The emergence of metallo-β-lactamase (MBL)-producing pathogens is an increasing therapeutic problem. These enzymes have a broad-substrate spectrum; they hydrolyze all β-lactams except for the monobactam aztreonam. At the present time, there is no clinically useful inhibitor available. Four distinct types of MBLs—IMP, VIM, SPM, and GIM enzymes—are known in Pseudomonas aeruginosa (3, 5, 9). Of the VIM MBL, 11 variants have been identified up to the present, constituting three main clusters, represented by VIM-1, VIM-2, and VIM-7 (http://www.lahey.org/Studies/). blaVIM genes are either chromosomally or plasmid located and have been described as parts of the variable region of class 1 integrons (8).
The multiresistant P. aeruginosa strain B63230 with resistance to carbapenems was investigated for the presence of MBLs. The strain was isolated in Berlin, Germany, in October 2003, from a blood culture of a 70-year-old male cancer patient during an episode of febrile neutropenia that followed a course of anticancer therapy. Antimicrobial susceptibility testing was performed using the broth microdilution method recommended by the NCCLS. The MICs of imipenem, meropenem, ceftazidime, cefepime, piperacillin, piperacillin-tazobactam, aztreonam, amikacin, gentamicin, ciprofloxacin, and levofloxacin were 128 mg/liter, ≥32 mg/liter, ≥64 mg/liter, ≥32 mg/liter, ≥128 mg/liter, ≥128 mg/liter, 8 mg/liter, 16 mg/liter, ≥64 mg/liter, ≥16 mg/liter, and 32 mg/liter, respectively. Sequential treatment with piperacillin-tazobactam and gentamicin for 4 days, meropenem-vancomycin for 1 day, ceftazidime-ciprofloxacin intravenous for 4 days, and ciprofloxacin given orally for 4 days was carried out. During the therapy with ceftazidime-ciprofloxacin intravenous, regeneration of the leukocytes was detected and the fever diminished. Considering the order of events and the susceptibility pattern of the isolate, the recovery of the patient seems to be mostly related to the end of the neutropenic episode.
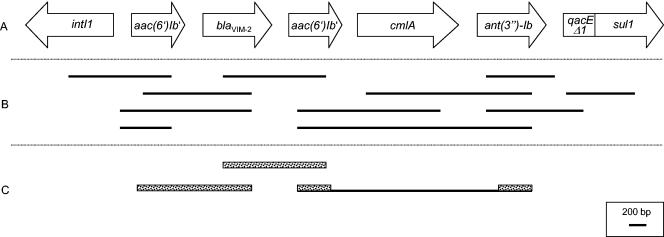
The presence of an MBL was proven using the EDTA-phenanthroline-imipenem microdilution test (6). Repeated attempts to transfer the MBL gene by conjugation, using a rifampin-resistant mutant of Escherichia coli W3110 as the recipient, and filter mating following a previously described method failed (10). PCR experiments were performed using consensus primers for the detection of blaVIM and blaIMP genes (11). Additionally, a PCR screening for the SHV, TEM, PSE, OXA-1-, OXA-2-, and OXA-10-group β-lactamase genes was conducted. blaSHV, blaTEM, and blaOXA PCRs were carried out as described previously (1, 2, 7). A PCR for the detection of blaPSE was performed with primers PSE-f (5′-AAAACAATAGCTTGCGCTAAA-3′) and PSE-r (5′-TCAGCGCGACTGTGATGTATA-3′). Positive results were obtained with the VIM and the PSE primers. To determine the blaVIM and blaPSE variants, the complete genes were sequenced on both strands. The sequences corresponded to blaVIM-2 and blaPSE-1. The genetic context of the detected blaVIM gene was further investigated by PCR mapping and partial sequencing (Fig. 1) (4). These experiments revealed that the detected blaVIM gene was part of a novel class 1 integron containing five gene cassettes. Furthermore, partial sequences of both aac(6′)-I genes and the ant(3′′)-I gene showed 100% nucleotide sequence identity with aac(6′)-Ib′ and ant(3′′)-Ib, respectively.
FIG. 1.
Characterization of the gene cassette array of a novel class 1 integron containing blaVIM-2. (A) Region characterized by PCR mapping. Genes are indicated by arrows showing their transcriptional orientations. (B) Lengths of PCR mapping products. (C) Sequenced regions of the class 1 integron indicated by structured bars.
To our knowledge, this is the first isolation of a VIM MBL in Germany. The detected enzyme VIM-2 has previously been found in several species isolated in Asian and European countries. This novel integron further elucidates the variable genetic context of blaVIM-2. Our findings are an additional indication of the emergence of MBLs in Europe and underline the need for intensified epidemiological surveillance.
Acknowledgments
This work was supported by Studienstiftung des deutschen Volkes (German National Academic Foundation) and AstraZeneca.
We thank Jutta Wagner, Freie Universität Berlin, Berlin, Germany, for providing P. aeruginosa B63230. The technical assistance of Nuria Riera is gratefully acknowledged.
REFERENCES
- 1.Arlet, G., G. Brami, D. Decre, A. Flippo, O. Gaillot, P. H. Lagrange, and A. Philippon. 1995. Molecular characterisation by PCR-restriction fragment length polymorphism of TEM β-lactamases. FEMS Microbiol. Lett. 134:203-208. [DOI] [PubMed] [Google Scholar]
- 2.Bert, F., C. Branger, and N. Lambert-Zechovsky. 2002. Identification of PSE and OXA β-lactamase genes in Pseudomonas aeruginosa using PCR-restriction fragment length polymorphism. J. Antimicrob. Chemother. 50:11-18. [DOI] [PubMed] [Google Scholar]
- 3.Castanheira, M., M. A. Toleman, R. N. Jones, F. J. Schmidt, and T. R. Walsh. 2004. Molecular characterization of a β-lactamase gene, blaGIM-1, encoding a new subclass of metallo-β-lactamase. Antimicrob. Agents Chemother. 48:4654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Livermore, D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634-640. [DOI] [PubMed] [Google Scholar]
- 6.Migliavacca, R., J. D. Docquier, C. Mugnaioli, G. Amicosante, R. Daturi, K. Lee, G. M. Rossolini, and L. Pagani. 2002. Simple microdilution test for detection of metallo-β-lactamase production in Pseudomonas aeruginosa. J. Clin. Microbiol. 40:4388-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nuesch-Inderbinen, M. T., H. Hachler, and F. H. Kayser. 1996. Detection of genes coding for extended-spectrum SHV β-lactamases in clinical isolates by a molecular genetic method, and comparison with the E test. Eur. J. Clin. Microbiol. Infect. Dis. 15:398-402. [DOI] [PubMed] [Google Scholar]
- 8.Poirel, L., T. Lambert, S. Turkoglu, E. Ronco, J. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toleman, M. A., A. M. Simm, T. A. Murphy, A. C. Gales, D. J. Biedenbach, R. N. Jones, and T. R. Walsh. 2002. Molecular characterization of SPM-1, a novel metallo-β-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance programme. J. Antimicrob. Chemother. 50:673-679. [DOI] [PubMed] [Google Scholar]
- 10.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yatsuyanagi, J., S. Saito, S. Harata, N. Suzuki, Y. Ito, K. Amano, and K. Enomoto. 2004. Class 1 integron containing metallo-β-lactamase gene blaVIM-2 in Pseudomonas aeruginosa clinical strains isolated in Japan. Antimicrob. Agents Chemother. 48:626-628. [DOI] [PMC free article] [PubMed] [Google Scholar]