Abstract
Analysis of Candida albicans cells using antibodies directed against Gas1p/Ggp1p, Saccharomyces cerevisiae homolog of Phr1p, revealed that Phr1p is a glycoprotein of about 88 kDa whose accumulation increases with the rise of external pH. This polypeptide is present both in the yeast form and during germ tube induction. In the Phr1− cells at pH 8 the solubility of glucans in alkali is greatly affected. In the parental strain the alkali-soluble/-insoluble glucan ratio shows a 50% decrease at pH 8 with respect to pH 4.5, whereas in the null mutant it is unchanged, indicating the lack of a polymer cross-linker activity induced by the rise of pH. The mutant has a sixfold increase in chitin level and is hypersensitive to calcofluor. Consistently with a role of chitin in strengthening the cell wall, Phr1− cells are more sensitive to nikkomycin Z than the parental strain.
PHR1 is a pH-regulated gene required for correct morphogenesis in Candida albicans. This gene codes for a protein of 548 amino acids 56% identical to the Gas1p/Ggp1p/Cwh52p of Saccharomyces cerevisiae and with no homology to other proteins with known function (17). Phr1p and Gas1p play a common function since PHR1 can replace the GAS1 gene and fully complements the morphological defects of an S. cerevisiae gas1 null mutant (20). In C. albicans it has been shown by Northern analysis that the PHR1 RNA transcript is regulated by the external pH. It is not detectable at pH 4.5 or 5 and becomes detectable at pH 5.5, then steadily increasing with the increase of the external pH, reaching a level at pH 8 ninefold higher than at pH 5.5 (17). The lack of Phr1p leads to several morphogenetic defects which become more evident and severe as the external pH rises to alkaline values, consistent with the pattern of expression of the gene (17). In this work we tested the reactivity of C. albicans extracts to antibodies directed against Gas1p. In order to gain insight into the role of Phr1p, we analyzed if the morphogenetic defects of the null mutant could be related to changes in the cell wall matrix.
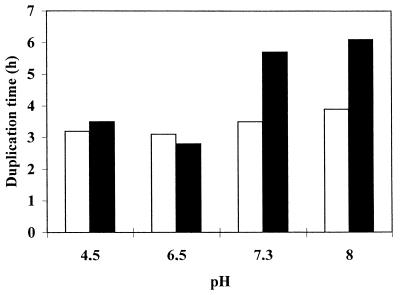
The Phr1+ parental CAF3-1 (PHR1/PHR1 Δura3::Δimm434/Δura3::Δimm434) and the Phr1− CAS8 (Δphr1/Δphr1 Δura3::Δimm434/Δura3::Δimm434) C. albicans strains were grown in YPD (1% peptone, 2% yeast extract, and 2% dextrose) medium supplemented with 50 mg of uridine per liter, 1/20 of the final volume of 1 M N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) (final concentration, 50 mM), and buffered with NaOH or HCl to different pH values before sterilization by filtration. Cells were grown at 25°C, a temperature at which C. albicans shows the typical yeast morphology. Growth, monitored as increase in cell number (21), was monitored for several hours. Duplication times (TD) calculated at the different pHs are given in Fig. 1. In YPD-HEPES medium, the TD of Phr1+ and Phr1− are rather similar at pH 4.5 and 6.5 whereas at pH 7.3 and 8 the mutant strain exhibits a TD longer than the parental one. At pH 7.3 and more so at pH 8, mutant cells lost the highly polarized shape typical of Phr1+ cells, became round and large, and often carried more than one bud with a smaller size, indicating defects in bud maturation. This is in agreement with the reported phenotype and its dependence upon the external pH (17).
FIG. 1.
Effect of different pHs on growth of C. albicans. The parental strain CAF3-1 (open bars) or the Phr1− mutant CAS8 (solid bars) were grown at 25°C in YPD containing 50 mM HEPES and buffered at the desired pH. The pH of the medium was monitored during growth and never changed by more than 0.1. Growth was monitored as increase in cell number.
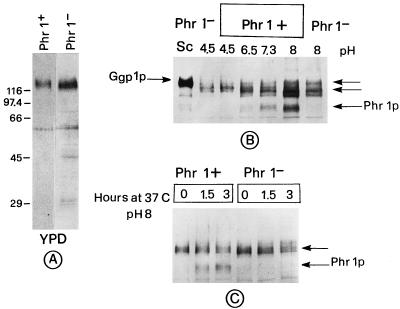
We used the technique of immunoblotting with polyclonal antibodies directed against S. cerevisiae Gas1p/Ggp1p (13) to probe the reactivity of C. albicans protein extracts, prepared as described elsewhere (19). The immunoblot analysis shows that the anti-Ggp1p/Gas1p antibodies detect a major, rather diffuse band of about 120 kDa in cells grown at 25°C in unbuffered YPD both in Phr1+ and Phr1− cells (Fig. 2A). To monitor the appearance of the PHR1 gene product, cells were grown in YPD-HEPES medium buffered at various pHs. In Phr1+ cells at pH 6.5 a band of about 88 kDa begins to be detectable. Its level steadily increases with the increase of the external pH to 7.3 and 8 (Fig. 2B). In Phr1− cells at pH 8, the 88-kDa band is not detected (Fig. 2B). Thus, since the 88-kDa polypeptide has a behavior that parallels that of the PHR1 mRNA (17) and is absent in Phr1− cells, it corresponds to the product of PHR1.
FIG. 2.
Immunoblot analysis of C. albicans extracts. About 60 μg of total protein was analyzed by immunoblotting with antibodies directed against Gas1p/Ggp1p and the ECL detection system (20). (A) Extracts from cells grown at 25°C in unbuffered YPD. (B) Extracts from cells grown at 25°C in YPD–50 mM HEPES. An extract (20 μg) of S. cerevisiae (Sc) was also loaded and deliberately overexposed. (C) Extracts from cells grown at 25°C in unbuffered YPD, filtered, and shifted at zero time to YPD pH 8 at 37°C. Arrows, immunoreactive bands detected by the anti-Ggp1p antibodies.
Moreover, anti-Ggp1p antibodies detect other bands of higher molecular weight. Beyond a band corresponding to a polypeptide of about 120 kDa present at acidic pH in both strains in unbuffered and buffered media, other diffuse bands of about 115 to 120 kDa are present at pH 6.5, 7.3, and 8 in both strains (Fig. 2B).
We monitored the appearance of the Phr1p during germ tube formation. A temperature of 37°C is optimal for germ tube emergence, and also, pH is known to influence this process (5). Cells exponentially growing at 25°C in unbuffered YPD medium were inoculated into fresh YPD medium buffered at pH 8 and incubated at 37°C. Extracts were prepared at the moment of the shift and 1.5 and 3 h later. At 1.5 and 3 h the p88 band was present in Phr1+ cells and absent in Phr1− cells (Fig. 2C). This indicates that PHR1 is expressed also during hyphal induction. In this blot the polypeptide of about 120 kDa is also present at time zero. Its persistence after the shift to alkaline pH and 37°C may suggest that it is still synthesized or its synthesis is switched off but the level is unchanged due to the stability of the protein. During the time course of this experiment the 115- to 120-kDa polypeptides did not appear.
The rather diffuse aspect of migration of Phr1p suggests that it is a glycoprotein. Extracts were treated with endo-β-N-acetylglucosaminidase F (EndoF), which removes N-linked oligosaccharide chains, and an immunoblot analysis was performed. The 88-kDa polypeptide was found to be EndoF sensitive. The 120- and the 115-kDa polypeptides were also EndoF sensitive, indicating that they are N-glycosylated proteins (data not shown). This could explain their diffuse migration in sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
To characterize changes occurring in the cell wall of the phr1 null mutant, we determined the levels of glucans and chitin in cells exponentially growing in YPD medium buffered at pH 4.5 or 8. As shown in Table 1, at pH 4.5 the levels of alkali-soluble and insoluble glucan and the corresponding ratio are about the same in the parental strain and in the mutant. At pH 8 in the parental strain the level of alkali-insoluble glucan increases and the alkali-soluble/-insoluble glucan ratio undergoes a 50% decrease, suggesting formation of more cross-links among the polymers. In contrast, in the mutant at pH 8 the alkali-insoluble glucan does not show an increase while the alkali-soluble/-insoluble ratio is almost unchanged compared to pH 4.5. Moreover, at pH 8 the level of 1,6-β-d-glucan is 25% less in the mutant than in the parental strain. Thus, in the absence of Phr1p the increase of polymer connections brought about by the pH up-shift does not occur. Chitin level was also estimated in the fraction containing the undigestible material obtained after zymolyase treatment (15). The amount of chitin in the mutant is about sixfold higher than in the control strain at pH 8 (Table 1). The mutant strain also appeared brighter than the control after calcofluor staining, suggesting an increase and delocalization of chitin (data not shown), as previously observed for the ggp1 null mutant (14).
TABLE 1.
Glucan and chitin levels in phr1Δ cellsa
| Strain | Amt (μg/mgb)
|
Alks/Alki | Chitind (μg/mgb) | ||
|---|---|---|---|---|---|
| Alkic
|
Alksc, 1,3- + 1,6β-Glucan | ||||
| 1,3- + 1,6β-Glucan | 1,6β-Glucan | ||||
| pH 4.5 | |||||
| CAF3-1 | 183 | ND | 165 | 0.9 | ND |
| CAS8 | 176 | ND | 140 | 0.8 | ND |
| pH 8 | |||||
| CAF3-1 | 225 | 65 | 112 | 0.5 | 7.5 |
| CAS8 | 169 | 49 | 163 | 1 | 41.5 |
CAF3-1 is Phr1+; CAS8 is Phr1−. Alki and Alks, alkali insoluble and alkali soluble, respectively. ND, not determined.
Dry weight of cells.
Carbohydrate content (measured by the sulfuric acid method).
Glucosamine content in the alkali-insoluble zymolyase-undigestible fractions.
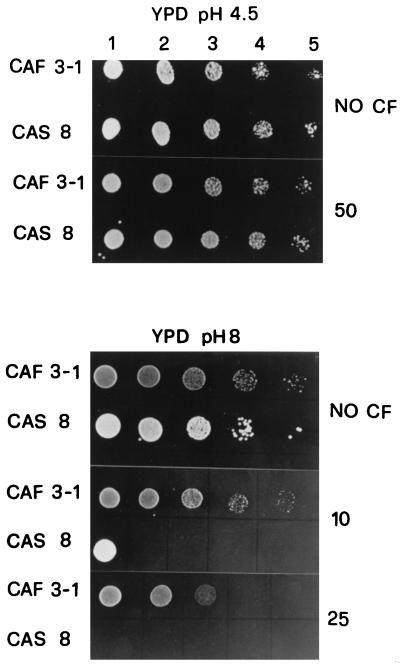
The phenotype of the Phr1− mutant was further investigated. Since the ggp1Δ mutant of S. cerevisiae is sensitive to Calcofluor White (CF) (Sigma, St. Louis, Mo.), a fluorescent stain that binds to nascent chitin molecules and prevents their assembly into fibrils (2), we tested if the phr1Δ mutant had similar behavior with respect to this compound. At pH 4.5 and 25°C no differences in the sensitivities to CF of the two strains were observed (Fig. 3). Instead, at pH 8, Phr1− cells exhibited a marked increase of sensitivity to CF with respect to control cells, being unable to grow in the presence of 25 μg/ml. In this experiment it is also possible to observe that the Phr1+ strain is more sensitive to CF at pH 8 than at 4.5, suggesting that the synthesis of chitin increases at the alkaline pH value.
FIG. 3.
Sensitivity to Calcofluor White of C. albicans Phr1+ and Phr1− cells. At a cell density of 5 × 106/ml, 5 μl of a concentrated suspension (lanes 1) of cells grown in YPD at pH 4.5 or 8 and of 10× (lanes 2), 100× (lanes 3), 1,000× (lanes 4), and 10,000× (lanes 5) dilution was spotted on a solid plate of YPD at the same pH. Amounts of CF present in the plate are indicated on the right in micrograms per milliliter.
It is known that cells defective in chitin synthesis tend to be resistant to CF (16). A marked increase of cellular chitin could cause an increase of binding of CF molecules that could significantly interfere with cell wall assembly. Since CF addition to C. albicans growing cells does not increase chitin synthesis (2), we propose that the hypersensitivity of the phr1 mutant to CF is directly related to the increase of chitin brought about by the PHR1 gene inactivation.
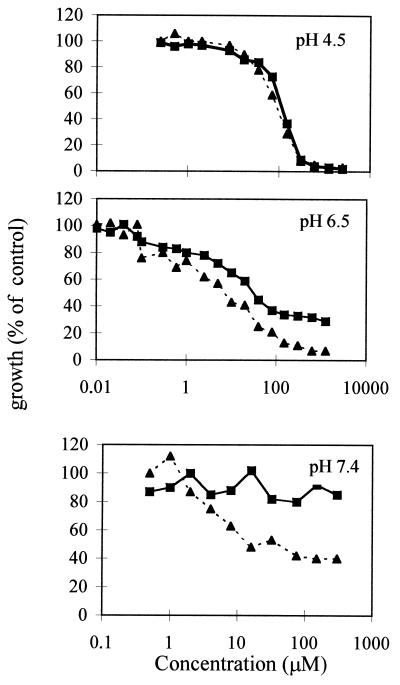
In order to assess if Phr1− cells could be more sensitive to the action of nikkomycin Z, a competitive inhibitor of chitin synthases both in vivo and in vitro (1, 3), a liquid broth microdilution assay was performed as previously described except that the microtiter plates were incubated at 25°C for 48 h (15). Nikkomycin Z is a peptide-nucleoside drug whose uptake is mediated by a peptide transport system in C. albicans (9); thus, YPD cannot be used due to peptides present in this medium which can compete with the drug (22). The assay of susceptibility to nikkomycin Z was performed in YNB-glucose medium (6.7-g/liter Difco yeast nitrogen base without amino acids and 2% glucose, supplemented with 50 mg of uridine per liter) buffered at the desired pH with 50 mM HEPES. As expected no differences were detected during growth at pH 4.5, whereas at pH 6.5 Phr1− cells were more sensitive to nikkomycin Z (Calbiochem, La Jolla, Calif.) than Phr1+ cells (Fig. 4, upper and middle panels). Since salts in YNB precipitate at pH >7, in order to test the effects at higher pH, which lead to a more severe phenotype, we used another synthetic medium, RPMI 1640 (Sigma), supplemented with 1% glucose, 50 mg of uridine per liter, and 35 g of 3-(N-morpholino)propanesulfonic acid (MOPS) per liter, and buffered with NaOH at pH 7.4. As shown in Fig. 4 (lower panel), Phr1− cells are much more sensitive to nikkomycin Z than control cells.
FIG. 4.
Sensitivity of Phr1− cells to nikkomycin Z. Phr1+ (strain CAF3-1) (▪) and Phr1− (strain CAS8) (▴) cells exponentially growing at pH 4.5 or 6.5 in YNB or at pH 7.4 in RPMI 1640 at 25°C were tested for sensitivity to nikkomycin Z by microdilution assay.
Our results indicate that anti-Ggp1p antibodies recognize Phr1p as a glycosylated polypeptide of about 88 kDa that, as expected, is absent in the phr1 null mutant and is detectable at progressively higher levels from pH 6.5 to pH 8, in agreement with the expression pattern of the PHR1 gene. Phr1p appears to be present during both yeast growth at 25°C and germ tube induction at 37°C.
Other polypeptides are detected by the anti-Ggp1p antibodies and could be the products of other homologs of the PHR1 gene. One of these, PHR2, has been isolated recently and encodes a protein 54% identical to Phr1p. The pattern of expression of the PHR2 gene is the inverse of the pattern described for PHR1, being maximal at an acidic pH (11, 12). However in cultures grown at different pH values at 25°C the situation of the high-molecular-weight polypeptides recognized by the anti-Ggp1p antiserum is rather complex. Beyond the 120-kDa polypeptide, other polypeptides of 115 to 120 kDa are detected at pH 6.5 and higher. The heterogeneous migration of these polypeptides could also arise from different degrees of N glycosylation of a single polypeptide chain. Further experiments will be necessary to clear up the nature of these bands. The availability of phr2Δ or phr1Δ phr2Δ mutants and the search through a molecular approach for other homologs will allow the correct assignment of these polypeptides. Gene redundancy is very well documented in S. cerevisiae and could also be common in C. albicans, in particular for cell wall-related genes, in order to allow a better adaptation of this organism to various environmental conditions. In the S. cerevisiae genome four homologs of GAS1 are present, but it is not yet known if they are expressed.
The apparent molecular mass of about 88 kDa of Phr1p as detected in C. albicans is higher than that reported for the protein expressed in S. cerevisiae (about 75 to 80 kDa) (20). This could be ascribed to slight differences in protein glycosylation between the two microorganisms.
Our results also demonstrate that the morphogenetic effects reported for Phr1− cells could be related to modification in cell wall organization. In fact, the total glucan level (alkali-soluble plus -insoluble fractions) does not change, whereas glucan solubility in alkali is highly affected. In particular, at pH 8, the alkali-soluble/-insoluble glucan ratio decreases from 0.9 to 0.5; conversely, it is almost unaffected in the mutant cells. Thus, in C. albicans Phr1p could catalyze the formation of cross-links between the glucans (for example, the linkage between nascent 1,3-β-d-glucan chains or the linkage between them and 1,6-β-d-glucan) rather than being involved in their biosynthesis as we previously postulated for Gas1p of S. cerevisiae (15). This is also in agreement with the lack of any differences between Phr1+ and Phr1− cells in sensitivity to L-733,560 (8), an inhibitor of 1,3-β-d-glucan synthase, both at pH 4.5 and at pH 6.5 (data not shown). Further biochemical studies will be necessary to shed some light on the enzymatic activity of this protein. It is worthwhile noting that the phr1 null mutant is less virulent than its isogenic parental strain (4), suggesting that subtle changes in the cell wall structure can play an important role in the virulence mechanism of this fungal pathogen.
Finally, our data indicate that in C. albicans, as already shown in gas1Δ mutants of S. cerevisiae, the increase of chitin could be activated in order to counteract a weakening of the cell wall. This response is uncoupled from an increase in the alkali-insoluble glucan, as would be expected on the basis of the reported data on the cross-links between chitin, glucans, and mannoproteins (6, 7). One possible explanation is that Phr1p itself participates in the formation of the cross-links responsible for the conversion of the alkali-soluble glucan to the insoluble one. Alternatively, it can be envisaged that the increase in chitin synthesis unbalances a mechanism that coordinates the synthesis of the cell wall polymers and their interconnections.
The induction of chitin synthesis could be a more general mechanism of yeast and fungi to counter a specific stress of the cell wall. In fact, recently the increase in sensitivity to nikkomycin Z was also observed in C. albicans bgl2 mutants (18), indicating that loss of function of some genes involved in the cell wall assembly renders cells more dependent on chitin as a structural element essential for cell integrity. Three chitin synthase genes, CHS1, CHS2, and CHS3, have been characterized in C. albicans (10), and this will allow further investigation of their involvement in the compensatory response.
Acknowledgments
We are very indebted to W. A. Fonzi for the CAF3-1 and CAS8 strains and for the helpful exchange of information. We thank Myra Kurtz for providing the inhibitor of 1,3-β-d-glucan synthase, L-733,560. We are indebted to Alfonso-Javier Carillo-Munoz for advice on nikkomycin Z assay. We are grateful to Antonio Grippo for preparing the figures.
This work was partially supported by grants from Ministero dell’Università e della Ricerca Scientifica e Tecnologica (art.65 DPR 382/80) to Lilia Alberghina.
REFERENCES
- 1.Choi W J, Santos B, Duran A, Cabib E. Are yeast chitin synthases regulated at the transcriptional or post-translational level? Mol Cell Biol. 1994;14:7685–7694. doi: 10.1128/mcb.14.12.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elorza M V, Rico H, Sentandreu R. Calcofluor White alters the assembly of chitin fibrils in Saccharomyces cerevisiae and Candida albicans cells. J Gen Microbiol. 1994;129:1577–1582. doi: 10.1099/00221287-129-5-1577. [DOI] [PubMed] [Google Scholar]
- 3.Gaughran J P, Lai M H, Kirsch D R, Silverman S J. Nikkomycin Z is a specific inhibitor of Saccharomyces cerevisiae chitin synthase isozyme Chs3 in vitro and in vivo. J Bacteriol. 1994;176:5857–5860. doi: 10.1128/jb.176.18.5857-5860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghannoum M A, Spellberg B, Saporito-Irwin S M, Fonzi W A. Reduced virulence of Candida albicans PHR1 mutants. Infect Immun. 1995;63:4528–4530. doi: 10.1128/iai.63.11.4528-4530.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gow N A R. Growth and guidance of the fungal hypha. Microbiology. 1994;140:3193–3205. doi: 10.1099/13500872-140-12-3193. [DOI] [PubMed] [Google Scholar]
- 6.Kollar R, Reinhold B B, Petrakova E, Yeh H J C, Ashwell G, Drgonova J, Kaptayn J C, Klis F M, Cabib E. Architecture of the yeast cell wall. J Biol Chem. 1997;272:17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- 7.Kollar R, Petrakova E, Ashwell G, Robbins P W, Cabib E. Architecture of the cell wall. J Biol Chem. 1995;270:1170–1178. doi: 10.1074/jbc.270.3.1170. [DOI] [PubMed] [Google Scholar]
- 8.Kurtz M B, Douglas C, Marrinan J, Nollstadt K, Onishi J, Dreikorn S, Milligan J, Mandala S, Thompson J, Balkovec J M, Bouffard F A, Dropinski J F, Hammond M L, Zambias R A, Abruzzo G, Bartizal K, McManus O B, Garcia M L. Increased antifungal activity of L-733,560, a water-soluble, semisynthetic pneumocandin, is due to enhanced inhibition of cell wall synthesis. Antimicrob Agents Chemother. 1994;38:2750–2757. doi: 10.1128/aac.38.12.2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarthy P J, Troke P F, Gull K. Mechanism of action of nikkomycin and the peptide transport system of Candida albicans. J Gen Microbiol. 1985;131:775–780. doi: 10.1099/00221287-131-4-775. [DOI] [PubMed] [Google Scholar]
- 10.Mio T, Yabe T, Sudoh M, Satoh Y, Nakajima N, Arisawa M, Yamada-Okabe H. Role of three chitin synthase genes in the growth of Candida albicans. J Bacteriol. 1996;178:2416–2419. doi: 10.1128/jb.178.8.2416-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muhlschlegel F, Fonzi W A. ASM Conference of Candida and Candidiasis: Biology, Pathogenesis, and Management. Washington, D.C: American Society for Microbiology; 1996. PHR2 is a pH-regulated gene of Candida albicans with an expression pattern that is the inverse of its homolog PHR1, abstr. A46. [Google Scholar]
- 12.Muhlschlegel F, Fonzi W A. Abstracts of the 13th Congress of the International Society for Human and Animal Mycology, Salsomaggiore Terme, Italy. 1997. PHR2 encodes a functional homolog of the C. albicans PHR1 gene with pH-dependent expression counterbalancing that of PHR1, abstr. S72; p. 58. [Google Scholar]
- 13.Popolo L, Grandori R, Vai M, Lacanà E, Alberghina L. Immunochemical characterization of gp115, a yeast glycoprotein modulated by the cell cycle. Eur J Cell Biol. 1988;47:173–180. [PubMed] [Google Scholar]
- 14.Popolo L, Vai M, Gatti E, Porello S, Bonfante P, Balestrini R, Alberghina L. Physiological analysis of mutants indicates involvement of the Saccharomyces cerevisiae GPI-anchored protein gp115 in morphogenesis and cell separation. J Bacteriol. 1993;175:1879–1885. doi: 10.1128/jb.175.7.1879-1885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popolo L, Gilardelli D, Bonfante P, Vai M. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1Δ mutant of Saccharomyces cerevisiae. J Bacteriol. 1997;179:463–469. doi: 10.1128/jb.179.2.463-469.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roncero C, Valdivieso M H, Ribas J C, Duran A. Isolation and characterization of Saccharomyces cerevisiae mutants resistant to Calcofluor white. J Bacteriol. 1988;170:1950–1954. doi: 10.1128/jb.170.4.1950-1954.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saporito-Irwin S, Birse C E, Sypherd P S, Fonzi W A. PHR1, a pH regulated gene of Candida albicans, is required for morphogenesis. Mol Cell Biol. 1995;15:601–613. doi: 10.1128/mcb.15.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarthy A V, McGonigal T, Coen M, Frost D J, Meulbroek J A, Goldman R C. Phenotype in Candida albicans of a disruption of the BGL2 gene encoding a 1,3-β-glucosyltransferase. Microbiology. 1997;143:367–376. doi: 10.1099/00221287-143-2-367. [DOI] [PubMed] [Google Scholar]
- 19.Vai M, Popolo L, Alberghina L. Immunological cross-reactivity of fungal and yeast plasma membrane H+-ATPase. FEBS Lett. 1986;206:135–141. doi: 10.1016/0014-5793(86)81355-2. [DOI] [PubMed] [Google Scholar]
- 20.Vai M, Orlandi I, Cavadini P, Alberghina L, Popolo L. Candida albicans homologue of GGP1/GAS1 is functional in Saccharomyces cerevisiae and contains the determinants for glycosylphosphatidyl-inositol attachment. Yeast. 1996;12:361–368. doi: 10.1002/(SICI)1097-0061(19960330)12:4%3C361::AID-YEA920%3E3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 21.Vanoni M, Vai M, Popolo L, Alberghina L. Structural heterogeneity in populations of the budding yeast Saccharomyces cerevisiae. J Bacteriol. 1983;156:1282–1291. doi: 10.1128/jb.156.3.1282-1291.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yadan J C, Gonneau M, Sarthou P, Le Goffic F. Sensitivity to Nikkomycin Z in Candida albicans: role of peptide permeases. J Bacteriol. 1984;160:884–888. doi: 10.1128/jb.160.3.884-888.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]