Abstract
We have generated a mutant form of the OmpR regulatory protein, OmpRD55E, that is active independent of the EnvZ kinase. Notably, the pattern of OmpF and OmpC expression can be altered simply by changing the level of this mutant protein in the cell. This result supports a key prediction of the current model of porin regulation, which states that the differential regulation of OmpF and OmpC is a direct consequence of the cellular level of the active form of OmpR.
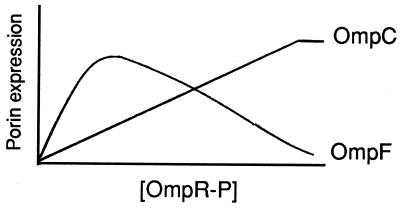
The major porin proteins of Escherichia coli K-12, OmpF and OmpC, are differentially expressed in response to several environmental signals, including changes in medium osmolarity (for recent reviews, see references 6 and 21). This expression is regulated at the transcriptional level by the two-component regulatory proteins EnvZ and OmpR. To accomplish this regulation, the sensor EnvZ is thought to control the activity of the transcriptional factor OmpR in two ways. First, EnvZ is a histidine kinase (1, 7, 12, 14) that responds to environmental stress by converting OmpR from an inactive form to an active protein via phosphorylation. Second, in the absence of environmental stress, EnvZ is able to dephosphorylate OmpR phosphate (OmpR-P) (1, 2, 13). The tension between the kinase and phosphatase activities of EnvZ therefore controls the level of OmpR-P in the cell. Based on the current model, the cellular level of OmpR-P is chiefly responsible for the differential expression of ompF and ompC (8, 21, 22, 25). A diagram of this model (adapted from reference 21) is presented in Fig. 1. In this study, we directly tested this model by generating a mutant OmpR protein that is active in the absence of the EnvZ kinase.
FIG. 1.
The current model of ompF and ompC regulation. According to this model, the differential expression of ompF and ompC is a direct consequence of the intracellular concentrations of the phosphorylated form of OmpR, OmpR-P. Under low-osmolarity conditions, low levels of OmpR-P are present in the cell, resulting in an OmpF+ OmpC− phenotype. Under high-osmolarity conditions, high levels of OmpR-P are present in the cell, resulting in an OmpF− OmpC+ phenotype. The relationship between porin expression and the concentration of OmpR-P is based on the mathematical modeling of Russo and Silhavy (22). This figure has been adapted from reference 21.
Our approach for generating the active OmpR mutant was based on studies of the two-component regulatory protein NtrC, which regulates nitrogen utilization in enteric bacteria (26). Changing the amino acid at the phosphorylation site of NtrC from aspartate to glutamate resulted in a constitutively active protein (15). Taking advantage of the homology between the two-component regulatory proteins, we used sequential PCR to generate a 1,050-bp DNA fragment which contains the analogous mutation in ompR, ompRD55E (3). Changing codon 55 of ompR from GAT to GAA results in the conversion of the conserved aspartate residue to glutamate. As a control, we also used PCR to generate the 1,050-bp DNA fragment containing the ompR+ allele. Both PCR products were cloned into pUC19, creating the plasmids pLAN801, which contains the ompR+ allele, and pLAN802, which contains the ompRD55E allele (Table 1). The DNA sequences of the PCR-generated inserts were then verified by DNA sequence analysis.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and description | Source and/or reference |
|---|---|---|
| Strains | ||
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | 4 |
| MH1160 | MC4100 ompR101 | 9 |
| LM101 | MC4100 Δ(ompR-envZ)::Kanr | 18 |
| FR247 | MC4100 ompR101 Φ(ompC′-lacZ+)10–25 envZ247 zhf37::Tn10 (λpSG10) | 22 |
| CYL304 | MC4100 ompR101 envZ247 zhf37::Tn10 | This study |
| MS10095 | JM107 zad::Tn10 pcnB80 | 17; M. Singer |
| CYL302 | MH1160 zad::Tn10 pcnB80 | This study |
| CYL303 | LM101 zad::Tn10 pcnB80 | This study |
| CYL305 | CYL304 zad::Tn10 pcnB80 | This study |
| Plasmids | ||
| pACYC177 | 5 | |
| pUC19 | 27 | |
| pLAN701 | ompR+ cloned into pACYC177a | This study |
| pLAN702 | ompRD55E cloned into pACYC177b | This study |
| pLAN801 | ompR+ cloned into pUC19a | This study |
| pLAN802 | ompRD55E cloned into pUC19b | This study |
A 1,050-bp EcoRI-HindIII PCR-generated DNA fragment containing the ompR+ sequence from position −127 to +923 from the start point of ompR transcription.
A 1,050-bp EcoRI-HindIII PCR-generated DNA fragment containing the ompR+ sequence with the codon at position 55 changed from GAT to GAA to generate the ompRD55E allele.
We first established that the OmpRD55E mutant protein was active independent of the EnvZ kinase. For this analysis, the DNA fragments containing the ompR+ and ompRD55E alleles were cloned into the ColE1-related plasmid pACYC177, creating the plasmids pLAN701 (ompR+) and pLAN702 (ompRD55E) (Table 1). These plasmids were transformed into two strains: CYL302, which contains an ompR null mutation and the envZ+ gene, and CYL303, which contains a deletion of both ompR and envZ. These strains also contain a pcnB mutation which lowers the copy number of ColE1-related plasmids by as much as 15-fold (16, 17). As shown in Table 2, the OmpRD55E mutant exhibited an OmpF+ OmpC− phenotype both in the presence and in the absence of a functional EnvZ protein. These results indicate that the phenotype of the OmpRD55E mutant is not dependent on the EnvZ kinase. In contrast, the phenotype conferred by the ompR+ allele on this plasmid is completely dependent on the EnvZ kinase. In the presence of EnvZ, the phenotype was OmpF+ OmpC+, whereas in the absence of EnvZ, the phenotype was OmpF−/+ OmpC−. Thus, unlike that conferred by wild-type OmpR, the phenotype conferred by OmpRD55E is not affected by the presence or absence of the EnvZ kinase, suggesting that this mutant protein bypasses the requirement for protein phosphorylation.
TABLE 2.
Phenotypes of the ompR alleles in the presence and absence of the EnvZ kinase
Because the OmpRD55E mutant was active in the absence of EnvZ, we were able to use this mutant to examine whether the cellular level of an active form of OmpR controls the differential expression of ompF and ompC. For these experiments, the phenotypes conferred by the ompR+ and ompRD55E alleles were determined at three different levels of expression. First, to generate the low-level expression condition, the pACYC177-derived plasmids pLAN701 (ompR+) and pLAN702 (ompRD55E) were transformed into the pcnB mutant strain (CYL303). In this strain, the chromosomal copy of ompR had been deleted and the only copy of the ompR gene was provided by the incoming plasmid. Therefore, in the pcnB mutant strain, the ompR gene should be present at a low copy number. Second, to generate the intermediate-level expression condition, pLAN701 and pLAN702 were transformed into the pcnB+ strain (LM101), which also contains the chromosomal ompR deletion. In this strain, the ompR gene should be expressed at an intermediate copy number. Finally, to generate the high-copy-number condition, the pUC19-derived plasmids pLAN801 (ompR+) and pLAN802 (ompRD55E) were transformed into the pcnB+ strain (LM101).
To confirm that the increased copy number of the ompR gene resulted in an increase in the cellular level of the OmpR protein, we performed Western immunoblot analysis. In these assays, sample sizes were adjusted such that equal amounts of total protein were present in all samples. We also varied the amount of total protein assayed in a series of independent experiments. A representative gel is shown in Fig. 2. The levels of OmpR protein under low-copy-number conditions (lanes 3 and 6) were close to those of the wild-type MC4100 strain, in which OmpR is present in a single copy (lane 1). In addition, increasing the copy number of the ompR gene led to increased levels of OmpR protein (lanes 3 to 5 and 6 to 8). These experiments confirmed that increasing the copy number of the two ompR alleles results in an increase in the amount of protein produced. Whether the increase in the total amount of wild-type OmpR also results in an increase in the amount of active OmpR (i.e., OmpR-P) in the cell cannot be determined with this assay. However, increasing the amount of OmpRD55E should correlate with an increasing amount of active OmpR in the cell. To test this hypothesis, we used these sets of conditions to examine the effect of the cellular level of active OmpR on porin expression.
FIG. 2.
Western immunoblot analysis of OmpR levels in the cells. Cells were grown overnight in LB medium without (lanes 1 and 2) or with (lanes 3 to 8) ampicillin (50 μg/ml), subcultured into 40 ml of the same medium, and grown to mid-log phase. Total cellular protein was prepared as described previously (10). One hundred micrograms of total protein from each sample was separated by SDS–10% PAGE and then transferred to a polyvinylidene difluoride membrane. Western blot analysis was then performed with rabbit anti-wild-type OmpR antiserum (1:5,000) and alkaline phosphatase-conjugated goat anti-rabbit antibody (1:10,000). The immune complexes were detected with the Vistra ECF substrate (Amersham Life Science Inc.) and visualized with a FluorImager Storm 840 system (Molecular Dynamics). Lanes: 1, MC4100; 2, LM101; 3, pLAN701 in CYL303; 4, pLAN701 in LM101; 5, pLAN801 in LM101; 6, pLAN702 in CYL303; 7, pLAN702 in LM101; 8, pLAN 802 in LM101.
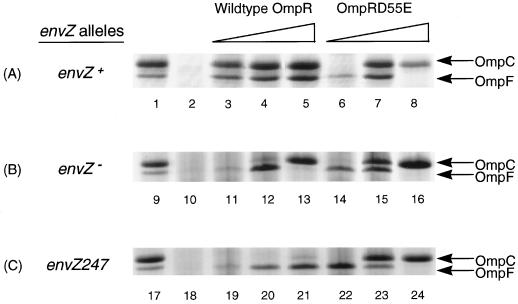
We examined the effects of the different levels of wild-type OmpR and OmpRD55E on OmpF and OmpC expression in the presence of three different envZ alleles: an envZ+ allele, an envZ null mutation, and the envZ247 mutation, which eliminates EnvZ kinase activity but does not affect its phosphatase activity. In these experiments, cells were grown in Luria broth (LB) medium to mid-log phase. The cellular envelopes were isolated and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on an 11% polyacrylamide gel containing 4 M urea. This analysis established that the pattern of OmpF and OmpC expression conferred by OmpRD55E is not dependent on phosphorylation by EnvZ or other cellular phosphate donors, such as nonpartner kinases or acetyl phosphate. In contrast, the pattern conferred by wild-type OmpR completely depends on the envZ allele that is present in the strain. The experiments that support these conclusions are presented in Fig. 3.
FIG. 3.
Effects of the level of OmpRD55E and wild-type OmpR on porin expression in the presence of three different alleles of envZ. Plasmids containing the ompR+ or ompRD55E allele were introduced into strains containing the envZ+ allele (A), an envZ null allele (B), or the envZ247 allele (C). Cells were grown overnight in LB medium with or without ampicillin (50 μg/ml), subcultured in 20 ml of the same medium, and grown to mid-log phase. The cellular envelopes were isolated as previously described (20). The samples were analyzed by electrophoresis on an SDS–11% polyacrylamide gel containing 4 M urea and identified by staining with Coomassie brilliant blue R-250 (Kodak). The positions of the outer membrane proteins OmpC and OmpF are indicated on the right. The cellular envelopes were prepared from the following strains, which are described in Table 1. Lanes: 1, MC4100; 2, MH1160; 3, pLAN701 in CYL302; 4, pLAN701 in MH1160; 5, pLAN801 in MH1160; 6, pLAN702 in CYL302; 7, pLAN702 in MH1160; 8, pLAN802 in MH1160; 9, MC4100; 10, LM101; 11, pLAN701 in CYL303; 12, pLAN701 in LM101; 13, pLAN801 in LM101; 14, pLAN702 in CYL303; 15, pLAN702 in LM101; 16, pLAN802 in LM101; 17, MC4100; 18, CYL304; 19, pLAN701 in CYL305; 20, pLAN701 in CYL304; 21, pLAN801 in CYL304; 22, pLAN702 in CYL305; 23, pLAN702 in CYL304; 24, pLAN802 in CYL304.
We first examined the phenotypes conferred by the ompR+ and ompRD55E alleles in the presence of EnvZ under the three sets of expression conditions (Fig. 3A). The ompR+ allele confers the same pattern of OmpF and OmpC expression regardless of its copy number (lanes 3 to 5). This result is consistent with those of previous studies (19, 23), which suggested that increasing the total amount of OmpR in the cell does not change the total amount of active OmpR (OmpR-P) in the cell when EnvZ is present. In contrast, the phenotype conferred by ompRD55E was markedly different under the three sets of expression conditions. At low levels of OmpRD55E, ompF expression is activated (lane 6). Then, as the level of OmpRD55E protein increases, expression of both ompF and ompC is activated (lane 7). Finally, at high levels of OmpRD55E, ompF expression is repressed and ompC expression is activated (lane 8). These observations indicate that the phenotype conferred by the ompRD55E mutation is highly dependent on the copy number and support the hypothesis that different levels of the active form of OmpR can result in the differential expression of OmpF and OmpC.
To further test this hypothesis, we also examined the phenotypes conferred by the ompR+ and ompRD55E alleles in an envZ null mutant under the three sets of expression conditions. As shown in Fig. 3B, the pattern of OmpF and OmpC expression conferred by the ompRD55E allele in the envZ null mutant matches the overall pattern observed in the presence of the envZ+ allele, supporting our supposition that the phenotype conferred by the ompRD55E allele is EnvZ independent. However, this experiment does not establish that the phenotype conferred by OmpRD55E is phosphorylation independent. Previous studies indicate that in the absence of EnvZ, the wild-type OmpR protein can be phosphorylated by other cellular phosphate donors, such as nonpartner kinases or acetyl phosphate, and is influenced by the copy number (8, 11, 22–24). At low levels of wild-type OmpR, low levels of OmpF expression are observed (lane 11). Then, as the level of wild-type OmpR increases, the expression of both ompF and ompC is activated (lane 12). Finally, at high levels of wild-type OmpR, ompF expression is repressed and ompC expression is activated (lane 13). These observations indicate that the phenotype conferred by the wild-type OmpR protein is highly dependent on the copy number in the absence of EnvZ and is similar to the phenotype conferred by the OmpRD55E mutant in the presence (Fig. 3A, lanes 6 to 8) and in the absence (Fig. 3B, lanes 14 to 16) of EnvZ. This similarity gave rise to the possibility that the phenotype conferred by OmpRD55E was still dependent on phosphorylation by other cellular phosphate donors even though it was not dependent on the kinase activity of EnvZ.
To rule out this possibility, we examined the phenotype conferred by the ompR+ and ompRD55E alleles in the envZ247 mutant under the three sets of expression conditions (Fig. 3C). The EnvZ247 mutant protein has lost its kinase activity but can still dephosphorylate OmpR-P generated either by EnvZ itself or by other cellular phosphate donors (22). As shown in Fig. 3C, the pattern of OmpF and OmpC expression conferred by the ompRD55E allele in the envZ247 mutant matches the overall pattern observed in the presence of the envZ+ and envZ null alleles, supporting our supposition that the phenotype conferred by the ompRD55E allele is not dependent on phosphorylation by EnvZ or other cellular phosphate donors. In contrast, our analysis using the ompR+ allele in the envZ247 mutant under the different sets of expression conditions illustrates the dependence of wild-type OmpR on phosphorylation by either EnvZ or other phosphate donors. The pattern of OmpF and OmpC expression in the envZ247 mutant is extremely different from the pattern observed in the presence of either the envZ+ or the envZ null allele (compare lanes 19 to 21 to lanes 3 to 5 and 11 to 13). The fact that the phenotype conferred by the ompR+ allele is different with these three envZ alleles supports the hypothesis that OmpR can be phosphorylated by other phosphate donors in the absence of EnvZ. These results also highlight the importance of EnvZ in controlling the level of OmpR-P in the cell.
In conclusion, we have isolated a mutant form of OmpR that is active independent of EnvZ and have used this mutant protein to examine the effects of different levels of active OmpR in the cell. Our results indicate that the pattern of OmpF and OmpC expression can be dramatically altered simply by changing the level of this mutant protein. Therefore, this study provides strong evidence supporting the current model, which states that the level of the active form of OmpR, OmpR-P, is responsible for the differential regulation of ompF and ompC.
Acknowledgments
We thank Miaw-Sheue Tsai and Jinling Li for assistance in the initial stages of this study.
This research was supported in part by Public Health Service grant GM48591 to M.M.I. from the National Institutes of Health.
REFERENCES
- 1.Aiba H, Mizuno T, Mizushima S. Transfer of phosphoryl group between two regulatory proteins involved in osmoregulatory expression of the ompF and ompC genes in Escherichia coli. J Biol Chem. 1989;264:8563–8567. [PubMed] [Google Scholar]
- 2.Aiba H, Nakasai F, Mizushima S, Mizuno T. Evidence for the physiological importance of the phosphotransfer between the two regulatory components, EnvZ and OmpR, in osmoregulation in Escherichia coli. J Biol Chem. 1989;264:14090–14094. [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1989. [Google Scholar]
- 4.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 5.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egger L A, Park H, Inouye M. Signal transduction via histidyl-aspartyl phosphorelay. Genes Cells. 1997;2:167–184. doi: 10.1046/j.1365-2443.1997.d01-311.x. [DOI] [PubMed] [Google Scholar]
- 7.Forst S, Delgado J, Inouye M. Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:6052–6056. doi: 10.1073/pnas.86.16.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forst S, Delgado J, Rampersaud A, Inouye M. In vivo phosphorylation of OmpR, the transcription activator of the ompF and ompC genes in Escherichia coli. J Bacteriol. 1990;172:3473–3477. doi: 10.1128/jb.172.6.3473-3477.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall M, Silhavy T J. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K-12. J Mol Biol. 1981;146:23–43. doi: 10.1016/0022-2836(81)90364-8. [DOI] [PubMed] [Google Scholar]
- 10.Hicks K A, Grossman A D. Altering the level and regulation of the major sigma subunit of RNA polymerase affects gene expression and development in Bacillus subtilis. Mol Microbiol. 1996;20:201–212. doi: 10.1111/j.1365-2958.1996.tb02501.x. [DOI] [PubMed] [Google Scholar]
- 11.Hsing W, Silhavy T J. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J Bacteriol. 1997;179:3729–3735. doi: 10.1128/jb.179.11.3729-3735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Igo M M, Ninfa A J, Silhavy T J. A bacterial environmental sensor that functions as a protein kinase and stimulates transcriptional activation. Genes Dev. 1989;3:598–605. doi: 10.1101/gad.3.5.598. [DOI] [PubMed] [Google Scholar]
- 13.Igo M M, Ninfa A J, Stock J B, Silhavy T J. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 1989;3:1725–1734. doi: 10.1101/gad.3.11.1725. [DOI] [PubMed] [Google Scholar]
- 14.Igo M M, Silhavy T J. EnvZ, a transmembrane environmental sensor of Escherichia coli K-12, is phosphorylated in vitro. J Bacteriol. 1988;170:5971–5973. doi: 10.1128/jb.170.12.5971-5973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klose K E, Weiss D S, Kustu S. Glutamate at the site of phosphorylation of nitrogen-regulatory protein NtrC mimics aspartyl-phosphate and activates the protein. J Mol Biol. 1993;232:67–78. doi: 10.1006/jmbi.1993.1370. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Parkinson J S. Genetics and sequence analysis of the pcnB locus, an Escherichia coli gene involved in plasmid copy number control. J Bacteriol. 1989;171:1254–1261. doi: 10.1128/jb.171.3.1254-1261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopilato J, Bortner S, Beckwith J. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 18.McDowell, L., and M. M. Igo. Unpublished data.
- 19.Mizuno T, Wurtzel E T, Inouye M. Cloning of the regulatory genes (ompR and envZ) for the matrix proteins of the Escherichia coli outer membrane. J Bacteriol. 1982;150:1462–1466. doi: 10.1128/jb.150.3.1462-1466.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morona R, Reeves P. The tolC locus of Escherichia coli affects the expression of three major outer membrane proteins. J Bacteriol. 1982;150:1016–1023. doi: 10.1128/jb.150.3.1016-1023.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pratt L A, Silhavy T J. Porin regulon of Escherichia coli. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 105–127. [Google Scholar]
- 22.Russo F D, Silhavy T J. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J Mol Biol. 1991;222:567–580. doi: 10.1016/0022-2836(91)90497-t. [DOI] [PubMed] [Google Scholar]
- 23.Russo F D, Silhavy T J. The essential tension: opposed reactions in bacterial two-component regulatory systems. Trends Microbiol. 1993;1:306–310. doi: 10.1016/0966-842x(93)90007-e. [DOI] [PubMed] [Google Scholar]
- 24.Skarphol K, Waukau J, Forst S A. Role of His243 in the phosphatase activity of EnvZ in Escherichia coli. J Bacteriol. 1997;179:1413–1416. doi: 10.1128/jb.179.4.1413-1416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slauch J M, Silhavy T J. Genetic analysis of the switch that controls porin gene expression in Escherichia coli K-12. J Mol Biol. 1989;210:281–292. doi: 10.1016/0022-2836(89)90330-6. . (Erratum, 212:429, 1990.) [DOI] [PubMed] [Google Scholar]
- 26.Weiss D S, Klose K E, Hoover T R, North A K, Porter S C, Wedel A B, Kustu S. Prokaryotic transcriptional enhancers. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 667–694. [Google Scholar]
- 27.Yanisch-Perron J C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]