Abstract
The outer membrane proteins responsible for the influx of carbapenem β-lactam antibiotics in the nonfermentative gram-negative pathogen Acinetobacter baumannii are still poorly characterized. Resistance to both imipenem and meropenem in multidrug-resistant clinical strains of A. baumannii is associated with the loss of a heat-modifiable 29-kDa outer membrane protein, designated CarO. The chromosomal locus containing the carO gene was cloned and characterized from different clinical isolates. Only one carO copy, present in a single transcriptional unit, was found in the A. baumannii genome. The carO gene encodes a polypeptide of 247 amino acid residues with a typical N-terminal signal sequence and a predicted transmembrane β-barrel topology. Its absence from different carbapenem-resistant clinical isolates of A. baumannii resulted from the disruption of carO by distinct insertion elements. The overall data thus support the notion that CarO participates in the influx of carbapenem antibiotics in A. baumannii. Moreover, database searches identified the presence of carO homologs only in species of the genera Acinetobacter, Moraxella, and Psychrobacter, disclosing the existence of a novel family of outer membrane proteins restricted to the family Moraxellaceae of the class γ-Proteobacteria.
The emergence of antibiotic resistance among both pathogenic and opportunistic microbes resident in hospitals represents a serious and recurrent problem for the treatment of infections (22). It generates a continuous demand for new antimicrobial agents, whose application feeds the undesired vicious circle of selection and dissemination of new patterns of antibiotic resistance. The otherwise bizarre but still elegant mechanisms that underlie some of these patterns of resistance demonstrate that the potential of microbes to challenge eradication attempts remains almost unexhausted (26). Yet, attempts to reduce the dissemination of rapidly evolving antibiotic-resistant pathogens are best based on a detailed knowledge of the causes that promote these patterns of resistance (26).
The genus Acinetobacter, recently reassigned to the family Moraxellaceae in the class γ-Proteobacteria (30), is constituted by gram-negative, pleomorphic aerobic species commonly isolated from many sources in the environment, including drinking and static water, soil, sewage, food, and the skin of humans and animals (5). Certain strains of a particular species of the genus, Acinetobacter baumannii, now account for a large percentage of nosocomial infections, including pneumonia, bacteremia, skin and wound infections, and urinary tract infections. These strains are almost invariably multidrug resistant, having successfully resisted eradication attempts by the use of penicillins, aminoglycosides, cephalosporins, and even fluoroquinolones (5). It is due to this outstanding ability to rapidly respond to the challenge of new antibiotics that the emerging resistance to carbapenems among nosocomial strains of A. baumannii represents a major concern (22).
The molecular bases of resistance to carbapenems, which have been best characterized in Pseudomonas aeruginosa, include the recruitment of new β-lactamases or modifications to the specificities of existing enzymes, alterations in outer membrane (OM) permeability due to the reduced content of specific protein channels such as OprD, increased expression of efflux pumps, and/or modifications in the contents of penicillin-binding proteins (PBPs) (13, 28, 29, 34). In A. baumannii, carbapenem resistance has mostly been ascribed to the acquisition of carbapenemases (3, 29) or to synergistic effects between β-lactamases with the ability to hydrolyze carbapenems and the decreased expression of certain PBPs (9, 10). The contribution of OM impermeability to this resistance has been less well characterized, although a number of reports have noted the absence of different OM proteins in a number of carbapenem-resistant A. baumannii strains of clinical origin (6, 7, 9, 33). It is worth noting here that our knowledge of the proteins responsible for the influx of β-lactam antibiotics through the OM of this pathogen is still limited (11).
We previously demonstrated (19) that imipenem resistance is associated with the loss of a 29-kDa OM protein in clinical isolates of A. baumannii in which no imipenemase activity could be detected. We report here on the cloning and characterization of a chromosomal locus containing the gene encoding this polypeptide. Our results indicate that this protein, designated CarO (for carbapenem resistance-associated outer membrane protein), is a member of a novel family of β-barrel OM proteins apparently restricted to the family Moraxellaceae of the class γ-Proteobacteria. The insertional disruption of carO by previously uncharacterized insertion elements was responsible for the loss of this protein in carbapenem-resistant clinical isolates of A. baumannii, supporting the notion that CarO participates in the influx of these antibiotics in this pathogen.
MATERIALS AND METHODS
Bacterial strains and susceptibility testing.
A. baumannii clinical isolates were obtained from the Bacteriology Section of the Hospital de Emergencias Clemente Alvarez, Rosario, Argentina. Isolates were routinely considered multiresistant when they were simultaneously resistant to at least two β-lactams (including ampicillin-sulbactam, ceftazidime, cefotaxime, piperacillin, and piperacillin-tazobactam), gentamicin or amikacin, ciprofloxacin, and trimethoprim-sulfamethoxazole.
Genomic relationships between isolates were determined from the profiles obtained by three different methods: PCR with degenerate primers, repetitive extragenic palindromic PCR, and pulsed-field gel electrophoresis (20). A particular subgroup of seven clonally related isolates, which included three carbapenem-resistant strains (20), was used for the present study. The antibiotic sensitivity profiles of the A. baumannii strains analyzed here are described in Table 1. The resistance to carbapenems in these strains could not be attributed to the presence of carbapenem-hydrolyzing enzymes, as judged by spectrophotometric analysis of bacterial extracts with imipenem as a substrate; PCR amplification with primers specific for blaIMP, blaVIM, and blaOXA; and DNA hybridization studies with probes specific for OXA-23, IMP-7, and VIM-2 (data not shown). Carbapenem-resistant isolate Ab125, also incorporated in this study (see below), belongs to a more comprehensive collection of isolates present at the Malbrán Institute (Buenos Aires, Argentina).
TABLE 1.
Antibiotic susceptibilities of the A. baumannii strains used in this work
| Strain | MIC (μg/ml)a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| IPM | MRP | CAZ | CTX | AMK | GEN | KAN | ||
| Ab244 | 1 | 1 | 256 | 256 | 16 | 1,024 | 1,024 | |
| Ab242 | 16 | 16 | 256 | 256 | 128 | 1,024 | 1,024 | |
| Ab825 | 16 | 16 | 256 | 256 | 128 | 1,024 | 512 | |
| Ab125 | 16 | 16 | 512 | 256 | 64 | 512 | ND | |
| Ab244R1 | 16 | 16 | 512 | 256 | 128 | ND | ND | |
| Ab244R2 | 16 | 16 | 512 | 256 | 128 | ND | ND | |
The MICs were determined by the macrodilution method, in accordance with the procedures recommended by the National Committee for Clinical Laboratory Standards (27). Abbreviations: IPM, imipenem; MRP, meropenem; CAZ, ceftazidime; CTX, cefotaxime; AMK, amikacin; GEN, gentamicin; KAN, kanamycin; ND, not determined.
Bacteria were routinely grown at 30°C in Luria-Bertani (LB) or Mueller-Hinton broth. MICs were determined by the micro- and macrodilution methods in accordance with the procedures recommended by the National Committee for Clinical Laboratory Standards (27).
Carbapenem-resistant clones of clinical isolate Ab244 were obtained by the successive selection of resistant bacteria in media containing increasing concentrations (1, 2, 4, 8, and 16 μg/ml, respectively) of imipenem, basically by the procedures described previously (19). Two selected clones resistant to both imipenem and meropenem, designated Ab244R1 and Ab244R2 (Table 1), were chosen for further characterization (see Fig. 8).
FIG. 8.
Loss of CarO in the OMs of A. baumannii strains selected for carbapenem resistance in vitro. Carbapenem-resistant strains Ab244R1 and Ab244R2 were derived from carbapenem-sensitive clinical strain Ab244 by selection in media containing successively increasing concentrations of imipenem. Bacterial OM fractions, each of which corresponds to 30 μg of protein, were analyzed by SDS-PAGE on a 12.5% polyacrylamide gel and immunoblotting. All procedures are described in Materials and Methods. (A) Coomassie blue staining. Lane 1, strain Ab244; lane 2, strain Ab244R1; lane 3, strain Ab244R2. An arrowhead indicates the position of CarO. The molecular mass and position of each of the different size markers (lane m) are also indicated. (B) Immunoblot analysis of the samples described for panel A by using antibodies directed against Ab244 CarO.
Preparation of bacterial OM.
The OM fractions of the different bacterial strains studied here were prepared by the N-lauroyl sarcosinate method (19). Briefly, the bacterial cells were grown overnight at 30°C in LB broth, harvested by centrifugation at 7,000 × g for 10 min, and washed once with ice-cold phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4 [pH 7.4]). The cells were resuspended in fresh PBS containing 0.1 mM phenylmethylsulfonyl fluoride and 1 mM dithiothreitol and were disrupted by ultrasonic disintegration with a Vibra-cell VCX-600 ultrasonic processor (Sonics & Materials). The resulting extracts were clarified by centrifugation at 5,000 × g for 10 min at 4°C, and the supernatants were collected. N-Lauroyl sarcosinate (sodium salt) was added to a final concentration of 2.2% (wt/vol), and the mixture was incubated for 30 min at 20°C. OM fractions were collected by centrifugation at 100,000 × g for 1 h at 4°C, washed once with 2.2% (wt/vol) sodium N-lauroyl sarcosinate, collected as described above, and finally resuspended in 20 mM Tris-HCl (pH 8.0)-0.1 mM EDTA-1% (wt/vol) sodium dodecyl sulfate (SDS) for further analyses (see below).
SDS-PAGE and immunoblotting.
The OM protein compositions of the fractions described above were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) with 12.5% (wt/vol) polyacrylamide gels and Coomassie blue staining by standard protocols (17). All samples were incubated for 5 min in a boiling water bath before they were subjected to electrophoresis. The resolved proteins were also analyzed by immunoblotting (8) with specific anti-CarO antibodies (see below) raised in rabbits. After incubation with goat anti-rabbit immunoglobulin G-alkaline phosphatase second antibody (Bio-Rad), immunodecoration was done with 5-bromo-4-chloro-3-indolylphosphate and p-nitroblue tetrazolium by standard techniques.
Determination of N-terminal sequence of mature CarO.
Strain Ab244 OM proteins were resolved by SDS-PAGE as described above and electrotransferred onto polyvinylidene difluoride membranes (19). The N-terminal sequence of CarO was determined by Edman degradation at the Interdisciplinary Center for Biotechnology Research, Protein Chemistry Core Facility (University of Florida, Gainesville).
PCR amplification of carO gene.
The DNA sequence encoding the mature form of CarO was obtained by PCR amplification of strain Ab244 genomic DNA. The forward primer (5′-CCATGGCTGACGAWGCAGTCGTACATGA-3′, where W is A or T) was designed after the first 7 amino acids of the N terminus of the mature protein determined as described above and contains, in addition, an NcoI tail. The reverse primer (5′-CCATGGCAAAAGTATTAAAAGTTTTAGCAGT-3′) corresponded to the 3′ end of a predicted carO homolog present in the genome of Acinetobacter sp. strain ADP1 and contains, in addition, a BamHI tail. To identify the gene homologous to A. baumannii carO in Acinetobacter sp. strain ADP1, a search for the best alignment with the 27 amino acids of the N terminus of the mature form of A. baumannii CarO (19) was done by using the Clustal W program (version 1.7) (35) with the contigs of the genome of Acinetobacter sp. strain ADP1 (available at www.genoscope.com), all possible open reading frames (ORFs) of which have previously been translated. While this report was in preparation, the complete annotated genome of Acinetobacter sp. strain ADP1 was released in the GenBank database under accession number CR543861. The predicted carO homolog in this genome can be located under accession number YP_047181.
PCRs were done with a 50-μl reaction mixture containing 10 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 1 μM each forward and reverse primers (see above), 50 ng of template DNA from strain Ab244, and 1 U of Taq polymerase (Invitrogen). An initial denaturation step at 94°C for 3 min was followed by 30 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C. A final extension step of 10 min at 72°C was performed. Only one fragment of the expected size (about 700 bp) was amplified, and this fragment was ligated to the pGem-T Easy vector (Promega) and transformed into Escherichia coli DH5α. After selection for ampicillin resistance, the plasmids were isolated for DNA sequence analysis.
Southern blot analysis and cloning of the A. baumannii genomic locus containing carO.
DNA samples from strain Ab244 or selected carbapenem-resistant strains were separately digested with BstXI, ClaI, EcoRI, HindIII, and XbaI. The resulting fragments were separated by electrophoresis in 0.7% (wt/vol) agarose, transferred to Hybond nylon membranes (Amersham), and hybridized with a [α-32P]ATP-labeled carO probe (see above) by the procedures recommended by the membrane supplier.
To clone the carO gene and neighboring regions from strain Ab244, the digests obtained with ClaI and HindIII were separately ligated into the corresponding sites in pBluescript SK− (Stratagene). After transformation into E. coli DH5α and selection for ampicillin resistance, the resulting colonies were screened by DNA hybridization with [α-32P]ATP-labeled carO, as described above. Different plasmids isolated from positive clones were found to contain the expected inserts of 3.7 kbp (HindIII digests) or 3.4 kbp (ClaI digests). Sequence analyses indicated that these inserts overlapped in a 1.3-kbp region which contained the complete carO-coding region present in Ab244.
Cloning of complete carO-coding sequence from different A. baumannii isolates.
The sequence data obtained as described above were used to design primers to specifically amplify the complete coding sequence of carO from the different A. baumannii strains described in Table 1. The forward primer (5′-CATATGAAAGTATTACGTGTTTTAGTG-3′) covered the DNA sequence corresponding to the initiation methionine and the subsequent 7 amino acids of the transit sequence and included an additional NdeI site at its 5′ end. The reverse primer (5′-GGTACCTTACCAGTAGAAGTTTACACC-3′) covered the DNA sequence corresponding to the last 6 amino acids and the TAA stop codon and included an additional KpnI site at its 3′ end.
PCRs were done as described above. The amplification products were ligated into the pGem-T Easy vector (Promega) and transformed into E. coli DH5α. After selection for ampicillin resistance, plasmids were isolated for DNA sequence analysis.
Expression of carO in E. coli.
The complete coding sequence of the strain Ab244 carO was amplified as described above with High Fidelity Taq polymerase (Invitrogen). The amplification product was digested with NdeI and KpnI and ligated into the equivalent sites of plasmid pTrx6. The ligation mixture was transformed into E. coli DH5α, and colonies were selected in LB agar plates containing 15 μg of chloramphenicol per ml. Plasmids from different clones were isolated and sequenced to verify the presence of the desired construction, and one of them, designated p-preCarO, was used for expression purposes.
To study carO expression in E. coli DH5α, the transformed cells were grown at 30°C in LB liquid medium containing 15 μg of chloramphenicol per ml. When the optical density at 600 nm of the cultures reached 0.5, l-arabinose was added to a final concentration of 0.01% (wt/vol), and incubation was continued for an additional 2 h. The cells were harvested and washed once with ice-cold PBS, and the OM fraction was analyzed by SDS-PAGE and immunoblotting with anti-CarO as described above.
Other procedures.
Protein contents were determined by a modified procedure of Lowry et al. (23). Antibodies were elicited in rabbits with CarO purified from A. baumannii Ab244 by preparative SDS-PAGE (12). The antibodies were further affinity purified by using nitrocellulose membranes to which purified CarO had been electrophoretically attached (32).
DNA manipulations were conducted by the protocols described elsewhere (32). All DNA sequencing was done at the DNA Sequencing Facility of the University of Maine, Orono.
Database searches and computer analyses.
Searches for the similarities of the sequences obtained with protein and DNA sequences in the GenBank database were performed by using the BLAST program (2).
Prediction of the transmembrane β strands on mature CarO and homologous proteins were done by the hidden Markov model method (4), available at http://bioinformatics.biol.uoa.gr/PRED-TMBB. Predictions of the CarO functional category and the putative presence of transmembrane α-helical segments were done with the ProtFun (version 2.2) and TMHMM programs (15, 16), respectively (both available at http://www.cbs.dtu.dk/services/). Predictions of consensus bacterial promoter sequences were conducted at http://www.softberry.com.
Nucleotide sequence accession numbers.
The 5,731-bp A. baumannii Ab244 genomic sequence containing carO and surrounding regions was deposited in the GenBank database under accession number AY684798. The 1,723-bp DNA sequence of the PCR fragment corresponding to the ISAba825-interrupted carO gene was deposited in the GenBank database under accession number AY751532. The 1,840-bp sequence corresponding to the ISAba125-interrupted carO gene received accession number AY751533.
RESULTS AND DISCUSSION
Carbapenem resistance is associated with the loss of a 29-kDa OM protein in multidrug-resistant clinical strains of A. baumannii.
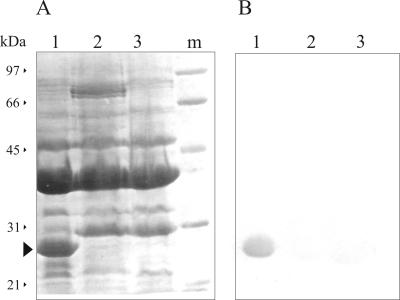
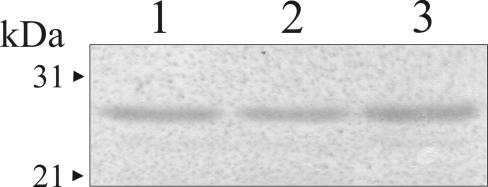
The OM protein compositions of seven clonally related multiresistant A. baumannii strains isolated at the Hospital de Emergencias Clemente Alvarez from different patients after the introduction of imipenem therapy (20) were analyzed by SDS-PAGE. Three different OM protein profiles were observed among these strains and are shown in Fig. 1A. Strains showing carbapenem sensitivity produced similar profiles, exemplified in Fig. 1 by strain Ab244 (lane 1). In turn, two profiles were distinguished among strains that were also resistant to imipenem and meropenem (Table 1), which are exemplified in Fig. 1 by strains Ab242 (lane 2) and Ab825 (lane 3). The OM protein pattern of strain Ab125, a carbapenem-resistant isolate obtained from the collection of A. baumannii strains of the Malbrán Institute at Buenos Aires, is also shown in Fig. 1. The relevance of this isolate is discussed below (see Fig. 5 and 7). As seen in Fig. 1 and in agreement with previous results (19), carbapenem resistance was associated with the loss of a 29-kDa OM protein. Determination of the N-terminal amino acid sequence of this protein isolated from strain Ab244 indicated complete identity (DEAVVHDSY) with the same region of the 29-kDa OM protein described previously (19) for isolate Ab288. Moreover, immunoblot analysis with a specific antiserum confirmed the absence of this 29-kDa OM protein in all carbapenem-resistant clinical strains (Fig. 1B). This protein was subsequently designated CarO, for carbapenem resistance-associated OM protein.
FIG. 1.
Loss of a 29-kDa OM protein in carbapenem-resistant clinical strains of A. baumannii. OM fractions of the different isolates, each of which contained 30 μg of protein, were analyzed by SDS-PAGE on 12.5% polyacrylamide gels and immunoblotting, as described in Materials and Methods. (A) Coomassie blue staining. Lane 1, carbapenem-sensitive strain Ab244; lane 2, carbapenem-resistant strain Ab242; lane 3, carbapenem-resistant strain Ab825; lane 4, carbapenem-resistant strain Ab125. The arrowhead indicates the final position of the 29-kDa OM protein (CarO) in the gel. The molecular mass and position of each of the different size markers (lane m) are shown on the left. (B) Immunoblot analysis of the same samples described for panel A with antibodies directed against the 29-kDa OM protein from strain Ab244.
FIG. 5.
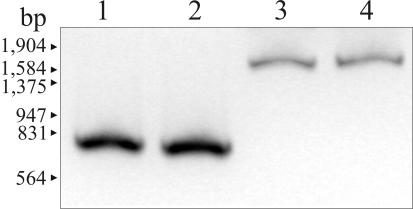
PCR amplification of the carO genes in the A. baumannii strains studied. Lane 1, carbapenem-sensitive strain Ab244; lane 2, carbapenem-resistant strain Ab242; lane 3, carbapenem-resistant strain Ab825; lane 4, carbapenem-resistant strain Ab125. The positions of the size markers (EcoRI-HindIII-digested bacteriophage λ DNA) are indicated on the left. For details, see Materials and Methods.
FIG. 7.
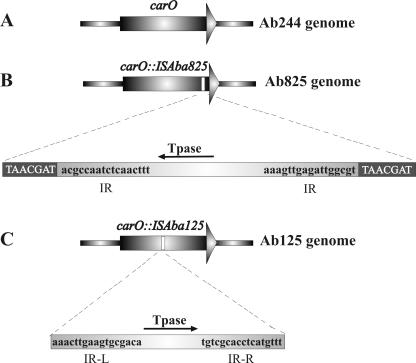
Schematic representation of IS-mediated inactivation of carO in the carbapenem-resistant A. baumannii strains studied in this work. (A) carO allele in carbapenem-sensitive strain Ab244; (B) carO-disrupted allele in strain Ab825. The duplicated carO sequences at the insertion site of ISAba825 are shown in white capital letters. The inverted repeats (IR) are shown in black lowercase letters at both sides of the transposase (Tpase) gene. (C) carO-disrupted allele in strain Ab125. The imperfect inverted repeats (IR-L and IR-R) of ISAba125 bordering the transposase gene are shown. No direct repeats could be recognized in this case. The directions of transcription of the transposase genes are indicated by arrows. The picture is not drawn to scale. The complete nucleotide sequences of the carO-disrupted genes and the corresponding ISs are present in the GenBank database under accession numbers AY751532 and AY751533, respectively.
Identification and characterization of a genomic locus containing the A. baumannii carO gene.
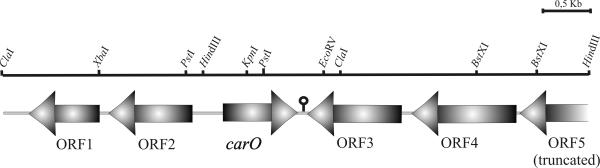
The information derived from the N-terminal sequence (see above) was used to identify and clone a 5.7-kbp chromosomal fragment from strain Ab244 containing the complete carO gene and surrounding regions (Fig. 2). Southern blot analysis indicated the presence of only one locus containing carO in Ab244 (data not shown).
FIG. 2.
Genetic organization of the A. baumannii chromosomal locus containing carO and flanking regions. A 5,731-bp chromosomal fragment from A. baumannii Ab244 containing the carO gene was cloned and characterized as described in Materials and Methods. ORFs (Table 2) identified in databases are indicated by arrows, which depict the direction of transcription. The stem-loop structure identified immediately downstream of the carO gene is shown. The complete nucleotide sequence of this region can be found in the GenBank database under accession number AY684798.
Database searches and comparisons (Fig. 2 and Table 2) indicated that carO shares a locus in the A. baumannii chromosome with genes that encode proteins involved in the diaminopimelate-lysine pathway of cell wall biosynthesis (ORF2), a LysR-type transcriptional regulator (ORF3), and sulfate import (ORF4 and ORF5). The carO gene consists of a single transcriptional unit in which the coding sequence covers a total of 744 nucleotides. A total of 294 nucleotides separate the initiation codon of carO from the dapD gene, and the two genes are transcribed in opposite directions (Fig. 2). A ribosome binding site (AGGAG) was identified approximately 7 bp upstream of the translational start codon of carO, and a 16-bp inverted repeat with characteristics of a rho-independent transcription terminator was located downstream of the termination codon (Fig. 2). Putative −35 (TTGAAA) and −10 (TTGTATCAA) promoter boxes were identified from 146 to 141 bp and from 122 to 114 bp, respectively, upstream of the carO initiation codon.
TABLE 2.
Predicted ORFs located in the vicinity of carO in the A. baumannii genome
| ORF | Size (no. of amino acids) | GenBank accession no. | E valuea | Proposed functionb |
|---|---|---|---|---|
| CarO | 247 | AY684798 | OM protein (this work) | |
| ORF1 | 236 | ZP_00145817 | 4e−80 | Organic radical-activating enzyme |
| ORF2 | 273 | NP_414708 | 1e−101 | 2,3,4,5-Tetrahydropyridine-2-carboxylate, N-succinyltransferase, lysine-diaminopimelate synthesis |
| ORF3 | 307 | NP_840656 | 1e−104 | LysR family type, transcriptional regulator |
| ORF4 | 353 | ZP_00023858 | 1e−105 | ABC-type sulfate and molybdate transporter, ATPase component |
| ORF5 | 232c | ZP_00029254 | 1e−82 | ABC-type sulfate transporter, permease component |
The expected (E) value was obtained as described previously (2).
Functions were assigned on the basis of amino acid similarities with proteins present in databases.
Truncated.
carO encodes a polypeptide of 247 amino acid residues with an estimated molecular mass of 26,418 Da. The first 21-amino-acid region represents a typical prokaryotic signal sequence (31): it contains two basic amino acid residues at the N terminus (residues 1 to 5), followed by a hydrophobic core from residues 6 through 21 and a peptidase I cleavage site (Ala-Met-Ala). In agreement, the experimentally determined N terminus of the mature polypeptide starts at aspartic acid residue 22 (Fig. 3) (see also reference 19). These data indicate that mature CarO is 226 amino acid residues in length and has a final estimated molecular mass of 24,334 Da.
FIG. 3.
Alignments and prediction of the structures of A. baumannii CarO and homolog proteins identified in databases. Homolog proteins were identified after nucleotide databases were searched with the BLAST program (2) by using the translated carO gene sequence as the query sequence. Sequence alignments were performed by using the Clustal W (version 1.7) program (35) and were refined by visual inspection. Identical residues in all sequences are indicated below the alignments by asterisks, and conservative substitutions are indicated by colons. The experimentally determined processing site for A. baumannii CarO is indicated above the sequence by an arrowhead. The different β-strand membrane-spanning regions predicted for A. baumannii CarO are indicated above the alignments. Predicted transmembrane-spanning regions present at similar locations in at least two of these polypeptides are indicated in shaded boxes. The PRED-TMBB program, which uses the posterior decoding method with the dynamic programming algorithm (4), was used. The GenBank accession numbers of the sequences are as follows: A. baumannii carO, AY684798; Acinetobacter sp. strain ADP1, YP_047181; M. catarrhalis G1b, AAS21230; Psychrobacter sp. strain 273-4, ZP_00147002.
The apparent molecular mass of CarO in SDS-polyacrylamide gels when OM samples were pretreated at 95°C for 5 min was 29 kDa (Fig. 1). Omission of the heating step prior to electrophoresis, however, showed a significantly faster migration rate for this polypeptide, which changed from 29 to 24 kDa (data not shown). This indicates that CarO represents a member of the so-called heat-modifiable proteins, a group formed by many β-barrel monomeric channels present in the OMs of different gram-negative bacteria (11, 28).
Densitometric estimates of the relative contents of CarO in strain Ab244 indicated that this protein represents between 20 and 25% of the total OM (Fig. 1). No substantial modifications in the relative contents of this protein in the OM were found in bacteria growing in different media (Mueller-Hinton or LB medium), at different temperatures (20, 30, or 37°C), in medium supplemented with subinhibitory concentrations of imipenem (0.25 μg/ml), or in medium of high osmolarity (data not shown). Therefore, the carO gene seems to be constitutively expressed under all of these conditions. Whether the content of CarO varies depending on the carbon or carbon and nitrogen sources is under scrutiny.
Expression of carO in E. coli.
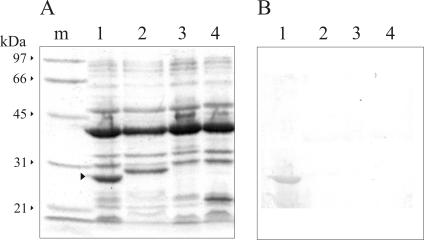
The complete carO gene, including the transient sequence-coding region, was subcloned in an E. coli expression vector and expressed in DH5α cells (Fig. 4). Analysis of the bacterial OM fractions by SDS-PAGE and immunoblotting with antisera against the 29-kDa OM protein of A. baumannii not only confirmed the identity of this polypeptide but also indicated that it has all the information needed to direct its secretion to the bacterial OM.
FIG. 4.
Expression of A. baumannii carO in E. coli directs the protein to the outer bacterial membrane. The complete carO gene of strain Ab244, including the transit sequence-coding region, was subcloned in expression plasmid pTrx6 and expressed in E. coli DH5α. Whole-cell extracts and OM fractions were analyzed by SDS-PAGE and immunoblotting with anti-CarO, as described in Materials and Methods. Lane 1, whole-cell lysates, 30 μg of total protein; lane 2, E. coli OM fraction, 10 μg of total protein; lane 3, A. baumannii Ab244 OM fraction, 10 μg of total protein. The positions and molecular masses of two size markers are indicated by the arrowheads on the left.
Predictions of CarO structure.
Integral membrane proteins can be divided into two distinct structural classes: α-helical and β-barrel proteins (4). Some of the attributes of CarO are characteristic of bacterial OM β-barrel proteins: heat modifiability (see above), a glycine-rich content (12.5%), and an absence of cysteine residues (28). In agreement, while no α-helix membrane-spanning regions were predicted by commonly used algorithms, the PRED-TMBB program indicated that CarO represents a β-barrel OM protein (score, 2.886) with up to 10 membrane-spanning regions (indicated in Fig. 3).
In turn, function predictions (ProtFun software, version 2.2) gave CarO the highest score (0.764) in the category transport and binding.
CarO belongs to an emerging novel family of bacterial OM proteins.
Databases similarity searches, including searches of the genomes of species of the major branches of the class Proteobacteria deposited in databases, identified only a very restricted number of putative polypeptides whose sizes, amino acid sequences, and predicted topologies were similar to those of A. baumannii CarO (Fig. 3). In fact, sequence comparisons indicated the closest homology to a putative homolog present in the soil species Acinetobacter sp. strain ADP1 (68% amino acid identity, 84% overall similarity). Much lower but still significant homology was found between A. baumannii CarO and a set of highly homologous OM proteins of Moraxella catarrhalis (29% identity and 55% overall similarity in the example given in Fig. 3; see reference 1 for details) and a predicted protein of Psychrobacter sp. strain 273-4 (27% identity, 53% overall similarity). All of these polypeptides contained predictable secretion signal sequences at their N-terminal regions (Fig. 3), scores compatible with those of β-barrel OM proteins (data not shown), and similar locations of the predicted β-strand-spanning regions along their primary sequences (Fig. 3). Remarkably, the C-terminal residues of all of these polypeptides are represented by either a tryptophan (Acinetobacter species) or a phenylalanine (Moraxella and Psychrobacter species) residue, a feature almost invariably found in OM protein channels (28). However, no close homology between CarO and any member of the different families of OM protein channels that have been described (28) could be detected at the sequence level.
It is worth noting that the genera Acinetobacter, Moraxella, and Psychrobacter compose the family Moraxellaceae of the class γ-Proteobacteria (30). Thus, the overall results provide evidence for the existence of a novel family of OM proteins restricted so far to this proteobacterial family. Several highly conserved short amino acid stretches could be observed in the sequence comparisons shown in Fig. 3 and may be characteristic of these proteins. In particular, the motif AEVGTXGYG (where X is T or L), which is located near their N-terminal regions and which is not found in other OM protein families (28), may represent a characteristic signature of this novel family.
Naturally occurring insertional inactivation of carO in different carbapenem-resistant clinical strains of A. baumannii.
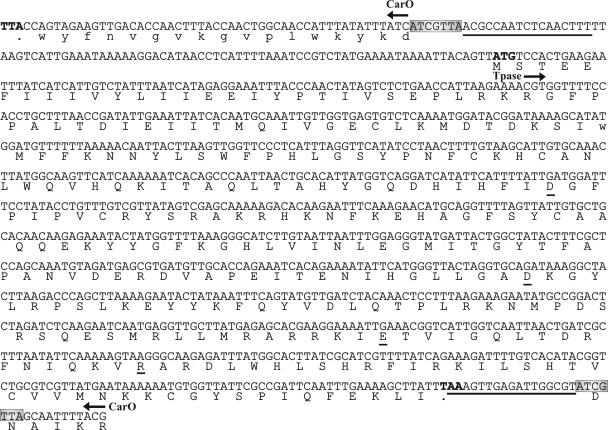
We next analyzed the possible causes of the loss of CarO in carbapenem-resistant A. baumannii clinical strains. A PCR amplification analysis with carO-specific primers indicated single expected fragments of about 750 bp in the case of carbapenem-sensitive strain Ab244 and carbapenem-resistant isolate Ab242 (Fig. 5, lanes 1 and 2, respectively). On the other hand, a single amplification fragment of 1,700 bp was obtained in the case of the carbapenem-resistant strain Ab825 (lane 3). Cloning and sequencing of this fragment indicated the presence of an extra insertion of about 975 bp near the 3′ end of the carO allele present in this strain (Fig. 6 and 7B). This extra fragment displayed all characteristics of an insertion element (Fig. 6): it generated a 7-bp duplication (TAACGAT) at the predicted insertion site, is bounded by a perfect 17-bp inverted repeat, and contains a 876-bp ORF whose product shows the characteristic signatures of a DDE-type transposase (25). Database searches (data not shown) indicated the closest homologies between this putative transposase and predicted proteins present in Psychrobacter sp. strain 273-4 (64% amino acid identity, 80% overall similarity) and the cyanobacterium Nostoc sp. strain PCC 7120 (47% identity, 66% overall similarity). No homology between this element and any member of the described insertion sequence (IS) families (http://www-is.biotoul.fr) was found at either the inverted repeat or transposase level. Thus, we designated this novel element ISAba825 after its immediate source. Southern hybridization analysis indicated that it occurs at two to three copies per genome in the carbapenem-resistant A. baumannii isolates analyzed here (data not shown). It is worth noting that the G+C content of ISAba825 (34.0%) differed significantly from that of the A. baumannii 5.7-kbp chromosomal fragment described above (39.8%), suggesting an exogenous origin for this new IS element.
FIG. 6.
carO is disrupted by an IS element in A. baumannii Ab825. The nucleotide sequence of a relevant region of the PCR fragment showing the ISAba825-disrupted carO allele is displayed. Note that the carO gene and the transposase (Tpase) gene are located on opposite strands. For schematic purposes, only the nucleotide sequence corresponding to the 6 amino acid residues (RKIAND) of CarO located immediately upstream of the insertion site is shown. The carO region (the first 54 nucleotides in the sequence) that encoded the last 17 amino acid residues present at the C-terminal region of the protein (in lowercase letters) is also shown. The termination codon of carO (the first three nucleotides in the scheme) is indicated in boldface. Gray boxes indicate the carO region (ATCGTTA) which was duplicated during transposition. The inverted repeats bordering ISAba825 are underlined. The initiation (ATG) and termination (TAA) codons of the transposase are also indicated in boldface. The amino acid residues conforming to the DDE triad and the accompanying R residue are underlined. The complete 1,723-nucleotide sequence was deposited in the GenBank database under accession number AY751532.
The results presented above predict the presence of different IS-mediated disruptive events targeting the carO gene in other carbapenem-resistant clinical isolates of A. baumannii. In fact, PCR analysis of a more comprehensive collection of clinical isolates obtained at a different health care center (Malbrán Institute, Buenos Aires, Argentina) revealed another carbapenem-resistant strain, Ab125, in which the amplification band corresponding to carO was larger than expected (Fig. 5, lane 4). Sequence analysis of this fragment indicated the presence of a different IS element disrupting carO at a different position in this strain (Fig. 7C). This previously uncharacterized IS, designated ISAba125, showed significant homology at both the imperfect terminal repeats and the transposase sequence with members of the widely distributed IS30 family (25) (Fig. 7C and data not shown).
CarO is lost from the OMs of bacteria selected for carbapenem resistance in vitro.
Clones derived from sensitive strain Ab244 were selected for resistance to 16 μg of imipenem per ml by growing the bacteria in media containing successively increasing concentrations of this antibiotic. Two clones, Ab244R1 and Ab244R2, which had evolved resistance to both imipenem and meropenem after this procedure were studied (Table 1). Analysis of the OM protein patterns of both clones indicated some modifications in the relative contents of some polypeptides as a result of the selection (Fig. 8A). However, the most noticeable change consisted of the complete loss of the CarO protein from their OM fractions (Fig. 8A and B). Thus, these results provided further support to the evidence presented above indicating that carbapenem resistance resulted from the inactivation of the carO gene in the clinical isolates of A. baumannii studied here.
It is worth noting that both PCR and Southern blot analysis with DNA from the in vitro-selected carbapenem-resistant strains described above provided no evidence for insertional disruption events on either the structural carO gene or its immediate upstream region that may account for the absence of CarO in these bacteria (data not shown). Remarkably, a similar observation was also made with carbapenem-resistant clinical isolate Ab242 (Fig. 1 and 5). Thus, a mechanism other than the insertional disruption event described for isolates Ab825 and Ab125 may also account for the loss of CarO in both clinical and laboratory-selected carbapenem-resistant strains of A. baumannii. This alternative mechanism is under study in our laboratory.
Concluding remarks.
The overall results of this work are consistent with the notion that CarO participates in the influx of carbapenems in A. baumannii (19). The acquisition of β-lactam resistance resulting from IS-mediated events targeting genes encoding OM channels has previously been described on only a few occasions, such as cefoxitin resistance in Klebsiella pneumoniae mediated by ompK36 disruption (14) and carbapenem resistance in P. aeruginosa mediated by oprD disruption (37). The results presented here provide another example of the challenges that bacterial mobile elements pose to efforts directed toward the prevention of the emergence of antibiotic resistance.
To our knowledge, specific OM channels for carbapenems have been described only in the case of the OprD of P. aeruginosa, another nonfermentative pathogen ascribed to the family Pseudomonadaceae of the class γ-Proteobacteria (21). OprD participates in the influx of basic amino acids and certain peptides, and mutations that inactivate oprD constitute one of the main mechanisms of carbapenem resistance among clinical strains of P. aeruginosa (21, 37). Still, A. baumannii CarO has no recognizable homology to P. aeruginosa OprD in terms of either its size or primary sequence (this work). Moreover, OM proteins whose sequences are related to OprD are seemingly absent in Acinetobacter species, as judged by immunoblot analysis of the OM fractions of different A. baumannii strains with an antiserum directed against P. aeruginosa OprD (18) and by the absence of oprD homologs in the recently released genome of Acinetobacter sp. strain ADP1 (GenBank accession number CR543861). Thus, CarO may well act as a functional OprD analog in A. baumannii, allowing the influx of carbapenems in this pathogen. Whether CarO performs this function by forming complexes with other OM proteins and the physiological roles of this protein in A. baumannii are under evaluation.
Finally, the apparent restriction of carO homolog genes to a single family of the class γ-Proteobacteria, the Moraxellaceae (Fig. 3), provides some restraints on the general dissemination of the mechanism of carbapenem resistance proposed here. In fact, several species of this family are either known or suspected human pathogens (1, 24, 36). Whether they can eventually evolve carbapenem resistance by a similar mechanism represents a distressing possibility that requires further awareness and study.
Acknowledgments
We are indebted to F. Pasterán and M. Galas for the generous gift of the A. baumannii isolates of the collection of the Malbrán Institute. We are also indebted to D. de Mendoza and N. Carrillo for critical reading of the manuscript. Plasmid pTrx6 was a generous gift of K. Nishihara, Kyoto, Japan. Rabbit antibodies against P. aeruginosa OprD were kindly provided by T. Nakae and H. Yoneyama, Tokai University, Kanagawa, Japan.
This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) of Argentina; Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET); the Subsecretaría de Investigación y Tecnología, Ministerio de Salud de la Nación Argentina; and the Departamento de Salud Pública, Municipalidad de Rosario. A.M.V. is a career investigator of CONICET, and M.A.M. is a fellow of the same institution. A.S.L. is a researcher of the National University of Rosario.
REFERENCES
- 1.Adlowitz, D. G., T. Hiltke, A. J. Lesse, and T. F. Murphy. 2004. Identification and characterization of outer membrane proteins G1a and G1b of Moraxella catarrhalis. Vaccine 22:2533-2540. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amyes, S. G. B., and H. K. Young. 1996. Acinetobacter: microbiology, epidemiology, infections, management, p. 185-223. CRC Press, Inc., New York, N.Y.
- 4.Bagos, P. G., T. D. Liakopoulos, I. C. Spyropoulos, and S. J. Hamodrakas. 2004. A Hidden Markov Model method, capable of predicting and discriminating beta-barrel outer membrane proteins. BMC Bioinformatics 5:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergogne-Berezin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bou, G., G. Cervero, M. A. Dominguez, C. Quereda, and J. Martinez-Beltran. 2000. Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: high-level carbapenem resistance in A. baumannii is not due solely to the presence of beta-lactamases. J. Clin. Microbiol. 38:3299-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark, R. B. 1996. Imipenem resistance among Acinetobacter baumannii: association with reduced expression of a 33-36 kDa outer membrane protein. J. Antimicrob. Chemother. 38:245-251. [DOI] [PubMed] [Google Scholar]
- 8.Dionisi, H. M., S. K. Checa, and A. M. Viale. 1995. Protein immunoblotting of stained gels. BioTechniques 19:348-350. [PubMed] [Google Scholar]
- 9.Fernandez-Cuenca, F., L. Martinez-Martinez, M. C. Conejo, J. A. Ayala, E. J. Perea, and A. Pascual. 2003. Relationship between beta-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 51:565-574. [DOI] [PubMed] [Google Scholar]
- 10.Gehrlein, M., H. Leying, W. Cullmann, S. Wendt, and W. Opferkuch. 1991. Imipenem resistance in Acinetobacter baumannii is due to altered penicillin-binding proteins. Chemotherapy 37:405-412. [DOI] [PubMed] [Google Scholar]
- 11.Gribun, A., Y. Nitzan, I. Pechatnikov, G. Hershkovits, and D. J. Katcoff. 2003. Molecular and structural characterization of the HMP-AB gene encoding a pore-forming protein from a clinical isolate of Acinetobacter baumannii. Curr. Microbiol. 47:434-443. [DOI] [PubMed] [Google Scholar]
- 12.Hames, B. D. 1982. An introduction to polyacrylamide gel electrophoresis, p. 65-70. In B. D. Hames and D. Rickwood (ed.), Gel electrophoresis of proteins: a practical approach. IRL Press, Oxford, United Kingdom.
- 13.Hancock, R. E., and F. S. Brinkman. 2002. Function of Pseudomonas porins in uptake and efflux. Annu. Rev. Microbiol. 56:17-38. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez-Alles, S., V. J. Benedi, L. Martinez-Martinez, A. Pascual, A. Aguilar, J. M. Tomas, and S. Alberti. 1999. Development of resistance during antimicrobial therapy caused by insertion sequence interruption of porin genes. Antimicrob. Agents Chemother. 43:937-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen, L., R. Gupta, N. Blom, D. Devos, J. Tamames, C. Kesmir, H. Nielsen, H. Stærfeldt, K. Rapacki, C. Workman, C. Andersen, S. Knudsen, A. Krogh, A. Valencia, and S. Brunak. 2002. Prediction of human protein function from post-translational modifications and localization features. J. Mol. Biol. 319:1257-1265. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, L. J., R. Gupta, H. H. Staerfeldt, and S. Brunak. 2003. Prediction of human protein function according to gene ontology categories. Bioinformatics 19:635-642. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Limansky, A. S. 2001. Ph.D. thesis. National University of Rosario, Rosario, Argentina.
- 19.Limansky, A. S., M. A. Mussi, and A. M. Viale. 2002. Loss of a 29-kilodalton outer membrane protein in Acinetobacter baumannii is associated with imipenem resistance. J. Clin. Microbiol. 40:4776-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Limansky, A. S., M. I. Zamboni, M. C. Guardati, G. Rossignol, E. Campos, and A. M. Viale. 2004. Evaluation of phenotypic and genotypic markers for clinical strains of Acinetobacter baumannii. Medicina (Buenos Aires) 64:306-312. [PubMed] [Google Scholar]
- 21.Livermore, D. M. 2001. Of Pseudomonas, porins, pumps and carbapenems. J. Antimicrob. Chemother. 47:247-250. [DOI] [PubMed] [Google Scholar]
- 22.Livermore, D. M. 2003. The threat from the pink corner. Ann. Med. 35:226-234. [DOI] [PubMed] [Google Scholar]
- 23.Lowry, O. H., N. J. Rosenbrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 24.Lozano, F., C. Florez, F. J. Recio, F. Gamboa, J. M. Gomez-Mateas, and E. Martin. 1994. Fatal Psychrobacter immobilis infection in a patient with AIDS. AIDS 8:1189-1190. [DOI] [PubMed] [Google Scholar]
- 25.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGowan, J. E., and F. C. Tenover. 2004. Opinion—anti-infectives: confronting bacterial resistance in healthcare settings: a crucial role for microbiologists. Nat. Rev. Microbiol. 2:251-258. [DOI] [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 28.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 30.Pettersson, B., A. Kodjo, M. Ronaghi, M. Uhlen, and T. Tonjum. 1998. Phylogeny of the family Moraxellaceae by 16S rDNA sequence analysis, with special emphasis on differentiation of Moraxella species. Int. J. Syst. Bacteriol. 48(Pt 1):75-89. [DOI] [PubMed] [Google Scholar]
- 31.Pugsley, A. P. 1993. Processing and methylation of PuIG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Mol. Microbiol. 9:295-308. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sato, K., and T. Nakae. 1991. Outer membrane permeability of Acinetobacter calcoaceticus and its implication in antibiotic resistance. J. Antimicrob. Chemother. 28:35-45. [DOI] [PubMed] [Google Scholar]
- 34.Spratt, B. G. 1994. Resistance to antibiotics mediated by target alterations. Science 264:388-393. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vela, A. I., M. D. Collins, M. V. Latre, A. Mateos, M. A. Moreno, R. Hutson, L. Dominguez, and J. F. Fernandez-Garayzabal. 2003. Psychrobacter pulmonis sp. nov., isolated from the lungs of lambs. Int. J. Syst. Evol. Microbiol. 53:415-419. [DOI] [PubMed] [Google Scholar]
- 37.Wolter, D. J., N. D. Hanson, and P. D. Lister. 2004. Insertional inactivation of oprD in clinical isolates of Pseudomonas aeruginosa leading to carbapenem resistance. FEMS Microbiol. Lett. 236:137-143. [DOI] [PubMed] [Google Scholar]