Abstract
Low vitamin D (vitD) levels have been consistently reported in schizophrenia (SCZ) suggesting a role in the etiopathology. However, little is known about the role of underlying shared genetic mechanisms. We applied a conditional/conjunctional false discovery rate approach (FDR) on large, nonoverlapping genome-wide association studies for SCZ (N cases = 53 386, N controls = 77 258) and vitD serum concentration (N = 417 580) to evaluate shared common genetic variants. The identified genomic loci were characterized using functional analyses and biological repositories. We observed cross-trait SNP enrichment in SCZ conditioned on vitD and vice versa, demonstrating shared genetic architecture. Applying the conjunctional FDR approach, we identified 72 loci jointly associated with SCZ and vitD at conjunctional FDR < 0.05. Among the 72 shared loci, 40 loci have not previously been reported for vitD, and 9 were novel for SCZ. Further, 64% had discordant effects on SCZ-risk and vitD levels. A mixture of shared variants with concordant and discordant effects with a predominance of discordant effects was in line with weak negative genetic correlation (rg = −0.085). Our results displayed shared genetic architecture between SCZ and vitD with mixed effect directions, suggesting overlapping biological pathways. Shared genetic variants with complex overlapping mechanisms may contribute to the coexistence of SCZ and vitD deficiency and influence the clinical picture.
Keywords: GWAS, genetic overlap, conditional, conjunctional false discovery rate, polygenic architecture
Introduction
Schizophrenia (SCZ) is a severe mental disorder characterized by psychotic symptoms with onset in early adulthood and large global disease burden.1 SCZ has a high heritability. However, the underlying disease mechanisms are not yet fully understood.1 A role of vitamin D (vitD) in SCZ pathophysiology has been supported by 2 decades of research.2 The primary hypothesis, proposing low vitD level as a risk factor for SCZ, was based on epidemiological evidence linking SCZ with winter births (low maternal vitD), urban upbringing, and migration to high latitudes causing reduced sun exposure.2–4 Moreover, neonatal vitD deficiency was associated with higher risk for SCZ5,6 and low vitD level has been linked to poor prognostic markers in individuals with a psychotic disorder, including more severe negative symptoms7,8 and poorer cognitive functioning.9
The potential role of vitD in the pathogenesis of SCZ is further supported by its involvement in brain development and function.2,10 The main source (~90%) of vitD is production of its D3 form (cholecalciferol) from 7-dehydrocholesterol in the skin by exposure to ultraviolet light.11,12 Additionally, vitD can be derived from some dietary sources such as fish liver oil (eg, cod), fatty fish (eg, mackerel), egg yolks, beef (eg, kidney), pork (eg, liver), mushrooms (eg, shiitake) as well as fortified orange juice and dairy products.13 VitD binds to its cytoplasmatic receptor (VDR) which subsequently forms a heterodimer with the retinoic acid receptor and, after translocation to the nucleus, regulates the expression of target genes.2 Regarding CNS-specific effects, vitD modulates prefrontal cortex development via non-genomic mechanisms, namely through modulation of L-type voltage-gated calcium channels.14,15 Animal models have indicated that vitD may also exert anti-proliferative and pro-apoptotic effects in the developing brain2 which is associated with altered expression of cyclin-dependent kinases and cyclin D1.16 Developmental vitD deficiency results in abnormal architecture and/or size of rodent brain structures including the hippocampus and lateral ventricles.2 It has been reported that developmental vitD deficiency downregulates some neurotrophic factors (eg, nerve growth factor and glial-derived neurotropic factor) which play a critical role in neurite outgrowth and dendritic arborization.2,17 Moreover, vitD modulates the inflammatory response and the glucocorticoid system, as evidenced by elevated maternal inflammatory cytokines and glucocorticoid levels in rodent models of vitD deficiency.2,18 On a behavioral level, developmental vitD deficiency in rats is associated with impaired mother-offspring interaction, disturbed ultrasonic vocalization, stereotyped behavior, sensitivity to psychomimetics, and impaired learning and memory.2
Several lines of evidence implicate vitD in a plethora of developmental processes linked with the pathogenesis of SCZ.2 Genetic variants may impact these neurobiological processes directly, and indirectly via systemic vitD levels (eg, through the gut vitD absorption feedback loop or the tan response), sensitivity to environmental factors (eg, a low sun exposure), as well as the resistance of the developing brain to vitD deficiency. Thus, it is important to scrutinize potential shared genetic architecture. SCZ is highly heritable, with broad-sense heritability estimated to be 0.64–0.81.19,20 Most of the genetic risk is attributed to thousands of common genetic variants with small effect sizes.21 While the most recent GWAS of SCZ identified 270 loci, the majority of genetic risk variants have not yet been discovered.22 The genetic loci associated with SCZ implicate processes related to brain development, signal transduction, and inflammatory response in the etiology of SCZ.20 The heritability of vitD levels has been estimated in twin and familial studies to be within a broad range of 0%–85%.23 A recent GWAS of vitD identified 143 loci associated with vitD concentration implicating among others genes associated with lipid metabolism, and estimated the SNP heritability to be 0.32.24 The potential genetic link between vitD and SCZ has been investigated using genetic correlation, which was estimated to be −0.086.24 However, genetic correlation fails to quantify mixtures of effect directions across variants since it is unable to differentiate scenarios of no genetic overlap from overlap with a balance of shared variants with concordant (a given variant has same direction of effect in both traits) and discordant effects (a given variant increases the risk of 1 trait while decreasing the risk of the second).25 This limitation is overcome by the conjunctional false discovery rate (conjFDR) approach which build on an empirical Bayesian statistical framework.26–28
In the current study, we applied the conjFDR method to large nonoverlapping GWASs of SCZ22 and vitD levels24 to investigate their shared polygenic architecture. An improved understanding of the overlapping genetic architecture and jointly associated loci between SCZ and vitD regulation may implicate pathophysiological processes of SCZ and may form the basis of preventive measures and identify new targets for drug development.
Methods and Materials
GWAS Samples
GWAS summary statistics for SCZ were obtained from the Psychiatric Genomics Consortium (53 386 cases and 77 258 controls),22 and for vitD serum concentration (417 580 individuals) from https://cnsgenomics.com/content/data. See Supplementary Materials for more details. GWASs investigated in the study were approved by the ethics committees, and informed consent was obtained from all participants.
Statistical Analyses
To estimate the polygenicity (number of casual variants) and discoverability (expected squared effect size of a given causal variant) for SCZ and vitD we applied univariate MiXeR.29 Univariate MiXeR is based on an analysis of the linkage disequilibrium (LD) structure from a detailed reference panel to estimate polygenicity and discoverability for a given trait from GWAS summary statistics25,29 and has been successfully applied for human phenotypes, including psychiatric disease and brain architecture.30,31 Consistent with previous studies, polygenicity is expressed as the number of causal variants with strongest effects required to explain 90% SNP heritability.
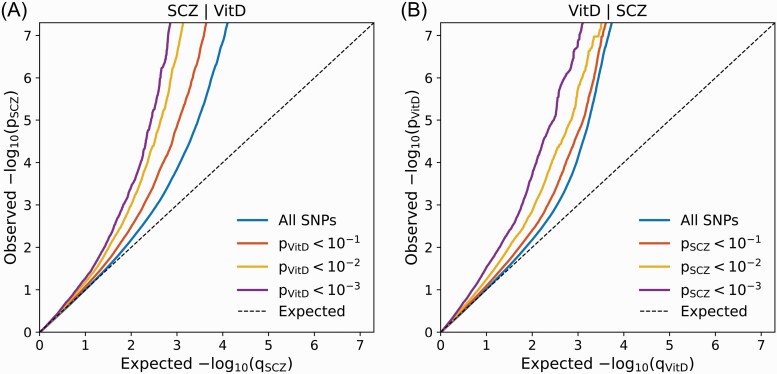
We visualized cross-trait SNP enrichment by generating conditional QQ plots which display the distribution of P-values for the primary trait conditioned on significance levels in a secondary trait (ie, SCZ trait conditioned on significance levels in vitD serum concentration phenotype and vice versa). The QQ plots were constructed after random pruning averaged over 500 iterations to control for spurious enrichment. In every iteration, 1 SNP was randomly selected for each LD block (r2 > 0.1) and corresponding P-values were used to construct the empirical cumulative distribution functions. To avoid potential bias associated with complex LD structure, we excluded SNPs within 2 regions (major histocompatibility complex region 6:25119106-33854733; 8:7200000-12500000). An empirical cumulative distribution of nominal P-values for a given primary trait was computed for all SNPs and for SNPs with significance levels below the indicated cutoffs for the secondary trait (corresponding to P < .1, P < .01, P < .001). Statistical enrichment of the given primary trait (cross-trait enrichment) is indicated by successive leftward deflection in the conditional QQ plot as levels of association with the secondary trait increase.28,32,33
To assess genetic overlap between phenotypes and to boost SNPs discovery rate, we used conditional FDR (condFDR) and conjunction FDR (conjFDR) approaches. The conjFDR method identifies loci with statistical pleiotropy.28 The method builds on an empirical Bayes mixture model,34 with 2 distributions (null and non-null effect loci) and has a higher replication rate compared to methods based on the infinitesimal model (all loci have an effect34). This approach has been applied previously to identify novel genetic loci for a series of psychiatric and somatic phenotypes, as well as to detect shared genetic loci between diverse phenotypes.35–39 CondFDR is defined as the posterior probability that a SNP of interest is null for the primary trait given that the P-values for both traits are as small or smaller than the observed P-values. ConjFDR identifies shared genetic loci and is defined as the posterior probability that a SNP is null for either 1 trait or both traits simultaneously, given the P-value for traits are equal to or smaller that the observed P-values. A conservative estimate of the conjFDR is defined by the maximum of 2 condFDR values (trait 1 conditioned on trait 2 and vice versa).40 We used FDR significance cutoffs for condFDR and conjFDR in line with previous studies, at 0.01 and 0.05, respectively.27,31 Due to extensive LD within these regions, we excluded SNPs located within the MHC and 8p23.1 inversion before fitting the cond/conjFDR model to avoid inflation of the estimated parameters.35 Although previous studies demonstrated that conjFDR findings for human phenotypes are well replicated in independent samples,41,42 in-depth experimental analyses are required to determine the mechanisms involved.43,44
We applied the FUMA protocol to identify independent genomic loci.45 We defined independent significant SNPs as those which reached a conjFDR < 0.05 and were independent from each other at r2 < 0.60. To create a subset of lead SNPs, we selected independent significant SNPs with LD with each other at r2 < 0.10. The range of each genomic locus was limited to SNPs in LD (r2 ≥ 0.60) with any of the independent significant SNPs within the locus. Subsequently, we merged all loci that were less than 250 kb apart to define distinct genomic loci and marked SNPs with the lowest P-value as a lead SNP of the locus. We used the 1000 Genomes Project European ancestry haplotype reference panel to annotate LD information.
Functional Annotation
Novel loci were defined as loci which were not physically overlapping with findings from the original GWAS, GWAS Catalog by National Human Genome Research Institute-European Bioinformatics (NHGRI-EBI46) and previous cond/conjFDR studies.31 We applied positional mapping by physical position (10-kb window) and expression quantitative trait locus association (eQTL) as gene-mapping strategies for candidate SNPs with cond/conjFDR ≤ 0.10.45 The direction of allelic effects in SNPs identified by conjFDR were investigated by comparing the z-scores in SCZ original GWAS against the z-scores in vitD levels original GWAS. Only genes identified by both strategies, after exclusion of those located within regions of complex LD, were provided for subsequent gene-set analysis. Gene-set enrichment analysis was performed using FUMA GENE2FUNC functionality.
Results
Univariate MiXeR
Univariate MiXeR analyses established a higher heritability (SCZ = 0.36; vitD = 0.076), higher polygenicity (SCZ = 8700 SNPs accounting for 90% of the heritability; vitD = 400), and lower discoverability (SCZ = 6.48e-05; vitD = 3.34e-04) for SCZ than for vitD levels. Moreover, it was estimated that the SNPs which were genome-wide significant in the original GWASs explained 4.5% (SCZ) and 52.9% (vitD) of the trait variance associated with SNPs.
QQ Plots of SCZ SNPs Conditioned on Association with vitD and Vice Versa
The conditional QQ plot for SCZ GWAS conditioned on vitD levels (figure 1A) demonstrated enrichment across different levels of significance for vitD. The earlier leftward deflection from the dashed line (no enrichment) indicated a greater proportion of true associations for a given nominal SCZ P-value. Successive leftward shifts for decreasing nominal vitD P-value thresholds demonstrated that the proportion of non-null effects in SCZ increases as the strength of the association with vitD increases. Similarly, the conditional QQ plot displayed strong polygenic enrichment for vitD given the strength of association with SCZ (figure 1B).
Fig. 1.
Conditional QQ plots illustrating cross-phenotype polygenic enrichment between schizophrenia (SCZ) and vitamin D levels (vitD). (A) SCZ trait is conditioned on vitD (B) vitD is conditioned on SCZ.
Applying the condFDR approach (condFDR < 0.01), a total of 286 distinct genomic loci were associated with SCZ conditional on their association with vitD (Supplementary figure 1 and Supplementary table 5). Sixteen of these loci have not been identified in the original SCZ GWAS,22 the NHGRI-EBI catalog46 or the previous cond/conjFDR studies (Supplementary table 1). At condFDR < 0.01, we identified 240 loci associated with vitD levels conditional on SCZ (Supplementary figure 2, Supplementary table 6). Of these, 97 loci have not been identified earlier, neither in the original vitD GWAS,24 nor in the NHGRI-EBI catalog46 (Supplementary table 2).
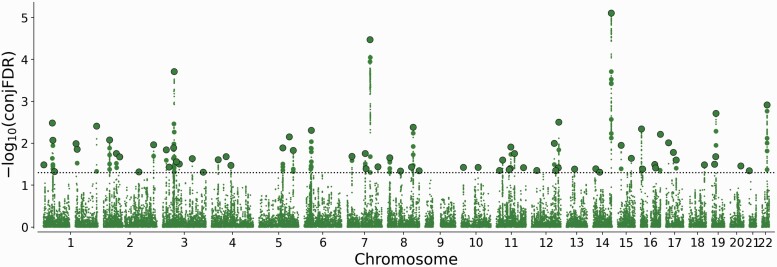
Applying the conjFDR approach, a total of 72 loci were jointly associated with SCZ and vitD at conjFDR < 0.05 (Figure 2, Supplementary table 4). Evaluation of the allelic effect direction for the 72 lead SNPs showed that 46 (64%) had discordant effect directions on SCZ-risk and vitD levels, ie, increased the risk for SCZ while reducing vitD levels or vice versa.
Fig. 2.
“ConjFDR Manhattan plot” showing common genetic variants jointly associated with schizophrenia and vitamin D levels at conjunctional false discovery rate (conjFDR) < 0.05. The y-axis shows the −log10 transformed conjFDR. Chromosomal position is presented along the x-axis. The threshold for significant shared associations (conjFDR < 0.05) is represented by the horizontal dotted line. Independent SNPs are indicated by a black perimeter.
Our conjFDR analysis displayed 9 novel loci for SCZ (table 1), not previously identified.22,46 Among the novel SCZ loci, 1 locus (lead SNP rs112936817, 3:186075556-186079990) was included in the GWAS catalog due to reported associations with chronotype47 and morningness.48 The other loci (lead SNP rs12546328, 8:59321609-59384675) were associated with narcolepsy with cataplexy.49 Moreover, we identified 40 novel loci for vitD levels (table 2), many of which were reported in the GWAS catalog for traits associated with vitD (eg, lead SNP rs6779146, 3:135798658-136507818,50–52 lead SNP rs78004870, 16:89736352-8988639253; table 3).
Table 1.
Novel Schizophrenia (SCZ) Genomic Loci also Associated with Vitamin D Levels (vitD) at Conjunctional FDR < 0.05
| Locus | Chr | Lead SNP | A1/A2 | Nearest gene | Functional category | P value SCZ | Odds Ratio SCZ | P value vitD | Beta vitD | conjFDR |
|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 2 | rs56393175* | T/G | TM4SF20 | intergenic | 3.04E-06 | 1.05 | 3.18E-05 | −1.12E-02 | 1.09E-02 |
| 20 | 3 | rs112936817* | T/G | DGKG | intronic | 1.16E-04 | 0.95 | 3.13E-05 | 1.33E-02 | 4.90E-02 |
| 23 | 4 | rs10516786 | G/A | AFF1 | intronic | 5.66E-05 | 0.96 | 2.52E-05 | −1.12E-02 | 3.37E-02 |
| 34 | 8 | rs12546328 | T/C | UBXN2B | intronic | 1.03E-04 | 1.04 | 7.97E-05 | 8.40E-03 | 4.60E-02 |
| 35 | 8 | rs4734176* | G/A | EMC2 | intronic | 6.63E-05 | 1.04 | 1.98E-04 | −8.13E-03 | 3.66E-02 |
| 39 | 10 | rs11001489* | G/A | C10orf11:RP11-367B6.2 | ncRNA_intronic | 3.41E-05 | 0.94 | 2.24E-04 | 1.20E-02 | 3.73E-02 |
| 40 | 11 | rs1950039 | T/C | SPON1 | ncRNA_intronic | 9.67E-05 | 0.97 | 6.68E-21 | 1.91E-02 | 4.47E-02 |
| 54 | 14 | rs79621605 | C/T | CDKL1 | intronic | 1.17E-04 | 0.91 | 2.13E-06 | −2.62E-02 | 4.92E-02 |
| 68 | 19 | rs72999390 | A/G | UPF1 | intronic | 3.79E-06 | 0.90 | 8.80E-05 | −1.81E-02 | 2.09E-02 |
| 70 | 20 | rs6020924* | T/G | KCNG1 | intergenic | 5.97E-05 | 0.95 | 5.90E-06 | 1.30E-02 | 3.46E-02 |
Note: The most strongly associated SNPs in independent genomic loci shared between SCZ and vitD at conjFDR < 0.05. Chromosomal position (Chr), nearest gene, and functional category, as well as P values and effect sizes (Odds ratio and Beta for SCZ and Vitamin D, respectively) from original summary statistics. The effect allele is A1. Novel for vitD levels marked with *.
Table 2.
Novel Vitamin D Levels Genomic Loci also Associated with Schizophrenia (SCZ) at Conjunctional FDR < 0.05
| Locus | Chr | Lead SNP | A1/A2 | Nearest gene | Functional category | P value Vitamin D | Beta Vitamin D | P value SCZ | Odds Ratio SCZ | conjFDR |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 1 | rs2842198 | A/G | HYI-AS1 | intergenic | 2.13E-05 | -8.68E-03 | 1.51E-07 | 0.95 | 8.40E-03 |
| 4 | 1 | rs12567744 | G/A | CDKN2C | intergenic | 3.31E-04 | −7.68E-03 | 1.03E-04 | 1.04 | 4.71E-02 |
| 7 | 1 | rs12743378 | A/G | SDCCAG8 | intronic | 6.55E-06 | 1.28E-02 | 5.10E-07 | 0.94 | 3.91E-03 |
| 10 | 2 | rs999494 | T/C | EMX1 | intronic | 8.85E-05 | −9.90E-03 | 2.61E-07 | 0.95 | 2.10E-02 |
| 11 | 2 | rs13024749 | T/G | LY75:LY75-CD302 | intronic | 3.38E-04 | −7.24E-03 | 3.97E-05 | 1.04 | 4.77E-02 |
| 12 | 2 | rs56393175* | T/G | TM4SF20 | intergenic | 3.18E-05 | −1.12E-02 | 3.04E-06 | 1.05 | 1.09E-02 |
| 13 | 3 | rs61182735 | A/G | AC091493.2 | intergenic | 4.85E-05 | −8.11E-03 | 1.16E-07 | 1.05 | 1.43E-02 |
| 14 | 3 | rs9831428 | A/C | RBMS3 | intronic | 2.02E-04 | 7.52E-03 | 6.77E-05 | 0.97 | 3.70E-02 |
| 17 | 3 | rs2241823 | A/C | ATXN7 | intronic | 1.38E-04 | 8.30E-03 | 1.26E-07 | 1.05 | 2.76E-02 |
| 18 | 3 | rs2088176 | G/A | ROBO2 | intronic | 1.66E-04 | 7.56E-03 | 1.57E-05 | 0.96 | 3.11E-02 |
| 19 | 3 | rs6779146 | C/T | MSL2 | intronic | 1.04E-04 | −9.21E-03 | 1.87E-06 | 1.05 | 2.32E-02 |
| 20 | 3 | rs112936817* | T/G | DGKG | intronic | 3.13E-05 | 1.33E-02 | 1.16E-04 | 0.95 | 4.90E-02 |
| 21 | 4 | rs80279461 | T/G | PCDH7 | intergenic | 1.15E-04 | −1.09E-02 | 1.55E-05 | 0.95 | 2.47E-02 |
| 25 | 5 | rs904612 | A/G | CTB-35F21.1 | ncRNA_intronic | 2.54E-06 | −1.05E-02 | 4.05E-06 | 1.05 | 7.06E-03 |
| 27 | 6 | rs9273493 | T/C | HLA-DQB1:HLA-DQB1-AS1:XXbac-BPG254F23.6 | ncRNA_exonic | 8.21-06 | −9.10E-03 | 2.35E-06 | 1.05 | 4.94E-03 |
| 30 | 7 | rs274632 | A/C | GRM3 | intergenic | 2.24E-04 | −7.43E-03 | 7.98E-05 | 1.04 | 4.03E-02 |
| 32 | 7 | rs568471 | T/C | RNU4-74P | upstream | 1.82E-04 | 1.15E-02 | 6.57E-05 | 0.95 | 3.64E-02 |
| 35 | 8 | rs4734176* | G/A | EMC2 | intronic | 1.98E-04 | −8.13E-03 | 6.63E-05 | 1.04 | 3.66E-02 |
| 37 | 8 | rs10875482 | C/T | TSNARE1 | intronic | 3.05E-04 | −7.40E-03 | 9.96E-12 | 0.94 | 4.49E-02 |
| 39 | 10 | rs11001489* | G/A | C10orf11:RP11-367B6.2 | ncRNA_intronic | 2.24E-04 | 1.20E-02 | 3.41E-05 | 0.94 | 3.73E-02 |
| 41 | 11 | rs2582894 | G/T | RP11-22P4.1 | intergenic | 1.18E-04 | −1.03E-02 | 1.78E-05 | 0.95 | 2.51E-02 |
| 42 | 11 | rs6591434 | G/A | RP11-734C14.2 | intergenic | 2.76E-04 | −7.78E-03 | 8.61E-08 | 1.05 | 4.24E-02 |
| 43 | 11 | rs484201 | T/C | MACROD1 | intronic | 2.53E-04 | −7.41E-03 | 4.44E-06 | 1.04 | 4.02E-02 |
| 45 | 11 | rs7948931 | C/T | DLG2 | intronic | 6.60E-05 | 8.08E-03 | 1.44E-05 | 1.04 | 1.74E-02 |
| 46 | 11 | rs61123830 | A/G | GRAMD1B | intergenic | 4.25E-05 | 8.90E-03 | 7.09E-05 | 0.96 | 3.80E-02 |
| 48 | 12 | rs10860964 | C/T | RP11-328J6.1 | intergenic | 2.82E-05 | 8.59E-03 | 6.39E-09 | 0.95 | 1.01E-02 |
| 49 | 12 | rs7313797 | C/T | KCTD10 | UTR5 | 1.55E-04 | −7.58E-03 | 9.48E-05 | 0.97 | 4.42E-02 |
| 50 | 12 | rs2464196 | A/G | HNF1A | exonic | 2.40E-04 | 8.05E-03 | 7.43E-05 | 1.04 | 3.89E-02 |
| 51 | 12 | rs79478560 | A/G | DNAH10 | intronic | 4.63E-06 | −9.53E-03 | 3.89E-07 | 1.05 | 3.11E-03 |
| 53 | 14 | rs17522122 | T/G | AKAP6 | intergenic | 2.59E-04 | −7.34E-03 | 1.08E-10 | 1.06 | 4.08E-02 |
| 56 | 15 | rs8035957 | C/T | RASGRP1 | intronic | 1.63E-05 | −9.85E-03 | 8.35E-06 | 1.04 | 1.12E-02 |
| 57 | 15 | rs12905223 | C/T | UBE2Q2P1:LINC00933 | ncRNA_exonic | 1.01E-04 | −8.68E-03 | 1.69E-09 | 0.94 | 2.28E-02 |
| 60 | 16 | rs116001925 | T/C | RP11-439I14.3 | intergenic | 1.05E-05 | 1.88E-02 | 5.17E-05 | 0.92 | 3.21E-02 |
| 61 | 16 | rs61733486 | T/C | PRMT7 | exonic | 2.40E-04 | −1.58E-02 | 2.87E-06 | 1.09 | 3.89E-02 |
| 62 | 16 | rs78004870 | T/G | FANCA | exonic | 1.40E-06 | 1.05E-02 | 3.27E-06 | 0.96 | 6.13E-03 |
| 63 | 17 | rs6502219 | C/T | ARHGAP44 | intronic | 1.44E-05 | 9.38E-03 | 6.60E-06 | 1.04 | 9.66E-03 |
| 64 | 17 | rs11263770 | A/G | MYO19 | intergenic | 5.99E-05 | −8.03E-03 | 4.83E-06 | 0.96 | 1.64E-02 |
| 65 | 17 | rs6504584 | T/G | CALCOCO2 | UTR3 | 6.16E-05 | 8.42E-03 | 3.31E-05 | 0.96 | 2.51E-02 |
| 67 | 19 | rs8103241 | G/A | NFIX | intronic | 2.36E-05 | 8.43E-03 | 5.07E-05 | 0.97 | 3.17E-02 |
| 70 | 20 | rs6020924* | T/G | KCNG1 | intergenic | 5.90E-06 | 1.30E-02 | 5.97E-05 | 0.95 | 3.46E-02 |
Note: The most strongly associated SNPs in independent genomic loci shared between SCZ and vitD at conjFDR < 0.05. Chromosomal position (Chr), nearest gene, and functional category, as well as P values and effect sizes (Odds ratio and Beta for SCZ and Vitamin D, respectively) from original summary statistics. The effect allele is A1. Novel for SCZ marked with *.
Table 3.
Novel for Vitamin D Levels Loci—the GWAS Catalog Traits Associated with Vitamin D Metabolism
| Locus | Lead SNP | Genomic position | Previously reported to GWAS catalog trait(s) |
|---|---|---|---|
| 17 | rs2241823 | 3: 63862842-64003983 | Glomerular filtration rate54 |
| 19 | rs6779146 | 3: 135798658-136507818 | Body mass index,50 Liver enzyme levels,51 Waist-hip ratio52 |
| 25 | rs904612 | 5: 139053438-139095473 | Fat-free mass55 |
| 45 | rs7948931 | 11: 82987208-83335764 | Oily fish consumption56 |
| 49 | rs7313797 | 12: 109849297-110042348 | Oily fish consumption,56 Total cholesterol levels57 |
| 50 | rs2464196 | 12: 121384495-121471337 | Gamma glutamyl transferase levels,58 Total cholesterol levels58 |
| 51 | rs79478560 | 12: 124388511-124496316 | Waist-hip index,59 Body mass index60 |
| 53 | rs17522122 | 14: 33292743-33309495 | Body mass index52 |
| 57 | rs12905223 | 15: 84672181-85280792 | Waist circumference adjusted for body mass index50 |
| 62 | rs78004870 | 16: 89736352-89886392 | Low tan response53 |
| 64 | rs11263770 | 17: 34825861-34952964 | Body mass index52 |
Note: Genomic position: Chromosome: lead SNP pair base lower threshold position—upper threshold position.
Functional Analyses
Functional annotation of all shared lead SNPs identified in conjFDR analysis displayed that the majority (81%) were intergenic or intronic, while 3 were exonic and 6 were associated with noncoding RNA (Supplementary table 1). Seven lead SNPs (rs2464196, rs2267791, rs72999390, rs61182735, rs10514040, rs13024749, rs112936817) had a CADD score above the threshold score of 12.37, indicative of deleteriousness.61 Among those 7 SNPs with CADD scores above 12.37, 5 had opposite effects on SCZ-risk and vitDlevels. Moreover, 4 SNPs among those 72 lead SNPs shared between SCZ and vitD (conjFDR) displayed low RDB score (1b and 2b), suggestive of regulatory functionality.62 Two shared loci were located within genomic regions with complex LD, namely the extended MHC region (lead SNP rs9273493, 6:32628247-32501395) and chromosomal region 8p23.1 (lead SNP rs6999466, 8:9390529-10283748). Given the effect of complex LD, these loci are evidence of the involvement of the MHC and 8p23.1 regions in the studied traits rather than of any specific gene.
Functional annotation of all 10556 SNPs in the loci shared between SCZ and vitD (conjFDR <0. 10) displayed that 84.7% of them were intronic or intergenic, while 1.3% were exonic. Having excluded regions with complex LD (MHC and 8p23.1 loci) we applied FUMA to link the 10196 candidate SNPs in the 70 shared loci to 858 genes. Positional mapping aligned the SNPs to 627 genes while cis-eQTL mapping implicated 528 genes. Two hundred and 97 genes were implicated by both gene-mapping procedures. Seven of those genes are located within newly identified loci, specifically TM4SF20 and AGFG1 at chromosome 2, C4orf36, RP11-397E7.4, and AFF1 at chromosome 4, UBXN2B and EMC2 at chromosome 8.
Gene Ontology gene-set analysis for genes indicated by both positional mapping and eQTL indicated 2 significantly associated processes, namely, “hyaluronan metabolic process” and “synapse organization.” Further, the genes were significantly associated with “platelet dense granule lumen” cellular components and with 3 molecular functions gene sets, namely, “adenyl nucleotide biding,” “ribonucleotide binding,” and “drug binding” (Supplementary table 7).
Discussion
In the present study, we found that the genetic architecture of SCZ consists of approximately 25 times more variants than vitD levels, probably reflecting a more complex genetic etiology of SCZ.31 The SNPs associated with SCZ and vitD levels are mutually enriched (QQ plots) demonstrating shared genetic architecture. We found a total of 72 loci jointly associated with SCZ and vitD level, 9 novel for SCZ, and 40 novel for vitD (conjFDR). Of the 72 loci shared between SCZ and vitD, 64% had discordant effects on SCZ and vitD levels. Further, functional analyses of the novel shared loci identified 2 genes (DGKG, EMC2), suggesting links between lipid metabolism, involved in 25OHD pathways and brain development. Together, the findings provide a better understanding of the overlapping genetic architecture and jointly associated loci between SCZ and vitD regulation beyond genetic correlation. These findings implicate pathophysiological processes of SCZ linked to vitD regulation which may form the basis of preventive measures and identify new targets for drug development.
Our finding highlights the complexity of the genetic relationship between SCZ and vitD levels, consistent with previously reported weak negative genetic correlation between the 2 traits.24 Weak negative correlation is consistent with genetic overlap if there is a mixture of concordant and discordant effect directions, since genetic correlation is a summary measure of the correlation of effect sizes. The sum of concordant and discordant effect directions therefore cancel each other out, resulting in a decrease in the absolute value of the genetic correlation estimate.63 The observed distribution of directional effects among the shared loci supports epidemiological findings that suggest co-occurrence of low vitD levels with SCZ.2,64 Moreover, in connection with previous findings, this may be consistent with subpopulations of patients with SCZ in which vitD levels might frame clinical manifestation of psychosis and CVD comorbidity.65,66 Consequently, even though a Mendelian randomization analysis did not support a causal role for vitD in SCZ, it might be beneficial for future studies to assess specific subgroups of patients with SCZ with a high SNP concordance/discordance rate within genes implicated in both traits.24
Further, our study supports the notion that genetic pleiotropy, ie, the association of a single gene or variant with more than 1 distinct phenotype plays a role in a range of human phenotypes. Extensive pleiotropy has been shown across brain-related traits and disorders, such as mental and cognitive traits, personality, and mental disorders.44 Similarly, polygenic pleiotropy between SCZ and somatic traits including CVD risk factors has also been reported.26,31 Here, we provide evidence of pleiotropy between SCZ and vitD levels, suggesting novel molecular pathways related to SCZ. Of particular interest, the novel loci shared between SCZ and vitD levels were also found at or in proximity to the genes DGKG, EMC2, and KCNG1.
DGKG encodes diacylglycerol kinase γ which is a member of the type I subfamily of diacylglycerol kinases, known to phosphorylate diacyloglycerol in the production of phosphatidic acid.67 Phosphatidic acid, a product of the reaction catalyzed by diacylglycerol kinases, plays an important role in cell proliferation and survival, the processes that have been linked to both vitD deficiency in a developing brain and SCZ.2,68 Interestingly, both diacylglycerol and vitD may directly impact the activation of Ca2+-permeable cation channels in neurons.14,69 Moreover, diacylglycerol kinase γ may influence the synthesis of phosophatidylinositol that has been linked to the pathogenesis of SCZ and other neurodevelopmental disorders, eg, autism spectrum disorders (please note that diacylglycerol kinases ε is the predominant enzyme of the phosphatidylinositol cycle).68,70,71 Diacylglycerol kinases provide precursors for the synthesis of cardiolipins, and an increased activity of anti-cardiolipin antibody accompanied by a higher activity of MMP9 has been detected in serum from patients with SCZ.72,73Dgkg has been shown to be highly expressed in GABAergic interneurons (especially somatostatin expressing) and to regulate neurite outgrowth (maturation) of GABAergic interneurons in mice.74 This is of particular interest in the context of the hypothesized role of inhibitory local circuit neurons in the pathogenesis of SCZ.75,76 Diacylglycerol kinases may impact on vitD serum level through inhibiting lipid droplet formation by removing the substrate and lipid signal that triggers this process.68 These processes may play an important role in vitD homeostasis since vitD receptors and key vitD enzymes (1α-hydroxylase and CYP 24) are accumulated within the membrane of lipid droplets.77
EMC2 encodes one of the cytosolic subunits of the endoplasmic reticulum (ER) membrane protein complex, participating in the insertion and folding of protein into the ER,78 and has been linked to SCZ79,80 (also DISC181–84). Misfolded ER proteins can induce ER stress—a cell death pathway.85 A recent study emphasized a protective role of vitD against ER stress in hippocampal and cortical neurons.86 Previous studies indicated that vitD might regulate lipid metabolism by inducing degradation of SREBP/SCAP complex located in ER.87 ER stress mediates the effect of vitD on intestinal epithelial cells in the small intestine (the main site of vitD absorption) and may play an important role in the vitD level feedback loop.88KCNG1 encodes potassium voltage-gated channel modifier subfamily G member 1. Expression of this ion channel has been associated with regions of the brain dedicated to cognitive functions, emotions, biological needs, and internally driven rhythms (eg, circadian rhythm).89
Two of the candidate SNPs among the 9 novel SCZ loci jointly associated with vitD levels, rs4643531 (2:228243905) and rs79621605 (14:50655307) were nonsynonymous exonic variants within TM4SF20 and SOS2, respectively. Nonsynonymous exonic variants are of particular interest as they potentially disrupt protein function by changing the amino acid sequence of the protein. TM4SF20 encodes a 4-transmembrane protein, and single-exon deletion of TM4SF20 has been associated with language delay and white matter hyperintensities.90SOS2 encodes a protein involved in signal transduction pathways. Two SNPs have been associated with late-onset Alzheimer’s disease.91 Moreover, variants in SOS2 have been identified as a very rare cause of Noonan syndrome, in which intellectual/learning disabilities and skin abnormalities might be expressed among others symptoms.92 A functional rare variant in SOS2 has also been associated with estimated glomerular filtration rate,93 while a study in zebrafish Sos2-knockdowns have shown changes in renal development and function93 that in consequence might modify vitD metabolisms.
The novel SCZ-risk loci positionally mapped to AFF1, UBXN2B, CDKL1, and UPF1 were also found to be shared between SCZ and vitD levels. AFF1 encodes AF4/FMR2 family, member 1 (AFF-1). AFF-1 protein has been demonstrated to play a role in dendrite reconnection following injury and to maintain regenerative potential during aging in C. elegans model.94 Prior GWASs have associated a different SNP at AFF-1 with risk of Alzheimer’s disease95 and systemic lupus erythematosus.96UBXN2B encodes UBX domain protein 2B, an adaptor protein involved in mitotic spindle orientation, Golgi, and ER biogenesis. Further, a SNP at UBXN2B has been associated with the age of onset of excessive daytime sleepiness in narcolepsy.49CDKL1 encodes cyclin-dependent kinase like 1 (CDKL1). In vivo studies in rats have shown that this protein is mainly expressed in glial cells, is upregulated in gliosis, and is probably involved in hyperphosphorylation of tau protein in Alzheimer’s diseases.97UPF1 encodes UPF1 RNA helicase and ATPase (UPF1) a part of multiprotein complex controlling mRNA nuclear export and surveillance. Which seems particularly interesting in view of a recent study assessing the role of RNA-binding protein (including UPF1) in the pathogenesis of SCZ.98
There were limitations to the present study. The conjFDR analysis, applied in our study, identifies loci with a statistical pleiotropy, and further in-depth experimental analyses are required to investigate if detected signals directly influence studied phenotypes and which causal mechanisms are involved.43,44 Another limitation of our study is that both GWASs applied in the current study has been performed using European ancestry population.
In conclusion, the current study demonstrates extensive genetic overlap between SCZ and vitD level beyond genetic correlations and identifies several novel polygenic effects. The current findings provide insights into potential mechanistic relationships underlying the epidemiological and clinical associations between these 2 phenotypes. Interestingly, the novel loci shared between SCZ and vitD levels were found in the proximity of genes (DGKG, EMC2) which are connected to pathogenic neurodevelopmental processes, and to autism spectrum disorders.99,100 Moreover, our results suggest that vitD may influence the resilience of the developing brain to adverse factors. This is a potential direction for future animal studies and research in large birth cohorts.
Supplementary Material
Acknowledgments
We thank Dr Wesley Thompson for statistical advice. This work was performed on resources provided by Tjeneste for Sensitive Data facilities. O.A.A. is a consultant for CorTechs.ai and has received speaker’s honoraria from Lundbeck and Sunovion. S.D. has received speaker’s honoria from Lundbeck. A.M.D. is a founder, holds equity, and serves on scientific-advisory board of CorTechs Labs, is a member of scientific-advisory board of Human Longevity, and receives research funding from General Electric Healthcare and Medtronic.
Contributor Information
Piotr Jaholkowski, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway.
Guy F L Hindley, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, UK.
Alexey A Shadrin, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway; KG Jebsen Centre for Neurodevelopmental Disorders, University of Oslo and Oslo University Hospital, Oslo, Norway.
Markos Tesfaye, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Department of Psychiatry, St. Paul’s Hospital Millennium Medical College, Addis Ababa, Ethiopia.
Shahram Bahrami, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway.
Mari Nerhus, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Department of Special Psychiatry, Akershus University Hospital, Lørenskog, Norway; Division of Health Services Research and Psychiatry, Institute of Clinical Medicine, Campus Ahus, University of Oslo, Oslo, Norway.
Zillur Rahman, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway.
Kevin S O’Connell, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway.
Børge Holen, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway.
Nadine Parker, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway.
Weiqiu Cheng, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway.
Aihua Lin, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway.
Linn Rødevand, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway.
Naz Karadag, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway.
Oleksandr Frei, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Center for Bioinformatics, Department of Informatics, University of Oslo, Oslo, Norway.
Srdjan Djurovic, Department of Medical Genetics, Oslo University Hospital, Oslo, Norway; NORMENT Centre, Department of Clinical Science, University of Bergen, Bergen, Norway.
Anders M Dale, Department of Radiology, University of California, San Diego, La Jolla, CA; Multimodal Imaging Laboratory, University of California San Diego, La Jolla, CA; Department of Psychiatry, University of California, San Diego, La Jolla, CA; Department of Neurosciences, University of California San Diego, La Jolla, CA.
Olav B Smeland, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway.
Ole A Andreassen, NORMENT, Centre for Mental Disorders Research, Division of Mental Health and Addiction, Oslo University Hospital, and Institute of Clinical Medicine, University of Oslo, Oslo, Norway; KG Jebsen Centre for Neurodevelopmental Disorders, University of Oslo and Oslo University Hospital, Oslo, Norway.
Funding
This work was supported by the Research Council of Norway ( 223273, 273291, 273446, 296030, 300309, 326813), The European Economic Area and Norway Grants (EEA-RO-NO-2018-0535, EEA-RO-NO-2018-0573), EU’s Horizon 2020 Research and Innovation (847776, 801133), and US NIMH (1R01 MH124839).
References
- 1. Jauhar S, Johnstone M, McKenna PJ.. Schizophrenia. Lancet. 2022;399:473–486. doi: 10.1016/S0140-6736(21)01730-X. [DOI] [PubMed] [Google Scholar]
- 2. Cui X, McGrath JJ, Burne THJ, Eyles DW.. Vitamin D and schizophrenia: 20 years on. Mol Psychiatry. 2021;26:2708–2720. doi: 10.1038/s41380-021-01025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McGrath JH. Hypothesis: is low prenatal vitamin D a risk-modifying factor for schizophrenia? Schizophr Res. 1999;40:173–177. doi: 10.1016/S0920-9964(99)00052-3. [DOI] [PubMed] [Google Scholar]
- 4. McGrath JJ, Burne TH, Féron F, MacKay-Sim A, Eyles DW.. Developmental vitamin D deficiency and risk of schizophrenia: a 10-year update. Schizophr Bull. 2010;36(6):1073–1078. doi: 10.1093/schbul/sbq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eyles DW, Trzaskowski M, Vinkhuyzen AAE, et al. The association between neonatal vitamin D status and risk of schizophrenia. Sci Rep. 2018;8(1). doi: 10.1038/s41598-018-35418-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McGrath JJ, Eyles DW, Pedersen CB, et al. Neonatal vitamin D status and risk of schizophrenia: a population-based case-control study. Arch Gen Psychiatry. 2010;67:889. doi: 10.1001/archgenpsychiatry.2010.110. [DOI] [PubMed] [Google Scholar]
- 7. Graham KA, Keefe RS, Lieberman JA, Calikoglu AS, Lansing KM, Perkins DO.. Relationship of low vitamin D status with positive, negative and cognitive symptom domains in people with first-episode schizophrenia. Early Interv Psychiatry. 2015;9:397–405. doi: 10.1111/eip.12122. [DOI] [PubMed] [Google Scholar]
- 8. Nerhus M, Berg AO, Kvitland LR, et al. Low vitamin D is associated with negative and depressive symptoms in psychotic disorders. Schizophr Res. 2016;178:44–49. doi: 10.1016/j.schres.2016.08.024 [DOI] [PubMed] [Google Scholar]
- 9. Nerhus M, Berg AO, Simonsen C, et al. Vitamin D deficiency associated with cognitive functioning in psychotic disorders. J Clin Psychiatry. 2017;78:e750–e757. doi: 10.4088/JCP.16m10880. [DOI] [PubMed] [Google Scholar]
- 10. Schwarzenberg SJ, Georgieff MK, NUTRITION CON, et al. Advocacy for improving nutrition in the first 1000 days to support childhood development and adult health. Pediatrics. 2018;141:e20173716. doi: 10.1542/peds.2017-3716. [DOI] [PubMed] [Google Scholar]
- 11. Jones G, Strugnell SA, DeLuca HF.. Current understanding of the molecular actions of vitamin D. Physiol Rev. 1998;78(4):1193–1231. doi: 10.1152/physrev.1998.78.4.1193. [DOI] [PubMed] [Google Scholar]
- 12. Makida K, Nishida Y, Morita D, et al. Low energy irradiation of narrow-range UV-LED prevents osteosarcopenia associated with vitamin D deficiency in senescence-accelerated mouse prone 6. Sci Rep. 2020;10(1). doi: 10.1038/s41598-020-68641-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Charoenngam N, Shirvani A, Holick MF.. Vitamin D for skeletal and non-skeletal health: what we should know. J Clin Orthop Trauma. 2019;10(6):1082–1093. doi: 10.1016/j.jcot.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gooch H, Cui X, Anggono V, et al. 1,25-Dihydroxyvitamin D modulates L-type voltage-gated calcium channels in a subset of neurons in the developing mouse prefrontal cortex. Transl Psychiatry. 2019;9(1). doi: 10.1038/s41398-019-0626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nanou E, Catterall WA.. Calcium channels, synaptic plasticity, and neuropsychiatric disease. Neuron. 2018;98:466–481. doi: 10.1016/j.neuron.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 16. Ko P, Burkert R, McGrath J, Eyles D.. Maternal vitamin D3 deprivation and the regulation of apoptosis and cell cycle during rat brain development. Dev Brain Res. 2004;153:61–68. doi: 10.1016/j.devbrainres.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 17. Eyles D, Brown J, Mackay-Sim A, McGrath J, Feron F.. Vitamin d3 and brain development. Neuroscience. 2003;118:641–653. doi: 10.1016/S0306-4522(03)00040-X [DOI] [PubMed] [Google Scholar]
- 18. Ali A, Cui X, Alexander S, Eyles D.. The placental immune response is dysregulated developmentally vitamin D deficient rats: relevance to autism. J Steroid Biochem Mol Biol. 2018;180:73–80. doi: 10.1016/j.jsbmb.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 19. Charlson FJ, Ferrari AJ, Santomauro DF, et al. Global epidemiology and burden of schizophrenia: findings from the global burden of disease study 2016. Schizophr Bull. 2018;44(6):1195–1203. doi: 10.1093/schbul/sby058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smeland OB, Frei O, Dale AM, Andreassen OA.. The polygenic architecture of schizophrenia — rethinking pathogenesis and nosology. Nat Rev Neurol. 2020;16:366–379. doi: 10.1038/s41582-020-0364-0. [DOI] [PubMed] [Google Scholar]
- 21. Zeng J, Xue A, Jiang L, et al. Widespread signatures of natural selection across human complex traits and functional genomic categories. Nat Commun. 2021;12(1). doi: 10.1038/s41467-021-21446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Trubetskoy V, Pardiñas AF, Qi T, et al. ; Indonesia Schizophrenia Consortium. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–508. doi: 10.1038/s41586-022-04434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang X, Kiel DP, Kraft P.. The genetics of vitamin D. Bone. 2019;126:59–77. doi: 10.1016/j.bone.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 24. Revez JA, Lin T, Qiao Z, et al. Genome-wide association study identifies 143 loci associated with 25 hydroxyvitamin D concentration. Nat Commun. 2020;11(1):1647. doi: 10.1038/s41467-020-15421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frei O, Holland D, Smeland OB, et al. Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat Commun. 2019;10(1):2417. doi: 10.1038/s41467-019-10310-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andreassen OA, Thompson WK, Schork AJ, et al. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 2013;9(4). doi: 10.1371/journal.pgen.1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Andreassen OA, Desikan RS, Wang Y, et al. Abundant genetic overlap between blood lipids and immune-mediated diseases indicates shared molecular genetic mechanisms. PLoS One. 2015;10:e0123057. doi: 10.1371/journal.pone.0123057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smeland OB, Frei O, Shadrin A, et al. Discovery of shared genomic loci using the conditional false discovery rate approach. Hum Genet. 2020;139(1):85–94. doi: 10.1007/s00439-019-02060-2. [DOI] [PubMed] [Google Scholar]
- 29. Holland D, Frei O, Desikan R, et al. Beyond SNP heritability: polygenicity and discoverability of phenotypes estimated with a univariate Gaussian mixture model. PLoS Genet. 2020;16:e1008612. doi: 10.1371/journal.pgen.1008612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O’Connell KS, Frei O, Bahrami S, et al. Characterizing the genetic overlap between psychiatric disorders and sleep-related phenotypes. Biol Psychiatry. 2021;90:621–631. doi: 10.1016/j.biopsych.2021.07.007. [DOI] [PubMed] [Google Scholar]
- 31. Cheng W, Frei O, Van Der Meer D, et al. Genetic association between schizophrenia and cortical brain surface area and thickness. JAMA Psychiatry. 2021;78(9):1020. doi: 10.1001/jamapsychiatry.2021.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Andreassen OA, Thompson WK, Dale AM.. Boosting the power of schizophrenia genetics by leveraging new statistical tools. Schizophr Bull. 2014;40(1):13–17. doi: 10.1093/schbul/sbt168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Andreassen OA, Djurovic S, Thompson WK, et al. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet. 2013;92(2):197–209. doi: 10.1016/j.ajhg.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thompson WK, Wang Y, Schork AJ, et al. An empirical Bayes mixture model for effect size distributions in genome-wide association studies. PLoS Genet. 2015;11:e1005717. doi: 10.1371/journal.pgen.1005717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smeland OB, Shadrin A, Bahrami S, et al. Genome-wide association analysis of Parkinson’s disease and schizophrenia reveals shared genetic architecture and identifies novel risk loci. Biol Psychiatry. 2021;89:227–235. doi: 10.1016/j.biopsych.2020.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smeland OB, Bahrami S, Frei O, et al. Genome-wide analysis reveals extensive genetic overlap between schizophrenia, bipolar disorder, and intelligence. Mol Psychiatry. 2020;25(4):844–853. doi: 10.1038/s41380-018-0332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andreassen OA, Zuber V, Thompson WK, et al. Shared common variants in prostate cancer and blood lipids. Int J Epidemiol. 2014;43(4):1205–1214. doi: 10.1093/ije/dyu090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Broce IJ, Tan CH, Fan CC, et al. Dissecting the genetic relationship between cardiovascular risk factors and Alzheimer’s disease. Acta Neuropathol. 2019;137(2):209–226. doi: 10.1007/s00401-018-1928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rødevand L, Bahrami S, Frei O, et al. Extensive bidirectional genetic overlap between bipolar disorder and cardiovascular disease phenotypes. Transl Psychiatry. 2021;11(1):407. doi: 10.1038/s41398-021-01527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nichols T, Brett M, Andersson J, Wager T, Poline JB.. Valid conjunction inference with the minimum statistic. Neuroimage. 2005;25:653–660. doi: 10.1016/j.neuroimage.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 41. Karadag N, Shadrin AA, O’Connell K, et al. Identification of novel genomic risk loci shared between common epilepsies and psychiatric disorders. Brain. 2023:awad038. doi: 10.1093/brain/awad038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wiström ED, O’Connell KS, Karadag N, et al. Genome-wide analysis reveals genetic overlap between alcohol use behaviours, schizophrenia and bipolar disorder and identifies novel shared risk loci. Addiction. 2022;117(3):600–610. doi: 10.1111/add.15680. [DOI] [PubMed] [Google Scholar]
- 43. Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW.. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 2013;14:483–495. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Watanabe K, Stringer S, Frei O, et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet. 2019;51(9):1339–1348. doi: 10.1038/s41588-019-0481-0. [DOI] [PubMed] [Google Scholar]
- 45. Watanabe K, Taskesen E, Van Bochoven A, Posthuma D.. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Buniello A, Macarthur JAL, Cerezo M, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D1012. doi: 10.1093/nar/gky1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jones SE, Lane JM, Wood AR, et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat Commun. 2019;10(1):343. doi: 10.1038/s41467-018-08259-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jansen PR, Watanabe K, Stringer S, et al. Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat Genet. 2019;51(3):394–403. doi: 10.1038/s41588-018-0333-3. [DOI] [PubMed] [Google Scholar]
- 49. Luca G, Haba-Rubio J, Dauvilliers Y, et al. Clinical, polysomnographic and genome-wide association analyses of narcolepsy with cataplexy: a European Narcolepsy Network study. J Sleep Res. 2013;22:482–495. doi: 10.1111/jsr.12044. [DOI] [PubMed] [Google Scholar]
- 50. Zhu Z, Guo Y, Shi H, et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J Allergy Clin Immunol. 2020;145:537–549. doi: 10.1016/j.jaci.2019.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pazoki R, Vujkovic M, Elliott J, et al. Genetic analysis in European ancestry individuals identifies 517 loci associated with liver enzymes. Nat Commun. 2021;12(1):2579. doi: 10.1038/s41467-021-22338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pulit SL, Stoneman C, Morris AP, et al. Meta-Analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet. 2019;28(1):166–174. doi: 10.1093/hmg/ddy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Visconti A, Duffy DL, Liu F, et al. Genome-wide association study in 176,678 Europeans reveals genetic loci for tanning response to sun exposure. Nat Commun. 2018;9(1):1684. doi: 10.1038/s41467-018-04086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wuttke M, Li Y, Li M, et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet. 2019;51(6):957–972. doi: 10.1038/s41588-019-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hübel C, Gaspar HA, Coleman JRI, et al. Genomics of body fat percentage may contribute to sex bias in anorexia nervosa. Am J Med Genet Part B Neuropsychiatr Genet. 2019;180(6):428–438. doi: 10.1002/ajmg.b.32709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Niarchou M, Byrne EM, Trzaskowski M, et al. Genome-wide association study of dietary intake in the UK biobank study and its associations with schizophrenia and other traits. Transl Psychiatry. 2020;10(1):51. doi: 10.1038/s41398-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Klarin D, Damrauer SM, Cho K, et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet. 2018;50(11):1514–1523. doi: 10.1038/s41588-018-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sinnott-Armstrong N, Tanigawa Y, Amar D, et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat Genet. 2021;53(2):1622–1622. doi: 10.1038/s41588-020-00757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Christakoudi S, Evangelou E, Riboli E, Tsilidis KK.. GWAS of allometric body-shape indices in UK Biobank identifies loci suggesting associations with morphogenesis, organogenesis, adrenal cell renewal and cancer. Sci Rep. 2021;11(1):10688. doi: 10.1038/s41598-021-89176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kichaev G, Bhatia G, Loh PR, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet. 2019;104(1):65–75. doi: 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kircher M, Witten DM, Jain P, O’roak BJ, Cooper GM, Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hindley G, Frei O, Shadrin AA, et al. Charting the landscape of genetic overlap between mental disorders and related traits beyond genetic correlation. Am J Psychiatry. 2022;179:833–843. doi: 10.1176/appi.ajp.21101051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Valipour G, Saneei P, Esmaillzadeh A.. Serum vitamin D levels in relation to schizophrenia: a systematic review and meta-analysis of observational studies. J Clin Endocrinol Metab. 2014;99:3863–3872. doi: 10.1210/jc.2014-1887. [DOI] [PubMed] [Google Scholar]
- 65. Tsiglopoulos J, Pearson N, Mifsud N, Allott K, O’Donoghue B.. The association between vitamin D and symptom domains in psychotic disorders: a systematic review. Schizophr Res. 2021;237:79–92. doi: 10.1016/j.schres.2021.08.001. [DOI] [PubMed] [Google Scholar]
- 66. Lally J, Gardner-Sood P, Firdosi M, et al. Clinical correlates of vitamin D deficiency in established psychosis. BMC Psychiatry. 2016;16(1):76. doi: 10.1186/s12888-016-0780-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang X, Devaiah SP, Zhang W, Welti R.. Signaling functions of phosphatidic acid. Prog Lipid Res. 2006;45:250–278. doi: 10.1016/j.plipres.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 68. Shulga Y V, Topham MK, Epand RM.. Regulation and functions of diacylglycerol kinases. Chem Rev. 2011;111:6186–6208. doi: 10.1021/cr1004106. [DOI] [PubMed] [Google Scholar]
- 69. Lucas P, Ukhanov K, Leinders-Zufall T, Zufall F.. A diacylglycerol-gated cation channel in vomeronasal neuron dendrites is impaired in TRPC2 mutant mice: mechanism of pheromone transduction. Neuron. 2003;40:551–561. doi: 10.1016/S0896-6273(03)00675-5. [DOI] [PubMed] [Google Scholar]
- 70. Clayton EL, Minogue S, Waugh MG.. Phosphatidylinositol 4-kinases and PI4P metabolism in the nervous system: roles in psychiatric and neurological diseases. Mol Neurobiol. 2013;47(1):361–372. doi: 10.1007/s12035-012-8358-6. [DOI] [PubMed] [Google Scholar]
- 71. Cuscó I, Medrano A, Gener B, et al. Autism-specific copy number variants further implicate the phosphatidylinositol signaling pathway and the glutamatergic synapse in the etiology of the disorder. Hum Mol Genet. 2009;18(10):1795–1804. doi: 10.1093/hmg/ddp092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chang SH, Chiang SY, Chiu CC, et al. Expression of anti-cardiolipin antibodies and inflammatory associated factors in patients with schizophrenia. Psychiatry Res. 2011;187:341–346. doi: 10.1016/j.psychres.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 73. Lepeta K, Kaczmarek L.. Matrix metalloproteinase-9 as a novel player in synaptic plasticity and schizophrenia. Schizophr Bull. 2015;41(5):1003–1009. doi: 10.1093/schbul/sbv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fukumoto K, Tamada K, Toya T, Nishino T, Yanagawa Y, Takumi T.. Identification of genes regulating GABAergic interneuron maturation. Neurosci Res. 2018;134:18–29. doi: 10.1016/j.neures.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 75. Chu J, Anderson SA.. Development of cortical interneurons. Neuropsychopharmacology. 2015;40(1):16–23. doi: 10.1038/npp.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Van Derveer AB, Bastos G, Ferrell AD, et al. A role for somatostatin-positive interneurons in neuro-oscillatory and information processing deficits in schizophrenia. Schizophr Bull. 2021;47(5):1385–1398. doi: 10.1093/schbul/sbaa184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Filipović N, Bočina I, Restović I, et al. Ultrastructural characterization of vitamin D receptors and metabolizing enzymes in the lipid droplets of the fatty liver in rat. Acta Histochem. 2020;122(2):151502. doi: 10.1016/j.acthis.2020.151502. [DOI] [PubMed] [Google Scholar]
- 78. O’donnell JP, Phillips BP, Yagita Y, et al. The architecture of EMC reveals a path for membrane protein insertion. Elife. 2020;9:e57887. doi: 10.7554/eLife.57887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kim P, Scott MR, Meador-Woodruff JH.. Abnormal ER quality control of neural GPI-anchored proteins via dysfunction in ER export processing in the frontal cortex of elderly subjects with schizophrenia. Transl Psychiatry. 2019;9(1):6. doi: 10.1038/s41398-018-0359-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Britzolaki A, Saurine J, Klocke B, Pitychoutis PM.. A role for SERCA pumps in the neurobiology of neuropsychiatric and neurodegenerative disorders. Adv Exp Med Biol. 2020;1131:131–161. doi: 10.1007/978-3-030-12457-1_6. [DOI] [PubMed] [Google Scholar]
- 81. Rittenhouse AR, Ortiz-Miranda S, Jurczyk A.. Mutations in DISC1 alter IP3R and voltage-gated Ca2+ channel functioning, implications for major mental illness. Neuronal Signal. 2021;5(4):NS20180122. doi: 10.1042/ns20180122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Park SJ, Lee SB, Suh Y, et al. DISC1 modulates neuronal stress responses by gate-keeping ER-mitochondria Ca2+ transfer through the MAM. Cell Rep. 2017;21(10):2748–2759.. doi: 10.1016/j.celrep.2017.11.043. [DOI] [PubMed] [Google Scholar]
- 83. Park SJ, Jeong J, Park YU, et al. Disrupted-in-schizophrenia-1 (DISC1) regulates endoplasmic reticulum calcium dynamics. Sci Rep. 2015;5:8694. doi: 10.1038/srep08694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Norkett R, Modi S, Birsa N, et al. DISC1-dependent regulation of mitochondrial dynamics controls the morphogenesis of complex neuronal dendrites. J Biol Chem. 2016;291:613–629. doi: 10.1074/jbc.M115.699447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Xu C, Bailly-Maitre B, Reed JC.. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115(10):2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jayachandran P, Koshy L, Sudhakaran PR, Nair GM, Gangaprasad AN, Nair AJ.. 1, 25-(OH)2D3 protects against ER stress and miRNA dysregulation in Mus musculus neurons. Genes Genom. 2022;44(12):1565–1576. doi: 10.1007/s13258-022-01256-7. [DOI] [PubMed] [Google Scholar]
- 87. Asano L, Watanabe M, Ryoden Y, et al. Vitamin D metabolite, 25-hydroxyvitamin D, regulates lipid metabolism by inducing degradation of SREBP/SCAP. Cell Chem Biol. 2017;24(2):207–217. doi: 10.1016/j.chembiol.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 88. Sittipo P, Kim HK, Han J, Lee MR, Lee YK.. Vitamin D3 suppresses intestinal epithelial stemness via ER stress induction in intestinal organoids. Stem Cell Res Ther. 2021;12(1):285. doi: 10.1186/s13287-021-02361-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cioli C, Abdi H, Beaton D, Burnod Y, Mesmoudi S.. Differences in human cortical gene expression match the temporal properties of large-scale functional networks. PLoS One. 2014;9:e115913. doi: 10.1371/journal.pone.0115913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wiszniewski W, Hunter JV, Hanchard NA, et al. TM4SF20 ancestral deletion and susceptibility to a pediatric disorder of early language delay and cerebral white matter hyperintensities. Am J Hum Genet. 2013;93(2):197–210. doi: 10.1016/j.ajhg.2013.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hamilton G, Proitsi P, Jehu L, et al. Candidate gene association study of insulin signaling genes and Alzheimer’s disease: evidence for SOS2, PCK1, and PPARγ as susceptibility loci. Am J Med Genet Part B Neuropsychiatr Genet. 2007;144(4):508–516. doi: 10.1002/ajmg.b.30503 [DOI] [PubMed] [Google Scholar]
- 92. Lissewski C, Chune V, Pantaleoni F, et al. Variants of SOS2 are a rare cause of Noonan syndrome with particular predisposition for lymphatic complications. Eur J Hum Genet. 2021;29(1):51–60. doi: 10.1038/s41431-020-00708-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Li M, Li Y, Weeks O, et al. SOS2 and ACP1 loci identified through large-scale exome chip analysis regulate kidney development and function. J Am Soc Nephrol. 2017;28(3):981–994. doi: 10.1681/ASN.2016020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kravtsov V, Oren-Suissa M, Podbilewicz B.. The fusogen AFF-1 can rejuvenate the regenerative potential of adult dendritic trees by self-fusion. Development. 2017;144(13):2364–2374. doi: 10.1242/dev.150037. [DOI] [PubMed] [Google Scholar]
- 95. Sherva R, Tripodis Y, Bennett DA, et al. Genome-wide association study of the rate of cognitive decline in Alzheimer’s disease. Alzheimers Dement. 2014;10(1):45–52. doi: 10.1016/j.jalz.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Okada Y, Shimane K, Kochi Y, et al. A genome-wide association study identified AFF1 as a susceptibility locus for systemic lupus eyrthematosus in Japanese. PLoS Genet. 2012;8:e1002455. doi: 10.1371/journal.pgen.1002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yen S‐H, Kenessey A, Lee SC, Dickson DW.. The distribution and biochemical properties of a Cdc2‐related kinase, KKIALRE, in normal and Alzheimer brains. J Neurochem. 1995;65(6):2577–2584. doi: 10.1046/j.1471-4159.1995.65062577.x. [DOI] [PubMed] [Google Scholar]
- 98. Park CY, Zhou J, Wong AK, et al. Genome-wide landscape of RNA-binding protein target site dysregulation reveals a major impact on psychiatric disorder risk. Nat Genet. 2021;53(2):166–173. doi: 10.1038/s41588-020-00761-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Amestoy A, Baudrillard C, Briot K, Pizano A, Bouvard M, Lai M-C.. Steroid hormone pathways, vitamin D and autism: a systematic review. J Neural Transm. 2023;130:207–241. doi: 10.1007/s00702-022-02582-6. [DOI] [PubMed] [Google Scholar]
- 100. Siracusano M, Riccioni A, Abate R, Benvenuto A, Curatolo P, Mazzone L.. Vitamin D deficiency and autism spectrum disorder. Curr Pharm Des. 2020;26:2460–2474. doi: 10.2174/1381612826666200415174311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.