Abstract
During the last decade, there has been great interest in elucidating the biological role of extracellular vesicles (EVs), particularly, their hormone-like role in cell-to-cell communication. The field of endocrinology is uniquely placed to provide insight into the functions of EVs, which are secreted from all cells into biological fluids and carry endocrine signals to engage in paracellular and distal interactions. EVs are a heterogeneous population of membrane-bound vesicles of varying size, content, and bioactivity. EVs are specifically packaged with signaling molecules, including lipids, proteins, and nucleic acids, and are released via exocytosis into biofluid compartments. EVs regulate the activity of both proximal and distal target cells, including translational activity, metabolism, growth, and development. As such, EVs signaling represents an integral pathway mediating intercellular communication. Moreover, as the content of EVs is cell-type specific, it is a “fingerprint” of the releasing cell and its metabolic status. Recently, changes in the profile of EV and bioactivity have been described in several endocrine-related conditions including diabetes, obesity, cardiovascular diseases, and cancer. The goal of this statement is to highlight relevant aspects of EV research and their potential role in the field of endocrinology.
Keywords: extracellular vesicle, exosome, microvesicle, apoptotic body, migrasome, oncosome, cellular messenger, ectosome, signaling, biogenesis
The constructs that define explicit knowledge of endocrinology have their origins in the works of Ernest Starling and Edward Sharpey-Schäfer, from over 100 years ago. The former introduced the term “hormone” in the Croonian Lecture delivered on June 20, 1905. “These chemical messengers, however, or ‘hormones’, as we might call them, have to be carried from the organ where they are produced to the organ which they affect by means of the blood stream and the continually recurring physiological needs of the organism must determine their repeated production and circulation through the body” (1). The latter classified these chemical messengers into endocrine and exocrine mediators (differentiating them based on their mechanism of release) and collectively referred to them as autacoids. These constructs underpin the conceptual framework by which research endeavor, characterization of physiological systems and clinical practice have advanced, including our contemporary understanding of reproductive endocrinology and the development of biofluid-based prognostic and diagnostic modalities. Indeed, much of reproductive medicine is predicated on the action or measurement of autacoids and how cells employ such mediators to communicate with each other.
Until recently it was not often appreciated that cells release a cloud of nanovesicles into extracellular compartments as a part of normal homoeostasis (2-4). Active macro-molecules including nucleic acids, proteins, and lipids are associated with these vesicles, exposed on the vesicle surface, intercalated in the bilipid layer membrane, or encapsulated within their lumen (5-11). When homoeostasis is challenged, acute adaptive responses may be accompanied by the increased release of bioactive nanovesicles (12-20). Extracellular vesicles (EVs) may act proximally or distally, are distributed in the extracellular fluid compartment, and may traverse cell barriers via paracellular pathways or transcytosis [eg, as occurs at the blood-brain barrier (21, 22)]. In addition, EVs are present in exocrine secretions where they may further engage in physiologically relevant processes (23-29). EVs, thus, are melded autacoids; that is, they are bifunctional, fulfilling the classification criteria of both endocrine and exocrine factors. Decoding the role of this signaling pathway in physiological and pathophysiological events remains formative, particularly in endocrinology. Thus, the aim of this scientific statement is to elucidate the endocrine roles of EVs and to inform the codification EV signaling pathways in the routine practice of endocrinology.
Extracellular Vesicles’ Heterogeneity
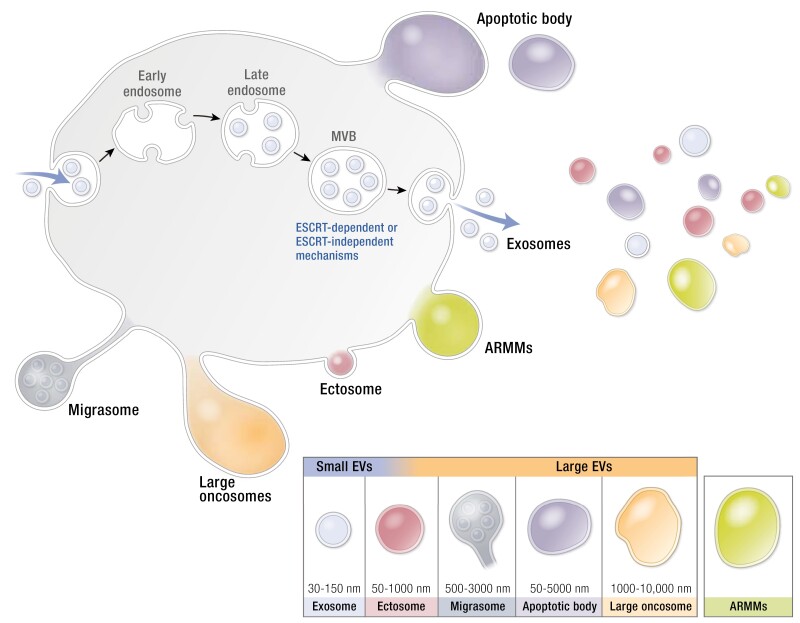
EVs are a heterogeneous population of membrane-bound particles of around 30 nm up to a few micrometers in diameter. The majority of these vesicles display a spherical, single bilayer morphology; however, vesicles with multiple membranes or with a tubular morphology also have been described (30). Based on their biogenesis and physical properties, EVs are often classified as small EVs (including exosomes) or large EVs (including ectosomes or microvesicles, migrasomes, apoptotic bodies) and large oncosomes (Table 1). The term “exosome” has been used to refer to EVs of ~30 to 150 nm in diameter that are formed via the inward membrane budding of multivesicular bodies (MVBs). Upon fusion of MVBs with the plasma membrane, vesicles are released into the extracellular space and are subsequently referred to as exosomes. The content of such EVs may be regulated by endosomal sorting complexes required for transport–dependent and –independent mechanisms and further contribute to vesicle heterogeneity (31, 32). The terms “ectosome,” “microvesicle,” and “microparticle” have been used to characterize EVs that are formed through direct budding from the plasma membrane. Formation of microvesicles involves Ca2+ influx and contraction of cortical actin (33). Finally, vesicular apoptotic bodies (up to a few microns) are formed when cells release membrane extrusions as part of the apoptotic process. They may contain nuclear and cytosolic fragments and even intact organelles. While apoptotic bodies are often regarded as unwanted contaminants of EV preparations, some argue that apoptotic bodies can also facilitate intercellular communication and may have potential as therapeutic modalities (34) (Fig. 1).
Table 1.
Classification of extracellular vesicles
| EV subtypes | Size | Biogenesis | Cargo | References |
|---|---|---|---|---|
| Exosomes | 30-150 nm | Originate in the endosomal pathway in the multi-vesicular bodies (MVBs) and are released upon fusion of MVBs with the plasma membrane | Proteins of the endosomal pathway and endosomal sorting complexes required for transport complex (Alix, TSG101, HSP70) and members of tetraspanin family (CD62, CD9, CD81) | (35-40) |
| Ectosomes (or microvesicles, microparticles) | 50-1000 nm | Released by direct budding from the plasma membrane | Proteins annexin A1, integrins, selectins, CD40 | (30, 41-43) |
| Migrasomes | 500-3000 nm | Released from migrating cells, dependent onactin polymerization | Protein TSPAN4 | (44-46) |
| Apoptotic bodies | 50-5000 nm | Released from apoptotic cells | Protein Annexin V, lipid phosphatidyl serine | (42, 47, 48) |
| Large oncosomes | 1000-10000 nm | Released from amoeboid cancer cells | Protein cytokeratin 18 | (49-51) |
| ARMMs (arrestin domain-containing protein 1 (ARRDC1)-mediated microvesicles) | Released via ARRDC1-driven outward budding of plasma membrane | Protein ARRDC1 | (52, 53) |
Figure 1.
Extracellular vesicle (EV) heterogeneity. EVs can be categorized according to their size as small and large EVs. Small EV including exosomes and ectosomes and large EV including ectosomes (some EV overlap with exosomes), migrasome, apoptotic bodies, and large oncosomes. Recently, arrestin domain-containing protein 1 (ARRDC1)-mediated microvesicles (ARMMs) have been identified.
Recent evidence suggests that even within these EV categories, subpopulations of vesicles exist. For example, crude exosome preparations can be further subdivided and separated into exosome subpopulations based on differences in size (54), surface proteome (55), or membrane lipids (56, 57). Some cancer cells secrete a distinct population of microvesicles termed “large oncosomes” of 1 to 10 µm in size (58), while other cell types release arrestin domain-containing protein 1 (ARRDC1)-mediated microvesicles (ARMMs) that require ARRDC1 for budding (52). A more in-depth understanding of the biological mechanisms underlying EV biogenesis is needed to better inform the classification of EV subpopulations.
In addition to biogenesis, the biological environment of EV-releasing cells also contributes to EV heterogeneity. For example, local changes in concentrations of growth factors may shift the balance between release of exosomes vs microvesicles (59). In addition, EV cargo is affected by changes in gene expression resulting from environmental cues such as oxygen levels (60), inflammation (61), and shear (62). Even when EVs are isolated from a single cell source (eg, from cells cultured in vitro), spatial and temporal changes in confluency, cell cycle stage, stress, and phase of circadian rhythm may contribute to the observed heterogeneity.
Stochasticity of gene expression and both localized and overall protein concentration in endosomal and plasma membranes also may play an important role in heterogeneity of EV cargo composition. Since at least part of the inclusion of EV cargo appears to be random, driven by local concentrations of biological molecules at sites of biogenesis, it can be envisioned that each individual EV carries a unique cargo repertoire (63).
Despite an increased appreciation of the complexity and variability of EV biogenesis and cargo loading, the functional relevance of EV heterogeneity remains largely unknown. While originally considered “waste bins” (64), contributing to, for example, protein quality control, EVs are now also being recognized as important mediators of intercellular communication via transfer of biological cargo. Since it seems highly unlikely that a single EV subpopulation can have these 2 functions at the same time, it is axiomatic to conclude that EV subpopulations serve different biological functions. Indeed, EV subpopulations separated based on size or surface protein profile have distinct protein and RNA profiles (35, 54, 56, 65).
Concordantly, EV subpopulations have been shown to display different functional properties. For example, small and large EVs derived from immature dendritic cells differ in their capacity to promote T helper cell responses (66) and EV subpopulations from encephalomyocarditis virus–infected cells vary in their ability to transfer virus infection (67). In addition, endothelial cell–derived intercellular adhesion molecule-1 (ICAM-1)-carrying EVs are more prone to induce expression of ICAM-1 in vascular endothelial cells and promote monocyte migration than ICAM-1–negative EVs (61). Nevertheless, in most studies to date, functional differences between EV subpopulations were found to be merely qualitative. Whether this is due to incomplete separation between these subtypes remains unclear. Thus, the highly relevant question of whether EV subpopulations are truly functionally distinct remains to be answered and may require technical developments in isolation and separation procedures.
Overall, despite significant progress in our understanding of the biology underlying EV biogenesis, many aspects of EV heterogeneity remain poorly understood. Since consensus on nomenclature has yet to be reached and it remains challenging to assign a biogenic origin to EVs after their isolation, the International Society of Extracellular Vesicles (ISEV) has suggested “extracellular vesicle” as the generic term for particles delimited by a lipid bilayer that cannot replicate. When relevant, subtypes of EVs could be further described using characteristics such as size (eg, small vs large EVs), density (eg, low density vs high density), or composition (eg, CD63+ve EVs vs CD9+ve EVs) (40). During the formative phase of EV research, the term “exosome” has been used to reference very different preparations of EVs. Its nonspecific usage, however, confounds meaningful comparative analysis of data. To facilitate data interpretation, the term “exosome” should be reserved to reference only EVs that can be proven to be formed within MVB of late endosomal origin. The term “extracellular vesicle” should be used where such precision (eg, a unique antigenic phenotype) cannot be provided.
Highlights
EVs represent a heterogeneous population of membrane-bound particles of 30 nm up to a few micrometers in size.
Based on biogenesis and physical and morphological features, EVs are often classified as exosomes or small EVs, ectosomes/microvesicles, or large vesicles and apoptotic bodies.
EVs are recognized as important mediators of intercellular communication via transfer of biological cargo.
Adoption of precise protocols that enable the reproducible isolation and characterization of vesicle populations released from cells should enable data comparison and advancement in understanding.
The term “extracellular vesicle” should be used to reference vesicle preparations where homogeneity of biogenesis is not evidenced. The use of other terms, including exosome and ectosome/microvesicle should be used where homogeneity of biogenesis is unequivocally established.
Isolation and Characterization Methods
EVs are secreted from all cells that have been studied to date, and they are present in large amounts in all biofluids. EVs have been isolated from diverse sources, including cell-conditioned media and biological fluids including plasma, serum, urine, saliva, and milk. EV preparations are used for equally diverse purposes, such as EV analysis, EV-based diagnostic markers, and therapeutic applications. Most techniques used to prepare EVs are predicated on biophysical properties. Preparations of EVs using such techniques are essentially enrichments of entities that have the targeted size or density range and not of lipid membrane-bound entities. Bodily fluids and culture medium are known to contain many nonvesicular macromolecules that have biophysical properties similar to or overlapping with those of EVs. For example, many lipoprotein complexes, such as high- and low-density lipoproteins (HDL and LDL, respectively) have sizes and densities that overlap with many different classes of EVs (68). The choice of EV isolation methodology and the characteristics of the EV preparation will be influenced by the starting material and intended use of the preparation. Independent of the isolation methods, however, all EV preparation must minimally exhibit key defining characteristics of EVs.
In 2018, ISEV issued a position paper known as the “Minimal Information for Studies of Extracellular Vesicles 2018” (MISEV2018) to address challenging issues in the study of EVs (40). Isolation and characterization of EVs were identified as some of the major challenges.
The following discussion provides a summary of the key points from MISEV2018 on the isolation and characterization of EVs. Readers are referred to MISEV2018 for more details (40). The reader should also be cautioned that the isolation method and nature of the starting materials (such as conditioned culture medium or biological fluids) will have an impact on EV isolation and the isolated EV preparation.
EV Isolation Techniques
At present, there is no practical technology to isolate EVs completely from other non-EV components (eg, soluble molecules) of the matrix, such as conditioned medium, biofluid, tissue, etc, or different types of EVs from each other. Many isolation techniques have been employed in EV isolation. They include ultracentrifugation, density gradient ultracentrifugation, precipitation, filtration, size exclusion chromatography, and immuno-isolation. Each technique has advantages and limitations in terms of recovery and specificity (ie, relative ratio of EV to non-EVs; eg, soluble molecules). Most researchers use a combination of techniques to maximize recovery and specificity. The following discussion provides a summary of the commonly used EV isolation techniques and their pros and cons (Table 2). MISEV2018 has categorized techniques according to EV recovery and specificity.
Table 2.
Comparison of the key features in commonly used extracellular vesicle enrichment techniques
| EV enrichment techniques | Time | Cost | Scalability | Recovery | Specificity |
|---|---|---|---|---|---|
| PEG precipitation | +++ | ++++ | ++++ | ++++ | + |
| Size exclusion chromatography | + | + | + | + | +++ |
| High MW centrifugal filters | ++++ | +++ | ++++ | +++ | ++ |
| Differential ultracentrifugation | + | ++ | + | + | ++ |
| Tangential flow filtration | +++ | ++ | ++++ | +++ | +++ |
| Affinity chromatography | ++ | + | ++ | ++ | ++++ |
| Immunomagnetic bead capture | ++++ | +++ | +++++ | +++ | ++++ |
The key features are length of operation time (time), cost of the equipment and consumables (cost), the ease of scaling the technique to process large volumes of fluids (scalability), the percentage of EVs in fluids that could be extracted (recovery) and the ratio of EVs extracted relative to total protein with a higher ratio being more specific (specificity). + denotes the desirability of the feature(++++: most desirable; +: least desirable).
Abbreviations: EV, extracellular vesicle; MW, molecular weight; PEG, polyethylene glycol.
High recovery and low specificity
Precipitation using high molecular weight polymer such as polyethylene glycol or centrifugation with low molecular weight cutoff filter generally results in high EV recovery but also high contamination from non-EV material (ie, low specificity). An advantage of these techniques is their scalability, speed, and low cost in processing large volumes of starting material.
Intermediate recovery and intermediate specificity
Techniques such as size exclusion chromatography, high molecular weight centrifugal filters, multistep differential ultracentrifugation, tangential flow filtration, and affinity chromatography columns, where the separation parameters are more stringent, generally have lower EV recovery, but this is compensated by an increase in specificity where there is more EV material than non-EV material. These techniques, however, have reduced scalability, slower speed, and higher cost. To circumvent these shortcomings, these techniques are often used on EV preparations that had been processed by precipitation or centrifugation with low molecular weight cutoff filters.
Low recovery and high specificity
Achieving high EV specificity is possible with a combination of isolation techniques but the use of multiple isolation processes will inevitably lead to a low recovery. To circumvent this, the strategy for isolating EV preparations with high specificity is to first enrich for EVs using a high recovery and low specificity technique and/or intermediate recovery and intermediate specificity techniques. The debulked EV preparation can then be further fractionated into narrower ranges of size, density, pH or surface charge. Alternatively, they could be further fractionated according to the protein/sugar/lipid composition on the surface of EVs. Some of the higher resolution fractionation techniques include fast protein/high performance liquid chromatography, using either size exclusion columns or ion exchange columns, microfluidic devices, and immune-affinity or other affinity isolation techniques. A more comprehensive list can be found in MISEV2018 (40).
EV Characterization
Each EV preparation should, independent of the isolation methodology, be characterized and quantified for key EV features to establish their identity as EV preparations and to facilitate comparison to other EV preparations. In this regard, the major MISEV2018’s recommendation on the global characterization of EV preparations is highlighted in the following discussion.
EV Source Quantification
As the biofluids and cell culture supernatant used in EV isolations are diverse and are prepared in many different ways, the biofluids (eg, conditioned culture medium or biological fluid) constitute a key feature of EV characterization. The conditions to which cells (as source of EVs) are exposed and clinical characteristics of the patients from whom the biofluids are derived need to be defined sufficiently precisely to allow replication by others. For conditioned culture medium, the description could include the volume of conditioned medium, number of cells or mass of tissue used for conditioning, the harvest procedure, and composition of the culture medium. Importantly, serum contains high amounts of EV, so information about EV-free serum or growth media without serum must be clearly described. In the case of biological fluids, parameters such as collection of fluid, processing of fluid, time after collection, storage conditions (eg, temperature and numbers of freezing-thawing cycles) and volume of processed fluid used in EV isolation should be included and described quantitatively if possible.
EV Abundance
Currently, there is not a gold standard method to quantify the number of EVs in a preparation. Instead, EVs are usually quantified indirectly by particulate features or components such as proteins, lipids, nucleic acids, and other biomolecules. The most commonly used parameters for EV quantification are protein, lipid, RNA content, or particle number. Using at least 2 different methods to identify the purified population can be highlighted as good practice.
Vesicular Identity
EV preparation may be further characterized by the presence of specific biomarkers, including proteins, lipids, and nucleic acids. Of these biomolecules, proteins (eg, transmembrane or glycosylphosphatidylinositol-anchored proteins associated with the plasma membrane and/or endosomes) are commonly used as EV markers, including tetraspanins [eg, cluster of differentiation (CD) 63, 81, 82], other multipass membrane proteins (CD47) and heterotrimeric G proteins, major histocompatibility complex class I, integrins, transferrin receptor, heparan sulfate proteoglycans, complement-binding proteins CD55 and CD59, and sonic hedgehog (40). MISEV2018 (40) recommends the determination of at least 3 positive protein markers including at least 1 transmembrane/lipid-bound protein or cytosolic protein to establish the presence of EVs. The rationale to include a transmembrane/lipid-bound protein or cytosolic protein is to determine whether lipid membranes are present in the preparation.
Highlights
EVs have been isolated from biofluids and cell-conditioned media, including cell-conditioned culture medium and bodily fluids such as plasma, serum, urine, saliva, and milk.
EV preparations are used for diverse purposes, including EV analysis, EV-based diagnostic markers, and therapeutic applications. Most techniques used to isolate EVs are predicated on biophysical properties such as size or density rather than specific properties unique to EVs or a specific EV type.
There are several methods to enrich EVs and all of them obtain heterogeneous populations of EVs (varies depending on the method). Method choice should be appropriate and fit for purpose to resolve the hypothesis being tested.
In 2018, the ISEV issued a position paper known as the MISEV2018 to address challenging issues in the study of EVs.
EV Composition
Most of the studies of composition of EVs have identified a wide range of bioactive molecules, including proteins, lipids, and nucleic acids. We summarize the current state of the literature to identify EV-associated molecules.
Proteins
As evidenced by the increasing number of studies, EVs play intriguing roles in intercellular communication and other physiological functions with cargo including proteins, nucleic acids, and metabolites (32). Among them, proteins are emphasized in the literature, as proteins represent actual functional molecules in the cell and most cellular functions are carried out by proteins. Several publicly available EV protein databases, such as ExoCarta (69), EVpedia, and Vesiclepedia (70), have been developed over the years and are continuously evolving with the assistance of high throughput mass spectrometry. Current studies on EV proteins mainly focus on 3 aspects: (1) molecular characterization of EVs by their protein composition; (2) protein markers to detect and monitor disease progression; and (3) surface functionalization of EVs for therapeutics.
Characterization of EVs by Protein Composition
While there is a dramatic increase in the number of scientific studies to explore physiological and pathological functions of EVs, we also face the challenges of the heterogeneity of EVs—different types, sizes, and cellular origins—and the need for definitive characterization of EVs. Protein composition may be different depending on the specific species (eg, human, mouse, or other organism), cell type, and experimental conditions. Early work with homogenous cell culture systems identified protein markers common to all EVs or specific for EV subtypes (40). These proteins have been used as positive markers to assess the yield and purity of EV isolation. They include transmembrane surface markers such as tetraspanins (CD9, CD63, CD81, etc.), and cytosolic proteins such as heat shock proteins HSC70, tumor susceptibility gene 101 (TSG101), and programmed cell death 6-interating protein (ALIX) and cytoskeleton proteins (eg, actin and tubulin) that may be present in EVs. In addition, due to the lack of specific EV isolation techniques, contamination is common, in particular, if EVs are isolated from biofluids such as plasma or serum in which albumin, immunoglobulins, and lipoproteins are highly abundant. These major components in biofluids, along with proteins typically associated with intracellular compartments (eg, Golgi bodies and mitochondria) are proposed as negative controls to examine EV purity. With EV biology attracting widespread interest and enthusiasm well beyond the EV research community our understanding of EV science continues to progress. The promotion of rigorous EV research remains paramount. A recent large-scale, proteome-wide profiling of EVs from 497 human- and murine-derived samples including cell lines, tissues, and biofluids revealed that a majority of commonly used EV protein markers were not detected in more than 50% of biofluids (71). For example, CD63 was commonly present in EVs from murine cell-line samples but rarely identified in EVs from human or murine biofluids. At the same time, the study using high throughput mass spectrometry identified a panel of more than 10 proteins that are present at high frequency in human-derived samples.
Protein Markers to Detect and Monitor Disease Progression
Early, accurate diagnosis and then triage to efficacious treatment and disease monitoring through noninvasive tests using biofluids such as blood and urine (liquid biopsy) are the aspiration of medical diagnostics. Successful applications based on protein biomarkers, however, remain limited, in particular, with regard to early screening for disease. This is, in large part, due to the complexity of biofluids, which have an extremely wide dynamic range and are typically dominated by a few highly abundant proteins, while protein biomarkers for diseases are in low abundance. This prevents the discovery and development of novel disease biomarkers. Available data indicate that EVs have great potential as 1 of the major components in liquid biopsy (71, 72). Identification of EV protein markers prior to the onset of symptoms or physiological detection of a tumor suggest that they could be used for detection of early-stage cancer and other diseases (73, 74). If EVs can be efficiently isolated with low contamination, focusing on EV proteins effectively overcomes the issue presented by highly abundant proteins in biofluids. In addition, EVs are membrane-encapsulated nano- or microparticles and thus shield their internal contents from external proteases and other enzymes in biofluids, making them highly stable in a biofluid for extended periods of time (75-77). These features, along with clinical convenience to access, compared to tissue biopsy, present protein markers in EVs as extremely appealing candidates to guide therapy and monitor disease progression. Compared to genomic analysis, the ability to detect proteins—genome output—can offer actual real-time information about the organism’s physiological functions and disease progression.
In most biological processes, posttranslational modification (PTM) finely tunes the cellular functions of each protein, such as regulation of gene expression, cellular differentiation, subcellular location, signaling and regulatory processes, and protein-protein interactions. Profiling proteins with PTMs in EVs, therefore, may provide snapshots of the process (78). For instance, EV surface proteins are largely glycosylated. Previous studies have reported that glycosylation effects the export and uptake in EV (79). Phosphorylation, 1 of the most important and ubiquitous PTMs, was discovered to be as widespread in circulating EVs as in cells (72). Protein phosphorylation is a key control mechanism for cellular regulatory pathways and one often targeted by drug developers to create inhibitors that block signaling pathways involved in cancer and other diseases. Due to active phosphatases in biofluids, however, there are few detectable phosphoproteins available for disease status analysis. Measuring a phosphorylation event through EVs in biofluids can provide unparalleled capability and accessibility to monitor the status of disease. Another known protein modification, by ubiquitin and ubiquitin-like modifiers, has been reported to regulate protein loading into EVs (80). Although the detailed mechanisms are still not very clear, several studies showed that cargo proteins undergo ubiquitination and deubiquitination sequentially.
Analysis of unknown protein PTMs in EVs can be challenging, particularly from biofluids with limited quantity, and it requires a sensitive analytical pipeline to enrich specific modified peptides in EVs from biofluids with high efficiency and low contamination. A general procedure for EV PTM analysis by mass spectrometry is illustrated in Figure 3. The importance of PTMs in cells highlights the need for a better understanding of EV PTMs and the need for deeper, comprehensive analyses of PTMs in EVs. Protocols to sequentially isolate phosphopeptides and N-glycopeptides have been introduced, enabling multiple PTM analyses of the same clinical samples (81).
Figure 3.
Extracellular vesicle uptake and interaction with target cells.
Adapted with permission from Moller A and Lobb RJ. Nat Rev Cancer, 2021; 20(12) © Springer Nature Limited.
Figure 2 provides an illustration of analytical workflow to analyze phosphorylated and glycosylated proteins in plasma EVs.
Figure 2.
Illustration of analytical workflow to analyze phosphorylated and glycosylated proteins in plasma EVs.
Surface Functionalization of EVs for Therapeutic Tools
While we have mainly considered proteins as EV cargo, proteins on the EV surface deserve particular attention. This is especially true when EVs are used for therapeutic purposes (82). Multiple proteins can be present on the EV surface, through which EVs execute specific functions such as antigen presentation, immune activation and suppression, and tumor growth and metastasis (83). Early work using EVs isolated from tumor cells demonstrated that these EVs have antigen and MHC class I molecules on their surface and, therefore, could modulate the immune system, which could be used to stimulate antitumor immune response (84). On the other hand, EVs prepared from immune cells play an important role in regulating the immune response in many events, which has been exploited for cancer therapeutics. Immune cell–derived vesicles are enriched in surface proteins with immune-modulating functions such as major histocompatibility complex (MHC) proteins, costimulatory proteins (CD86), and adhesion proteins (eg, CD11b and CD54). EVs derived from antigen-presenting cells and other immunocytes (85), which express antigen-presenting molecules (MHC class I and II) on their surface, present antigen to T cells and activate the immune response (86). Finally, there has been considerable effort to engineer therapeutic EVs through effective presentation of membrane proteins to improve targeted biodistribution and thus therapeutic efficacy (87, 88).
Highlights
Current studies on EV proteins mainly focus on 3 aspects: (1) molecular characterization of EVs by their protein composition; (2) protein markers to detect and monitor disease progression; and (3) surface functionalization of EVs for therapeutics.
Surface markers such as tetraspanins (CD9, CD63, CD81, etc) and cytosolic proteins such as heat shock proteins HSC70, TSG101, and ALIX are associated with EVs.
Due to the lack of specific EV isolation techniques, contamination is quite common, in particular if EVs are isolated from biofluids such as plasma or serum in which albumin, immunoglobulins, and lipoproteins are highly abundant.
Multiple proteins can be present on the EV surface, through which EVs execute specific functions such as antigen presentation, immune activation and suppression, and tumor growth and metastasis.
Lipids
Extracellular Vesicle Lipids
Lipids are defined as organic compounds that are soluble in nonpolar solvents (89). This chemical characteristic is present in an extremely heterogeneous class of bioactive mediators that have the potential to generate up to 106 structurally different molecular species (90-92). Included within this class of biomolecules are fatty acids (eg, eicosanoids derived from arachidonic acid, eicosapentaenoic acid), glycerolipids (eg, triglycerides), glycerophospholipids (eg, phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine), polyketides (eg, erythromycins, tetracyclines), prenol lipids (eg, carotenoids, vitamin E, vitamin K), saccharolipids (eg, acylated glucosamine precursors of the lipid A component of Gram-negative bacteria lipopolysaccharides), sphingolipids (eg, phosphosphingolipids, neutral and acidic glycosphingolipids), and sterol lipids (eg, cholesterol) (90).
Lipids are obligate components of all EVs, independent of their mode of biogenesis or release. The most abundant lipids in eukaryotic cell membranes are the amphipathic phospholipids: phosphatidylserine, phosphatidylcholine, phosphatidyl ethanolamine, and sphingomyelin. Together with cholesterol and glycolipids, they determine the structure, rigidity, and curvature of cell membranes (93). As EVs are derived from the cell’s bilipid membranes, it is axiomatic that these lipids are most abundant in EVs and that they have been most often studied and reported in the literature. Other classes of bioactive EV lipids, however, have been identified and may play a significant role in EV signaling pathways.
Lipids not only form the physical structure of extracellular vesicles and encapsulate their cargo but also function as first and secondary signaling messengers (94). As first messengers, extracellular lipids may interact with cell-surface receptors triggering diverse signaling pathways. For example, endocannabinoids bind to G-protein-coupled cell surface receptors to trigger multiple second messenger pathways (95). Lipid second messengers are generated via the activation of cell-surface receptors including N-methyl-D-aspartate receptor activation of the arachidonic acid cascade, which generates prostaglandins, lipoxins, and leukotrienes (96) and T-cell receptor–mediated activation of diacylglycerol and phosphatidic acid signaling pathways (97). Lipid biomolecules that are associated with EVs thus may affect target cell function via direct activation of cell surface receptors or as secondary messengers following endocytosis. In addition, fusion of EVs with target cell membranes may alter membrane fluidity, permeability, and receptor responsiveness (98).
Consistent with the original definition of a hormone, EV-associated lipids are released from cells into biofluids and are transported to distant tissues where they elicit specific effects. In addition, EVs are present in exocrine secretions where they may further engage in physiologically relevant processes (23-29). As a consequence of the enormous diversity of lipids, systematic and comprehensive analysis of their role in EV signaling remains formative and is a fertile area for future study.
Given the enormous chemical diversity of lipids, it is not surprising that only a small number have been associated with EVs. Recent reviews catalogue more than 1900 lipid molecules identified in EV isolates obtained from multiple species, cells, and biological fluids (11, 99-106). The composition and distribution of lipids in bilipid membranes is dynamic, often asymmetric between the inner and outer layer and varies across subcellular organelles and cell types (107). Lipids associated with EVs similarly may vary between cell of origin and/or selective loading and enrichment of specific lipids (107, 108).
EV Lipids as Endocrine Mediators
Decoding the biological role of EV-associated lipids in physiological and pathophysiological events remains formative and has been frequently inferred from the well-known bioactivities of nonvesicle-associated lipid mediators. One of the first documented examples of the biological activity of EVs is that of membrane-bound, extracellular nanovesicles released by chondroblasts and osteoblasts to promote cartilage calcification (109). These vesicles are enriched in acidic phospholipids (eg, phosphatidylserine) that function as nucleation sites for apatite deposition and promote calcification (110). In the intervening 40 years, data consistent with the biological role of EV-associated lipids have been obtained and reviewed in detail (11, 102, 111). To unequivocally establish lipid-specific effects of EVs, however, the effects of lipid-deplete vs lipid-replete EVs on the experimental endpoint under test must be compared. Currently, there is a paucity of data that have been obtained using this experimental design. Several experimental approaches that satisfy the previously discussed requirements are available, including the use of endogenous EVs isolated from selective lipid-synthesizing enzyme knockdown or overexpression models [eg, viral (112) or episomal (113) vectors, CRISPR/Cas 9 gene editing (114)] and selective lipid loading of nanovesicles (115). The application of such experimental designs is requisite to further understanding the endocrine role and clinical application of EV-associated lipids.
Highlights
EVs are particles naturally released from cells that are delimited by a lipid bilayer and cannot replicate. As such, lipids are obligate components of all EVs, independent of their mode of biogenesis or release.
As EVs are derived from the cellular bilipid membranes, it is axiomatic that these lipids are the most abundant in EVs and that they have been most often studied and reported in the literature.
Lipids not only form the physical structure of EVs and encapsulate their cargo but also function as first and secondary signaling messengers.
Decoding the biological role of EV-associated lipids in physiological and pathophysiological events remains formative and has been frequently inferred from the well-known bioactivities of nonvesicle-associated lipid mediators.
Nucleic Acids
RNA Carriers in Biofluids
Initial observations that the cargo of EVs contain RNAs (7, 116) launched the field of extracellular RNAs (exRNAs), as investigators sought to characterize the role of these novel entities both as biomarkers of disease as well as mediators of intercellular signaling. Studies that demonstrated the association of RNAs in biofluids with ribonucleoproteins (notably Ago 2) (117, 118) as well as lipoproteins, added additional complexity to this nascent field. Using methodologies to segregate these different RNA carrier subtypes (including selective immunoprecipitation of carriers like Ago2) yielded the finding that classes of small RNAs associated with each of these carrier subtypes may be different (117, 119, 120). There was therefore growing realization that the lack of gold-standard methodologies to comprehensively separate these carrier subtypes may be a major source of variability across different studies and therefore contribute in part to problems with rigor and reproducibility in the field (121).
Recognition of the potential sources of variability in the characterization of the RNA content of different biofluids led to concerted efforts across the field to better define methodologies and approaches that could serve to assist investigators in addressing these variabilities. In this regard, the ISEV convened working groups and statements forging minimal scientific requirements for the characterization of EVs (40, 122) and EV-RNAs and the Extracellular RNA Communication Consortium facilitated the completion of several studies across multiple sites to address sources of variability in measurement of exRNAs. Using large data sets from multiple studies, the Extracellular RNA Communication Consortium investigators were able to develop computational pipelines that could allow for deconvolution of plasma RNAs into their carrier subtypes (119). In a complementary study, the investigators also suggested that the biases in the RNA isolation methodology for exRNA carrier subtypes could explain a significant source of variability across studies (120). In particular, the latter study defined microRNA (miRNA) profiles associated with key exRNA carrier subtypes and devised a tool (miRDAR, available at exRNA.org) that could assist the investigator in choosing the appropriate RNA isolation methodology for their miRNA of interest.
This growing recognition of the heterogeneity of EVs themselves has spurred several studies that seek to define whether EV subtypes themselves are associated with unique RNA profiles.
Fractionation of small EVs secreted by mesenchymal stromal cells using specific membrane lipid binding proteins, namely cholera toxin B chain, Shiga toxin, and annexin V revealed that RNA was present in Shiga toxin–binding EVs and not in cholera toxin B chain or annexin V–binging vesicles (123). Using high-resolution density gradient centrifugation and immunoaffinity capture methodology, Jeppesen et al defined the distinct cargo of exosomes, microvesicles, and other nonvesicular components (41). Other methodologies recently adapted to study EVs, such as asymmetric flow field-flow fractionation that separate particles based on their density and hydrodynamic properties have led to the discovery of nonmembranous nanoparticles termed “exomeres” in addition to large and small exosomes, each with distinct RNA profiles (124). Importantly, careful proteomic analysis of these distinct particles yielded key protein markers that characterize these EV subpopulations. Together, the refinement of markers for EV subpopulations and the development of computational platforms for deconvolution will allow investigators to define RNAs characteristic for each EV subtype and to study how these are altered in disease states.
Composition of EV-RNAs
Based on the initial studies of EV-RNAs that showed a predominance of small RNAs, particularly miRNAs, subsequent investigations to characterize the RNA contents of EVs focused on profiling of small RNAs using a variety of techniques, including next-generation RNA sequencing (125, 126) and a variety of other platforms that were based on either hybridization of specific probes or sequencing based detection of probes (against RNAs of interest) that incorporated bar codes. Challenges that were noted from RNA sequencing experiments were the extremely low amount of input RNA present in samples, leading to high variability in biological replicates and the inherent protocol and sequence-specific biases from RNA sequencing methodologies. Investigators, however, also noted that these biases could be mitigated by choosing adaptors for RNA sequencing that had degenerate bases (126). These studies were confirmed by other studies, which also noted that concordance of results across different platforms were most dependent on the level of RNA expression in the biofluids, highlighting the difficulties in reproducibility of studies across different platforms for RNA species that had low expression (127). Platform-specific differences in measurement of specific RNAs (128) also suggest that careful assessment of the technology for the purpose of measuring specific RNAs of interest may be warranted prior to large-scale studies using that technology.
Initial focus on the miRNA content of EVs has also shifted to the profiling of other small RNAs as well as messenger RNAs (mRNAs) and long noncoding RNAs (lncRNAs). Interestingly, studies in EVs secreted by cancer cell lines showed varied presence of different species of RNAs, depending on the biophysical characteristics of EVs. For example, small EVs were noted to contain a large number of fragments derived from transfer RNAs (tRNAs), while medium-sized EVs contained mRNAs that closely reflected the transcriptome of the parent cell type (129). Study of these other species of RNAs have revealed that for some cell types, such as T cells, there is selective enrichment of tRNA fragments into MVBs and, eventually, into exosomes (130). Interestingly, the release of these RNAs into EVs serves to remove tRNA fragments that functionally repress T-cell activation. Similarly, for cardiosphere-derived cells, a large component of the EV cargo comprises a Y RNA fragment that appears to confer cardioprotection by modulating the immune system (131). The detection of large RNA molecules, including fragments of mRNAs and lncRNAs in biofluids and cell culture supernatants, has more recently focused attention on whether the long RNA transcriptome of EVs may serve to better reflect changes in disease trajectory, as these RNA species may more closely approximate their cell of origin (132). While there is burgeoning interest in the long RNA transcriptome of EVs, whether they truly reflect cellular transcriptome with greater fidelity, and how they may change with stress signals, is not completely clear.
Sorting of RNAs Into EVs
Complementary to the investigation of RNA cargoes of EVs, researchers also sought to determine mechanisms of RNA sorting into EVs. Experiments that compared the EV RNAs with the RNA transcriptome of the parent cell have shown selective enrichment of specific RNAs in the EVs (133-136). These studies have identified RNA binding proteins such as hnRNPA2B1 or Y-box protein-1, which may bind to specific sequence motifs to direct trafficking of miRNAs into exosomes, although some of these mechanisms may be specific to certain cell types and perhaps not generalizable. Similarly, proteins such as K-ras, which direct trafficking of specific miRNAs into exosomes, may play a role specifically in cancer cells. Recent interesting studies have touched on other mechanisms such as inflammasome activation that can regulate loading of miRNAs into exosomes, based on motif-specific binding to the FMR1 RNA binding protein (137). It is important to keep in perspective that the mechanisms of miRNA export into EVs may be context- and cell-specific. In contrast to these mechanistic studies investigating sorting of miRNAs into exosomes, there is far less mechanistic insight into how other RNA species, particularly mRNAs and lncRNAs, get transported into EVs, whether these are all fragmented, and whether there are any sequence-specific motifs that are enriched in the EV cargo.
EV-RNAs Metabolic Disorders
The role of EVs and their molecular cargo as potential biomarkers and mediators of intercellular signaling in obesity and metabolic disorders is described in detail in other sections of this statement. EV-RNAs and may play an important role as “functional” biomarkers—possible prognosticators of disease as well as causal mediators of disease pathogenesis (138). Most studies of ex-RNAs as biomarkers in diabetes and obesity have profiled total plasma and serum from human subjects (rather than RNAs associated with specific carrier subtypes) and hence may be subject to the kind of methodological biases as described in the previous discussion. Nonetheless, these studies describe miRNAs that are differentially expressed in plasma or serum from patients across the spectrum of cardiometabolic disorders (139, 140). Interestingly, there are consistent findings across several of these studies: miR-122 appears to be associated with insulin resistance, altered in a similar direction in murine models of obesity, and implicated in regulation of key metabolic pathways (139, 140). Notably, it appears that profiling of EV miRNAs (as opposed to total plasma RNAs) yielded a stronger signal for miRNAs that were differentially expressed in type 2 diabetes mellitus (T2DM) compared to subjects with normal glucose tolerance (141). A second study noted differences in expression of miR-21-5p in EVs released from beta cells subjected to inflammatory stress, consistent with observations in human subjects (142). There are far fewer studies identifying other RNA species in EVs that can discriminate across the spectrum of metabolic disorders. While the lack of consistent results across the multiple studies may arise from the methodological sources of variation detailed in the previous sections, the more compelling data for a functional role for some of these EV-RNAs in cellular processes important in diabetes pathogenesis provide some level of confidence. In this regard, 2 compelling studies described an important role for miRNAs contained within EVs derived from adipose cells or adipose tissue macrophages (ATMs) in the pathogenesis of insulin resistance (143, 144). Ultimately, while these early studies show some promise of EV-RNAs as potential prognostic biomarkers for diseases across the spectrum of cardiometabolic disorders, future studies that pay careful attention to standardized methodologies to improve rigor and reproducibility are needed to advance them into the clinical arena. Nonetheless, these studies provide much needed insight into novel areas of signaling that pertain to the pathogenesis of insulin resistance and diabetes, paving the way for future identification of therapeutic targets.
Highlights
Initial studies of EV-RNAs showed a predominance of small RNAs, with initial focus on miRNAs as mediators of EV-mediated intercellular communication.
Different RNA carrier subtypes may have different RNA cargoes; RNA cargo is likely cell- and context-specific.
More recent studies have started uncovering novel small RNA species, mRNA, and lncRNA components of EVs.
Experiments that compared the EV RNAs with the RNA transcriptome of the parent cell suggest selective enrichment of specific RNAs in the EVs and a possible functional role for EV-RNAs in the pathogenesis of metabolic diseases.
The role of other RNA species, such as tRNA-derived small RNAs, Y RNAs, small nucleolar RNAs, and fragments of lncRNAs or mRNAs as mediators of signaling have not been investigated in detail.
Biodistribution and Interaction With Target Cells
Ways That EVs Interact With Cells
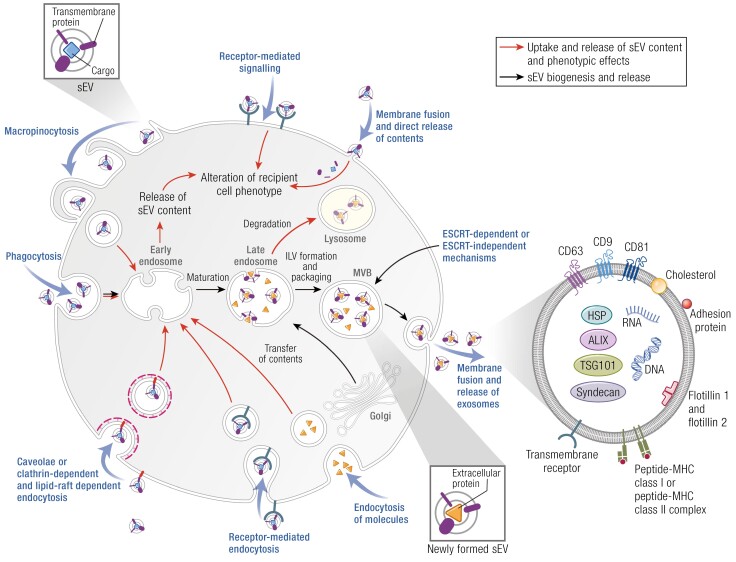
EVs can interact with recipient cells in both autocrine and paracrine manners. Although theoretically EVs could be taken up nonspecifically by cells, there are many reports of EVs binding to cells in a specific manner, using ligand-receptor interactions (98, 145). In fact, EVs carry many types of adhesion molecules that have been shown to enhance binding to recipient cells. This includes ephrin-Eph interactions (146, 147), integrin-ligand interactions (148-150), and peptide-MHC class II complexes to T-cell receptors (85). In addition, EVs are enriched in glycoproteins, such as heparan sulfate proteoglycans and proteins containing a variety of glycosylations, including sialic acid and mannose, in a cell-type and cell state–specific manner (151, 152). In some cases, these sugar groups may increase EV binding and uptake via lectins, such as dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin, on recipient cells (98, 153-155). Conversely, cleavage of sugar groups has also been shown to reduce uptake in other systems, possibly by altering the charge of the EVs. In addition, heparan sulfate proteoglycans on recipient cells have been shown to enhance uptake of EVs (156, 157).
Once bound to recipient cells, EVs can deliver biological signals in multiple ways. Thus, ligand-receptor interactions between EVs and cells may directly induce cellular signaling by both proteins and bioactive lipids. EVs may also fuse with cellular membranes to deliver internal cargoes to recipient cell cytoplasm. EV fusion would also deliver transmembrane and membrane-linked molecules into recipient cell membranes. Due to the way that EV biogenesis occurs, the internal content of EVs is derived from molecules and organelles found in the cytoplasm of cells (32, 158). Thus, internal cargoes of EVs may include RNAs and RNA-binding proteins, metabolic enzymes and metabolites, and other normally cytoplasmic constituents. Such cargoes could affect the cellular phenotype in a variety of ways, including via altering gene expression by miRNA and mRNA delivery (159).
Biodistribution of EVs
EVs not only interact with cells locally but can also transit through blood and lymph vessels and travel to distant tissues. The biodistribution of EVs may influence the course of various diseases. For example, in cancer, small EVs have been shown to seed premetastatic niches, and the integrin cargo content of those EVs may influence the site of metastatic spread by binding to the extracellular matrix at those sites (160-162). Interestingly, different subpopulations of cancer EVs injected into the bloodstream have different biodistributions (124), suggesting that anatomical factors do not solely affect EV distribution.
Methods to Visualize EV Uptake by Tissues and Cells
There are a variety of methods to detect EV uptake and biodistribution (Fig. 3). Purified EVs can be labeled with lipophilic dyes or even purified from cells that are themselves labeled with lipophilic dyes, with the caveat that dye aggregates can resemble EVs, necessitating careful controls (163-169). Cells can also be engineered to express EV-targeting labels, including luciferase (170-173) and fluorescent tags. Typically, this labeling approach either utilizes tagging of EV marker proteins, such as tetraspanins, or linking luciferase or fluorescent protein labels to lipid anchoring sequences (87, 174-177). Depending on which protein or lipid anchor is utilized, the tag may be targeted more to the plasma membrane or to endosomes. This differential targeting can lead to preferential labeling of plasma membrane–derived microvesicles or endosome-derived exosomes. A final approach has been to label the endosome-targeted tetraspanin CD63 with a pH-sensitive green florescent protein called pHluorin (178-181). These constructs are nonfluorescent within acidic late endosomal EVs but fluoresce upon fusion with the plasma membrane since the extracellular environment is at neutral pH. These constructs are especially helpful for tracking high-resolution exosome secretion events and also allow visualization of exocytosed endosomes.
Highlights
EVs can interact with recipient cells in both autocrine and paracrine manners.
EVs not only interact with cells locally but can also transit through blood and lymph vessels and travel to distant tissues.
Different classes of cancer EVs injected into the bloodstream have different biodistributions.
There are a variety of methods to detect EV uptake and biodistribution.
EVs and Endocrine Disorders
Obesity and Insulin Resistance
As discussed in earlier sections, it is important to employ methods that rigorously enrich EVs using standardized methodologies and then to characterize the resulting preparations with respect to particle size and well-accepted EV-associated markers. Several papers in the field of obesity report studies using preparations that contain substantial amounts of a heterogenic population of EVs such as exosomes, microvesicles, apoptotic bodies, etc (see the previous section Extracellular Vesicles’ Heterogeneity).
Since increased adipose tissue is the hallmark of obesity, most studies in the obesity field have focused on small EVs or exosomes derived from this tissue (182). ATMs and adipocytes, as well as other cell types within the adipose tissue, all produce EVs. In addition, adipose tissue explant–derived EVs from pregnant women from normal and gestational diabetes mellitus (GDM) have been characterized (183), and the total number of EVs present in maternal circulation strongly correlated with maternal BMI (184). A number of biological effects have been ascribed to adipose tissue EVs, which are mostly, but not exclusively, mediated by their miRNA cargo. In the context of obesity, ATM and adipocyte-derived EVs have been the most extensively studied. Studies of small EVs or exosomes in obesity have centered on either the use of EV cargo as a biomarker or on their biological effects.
Biomarkers
In the case of obesity, while it might not be necessary to identify a circulating biomarker to diagnose obesity, it would be useful to have biomarkers that are predictive of grade of inflammation and obesity development, success rates for weight loss, or predicters of recidivism (weight regain). Such biomarker studies focus on circulating exosomes, but an important caveat must be acknowledged. Circulating EVs represent the composite of EV released from a large variety of different cell types, and in a given disease, EVs from the relevant cell type or tissue may be masked or confounded by the contribution of EV cargo from multiple other cell types.
Several papers have assessed circulating miRNA content in normal vs obese subjects (139, 185-195), and several different miRNAs were reported as differentially expressed between the 2 groups. However, within these papers the specific differentially expressed miRNAs are not concordant and different patterns were observed. In addition, weight loss measures such as bariatric surgery, exercise, and low-calorie diets all lead to changes in the circulating EV miRNA profiles, but, again, specific miRNA signatures across these studies differ (196-198). At this point, no consensus miRNAs have consistently emerged. Technical issues might also be at work across the different reports, since some studies have examined blood miRNAs and others have assessed miRNAs contained within EVs, with a few focusing on miRNAs within exosomes. The cargo across these different components could differ, resulting in differential results. In addition, identifying the cellular source of potential circulating miRNA biomarkers is a key goal.
Clearly, the potential of circulating miRNAs as biomarkers for metabolic diseases is of great interest and additional work is needed to define consistent patterns that differentiate normal vs obese states. In particular, it would be important to identify exosomal miRNA biomarkers that predict the onset of obesity in susceptible individuals or their responses to nutritional or pharmacological treatment. In addition, recidivism after successful weight loss is an enormous clinical problem, and biomarkers that would have some predictive value on this phenomenon would be important.
It would be of great value if validated standardized methodologies were used for EV preparation and miRNA detection across multiple studies. If such studies were performed in adequately sized cohorts over diverse demographic characteristics, this would greatly aid in the search for circulating EV-based biomarkers.
Biological Effect of EVs
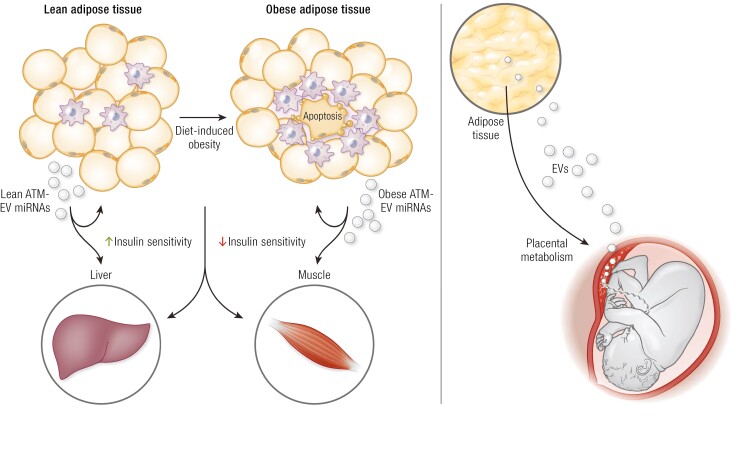
With respect to biological effects, EVs derived from adipose tissue have been well studied in mice, with more limited studies in humans. For example, Ferrante et al showed that EV preparations from human adipose tissue contained a number of miRNAs that were more highly expressed in obese vs lean EVs and reported that a subset of these miRNAs can regulate intracellular signaling pathways important for insulin action (199). Using EVs prepared from adipose tissue from obese vs lean mice, Dang et al also reported a deleterious effect of obese adipose tissue-derived EVs on insulin sensitivity (200). They found that a deficiency of miR-141-3p in the obese EVs contributes to insulin resistance. In this study, as well as in other studies, EVs were extracted from intact adipose tissue. It is not possible, therefore, to know from which cell type they were derived. Using blood as a source of circulating EVs, lean mice treated with circulating EVs from obese mice develop glucose intolerance and insulin resistance (186). The authors of this study also found increased expression of miR-122, -192, -27a-3p, and -27b-3p in those “obese” EVs, and using mimics of these miRNAs, they were able to reproduce the adverse effects on glucose tolerance. With respect to adipocytes, obesity leads to an increase in miR-222 expression that causes impaired insulin sensitivity by inhibiting Glut4, IRS1, and ER2 expression (201). In addition, Thomou et al published an extensive paper probing the circulating exosomal cargo derived specifically from adipocytes (143). Using a combination of techniques, including studies of adipocyte-specific Dicer knockout mice, which do not incorporate miRNAs into EVs, they found that brown adipocytes release EVs containing high levels of miR-99b, which travel to the liver and inhibit FGF21 expression. The subsequent changes in circulating FGF21 levels may contribute to the metabolic dysfunction in obesity.
Obesity is characterized by a substantial accumulation of proinflammatory macrophages in visceral adipose tissue, and numerous studies have shown that these ATMs are major contributors to the insulin resistant state (144, 202-205). Therefore, studies of exosomes specifically derived from ATMs have important implications for obesity-mediated metabolic disease. Thus, exosomes harvested from ATMs in obese mice cause insulin resistance in adipocytes, myocytes, and primary hepatocytes after in vitro treatment, demonstrating direct effects (144). When “obese” ATM exosomes are given to lean mice by intravenous injection, the lean recipient mice develop glucose intolerance, hyperinsulinemia, and insulin resistance, comparable to the obese state, despite the fact that treatment with these exosomes did not cause changes in eating behavior or body weight. In contrast, exosomes obtained from lean mouse ATMs lead to an opposite phenotype (144). Thus, treatment of adipocytes, myocytes, and primary hepatocytes in vitro with “lean” ATM exosomes leads to increased cellular insulin sensitivity and in vivo treatment of obese mice with these preparations produces improved glucose tolerance and decreased insulin resistance (Fig. 4). Using cultured bone marrow-derived macrophages differentiated toward the M2 state in culture as a platform for harvesting exosomes in vitro, a recent study has shown that M2 exosomal miR-690 is the major driver of increased insulin sensitivity induced by these preparations (25, 206). Treatment with a miR-690 mimic leads to increased insulin sensitivity in vitro in adipocytes, muscle cells, and primary hepatocytes and decreases the inflammatory tone of a variety of macrophage preparations. When given in vivo to obese mice, the miR-690 mimic cause a marked improvement in glucose tolerance and insulin sensitivity with no change in body weight. Treatment with a miR-690 antagomir reverses these effects. Other studies have produced results consistent with the concepts summarized in Figure 1. For example, adipose tissue-derived exosomes can enhance macrophage-mediated inflammation and miR-223;-155, and -27a promote proinflammatory signaling in macrophages (207, 208).
Figure 4.
Extracellular vesicles (EVs; eg, small EVs such as exosomes) harvested from adipose tissue macrophages (ATMs) in obese mice promote insulin resistance while exosomes preparations from ATMs in lean mice induce insulin sensitivity and might be involved in fetal growth in gestational diabetes mellitus.
Hepatocyte-derived exosomes can also contribute to metabolic regulation in obesity (209). Thus, in the early stages of obesity, hepatocytes secrete exosomes that cause improved glucose tolerance and insulin sensitivity, and miR-3075 is the dominant, and perhaps only, miRNA that causes this phenotype. FA2H as a major target of miR-3075 and hepatocyte exosomes from early onset obese mice (4-week high-fat diet) promote insulin sensitivity through miR-3075 and inhibition of its target FA2H, both in vitro and in vivo. Conversely, in chronic obesity (16-week high-fat diet), this entire situation is reversed, and miR-3075 is no longer expressed in hepatocytes, or at least at very low levels, and, instead, the hepatocytes secrete pathogenic exosomes that promote insulin resistance. They do not do this by directly causing insulin resistance in adipocytes, myocytes, or primary hepatocytes, but, rather, they promote macrophage-mediated inflammation, and this is the way they cause decreased insulin sensitivity. Interestingly, EVs produced by adipose tissue function as an endocrine link during pregnancy, facilitating the communication between mother and fetus by regulating placental metabolism in GDM that might lead to changes in fetal growth (Fig. 4). EVs have been implicated in the molecular crosstalk at the level of intercellular and interorgan signaling and play pivotal roles in regulation of metabolism (210).
In summary, as a general pattern, exosomes derived from blood, hepatocytes, whole adipose tissue, adipocytes, or ATMs from obese mice or humans can all participate in the etiology of insulin resistance and glucose intolerance in obesity. Furthermore, exosomes prepared from lean mouse ATMs might have therapeutic value, and the current literature supports this concept. As in the biomarker field, the specific miRNAs or groups of mRNAs that lead to detrimental or beneficial effects are multiple and not consistent across different studies. miR-155, -27b-3p, and -122 are ones in which more than 1 study has shown effects either causing or preventing metabolic dysfunction.
Since obesity is a dominant characteristic of patients with T2DM, it is likely that many of these principles derived from studies of obesity will also pertain to T2DM. The reader is referred to a number of recent review articles on this subject (208, 210-213) Eventually methods of delivery of miRNAs within exosomes or liposomes or other vesicles will be established to provide therapeutic miRNAs or miRNA inhibitors, and there are already studies progressing through clinical development in other diseases that might serve as useful guides.
Highlights
Since increased adipose tissue is the hallmark of obesity, most studies in the obesity field have focused on small EVs or exosomes derived from this tissue.
Circulating EVs represent the composite of EVs released from a large variety of different cell types, and in a given disease, EVs from the relevant cell type or tissue may be masked or confounded by the contribution of exosomal cargo from multiple other cell types.
At this point, no consensus miRNAs associated with obesity have consistently emerged.
EVs/exosomes derived from blood, whole adipose tissue, adipocytes, or ATMs from obese mice or humans can all participate in the etiology of insulin resistance and glucose intolerance in obesity.
Diabetes
As a prevalent endocrine disease, diabetes mellitus is characterized by abnormal elevation of glucose levels in blood and/or urine, which results from impaired uptake and utilization of glucose by tissue cells. Severe diabetes can cause complications, including diabetic nephropathy, cardiomyopathy, and impaired wound healing. Pancreatic islets secrete insulin to promote glucose uptake and utilization by the brain, muscles, liver, and other organs. Insulin deficiency, largely in part resulting from destruction of islet cells, can lead to type 1 diabetes, even in the early stages of life. Resistance to insulin action (sometimes accompanied by β-cell dysfunction and relative insulin insufficiency), which is closely associated with obesity, may result in the development of T2DM. In addition, glucose intolerance during pregnancy is defined as GDM (214). EVs mediate intercellular communication within or among the endocrine organs under physiological and pathophysiological conditions, including pregnancy (215, 216). The alteration of EV number or content in blood or urine from patients with diabetes has been verified by several studies, indicating the mutual interaction between metabolic disorders and EV characteristics (141, 142, 217-222).
EVs in the Pathogenesis of Diabetes
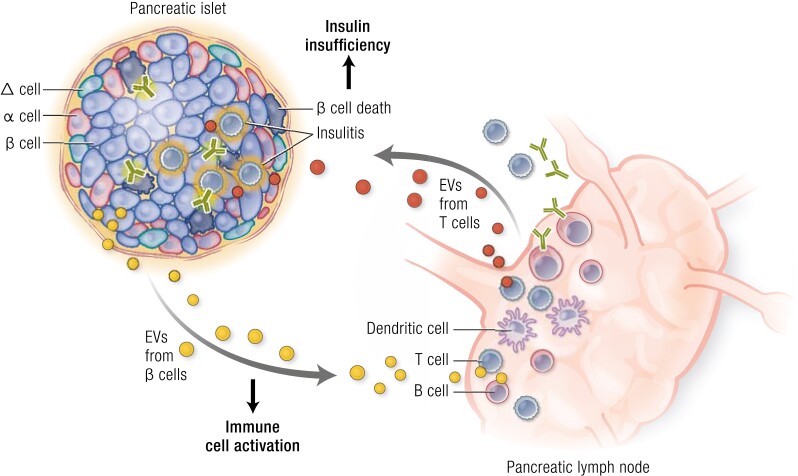
It is largely undefined how EVs participate in the physiological functions of endocrine organs such as the pancreas and liver under healthy conditions; however, an active involvement of EVs in the pathological processes of diabetes is being revealed. Type 1 diabetes is an autoimmune disease in which both innate and adaptive immune cells are involved. EVs regulate the crosstalk between immune cells and insulin-producing cells during the development of diabetes. Either small or large EVs from human or rodent islet β cells or insulinoma cells contribute to insulitis by stimulating antigen-presenting cells, T cells or B cells. Several intracellular autoantigens such as GAD65 and IA-2 have been found in β-cell–derived EVs, which may exert immunostimulatory effects (223-227). The release of EVs may be increased while their contents become more pathogenic upon stress stimuli such as inflammation or hypoxia (142, 225, 228). Conversely, lymphocytes can also release small EVs such as exosomes to act on β cells. Exosomes released by human or NOD mice T cells contained abundant specific miRNA involved in diabetes progression, possessing an ability to promote β-cell apoptosis (229) (Fig. 5). Direct in vivo evidence supporting the contribution of these exosomes to type 1 diabetes, however, remain to be found. Furthermore, horizontal transfer of exosomal miRNA between β cells was also observed upon inflammatory stress (230). Insulin resistance is the primary cause for T2DM in which EVs participate in the crosstalk between adipocytes, hepatocytes, muscle cells, and/or immune cells. Circulating EVs including exosomes from patients with T2DM show an alteration in miRNA or protein signature and can impair the insulin action in skeletal muscle cells and affect the functions of leukocytes (141, 220). To delineate the contribution of EVs to T2DM, it is necessary to identify the complex origin and various functions of theses EVs. Currently, several studies have characterized some EVs from adipose tissue, including adipocytes and macrophages, as well as their effects on insulin action. EVs released from adipose tissue of some patients with T2DM showed direct impairment on insulin action in hepatocytes or myotubes, while another animal study showed the indirect effect of EVs from obese adipose tissue on insulin resistance through activating macrophages (231, 232). EVs from stressed adipocytes may play an important role in reducing insulin action in direct or indirect manners (205, 233-237). Meanwhile, EVs from stressed macrophages may also blunt insulin actions in adipocytes, hepatocytes, and muscle cells (144). In pregnant women, changes in EV characteristics, including their origin and components, have been found in either blood or urine with the development of GDM. Several specific miRNAs upregulated in serum EVs were related to glucose metabolism, while placenta-derived EVs, such as exosomes present in both maternal circulation and urine, possessed distinct protein or miRNA profiles related to inflammation or metabolism (219, 238-241). Using cellular or animal experiments, several studies provided evidence for the adverse effects of EVs from plasma, placenta, or adipose tissue of GDM patients on glucose metabolism, indicating the pathological roles of EVs in GDM progression (183, 222, 242). Recently, small EVs from GDM have been demonstrated to induce glucose intolerance in vivo using an elegant system involving the continuous chronic infusion using miniosmotic pumps of EV that is likely to mimic physiological conditions (183, 222, 242). In addition, EVs are also involved in the pathologies of various diabetic complications including diabetic nephropathy, cardiomyopathy, or retinopathy, which are not described in detail here. These findings provide the possibility to use EVs as biomarkers in monitoring the development of diabetes mellitus. EVs in body fluid may be promising candidates because of convenient collection and easy manipulation. Several studies have shown that EVs in circulation or urine display unique features during different types of diabetes. Taking exosomal miRNA as the example, miRNA-16-5p was downregulated in plasma exosomes from patients with long-term type 1 diabetes but upregulated in urine exosomes from subjects with GDM (217, 239). However, there is still a long way to translate these observations into diabetes-associated EV biomarkers in the clinic. In this field, large amounts of work on standardization and validation remain to be established.
Figure 5.
The contribution of extracellular vesicles (EVs) to the pathogenesis of type 1 diabetes. Islet β cells, especially upon inflammatory stimuli, can produce EVs to activate antigen-presenting cells and T and B lymphocytes, causing insulitis and β-cell destruction. In addition to direct immune attack, T cells can also release EVs to induce β-cell apoptosis, thereby aggravating the development of type 1 diabetes.
EVs in the Therapy of Diabetes
As an effective vesicle to transport bioactive components to target tissues or cells, EVs possess predominant advantages in disease treatment. Emerging evidence has shown the potential of EVs, particularly exosomes from mesenchymal stem cells (MSCs), in treating diabetes mellitus and associated complications. In mouse models of type 1 diabetes induced by streptozotocin, delivery of exosomes from adipose tissue–derived MSCs ameliorated the diabetic symptoms, possibly by suppressing inflammatory T-cell responses (243). Infusion of exosomes from human umbilical cord MSCs also exhibited desired effects on treating T2DM in a rat model induced by a high-fat diet combined with streptozotocin, largely by reducing insulin resistance and inhibiting β-cell apoptosis (244). There are also some animal studies showing the therapeutic effects of MSC-derived exosomes on diabetic complications, particularly diabetic wounds (245-247). Thus far, only a few clinical trials using EV therapies for diabetes have been reported.
In summary, EVs released from cells may be changed in terms of concentration or content, depending on age, sex, genetic background, and physiological or pathological conditions. To clarify the physiological or pathological roles of EVs in the development of diabetes mellitus, further exploration of their cell origins, components, target cells, and action pathways is required. To propose specific EVs as diagnostic biomarkers for diabetes or its complications, standardization of the physiological characteristics of EV profiles, based on large scales of cohorts that cover age, sex, and so on would be necessary. Furthermore, identification of abnormal EV indicators for pathologies in different types or stages may require dynamic monitoring, in which the EV signature needs to be specific enough to exclude other background signals. Regarding the potential EV therapy for diabetes mellitus and associated complications, all processes, including EV preparation and identification, evaluation of EV efficiency and side effects should be strictly determined, through a series of in vitro and in vivo studies including animal and human trials. It should be noted that these future prospects for EV application in diabetes therapy are only based on a few animal experiments. Currently, it is still an open question to elucidate the effectiveness and practicality of EVs in treating diabetes. If so, exact molecular mechanisms of EV action need to be clarified and appropriate therapeutic window should be evaluated.
Highlights
EVs regulate the crosstalk between immune cells and insulin-producing cells during the development of diabetes.
Circulating EVs including exosomes from patients with T2DM show an alteration in miRNA or protein signature and can impair the insulin action in skeletal muscle cells and affect the functions of leukocytes.
In pregnant women, the change of EV characteristics including origins or components has been found in either blood or urine with the development of GDM.
Emerging experimental evidence has shown the potential of EVs, particularly exosomes from MSCs, in treating diabetes mellitus.
Cancer
The impact of EVs and evaluation of their cargo not only apply to the diagnostic and prognostic challenges of cancer progression but also to the challenges in understanding the mechanisms of tumor growth, invasion, metastatic disease, and resistance to therapy (82, 248-251). In the context of endocrine-related cancers, a potential role of EV in pituitary adenoma have been proposed (252-254). Hydrocortisone increased the secretion and altered the RNA profile of EVs from pituitary cells, suggesting a stress-associated response (252). Interestingly, growth hormone-secreting pituitary adenoma EVs stimulates osteoblast proliferation via increased cell viability and DNA replication, associated with changes in the bioactivity of miR-21-5p within EVs (253). lncRNA H19 inhibits the growth of distal pituitary tumors by suppressing the eukaryotic translation initiation factor 4E-binding protein 1 phosphorylation. Interestingly, the level of EV-associated lncRNA H19 in the patients with all subtypes of pituitary tumors was significantly lower compared to healthy controls, suggesting that this molecule within EV might be used as a biomarker for predicting responses of patients with prolactinomas (254).
The role of EVs as cancer biomarkers is evolving (255, 256). EVs have been enriched from all biological fluids and are most often investigated in the blood. Regardless of the biological fluid under study, the goal underlying the use of EVs as biomarkers of pathology is to capture discernible molecular changes that are specific to that pathology, whether symptomatic or not. Should EVs represent the cells they originate from and should circulating EVs represent the entire system’s molecular homeostasis, a cancer lesion would thus shed EVs with unique features that distinguish them from EVs shed by noncancerous lesions or normal parenchyma. Although these assumptions frame the studies of EVs as cancer biomarkers, they present valid challenges in the field of EVs as cancer diagnostic or prognostic tools. First, in a given pool of EVs collected from the circulation, the relatively low frequency of cancer-derived EVs may severely limit the detection of cancer-specific signals. This is particularly problematic when biomarkers are studied for early cancer detection, when small lesions would contribute even less to the EV pool in circulation and when symptomology and imaging cannot decisively assist. Second, EVs are not only inherently diverse but also are enriched using methodologies that are still evolving and that present respective limitations in obtaining distinct purity and quality controls for a defined EV subpopulation. The technological limitations are compounded by a dynamic and rapidly expanding field of research that may disagree on the relative abundance of EVs’ given cargo of interest (248). Nonetheless, EVs as cancer biomarkers are likely to continue to expand and find utility in clinical oncology. Despite the issue of signal to noise (common to all biomarkers), EVs give a unique advantage: they present with the potential of interrogating more than 1 molecular species. EVs have been dubbed a “multicomponent biomarker platform” in cancer (255), since their purification and study from biological fluid(s) enable not only profiling of proteins but also other biologicals. The cargo of EVs is indeed complex and rich in distinct biologicals, possibly preserved from enzymatic degradation in circulation, and include not only proteins but also nucleic acids, metabolites, and lipids (248) (see the previous section EV Composition). Surface proteins on EVs may be used to enhance signal to noise, adding power to tests detecting cancer-specific sequences in the DNA, mRNA, miRNA, and other RNA species found in EVs. The combination of multiple biological readouts may also offer a mean to distinguish cancer from other pathologies, with shared vs uniquely assigned EV cargos. This may prove useful in aiding the interpretation of medical imaging without the need for invasive tissue biopsy. Finally, EVs may shield and thus capture nucleic acids with distinct properties (eg, short fragments, transcriptome rather than genome), which may offset sequencing limitations associated with rare or fragile species of nucleic acids in the circulation (257). As alluded to in the previous discussion, EV cargo with distinct cargo enrichment when compared to the cells they originate from may also give biomarker studies an additional advantage, with “cancer-specific cargo” impacting the detection of cancer-specific EVs in the pool of circulating EVs (82). The role of EVs as cancer biomarker may also extend to predicting response to therapy (255, 256, 258), with recent findings indicating that the frequency of EVs with surface expression of immunomodulatory molecules (eg, PD-L1) may inform on response to immune checkpoint blockade (259).
Circulating EVs not only inform on cancer cell biology (cancer cell–derived EVs) but also on the host response (stromal cell–derived EVs), including immune cells (248, 249). Transmembrane proteins and associated extracellular proteins can act as ligands in cell signaling, activating or suppressing pathways involved in various biological responses, including resistance to cancer therapy. The role of EVs in cancer has largely been associated with protumorigenic functions, although it is noted that these may reflect predominantly the role of cancer cell–derived EVs. Cancer cell–derived EVs have been reported to play a role in metastasis when mice are administered bolus injections of purified EVs (160, 260, 261). EVs may also modulate cancer progression by regulating the tumor microenvironment and generating a more tumor permissive environment (251, 262, 263). The complexity of EV intercellular communication is enormous, and intercellular communication is particularly challenging to study in vivo due to its dynamic component. Although it is possible that EVs are efficiently exchanged between a limited number of cell types, it is more likely that the exchange of EVs is more promiscuous and EVs can be exchanged as part of an endless combination of intercellular communication highways. Several such highways in EV exchange that modify the tumor microenvironment include cancer cell–derived EVs impacting stromal cells (fibroblasts, immune cells, endothelial cells etc), and vice-versa. The effect of EV exchange includes transcriptomic, signaling, and metabolic changes (248, 264-266). It is intriguing that EVs (studied in the context of exogenous administration of EVs in animals) demonstrate tropism for some organs (267, 268), which suggests a role for EVs in organ systems communication and interorgan communication (269, 270). The study of how such “preferred” EV exchange routes are used, highjacked, or hindered in cancer will likely guide our understanding of cancer as a systemic disease. In this regard, the role of EVs in promoting cancer cachexia is also emerging (271), although more studies are needed to determine whether they indeed participate in this cancer-associated metabolic syndrome in a rate-limiting fashion. The future studies of EVs in cancer may also reveal an antitumor function of EVs, with a role in limiting tumor angiogenesis, for example (272). It is likely that EVs function to both promote and limit cancer progression, given their cell of origin, microenvironmental milieu, and cancer type and stage of progression.
Since the discovery of the biological impact of the exchange of EV cargos between cells, the role of EVs as natural carriers of therapeutic payload has gained traction toward clinical applications (82, 248, 262, 273). Selecting EVs as nanovesicles of therapeutic payloads over synthetic nanoparticles was motivated by their “physiological camouflage,” where in exogenous administration of EVs, even if modified, benefit from their cell-like exterior and physiological predisposition for efficient cellular uptake (274). This may aid EVs as therapeutic agents by preserving their cargo from degradation during transit, limiting their clearance by immune cells, and minimizing toxic (adverse) reactions that synthetic nanoparticles can elicit (256, 258, 275). By harnessing the tools to modify cells in vitro, it is possible to engineer EVs produced by these cells to present specific ligands or receptors on their surface, the conformation of which, on EVs’ lipid bilayer, may enhance the desired signaling event. This approach was used to harness the antitumor role of dendritic cell–derived EVs in lung cancer therapy (262, 276). As nanoparticles, EVs can also be “loaded” (eg, by electroporation) with a desired therapeutic payload. Therapeutic payloads include small interfering RNAs to regulate target cell transcriptome, enabling the downregulation of otherwise undruggable targets (267, 277). Chemotherapeutic payloads of EVs may enhance the benefit of the encapsulated drug by controlling their delivery to target organs (cancer lesions) and limiting off-target effects. The role of EVs in cancer therapeutics is rapidly evolving, informed from a growing number of preclinical studies and emerging clinical trials (248).
EVs hold several key possible roles in our study of cancer, from their use as biomarkers to their development as a novel class of therapeutics. Their study will likely inform us about cancer as a systemic disease and will likely continue to give new insights into harnessing the host’s response to control cancer progression.
Highlights
In the context of cancer, the impact of EVs and evaluation of their cargo not only apply to the diagnostic and prognostic challenges of cancer progression but also to the challenges in understanding the mechanisms of tumor growth, invasion, metastatic disease, and resistance to therapy.
EVs are not only inherently diverse but also are enriched using methodologies that are still evolving and that present different limitations in obtaining distinct purity and quality controls for a defined EV subpopulation.
The role of EVs as cancer biomarker may also extend to predicting response to therapy.
Selecting EVs as nanovesicles of therapeutic payloads over synthetic nanoparticles was motivated by their “physiological camouflage,” wherein exogenous administration of EVs, even if modified, benefits from EVs’ cell-like exterior and physiological predisposition for efficient cellular uptake.
Cardiovascular Disease
EVs, Cardiovascular Disease, and Endocrine Disorder
Normal endocrine function is essential for cardiovascular health. Disorders of the endocrine system impact metabolic syndrome, adiposity, insulin resistance, high blood pressure (BP), hormonal changes, obesity, and diabetes, which are well-known comorbidities that can significantly elevate the risk of death from cardiovascular disease (CVD). Tissue-specific EVs released from diverse cell types, including adipose tissues, immune systems, liver, muscles, etc, to the central circulation may carry and transfer functional RNA, proteins, and lipids to regulate the metabolic function of the target cells and tissues related to CVD. Changes in the EV profile and bioactivity have been described in several endocrine-related conditions in CVDs. Here, we discuss the composition and function of EVs in regulation of various cardiac manifestations secondary to endocrine dysfunction, and vice versa, and their possible clinical use as blood-based biomarkers.
EVs and EV lncRNAs in Cardiometabolic Diseases, Including Endothelial Dysfunction, Diabetes, and Atherosclerosis
Metabolic syndrome is a clustering of risk factors that increases susceptibility to serious cardiometabolic complications, including T2DM and myocardial infarction. The clustering of several cardiometabolic disease traits [eg, abdominal obesity, atherogenic dyslipidemia, hyperglycemia, insulin resistance, and/or hypertension (HTN)] strikingly elevates the risk of developing T2DM or overt CVD to a much greater extent than having only 1 disease trait. One of the prominent features of cardiometabolic diseases is endothelial dysfunction. It has been demonstrated that endothelial cell homeostasis and response to pathological stimuli are critically impacted by regulatory molecules carried by circulating EVs such as noncoding RNAs and proteins. Interestingly, one of the strongest genetic associations in the development of coronary artery disease (CAD), cholesterol metabolism, and T2DM is with 9p21, a lncRNA known as ANRIL (278) that has been shown to be secreted in association with EVs (279, 280). Similar to ANRIL, polymorphisms in lncRNAs, which are detected extracellularly in circulation in association with EVs, in addition to intracellular expression, such as MALAT-1 (281), H19 (282), and p21 (283) have also been shown to be associated with an increased risk of CAD, hypertrophy, myocardial ischemia, and contractile dysfunction. Compared with healthy volunteers, patients with atherosclerosis, CAD, myocardial ischemia, and heart failure have been shown to have higher serum levels of H19, LIPCR, and HIF1A-AS1 (282, 284) and lower serum levels of ANRIL, KCNQ1OT1, and MALAT1. However, serum levels of some of these lncRNAs were shown to be poor predictors of left ventricular dysfunction (283). Of note, studies using human cultured cells in vitro, as well as those using patient samples, have shown that many of these lncRNAs are associated with secreted exosomes or EVs in circulation (285). Additional controlled studies are needed before EV-associated circulating lncRNAs can be used reliably as biomarkers for atherosclerotic diseases.
EV-associated Circulating miRNAs in Cardiometabolic Diseases and Endothelial Cell Dysfunction
Circulating miRNAs can be modulated by many of the characteristic features of cardiometabolic diseases, including HTN, hyperglycemia, insulin resistance, obesity, and dyslipidemia [reviewed in (286)]. When these comorbidities are combined in patients with metabolic syndrome, a distinct set of circulating miRNAs, including miRNAs enriched in the endothelium, is dysregulated.
Interestingly, several of the identified circulating miRNAs are claimed to have tissue specificity. Many circulating miRNAs that are present at lower concentrations in the blood of patients with CAD compared with healthy controls (such as miR-126, -17, and -92a) are known to be highly expressed in the vascular wall, especially in the endothelium (287). Others are enriched in cardiac muscle (miR-208, -199, -133a, -1, and -499), in vascular smooth muscle cells (e.g., miR-145 and -143) and in inflammatory cells (miR-155) (287-289). Moreover, another study using human, murine, and cell-culture models suggested that circulating and tissue expression of ex-RNAs are temporally regulated and are associated with the pathological state of the heart or other tissues that they are released from (290). Presence of circulating miRNAs, often studied in the context of biomarkers, is controversial in relation to their presence in different plasma components, such as association with circulating EVs, Argonaute complexes, LDLs, HDLs, etc (119, 291, 292). It is widely established, however, that multiple different types of RNA species are transported by EVs from different cellular origins.
Other EV-associated Biomolecules and Factors in Cardiovascular Diseases
Interestingly, large EVs, but not small EVs, from cardiomyocyte origin were shown to express cardiac troponin T (293), a well-known and sensitive biomarker that can detect a small degree of damage to the heart. EVs originating from cardiomyocytes and myocardial tissue were shown to be rapidly taken up by infiltrating monocytes and to regulate local inflammatory processes. Another noteworthy clinical study investigating protein epitope profile has demonstrated that EV levels of CD62p, CD42a, CD41b, CD31, and CD40 increased in acute myocardial infarction (ST-elevation myocardial infarction and angina patients) (294), some of which have high potential as biomarkers.
A number of studies have reported that the numbers of circulating EVs is increased in several pathological conditions including insulin-resistant patients (295) and in patients with T2DM and microvascular complications, including CVD, HTN (296), atherosclerosis (297, 298), obesity (299), stroke, and myocardial infarction (293, 300), and postsurgical interventions of the heart (301), whereas many studies report significant reductions in EV numbers after calorific restriction or bariatric surgery (302). These studies provide evidence of trafficking of EVs from the heart or from other tissues to peripheral circulation carrying biomolecules that may serve as biomarkers or potential disseminators of disease.
EVs in Cardio-endocrine System
The same factors that increase the risk of cardiometabolic disease are also risk factors for several endocrine disorders, such as polycystic ovary syndrome (PCOS) (303, 304), which leads to reduced fertility. Several studies have now shown that in accordance with these increased risk factors, PCOS patients have increased circulating levels of EVs, particularly procoagulant and proinflammatory platelet EVs (305, 306), although a causal relationship has not yet been established between EVs and the other symptoms of PCOS. Nevertheless, these studies implicate that EVs can play an important role in the pathogenesis of cardiometabolic and cardio-endocrine disorders and that EV-associated biomolecules could be important biomarkers for predicting risk. In addition, inflammation could be at the center of CVD and endocrine disorders, connecting adipose tissue, liver, skeletal muscle, pancreas, heart, and other organs via circulating EVs that contribute to the development of obesity, insulin resistance, diabetes, and CVD.
Highlights
Changes in the EV profile and bioactivity have been described in several endocrine-related conditions in CVDs.
Presence of circulating miRNAs, often studied in the context of biomarkers, is controversial in relation to their presence in different plasma components, such as association with circulating EVs, Argonaute complexes, LDLs, HDLs, etc.
EVs originating from cardiomyocytes and myocardial tissue were shown to be rapidly taken up by infiltrating monocytes and shown to regulate local inflammatory processes.
EVs can play an important role in the pathogenesis of cardiometabolic and cardio-endocrine disorders and EV-associated biomolecules could be important biomarkers for predicting risk.
Hypertension
Arterial Hypertension
Multiple systems contribute to BP homeostasis, including the vasculature, nervous, and immune systems, along with the kidney and the various hormonal regulators (307). HTN remains a major cause of premature death worldwide, and therefore developing a better understanding of the molecular pathogenesis of HTN is crucial (308). EVs have already gained significant attention as potential new surrogate biomarkers for endothelial dysfunction and vascular damage and may serve as novel biomarkers and bioregulators in primary and secondary HTN (309-311).
EVs as Biomarkers
In clinical studies, plasma-derived EV levels of endothelial origin were found to be higher in patients with primary HTN but could be lowered with antihypertensive treatments. Most studies, however, focused on studying EV surface and protein markers of endothelial cell origin using flow cytometry (eg, E-selectin, platelet endothelial cell adhesion molecule, vascular endothelial-cadherin, S-endoglin) (312-314). A few studies from rodent models of HTN and human HTN included other circulating EV subtypes and identified leukocyte- and T-cell (CD3+)-derived EVs as a major subset of EVs (315, 316). These EVs correlated significantly with BP severity, defining them as clinically relevant biomarkers. Interestingly, no correlations with BP levels were observed with levels of endothelial-derived EVs in these models. This finding is consistent with the paradigm that the immune system and, in particular, T cells play a significant role in HTN and support a functional role of EVs in HTN pathogenesis (317). EV-RNA cargo was also studied as biomarkers in HTN. Comparing the RNA cargo of plasma derived exosomes from spontaneously hypertensive rats (SHR) to normotensive Wistar Kyoto (WKY) rats, the SHR EVs carried more unique miRNAs (318). Further analysis of EVs’ origin and EV cargo opens up discovery for novel pathologic pathways and treatment targets. In addition, EVs might play a role as early biomarkers for end-organ damage in HTN. HTN is a silent disease and current markers of end-organ damage such as left ventricular hypertrophy or hypertensive retinopathy are indicating already advanced organ damage. As an example, for early and more sensitive and specific EV biomarkers, EVs deriving from peritubular capillaries in the kidney were identified by flow cytometry as early markers for hypertensive kidney damage (319). Using proteomics for urinary EV (uEV) cargo characterization, an altered protein pattern was also found in a group of 40 nondiabetic hypertensive patients with and without albuminuria (320).
Beside the diagnostic role of EVs in HTN, a prognostic role has also been investigated in 1 large clinical study of 844 individuals from the Framingham Offspring cohort, who did not have HTN at study entry. Levels of endothelial-derived EVs (CD144+) were identified as a cardiovascular risk factor and was significantly associated with the development of HTN (311). More large clinical studies are crucially needed to confirm the diagnostic and prognostic role of circulating or uEVs in HTN.
Moreover, EVs have been studied as biomarkers in secondary HTN, which represents about 4% to 5% of cases with elevated BP due to a known cause (308). Kidney-derived EVs from podocytes (podocalyxin and nephrin positive) were significantly elevated in renovascular HTN and showed an inverse correlation with renal blood flow (321). Primary hyperaldosteronism (PA), the most common endocrine cause for secondary HTN, is characterized by the inappropriate production of aldosterone and overstimulation of the mineralocorticoid receptor, leading to hypokalemia and increase in BP. Microarray analysis of EV mRNA showed that 19 genes were differentially expressed, of which the endothelin 1 gene (EDN1) was downregulated and only detected in PA patients, representing a potential biomarker (322). EVs have also been examined extensively in preeclampsia (323), a secondary cause for severe HTN in pregnancy. Podocyte-derived EVs were identified in humans with preeclampsia and associated with altered podocyte expression (324).
Bioactivity
As EVs carry proteins, metabolites, and RNAs, they have gained significant attention as possible vascular effectors in HTN, promoting intercellular and interorgan communication (325). In particular, they have been found to carry angiotensin type 1 receptor (AT1 R) (326), endothelial nitric oxide (NO) synthases (eNOS), and nicotinamide adenine dinucleotide phosphate subunits that impair NO release (327) or the transfer of chemokines and adhesion molecules to endothelial cells (327). Pironti et al demonstrated in a refined in vivo model that AT1 R–enriched exosomes are released from the heart undergoing a cardiac pressure load. These EVs are functional as exogeneous transfer of AT1 R–enriched exosomes increased BP in AT1 R knockout mice. The EV transfer improved their response to angiotensin II (326). Another example of EVs’ interaction with the renin-angiotensin system (RAS) is work by Ren et al (328) and Tong et al (329). Angio-converting enzyme (ACE) content was found to be higher in adventitial fibroblast derived EVs in SHR compared to WKY. Increased ACE in EVs from SHR increased angiotensin II levels, activated AT1R, and promoted vascular smooth muscle migration, likely through miR155-5p. In addition, monocyte miR-27a in EVs decreases Mas receptor expression, impairing Ang1-7 vasodilation and thus causing HTN (330). Thus, EVs can be seen as novel messengers of the RAS through transfer of molecules from proteins to RNA. This effect is bidirectional as RAS increases the formation of EVs as well.
EVs’ direct vascular function has been largely tested in ex vivo studies using pressure myography of small arteries. Most of the EVs investigated were generated in cell culture in vitro (327, 331, 332). These studies demonstrated that endothelial- and T-cell–derived EVs reduce endothelial dependent vasodilation in resistance arteries from mice, likely via a NO-dependent mechanism. Platelet-derived EVs resulted in thromboxane A2 receptor–mediated vasoconstriction (331, 332). These results were confirmed by studies using EVs generated from in vivo models, SHRs, and humans. Interestingly, EVs’ vasoactive effects occur during or after the development of HTN in these models (333). There is also evolving evidence for functional EV-RNA cargo in HTN. Monocyte miR-27a in EVs has been found to decrease Mas receptor expression and eNOS phosphorylation, impairing angiotensin 1-7 vasodilation, which can lead to elevated BP (330).
A possibly therapeutic effect of EVs on BP regulation in in vivo models has only been reported by a few groups (334, 335). For instance, Otani et al isolated circulating EVs from normotensive WKY rats and administered them intraperitoneally weekly, over 6 weeks, into SHR rats and measured BP by tail cuff (334). BP in hypertensive animals could be lowered. However, the dose effect of these functional EVs, their cellular origin, their exact vasoactive factor(s) (EV cargo of protein/metabolites/RNA), and cellular target(s) are still unknown. The therapeutic role of MSC-derived EVs was tested in HTN by injecting them into aging mice intravenously. Aging-related vascular stiffness and HTN were mitigated (336).
Furthermore, EVs of tubular origin have been investigated in HTN. These EVs carry functional units including sodium transporters (eg, epithelial sodium channel, thiazide-sensitive sodium chloride cotransporter) and water channels (aquaporin) (337). Evolving evidence is demonstrating that these EVs of tubular origin might play an important role in BP and volume regulation, which can be of importance in salt-sensitive individuals or patients with diuretic resistance.
In summary, EVs not only represent novel biomarkers of HTN severity and end-organ damage but also play a mechanistic role in HTN pathogenesis as bioregulators. Understanding EV biology and EV cargo in HTN can open up discoveries of novel and needed treatment targets (Fig. 6). In addition, circulating or uEVs have the potential to better characterize the different phenotypes of HTN. As the optimal BP threshold for initiating BP treatment is still controversial, EVs might represent easy (plasma EVs) and noninvasive (uEVs) clinical biomarkers for detection of early end-organ damage, which can guide effective treatment in HTN and lower cardiovascular risk. To advance the field of EVs in HTN, more rigor and transparency in reporting per guidelines of the EV societies are needed.
Figure 6.
Functional role of extracellular vesicles (EVs) in essential hypertension.
Adapted with permission from La Salvia S et al. Curr Hypertens Rep, 2020; 22(10) © Springer Science Business Media, LLC, part of Springer Nature.
Highlights
EVs have already gained significant attention as potential new surrogate biomarkers for endothelial dysfunction and vascular damage and may serve as novel biomarkers and bioregulators in primary and secondary HTN.
EVs might play a role as early biomarkers for end-organ damage in HTN.
EVs have also been examined extensively in preeclampsia and PA as secondary cause for HTN.
Endothelial- and T-cell–derived EVs reduce endothelial dependent vasodilation in resistance arteries from mice, likely via a NO-dependent mechanism.
EVs are novel messengers of the RAS, which also contributes to release of EVs.
The therapeutic and BP-lowering role of EVs from stem cells and normotensive animals was tested in HTN by injecting them into aging mice and the spontaneously hypertensive rat intravenously.
Final Remarks
“Nevertheless, the history of science has repeatedly shown how the introduction of a new word can act as a catalyst for research—just consider the words ‘radioactivity’, ‘chromosome’, ‘antibiotic’, ‘apoptosis’ and, of course, ‘molecular biology’. When Starling first introduced ‘hormone’ a hundred years ago, virtually nothing was known about the nature of hormones or chemical messengers. Biochemistry was then still in its infancy, but it soon became obvious to many physiologists that a chemical approach was needed to understand the nature and actions of hormones” (338). The quote from Tata’s 2005 review is equally applicable to the field of endocrinology, codified by the term “extracellular vesicle.” EV is now ensconced in the lexicon of endocrinology. As it was for hormones 120 years ago, our understanding of the role of EV signaling pathways in physiology and pathophysiology, as biomarkers of disease and their potential as therapeutic agents, is formative. Advances in understanding and application will be enabled by the use of a standardized EV taxonomy, in which subtypes of vesicles are better defined. The current lack of standardization is acknowledged and represents a significant caveat to data interpretation and limits progress. Precise definition of the molecular cargo of EVs may represent a more effective approach for assigning specific functions to EVs, rather than size, density, or biogenic mechanism.
The following statement is issued as an aid in standardizing EV research practice within the field of endocrinology.
EVs are vesicular mediators released by cells into biofluid compartments;
EVs are carriers of diverse molecular cargoes, including protein, nucleic acid, and lipids;
EVs are released by cells to mediate autocrine, paracrine, endocrine, and exocrine activities;
EVs are released both under normal conditions and in response to challenges to cellular homoeostasis;
EVs are transported within biofluids to target cells where they affect a specific cellular response;
EVs represent non- or less-invasive diagnostics and next-generation drug delivery vehicles;
EVs are composed of different subtypes with distinct biophysical properties and biogenesis pathways and distinct surface markers and cargo;
EVs are potential novel biomarkers associated with disease phenotypes across a range of endocrine and metabolic disorders;
When state-of-the-art methods, as described by the ISEV and in the section Isolation and Characterization Methods, are followed, including characterization of exosomal protein markers, then the use of the term “exosome” is appropriate in defining such vesicles; otherwise, the term “ extracellular vesicle” is preferred.
Characterization of EV phenotype, cargo, and function should align with the recommendations of the EV societies to provide the necessary rigor, reproducibility, and interoperability in EV research (MISEV, EV-Track, MiCytFlowEV, uEV position paper).
Acknowledgments
The authors thank E. Dale Abel, Aditi Bhargava, and Robert M. Carey for carefully reading the manuscript.
Contributor Information
Carlos Salomon, Exosome Biology Laboratory, Centre for Clinical Diagnostics, University of Queensland Centre for Clinical Research, Royal Brisbane and Women’s Hospital, Faculty of Medicine, The University of Queensland, Brisbane, Australia.
Saumya Das, Cardiovascular Research Center of Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA.
Uta Erdbrügger, Department of Medicine, Nephrology Division, University of Virginia, Charlottesville, VA, USA.
Raghu Kalluri, Department of Cancer Biology, Metastasis Research Center, University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Sai Kiang Lim, Institute of Molecular and Cell Biology, Agency for Science, Technology and Research, Singapore.
Jerrold M Olefsky, Department of Medicine, University of California-San Diego, La Jolla, CA, USA.
Gregory E Rice, Inoviq Limited, Notting Hill, Australia.
Susmita Sahoo, Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
W Andy Tao, Department of Biochemistry, Purdue University, West Lafayette, IN, USA.
Pieter Vader, CDL Research, Division LAB, UMC Utrecht, Utrecht, the Netherlands Faculty of Medicine, Utrecht University, Utrecht, the Netherlands; Laboratory of Experimental Cardiology, UMC Utrecht, Utrecht, The Netherlands.
Qun Wang, Key Laboratory of Infection and Immunity of Shandong Province, Department of Immunology, School of Basic Medical Sciences, Cheeloo College of Medicine, Shandong University, Jinan, China.
Alissa M Weaver, Department of Cell and Developmental Biology, Vanderbilt University School of Medicine, Nashville, TN, USA; Department of Pathology, Microbiology and Immunology, Vanderbilt University Medical Center, Nashville, TN, USA.
Disclosures
C.S., U.E., J.M.O., S.S., Q.W., and A.M.W. declare no conflicts. S.D. is a founding member and holds equity in Switch Therapeutics. MD Anderson Cancer Center and R.K. hold patents in the area of exosome biology that are licensed to Codiak Biosciences, Inc. MD Anderson Cancer Center and R.K. are stock equity holders in Codiak Biosciences, Inc. R.K. is a consultant and scientific adviser for Codiak Biosciences, Inc. L.S.K. owns founder shares in Paracrine Therapeutics. GER was appointed Chief Scientific Officer of INOVIQ Limited Australia, a publicly listed diagnostic company, in September 2021. W.A.T. is the cofounder of Tymora Analytical Operation. P.V. serves on the scientific advisory board of Evox Therapeutics.
Data Availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
References
- 1. Starling EH. The Croonian Lecture; the chemical correlation of the functions of the body. Lancet. 1905;166:339-341. [Google Scholar]
- 2. Takahashi A, Okada R, Nagao K, et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun. 2017;8:15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Salomon C, Rice GE. Role of exosomes in placental homeostasis and pregnancy disorders. Prog Mol Biol Transl Sci. 2017;145:163-179. [DOI] [PubMed] [Google Scholar]
- 4. Mincheva-Nilsson L. Placental exosome-mediated immune protection of the fetus: feeling groovy in a cloud of exosomes. Expert Rev Obstet Gynaecol. 2010;5:619-634. [Google Scholar]
- 5. Théry C, Boussac M, Véron P, et al. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166(12):7309-7318. [DOI] [PubMed] [Google Scholar]
- 6. Johnstone RM, Mathew A, Mason AB, Teng K. Exosome formation during maturation of mammalian and avian reticulocytes: evidence that exosome release is a major route for externalization of obsolete membrane proteins. J Cell Physiol. 1991;147(1):27-36. [DOI] [PubMed] [Google Scholar]
- 7. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654-659. [DOI] [PubMed] [Google Scholar]
- 8. Taylor DD, Zacharias W, Gercel-Taylor C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol Biol. 2011;728:235-246. [DOI] [PubMed] [Google Scholar]
- 9. Chen HS, Tong HS, Zhao Y, Hong CY, Bin JP, Su L. Differential expression pattern of exosome long non-coding RNAs (lncRNAs) and MicroRNAs (miRNAs) in vascular endothelial cells under heat stroke. Med Sci Monit. 2018;24:7965-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Llorente A, Skotland T, Sylvänne T, et al. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim Biophys Acta. 2013;1831(7):1302-1309. [DOI] [PubMed] [Google Scholar]
- 11. Skotland T, Hessvik NP, Sandvig K, Llorente A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J Lipid Res. 2019;60(1):9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mayo JN, Bearden SE. Driving the hypoxia-inducible pathway in human pericytes promotes vascular density in an exosome-dependent manner. Microcirculation. 2015;22(8):711-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harischandra DS, Ghaisas S, Rokad D, et al. Environmental neurotoxicant manganese regulates exosome-mediated extracellular miRNAs in cell culture model of Parkinson’s disease: Relevance to α-synuclein misfolding in metal neurotoxicity. Neurotoxicology. 2018;64:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giusti I, D’Ascenzo S, Millimaggi D, et al. Cathepsin B mediates the pH-dependent proinvasive activity of tumor-shed microvesicles. Neoplasia. 2008;10(5):481-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lobos-González L, Bustos R, Campos A, et al. Exosomes released upon mitochondrial ASncmtRNA knockdown reduce tumorigenic properties of malignant breast cancer cells. Sci Rep. 2020;10(1):343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Truong G, Guanzon D, Kinhal V, et al. Oxygen tension regulates the miRNA profile and bioactivity of exosomes released from extravillous trophoblast cells—liquid biopsies for monitoring complications of pregnancy. PloS One. 2017;12(3):e0174514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rice GE, Scholz-Romero K, Sweeney E, et al. The effect of glucose on the release and bioactivity of exosomes from first trimester trophoblast cells. J Clin Endocrinol Metab. 2015;100(10):E1280-E1288. [DOI] [PubMed] [Google Scholar]
- 19. Salomon C, Kobayashi M, Ashman K, Sobrevia L, Mitchell MD, Rice GE. Hypoxia-induced changes in the bioactivity of cytotrophoblast-derived exosomes. PloS One. 2013;8(11):e79636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salomon C, Ryan J, Sobrevia L, et al. Exosomal signaling during hypoxia mediates microvascular endothelial cell migration and vasculogenesis. PloS One. 2013;8(7):e68451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen CC, Liu L, Ma F, et al. Elucidation of exosome migration across the blood-brain barrier model in vitro. Cell Mol Bioeng. 2016;9(4):509-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Betzer O, Perets N, Angel A, et al. In vivo neuroimaging of exosomes using gold nanoparticles. ACS Nano. 2017;11(11):10883-10893. [DOI] [PubMed] [Google Scholar]
- 23. Wu CX, Liu ZF. Proteomic profiling of sweat exosome suggests its involvement in skin immunity. J Invest Dermatol. 2018;138(1):89-97. [DOI] [PubMed] [Google Scholar]
- 24. Lässer C, O’Neil SE, Ekerljung L, Ekström K, Sjöstrand M, Lötvall J. RNA-containing exosomes in human nasal secretions. Am J Rhinol Allergy. 2011;25(2):89-93. [DOI] [PubMed] [Google Scholar]
- 25. Lässer C, Alikhani VS, Ekström K, et al. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011;9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nocera AL, Mueller SK, Stephan JR, et al. Exosome swarms eliminate airway pathogens and provide passive epithelial immunoprotection through nitric oxide. J Allergy Clin Immunol. 2019;143(4):1525-1535.e1. [DOI] [PubMed] [Google Scholar]
- 27. Mueller SK, Nocera AL, Bleier BS. Exosome function in aerodigestive mucosa. Nanomedicine. 2018;14(2):269-277. [DOI] [PubMed] [Google Scholar]
- 28. Grigor’eva AE, Tamkovich SN, Eremina AV, et al. Exosomes in tears of healthy individuals: isolation, identification, and characterization. Biochem (Mosc) Suppl B: Biomed Chem. 2016;10:165-172. [Google Scholar]
- 29. Klingeborn M, Dismuke WM, Bowes Rickman C, Stamer WD. Roles of exosomes in the normal and diseased eye. Prog Retin Eye Res. 2017;59:158-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arraud N, Linares R, Tan S, et al. Extracellular vesicles from blood plasma: determination of their morphology, size, phenotype and concentration. J Thromb Haemost. 2014;12(5):614-627. [DOI] [PubMed] [Google Scholar]
- 31. Edgar JR, Eden ER, Futter CE. Hrs- and CD63-dependent competing mechanisms make different sized endosomal intraluminal vesicles. Traffic. 2014;15(2):197-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213-228. [DOI] [PubMed] [Google Scholar]
- 33. Meldolesi J. Exosomes and ectosomes in intercellular communication. Curr Biol. 2018;28(8):R435-R444. [DOI] [PubMed] [Google Scholar]
- 34. Phan TK, Ozkocak DC, Poon IKH. Unleashing the therapeutic potential of apoptotic bodies. Biochem Soc Trans. 2020;48(5):2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113(8):E968-E977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation: association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987;262(19):9412-9420. [PubMed] [Google Scholar]
- 37. Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116-125. [DOI] [PubMed] [Google Scholar]
- 38. Dragovic RA, Gardiner C, Brooks AS, et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine. 2011;7(6):780-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van der Vlist EJ, Nolte-‘t Hoen EN, Stoorvogel W, Arkesteijn GJ, Wauben MH. Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat Protoc. 2012;7(7):1311-1326. [DOI] [PubMed] [Google Scholar]
- 40. Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jeppesen DK, Fenix AM, Franklin JL, et al. Reassessment of exosome composition. Cell. 2019;177(2):428-445.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yáñez-Mó M, Siljander PRM, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4(1) 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dragovic RA, Southcombe JH, Tannetta DS, Redman CW, Sargent IL. Multicolor flow cytometry and nanoparticle tracking analysis of extracellular vesicles in the plasma of normal pregnant and pre-eclamptic women. Biol Reprod. 2013;89(6):151. [DOI] [PubMed] [Google Scholar]
- 44. da Rocha-Azevedo B, Schmid SL. Migrasomes: a new organelle of migrating cells. Cell Res. 2015;25(1):1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tavano S, Heisenberg CP. Migrasomes take center stage. Nat Cell Biol. 2019;21(8):918-920. [DOI] [PubMed] [Google Scholar]
- 46. Ma L, Li Y, Peng J, et al. Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res. 2015;25(1):24-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Caruso S, Poon IKH. Apoptotic cell-derived extracellular vesicles: more than just debris. Front Immunol. 2018;9:1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Suzuki J, Denning DP, Imanishi E, Horvitz HR, Nagata S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science. 2013;341(6144):403-406. [DOI] [PubMed] [Google Scholar]
- 49. Minciacchi VR, You S, Spinelli C, et al. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget. 2015;6(13):11327-11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Al-Nedawi K, Meehan B, Micallef J, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10(5):619-624. [DOI] [PubMed] [Google Scholar]
- 51. Di Vizio D, Kim J, Hager MH, et al. Oncosome formation in prostate cancer: association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res. 2009;69(13):5601-5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci U S A. 2012;109(11):4146-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang Q, Lu Q. Plasma membrane-derived extracellular microvesicles mediate non-canonical intercellular NOTCH signaling. Nat Commun. 2017;8(1):709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Willms E, Johansson HJ, Mäger I, et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci Rep. 2016;6:22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Matsuda A, Kuno A, Yoshida M, et al. Comparative glycomic analysis of exosome subpopulations derived from pancreatic cancer cell lines. J Proteome Res. 2020;19(6):2516-2524. [DOI] [PubMed] [Google Scholar]
- 56. Lai RC, Tan KH, Lim SK. Membrane lipid binding molecules for the isolation of bona fide extracellular vesicle types and associated biomarkers in liquid biopsy. J Cancer Metastasis Treat. 2019;5;65. [Google Scholar]
- 57. Rodrigues-Junior DM, Tan SS, de Souza Viana L, et al. A preliminary investigation of circulating extracellular vesicles and biomarker discovery associated with treatment response in head and neck squamous cell carcinoma. BMC Cancer. 2019;19(1):373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Di Vizio D, Morello M, Dudley AC, et al. Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. Am J Pathol. 2012;181(5):1573-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lopatina T, Bruno S, Tetta C, Kalinina N, Porta M, Camussi G. Platelet-derived growth factor regulates the secretion of extracellular vesicles by adipose mesenchymal stem cells and enhances their angiogenic potential. Cell Commun Signal. 2014;12:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kucharzewska P, Christianson HC, Welch JE, et al. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc Natl Acad Sci U S A. 2013;110(18):7312-7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hosseinkhani B, van den Akker NMS, Molin DGM, Michiels L. (Sub)populations of extracellular vesicles released by TNF-α-triggered human endothelial cells promote vascular inflammation and monocyte migration. J Extracell Vesicles. 2020;9(1):1801153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hergenreider E, Heydt S, Tréguer K, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14(3):249-256. [DOI] [PubMed] [Google Scholar]
- 63. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487-514. [DOI] [PubMed] [Google Scholar]
- 64. Wolf P. The nature and significance of platelet products in human plasma. Br J Haematol. 1967;13(3):269-288. [DOI] [PubMed] [Google Scholar]
- 65. Crescitelli R, Lasser C, Szabo TG, et al. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles. 2013;2. doi:10.3402/jev.v2i0.20677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tkach M, Kowal J, Zucchetti AE, et al. Qualitative differences in T-cell activation by dendritic cell-derived extracellular vesicle subtypes. Embo J. 2017;36(20):3012-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. van der Grein SG, Defourny KAY, Rabouw HH, et al. Picornavirus infection induces temporal release of multiple extracellular vesicle subsets that differ in molecular composition and infectious potential. PloS Pathog. 2019;15(2):e1007594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yuana Y, Levels J, Grootemaat A, et al. Co-isolation of extracellular vesicles and high-density lipoproteins using density gradient ultracentrifugation. J Extracellular Vesicles. 2014;3: doi: 10.3402/jev.v3.23262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mathivanan S, Simpson RJ. ExoCarta: a compendium of exosomal proteins and RNA. Proteomics. 2009;9(21):4997-5000. [DOI] [PubMed] [Google Scholar]
- 70. Kalra H, Simpson RJ, Ji H, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PloS Biol. 2012;10(12):e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hoshino A, Kim HS, Bojmar L, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182(4):1044-1061.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen IH, Xue L, Hsu CC, et al. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc Natl Acad Sci U S A. 2017;114(12):3175-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin Appl. 2015;9(3-4):358-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Saraswat M, Joenväära S, Musante L, Peltoniemi H, Holthofer H, Renkonen R. N-linked (N-) glycoproteomics of urinary exosomes [Corrected]. Mol Cell Proteomics. 2015;14(2):263-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sokolova V, Ludwig AK, Hornung S, et al. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf B Biointerfaces. 2011;87(1):146-150. [DOI] [PubMed] [Google Scholar]
- 76. Palmisano G, Jensen SS, Le Bihan MC, et al. Characterization of membrane-shed microvesicles from cytokine-stimulated β-cells using proteomics strategies. Mol Cell Proteomics. 2012;11(8):230-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25(6):364-372. [DOI] [PubMed] [Google Scholar]
- 78. Zhang Y, Wu X, Andy Tao W. Characterization and applications of extracellular vesicle proteome with post-translational modifications. Trends Analyt Chem. 2018;107:21-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chen IH, Aguilar HA, Paez Paez JS, et al. Analytical pipeline for discovery and verification of glycoproteins from plasma-derived extracellular vesicles as breast cancer biomarkers. Anal Chem. 2018;90(10):6307-6313. [DOI] [PubMed] [Google Scholar]
- 80. Li DT, Habtemichael EN, Julca O, et al. GLUT4 storage vesicles: specialized organelles for regulated trafficking. Yale J Biol Med. 2019;92(3):453-470. [PMC free article] [PubMed] [Google Scholar]
- 81. Andaluz Aguilar H, Iliuk AB, Chen IH, Tao WA. Sequential phosphoproteomics and N-glycoproteomics of plasma-derived extracellular vesicles. Nat Protoc. 2020;15(1):161-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Möller A, Lobb RJ. The evolving translational potential of small extracellular vesicles in cancer. Nat Rev Cancer. 2020;20(12):697-709. [DOI] [PubMed] [Google Scholar]
- 83. Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21(1):9-17. [DOI] [PubMed] [Google Scholar]
- 84. André F, Schartz NE, Chaput N, et al. Tumor-derived exosomes: a new source of tumor rejection antigens. Vaccine. 2002;20(suppl 4):A28-A31. [DOI] [PubMed] [Google Scholar]
- 85. Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183(3):1161-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lindenbergh MFS, Stoorvogel W. Antigen presentation by extracellular vesicles from professional antigen-presenting cells. Annu Rev Immunol. 2018;36:435-459. [DOI] [PubMed] [Google Scholar]
- 87. Lai CP, Kim EY, Badr CE, et al. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat Commun. 2015;6:7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hung ME, Leonard JN. Stabilization of exosome-targeting peptides via engineered glycosylation. J Biol Chem. 2015;290(13):8166-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. McNaught AD, Wilkinson A. Compendium of Chemical Terminology: IUPAC Recommendations. 2nd ed. Blackwell Scientific Publications; 1997. [Google Scholar]
- 90. Fahy E, Subramaniam S, Brown HA, et al. A comprehensive classification system for lipids. J Lipid Res. 2005;46(5):839-861. [DOI] [PubMed] [Google Scholar]
- 91. Li L, Han J, Wang Z, et al. Mass spectrometry methodology in lipid analysis. Int J Mol Sci. 2014;15(6):10492-10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010;11(8):593-598. [DOI] [PubMed] [Google Scholar]
- 93. Albert B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. 4th ed. Garland Science; 2002. [Google Scholar]
- 94. Rice GE. Secretory phospholipases and membrane polishing. Placenta. 1998;19(1):13-20. [DOI] [PubMed] [Google Scholar]
- 95. Gonsiorek W, Lunn C, Fan X, Narula S, Lundell D, Hipkin RW. Endocannabinoid 2-arachidonyl glycerol is a full agonist through human type 2 cannabinoid receptor: antagonism by anandamide. Mol Pharmacol. 2000;57(5):1045-1050. [PubMed] [Google Scholar]
- 96. Dumuis A, Sebben M, Haynes L, Pin JP, Bockaert J. NMDA receptors activate the arachidonic acid cascade system in striatal neurons. Nature. 1988;336(6194):68-70. [DOI] [PubMed] [Google Scholar]
- 97. Krishna S, Zhong XP. Regulation of lipid signaling by diacylglycerol kinases during T cell development and function. Front Immunol. 2013;4:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3. doi:10.3402/jev.v3.24641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Record M, Silvente-Poirot S, Poirot M, Wakelam MJO. Extracellular vesicles: lipids as key components of their biogenesis and functions. J Lipid Res. 2018;59(8):1316-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Haraszti RA, Miller R, Dubuke ML, et al. Serum deprivation of mesenchymal stem cells improves exosome activity and alters lipid and protein composition. Iscience. 2019;16:230-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Nishida-Aoki N, Izumi Y, Takeda H, et al. Lipidomic analysis of cells and extracellular vesicles from high- and low-metastatic triple-negative breast cancer. Metabolites. 2020;10(2):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Skotland T, Sandvig K, Llorente A. Lipids in exosomes: current knowledge and the way forward. Prog Lipid Res. 2017;66:30-41. [DOI] [PubMed] [Google Scholar]
- 103. Kreimer S, Belov AM, Ghiran I, et al. Mass-spectrometry-based molecular characterization of extracellular vesicles: lipidomics and proteomics. J Proteome Res. 2015;14(6):2367-2384. [DOI] [PubMed] [Google Scholar]
- 104. Choi DS, Kim DK, Kim YK, Gho YS. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013;13(10-11):1554-1571. [DOI] [PubMed] [Google Scholar]
- 105. Boilard E. Extracellular vesicles and their content in bioactive lipid mediators: more than a sack of microRNA. J Lipid Res. 2018;59(11):2037-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Egea-Jimenez AL, Zimmermann P. Lipids in exosome biology. Handb Exp Pharmacol. 2020;259:309-336. [DOI] [PubMed] [Google Scholar]
- 107. Haraszti RA, Didiot MC, Sapp E, et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J Extracell Vesicles. 2016;5:32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Subra C, Laulagnier K, Perret B, Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie. 2007;89(2):205-212. [DOI] [PubMed] [Google Scholar]
- 109. Anderson HC. Vesicles associated with calcification in the matrix of epiphyseal cartilage. J Cell Biol. 1969;41(1):59-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wu LN, Genge BR, Kang MW, Arsenault AL, Wuthier RE. Changes in phospholipid extractability and composition accompany mineralization of chicken growth plate cartilage matrix vesicles. J Biol Chem. 2002;277(7):5126-5133. [DOI] [PubMed] [Google Scholar]
- 111. Skotland T, Sagini K, Sandvig K, Llorente A. An emerging focus on lipids in extracellular vesicles. Adv Drug Deliv Rev. 2020;159:308-321. [DOI] [PubMed] [Google Scholar]
- 112. Verlekar D, Wei SJ, Cho H, Yang S, Kang MH. Ceramide synthase-6 confers resistance to chemotherapy by binding to CD95/Fas in T-cell acute lymphoblastic leukemia. Cell Death Dis. 2018;9(9):925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Yan FX, Dong GR, Qiang S, Niu YJ, Hu CY, Meng YH. Overexpression of △12, △15-desaturases for enhanced lipids synthesis in yarrowia lipolytica. Front Microbiol. 2020;11:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ercolano G, De Cicco P, Rubino V, et al. Knockdown of PTGS2 by CRISPR/CAS9 system designates a new potential gene target for melanoma treatment. Front Pharmacol. 2019;10:1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Li SP, Lin ZX, Jiang XY, Yu XY. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol Sin. 2018;39(4):542-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Skog J, Würdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Murillo OD, Thistlethwaite W, Rozowsky J, et al. exRNA Atlas analysis reveals distinct extracellular RNA cargo types and their carriers present across human biofluids. Cell. 2019;177(2):463-477.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Srinivasan S, Yeri A, Cheah PS, et al. Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell. 2019;177(2):446-462.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Das S, Ansel KM, Bitzer M, et al. ; Extracellular RNA Communication Consortium. The Extracellular RNA Communication Consortium: establishing foundational knowledge and technologies for extracellular RNA research. Cell. 2019;177(2):231-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Mateescu B, Kowal EJ, van Balkom BW, et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA—an ISEV position paper. J Extracell Vesicles. 2017;6(1):1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lai RC, Tan SS, Yeo RW, et al. MSC secretes at least 3 EV types each with a unique permutation of membrane lipid, protein and RNA. J Extracell Vesicles. 2016;5:29828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Zhang H, Freitas D, Kim HS, et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol. 2018;20(3):332-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Danielson KM, Rubio R, Abderazzaq F, Das S, Wang YE. High throughput sequencing of extracellular RNA from human plasma. PloS One. 2017;12(1):e0164644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Giraldez MD, Spengler RM, Etheridge A, et al. Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling. Nat Biotechnol. 2018;36(8):746-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Yeri A, Courtright A, Danielson K, et al. Evaluation of commercially available small RNASeq library preparation kits using low input RNA. BMC Genomics. 2018;19(1):331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Godoy PM, Barczak AJ, DeHoff P, et al. Comparison of reproducibility, accuracy, sensitivity, and specificity of miRNA quantification platforms. Cell Rep. 2019;29(12):4212-4222.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Gyuris A, Navarrete-Perea J, Jo A, et al. Physical and molecular landscapes of mouse glioma extracellular vesicles define heterogeneity. Cell Rep. 2019;27(13):3972-3987.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Chiou NT, Kageyama R, Ansel KM. Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Rep. 2018;25(12):3356-3370.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Cambier L, de Couto G, Ibrahim A, et al. Y RNA fragment in extracellular vesicles confers cardioprotection via modulation of IL-10 expression and secretion. EMBO Mol Med. 2017;9(3):337-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Rodosthenous RS, Hutchins E, Reiman R, et al. Profiling extracellular long RNA transcriptome in human plasma and extracellular vesicles for biomarker discovery. Iscience. 2020;23(6):101182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Cha DJ, Franklin JL, Dou Y, et al. KRAS-dependent sorting of miRNA to exosomes. Elife. 2015;4:e07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife. 2016;5:e19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Santangelo L, Giurato G, Cicchini C, et al. The RNA-binding protein SYNCRIP Is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016;17(3):799-808. [DOI] [PubMed] [Google Scholar]
- 136. Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Wozniak AL, Adams A, King KE, et al. The RNA binding protein FMR1 controls selective exosomal miRNA cargo loading during inflammation. J Cell Biol. 2020;219(10):e201912074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Li J, Salvador AM, Li G, et al. Mir-30d regulates cardiac remodeling by intracellular and paracrine signaling. Circ Res. 2021;128(1):e1-e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Jones A, Danielson KM, Benton MC, et al. miRNA signatures of insulin resistance in obesity. Obesity (Silver Spring). 2017;25(10):1734-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Shah R, Murthy V, Pacold M, et al. Extracellular RNAs are associated with insulin resistance and metabolic phenotypes. Diabetes Care. 2017;40(4):546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Katayama M, Wiklander OPB, Fritz T, et al. Circulating exosomal miR-20b-5p is elevated in type 2 diabetes and could impair insulin action in human skeletal muscle. Diabetes. 2019;68(3):515-526. [DOI] [PubMed] [Google Scholar]
- 142. Lakhter AJ, Pratt RE, Moore RE, et al. Beta cell extracellular vesicle miR-21-5p cargo is increased in response to inflammatory cytokines and serves as a biomarker of type 1 diabetes. Diabetologia. 2018;61(5):1124-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Thomou T, Mori MA, Dreyfuss JM, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542(7642):450-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Ying W, Riopel M, Bandyopadhyay G, et al. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell. 2017;171(2):372-384.e12. [DOI] [PubMed] [Google Scholar]
- 145. French KC, Antonyak MA, Cerione RA. Extracellular vesicle docking at the cellular port: extracellular vesicle binding and uptake. Semin Cell Dev Biol. 2017;67:48-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Gong J, Körner R, Gaitanos L, Klein R. Exosomes mediate cell contact-independent ephrin-Eph signaling during axon guidance. J Cell Biol. 2016;214(1):35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Sato S, Vasaikar S, Eskaros A, et al. EPHB2 carried on small extracellular vesicles induces tumor angiogenesis via activation of ephrin reverse signaling. JCI Insight. 2019;4(23):e132447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Altei WF, Pachane BC, Dos Santos PK, et al. Inhibition of αvβ3 integrin impairs adhesion and uptake of tumor-derived small extracellular vesicles. Cell Commun Signal. 2020;18(1):158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Carney RP, Hazari S, Rojalin T, et al. Targeting tumor-associated exosomes with integrin-binding peptides. Adv Biosyst. 2017;1(5):1600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Shimaoka M, Kawamoto E, Gaowa A, et al. Connexins and integrins in exosomes. Cancers (Basel). 2019;11(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Shimoda A, Sawada SI, Sasaki Y, Akiyoshi K. Exosome surface glycans reflect osteogenic differentiation of mesenchymal stem cells: profiling by an evanescent field fluorescence-assisted lectin array system. Sci Rep. 2019;9(1):11497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Williams C, Pazos R, Royo F, et al. Assessing the role of surface glycans of extracellular vesicles on cellular uptake. Sci Rep. 2019;9(1):11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Hao S, Bai O, Li F, Yuan J, Laferte S, Xiang J. Mature dendritic cells pulsed with exosomes stimulate efficient cytotoxic T-lymphocyte responses and antitumour immunity. Immunology. 2007;120(1):90-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Näslund TI, Paquin-Proulx D, Paredes PT, Vallhov H, Sandberg JK, Gabrielsson S. Exosomes from breast milk inhibit HIV-1 infection of dendritic cells and subsequent viral transfer to CD4+ T cells. AIDS. 2014;28(2):171-180. [DOI] [PubMed] [Google Scholar]
- 155. van Dongen HM, Masoumi N, Witwer KW, Pegtel DM. Extracellular vesicles exploit viral entry routes for cargo delivery. Microbiol Mol Biol Rev. 2016;80(2):369-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Christianson HC, Belting M. Heparan sulfate proteoglycan as a cell-surface endocytosis receptor. Matrix Biol. 2014;35:51-55. [DOI] [PubMed] [Google Scholar]
- 157. Christianson HC, Svensson KJ, van Kuppevelt TH, Li JP, Belting M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc Natl Acad Sci U S A. 2013;110(43):17380-17385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Maas SLN, Breakefield XO, Weaver AM. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. 2017;27(3):172-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. O’Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO, et al. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol. 2020;21(10):585-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Peinado H, Alečković M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Sato S, Weaver AM. Extracellular vesicles: important collaborators in cancer progression. Essays Biochem. 2018;62(2):149-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Collot M, Ashokkumar P, Anton H, et al. MemBright: a family of fluorescent membrane probes for advanced cellular imaging and neuroscience. Cell Chem Biol. 2019;26(4):600-614.e7. [DOI] [PubMed] [Google Scholar]
- 164. Dabrowska S, Del Fattore A, Karnas E, et al. Imaging of extracellular vesicles derived from human bone marrow mesenchymal stem cells using fluorescent and magnetic labels. Int J Nanomedicine. 2018;13:1653-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Macklin R, Wang H, Loo D, et al. Extracellular vesicles secreted by highly metastatic clonal variants of osteosarcoma preferentially localize to the lungs and induce metastatic behaviour in poorly metastatic clones. Oncotarget. 2016;7(28):43570-43587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Nicola AM, Frases S, Casadevall A. Lipophilic dye staining of Cryptococcus neoformans extracellular vesicles and capsule. Eukaryot Cell. 2009;8(9):1373-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Simonsen JB. Pitfalls associated with lipophilic fluorophore staining of extracellular vesicles for uptake studies. J Extracell Vesicles. 2019;8(1):1582237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Takov K, Yellon DM, Davidson SM. Confounding factors in vesicle uptake studies using fluorescent lipophilic membrane dyes. J Extracell Vesicles. 2017;6(1):1388731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Pužar Dominkuš P, Stenovec M, Sitar S, et al. PKH26 labeling of extracellular vesicles: characterization and cellular internalization of contaminating PKH26 nanoparticles. Biochim Biophys Acta Biomembr. 2018;1860(6):1350-1361. [DOI] [PubMed] [Google Scholar]
- 170. Danzer KM, Kranich LR, Ruf WP, et al. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol Neurodegener. 2012;7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Hikita T, Miyata M, Watanabe R, Oneyama C. Sensitive and rapid quantification of exosomes by fusing luciferase to exosome marker proteins. Sci Rep. 2018;8(1):14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Lai CP, Mardini O, Ericsson M, et al. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano. 2014;8(1):483-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Takahashi Y, Nishikawa M, Shinotsuka H, et al. Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J Biotechnol. 2013;165(2):77-84. [DOI] [PubMed] [Google Scholar]
- 174. Liu Q, Rojas-Canales DM, Divito SJ, et al. Donor dendritic cell-derived exosomes promote allograft-targeting immune response. J Clin Invest. 2016;126(8):2805-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Men Y, Yelick J, Jin S, et al. Exosome reporter mice reveal the involvement of exosomes in mediating neuron to astroglia communication in the CNS. Nat Commun. 2019;10(1):4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Neckles VN, Morton MC, Holmberg JC, et al. A transgenic inducible GFP extracellular-vesicle reporter (TIGER) mouse illuminates neonatal cortical astrocytes as a source of immunomodulatory extracellular vesicles. Sci Rep. 2019;9(1):3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Shen B, Wu N, Yang JM, Gould SJ. Protein targeting to exosomes/microvesicles by plasma membrane anchors. J Biol Chem. 2011;286(16):14383-14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Miesenböck G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394(6689):192-195. [DOI] [PubMed] [Google Scholar]
- 179. Sung BH, Ketova T, Hoshino D, Zijlstra A, Weaver AM. Directional cell movement through tissues is controlled by exosome secretion. Nat Commun. 2015;6:7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Sung BH, von Lersner A, Guerrero J, et al. A live cell reporter of exosome secretion and uptake reveals pathfinding behavior of migrating cells. Nat Commun. 2020;11(1):2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Verweij FJ, Bebelman MP, Jimenez CR, et al. Quantifying exosome secretion from single cells reveals a modulatory role for GPCR signaling. J Cell Biol. 2018;217(3):1129-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Gao X, Salomon C, Freeman DJ. Extracellular vesicles from adipose tissue-a potential role in obesity and type 2 diabetes? Front Endocrinol (Lausanne). 2017;8:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Jayabalan N, Lai A, Ormazabal V, et al. Adipose tissue exosomal proteomic profile reveals a role on placenta glucose metabolism in gestational diabetes mellitus. J Clin Endocrinol Metab. 2019;104(5):1735-1752. [DOI] [PubMed] [Google Scholar]
- 184. Elfeky O, Longo S, Lai A, Rice GE, Salomon C. Influence of maternal BMI on the exosomal profile during gestation and their role on maternal systemic inflammation. Placenta. 2017;50:60-69. [DOI] [PubMed] [Google Scholar]
- 185. Can U, Buyukinan M, Yerlikaya FH. The investigation of circulating microRNAs associated with lipid metabolism in childhood obesity. Pediatr Obes. 2016;11(3):228-234. [DOI] [PubMed] [Google Scholar]
- 186. Castaño C, Kalko S, Novials A, Párrizas M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc Natl Acad Sci U S A. 2018;115(48):12158-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Hsieh CH, Rau CS, Wu SC, et al. Weight-reduction through a low-fat diet causes differential expression of circulating microRNAs in obese C57BL/6 mice. BMC Genomics. 2015;16:699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Nunez Lopez YO, Coen PM, Goodpaster BH, Seyhan AA. Gastric bypass surgery with exercise alters plasma microRNAs that predict improvements in cardiometabolic risk. Int J Obes (Lond). 2017;41(7):1121-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Ortega FJ, Mercader JM, Catalán V, et al. Targeting the circulating microRNA signature of obesity. Clin Chem. 2013;59(5):781-792. [DOI] [PubMed] [Google Scholar]
- 190. Pescador N, Pérez-Barba M, Ibarra JM, Corbatón A, Martínez-Larrad MT, Serrano-Ríos M. Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PloS One. 2013;8(10):e77251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Villard A, Marchand L, Thivolet C, et al. Diagnostic value of cell-free circulating MicroRNAs for obesity and type 2 diabetes: a meta-analysis. J Mol Biomark Diagn. 2015;6(6):251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Wang R, Hong J, Cao Y, et al. Elevated circulating microRNA-122 is associated with obesity and insulin resistance in young adults. Eur J Endocrinol. 2015;172(3):291-300. [DOI] [PubMed] [Google Scholar]
- 193. Willeit P, Skroblin P, Moschen AR, et al. Circulating MicroRNA-122 is associated with the risk of new-onset metabolic syndrome and type 2 diabetes. Diabetes. 2017;66(2):347-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Wu Q, Li JV, Seyfried F, et al. Metabolic phenotype-microRNA data fusion analysis of the systemic consequences of Roux-en-Y gastric bypass surgery. Int J Obes (Lond). 2015;39(7):1126-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Iacomino G, Russo P, Stillitano I, et al. Circulating microRNAs are deregulated in overweight/obese children: preliminary results of the I.Family study. Genes Nutr. 2016;11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Bae YU, Kim Y, Lee H, et al. Bariatric surgery alters microRNA content of circulating exosomes in patients with obesity. Obesity (Silver Spring). 2019;27(2):264-271. [DOI] [PubMed] [Google Scholar]
- 197. Giardina S, Hernández-Alonso P, Díaz-López A, Salas-Huetos A, Salas-Salvadó J, Bulló M. Changes in circulating miRNAs in healthy overweight and obese subjects: effect of diet composition and weight loss. Clin Nutr. 2019;38(1):438-443. [DOI] [PubMed] [Google Scholar]
- 198. Safdar A, Saleem A, Tarnopolsky MA. The potential of endurance exercise-derived exosomes to treat metabolic diseases. Nat Rev Endocrinol. 2016;12(9):504-517. [DOI] [PubMed] [Google Scholar]
- 199. Ferrante SC, Nadler EP, Pillai DK, et al. Adipocyte-derived exosomal miRNAs: a novel mechanism for obesity-related disease. Pediatr Res. 2015;77(3):447-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Dang SY, Leng Y, Wang ZX, et al. Exosomal transfer of obesity adipose tissue for decreased miR-141-3p mediate insulin resistance of hepatocytes. Int J Biol Sci. 2019;15(2):351-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Shi Z, Zhao C, Guo X, et al. Differential expression of microRNAs in omental adipose tissue from gestational diabetes mellitus subjects reveals miR-222 as a regulator of ERα expression in estrogen-induced insulin resistance. Endocrinology. 2014;155(5):1982-1990. [DOI] [PubMed] [Google Scholar]
- 202. Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117(1):175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Xu H, Barnes GT, Yang Q, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112(12):1821-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Lackey DE, Olefsky JM. Regulation of metabolism by the innate immune system. Nat Rev Endocrinol. 2016;12(1):15-28. [DOI] [PubMed] [Google Scholar]
- 205. Zhang Y, Mei H, Chang X, Chen F, Zhu Y, Han X. Adipocyte-derived microvesicles from obese mice induce M1 macrophage phenotype through secreted miR-155. J Mol Cell Biol. 2016;8(6):505-517. [DOI] [PubMed] [Google Scholar]
- 206. Ying W, Gao H, Dos Reis FCG, et al. MiR-690, an exosomal-derived miRNA from M2-polarized macrophages, improves insulin sensitivity in obese mice. Cell Metab. 2021;33(4):781-790.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Yao F, Yu Y, Feng L, et al. Adipogenic miR-27a in adipose tissue upregulates macrophage activation via inhibiting PPARγ of insulin resistance induced by high-fat diet-associated obesity. Exp Cell Res. 2017;355(2):105-112. [DOI] [PubMed] [Google Scholar]
- 208. Kahn CR, Wang G, Lee KY. Altered adipose tissue and adipocyte function in the pathogenesis of metabolic syndrome. J Clin Invest. 2019;129(10):3990-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Ji Y, Luo Z, Gao H, et al. Hepatocyte-derived exosomes from early onset obese mice promote insulin sensitivity through miR-3075. Nat Metab. 2021;3(9):1163-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Isaac R, Reis FCG, Ying W, Olefsky JM. Exosomes as mediators of intercellular crosstalk in metabolism. Cell Metab. 2021;33(9):1744-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Mori MA, Ludwig RG, Garcia-Martin R, Brandão BB, Kahn CR. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. 2019;30(4):656-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Chen H, Lan HY, Roukos DH, Cho WC. Application of microRNAs in diabetes mellitus. J Endocrinol. 2014;222(1):R1-R10. [DOI] [PubMed] [Google Scholar]
- 213. Ji C, Guo X. The clinical potential of circulating microRNAs in obesity. Nat Rev Endocrinol. 2019;15(12):731-743. [DOI] [PubMed] [Google Scholar]
- 214. American Diabetes Association. (2) Classification and diagnosis of diabetes. Diabetes Care. 2015;38(suppl):S8-S16. [DOI] [PubMed] [Google Scholar]
- 215. Nair S, Ormazabal V, Lappas M, McIntyre HD, Salomon C. Extracellular vesicles and their potential role inducing changes in maternal insulin sensitivity during gestational diabetes mellitus. Am J Reprod Immunol. 2021;85(2):e13361. [DOI] [PubMed] [Google Scholar]
- 216. Nair S, Salomon C. Extracellular vesicles as critical mediators of maternal-fetal communication during pregnancy and their potential role in maternal metabolism. Placenta. 2020;98:60-68. [DOI] [PubMed] [Google Scholar]
- 217. Garcia-Contreras M, Shah SH, Tamayo A, et al. Plasma-derived exosome characterization reveals a distinct microRNA signature in long duration type 1 diabetes. Sci Rep. 2017;7(1):5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Kong Q, et al. Urinary exosome miR-424 and miR-218 as biomarkers for type 1 diabetes in children. Clin Lab. 2019;65(6). doi:10.7754/Clin.Lab.2018.180921 [DOI] [PubMed] [Google Scholar]
- 219. Salomon C, Scholz-Romero K, Sarker S, et al. Gestational diabetes mellitus is associated with changes in the concentration and bioactivity of placenta-derived exosomes in maternal circulation across gestation. Diabetes. 2016;65(3):598-609. [DOI] [PubMed] [Google Scholar]
- 220. Freeman DW, Noren Hooten N, Eitan E, et al. Altered extracellular vesicle concentration, cargo, and function in diabetes. Diabetes. 2018;67(11):2377-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Nair S, Guanzon D, Jayabalan N, et al. Extracellular vesicle-associated miRNAs are an adaptive response to gestational diabetes mellitus. J Transl Med. 2021;19(1):360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Nair S, Jayabalan N, Guanzon D, et al. Human placental exosomes in gestational diabetes mellitus carry a specific set of miRNAs associated with skeletal muscle insulin sensitivity. Clin Sci (Lond). 2018;132(22):2451-2467. [DOI] [PubMed] [Google Scholar]
- 223. Sheng H, Hassanali S, Nugent C, et al. Insulinoma-released exosomes or microparticles are immunostimulatory and can activate autoreactive T cells spontaneously developed in nonobese diabetic mice. J Immunol. 2011;187(4):1591-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Bashratyan R, Sheng H, Regn D, Rahman MJ, Dai YD. Insulinoma-released exosomes activate autoreactive marginal zone-like B cells that expand endogenously in prediabetic NOD mice. Eur J Immunol. 2013;43(10):2588-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Cianciaruso C, Phelps EA, Pasquier M, et al. Primary human and rat β-cells release the intracellular autoantigens GAD65, IA-2, and proinsulin in exosomes together with cytokine-induced enhancers of immunity. Diabetes. 2017;66(2):460-473. [DOI] [PubMed] [Google Scholar]
- 226. Rutman AK, Negi S, Gasparrini M, Hasilo CP, Tchervenkov J, Paraskevas S. Immune response to extracellular vesicles from human islets of Langerhans in patients with type 1 diabetes. Endocrinology. 2018;159(11):3834-3847. [DOI] [PubMed] [Google Scholar]
- 227. Hasilo CP, Negi S, Allaeys I, et al. Presence of diabetes autoantigens in extracellular vesicles derived from human islets. Sci Rep. 2017;7(1):5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Saravanan PB, Vasu S, Yoshimatsu G, et al. Differential expression and release of exosomal miRNAs by human islets under inflammatory and hypoxic stress. Diabetologia. 2019;62(10):1901-1914. [DOI] [PubMed] [Google Scholar]
- 229. Guay C, Kruit JK, Rome S, et al. Lymphocyte-derived exosomal MicroRNAs promote pancreatic β cell death and may contribute to type 1 diabetes development. Cell Metab. 2019;29(2):348-361.e6. [DOI] [PubMed] [Google Scholar]
- 230. Guay C, Menoud V, Rome S, Regazzi R. Horizontal transfer of exosomal microRNAs transduce apoptotic signals between pancreatic beta-cells. Cell Commun Signal. 2015;13:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Kranendonk ME, Visseren FL, van Herwaarden JA, et al. Effect of extracellular vesicles of human adipose tissue on insulin signaling in liver and muscle cells. Obesity (Silver Spring). 2014;22(10):2216-2223. [DOI] [PubMed] [Google Scholar]
- 232. Deng ZB, Poliakov A, Hardy RW, et al. Adipose tissue exosome-like vesicles mediate activation of macrophage-induced insulin resistance. Diabetes. 2009;58(11):2498-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Mleczko J, Ortega FJ, Falcon-Perez JM, et al. Extracellular vesicles from hypoxic adipocytes and obese subjects reduce insulin-stimulated glucose uptake. Mol Nutr Food Res. 2018;62(5):1700917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Kranendonk ME, Visseren FL, van Balkom BW, et al. Human adipocyte extracellular vesicles in reciprocal signaling between adipocytes and macrophages. Obesity (Silver Spring). 2014;22(5):1296-1308. [DOI] [PubMed] [Google Scholar]
- 235. Pan Y, Hui X, Hoo RLC, et al. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J Clin Invest. 2019;129(2):834-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Song M, Han L, Chen FF, et al. Adipocyte-derived exosomes carrying sonic hedgehog mediate M1 macrophage polarization-induced insulin resistance via Ptch and PI3K pathways. Cell Physiol Biochem. 2018;48(4):1416-1432. [DOI] [PubMed] [Google Scholar]
- 237. Camino T, Lago-Baameiro N, Bravo SB, et al. Vesicles shed by pathological murine adipocytes spread pathology: characterization and functional role of insulin resistant/hypertrophied adiposomes. Int J Mol Sci. 2020;21(6):2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Jayabalan N, Lai A, Nair S, et al. Quantitative proteomics by SWATH‐MS suggest an association between circulating exosomes and maternal metabolic changes in gestational diabetes mellitus. Proteomics. 2019;19(1-2):e1800164. [DOI] [PubMed] [Google Scholar]
- 239. Herrera-Van Oostdam AS, Toro-Ortíz JC, López JA, et al. Placental exosomes isolated from urine of patients with gestational diabetes exhibit a differential profile expression of microRNAs across gestation. Int J Mol Med. 2020;46(2):546-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Gillet V, Ouellet A, Stepanov Y, et al. miRNA profiles in extracellular vesicles from serum early in pregnancies complicated by gestational diabetes mellitus. J Clin Endocrinol Metab. 2019;104(11):5157-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Kandzija N, Zhang W, Motta-Mejia C, et al. Placental extracellular vesicles express active dipeptidyl peptidase IV; levels are increased in gestational diabetes mellitus. J Extracell Vesicles. 2019;8(1):1617000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. James-Allan LB, Rosario FJ, Barner K, et al. Regulation of glucose homeostasis by small extracellular vesicles in normal pregnancy and in gestational diabetes. FASEB J. 2020;34(4):5724-5739. [DOI] [PubMed] [Google Scholar]
- 243. Nojehdehi S, Soudi S, Hesampour A, Rasouli S, Soleimani M, Hashemi SM. Immunomodulatory effects of mesenchymal stem cell-derived exosomes on experimental type-1 autoimmune diabetes. J Cell Biochem. 2018;119(11):9433-9443. [DOI] [PubMed] [Google Scholar]
- 244. Sun Y, Shi H, Yin S, et al. Human mesenchymal stem cell derived exosomes alleviate type 2 diabetes mellitus by reversing peripheral insulin resistance and relieving β-cell destruction. ACS Nano. 2018;12(8):7613-7628. [DOI] [PubMed] [Google Scholar]
- 245. Chen CY, Rao SS, Ren L, et al. Exosomal DMBT1 from human urine-derived stem cells facilitates diabetic wound repair by promoting angiogenesis. Theranostics. 2018;8(6):1607-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Shi Q, Qian Z, Liu D, et al. GMSC-derived exosomes combined with a chitosan/silk hydrogel sponge accelerates wound healing in a diabetic rat skin defect model. Front Physiol. 2017;8:904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Wang M, Wang C, Chen M, et al. Efficient angiogenesis-based diabetic wound healing/skin reconstruction through bioactive antibacterial adhesive ultraviolet shielding nanodressing with exosome release. ACS Nano. 2019;13(9):10279-10293. [DOI] [PubMed] [Google Scholar]
- 248. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478):eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl). 2013;91(4):431-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-mediated metastasis: communication from a distance. Dev Cell. 2019;49(3):347-360. [DOI] [PubMed] [Google Scholar]
- 252. Zhou C, Shen S, Moran R, Deng N, Marbán E, Melmed S. Pituitary somatotroph adenoma-derived exosomes: characterization of nonhormonal actions. J Clin Endocrinol Metab. 2022;107(2):379-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Xiong Y, Tang Y, Fan F, et al. Exosomal hsa-miR-21-5p derived from growth hormone-secreting pituitary adenoma promotes abnormal bone formation in acromegaly. Transl Res. 2020;215:1-16. [DOI] [PubMed] [Google Scholar]
- 254. Zhang Y, Liu YT, Tang H, et al. Exosome-transmitted lncRNA H19 inhibits the growth of pituitary adenoma. J Clin Endocrinol Metab. 2019;104(12):6345-6356. [DOI] [PubMed] [Google Scholar]
- 255. LeBleu VS, Kalluri R. Exosomes as a multicomponent biomarker platform in cancer. Trends Cancer. 2020;6(9):767-774. [DOI] [PubMed] [Google Scholar]
- 256. Fitts CA, Ji N, Li Y, Tan C. Exploiting exosomes in cancer liquid biopsies and drug delivery. Adv Healthc Mater. 2019;8(6):e1801268. [DOI] [PubMed] [Google Scholar]
- 257. Kahlert C. Liquid biopsy: is there an advantage to analyzing circulating exosomal DNA compared to cfDNA or are they the same? Cancer Res. 2019;79(10):2462-2465. [DOI] [PubMed] [Google Scholar]
- 258. Barile L, Vassalli G. Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017;174:63-78. [DOI] [PubMed] [Google Scholar]
- 259. Chen G, Huang AC, Zhang W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560(7718):382-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Chronopoulos A, Kalluri R. Emerging role of bacterial extracellular vesicles in cancer. Oncogene. 2020;39(46):6951-6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Wee I, Syn N, Sethi G, Goh BC, Wang L. Role of tumor-derived exosomes in cancer metastasis. Biochim Biophys Acta Rev Cancer. 2019;1871(1):12-19. [DOI] [PubMed] [Google Scholar]
- 262. Kugeratski FG, Kalluri R. Exosomes as mediators of immune regulation and immunotherapy in cancer. FEBS J. 2021;288:10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. LeBleu VS, Kalluri R. Exosomes exercise inhibition of anti-tumor immunity during chemotherapy. Immunity. 2019;50(3):547-549. [DOI] [PubMed] [Google Scholar]
- 264. Bastos N, Ruivo CF, da Silva S, Melo SA. Exosomes in cancer: use them or target them? Semin Cell Dev Biol. 2018;78:13-21. [DOI] [PubMed] [Google Scholar]
- 265. Rajagopal C, Harikumar KB. The origin and functions of exosomes in cancer. Front Oncol. 2018;8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Yang H, Sun L, Mao Y. The role of exosomes in tumor immunity. Ann Transl Med. 2018;6(suppl 2):S116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Mendt M, Kamerkar S, Sugimoto H, et al. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight. 2018;3(8):e99263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Wiklander OP, Nordin JZ, O’Loughlin A, et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J Extracell Vesicles. 2015;4:26316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Kita S, Maeda N, Shimomura I. Interorgan communication by exosomes, adipose tissue, and adiponectin in metabolic syndrome. J Clin Invest. 2019;129(10):4041-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Castillo-Armengol J, Fajas L, Lopez-Mejia IC. Inter-organ communication: a gatekeeper for metabolic health. EMBO Rep. 2019;20(9):1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Chitti SV, Fonseka P, Mathivanan S. Emerging role of extracellular vesicles in mediating cancer cachexia. Biochem Soc Trans. 2018;46(5):1129-1136. [DOI] [PubMed] [Google Scholar]
- 272. Brossa A, Fonsato V, Bussolati B. Anti-tumor activity of stem cell-derived extracellular vesicles. Oncotarget. 2019;10(20):1872-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Xunian Z, Kalluri R. Biology and therapeutic potential of mesenchymal stem cell-derived exosomes. Cancer Sci. 2020;111(9):3100-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Liao W, Du Y, Zhang C, et al. Exosomes: the next generation of endogenous nanomaterials for advanced drug delivery and therapy. Acta Biomater. 2019;86:1-14. [DOI] [PubMed] [Google Scholar]
- 275. Ferguson SW, Nguyen J. Exosomes as therapeutics: the implications of molecular composition and exosomal heterogeneity. J Control Release. 2016;228:179-190. [DOI] [PubMed] [Google Scholar]
- 276. Gehrmann U, Näslund TI, Hiltbrunner S, Larssen P, Gabrielsson S. Harnessing the exosome-induced immune response for cancer immunotherapy. Semin Cancer Biol. 2014;28:58-67. [DOI] [PubMed] [Google Scholar]
- 277. Kamerkar S, LeBleu VS, Sugimoto H, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546(7659):498-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Holdt LM, Teupser D. Long Noncoding RNA ANRIL: Lnc-ing genetic variation at the chromosome 9p21 locus to molecular mechanisms of atherosclerosis. Front Cardiovasc Med. 2018;5:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Kathiresan S, Voight BF, Purcell S, et al. Myocardial Infarction Genetics Consortium. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41(3):334-3 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Abbastabar M, Sarfi M, Golestani A, Karimi A, Pourmand G, Khalili E. Tumor-derived urinary exosomal long non-coding RNAs as diagnostic biomarkers for bladder cancer. Excli J. 2020;19:301-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res. 2014;115(7):668-677. [DOI] [PubMed] [Google Scholar]
- 282. Zhang Z, Gao W, Long QQ, et al. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Sci Rep. 2017;7(1):7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Pierce JB, Feinberg MW. Long noncoding RNAs in atherosclerosis and vascular injury: pathobiology, biomarkers, and targets for therapy. Arterioscler Thromb Vasc Biol. 2020;40(9):2002-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Wang Y, Liang J, Xu J, et al. Circulating exosomes and exosomal lncRNA HIF1A-AS1 in atherosclerosis. Int J Clin Exp Pathol. 2017;10(8):8383-8388. [PMC free article] [PubMed] [Google Scholar]
- 285. Yuan Z, Huang W. New developments in exosomal lncRNAs in cardiovascular diseases. Front Cardiovasc Med. 2021;8:709169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Rottiers V, Näär AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13(4):239-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Fichtlscherer S, De Rosa S, Fox H, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107(5):677-684. [DOI] [PubMed] [Google Scholar]
- 288. Jansen F, Yang X, Proebsting S, et al. MicroRNA expression in circulating microvesicles predicts cardiovascular events in patients with coronary artery disease. J Am Heart Assoc. 2014;3(6):e001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Zampetaki A, Willeit P, Tilling L, et al. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol. 2012;60(4):290-299. [DOI] [PubMed] [Google Scholar]
- 290. Danielson KM, Shah R, Yeri A, et al. Plasma circulating extracellular RNAs in left ventricular remodeling post-myocardial infarction. Ebiomedicine. 2018;32:172-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Cheng L, Sharples A, Scicluna BJ, et al. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles. 2014;3. doi:10.3402/jev.v3.23743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Chevillet JR, Kang Q, Ruf IK, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014;111(41):14888-14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Loyer X, Zlatanova I, Devue C, et al. Intra-cardiac release of extracellular vesicles shapes inflammation following myocardial infarction. Circ Res. 2018;123(1):100-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Burrello J, Bolis S, Balbi C, et al. An extracellular vesicle epitope profile is associated with acute myocardial infarction. J Cell Mol Med. 2020;24(17):9945-9957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Jayachandran M, Litwiller RD, Lahr BD, et al. Alterations in platelet function and cell-derived microvesicles in recently menopausal women: relationship to metabolic syndrome and atherogenic risk. J Cardiovasc Transl Res. 2011;4(6):811-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Chen Y, Feng B, Li X, Ni Y, Luo Y. Plasma endothelial microparticles and their correlation with the presence of hypertension and arterial stiffness in patients with type 2 diabetes. J Clin Hypertens (Greenwich). 2012;14(7):455-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Diamant M, Nieuwland R, Pablo RF, Sturk A, Smit JW, Radder JK. Elevated numbers of tissue-factor exposing microparticles correlate with components of the metabolic syndrome in uncomplicated type 2 diabetes mellitus. Circulation. 2002;106(19):2442-2447. [DOI] [PubMed] [Google Scholar]
- 298. Feng B, Chen Y, Luo Y, Chen M, Li X, Ni Y. Circulating level of microparticles and their correlation with arterial elasticity and endothelium-dependent dilation in patients with type 2 diabetes mellitus. Atherosclerosis. 2010;208(1):264-269. [DOI] [PubMed] [Google Scholar]
- 299. Stepanian A, Bourguignat L, Hennou S, et al. Microparticle increase in severe obesity: not related to metabolic syndrome and unchanged after massive weight loss. Obesity (Silver Spring). 2013;21(11):2236-2243. [DOI] [PubMed] [Google Scholar]
- 300. D’Alessandra Y, Devanna P, Limana F, et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31(22):2765-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Emanueli C, Shearn AI, Laftah A, et al. Coronary artery-bypass-graft surgery increases the plasma concentration of exosomes carrying a cargo of cardiac microRNAs: an example of exosome trafficking out of the human heart with potential for cardiac biomarker discovery. PloS One. 2016;11(4):e0154274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Cheng V, Kashyap SR, Schauer PR, Kirwan JP, McCrae KR. Restoration of glycemic control in patients with type 2 diabetes mellitus after bariatric surgery is associated with reduction in microparticles. Surg Obes Relat Dis. 2013;9(2):207-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Daskalopoulos G, Karkanaki A, Piouka A, et al. Excess metabolic and cardiovascular risk is not manifested in all phenotypes of polycystic ovary syndrome: implications for diagnosis and treatment. Curr Vasc Pharmacol. 2015;13(6):788-800. [DOI] [PubMed] [Google Scholar]
- 304. Lawson C, Vicencio JM, Yellon DM, Davidson SM. Microvesicles and exosomes: new players in metabolic and cardiovascular disease. J Endocrinol. 2016;228(2):R57-R71. [DOI] [PubMed] [Google Scholar]
- 305. Willis GR, Connolly K, Ladell K, et al. Young women with polycystic ovary syndrome have raised levels of circulating annexin V-positive platelet microparticles. Hum Reprod. 2014;29(12):2756-2763. [DOI] [PubMed] [Google Scholar]
- 306. Carvalho LML, Ferreira CN, Sóter MO, et al. Microparticles: inflammatory and haemostatic biomarkers in polycystic ovary syndrome. Mol Cell Endocrinol. 2017;443:155-162. [DOI] [PubMed] [Google Scholar]
- 307. Coffman TM. Under pressure: the search for the essential mechanisms of hypertension. Nat Med. 2011;17(11):1402-1409. [DOI] [PubMed] [Google Scholar]
- 308. Oparil S, Acelajado MC, Bakris GL, et al. Hypertension. Nat Rev Dis Primers. 2018;4:18014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Erdbrügger U, Le TH. Extracellular vesicles as a novel diagnostic and research tool for patients with HTN and kidney disease. Am J Physiol Renal Physiol. 2019;317(3):F641-F647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Horn P, Cortese-Krott MM, Amabile N, et al. Circulating microparticles carry a functional endothelial nitric oxide synthase that is decreased in patients with endothelial dysfunction. J Am Heart Assoc. 2012;2(1):e003764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Amabile N, Cheng S, Renard JM, et al. Association of circulating endothelial microparticles with cardiometabolic risk factors in the Framingham Heart Study. Eur Heart J. 2014;35(42):2972-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Huang PH, Huang SS, Chen YH, et al. Increased circulating CD31+/annexin V+ apoptotic microparticles and decreased circulating endothelial progenitor cell levels in hypertensive patients with microalbuminuria. J Hypertens. 2010;28(8):1655-1665. [DOI] [PubMed] [Google Scholar]
- 313. Preston RA, Jy W, Jimenez JJ, et al. Effects of severe hypertension on endothelial and platelet microparticles. Hypertension. 2003;41(2):211-217. [DOI] [PubMed] [Google Scholar]
- 314. Sansone R, Baaken M, Horn P, et al. Endothelial microparticles and vascular parameters in subjects with and without arterial hypertension and coronary artery disease. Data Brief. 2018;19:495-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. La Salvia S, Musante L, Lannigan J, Gigliotti JC, Le TH, Erdbrügger U. T cell-derived extracellular vesicles are elevated in essential HTN. Am J Physiol Renal Physiol. 2020;319(5):F868-F875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Zu L, Ren C, Pan B, et al. Endothelial microparticles after antihypertensive and lipid-lowering therapy inhibit the adhesion of monocytes to endothelial cells. Int J Cardiol. 2016;202:756-759. [DOI] [PubMed] [Google Scholar]
- 317. Guzik TJ, Hoch NE, Brown KA, et al. Role of the T cell in the genesis of angiotensin II induced hypertension and vascular dysfunction. J Exp Med. 2007;204(10):2449-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Liu X, Yuan W, Yang L, Li J, Cai J. miRNA profiling of exosomes from spontaneous hypertensive rats using next-generation sequencing. J Cardiovasc Transl Res. 2019;12(1):75-83. [DOI] [PubMed] [Google Scholar]
- 319. Sun IO, Santelli A, Abumoawad A, et al. Loss of renal peritubular capillaries in hypertensive patients is detectable by urinary endothelial microparticle levels. Hypertension. 2018;72(5):1180-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Gonzalez-Calero L, Martínez PJ, Martin-Lorenzo M, et al. Urinary exosomes reveal protein signatures in hypertensive patients with albuminuria. Oncotarget. 2017;8(27):44217-44231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Kwon SH, Woollard JR, Saad A, et al. Elevated urinary podocyte-derived extracellular microvesicles in renovascular hypertensive patients. Nephrol Dial Transplant. 2017;32(5):800-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Burrello J, Gai C, Tetti M, et al. Characterization and gene expression analysis of serum-derived extracellular vesicles in primary aldosteronism. Hypertension. 2019;74(2):359-367. [DOI] [PubMed] [Google Scholar]
- 323. Salomon C, Guanzon D, Scholz-Romero K, et al. Placental exosomes as early biomarker of preeclampsia: potential role of exosomal microRNAs across gestation. J Clin Endocrinol Metab. 2017;102(9):3182-3194. [DOI] [PubMed] [Google Scholar]
- 324. Gilani SI, Anderson UD, Jayachandran M, et al. Urinary extracellular vesicles of podocyte origin and renal injury in preeclampsia. J Am Soc Nephrol. 2017;28(11):3363-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. La Salvia S, Gunasekaran PM, Byrd JB, Erdbrügger U. Extracellular vesicles in essential hypertension: hidden messengers. Curr Hypertens Rep. 2020;22(10):76. [DOI] [PubMed] [Google Scholar]
- 326. Pironti G, Strachan RT, Abraham D, et al. Circulating exosomes induced by cardiac pressure overload contain functional angiotensin II type 1 receptors. Circulation. 2015;131(24): 2120-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Burger D, Turner M, Munkonda MN, Touyz RM. Endothelial microparticle-derived reactive oxygen species: role in endothelial signaling and vascular function. Oxid Med Cell Longev. 2016;2016:5047954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Ren XS, Tong Y, Qiu Y, et al. MiR155-5p in adventitial fibroblasts-derived extracellular vesicles inhibits vascular smooth muscle cell proliferation via suppressing angiotensin-converting enzyme expression. J Extracell Vesicles. 2020;9(1):1698795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Tong Y, Ye C, Ren X-S, et al. Exosome-mediated transfer of ACE (angiotensin-converting enzyme) from adventitial fibroblasts of spontaneously hypertensive rats promotes vascular smooth muscle cell migration. Hypertension. 2018;72(4):881-888. [DOI] [PubMed] [Google Scholar]
- 330. Zou X, Wang J, Chen C, et al. Secreted monocyte miR-27a, via mesenteric arterial mas receptor-eNOS pathway, causes hypertension. Am J Hypertens. 2020;33(1):31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Pfister SL. Role of platelet microparticles in the production of thromboxane by rabbit pulmonary artery. Hypertension. 2004;43(2):428-433. [DOI] [PubMed] [Google Scholar]
- 332. Martin S, Tesse A, Hugel B, et al. Shed membrane particles from T lymphocytes impair endothelial function and regulate endothelial protein expression. Circulation. 2004;109(13):1653-1659. [DOI] [PubMed] [Google Scholar]
- 333. Good ME, Musante L, La Salvia S, et al. Circulating extracellular vesicles in normotension restrain vasodilation in resistance arteries. Hypertension. 2020;75(1):218-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Otani K, Yokoya M, Kodama T, et al. Plasma exosomes regulate systemic blood pressure in rats. Biochem Biophys Res Commun. 2018;503(2):776-783. [DOI] [PubMed] [Google Scholar]
- 335. Marrachelli VG, Mastronardi ML, Sarr M, et al. Sonic hedgehog carried by microparticles corrects angiotensin II-induced hypertension and endothelial dysfunction in mice. PloS One. 2013;8(8):e72861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336. Feng R, Ullah M, Chen K, Ali Q, Lin Y, Sun Z. Stem cell-derived extracellular vesicles mitigate ageing-associated arterial stiffness and hypertension. J Extracell Vesicles. 2020;9(1):1783869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Pathare G, Tutakhel OAZ, van der Wel MC, et al. Hydrochlorothiazide treatment increases the abundance of the NaCl cotransporter in urinary extracellular vesicles of essential hypertensive patients. Am J Physiol Renal Physiol. 2017;312(6):F1063-F1072. [DOI] [PubMed] [Google Scholar]
- 338. Tata JR. One hundred years of hormones. EMBO Rep. 2005;6(6):490-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.