Abstract
CTX-M-10 has been widely disseminated among multiple clones of several species of Enterobacteriaceae, harboring seemingly different plasmids, for over a decade in Ramón y Cajal University Hospital, Madrid, Spain. Cloning and sequencing of a 12.2-kb DNA fragment from plasmid pRYCE21 from Klebsiella pneumoniae strain KP4aC revealed a novel phage-related element immediately upstream of blaCTX-M-10 conserved among different CTX-M-10-producing strains. This is the first report showing an extended-spectrum-β-lactamase gene linked to a phage-related element.
In 1989 a new group of extended-spectrum β-lactamases (ESBL), not related to TEM or SHV enzymes, was described in Germany and was designated CTX-M-1 due to its preferential hydrolysis of cefotaxime (3). Simultaneously, another CTX-M enzyme, later called CTX-M-2, was found to be widely disseminated among Salmonella strains in Argentina (4). Nevertheless, the first-published CTX-M β-lactamase was actually FEC-1, described in Japan in 1986 (18), which was later found to be almost identical to CTX-M-3, characterized in Poland in 1996 (12), when both nucleotide sequences were available (5). In the last few years, there has been an explosive dissemination of CTX-M enzymes, and today they are probably the most widespread ESBL group (5).
Five different major groups of plasmid-mediated CTX-M β-lactamases have been recognized so far, all of which share high degrees of homology with the chromosomal β-lactamases of several species of the genus Kluyvera (5). Chromosomal β-lactamases of Kluyvera ascorbata and Kluyvera georgiana are almost identical to the plasmid-mediated enzymes from the CTX-M-2 and CTX-M-8 groups, respectively, whereas that of Kluyvera cryocrescens is closely related to those of the CTX-M-1 group, although it does not seem to be its direct ancestor (10, 15, 24). The surprisingly rapidly increasing recognition of clinical isolates containing different CTX-M β-lactamases worldwide has led to a growing interest in the investigation of the genetic elements responsible for their explosive emergence and spread. ISEcp1-like insertion sequences have been frequently found upstream of several blaCTX-M genes from different groups and from different geographical origins, and it is believed that they might play a role in CTX-M β-lactamase mobility and expression (8, 11, 16, 25, 29). Genes encoding CTX-M-9 and CTX-M-2 have also been found as part of class 1 integrons containing open reading frame 513 (ORF513) (1, 28).
CTX-M-10 was initially described in 2001 to occur in an Escherichia coli strain isolated in 1997 at Ramón y Cajal University Hospital in Madrid, Spain (20). Long-term molecular epidemiology studies of ESBL-producing Enterobacteriaceae in this institution revealed that this enzyme, present since at least 1990, was widely disseminated among strains of E. coli, Klebsiella pneumoniae, and different species of the genus Enterobacter (7, 9; T. M. Coque, M. C. Varela, A. Oliver, M. I. Morosini, F. Baquero, and R. Cantón, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstract C2-298, p. 118, 2001). Since blaCTX-M-10 had been detected for over a decade in multiple clones of these species and was found to be harbored by seemingly different transferable plasmids by restriction fragment length polymorphism analysis (7, 9), we decided to elucidate whether there was a common genetic environment that could explain the efficient dissemination of this gene among different plasmids and/or bacterial organisms in our institution.
The K. pneumoniae clinical strain KP4aC containing blaCTX-M-10 in a 60-kb plasmid (pRYCE21) (9), recovered from a urine sample in 1997, was used to characterize the genetic environment of this gene. Plasmid pRYCE21 was transferred to a rifampin-resistant mutant of the E. coli laboratory strain BM21 (BM21R) by conjugation with the filter mating method. Transconjugants were selected on MacConkey agar plates containing rifampin (100 μg/ml) plus cefotaxime (2 μg/ml). Plasmid pRYCE21 was isolated from a BM21R transconjugant with the Plasmid Midi Kit (QIAGEN, Hilden, Germany). Analysis of pRYCE21 DNA digested with BamHI or EcoRI showed the presence of three BamHI fragments (360 bp, 8.5 kb, and >40 kb) and 10 EcoRI fragments (ranging from 500 bp to 20 kb). DNA from pRYCE21 digested with either EcoRI or BamHI was ligated to pBGS18− (31), digested with the same enzymes, by using T4 DNA ligase at 16°C overnight. Recombinant plasmids were transformed into the E. coli strain XL1-Blue made competent with CaCl2 and were further selected in MacConkey agar plates containing 50 μg of kanamycin per ml with and without 2 μg of cefotaxime per ml. Transformants were checked by plasmid extraction and digestion with restriction enzymes to obtain a genetic library containing all 10 EcoRI fragments and the 360-bp and the 8.5-kb BamHI fragments described above (we failed to obtain a recombinant plasmid harboring the >40-kb BamHI fragment). Transformants selected on MacConkey agar plates containing kanamycin (50 μg/ml) plus cefotaxime (2 μg/ml) were used to select bacterial colonies containing the blaCTX-M-10 gene, which was found to be present in 7.2-kb EcoRI and 8.5-kb BamHI DNA fragments. The different EcoRI or BamHI DNA fragments were overlapped by PCR using specific primers from the ends of the sequences. A 12.2-kb region surrounding the blaCTX-M-10 gene was then fully sequenced. Comparisons of DNA and amino acid sequences were carried out with BLASTN and BLASTP programs, available at www.ncbi.nlm.nih.gov/BLAST. Multiple sequence alignments were done using the Clustal W 1.8 program (32) available at www.infobiogen.fr. Additionally, specific sets of primers (Table 1) were used to amplify this 12.2-kb region in two additional strains of K. pneumoniae (KP30C and KP36C) (9), three E. coli strains (EC16, EC22, and EC54) (T. M. Coque, M. C. Varela, A. Oliver, M. I. Morosini, F. Baquero, and R. Cantón, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-298, p. 118, 2001), two Enterobacter cloacae strains (ECL4 and ECL5), and one Enterobacter gergoviae strain (7). Strains in each species were selected from different clones harboring apparently different CTX-M-10-producing plasmids, by restriction fragment length polymorphism analysis, as described in their respective published studies.
TABLE 1.
Primers used for PCR amplification of different regions of the 12.2-kb DNA fragment containing blaCTX-M-10 from plasmid pRYCE21
| Primer | Sequence (5′→3′) | Positions | PCR amplification | PCR product size (kb) |
|---|---|---|---|---|
| RYCE21-F1 | CAGGACGCGGTATCACC | 335-351 | ||
| RYCE21-R1 | GGCTGGGATGTCGCGTAAC | 1785-1767 | RYCE21-F1-RYCE21-R1 | 1.5 |
| RYCE21-F2 | GACATTTCCATCGAAGAGCC | 2039-2058 | ||
| RYCE21-R2 | GCCGAGCGGATTAATCAGG | 4032-4014 | RYCE21-F2-RYCE21-R2 | 2.0 |
| RYCE21-F3 | CCCATGAGCCCGCTTACG | 3880-3897 | ||
| RYCE21-R3 | GAGCCACAAAGTGTAGCGC | 4944-4936 | RYCE21-F3-RYCE21-R3 | 2.1 |
| CTX-M-F8 | CCGCGCTACACTTTGTGGC | 4924-4942 | ||
| CTX-M-R3 | TTACAAACCGTCGGTGACG | 5885-5867 | CTX-M-F8-CTXM-R3 | 1.0 |
| RYCE21-F4 | CCCAACCTAAGGCAGAAAG | 5815-5833 | ||
| RYCE21-R4 | ATCGACAAGGTCATGCTGATG | 6425-6405 | RYCE21-F4-RYCE21-R4 | 0.6 |
| RYCE21-R5 | CCATGCTGTTTTCCGTAGTAC | 6828-6808 | RYCE21-F4-RYCE21-R5 | 1.0 |
| RYCE21-R6 | CTCGCTTACTAATTCCCAGC | 8030-8011 | RYCE21-F4-RYCE21-R6 | 2.2 |
| RYCE21-R7 | GGGTTCTGTCACCCTGAC | 8551-8534 | RYCE21-F4-RYCE21-R7 | 2.7 |
| RYCE21-F5 | GCTGAATGTGGACTATGTC | 8922-8940 | ||
| RYCE21-R8 | GCACGATCATTCCTGATAC | 11523-11505 | RYCE21-F5-RYCE21-R8 | 2.6 |
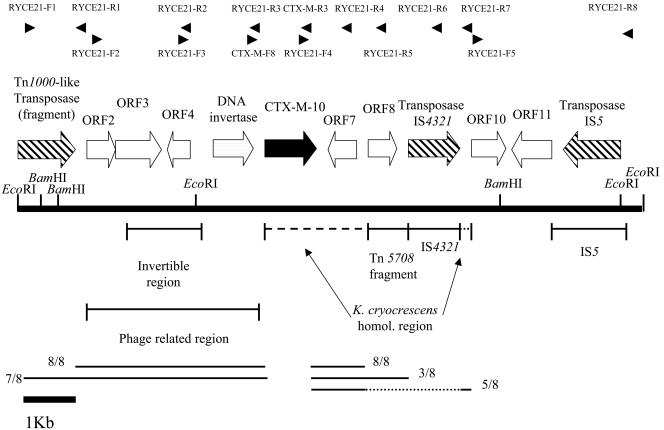
Figure 1 shows the 12.2-kb DNA fragment containing the blaCTX-M-10 gene from pRYCE21. The 3.5-kb fragment upstream from blaCTX-M-10 corresponded to four conserved phage-related ORFs, which were preceded by a Tn1000-like transposase. Three of these four ORFs (ORF2, ORF3, and ORF4) showed homology to conserved phage tail proteins, closest to those found in phage sequences from the genomes of Chromobacterium violaceum or Neisseria meningitidis (46 to 54% identity) (6, 21). Interestingly, a phage-related DNA invertase, 50 to 52% identical to different Pin, Cin, and Gin DNA invertases, preceded by a 1.5-kb invertible region was found 200 bp immediately upstream from the blaCTX-M-10 gene (Fig. 1). Figure 2 shows results from multiple sequence alignment of the plasmid pRYCE21 DNA invertase with other related enzymes. The orientation of the invertible sequence modified the nature of one of the phage tail proteins, making it similar to that found in other phage DNA invertases.
FIG. 1.
Representation of the 12.2-kb region of pRYCE21 harboring the blaCTX-M-10 gene. EcoRI and BamHI restriction sites used for cloning are represented with vertical lines. The locations of the primers used for the mapping of the 12.2-kb region by PCR amplification in different CTX-M-10-producing strains of Enterobacteriaceae are represented with black arrows. The conserved regions and the numbers of strains in which they are found are represented at the bottom of the figure. homol., homologous.
FIG. 2.
Clustal W multiple sequence alignments of DNA invertase (DNA INV) from plasmid pRYCE21 with related phage DNA invertases: Pin DNA invertase from lambdoid prophage (23), PinH DNA invertase from prophage CP-933H (27), Cin from Enterobacteriaceae phages P1 and P7 (14, 26), and Gin from Enterobacteriaceae phage Mu (22).
Downstream of the DNA invertase, a 1.5-kb DNA fragment (including blaCTX-M-10 and an unknown 483-bp ORF designated ORF7) was 90% identical to the Kluyvera cryocrescens chromosomal β-lactamase region. Interestingly, the Kluyvera cryocrescens-homologous region apparently contained two insertion sequences: first, a 700-bp fragment, 97% identical at the nucleotide level to Tn5708 (GenBank accession number AJ010745) containing the left inverted repeat and a 366-bp ORF (ORF8) with unknown function, and second, a complete copy of IS4321. A 38-bp fragment homologous to the K. cryocrescens chromosomal region contiguous to the above-described 1.5-kb fragment was also found (Fig. 1). Finally, the 12.2-kb fragment was completed by a 651-bp ORF (ORF10) coding for a protein that is 65% identical to a conserved hypothetical protein from E. coli, a 747-bp ORF (ORF11) coding for a nucleoprotein- or polynucleotide-associated enzyme that is 56% identical to YaiL from E. coli K-12, and a complete copy of IS5.
Conservation of the 12.2-kb genetic element identified in pRYCE21 from K. pneumoniae strain KP4aC among different clinical strains is represented in Fig. 1. The 5 kb upstream of blaCTX-M-10 including the phage-related region with the DNA invertase and its invertible region was found to be present in all the studied strains, with the exception of Kp36C, for which the fragment corresponding to the Tn1000-like transposase region was not amplified. The downstream region of blaCTX-M-10 was found to be more varied. All strains shared the 0.6-kb Kluyvera cryocrescens-homologous region downstream of blaCTX-M-10 (positive PCR amplification of RYCE21-F4 and RYCE21-R4) (Table 1; Fig. 1). On the other hand, only three additional strains (E. coli strains EC22 and EC54 and E. cloacae strain ECL4) had the Tn5708 fragment (positive PCR amplification with RYCE21-F4 and RYCE21-R5), but none of them were interrupted by IS4321 (negative PCR amplification with RYCE21-F4 and RYCE21-R6). The remaining strains had neither the Tn5708 fragment nor IS4321 inserted in the Kluyvera cryocrescens-homologous region. For these strains, a specific 0.7-kb band was obtained after RYCE21-F4-RYCE21-R7 amplification (Table 1; Fig. 1), as expected, when no additional sequences were inserted in the Kluyvera cryocrescens-homologous region. Sequencing of the 0.7-kb PCR products from two of the isolates confirmed this assumption. No temporal or spatial relationship of strains lacking or gaining the Tn5708 fragment was observed within the studied collection.
CTX-M-10 belongs to the CTX-M-1 group of CTX-M β-lactamases, differing from CTX-M-3 (12) in only two amino acids (Ala27Val and Arg38Gln). Despite this high homology, an important degree of polymorphism is observed when the nucleotide sequences of the genes coding for these closely related β-lactamases are compared. blaCTX-M-10 differs from blaCTX-M-3 in 21 nucleotides (2.4% of the coding sequence). This high degree of polymorphism suggests that both plasmid-mediated β-lactamase genes may be derived from an independent chromosome mobilization process occurring in different strains from the same species of the genus Kluyvera. CTX-M-10 has been successfully disseminated in our hospital for over a decade, despite the fact that blaCTX-M-10-containing plasmids, unlike integron-borne enzymes, do not harbor additional antibiotic resistance determinants. Nevertheless, the absence of CTX-M-10-producing strains reported in other studies until very recently suggested that the high dissemination in our institution was of only local dimensions. In a recent Spanish nationwide study of the prevalence of ESBL in a collection of 170 and 70 ESBL-producing E. coli and K. pneumoniae isolates collected in 2000, respectively, only one K. pneumoniae isolate and three E. coli isolates with CTX-M-10 were recovered, three of which (two E. coli isolates and one K. pneumoniae isolate) were found in two institutions not far from our hospital and the fourth of which (E. coli) was from northern Spain (13). In France, a single E. coli isolate was recently recognized to produce the CTX-M-10 enzyme (17). In contrast to the findings from our institution, ISEcp1 preceded blaCTX-M-10 in the French strain, as did many other CTX-M genes, suggesting that the blaCTX-M-10 gene has been captured in France by a genetic environment different from that of our strains. This is an excellent example of the influence of local genetic patterns on the local dissemination of resistance genes (2).
Whether the phage-like structure found is self-transferable (functional) into different plasmids or whether the high plasmid diversity in CTX-M-10-producing strains of Enterobacteriaceae is a consequence of frequent modifications of a single blaCTX-M-10-bearing plasmid remains to be elucidated. Nevertheless, the results of this work suggest that the transfer of blaCTX-M-10 from the chromosome of Kluyvera spp. to a transferable plasmid may have been mediated by transduction by a bacteriophage, highlighting the potential role of phages in the dissemination of resistance determinants. The first description of a β-lactamase gene transfer mediated by a bacteriophage was reported in 1972 (30). More-recent works show that bacteriophages found in sewage frequently harbor β-lactamase genes of the OXA and PSE type (19). Our study of the genetic environment of the gene coding for the CTX-M-10 β-lactamase illustrates the importance of the environmental reservoir of genetic elements that, as phages, may serve as tools for natural genetic engineering processes that eventually lead to the evolution and spread of antibiotic resistance.
Nucleotide sequence accession number.
The GenBank accession number for the 12.2-kb DNA fragment from pRYCE21 is AY598759.
Acknowledgments
This work was partially supported by the Red Española de Investigación en Patología Infecciosa (REIPI) from the Ministerio de Sanidad of Spain (C03-014), the Fondo de Investigaciones Sanitarias of the Ministerio de Sanidad of Spain (PI02043), the Ministerio de Ciencia y Tecnología of Spain (SAF 2003-09285), and the European Commission (LSHM-CT-2003-503335).
REFERENCES
- 1.Arduino, S. M., P. H. Roy, G. A. Jacoby, B. E. Orman, S. A. Pineiro, and D. Centron. 2002. blaCTX-M-2 is located in an unusual class 1 integron (In35) which includes Orf513. Antimicrob. Agents Chemother. 46:2303-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baquero, F. 2004. From pieces to patterns: evolutionary engineering in bacterial pathogens. Nat. Rev. Microbiol. 2:510-518. [DOI] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., H. Grimm, and S. Schweighart. 1990. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 18:294-298. [DOI] [PubMed] [Google Scholar]
- 4.Bauernfeind, A., J. M. Casellas, M. Goldberg, M. Holley, R. Jungwirth, P. Mangold, T. Rohnisch, S. Schweighart, and R. Wilhelm. 1992. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection 20:294-298. [DOI] [PubMed] [Google Scholar]
- 5.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brazilian National Genome Project Consortium. 2003. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc. Natl. Acad. Sci. USA 30:11660-11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantón, R., A. Oliver, T. M. Coque, M. C. Varela, J. C. Pérez-Díaz, and F. Baquero. 2002. Epidemiology of extended-spectrum β-lactamase-producing Enterobacter isolates in a Spanish hospital during a 12-year period. J. Clin. Microbiol. 40:1237-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, V., T. Lambert, and P. Courvalin. 2002. ColE1-like plasmid pIP843 of Klebsiella pneumoniae encoding extended-spectrum β-lactamase CTX-M-17. Antimicrob. Agents Chemother. 46:1212-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coque, T. M., A. Oliver, J. C. Pérez-Díaz, F. Baquero, and R. Cantón. 2002. Genes encoding TEM-4, SHV-2, and CTX-M-10 extended-spectrum β-lactamases are carried by multiple Klebsiella pneumoniae clones in a single hospital (Madrid 1989 to 2000). Antimicrob. Agents Chemother. 46:500-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decousser, J. W., L. Poirel, and P. Nordmann. 2001. Characterization of a chromosomally encoded extended-spectrum class A β-lactamase from Kluyvera cryocrescens. Antimicrob. Agents Chemother. 45:3595-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dotour, C., R. Bonnet, H. Marchandin, M. Boyer, C. Chanal, and D. Sirot. 2002. CTX-M-1, CTX-M-3, and CTX-M-14 β-lactamases from Enterobacteriaceae isolated in France. Antimicrob. Agents Chemother. 46:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gniadkowski, M., I. Schneider, A. Palucha, R. Jungwirth, B. Mikiewicz, and A. Bauernfeind. 1998. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 42:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernández, J. R., L. Martínez-Martínez, R. Cantón, T. M. Coque, A. Pascual, and the Spanish Group for Nosocomial Infection (GEIH). Nationwide study of Escherichia coli and Klebsiella pneumoniae producing extended-spectrum β-lactamases in Spain. Antimicrob. Agents Chemother., in press. [DOI] [PMC free article] [PubMed]
- 14.Hiestand-Nauer, R., and S. Iida. 1983. Sequence of the site-specific recombinase gene cin and of its substrates serving in the inversion of the C segment of bacteriophage P1. EMBO J. 2:1733-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humeniuk, C., G. Arlet, V. Gautier, P. Grimont, R. Labia, and A. Philippon. 2002. β-Lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46:3045-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karim, A., L. Poirel, S. Nagarajan, and P. Nordmann. 2001. Plasmid-mediated extended spectrum β-lactamase (CTX-M-3) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 17.Lartigue, M. F., L. Poirel, and P. Nordmann. 2004. Diversity of genetic environment of blaCTX-M genes. FEMS Microbiol. Lett. 234:20120-20127. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto, Y., F. Ikeda, T. Kamimura, Y. Yokota, and Y. Mine. 1988. Novel plasmid-mediated β-lactamase from Escherichia coli that inactivates oxymino-cephalosporins. Antimicrob. Agents Chemother. 32:1243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muniesa, M., A. García, E. Miró, B. Mirelis, G. Prats, J. Jofre, and F. Navarro. 2004. Bacteriophages and diffusion of β-lactamase genes. Emerg. Infect. Dis. 10:1134-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliver, A., J. C. Pérez-Díaz, T. M. Coque, F. Baquero, and R. Cantón. 2001. Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing β-lactamase isolated in Spain. Antimicrob. Agents Chemother. 45:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 22.Plasterk, R. H., A. Brinkman, and P. van de Putte. 1983. DNA inversions in the chromosome of Escherichia coli and in bacteriophage Mu: relationship to other site-specific recombination systems. Proc. Natl. Acad. Sci. USA 80:5355-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plasterk, R. H., and P. van de Putte. 1985. The invertible P-DNA segment in the chromosome of Escherichia coli. EMBO J. 4:237-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirel, L., P. Kampfer, and P. Nordmann. 2002. Chromosome-encoded Ambler class A beta-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 46:4038-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirel, L., J. W. Decousser, and P. Nordmann. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritthaler, W., and D. Kamp. 1988. DNA sequence of the site-specific recombination function cin of phage P7. Nucleic Acids Res. 16:6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rose, D. J., G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. Dimalanta, K. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattne. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 28.Sabaté, M., F. Navarro, E. Miro, S. Campoy, B. Mirelis, J. Barbe, and G. Prats. 2002. Novel complex sul1-type integron in Escherichia coli carrying blaCTX-M-9. Antimicrob. Agents Chemother. 46:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saladin, M., V. T. Cao, T. Lambert, J. L. Donay, J. L. Hermann, Z. Ould-Hocine, C. Verdet, F. Delisle, A. Philippon, and G. Arlet. 2002. Diversity of CTX-M β-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209:161-168. [DOI] [PubMed] [Google Scholar]
- 30.Smith, H. W. 1972. Ampicillin resistance in Escherichia coli by phage infection. Nat. New Biol. 238:205-206. [DOI] [PubMed] [Google Scholar]
- 31.Spratt, B. G., P. J. Hedge, S. T. Heesen, A. Edelman, and J. K. Broome-Smith. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8, and pEMBL9. Gene 41:337-342. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 11:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]