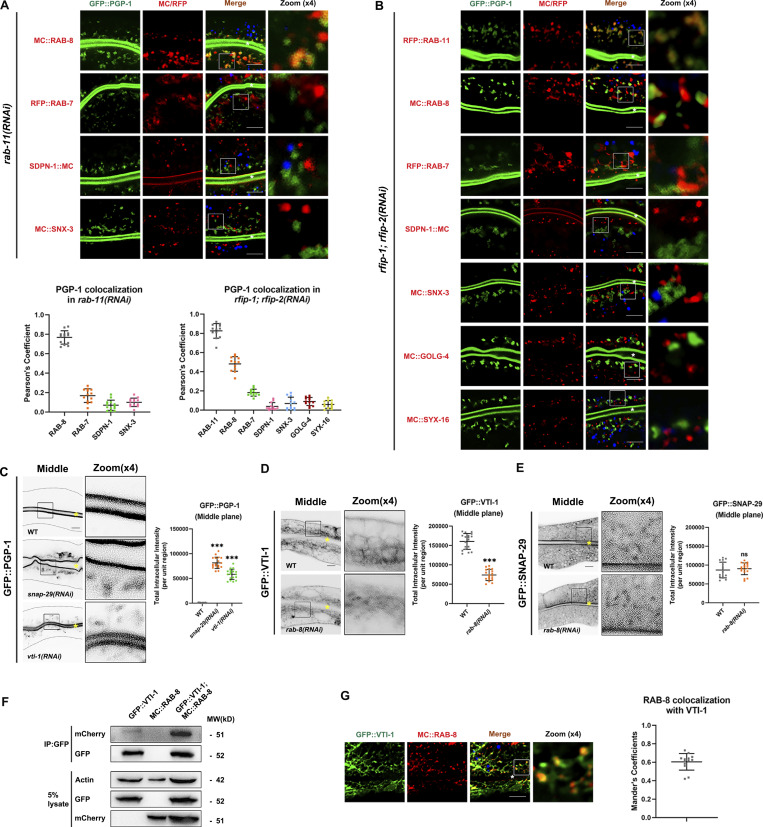
Based on the aforementioned data, we propose that RAB-8 could be deactivated on endosomes by RABI-8, which in turn is recruited by RAB-11. If this is the case, loss of RAB-11 would lead to retention of a certain amount of PGP-1 on RAB-8–positive endosomes. Indeed, apical endosomes labeled with GFP-tagged RAB-8 were grossly enlarged and displayed irregular shapes upon rab-11 knockdown (Fig. S5 G). The increased fluorescence of RAB-8 was similar to that in rabi-8 mutants (Fig. 2 A). Notably, we observed some PGP-1 was localized to subsets of enlarged RAB-8 puncta, but not to RAB-7–labeled late endosomes (Fig. 7 A). Furthermore, to examine the possibility that the aberrant PGP-1 accumulation of rab-11 knockdown animals was the consequence of defects in endocytic recycling, we tested whether PGP-1 was retained in early/sorting endosomes. No obvious overlapping between PGP-1 and two early/sorting endosome makers, SDPN-1 and SNX-3 (Tian et al., 2021; Gleason et al., 2016), was observed in rab-11 knockdown animals (Fig. 7 A). Taken together, these results suggest the absence of RAB-11 results in the retention of PGP-1 on RAB-8–positive endosomal structures.
Figure 7.
VTI-1 interacts with RAB-8 and is involved in unconventional apical protein transport. (A) Confocal images and quantification showing colocalization pattern between intracellular GFP::PGP-1 and several endosomal markers in rab-11(RNAi) knockdown animals. Intracellular GFP::PGP-1 puncta overlapped with subsets of MC::RAB-8. Intracellular GFP::PGP-1 puncta were separated from RAB-7–labeled late endosomes, and SDPN-1– and SNX-3–labeled early/sorting endosomes. (B) Confocal images and quantification showing colocalization pattern between intracellular GFP::PGP-1 and several endosomal markers in rfip-1; rfip-2 (RNAi) knockdown animals. Intracellular GFP::PGP-1 puncta overlapped with subsets of RFP::RAB-11 and MC::RAB-8. Intracellular GFP::PGP-1 puncta were separated from RAB-7–labeled late endosomes, SNX-3– and SDPN-1–labeled early/sorting endosomes, GOLG-4 and SYX-16–labeled trans-Golgi. In each set of images, broad-spectrum intestinal autofluorescent lysosome-like organelles can be seen in blue. Pearson’s correlation coefficients for GFP and MC/RFP signals were calculated (n = 12 animals). The signals from the apical membrane were avoided by manual ROI selection. (C) Confocal images and quantification showing the abnormal intracellular GFP::PGP-1 in vti-1(RNAi) and snap-29(RNAi) knockdown animals, in comparison with wild-type animals. For quantification, the signals from the apical membrane were avoided by manual ROI selection. Data are shown as mean ± SD (n = 18 each, six animals of each genotype sampled in three different unit regions of each intestine defined by a 100 × 100 [pixel2] box positioned at random). Statistical significance was determined using a one-way ANOVA followed by a post-hoc test (Dunn’s Multiple Comparison Test) for multiple comparisons. ***P < 0.001. Data distribution was assumed to be normal but this was not formally tested. (D) Confocal images and quantification showing the abnormal localization and decreased fluorescent intensity of GFP::VTI-1 in rab-8(RNAi) knockdown animals, in comparison with wild-type animals. Statistical significance was determined using a two-tailed, unpaired Student’s t test. ***P < 0.001. Data distribution was assumed to be normal but this was not formally tested. (E) Confocal images and quantification showing the intact localization of GFP::SNAP-29 in rab-8(RNAi) knockdown animals, in comparison with wild-type animals. Statistical significance was determined using a two-tailed, unpaired Student’s t test. ns, no significance. Data distribution was assumed to be normal but this was not formally tested. (F) GFP::VTI-1 precipitated MC::RAB-8 in a representative Co-IP assay. (G) Confocal images and quantification showing noticeable colocalization between GFP::VTI-1 and MC::RAB-8. Manders’ correlation coefficients for GFP and MC signals were calculated (n = 12 animals). Scale bars: 10 μm. White and yellow asterisks indicate intestinal lumen. Dashed lines indicate the outline of the intestine. Source data are available for this figure: SourceData F7.
Rab GTPases typically function by recruiting effector proteins to endosomal compartments (Stenmark, 2009). To further test the involvement of RAB-11 in the unconventional apical protein transport, we examined the role of RAB11 family interacting proteins (Rab11-FIPs) in the apical exocytosis of PGP-1. As the effectors of Rab11 GTPases, Rab11-FIPs modulate Rab11-dependent membrane traffic, including the apical targeting from ARE in polarized epithelial cells (Hales et al., 2001; Horgan and McCaffrey, 2009; Prekeris et al., 2000). RFIP-1 is the C. elegans ortholog of human Rab11-FIP3 and Rab11-FIP4 (Zheng et al., 2015), and RFIP-2 is the ortholog of human Rab11-FIP1 and Rab11-FIP5 by sequence blast. In intestinal cells with simultaneous knockdown of RFIP-1 and RFIP-2, the distinct RAB-11 puncta became much more diffuse, with smaller dots scattered in the cytosol (Fig. S5 H). This resembles the phenotype of mammal cells upon Rab11-FIP3 knockdown (Horgan et al., 2007) and supports the impact of effector proteins on the localization of Rab proteins. PGP-1 aberrantly accumulated in the cytosol of rfip-1; rfip-2 knockdown cells (Fig. 7 B). Notably, the accumulated PGP-1 colocalized well with RAB-11 puncta, partially colocalized with RAB-8 puncta, but not with RAB-7–labeled late endosomes, SNX-3– and SDPN-1–labeled early/sorting endosomes, or GOLG-4– and SYX-16–labeled trans-Golgi (Fig. 7 B), suggesting that upon Rab11-FIP deficiency, PGP-1 was retained in the RAB-11–positive endosomal compartments.
To identify potential RAB-8–interacting factors involved in PGP-1 secretion, we performed a small-scale candidate screen of C. elegans SNARE proteins, with initial attention to the abnormal intracellular PGP-1 signal. SNAP-29 served as a positive control, given it is required for organellar integrity of the whole endomembrane system (Sato et al., 2011). Next, we tested the effect of RAB-8 knockdown on the subcellular localization of these candidates based on the hypothesis that Rab GTPases could contribute to the compartmental specificity of some SNAREs (Ohya et al., 2009; Stenmark, 2009). As displayed in Fig. 7 C, PGP-1 accumulated abnormally in the cytosol of vti-1 knockdown animals. Besides, RAB-8 deficiency modestly disrupted the normal tubulovesicular distribution of GFP-tagged VTI-1 and decreased its fluorescent intensity but had little effect on SNAP-29 (Fig. 7, D and E). The Qb SNARE VTI-1 is the sole C. elegans homology to mammal Vti1a and Vti1b. Vti1a is proposed to mediate the homotypic fusion of endosomes and is required for retrograde endosome-to-TGN trafficking (Antonin et al., 2000; Brandhorst et al., 2006; Ganley et al., 2008; Mallard et al., 2002). In neurons, Vti1a/b regulates synaptic vesicle and dense core vesicle secretion (Walter et al., 2014; Emperador-Melero et al., 2018). Interestingly, Vti1a is also involved in unconventional traffic of KChIP1 and Kv4 potassium channels to the plasma membrane (Flowerdew and Burgoyne, 2009). To support the functional relevance of RAB-8 and VTI-1, we further performed the Co-IP assay to determine the interaction (Fig. 7 F). Consistent with the interaction, considerable colocalization between RAB-8 and VTI-1 was visible (Fig. 7 G). Together, these data reveal that VTI-1 could function as a potential RAB-8–interacting factor involved in unconventional apical protein transport.
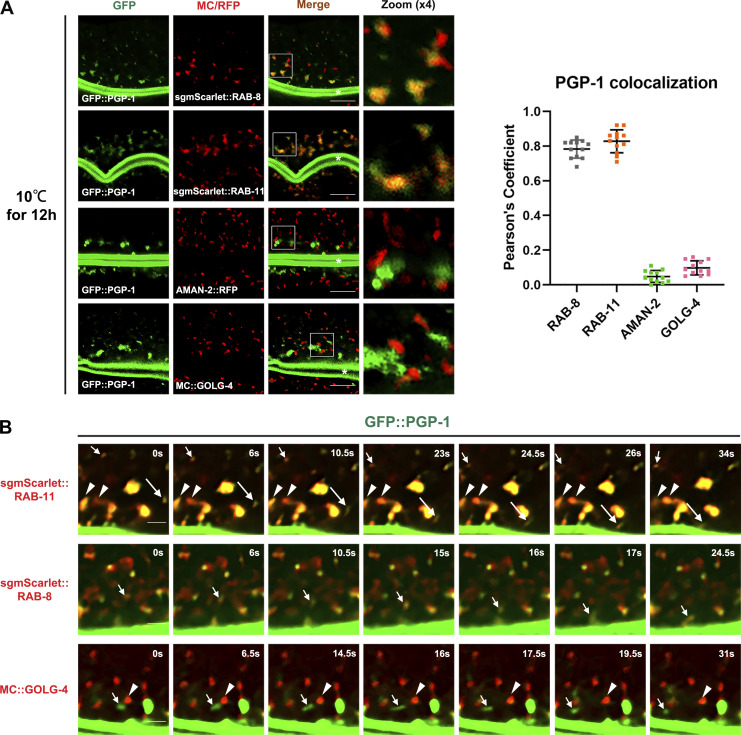
Due to the fast speed of PGP-1 transport of wild-type L4 animals under steady state, to confirm the transit of RAB-8– and RAB-11–positive endosomes by PGP-1, we further developed an assay to visualize the secretion. We transferred the worms which were kept at standard laboratory 20–10°C for 12 h and imaged the subcellular localization of PGP-1. Due to the low temperature, the secretion of PGP-1 was blocked. Interestingly, the intracellular accumulated PGP-1 showed good overlapping with RAB-8 and RAB-11 puncta (Fig. 8 A). By contrast, PGP-1 signals were separate from the AMAN-2–labeled cis-/medial-Golgi and GOLG-4–labeled trans-Golgi (Fig. 8 A). Further, animals were subsequently subjected to live-cell imaging after recovery for up to 30 min at 20°C. Fluorescence analysis revealed that populations of mobile mScarlet::RAB-11 or RAB-8–positive vesicular structures containing GFP::PGP-1 moved toward the apical side (Fig. 8 B and Videos 1 and 2). In particular, large quantities of RAB-11 signal either interacted with the RAB-11–positive endosomal subpopulation located along the apical surface or disappeared below the PGP-1–labeled apical membrane (Fig. 8 B), which possibly suggests a fusion event. PGP-1 puncta were barely observed to associate with Golgi marker GOLG-4 (Fig. 8 B and Video 3). Together, these results indicate RAB-8– and RAB-11–positive endosomes serve as intermediate carriers in the Golgi-bypassing unconventional apical protein transport.
Figure 8.
RAB-8– and RAB-11–positive endosomes serve as intermediate carries in the unconventional apical protein transport. (A) Confocal images and quantification showing colocalization pattern between intracellular GFP::PGP-1 with MC::RAB-8, RFP::RAB-11, Golgi marker AMAN-2::RFP, and MC::GOLG-4 in wild-type animals kept at 10°C for 12 h. Intracellular GFP::PGP-1 puncta overlapped with subsets of RAB-8 and RAB-11 and were separated from AMAN-2–labeled cis- and medial-Golgi and GOLG-4–labeled trans-Golgi. In each set of images, broad-spectrum intestinal autofluorescent lysosome-like organelles can be seen in blue. Pearson’s correlation coefficients for GFP and MC/RFP signals were calculated (n = 12 animals). The signals from the apical membrane were avoided by manual ROI selection. White asterisks indicate intestinal lumen. Scale bars: 10 μm. (B) Transgenic animals of GFP::PGP-1 and single-copy mScarlet::RAB-11 or mScarlet::RAB-8 or MC::GOLG-4 were imaged live by spinning disk confocal microscope at 20°C after a 12 h incubation at 10°C. Images were captured every 0.5 s for 2 min. Series of images that depict mScarlet::RAB-11– and mScarlet::RAB-8–labeled vesicles transporting GFP-labeled PGP-1 towards the apical surface are shown (see Videos 1 and 2). RAB-11 puncta can be observed to either interact with the RAB-11–positive endosomal subpopulation located along the apical surface (indicated by arrowheads) or disappear below the PGP-1–labeled apical membrane (indicated by long arrows). Image series show the movement of GFP::PGP-1 vesicles (indicated by arrows), with a neighboring MC::GOLG-4–labeled Golgi (indicated by arrowheads) remaining stationary at the time interval (see Video 3). Scale bars: 4 μm.
Video 1.
Transgenic animals of GFP::PGP-1 and single-copy mScarlet::RAB-11 were imaged live by spinning disk confocal microscope at 20°C after a 12-h incubation at 10°C. Intestinal cells were imaged every 0.5 s for 2 min at the middle focal plane to observe the apical membrane. The movie is compressed to 7 frames per second.
Video 2.
Transgenic animals of GFP::PGP-1 and single-copy mScarlet::RAB-8 were imaged live by spinning disk confocal microscope at 20°C after a 12-h incubation at 10°C. Intestinal cells were imaged every 0.5 s for 2 min at the middle focal plane to observe the apical membrane. The movie is compressed to 7 frames per second.
Video 3.
Transgenic animals of GFP::PGP-1 and MC::GOLG-4 were imaged live by spinning disk confocal microscope at 20°C after a 12-h incubation at 10°C. Intestinal cells were imaged every 0.5 s for 2 min at the middle focal plane to observe the apical membrane. The movie is compressed to 7 frames per second.
Discussion
The nematode C. elegans has proved to be a powerful genetic model for studying general mechanisms of membrane trafficking (Winter et al., 2012, Chen et al., 2006; Sato et al., 2014; Balklava et al., 2007). Our current work of unconventional apical protein transport has benefited from the investigation of exogenously expressed apical reporters such as P-glycoprotein PGP-1, which are predominantly delivered to apical surface from L4 larvae stage onward (Sato et al., 2007; Wang et al., 2022). Although not endogenously expressed, studying PGP-1’s transport has enabled us to discover the steady-state unconventional apical protein transport that is important for the physiology of C. elegans intestinal epithelia. Our prior studies have revealed that the absence of RAB-8 results in a decrease in PGP-1 delivery, thus making cells more susceptible to multiple environmental stressors (Wang et al., 2022). In this study, we provide evidence that hyperactive RAB-8 also leads to PGP-1 trafficking defects, and overexpression of PGP-1 significantly alleviates the reduced stressor resistance, no matter under the conditions of downregulated or upregulated RAB-8 activity. We propose that to ensure the progression of apical-directed Golgi-bypassing unconventional protein transport, RAB-8 needs to be deactivated appropriately, and this is achieved by inhibiting SMGL-1’s GEF activity with RABI-8, which is recruited by RAB-11 to endosomal structures. It is notable that upon rab-11 deficiency, intracellular accumulated PGP-1 is retained on RAB-8–positive endosomes, while upon deficiency of RAB-11 effector proteins, PGP-1 is accumulated on RAB-11–positive endosomes. Further, RAB-8– and RAB-11–postive endosomal structures containing PGP-1 move toward the apical surface. Our findings demonstrate the finely regulated RAB-8 activity is essential for the unconventional apical protein transport of C. elegans intestine and that RAB-8/RAB-11–positive endosomes are involved in the process.
Mammalian Rab8 and Rab10 belong to a Rab subfamily that includes Rab13 as well and are the closest relatives of yeast Sec4p (Pereira-Leal and Seabra, 2001). In MDCK cells, Rab10 is associated with the CRE and is implicated in basolateral recycling, probably regulating the transport from basolateral sorting endosomes to CRE (Babbey et al., 2006). In C. elegans intestine, in a similar way, RAB-10 mediates the basolateral recycling of clathrin-independent membrane cargo hTAC (Chen et al., 2006). Simultaneous knockdown of Rab10 and Rab8 in polarized MDCK cells produced more severe defects than individual knockdown on biosynthetic transport and epithelial polarization (Schuck et al., 2007), suggesting Rab8 and Rab10 may act cooperatively during the biosynthetic route. However, in our assays, the depletion of RAB-10 did not result in PGP-1 trafficking defects. In addition, the overexpression of RAB-10 had no rescue effects on PGP-1 accumulation in rab-8 mutants (Wang et al., 2022), which suggests that the RAB-10 and RAB-8 might function differently in the unconventional apical protein transport.
In yeast, Sec4p is transported to sites of polarized growth and constitutes the final Rab in the secretory pathway (Salminen and Novick, 1987; Ortiz et al., 2002). Ypt32 and Sec2p are yeast homologs of Rab11 and Rabin8, respectively, and there exists a Ypt32–Sec2p–Sec4p cascade as well (Ortiz et al., 2002). However, a study in mouse intestinal cells showed that Rab11a primarily localizes to the subapical region, whereas Rab8a primarily localizes to the perinuclear region (Sobajima et al., 2014). This is also the case in C. elegans intestinal cells, physically, Rab11 is closer to the apical membrane (Fig. S5 F). The differential localization indicates that Rab8 and Rab11 might function at different steps along the apical transport process in epithelial cells. Of note, immunofluorescence analysis of polarized MDCK in combination with immunoblots from apical post-Golgi carriers revealed a sequential passage of lactase-phlorizin hydrolase and sucrase-isomaltase through Rab-4, Rab-8, and Rab11-positive endosomes, before arriving at the apical membrane (Cramm-Behrens et al., 2008). In addition, by directly visualizing the biosynthetic trans-endosomal trafficking of apically targeted rhodopsin in polarized MDCK cells, it is reported that Rab11 present on the AREs was rapidly released upon the final fusion onset (Thuenauer et al., 2014), suggesting that Rab11-positive AREs are likely the final endosomal structures traversed by apical membrane cargoes. Consistently, our findings reveal that RAB-8 can be deactivated by RABI-8, which in turn is recruited by RAB-11. Furthermore, upon deficiency of RAB-11 effectors, PGP-1 accumulated on RAB-11 punctate structures, indicating PGP-1 transits RAB-11–positive endosomes en route to the apical surface.
Our results suggest a variance in the Rab11–Rabin8–Rab8 cascade, in which the localization and function of RABI-8/Rabin8 are Rab11 dependent as well. As the activation of RAB-8/Rab8/Sec4 is essential for polarized exocytosis, the deactivation of RAB-8 is also needed for unconventional apical protein transport. The exact reason why RABI-8 functions to attenuate rather than activate RAB-8 in the intestine of C. elegans is still unknown since RABI-8 promoted the release of MANT-GDP from RAB-8 in in vitro GEF assay (data not shown). Considering the loss of an obvious Rab8 GEF Rabin8/Sec2 homolog but retention of Rab8 in Drosophila (Lamber et al., 2019), combined with the discovery of SMGL-1 as the RAB-8 GEF in C. elegans, a simple explanation is the diversity of regulatory pathways controlling Rab proteins in various cell-specific and tissue-specific processes. The biophysical properties of RABI-8 might be tuned for its in vivo performance. Nevertheless, our results undoubtedly add substantial support to the concept that AREs constitute a central intermediate compartment along multiple intracellular biosynthetic trafficking routes in polarized cells, including secretory basolateral and apical protein targeting (Lock and Stow, 2005; Cresawn et al., 2007; Thuenauer et al., 2014), transcytosis in both directions (Apodaca et al., 1994; Pelissier et al., 2003), and unconventional apical protein transport as well.
A number of studies carried out in both polarized and non-polarized cells have demonstrated the key regulatory role of Rab11 in the membrane trafficking in and out of the recycling endosomes (Welz et al., 2014). In this study, apical Golgi-bypassing membrane cargoes PGP-1 and SID-2 displayed similar abnormal accumulation in the cytosol of both rab-8 mutants and rab-11 knockdown animals. However, the phenotype in rab-11 knockdown animals was actually much stronger than in rab-8 mutants, implying that RAB-11 appears to be a key molecular in the unconventional apical protein transport. It is tempting to speculate that more than one Golgi-bypassing UcPS pathway is at play. While cargoes such as PGP-1 use the RAB-8–dependent pathway, they might also be shipped through RAB-8–independent and RAB-11–dependent mechanisms. Furthermore, although we have revealed the unconventional apical protein transport in the C. elegans intestinal epithelia under steady state, it is believed that UcPS is mostly triggered by stress events. Considering the role of PGP-1 in the resistance of various stressors, future work will be needed to define and dissect specific roles of RAB-8 and RAB-11 under stress conditions.
Materials and methods
General methods and strains
All strains of C. elegans were derived from Bristol strain N2. The animals were cultured and maintained according to standard procedures unless indicated otherwise (Brenner, 1974). A complete list of strains used in this study can be found in Table S1.
RABI-8 antibody production
To generate the F54C9.11/RABI-8 antibodies, the sequence encoding amino acids of F54C9.11 was amplified from an N2 cDNA library and cloned into pGEX-2T for 6xHis tagging. His fusions were purified from bacteria and injected into two rats (GL Biochem). RABI-8 antibodies were affinity purified using the same antigen. Other antibodies used in this study include mouse anti-actin monoclonal antibody (sc-47778; Santa Cruz Biotechnology), rabbit anti-HA monoclonal antibody (C29F4; Cell Signaling Technology), rabbit anti-mCherry polyclonal antibody (ab167453; Abcam), and rabbit anti-GFP polyclonal antibody (ab290; Abcam).
RNAi treatment
RNAi-mediated interference was performed according to standard protocols (Timmons and Fire, 1998). The feeding constructs were from the Ahringer library or prepared by PCR from cDNA fragments and cloned into the RNAi vector L4440 (Timmons and Fire, 1998). For most experiments, four to five L4 larvae were plated onto the nematode growth media (NGM) plates containing 1 mM IPTG seeded with dsRNA bacterial clone. The F1 progeny were then examined. For essential genes such as rab-11, L1 larvae were plated onto the NGM plates and young adults were examined.
CRISPR/Cas9 knock-in strains
To generate the RABI-8::2xFLAG knock-in alleles, donor cocktails were used as repair templates, which can be synthesized using two reannealed fragments: one was FLAG sequence with 40 bp upstream and downstream homology arms of the guide cut site, and the other was FLAG sequence only, without any homology arms. A single gRNA (sgRNA) template containing T7 promoter (5′-AAGCTAATACGACTCACTATAGG-3′), sgRNA sequence (rabi-8 sgRNA: 5′-ATGGCGTATGTTGGAACGC-3′), and a transactivating CRISPR RNA sequence (5′-GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTT-3′) were prepared using PCR. The PCR products were used as the template for sgRNA transcription using the HiScribe Quick T7 High Yield RNA Synthesis Kit (E2050; New England Biolabs). sgRNAs were purified using a Monarch RNA Cleanup Kit (T2040; New England Biolabs). sgRNA and 0.5 μl Cas9 nuclease (10 μg/μl; Integrated DNA Technologies) were incubated at 37°C for 10 min, then repaired using a template donor cocktail (final 40 ng/μl), after which the selection marker pRF4[rol-6(su1006)] vectors (final 50 ng/μl) were added. Transgenic strains were obtained using standard microinjection techniques. The candidate F1 animals were identified by following a roller phenotype. The homozygote F2 progenies were identified by PCR and sequencing.
Reporter construction
For reporters under the intestine-specific vha-6 promoter, vectors were constructed using Gateway technology (Invitrogen). The cDNAs encoding RABI-8, RABI-8(1–200 aa), RABI-8(200–367 aa), SYX-16, VTI-1, and SNAP-29 were cloned into the entry vector pDONR221 via BP reaction and then transferred into intestinal expression vectors by LR reaction to generate fusion plasmids (Chen et al., 2006). Plasmids were coinjected with the selective marker Podr-1::gfp or Podr-1::rfp into the germline of N2 hermaphrodites. At least two stable transgenic lines were analyzed for each reporter. Insertion of single-copy arrays of GFP::RAB-11 (I), mScarlet::RAB-11 (II), and mScarlet::RAB-8 (II) at the specific genomic region on chromosomes was achieved by CRISPR/Cas9 (Xu et al., 2016). ttTi4348 and ttTi5605 genome locus were used for the insertion sites on chromosome I and II, respectively.
RNA isolation and quantitative real-time PCR
Young adults were collected using M9 buffer (20 mM KH2PO4, 40 mM Na2HPO4, 100 mM NaCl, and 1 mM MgSO4). Total RNA was extracted using an RNeasy Plus Mini Kit (QIAGEN) and reverse-transcribed using a Revert Aid RT Kit (Thermo Fisher Scientific). Quantitative PCR reactions were performed using iTaq Universal SYBR Green (Bio-rad) and a CFX Connect Real-Time PCR Detection System (Bio-Rad). Actin was used as the internal control. Quantitative PCR reactions were performed with the following primers: act-5 F: 5′-CAACATTCAGGCTGTGCTTT-3′, act-5 R: 5′-TGATGGATTGGTAGGTGGTCT-3′; pgp-1 F: 5′-CAATCTGGAAAGAGTAAAA-3′, and pgp-1 R: 5′-CGTAAGCATATCTCCAAAGT-3′. The mRNA level in mutant animals was normalized to the level observed in wild-type worms, which was set to 1. Error bars indicate the standard deviation (SD) of three independent experiments.
Protein purification
The cDNA of SMGL-1, SMGL-1 NT (1–399 aa), SMGL-1 CT (393–1,615 aa), and RABI-8 was cloned into the pGEX-2T vector (GE Healthcare). A cleavage site for the PreScission protease (LEVLFQGP) was added to the NT. The plasmid was then transformed into an Arctic Express strain of Escherichia coli (Stratagene) for protein expression. Bacterial pellets were suspended in lysis buffer containing 50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1 mM DTT, 1 mM PMSF, and complete protease inhibitor cocktail. Extracts were cleared by centrifugation and supernatants were incubated with 300–3,000 µl glutathione sepharose 4B beads (GE Healthcare) at 4°C 6 h. Beads were washed six times with the above buffer and three additional times with preScission cleavage buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, and 1 mM DTT). Cleavage of GST tag was achieved by PreScission protease overnight at 4°C. The resulting protein sample was further purified through size-exclusion chromatography (Superdex 200 or Superose 6 increase; GE Healthcare).
In vitro pull-down assay
cDNAs encoding RABI-8, SMGL-1 NT (1–399 aa), RAB-11(T25N), and RAB-11(Q70L) were cloned into a modified vector pcDNA3.1(+; Invitrogen) for 2xHA tagging. cDNAs encoding RABI-8, SMGL-1, SMGL-1 NT (1–399 aa), and SMGL-1 CT (393–1,615 aa) were cloned into a pGEX-2T vector (GE Healthcare) for GST tagging. The HA-tagged proteins were synthesized in vitro using the Quick Coupled Transcription/Translation System–coupled transcription-translation system (Promega). GST fusion proteins were expressed in the Arctic Express strain of E. coli (Stratagene). Bacterial pellets were lysed in 50 ml lysis buffer (50 mM Hepes [pH 7.5], 400 mM NaCl, 1 mM DTT, and 1 mM PMSF) with Complete Protease Inhibitor Mixture Tablets (Sigma-Aldrich). Extracts were cleared by centrifugation and the supernatants were incubated with glutathione sepharose 4B beads (GE Healthcare) at 4°C for 6 h. Beads were washed six times with cold STET buffer (10 mM Tris-HCl [pH 8.0], 300 mM NaCl, 1 mM EDTA, and 0.1% Tween-20). In vitro–synthesized HA-tagged proteins were incubated with glutathione sepharose 4B beads. After six washes with STET buffer, the eluted proteins were separated on SDS-polyacrylamide-gel electrophoresis and blotted to nitrocellulose. After blocking and washing with TBST buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 0.1% Tween-20), the blot was probed with an anti-HA antibody.
Co-IP assay
Young adult worms were collected with M9 buffer. About 100 μl worm pellet was resuspended in ice-cold lysis buffer (25 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM PMSF, 1 mM DTT, 1 mM Na3VO4, and 10 mM NaF) containing protease inhibitor cocktail (Sigma-Aldrich). Then, the worm pellet was lysed with an automatic grinding machine (Jingxin Inc.) at 65 Hz for 5 min (10-s intervals after every minute). The lysates were centrifuged at 12,000 g for 10 min at 4°C. The supernatant was collected and incubated with GFP-Trap Agarose (Chromotek) or Protein A+G agarose beads (Beyotime) that was preloaded with RABI-8 antibody for 2 h at 4°C. The precipitates were washed 5 × 10 min with lysis buffer and subjected to immunoblotting with anti-mCherry, anti-GFP, anti-RABI-8, and anti-actin antibodies.
Membrane fractionation assay
Wild-type and mutant animals expressing intestinal GFP fusion proteins were synchronized and collected as young adults (9-cm plates × 10). Animals were washed off with M9 buffer and the pellet was resuspended in 500 μl lysis buffer (50 mM Tris-HCl [pH 8.0], 20% sucrose, 10% glycerol, 1 mM DTT, and protease inhibitor). The worms were lysed and centrifuged at 1,000 g for 10 min at 4°C. 200 μl of the cleared lysate was centrifuged at 100,000 g for 1 h. Supernatants were collected and the pellets were reconstituted in the same volume of lysis buffer. Equal volumes of supernatants and pellets were subjected to immunoblotting with anti-GFP and anti-actin antibodies.
BFA microinjection
5 μM BFA (Beyotime) diluted in 0.2% DMSO were microinjected into young adult intestines. Worms were allowed to recover at 20°C for 2 h before examination.
ABC transporter-mediated xenobiotic transport assay
The PGP functional assay was adapted from a method previously described (Ménez et al., 2019). Briefly, synchronized L1 larvae were collected in M9 buffer containing E. coli strain OP50 and 10 μM rho123 (Beyotime) for 48 h at 20°C. After washing with M9 buffer, young adult worms were transferred to NGM plates to recover for 30 min at 20°C before microscopy.
In vitro GEF assay
Briefly, 50 μM MANT-GDP (Invitrogen) was incubated with 500 pmol 6xHis-RAB-8 in the preloading buffer (50 mM Tris-HCl [pH 8.0], 110 mM NaCl, 1 mM EDTA, 0.8 mM DTT, and 0.005% Triton X-100). After Rab preloading at 25°C for 60 min, 10 mM MgCl2 was added to stop the reaction. Then, 200 pmol preloaded RAB-8 was incubated with 100 pmol GST-tagged proteins in a GEF buffer (50 mM Tris [pH 8.0], 150 mM NaCl, and 0.5 mM MgCl2) at 25°C for 100 s. The nucleotide exchange reaction (the dissociation of MANT-GDP from Rab proteins) was triggered by adding the hydrolysis-resistant GTP analog Guanosine 5'-[β,γ-imido]triphosphate (Merck) at a final concentration of 0.1 mM and was recorded using a Synergy 2 Multi-Mode Reader (BioTek) at an excitation wavelength of 366 nm and an emission wavelength of 443 nm (Sakaguchi et al., 2015).
Bacterial resistance assay
PA14 culture was grown at 25°C overnight and spread on NGM plates. After spreading the bacterial culture, plates were incubated at 37°C for 24 h and then placed at room temperature for 8–12 h. 30 L4 larvae were transferred to these assay plates and incubated at 25°C. For essential genes such as rab-11, L1 larvae were fed on dsRNA-expressing bacteria and L4 larvae were transferred to plates with PA14 lawn. Mortality was scored after 24 h. Animals were considered dead if they failed to respond to a gentle tap on the head and tail with a platinum wire. Three independent assays were performed for each experiment.
Drug sensitivity assay
15 young adult hermaphrodites were plated onto the NGM plates containing 2 mM colchicine (TargetMol) and allowed to lay eggs for 24 h. The parents were then removed and the eggs were counted. The survival rate was calculated as the total number of adults divided by the number of eggs. Three independent assays were performed for each experiment.
Heavy metal exposure assay
30 L1 larvae were plated onto the NGM plates containing NaAsO2 (2 mM; Sigma-Aldrich) for 48 h. Animals were considered dead if they failed to respond to a gentle tap on the head and tail with a platinum wire. Three independent assays were tested for each experiment.
Microscopy and imaging analysis
Fluorescence images (GFP, mCherry, and DAPI channels) were obtained at 20°C using a C2 laser scanning confocal microscope (Nikon) equipped with a 100× NA 1.2 oil-immersion objective and captured using NIS-Elements AR 4.40.00 software. The Z-series of the optical sections were acquired using a 0.8-μm step size. Fluorescence data from the GFP channel were analyzed using Metamorph software version 7.8.0.0 (Universal Imaging). The “Integrated Morphometry Analysis” function of Metamorph was used to measure the fluorescent intensity (total intracellular intensity, a composite index of endosomal quantity, and mean intensity) and fluorescent area within unit regions. From a total of six animals of each genotype, “total intracellular intensity” and “total area” were sampled in three different unit regions of each animal defined by a 100 × 100 (pixel2) box positioned at random (n = 18 each). Signals from the apical membrane were avoided when using Metamorph for manual region of interest (ROI) selection. Colocalization images were quantified using Fiji (Image J) software. Pearson’s and Manders’ correlation coefficients for GFP and mCherry/RFP signals were calculated and the entire imaging area of 12 animals was analyzed for most samples. Signals from the apical membrane were deliberately avoided when using Metamorph for manual ROI selection. Live-cell imaging was performed on a Dragonfly spinning disk confocal microscope (Andor) equipped with a 63× (NA 1.4) oil objective. Dynamic images were captured using Fusion software (Andor).
Statistical analysis
The significance of differences and 95% confidence intervals were assessed and plotted using Prism software version 8.02 (GraphPad Software). SD was used as the y-axis error bars for bar charts plotted from the mean values. Two-tailed unpaired Student’s t test and one-way ANOVA followed by a post-hoc test (Dunn’s Multiple Comparison Test) were performed.
Online supplemental material
Fig. S1 shows the normal localization of some membrane cargoes upon BFA treatment and in RABI-8–depletion cells. Fig. S2 shows the intact pattern of some endocytic markers and stress reporters in rabi-8(tm2518) mutants. Also shown are the unaffected brood size and hatch rate. Fig. S3 shows the localization of RABI-8 and SMGL-1 and the association of SMGL-1 with RAB-8. Fig. S4 shows the oligomerization state of SMGL-1 NT and CT. Fig. S5 shows the subcellular localization of RABI-8, RAB-8, and RAB-11 in indicated genetic backgrounds. Table S1 contains information on C. elegans strains. Video 1 depicts the mobility of RAB-11– and PGP-1–labeled vesicles. Video 2 depicts the mobility of RAB-8– and PGP-1–labeled vesicles. Video 3 depicts the mobility of PGP-1–labeled vesicles and the relative static state of GOLG-4–labeled Golgi at the time interval.
Supplementary Material
contains information on C. elegans strains.
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 6.
is the source file for Fig. 7.
is the source file for Fig. S1.
is the source file for Fig. S3.
is the source file for Fig. S5.
Acknowledgments
We thank all lab members for their helpful discussions.
L. Lin was funded by the National Natural Science Foundation of China (92254302, 32270741, and 32070703). Y.R. Guo was funded by STI2030-Major Projects (2022ZD0206600).
Author contributions: Conceptualization: X. Li, A. Shi, and L. Lin. Data curation: X. Li, A. Shi, and L. Lin. Formal analysis: X. Li, Y.R. Guo, A. Shi, and L. Lin. Funding acquisition: Y.R. Guo and L. Lin. Investigation: X. Li, B. Liu, Y. Wen, and J. Wang. Methodology: X. Li, A. Shi, and L. Lin. Project administration: A. Shi and L. Lin. Resources: X. Li, B. Liu, Y. Wen, J. Wang, Y.R. Guo, A. Shi, and L. Lin. Software: X. Li, Y.R. Guo, A. Shi, and L. Lin. Supervision: A. Shi and L. Lin. Validation: X. Li, A. Shi, and L. Lin. Visualization: X. Li, Y.R. Guo, and L. Lin. Writing—original draft: X. Li, A. Shi, and L. Lin. Writing—review and editing: A. Shi and L. Lin.
Data availability
Data are available in the article itself and its supplementary materials.
References
- Ang, A.L., Taguchi T., Francis S., Fölsch H., Murrells L.J., Pypaert M., Warren G., and Mellman I.. 2004. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J. Cell Biol. 167:531–543. 10.1083/jcb.200408165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin, W., Holroyd C., Fasshauer D., Pabst S., Von Mollard G.F., and Jahn R.. 2000. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 19:6453–6464. 10.1093/emboj/19.23.6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apodaca, G., Gallo L.I., and Bryant D.M.. 2012. Role of membrane traffic in the generation of epithelial cell asymmetry. Nat. Cell Biol. 14:1235–1243. 10.1038/ncb2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apodaca, G., Katz L.A., and Mostov K.E.. 1994. Receptor-mediated transcytosis of IgA in MDCK cells is via apical recycling endosomes. J. Cell Biol. 125:67–86. 10.1083/jcb.125.1.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbey, C.M., Ahktar N., Wang E., Chen C.C., Grant B.D., and Dunn K.W.. 2006. Rab10 regulates membrane transport through early endosomes of polarized Madin-Darby canine kidney cells. Mol. Biol. Cell. 17:3156–3175. 10.1091/mbc.e05-08-0799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balklava, Z., Pant S., Fares H., and Grant B.D.. 2007. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat. Cell Biol. 9:1066–1073. 10.1038/ncb1627 [DOI] [PubMed] [Google Scholar]
- Bos, J.L., Rehmann H., and Wittinghofer A.. 2007. GEFs and GAPs: Critical elements in the control of small G proteins. Cell. 129:865–877. 10.1016/j.cell.2007.05.018 [DOI] [PubMed] [Google Scholar]
- Brandhorst, D., Zwilling D., Rizzoli S.O., Lippert U., Lang T., and Jahn R.. 2006. Homotypic fusion of early endosomes: SNAREs do not determine fusion specificity. Proc. Natl. Acad. Sci. USA. 103:2701–2706. 10.1073/pnas.0511138103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. 10.1093/genetics/77.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeks, A., Gerrard B., Allikmets R., Dean M., and Plasterk R.H.. 1996. Homologues of the human multidrug resistance genes MRP and MDR contribute to heavy metal resistance in the soil nematode Caenorhabditis elegans. EMBO J. 15:6132–6143. 10.1002/j.1460-2075.1996.tb01001.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeks, A., Janssen H.W., Calafat J., and Plasterk R.H.. 1995. A P-glycoprotein protects Caenorhabditis elegans against natural toxins. EMBO J. 14:1858–1866. 10.1002/j.1460-2075.1995.tb07178.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz, P., Pelegrín P., and Shao F.. 2020. The gasdermins, a protein family executing cell death and inflammation. Nat. Rev. Immunol. 20:143–157. 10.1038/s41577-019-0228-2 [DOI] [PubMed] [Google Scholar]
- Bryant, D.M., Datta A., Rodríguez-Fraticelli A.E., Peränen J., Martín-Belmonte F., and Mostov K.E.. 2010. A molecular network for de novo generation of the apical surface and lumen. Nat. Cell Biol. 12:1035–1045. 10.1038/ncb2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.C., Schweinsberg P.J., Vashist S., Mareiniss D.P., Lambie E.J., and Grant B.D.. 2006. RAB-10 is required for endocytic recycling in the Caenorhabditis elegans intestine. Mol. Biol. Cell. 17:1286–1297. 10.1091/mbc.e05-08-0787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils, J., and Zeghouf M.. 2013. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93:269–309. 10.1152/physrev.00003.2012 [DOI] [PubMed] [Google Scholar]
- Cramm-Behrens, C.I., Dienst M., and Jacob R.. 2008. Apical cargo traverses endosomal compartments on the passage to the cell surface. Traffic. 9:2206–2220. 10.1111/j.1600-0854.2008.00829.x [DOI] [PubMed] [Google Scholar]
- Cresawn, K.O., Potter B.A., Oztan A., Guerriero C.J., Ihrke G., Goldenring J.R., Apodaca G., and Weisz O.A.. 2007. Differential involvement of endocytic compartments in the biosynthetic traffic of apical proteins. EMBO J. 26:3737–3748. 10.1038/sj.emboj.7601813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan, K.W., and Bretscher A.. 2012. Myosin-V is activated by binding secretory cargo and released in coordination with Rab/exocyst function. Dev. Cell. 23:769–781. 10.1016/j.devcel.2012.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont, N., Jiang S., Pilli M., Ornatowski W., Bhattacharya D., and Deretic V.. 2011. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 30:4701–4711. 10.1038/emboj.2011.398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran, J.M., Anjard C., Stefan C., Loomis W.F., and Malhotra V.. 2010. Unconventional secretion of Acb1 is mediated by autophagosomes. J. Cell Biol. 188:527–536. 10.1083/jcb.200911154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejlerskov, P., Rasmussen I., Nielsen T.T., Bergström A.L., Tohyama Y., Jensen P.H., and Vilhardt F.. 2013. Tubulin polymerization-promoting protein (TPPP/p25α) promotes unconventional secretion of α-synuclein through exophagy by impairing autophagosome-lysosome fusion. J. Biol. Chem. 288:17313–17335. 10.1074/jbc.M112.401174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emperador-Melero, J., Huson V., van Weering J., Bollmann C., Fischer von Mollard G., Toonen R.F., and Verhage M.. 2018. Vti1a/b regulate synaptic vesicle and dense core vesicle secretion via protein sorting at the Golgi. Nat. Commun. 9:3421. 10.1038/s41467-018-05699-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S., Knödler A., Ren J., Zhang J., Zhang X., Hong Y., Huang S., Peränen J., and Guo W.. 2012. A Rab8 guanine nucleotide exchange factor-effector interaction network regulates primary ciliogenesis. J. Biol. Chem. 287:15602–15609. 10.1074/jbc.M111.333245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filaquier, A., Marin P., Parmentier M.L., and Villeneuve J.. 2022. Roads and hubs of unconventional protein secretion. Curr. Opin. Cell Biol. 75:102072. 10.1016/j.ceb.2022.02.006 [DOI] [PubMed] [Google Scholar]
- Flowerdew, S.E., and Burgoyne R.D.. 2009. A VAMP7/Vti1a SNARE complex distinguishes a non-conventional traffic route to the cell surface used by KChIP1 and Kv4 potassium channels. Biochem. J. 418:529–540. 10.1042/BJ20081736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gálvez-Santisteban, M., Rodriguez-Fraticelli A.E., Bryant D.M., Vergarajauregui S., Yasuda T., Bañón-Rodríguez I., Bernascone I., Datta A., Spivak N., Young K., et al. 2012. Synaptotagmin-like proteins control the formation of a single apical membrane domain in epithelial cells. Nat. Cell Biol. 14:838–849. 10.1038/ncb2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley, I.G., Espinosa E., and Pfeffer S.R.. 2008. A syntaxin 10-SNARE complex distinguishes two distinct transport routes from endosomes to the trans-Golgi in human cells. J. Cell Biol. 180:159–172. 10.1083/jcb.200707136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, J., Zhao L., Luo Q., Liu S., Lin Z., Wang P., Fu X., Chen J., Zhang H., Lin L., et al. 2020. An EHBP-1-SID-3-DYN-1 axis promotes membranous tubule fission during endocytic recycling. PLoS Genet. 16:e1008763. 10.1371/journal.pgen.1008763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee, H.Y., Noh S.H., Tang B.L., Kim K.H., and Lee M.G.. 2011. Rescue of ΔF508-CFTR trafficking via a GRASP-dependent unconventional secretion pathway. Cell. 146:746–760. 10.1016/j.cell.2011.07.021 [DOI] [PubMed] [Google Scholar]
- Gleason, A.M., Nguyen K.C.Q., Hall D.H., and Grant B.D.. 2016. Syndapin/SDPN-1 is required for endocytic recycling and endosomal actin association in the C. elegans intestine. Mol. Biol. Cell. 27:3746–3756. 10.1091/mbc.e16-02-0116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales, C.M., Griner R., Hobdy-Henderson K.C., Dorn M.C., Hardy D., Kumar R., Navarre J., Chan E.K., Lapierre L.A., and Goldenring J.R.. 2001. Identification and characterization of a family of Rab11-interacting proteins. J. Biol. Chem. 276:39067–39075. 10.1074/jbc.M104831200 [DOI] [PubMed] [Google Scholar]
- Horgan, C.P., and McCaffrey M.W.. 2009. The dynamic Rab11-FIPs. Biochem. Soc. Trans. 37:1032–1036. 10.1042/BST0371032 [DOI] [PubMed] [Google Scholar]
- Horgan, C.P., Oleksy A., Zhdanov A.V., Lall P.Y., White I.J., Khan A.R., Futter C.E., McCaffrey J.G., and McCaffrey M.W.. 2007. Rab11-FIP3 is critical for the structural integrity of the endosomal recycling compartment. Traffic. 8:414–430. 10.1111/j.1600-0854.2007.00543.x [DOI] [PubMed] [Google Scholar]
- Jin, M.S., Oldham M.L., Zhang Q., and Chen J.. 2012. Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature. 490:566–569. 10.1038/nature11448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, J., Kim J., Roh S.H., Jun I., Sampson R.D., Gee H.Y., Choi J.Y., and Lee M.G.. 2016. The HSP70 co-chaperone DNAJC14 targets misfolded pendrin for unconventional protein secretion. Nat. Commun. 7:11386. 10.1038/ncomms11386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerboeuf, D., and Guégnard F.. 2011. Anthelmintics are substrates and activators of nematode P glycoprotein. Antimicrob. Agents Chemother. 55:2224–2232. 10.1128/AAC.01477-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Gee H.Y., and Lee M.G.. 2018. Unconventional protein secretion - new insights into the pathogenesis and therapeutic targets of human diseases. J. Cell Sci. 131:jcs213686. 10.1242/jcs.213686 [DOI] [PubMed] [Google Scholar]
- Knödler, A., Feng S., Zhang J., Zhang X., Das A., Peränen J., and Guo W.. 2010. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl. Acad. Sci. USA. 107:6346–6351. 10.1073/pnas.1002401107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamber, E.P., Siedenburg A.C., and Barr F.A.. 2019. Rab regulation by GEFs and GAPs during membrane traffic. Curr. Opin. Cell Biol. 59:34–39. 10.1016/j.ceb.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Lock, J.G., and Stow J.L.. 2005. Rab11 in recycling endosomes regulates the sorting and basolateral transport of E-cadherin. Mol. Biol. Cell. 16:1744–1755. 10.1091/mbc.e04-10-0867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock, R., Kenific C.M., Leidal A.M., Salas E., and Debnath J.. 2014. Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov. 4:466–479. 10.1158/2159-8290.CD-13-0841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos, S., Tan M.W., Rahme L.G., and Ausubel F.M.. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 96:47–56. 10.1016/S0092-8674(00)80958-7 [DOI] [PubMed] [Google Scholar]
- Malhotra, V. 2013. Unconventional protein secretion: An evolving mechanism. EMBO J. 32:1660–1664. 10.1038/emboj.2013.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard, F., Tang B.L., Galli T., Tenza D., Saint-Pol A., Yue X., Antony C., Hong W., Goud B., and Johannes L.. 2002. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 156:653–664. 10.1083/jcb.200110081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménez, C., Alberich M., Courtot E., Guegnard F., Blanchard A., Aguilaniu H., and Lespine A.. 2019. The transcription factor NHR-8: A new target to increase ivermectin efficacy in nematodes. PLoS Pathog. 15:e1007598. 10.1371/journal.ppat.1007598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, M.P., and Goody R.S.. 2018. Molecular control of Rab activity by GEFs, GAPs and GDI. Small GTPases. 9:5–21. 10.1080/21541248.2016.1276999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya, T., Miaczynska M., Coskun U., Lommer B., Runge A., Drechsel D., Kalaidzidis Y., and Zerial M.. 2009. Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature. 459:1091–1097. 10.1038/nature08107 [DOI] [PubMed] [Google Scholar]
- Ortiz, D., Medkova M., Walch-Solimena C., and Novick P.. 2002. Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J. Cell Biol. 157:1005–1015. 10.1083/jcb.200201003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelissier, A., Chauvin J.P., and Lecuit T.. 2003. Trafficking through Rab11 endosomes is required for cellularization during Drosophila embryogenesis. Curr. Biol. 13:1848–1857. 10.1016/j.cub.2003.10.023 [DOI] [PubMed] [Google Scholar]
- Pereira-Leal, J.B., and Seabra M.C.. 2001. Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 313:889–901. 10.1006/jmbi.2001.5072 [DOI] [PubMed] [Google Scholar]
- Perez Bay, A.E., Schreiner R., Benedicto I., Paz Marzolo M., Banfelder J., Weinstein A.M., and Rodriguez-Boulan E.J.. 2016. The fast-recycling receptor Megalin defines the apical recycling pathway of epithelial cells. Nat. Commun. 7:11550. 10.1038/ncomms11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponpuak, M., Mandell M.A., Kimura T., Chauhan S., Cleyrat C., and Deretic V.. 2015. Secretory autophagy. Curr. Opin. Cell Biol. 35:106–116. 10.1016/j.ceb.2015.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris, R., Klumperman J., and Scheller R.H.. 2000. A Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol. Cell. 6:1437–1448. 10.1016/S1097-2765(00)00140-4 [DOI] [PubMed] [Google Scholar]
- Rabouille, C. 2017. Pathways of unconventional protein secretion. Trends Cell Biol. 27:230–240. 10.1016/j.tcb.2016.11.007 [DOI] [PubMed] [Google Scholar]
- Sakaguchi, A., Sato M., Sato K., Gengyo-Ando K., Yorimitsu T., Nakai J., Hara T., Sato K., and Sato K.. 2015. REI-1 is a guanine nucleotide exchange factor regulating RAB-11 localization and function in C. elegans embryos. Dev. Cell. 35:211–221. 10.1016/j.devcel.2015.09.013 [DOI] [PubMed] [Google Scholar]
- Salminen, A., and Novick P.J.. 1987. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell. 49:527–538. 10.1016/0092-8674(87)90455-7 [DOI] [PubMed] [Google Scholar]
- Sato, K., Norris A., Sato M., and Grant B.D.. 2014. C. elegans as a model for membrane traffic. WormBook. 1–47. 10.1895/wormbook.1.77.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M., Saegusa K., Sato K., Hara T., Harada A., and Sato K.. 2011. Caenorhabditis elegans SNAP-29 is required for organellar integrity of the endomembrane system and general exocytosis in intestinal epithelial cells. Mol. Biol. Cell. 22:2579–2587. 10.1091/mbc.e11-04-0279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, T., Mushiake S., Kato Y., Sato K., Sato M., Takeda N., Ozono K., Miki K., Kubo Y., Tsuji A., et al. 2007. The Rab8 GTPase regulates apical protein localization in intestinal cells. Nature. 448:366–369. 10.1038/nature05929 [DOI] [PubMed] [Google Scholar]
- Schotman, H., Karhinen L., and Rabouille C.. 2008. dGRASP-mediated noncanonical integrin secretion is required for Drosophila epithelial remodeling. Dev. Cell. 14:171–182. 10.1016/j.devcel.2007.12.006 [DOI] [PubMed] [Google Scholar]
- Schuck, S., Gerl M.J., Ang A., Manninen A., Keller P., Mellman I., and Simons K.. 2007. Rab10 is involved in basolateral transport in polarized Madin-Darby canine kidney cells. Traffic. 8:47–60. 10.1111/j.1600-0854.2006.00506.x [DOI] [PubMed] [Google Scholar]
- Schweinsberg, P.J., and Grant B.D.. 2013. C. elegans gene transformation by microparticle bombardment. WormBook. 1–10. 10.1895/wormbook.1.166.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, A., Chen C.C., Banerjee R., Glodowski D., Audhya A., Rongo C., and Grant B.D.. 2010. EHBP-1 functions with RAB-10 during endocytic recycling in Caenorhabditis elegans. Mol. Biol. Cell. 21:2930–2943. 10.1091/mbc.e10-02-0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobajima, T., Yoshimura S., Iwano T., Kunii M., Watanabe M., Atik N., Mushiake S., Morii E., Koyama Y., Miyoshi E., and Harada A.. 2014. Rab11a is required for apical protein localisation in the intestine. Biol. Open. 4:86–94. 10.1242/bio.20148532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son, S.M., Cha M.Y., Choi H., Kang S., Choi H., Lee M.S., Park S.A., and Mook-Jung I.. 2016. Insulin-degrading enzyme secretion from astrocytes is mediated by an autophagy-based unconventional secretory pathway in Alzheimer disease. Autophagy. 12:784–800. 10.1080/15548627.2016.1159375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark, H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525. 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- Thuenauer, R., Hsu Y.C., Carvajal-Gonzalez J.M., Deborde S., Chuang J.Z., Römer W., Sonnleitner A., Rodriguez-Boulan E., and Sung C.H.. 2014. Four-dimensional live imaging of apical biosynthetic trafficking reveals a post-Golgi sorting role of apical endosomal intermediates. Proc. Natl. Acad. Sci. USA. 111:4127–4132. 10.1073/pnas.1304168111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, G., Ropelewski P., Nemet I., Lee R., Lodowski K.H., and Imanishi Y.. 2014. An unconventional secretory pathway mediates the cilia targeting of peripherin/rds. J. Neurosci. 34:992–1006. 10.1523/JNEUROSCI.3437-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Y., Kang Q., Shi X., Wang Y., Zhang N., Ye H., Xu Q., Xu T., and Zhang R.. 2021. SNX-3 mediates retromer-independent tubular endosomal recycling by opposing EEA-1-facilitated trafficking. PLoS Genet. 17:e1009607. 10.1371/journal.pgen.1009607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons, L., and Fire A.. 1998. Specific interference by ingested dsRNA. Nature. 395:854. 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Walter, A.M., Kurps J., de Wit H., Schöning S., Toft-Bertelsen T.L., Lauks J., Ziomkiewicz I., Weiss A.N., Schulz A., Fischer von Mollard G., et al. 2014. The SNARE protein vti1a functions in dense-core vesicle biogenesis. EMBO J. 33:1681–1697. 10.15252/embj.201387549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Li X., Wang J., Wang J., Hu C., Zeng J., Shi A., and Lin L.. 2022. SMGL-1/NBAS acts as a RAB-8 GEF to regulate unconventional protein secretion. J. Cell Biol. 221:e202111125. 10.1083/jcb.202111125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz, O.A., and Rodriguez-Boulan E.. 2009. Apical trafficking in epithelial cells: Signals, clusters and motors. J. Cell Sci. 122:4253–4266. 10.1242/jcs.032615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz, T., Wellbourne-Wood J., and Kerkhoff E.. 2014. Orchestration of cell surface proteins by Rab11. Trends Cell Biol. 24:407–415. 10.1016/j.tcb.2014.02.004 [DOI] [PubMed] [Google Scholar]
- Westlake, C.J., Baye L.M., Nachury M.V., Wright K.J., Ervin K.E., Phu L., Chalouni C., Beck J.S., Kirkpatrick D.S., Slusarski D.C., et al. 2011. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. USA. 108:2759–2764. 10.1073/pnas.1018823108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, J.F., Höpfner S., Korn K., Farnung B.O., Bradshaw C.R., Marsico G., Volkmer M., Habermann B., and Zerial M.. 2012. Caenorhabditis elegans screen reveals role of PAR-5 in RAB-11-recycling endosome positioning and apicobasal cell polarity. Nat. Cell Biol. 14:666–676. 10.1038/ncb2508 [DOI] [PubMed] [Google Scholar]
- Xu, S., Wang Z., Kim K.W., Jin Y., and Chisholm A.D.. 2016. Targeted mutagenesis of duplicated genes in Caenorhabditis elegans using CRISPR-Cas9. J. Genet. Genomics 43:103–106. 10.1016/j.jgg.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M., Kenny S.J., Ge L., Xu K., and Schekman R.. 2015. Translocation of interleukin-1β into a vesicle intermediate in autophagy-mediated secretion. Elife. 4:e11205. 10.7554/elife.11205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M., Liu L., Lin X., Wang Y., Li Y., Guo Q., Li S., Sun Y., Tao X., Zhang D., et al. 2020. A translocation pathway for vesicle-mediated unconventional protein secretion. Cell. 181:637–652.e15. 10.1016/j.cell.2020.03.031 [DOI] [PubMed] [Google Scholar]
- Zhang, M., and Schekman R.. 2013. Cell biology. Unconventional secretion, unconventional solutions. Science. 340:559–561. 10.1126/science.1234740 [DOI] [PubMed] [Google Scholar]
- Zheng, C., Diaz-Cuadros M., and Chalfie M.. 2015. Hox genes promote neuronal subtype diversification through posterior induction in Caenorhabditis elegans. Neuron. 88:514–527. 10.1016/j.neuron.2015.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J., and Ge L.. 2022. Diverse cellular strategies for the export of leaderless proteins. Natl. Sci. Open. 1:20220018. 10.1360/nso/20220018 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
contains information on C. elegans strains.
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 6.
is the source file for Fig. 7.
is the source file for Fig. S1.
is the source file for Fig. S3.
is the source file for Fig. S5.
Data Availability Statement
Data are available in the article itself and its supplementary materials.