Abstract
Copy number variations at 7q11.23 cause neurodevelopmental disorders with shared and opposite manifestations. Deletion causes Williams-Beuren syndrome featuring hypersociability, while duplication causes 7q11.23 microduplication syndrome (7Dup), frequently exhibiting autism spectrum disorder (ASD). Converging evidence indicates GTF2I as key mediator of the cognitive-behavioral phenotypes, yet its role in cortical development and behavioral hallmarks remains largely unknown. We integrated proteomic and transcriptomic profiling of patient-derived cortical organoids, including longitudinally at single-cell resolution, to dissect 7q11.23 dosage–dependent and GTF2I-specific disease mechanisms. We observed dosage-dependent impaired dynamics of neural progenitor proliferation, transcriptional imbalances, and highly specific alterations in neuronal output, leading to precocious excitatory neuron production in 7Dup, which was rescued by restoring physiological GTF2I levels. Transgenic mice with Gtf2i duplication recapitulated progenitor proliferation and neuronal differentiation defects alongside ASD-like behaviors. Consistently, inhibition of lysine demethylase 1 (LSD1), a GTF2I effector, was sufficient to rescue ASD-like phenotypes in transgenic mice, establishing GTF2I-LSD1 axis as a molecular pathway amenable to therapeutic intervention in ASD.
Omics dissection of cortical development in 7q11.23 reveals defects in neuronal maturation and social behavior linked to GTF2I.
INTRODUCTION
Copy number variations (CNVs) at the 7q11.23 locus spanning 26 to 28 genes cause two rare neurodevelopmental disorders featuring shared and opposite multisystemic phenotypes. Heterozygous deletion causes Williams-Beuren syndrome [WBS; OMIM (Online Mendelian Inheritance in Man) 194050], featuring intellectual disability coupled with hypersociability, anxiety, and comparatively well-preserved language abilities (1). 7q11.23 heterozygous duplication (7Dup; OMIM 609757) shares intellectual disability and anxiety, with a contrasting impairment in expressive language and a high incidence of autism spectrum disorder (ASD) (2, 3). The multifaceted combination of shared and opposite clinical phenotypes, including for some in mirroring ways, suggests that the underlying molecular mechanisms might likewise be dichotomous or similar under both conditions. Hence, through their unique combination of neuropsychiatric manifestations and genetic lesions, 7q11.23 CNV syndromes offer unprecedented opportunities to dissect gene dosage-vulnerable circuits involved in social behavior and language competence (4). The relevance of gene dosage–phenotype associations is highlighted in recent studies using large human samples linking genome dosage sensitive regions with specific disorders, revealing CNVs properties and identifying thousands of highly dosage sensitive genes (5).
Convergent evidence from mouse and human studies, including from WBS individuals carrying atypical deletions, has pointed to general transcription factor I (GTF2I) as a key driver of the 7q11.23 CNV cognitive-behavioral manifestations (6, 7); however, its mechanistic underpinnings and specific roles during cortical development and in behavioral alterations remain elusive. GTF2I is a signal-induced transcription factor, and its target genes are involved in a variety of neuronal functions including axon guidance, calcium signaling, as well as cell cycle and maturation of gamma-aminobutyric acid (GABA) interneurons (INs) (8–10). Previously, we defined the GTF2I dose-dependent transcriptional dysregulation at the pluripotent state and traced its amplification upon differentiation into neural progenitors. Critically, we found that GTF2I represses transcription by binding the histone lysine demethylase 1 (LSD1 or KDM1A), whereas chemical and genetic LSD1 inhibition rescued the aberrant transcriptional repression brought about by increased GTF2I dosage in 7Dup lineages, affecting expression of key genes involved in intellectual disability and ASD and underlining the potential of LSD1 inhibition for therapeutic intervention (11).
The use of patient-derived pluripotent stem cell models, in particular, brain organoids in combination with single-cell transcriptomics, is a powerful platform to model neuropsychiatric disorders with unprecedented accuracy (12–14). Its application to study processes that reach unparalleled complexity in humans, such as the genetic circuits regulating cortical development and behavior, holds enormous potential to uncover hubs of genetic regulation susceptible to impairment during development. Testifying to the potential of these approaches, we and others recently showed that ASD-risk genes’ pathogenic impact converge on alterations of neuronal differentiation dynamics and neurodevelopmental trajectories (13, 15).
Here, we undertook a systematic dissection of the impact of 7q11.23 CNVs in cortical development, juxtaposing WBS and 7Dup transcriptional and phenotypic landscapes across multiple individuals, key developmental time windows, and experimental models. Our results, from integrating cortical organoids (COs) with single-cell transcriptomics in 13 induced pluripotent stem cell (iPSC) lines, revealed changes in the dynamics of progenitor proliferation and neuronal maturation as a result of 7q11.23 gene dosage imbalances. Changes in GTF2I dosage alone, either in COs or in transgenic mice, were sufficient to phenocopy key neuronal differentiation alterations observed in COs and caused ASD-like behaviors that could be rescued by inhibition of the GTF2I effector, LSD1.
RESULTS
7q11.23 CNVs cause progenitor proliferation and differentiation imbalances in COs
We generated patient-derived COs that recapitulate early to mid-fetal corticogenesis (16–18) from 13 iPSC lines reprogrammed from eight affected individuals and five healthy controls [four WBS, four 7Dup, and five CTL; Fig. 1A], including, respectively, one clone per individual following the design that we previously benchmarked (19), to avoid artificial inflation of spurious differential expression due to donors’ genetic backgrounds. Array comparative genomic hybridization confirmed that the 7q11.23 CNV is preserved upon reprogramming (fig. S1A). Likewise, the transcriptional readout of the locus dosage imbalance was confirmed by unattended clustering of the expression levels of the 7q11.23 genes in 7Dup, WBS, and CTL COs at days 18, 50, and 100 of differentiation (fig. S1, B and C).
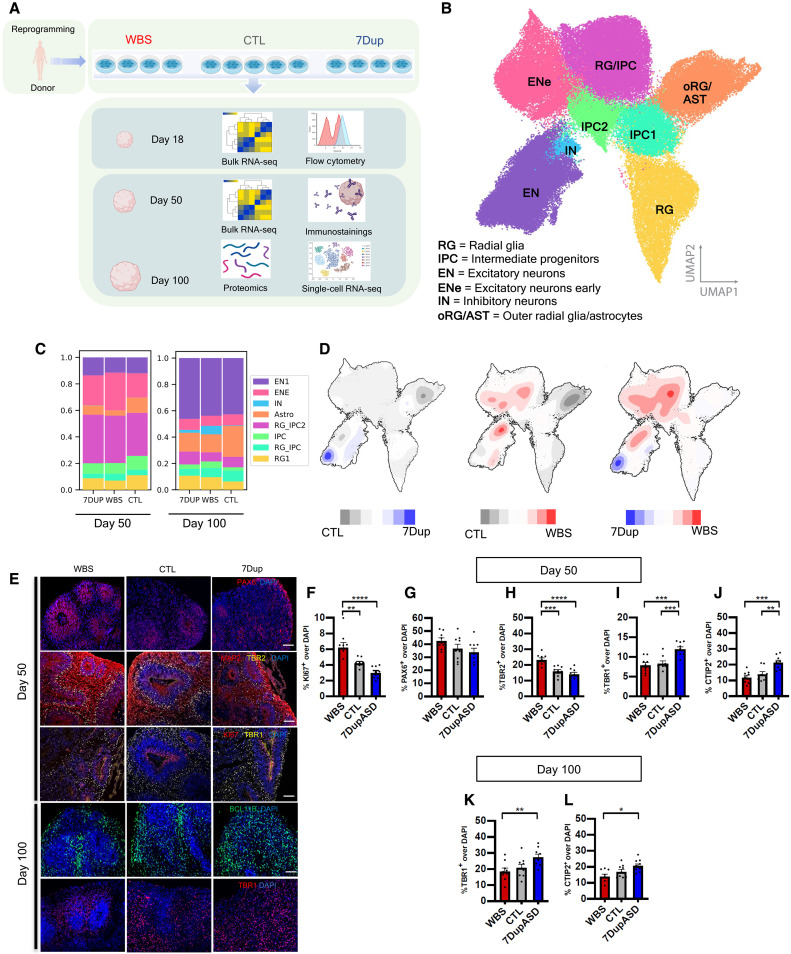
Fig. 1. 7q11.23 CNVs cause imbalances in cortical cell population composition in patient-derived COs.
(A) Experimental design: 13 reprogrammed iPSC lines from 13 different donors differentiated into COs and profiled for bulk transcriptomics, single-cell RNA sequencing (RNA-seq) and whole proteome at the specified time points. (B) Uniform manifold approximation and projection (UMAP) from (n = 97,108) single-cell transcriptomes. Leiden clustering algorithm identified eight distinct cell clusters (RG, RG/IPC, IPC1, IPC2, EN1, EN2, IN, and oRG/AST), composed by the following populations: RG, IPC, EN, IN, and oRG/AST. (C) Proportion of cells from each genotype in each cell cluster in COs at 50 and 100 days (three pooled individual organoids per genotype). (D) Density plots illustrating differences in (max-min) cell abundance in UMAP discriminated by condition; red indicates high abundance in WBS, blue indicates high abundance in 7Dup, and gray indicates high abundance in control. (E) Representative widefield fluorescence images from days 50 to 100 COs across genotypes (WBS/CTL/7Dup), immunostained with markers for different cortical populations: PAX6 (apical radial glia), microtubule-associated protein 2 (MAP2) (postmitotic neurons), TBR2 (intermediate progenitors), TBR1 (layer VI neurons), Ki67 (cycling cells), BCL11B (layer V toVI neurons). Scale bars, 50 um. (F to J) Quantification of the percentage cells positive for different population markers relative to total nuclei [4′,6-diamidino-2-phenylindole (DAPI)] at day 50 of differentiation. (K and L) Quantification of the percentage of layer V and VI neuron markers relative to total nuclei (DAPI) at day 100 of differentiation. Each data point is an individual section from five organoids per line, three independent human iPSC lines per genotype. All data are shown as means ± SEM (27 sections from three COs per cell line, three cell lines per genotype), one-way analysis of variance (ANOVA), post hoc Tukey test; *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001.
To investigate the impact of 7q11.23 CNVs on corticogenesis, we performed droplet-based transcriptional profiling of ~100.000 single cells from patient-derived COs at two time points (days 50 and 100) (Fig. 1A) that we previously showed to have a selective enrichment in ventricular radial glia intermediate progenitor cells (IPCs; day 50), excitatory neurons (ENs), and outer radial glia (oRG; day 100), a progenitor population essential for cortical expansion in humans (20–22). Leiden cluster annotation identified eight cell populations encompassing radial glial (RG) cells, IPC, oRG/astrocytes (oRG/ASTs), and ENs (Fig. 1B). Population annotation was achieved by tracing the expression pattern of known cortical population markers (fig. S2A), unattended determination of cluster-specific highly expressed genes (fig. S2B), and overlap with public fetal cortical single-cell datasets (fig. S2, C and D). We confirmed that control lines showed reproducible proportions of all identified populations (fig. S2E), as previously shown for this CO protocol (18, 23), and proceeded to ascertain that each line independently reproduced stereotypical developmental trajectories from radial glia progenitors to mature ENs and ASTs (fig. S2F).
We first looked at the distribution of cell cycle–related gene signatures between WBS and 7Dup COs at day 50, revealing reciprocally contrasting proportions of cells in S + G2M phases (WBS % = 33, CTL % = 29, and 7Dup % = 25) (fig. S3A). To evaluate whether this cycling imbalance was present at the progenitor stage, we dissociated and stained day 18 COs with propidium iodine (PI+) and used a Gaussian estimate of the percentage’s distribution across cell cycle phases by flow cytometry. We confirmed the presence of an early increase in S + G2M in WBS and a decrease in 7Dup (fig. S2B). Consistently, immunofluorescent staining with the cell cycling marker KI67 at day 50 showed an increase in active proliferation in WBS and reduction in 7Dup (Fig. 1, E and F), indicating the presence of a reciprocally opposite imbalance in cell proliferation across 7q11.23 genotypes during early corticogenesis.
We then examined the proportion of cortical populations across conditions through days 50 to 100, finding multiple changes in cluster size at both time points. Specifically, we found a twofold reduction in the number of oRG/AST cells, a 1.5-fold increase in the total number of ENs, and the appearance of a population rich in ventral telencephalon and IN markers distal-less homeobox 1/2 (DLX1/2) at day 100 in both WBS and 7Dup COs (Fig. 1C). Condition-specific changes included WBS-specific increase in RG/IPCs at day 50. While there were no significant differences at cluster level between WBS and 7Dup EN percentages, detailed mapping of cell abundances within clusters using a density plot revealed particular EN subpopulations specifically enriched under each condition in the area corresponding to the EN (Fig. 1D).
To quantitatively verify the variable abundance of progenitors and neuronal populations of the excitatory lineage in WBS and 7Dup, we immunostained and quantified population markers of early to mid-corticogenesis. We found a consistent increase in the proportion of paired-box 6 (PAX6)+ progenitors and T-box brain protein 2 (TBR2)+ intermediate progenitors in WBS compared with 7Dup COs (Fig. 1, G and H), while, conversely, postmitotic neurons positive for layer V to VI markers (TBR1+. BCL11B+) were increased in 7Dup COs compared with WBS (Fig. 1, G to I). These results establish that gene dosage imbalances within the 7q11.23 locus impact corticogenesis by altering the dynamics of progenitor proliferation and the subsequent output of neuronal populations through a combination of shared and reciprocally mirroring effects.
Transcriptomic and proteomic profiling of 7q11.23 CNV COs uncover alterations in proliferative and neurogenic programs in WBS and 7Dup
To identify the transcriptional dysregulation caused by 7q11.23 CNVs, we performed bulk transcriptomic profiling of COs at days 18, 50, and 100, interrogating the effect of the opposite gene dosages in developmental gene expression networks through a differential expression analysis (DEA) comparing 7Dup and WBS COs. First, we focused on the transcriptional changes that persisted throughout differentiation by differential analysis of the complete CO cohort, specifying the differentiation stage as covariable in the model. This analysis revealed 164 differentially expressed genes (DEGs) [false discovery rate (FDR) < 0.1, log2 fold change (log2FC) > 1 as absolute value] (Fig. 2A). 7Dup COs showed an up-regulation of the forebrain development regulator (FOXG1), as well as glutamatergic lineage drivers, including (NEUROD6) and (TBR1) (Fig. 2B), which regulate neuronal fate specification and differentiation during cortical development and are recurrently disrupted in ASD (12, 24, 25). In addition, 7Dup COs showed an up-regulation of members of the protocadherin family (i.e., PCDHGA3, PCDHGA4, PCDHB5, PCDHB15, and PCDHGB3) (Fig. 2B) known to regulate neuronal connectivity, extracellular matrix (ECM) remodeling, and synapse organization (26). Conversely, WBS organoids showed an up-regulation of transcription factors involved in anterior-lateral brain patterning (PHOX2A and PHOX2B) and the G protein–coupled receptor (GPR50), known for its association with bipolar affective disorder, major depressive disorder, and schizophrenia (27).
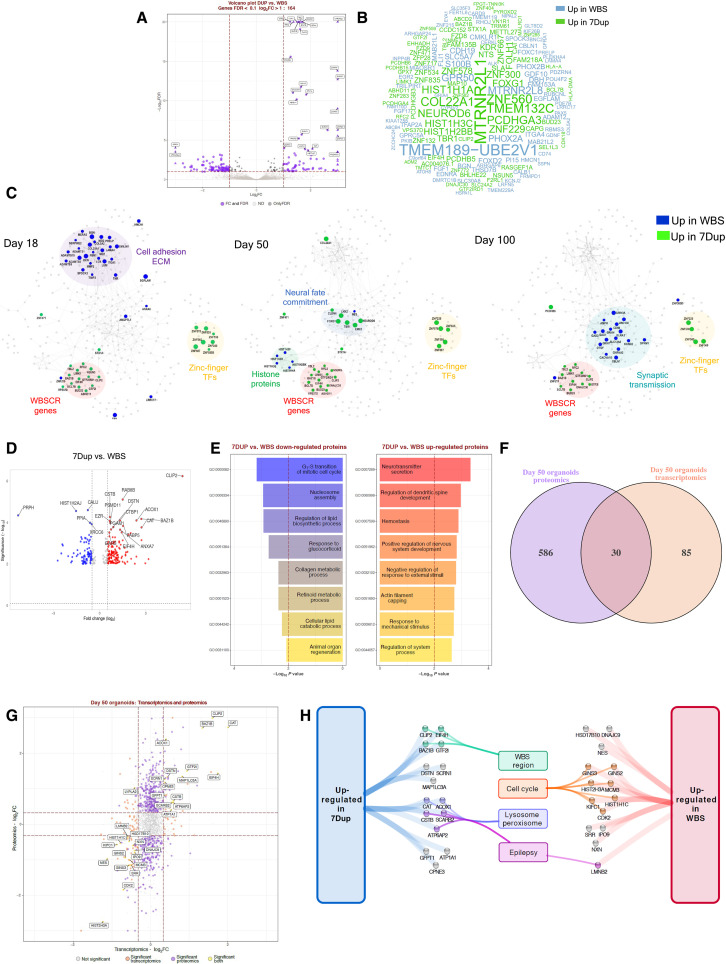
Fig. 2. Transcriptomic and proteomic profiling of 7q11.23 CNV COs reveal imbalances in proliferative vis-a-vis neurogenic programs.
(A) DEA for 7Dup versus WBS COs. Plots report tested genes as −log10 FDR and log2FC. Purple indicates significantly modulated genes (FDR < 0.1 and absolute log2FC > 1), black indicates genes passing FDR threshold, and gray indicates not significant. Top 30 genes are highlighted. (B) Word cloud reporting DEGs gene symbol; word size represents fold change magnitude, green represents up-regulated in 7Dup, and blue represents up-regulated in WBS. (C) STRING-based network reconstructed for modulated genes (FDR < 0.1, log2FC > 1) in at least one time point by stage-wise DEA. Each visualization highlights the most relevant findings for the reported time point. Node size and color: magnitude and direction of log2FC at the specified stage, green denotes up-regulation in 7Dup, and blue denotes up-regulation in WBS. Functionally relevant regions are highlighted. (D) Differentially expressed proteins (DEP) comparing 7Dup and WBS COs, (one CO per line and three lines per genotype in duplicate). DEP cutoff: FDR < 0.01 and FC > 1. (E) Top 8 enriched biological processes for up-regulated (P < 0.05, FC > 2) and down-regulated (P < 0.05, FC < −2) proteins from 7Dup versus WBS at day 50. (F) Overlap between transcriptomics and proteomics in 7Dup versus WBS at day 50. Features are selected as modulated with a P < 0.05 and absolute log2FC > log2(1.25). (G) Relationship of the fold change in 7Dup versus WBS in the proteome (y axis) and transcriptome (x axis) for genes tested by both approaches. Yellow indicates features modulated in both modalities; they are all modulated in the same direction except LYPLA2. Purple represents DEPs absent among DEGs, and orange represents DEGs absent among DEPs. (H) Manually curated functional annotation for genes modulated in the same direction at the transcript and protein level.
To identify the time dependency of these transcriptional changes, we applied DEA separately at each CO differentiation stage and used the genes differentially expressed at least at one stage (FDR < 0.1 and log2FC > 1 as absolute value) as input for the generation of stage-wise gene networks (fig. S4A). As expected, several genes from the 7q11.23 locus were consistently up-regulated in 7Dup at all stages (Fig. 2C). Other genes consistently up-regulated in 7Dup in more than one stage were a group of poorly characterized zinc finger transcription factors (e.g., ZNF229, ZNF300, ZNF560, and ZNF578). Focusing at each differentiation stage separately, the most outstanding observations included the following: (i) at day 18, a modulation of cell adhesion and ECM components, mostly up-regulated in WBS COs. Notably, several of these genes (e.g., COL12A, PCOLCE, BGN, NID2, and EMILI1) belong to a fetal cortex gene coexpression module characterized by nonmonotonic expression pattern during cortex development and functionally associated with ECM (16). (ii) At day 50, an up-regulation in WBS COs of histone protein genes and RG marker Nestin, consistent with the observed increase in proliferative cells in WBSs COs. This was accompanied at the same stage with the up-regulation in 7Dup COs of dorsal forebrain commitment drivers, including FOXG1, LHX2, NEUROD6, and TBR1, whose trend of modulation was conserved in fold change also at day 100 (fig. S4A). (iii) The up-regulation at day 100 of genes involved in synaptic transmission in WBS COs (e.g., GRIK1, GRIA4, and GRIN3A). Noteworthy, among the top up-regulated genes in 7Dup COs at day 50 was COL22A1, a recently identified marker of a subset of layer III glutamatergic neurons, linked with human-specific evolutionary protracted developmental programs (28, 29).
To pinpoint potential convergences between the transcriptional programs affected by 7q11.23 CNV and the causative genes of other neurodevelopmental disorders, we performed an overlap analysis of DEGs with public gene databases for neurodevelopmental syndromes [Simons Foundation Autism Research Initiative (SFARI) and AutismKB] and genetic diseases (OMIM, Orphanet, and Decipher). This analysis retrieved significant overlaps for several gene sets, revealing a notable convergence between the genes transcriptionally modulated by 7q11.23 CNV and disease-causing genes, among which ASD and other neurodevelopmental disorders were prominently featured (fig. S4B).
Last, reasoning that the neuronal imbalances observed at day 100 represented the output of earlier dysregulations during the mid-stage development, we focused proteome profiling on day 50 to uncover the upstream CNV-dependent protein expression changes that could account for later population imbalances. Analysis of differentially expressed proteins between 7Dup and WBS confirmed an up-regulation of proteins related to cell cycle and progenitor proliferation categories in WBS and, conversely, an up-regulation of neuronal maturation proteins in 7Dup (Fig. 2, D and E). Transcriptome/proteome overlap highlighted 30 common dysregulated genes/proteins (Fig. 2, F and G), including a subset of the 7q11.23 members (higher in 7Dup) and a set involved in cell cycle control up-regulated in WBS compared to 7Dup (Fig. 2H), further exposing the molecular underpinnings of the reciprocal imbalance between proliferative pathways in WBS and neurogenic programs in 7Dup.
7Dup COs exhibit anticipated production of ENs
To elucidate in detail the impact of 7q11.23 gene dosage on neuronal differentiation, we performed an in-depth inspection of the single-cell data, focusing on the composition of the postmitotic neuron cluster (EN), where we identified major differences between patient and control COs (Fig. 1D). To maximize the sensitivity of differential neurodevelopmental trajectory analysis between conditions, we used a force-directed graph layout (30) and applied Leiden clustering to identify the subpopulations composing the EN cluster (Fig. 3A). Benchmarking of the EN cluster against transcriptome from the human fetal cortex (31) revealed the presence of transcriptional signatures of early neurons, layer V to VI, layer II to III, prefrontal, occipital, and IN progenitors, highlighting the neuronal diversity that is recapitulated in COs (Fig. 3A). These subpopulations were further confirmed by observing the expression of canonical cortical plate markers (fig. S6A). A close inspection of the distribution of cells in this neuronal compartment at day 50 revealed marked differences between genotypes. Specifically, while cells from both WBS and CTL COs broadly overlap in areas with differentiating populations of excitatory and inhibitory neuronal lineages, 7Dup cells already occupy lower and upper layer identities at day 50, suggesting an anticipated acquisition of these fates (Fig. 3B).
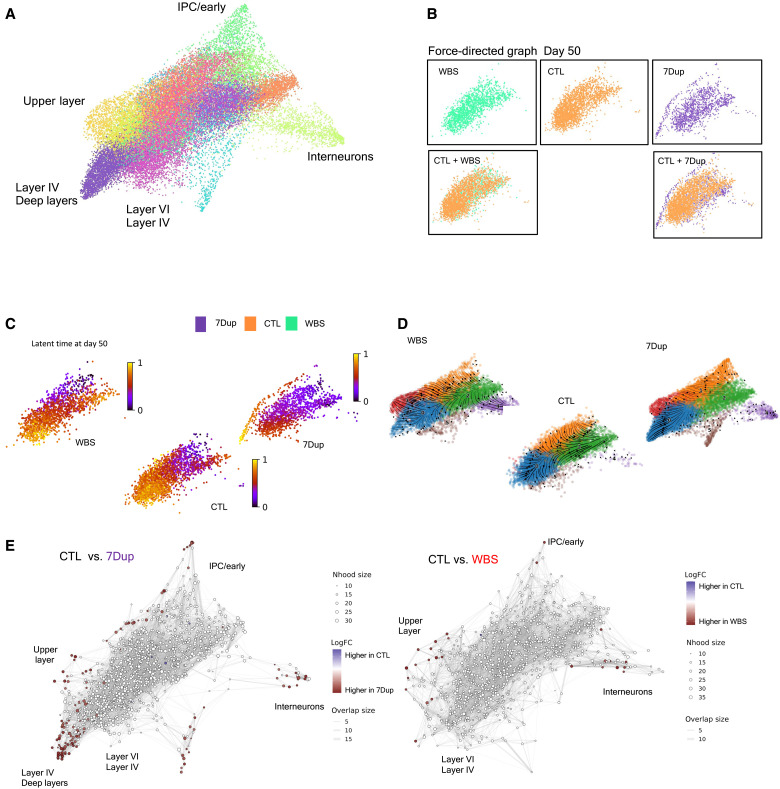
Fig. 3. Gene dosage imbalances at 7q11.23 accelerate neuronal differentiation in 7Dup COs.
(A) Force-directed graph drawing of only the EN2/IN populations (recalculating neighbors and distances subselecting only EN2/IN populations), divided in subclusters and manually annotated. This dimensionality reduction highlights the most differentiated cell population (the pointed extremity of the graph, manually annotated). (B) Differences in cell distribution in force-directed graph for day 50 organoids. WBS and CTL occupy the same space, while 7Dup has more mature cells. (C) Latent time calculated only on the EN2 and IN populations at day 50. The starting cell is identified considering the total population. (D) Representation of the EN2 and IN populations developmental path in the three genotypes in force-directed graph, overimposed with RNA velocity. (E) Differential abundance of cells in different areas of the force-directed graph. The nodes are neighborhoods of cells aggregated based on RNA expression similarity, while graph edges depict the number of cells connected between adjacent neighborhoods. White nodes are neighborhoods that are not detected as differentially abundant (FDR > 1%), red dots, on the other end, are significant (FDR < 1%).
To inquire whether the increase in postmitotic neurons in 7Dup organoids was indeed the result of an accelerated neuronal differentiation program, we applied to the postmitotic neuron cluster the CellRank algorithm (32) that combines three sources of information (pseudo-time, splicing, and transcriptomic distance) to determine the probability of each neurogenic trajectory for each genotype and calculated the latent time, a pseudo-time representation that highlights the developmental trajectory. This analysis revealed a selective bias of 7Dup COs toward a neuronal cluster enriched in postmitotic neurons, which were present already at day 50 in 7Dup but absent in WBS and CTL (Fig. 3C); this accelerated differentiation bias of 7Dup COs was further supported by the results of velocyto algorithm (33) (Fig. 3D), where the distribution of velocities (arrows) in the 7Dup versus WBS force-directed graph maps highlights a steady state of already differentiated cells in 7Dup compared to WBS and CTL. We then examined the differential abundance of cells in the force-directed graph representation (Fig. 3E). The analysis uncovered a higher abundance of 7Dup cells in nodes representing mature neuronal populations. Collectively, the developmental trajectory analysis, velocyto, and the density of cells revealed that cells from both WBS and 7Dup COs are enriched in clusters representing more differentiated neurons compared with CTL, including deeper and upper layer neurons and INs. Likewise, the exclusive presence of 7Dup cells in areas occupied by more mature neurons (i.e., extremities of the graph) at day 50 indicates an anticipated neuronal differentiation in 7Dup COs (Fig. 3, C to F).
Reduction of GTF2I levels rescues neuronal differentiation acceleration in 7Dup COs
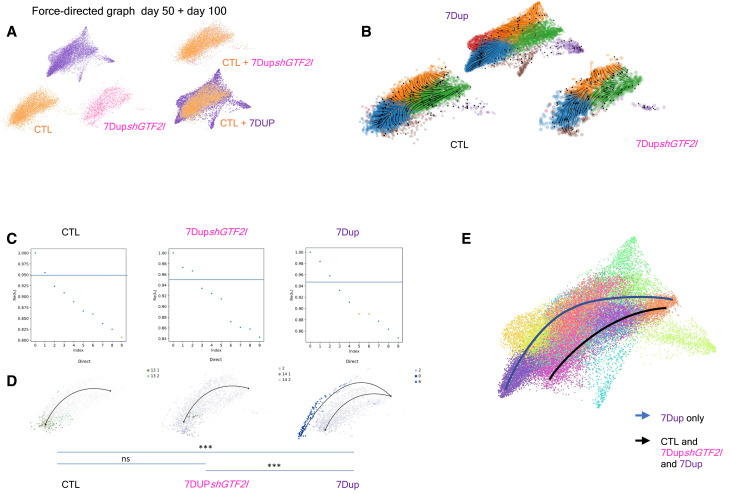
Given the pivotal role that GTF2I plays in the cognitive-behavioral manifestations in 7q11.23 CNV syndromes (6, 11, 34, 35) and its abundant expression in early and postmitotic neurons compared to its homologues GTF2RD1 and GTF2IRD2 in COs and primary fetal cortex (fig. S5, A to E), we interrogated the effect of down-regulating GTF2I in 7Dup COs. To this end, we transduced two different 7Dup iPSC lines with a lentivirus constitutively expressing short hairpin RNA (shRNA) against GTF2I and differentiated COs to 100 days. We tested two different hairpins against GTF2I (shA and shB) and chose shB as it showed knockdown effects comparable to 7q11.23 control dosage at the protein level (fig. S5, F and G). COs generated from 7Dup iPSC lines constitutively expressing an shRNA against GTF2I (7DupshGTF2I) showed reduced presence of mature neuronal populations, with a population composition resembling closely the one of control COs (Fig. 4A). Likewise, RNA velocity confirmed that GTF2I down-regulation in 7Dup COs leads to differentiation trajectories matching those of control COs (Fig. 4B), indicating that GTF2I dosage is critical for the accelerated production of postmitotic ENs in 7Dup individuals. We then once again applied CellRank to determine the probability of each neurogenic trajectory for each condition. First, we identified all possible end points for 7Dup, 7DupshGTF2I, and control and then plotted the distribution of trajectory end points with the attending probabilities (P > 0.95, P < 0.05) (Fig. 4, C and D, and fig. S6, A and B). This approach revealed a robust bias of 7Dup COs to produce neurons enriched in layer IV and deep layers markers at day 50. This bias was absent in COs with a constitutive knockdown of GTF2I (Fig. 4, C to E), thus confirming the central role of GTF2I in accelerated neuronal production in 7Dup COs.
Fig. 4. GTF2I duplication drives precocious neuronal differentiation in 7Dup COs.
(A) Force-directed graph stratified for genotype (CTL, 7Dup, and 7DupshGTF2I) from left to right, first line at day 50. The second line shows the differences in cell distribution among genotypes, highlighting areas in the EN2 population that are unique of 7Dup and WBS, overimposed on the right. (B) Eigenvalue of the transition matrix stratified by genotype (CTL, 7Dup, and 7DupshGTF2I). In the plot, each dot represents a potential differentiation trajectory, with y axis quantifying the likelihood of solving the transition matrix (probability to be a proper differentiation trajectory); the blue line identifies the imposed probability threshold (0.95) applied to all the conditions. (C) Force-directed graph over impose with RNA velocity, depicting the developmental path of different genotypes (CTL, 7Dup, and 7DupshGTF2I). (D) Force-directed graph with color code as the probability of a cell to go to a certain endpoint, defined by the graph in (C); ***P < 0.001. Given the clustering used, we have multiple clusters that identify the same populations. The significance is defined as the probability of going from the beginning, identified as the population with minimum of total pseudo-time, to the end points in 7Dup. ns, not significant. (E) Force-directed graph with a visual representation of the differentiation trajectory and populations that are involved in the process, only 7Dup passes in regions showing upper layer markers.
Increased Gtf2i dosage drives accelerated neuronal differentiation and ASD-related behaviors in the mouse
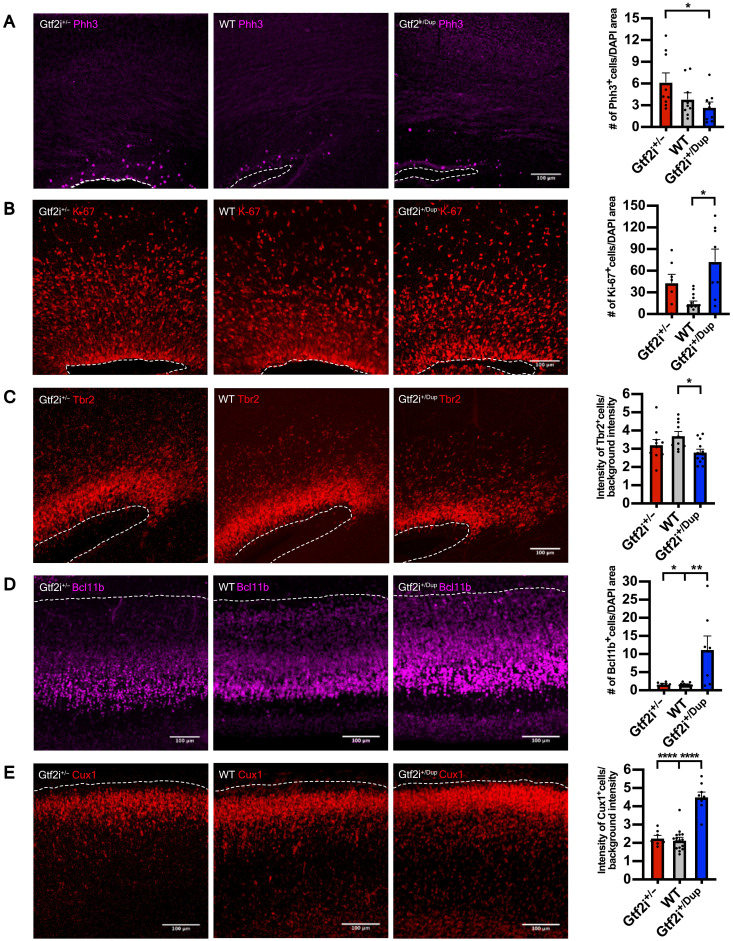
Having defined GTF2I dosage as the critical effector of the altered neurodevelopmental trajectories uncovered in 7Dup, we asked whether this specificity could be leveraged in vivo to rescue 7Dup-associated cognitive-behavioral manifestations. To this end, we resorted to murine models specifically recapitulating the GTF2I dosage of the two syndromes, i.e., carrying either one (Gtf2i+/−) or three (Gtf2i+/Dup) Gtf2i copies, alongside their wild-type (WT) controls, with the physiologic complement of two alleles (36). First, we probed the extent to which these murine models featured equivalent alterations to those observed in human corticogenesis, in terms of GTF2I dosage–dependent alterations of the proliferative and mature neuronal compartments. We thus quantified the expression of proliferative and neuronal markers in the cortices of transgenic mouse embryos at embryonic day 17.5 (E17.5). We found increased phospho-histone H3 (Phh3) in the Gtf2i+/− embryos indicating a higher proportion of neuronal progenitors undergoing mitosis (Fig. 5A), consistent with the findings from WBS COs. Ki67 was instead expressed more in Gtf2i+/Dup embryos (Fig. 5B). This contradiction points likely to species-specific differences and the well-established distinct features of cell cycle phases captured by these two markers, namely, with Phh3 expressed just before mitosis, while Ki67 marking the entire cell cycle and even expressed in nonactively proliferating cells (37, 38). Intermediate progenitor marker Tbr2 was significantly higher in WT compared with Gtf2i+/Dup embryos (Fig. 5C), whereas the increase in Gtf2i+/− genotype vis-a-vis the Gtf2i+/Dup did not reach significance. These discrepancies between organoids and mice are most likely due to species-specific differences, possibly compounded by the fact that mice differ only in Gtf2i gene dosage, while patients differ in the whole CNV (i.e., 26 to 28 genes), alongside the challenges inherent to exactly matching the age of organoids in vitro with the mouse embryonic stage in vivo. In terms of the postmitotic compartment, lower and upper cortical layer neuronal markers, Bcl11b and Cux1, respectively, were both robustly increased in Gtf2i+/Dup embryos at E17.5 compared with Gtf2i+/− and WT (Fig. 5, D and E).
Fig. 5. Gtf2i dosage drives accelerated neuronal differentiation in the mouse developing cortex.
(A) Left: Representative images of mitosis marker Phh3 expression in Gtf2i+/−, WT, and Gtf2i+/Dup E17.5 embryos. Right: Quantification of Phh3 expression at E17.5 in the three genotypes. Gtf2i+/−, n = 9 sections from six mice; WT, n = 9 sections from five mice; Gtf2i+/Dup, n = 9 sections from five mice. (B) Left: Representative images of cell cycle marker Ki-67 expression. Right: Scatter plot with bar quantifying the Ki-67 expression at E17.5. Gtf2i+/−, n = 7 sections from four mice; WT, n = 13 sections from eight mice; Gtf2i+/Dup, n = 8 sections from four mice. (C) Left: Representative images of intermediate progenitor marker Tbr2. Right: Scatter plot with bar quantifying Tbr2 expression at E17.5. Gtf2i+/−, n = 11 sections from six mice; WT, n = 10 sections from five mice; Gtf2i+/Dup, n = 12 sections from five mice. (D) Left: Representative images of lower layer cortical neuronal marker Bcl11b. Right: Scatter plot with bar quantifying Bcl11b expression at E17.5. Gtf2i+/−, n = 7 sections from four mice; WT, n = 16 sections from eight mice; Gtf2i+/Dup, n = 8 sections from four mice. (E) Left: Representative images of upper layer cortical neuronal markers Cux1. Right: Scatter plot with bar quantifying Cux1 expression at E17.5. Gtf2i+/−, n = 7 sections from four mice; WT, n = 16 sections from eight mice; Gtf2i+/Dup, n = 8 sections from four mice. All data are shown as means ± SEM. Statistical analyses were performed using one-way ANOVA, followed by Tukey’s multiple comparisons test. Significance level was set to P < 0.05. *P < 0.05, **P < 0.01, and ****P < 0.0001.
To further probe the Gtf2i dosage-sensitive corticogenesis phenotypes, we electroporated in utero pCAG–green fluorescent protein (GFP) at E14.5 and analyzed cortices at E17.5 or postnatal day 7.5 (P7.5) (fig. S7A). Neurons from Gtf2i+/Dup mice at E17.5 had an aberrant orientation and were stalled between subventricular zone (SVZ) and intermediate zone (IZ), failing to migrate into the cortical plate (fig. S7, B and C). We also examined the neuronal morphology postnatally at P7.5 mice (fig. S7D). Both mutants had an altered neuronal morphology compared with WT, with fewer dendritic branches and lower number of dendrites (fig. S7, E to G). Together, these results validate in vivo the robust impact of an increased Gtf2i dosage on the neuronal proliferation/differentiation dynamics during corticogenesis, while uncovering additional alterations in migration and branching (the latter shared with the hemideleted dosage) and underscoring how, even at the level of a single gene, the 7q11.23 CNV exerts composite effects spanning the range of shared versus reciprocally opposite endophenotypes.
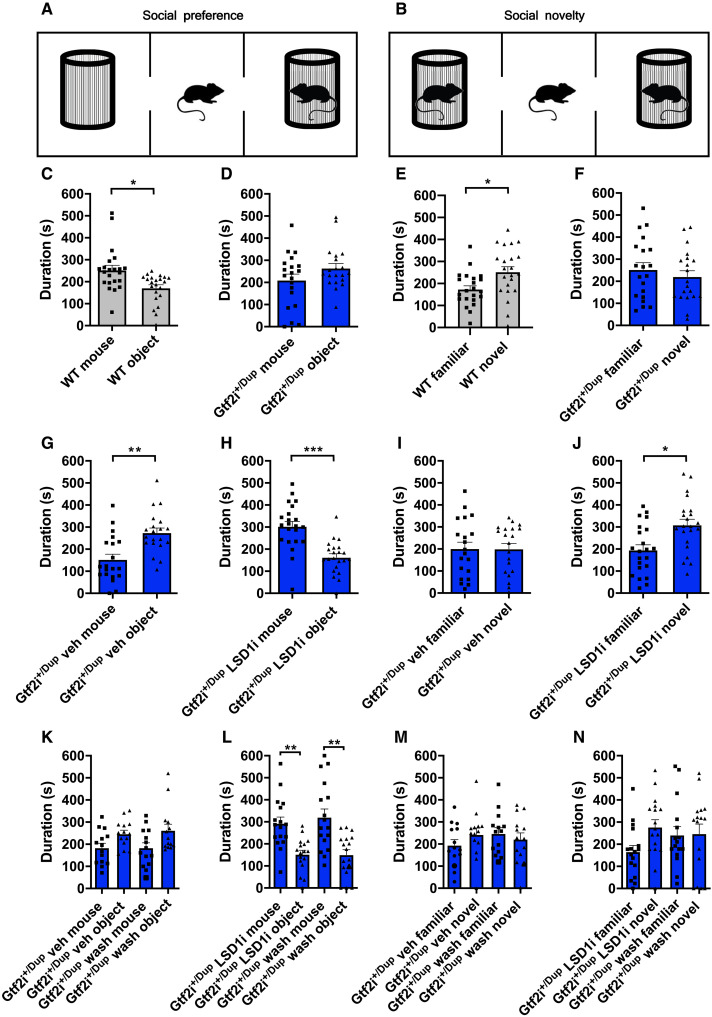
Next, having elucidated the impact of GTF2I dosage on human corticogenesis and validated partially equivalent phenotypes in murine models recapitulating its CNV, we set out to define its contribution to ASD-relevant behavioral phenotypes using the three-chamber sociability test (Fig. 6, A and B) (39). In the first part of the test, we quantified the time that male or female Gtf2i+/Dup and WT littermates spent with an object or a conspecific. In contrast to WT that spent more time with the conspecific, Gtf2i+/Dup showed either no preference for the object versus the conspecific or spent more time with the object (for male and female mice, respectively; Fig. 6, C and D, and fig. S8, A and B). During the second part of the test, mice had a choice between spending time with a familiar or a novel conspecific. As expected, WT mice spent significantly more time with a novel conspecific, whereas Gtf2i+/Dup showed no preference for the familiar versus the novel mouse (Fig. 6, E and F, and fig. S8, C and D). The robustness and significance of these phenotypes, particularly salient given the outbred nature of the GTF2I strains (CD-1 background), provided the rationale for testing rescue strategies, given the precedents of other syndromes such as Fragile X, in which the pharmacological rescue of behavioral phenotypes in adult animals had redefined the horizon of treatment options for neurodevelopmental disorders (40, 41).
Fig. 6. Inhibition of LSD1 rescues ASD-like phenotypes in Gtf2i+/Dup.
(A and B) Schematic representation of the three-chamber sociability apparatus for measuring social preference and social novelty, respectively, in male mice. (C to F) Bar plots with dots depicting the time spent with a conspecific versus object and with a novel versus familiar mouse at baseline, in WT (n = 22) and Gtf2i+/Dup (n = 20) mice. (G to J) Bar plots with dots depicting the time spent with a conspecific versus object and with a novel versus familiar mouse in Gtf2i+/Dup mice following four oral gavage administrations (two times per week over 2 weeks) of vehicle (n = 20) or LSD1 inhibitor (10 mg/kg; n = 22). (K to N) Bar plots with dots depicting social preference and social novelty results in Gtf2i+/Dup mice tested after the fourth administration of vehicle (n = 14) or LSD1 inhibitor (n = 17) and after a 2-week washout period. Data are shown as means ± SEM. Statistical analyses were performed using paired Student’s t test, followed by Holm-Bonferroni correction for multiple testing, except for (C), (I), and (M), where the data did not have a normal distribution and Wilcoxon signed rank test was used instead. Significance level P < 0.05. *P < 0.05, **P < 0.01, and ***P < 0.001.
LSD1 inhibition rescues ASD-like behaviors in Gtf2i+/Dup mice
We had previously established that GTF2I recruits a chromatin repressive complex featuring histone H3 lysine 4 demethylase LSD1 and that LSD1 inhibition relieves aberrant gene repression brought about by increased GTF2I dosages in differentiating 7Dup iPSC (11). We thus reasoned that LSD1 inhibition in vivo could be a viable strategy to rescue ASD-like GTF2I-dependent behavioral phenotypes. We, thus, targeted LSD1 with a specific inhibitor (42) that readily crosses the blood brain barrier (43) in Gtf2i+/Dup male mice, given the higher prevalence of ASD in males (44). As shown in (Fig. 6, G to J), administration of LSD1 inhibitor via oral gavage for 2 weeks rescued deficits in social preference and social novelty in Gtf2i+/Dup mice. Blood samples taken after LSD1 inhibitor administration confirmed that the treatment did not cause hematopoietic toxicity (fig. S9, A to C) (45). Next, we sought to test an interval treatment regimen that would have even greater translational relevance, especially for the prospect of chronic administration starting from a young age. Although we used an irreversible LSD1 inhibitor, it was in fact not known whether and for how long the beneficial impact on behavior would last. We thus devised a regimen in which, following LSD1 inhibitor treatment, we allowed mice 2 weeks of “rest” to remove the LSD1 inhibitor from the system and then tested them again in the three-chamber sociability apparatus. The beneficial impact of LSD1 inhibition on ASD-relevant behaviors was evident even after the 2-week “washout” period without detectable hematopoietic toxicity (Fig. 6, K to N, and fig. S9, D to F). Collectively, these results show that increased Gtf2i gene dosage underlies ASD-like behaviors and that its interference by LSD1 inhibition is a viable path for therapeutic intervention in 7Dup.
LSD1 inhibition alters transcriptional programs linked to neurodevelopment and synaptic organization in the cortex of Gtf2i+/Dup mice
Last, to define the molecular underpinnings of the efficacy of LSD1 inhibitor, we performed whole transcriptome analysis of Gtf2i+/Dup mice treated with either one or four doses of LSD1 inhibitor and collected the cortices: (i) 24 hours after for the single dose (acute) or (ii) 2 hours following the fourth dose and immediately after the behavioral test (chronic; fig. S10A). We focused our analysis on prefrontal and somatosensory cortices, two brain regions implicated in ASD pathophysiology (46, 47). As expected, inhibition of the transcriptional repressor LSD1 lead to an overall up-regulation among DEGs. In particular, acute treatment resulted in 114 DEGs (FDR < 0.1, 99 up-regulated and 15 down-regulated) (fig. S10, B and C). Prominent up-regulated genes are implicated in brain development and neurodevelopmental syndromes, including Ahi1, which is required for cerebellar and cortical development and whose mutation cause Joubert syndrome (48); Irf2bpl, which is associated with neurodevelopmental disorder with regression and plays a role in development of central nervous system via negative regulation of Wnt signaling (49); Doc2a, implicated in schizophrenia and ASD (50); and Arvcf linked to DiGeorge syndrome and schizophrenia (51). On the other hand, chronic LSD1 inhibition uncovered 54 DEGs (FDR < 0.1), 46 of which up-regulated (fig. S10, D and E). Among the dysregulated genes was a consistent up-regulation of protocadherin gene family members (i.e., Pcdhgb5, Pcdhb6, Pcdhb7, Pcdhb11, and Pcdhga5), as in 7Dup COs. Protocadherins are calcium-dependent cell adhesion protein, which establish and maintain synapse formation and neuronal connections (52), and their dysregulation has been strongly implicated in neurodevelopmental disorders (53). These findings suggest that inhibition of Gtf2i-LSD1 complex causes a two-tiered effect on the mouse’s cortex, with an up-regulation of genes critical for neurodevelopment manifested at short term, followed by a remodeling of synaptic organization evident upon sustained inhibition of the complex, enabled by increased expression of protocadherins that are, in turn, critical for sociability (53).
DISCUSSION
WBS and 7Dup syndromes are paradigmatic of the subset of neurodevelopmental disorders whose opposite CNVs are mirrored in both shared and reciprocally opposite phenotypes, thereby affording a unique glimpse into the dosage-sensitive mechanisms that underlie foundational aspects of the human condition such as sociability and language (4). This uniquely explanatory potential has remained, however, largely unfulfilled because of three main limitations. First, while genetic evidence provides the unequivocal causal links between gene dosage and high-order human phenotypes, the neurodevelopmental antecedents that mediate those links are still mostly unscrutinized. Hence, the neuroconstructivist framework that emphasized developmental constraints and the need to resolve both typical and atypical development in terms of the underlying trajectories and that owes so much of its articulation precisely to WBS is yet to be translated in molecular terms (54). Second, tracing the neurodevelopmental antecedents mediating those causal links requires the combination of multiple experimental systems, so as to bridge the distance between higher-order functions and the underlying gene dosage. However, previous studies used, separately, either animal models or two-dimensional (2D) in vitro cellular models profiled in bulk, thus precluding both the tracing of neurodevelopmental trajectories at high resolution and the cross-validation between experimental systems needed to bridge across endophenotypes unfolding at different layers of biological function. Third, the cis-epistasis of the 7q11.23 CNV, in which the simultaneous dosage imbalances of several genes within the CNV can potentially contribute to phenotypes, has made its modeling notoriously challenging. Thus, despite recent successes by us and others in delineating the roles of 7q11.23 locus’ genes in phenotypes relevant for disease pathophysiology (11, 55–58), the impact of 7q11.23 gene dosage alterations specifically on neurodevelopment and behavioral manifestation remains unknown. Here, we combine evidence from COs and transgenic mice to identify GTF2I and its axis as a key hub of neuronal maturation that underlies complex behavioral manifestations.
Analysis of single-cell transcriptome of neural progenitors and postmitotic neurons revealed a set of genotype-specific alterations, including an imbalance in cell cycle across patient WBS-7Dup COs and an early production of postmitotic neurons in 7Dup COs, while medial ganglionic eminence (MGE)-derived IN progenitors were similar in both genotypes. Definition of differentiation trajectories through RNA velocity revealed that these alterations in cortical neuronal fates are the result of accelerated neuronal differentiation in 7Dup. Critically, we found consistent abnormal neuronal differentiation dynamics in WBS and 7Dup using 2D neuronal cultures (59), corroborating the timing of neuronal differentiation as a critical and experimentally tractable phenotype across experimental models and systems. Asynchronous generation of specific neuronal populations is an emerging cellular mechanism underlying the convergent impact of autism genes on tractable neuronal phenotypes, as we and others have recently shown (12, 13, 15, 60). Here, we reveal that this cellular mechanism extends beyond high confidence autism risk genes to symmetrically opposite CNV, thereby providing a high-resolution cellular framework to annotate the neurodevelopmental trajectories of the sociability spectrum (4). This heterochronic neuronal differentiation was largely corroborated by our findings in mice, where we found an increased number of postmitotic neurons in Gtf2i+/Dup compared with Gtf2i+/− mice.
GTF2I preferentially binds promoters and drives both basal and signal-induced transcription (61), and it targets and interacts with critical genes for embryonic development and patterning, cell growth, and proliferation, including SMAD2, ERK, and JAK2 (62, 63). The formation of transcriptional suppressive complex with the well-established pluripotency regulator LSD1 (64), as we previously showed (11), lent further support to the notion that its effects most likely involve transcriptional programs critical for fate specification and neuronal differentiation. Previously, Gtf2i+/Dup mice pups showed increased ultrasonic vocalizations due to maternal separation anxiety (36), whereas, more recently, Gtf2i+/Dup showed no evidence of hyposociality in behavioral assays for ASD (65). Besides major differences in apparatuses, group sizes, and the tests for assessing social preference, the Gtf2i mice from these reports were in an inbred background (C57BL/6), in contrast to the current experiments for which we selected the more translationally relevant outbred strain (CD-1). Our findings now show that drugs targeting chromatin regulation and transcriptional control can be a viable strategy for neurodevelopmental disorders that have been so far impossible to treat in terms of core ASD symptoms. LSD1 inhibitors are under intense investigation for their promising efficacy in various human malignancies (66), and, more recently, LSD1 inhibitors, albeit different from the one we used here, proved efficacious in rescuing ASD-like behaviors in mouse models with deficient Shank3 and Cul3 (67). The current data, as well as our recent report on the efficacy of histone deacetylase inhibitors regulating GTF2I expression (68), highlight the potential of drug repurposing for neurodevelopmental disorders with extremely limited treatment options.
While the knockdown of GTF2I rescues the accelerated production of ENs in 7Dup, it had only a negligible impact on the imbalance of progenitor production, underscoring how, in the context of a complex cis-epistatic CNV, the longitudinal dissection of endophenotypes serves to define single-gene contributions that can be highly specific and time dependent. This also paves the way to further studies aimed at dissecting how other genes from the same locus, acting alone or in combination with GTF2I, bring about distinct aspects of the endophenotypes we have now charted.
Animal and population studies have attributed a critical role to GTF2I in sociability phenotypes (4, 69–72). Protocadherins, on the other hand, are synaptic cell adhesion molecules that play critical roles in synapse organization and function (73, 74). Expectedly, mutations in protocadherins lead to aberrant neuronal communication and function, which, in turn, affect higher cognitive processes, including sociability (75). Bulk RNA sequencing (RNA-seq) results revealed a modulation of protocadherins in 7Dup COs and Gtf2i+/Dup mice following LSD1 inhibition, suggesting a convergence across species and experimental models on a dosage-sensitive GTF2I -protocadherins molecular axis mediating sociability. Together, these data provide a comprehensive molecular characterization of the 7q11.23 gene dosage alterations impact on neurodevelopment and behavioral phenotypes and uncover the GTF2I-LSD1 axis as a central mediator of 7q11.23 disease pathophysiology with therapeutic potential.
MATERIALS AND METHODS
iPSC culture
We used 13 control and patient-derived iPSC lines that we generated previously and reported in (11, 68) (see also table S2). Relevant ethics approvals are referred to in the original publications reporting their first use and/or derivation.
iPSC lines were cultured on plates coated with human-qualified Matrigel [Corning; 1:40 dilution in Dulbecco’s modified Eagle’s medium (DMEM)–F12] in mTeSR (STEMCELL Technologies) or TeSR-E8 medium (STEMCELL Technologies). Cells were passaged with ReLeSR (STEMCELL Technologies) or with Accutase (Sigma-Aldrich). For single-cell culture, cells were resuspended in mTeSR or TeSR-E8 medium supplemented with 5 μM Y-27632 (ROCK inhibitor, Sigma-Aldrich).
GTF2I knockdown in iPSC lines
GTF2I knockdown was performed using a validated pLKO.1-Puro TRC vector, TRCN 0000364552 (referred to as shRNA A) and TRCN0000369208 (referred to as shRNA B) (11). Viral particles were produced using the psPAX2 and pMD2.G packaging vectors in human embryonic kidney 293T cells. Viral particles were collected at 24 and 36 hours after transfection and concentrated 250-fold by ultracentrifugation at 24,000g for 2 hours at 16°C. Two iPSC 7Dup lines were infected with different amounts of viral particles, and the amount that yielded <70% survival after 72 hours of treatment with puromycin (1 μg/ml) was selected for further experiments. Infected iPSC lines and derived COs were maintained in medium containing puromycin (1 μg/ml) throughout the expansion and differentiation process.
Quantitative reverse transcription polymerase chain reaction
A custom TaqMan kit (Invitrogen AM1729) was used to extract the RNA and perform reverse transcription to obtain cDNA, according to the manufacturer’s instructions. Three COs per condition were collected in sample tubes and received 100 μl of 1 × lysis solution, with diluted deoxyribonuclease I, samples were then incubated for 5 min at room temperature. Subsequently Stop solution (10 μl) was added, and the solution was incubated at room temperature for 2 min. Then, 30 μl of lysates was transferred to a polymerase chain reaction (PCR) plate with 40 μl of reverse transcription enzyme mix previously added to each well using GTF2I TaqMan probes (Thermo Fisher Scientific, Hs01073660_m1). The thermal cycling conditions were as follows: 60 min at 42°C and 5 min at 85°C. cDNA was diluted with 50 μl of water, and then a 10-μl aliquot of each cDNA reaction was added to 10 μl of each TaqMan master mix reaction into 96-well PCR plates. A QuantStudio 6 Flex Real-Time PCR system (Applied Biosystems) was used to determine the cycle threshold (Ct) values. Relative mRNA expression levels were normalized to housekeeping genes and analyzed through the comparative ΔΔCt method using the QBase Biogazelle software.
Generation of COs
Cortical brain organoids were generated by following the modified protocol described in (17, 22). Since we optimized the protocol to avoid the use of mouse embryonic fibroblast for stem cell culture, the following procedures were followed; when the human iPSC line reached 80% confluency in a 10-cm dish, colonies were dissociated with Accutase and centrifuged. After resuspension in TeSR/E8 medium supplemented with 5 μM ROCK inhibitor Y-27632 (Corning) cells were counted with a TC20 automated cell counter (Bio-Rad) and resuspended to get a final concentration of 2 × 105 cells/ml. Cell suspension (100 μl per well) was seeded into ultralow attachment PrimeSurface 96-well plates (SystemBio), and the plates were centrifuged at 850 rpm for 3 min to promote the formation of embryoid bodies (EBs). The day of the EB generation is referred to as day −2. On day 0, neural induction medium was added, consisting of 80% DMEM-F12 medium (1:1), 20% knockout serum (Gibco), 1 mM nonessential amino acids (Sigma-Aldrich), 0.1 mM cell culture grade 2-mercaptoethanol solution (Gibco), GlutaMAX (Gibco; 1:100), penicillin (100 U/ml), streptomycin (100 μg/ml), 7 μM dorsomorphin (Sigma-Aldrich), and 10 μM transforming growth factor–β inhibitor SB431542 (MedChemExpress) for promoting the induction of neuroectoderm. From day 0 to day 4, medium change was performed every day, while on day 5, neuronal differentiation medium was added, consisting of neurobasal medium (Gibco) supplemented with B-27 supplement without vitamin A (Gibco; 1:50), GlutaMAX (1:100), penicillin (100 U/ml), streptomycin (100 μg/ml), fibroblast growth factor 2 (20 ng/ml; Thermo Fisher Scientific) and epidermal growth factor (20 ng/ml; Thermo Fisher Scientific) until day 25. From days 25 to 43, the neuronal differentiation medium was supplemented with brain-derived neurotrophic factor (20 ng/ml) and neurotrophin-3 (NT3) (PeproTech). From day 43 onward, no growth or neuronal maturation factors were added to the medium.
Western blot
Organoids were snap-frozen in dry ice and lysed in SDS buffer [100 mM tris-HCl (pH 7.6), 20% glycerol, and 4.7% SDS] denatured at 95°C for 10 min and sonicated for 15 s. Protein extracts (30 to 50 μg) were mixed with Laemmli loading buffer, denatured for 5 min at 95°C, electrophoresed in 10% SDS–polyacrylamide gel electrophoresis gel and transferred to polyvinylidene difluoride membranes. The membranes were blocked in TBST [50 mM tris (pH 7.5), 150 mM NaCl, and 0.1% Tween 20] and 5% milk for 1 hour and incubated overnight with GTF2I (BD Biosciences) and glyceraldehyde-3-phosphate dehydrogenase (Millipore) primary antibodies (see table S1). Blots were developed with the ECL Prime Western Blotting Detection Reagents (Sigma-Aldrich), and bands imaged and quantified using the ChemiDoc system (Bio-Rad).
Bulk transcriptome analysis
At the designated time point, COs were washed once with phosphate-buffered saline (PBS) and snap-frozen in dry ice. Total RNA was isolated with the RNeasy Micro Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. RNA was quantified with Nanodrop, and then the integrity was evaluated with Agilent 2100 Bioanalyzer. A TruSeq Stranded Total RNA LT Sample Prep Kit (Illumina) was used for library preparation starting from 500 ng of total RNA for each sample. Sequencing was performed with the Illumina NovaSeq 6000 platform, with an average depth of 35 million 50–base pair paired-end reads per sample.
Cortical organoids
RNA-seq FASTQ data were quantified at the gene level using Salmon (version 0.8.2). GRCh38 Genecode 27 was used as reference for quantification and annotation. Protein-coding genes were selected for downstream investigations.
DEA comparing 7Dup versus WBS COs were performed by edgeR either on the complete organoid cohort (33 samples: three WBS, four CTL, and three 7Dup for day 18 organoids; four WBS, four CTL, and four 7Dup for day 50 organoids; three WBS, five CTL, and three 7Dup for day 100 organoids) or selecting by differentiation stage. For the complete cohort analysis, genes with expression levels higher than 2 cpm in at least four samples were tested for differential expression; the information about organoid differentiation stage was used as a covariate in the statistical model. DEGs were selected imposing a threshold of FDR < 0.1 and absolute log2FC > 1.
Similarly, stage-wise DEAs were performed comparing 7Dup versus WBS and selecting genes with an expression level higher than 2 cpm in at least three samples for day 18 and day 100 and in at least four samples for day 50. A total of 315 genes that were found significantly modulated (FDR < 0.1, absolute log2FC > 1) in at least one time point were selected for network reconstruction by retrieving STRING database connections (https://string-db.org/) and generating the network in Cytoscape (76). Node shape highlights the stage in which the genes are significantly modulated as follows: diamond for day 18, circle for day 50, square for day 100, and hexagon for genes significant in more than one stage. Identification of functionally relevant regions in the network was performed by manual curation of node biological function.
The following knowledge bases were considered for the overlap analysis: MIM (Mendelian Inheritance in Man), Orphanet, Decipher, SFARI Genes, and AutismKB. MIM, Orphanet, and Decipher genes were obtained by interrogating version 100 of Biomart (www.ensembl.org/biomart/martview/). SFARI genes and AutismKB were retrieved respectively from https://gene.sfari.org/database/human-gene/ and (77) in April 2020. Overlap between these gene sets and DEGs at day 18, day 50, or day 100 (either considered together or divided in up-regulated and down-regulated) was tested by GeneOverlap R library (version 1.14.0). Overlaps were considered significant with an odds ratio higher than 1 and P value lower than 0.1. Where not otherwise specified, analyses were performed in R, version 3.4.4.
Liquid chromatography–tandem mass spectrometry analysis and raw data processing
In all cases, sample preparation was performed according to the in-StageTip protocol (78). Briefly, samples (one CO per cell line, three cell lines per genotype in duplicate) were incubated in PreOmics lysis buffer (catalogue number P.O. 00001, PreOmics) for cysteine reduction and alkylation, followed by a protein denaturation step at 95°C for 10 min. Then, proteins from each sample were digested by adding the Trypsin/LysC mix (digestion buffer, PreOmics) at 1:50 enzyme-to-protein ratio at 37°C overnight. Proteolytic peptides were eluted from the solid-phase extraction material and dried out completely in a SpeedVac centrifuge (Eppendorf). Last, peptides were resuspended in 5 μl of load buffer (PreOmics) and analyzed by nano–reversed phase liquid chromatography–tandem mass spectrometry (LC-MS/MS) using an EASY-nLC 1200 (Thermo Fisher Scientific, catalog no. LC140) connected to a Q-Exactive HF instrument (Thermo Fisher Scientific) through a nano-electrospray ion source. In all cases, the nano-LC system was operated in one-column setup with an EasySpray PEPMAP RSLC C18 column (Thermo Fisher Scientific) kept at the constant temperature of 45°C. Solvent A was 0.1% formic acid (FA), and solvent B was 0.1% FA in 80% acetonitrile. Peptides were separated along a 64-min gradient of 3 to 30% solvent B, followed by a gradient of 30 to 60% for 10 min and 60 to 95% over 5 min, at a flow rate of 250 nl/min. The Q-Exactive was operated in the data-dependent acquisition mode to automatically switch between MS and MS/MS mode. The 15 most intense peptide ions with charge states ≥2 were sequentially isolated to a target value of 3 × 106 (top 15). Spray voltage was set to 1.7 kV, s-lens radio frequency level at 50, and heated capillary temperature at 275°C. Selected target ions were dynamically excluded for 20 s, and all experiments were acquired using positive polarity mode. The MS spectra [from mass/charge ratio (m/z) of 375 to 1550) were analyzed in the Orbitrap detector with resolution R = 60,000 at m/z 200. MS2 data were acquired at R = 15,000 resolution and an ion target value of 1 × 105. Higher-energy collisional dissociation fragment scans were acquired using 1.4 m/z isolation width and a normalized collision energy of 28. The maximum allowed ion accumulation times were 20 ms for full scans and 80 ms for MSMS.
The acquired raw MS data were analyzed using the integrated MaxQuant version 1.6.2.3 (79), using the Andromeda search engine (80), and the Human Fasta Database downloaded from UniprotKB (74470 Entries) was used. Carbamidomethylation of cysteine was set as a fixed modification. Enzyme specificity was set as C terminus to arginine and lysine as expected, using Trypsin and LysC as proteases. A maximum of two missed cleavages was allowed. Peptide identification was carried out with the Andromeda algorithm with an initial precursor mass deviation of up to 7 parts per million (ppm) and a fragment mass deviation of 20 ppm. FDR for both peptide and proteins was set to a maximum of 1% for identification. All proteins and peptides matching the reversed database were filtered out. The label-free quantitation (LFQ) intensity calculation was enabled, as well as the match between runs feature (81). The “protein groups” output file from MaxQuant was analyzed using Perseus software (80). Briefly, no imputation was used, and data were filtered to have at least three valid values in at least one group. To identify significantly regulated proteins, an FDR = 0.05 was imposed in the t test analysis.
Transcriptome-proteome comparison
Results from the proteomics profiling were compared with the transcriptional changes identified at day 50 (7Dup versus WBS DEA). The comparison between the two approaches was performed by selecting features (genes or proteins) measured by both techniques and discarding those with duplicated gene symbols. Significant modulation has been defined according to the following thresholds: P <0.05 and FC > 1.25 (absolute value); overlap was tested with a Fisher’s test and visualized by Venn diagram (R package Venn Diagram version 1.6.20, https://cran.r-project.org/package=VennDiagram) and by custom scatter plot.
Proteomic functional analysis
Gene ontology (GO) enrichment analysis for the Biological Process domain was performed on the proteins identified as changed in 7Dup versus WBS with a conventional P < 0.05 and an absolute log2FC value of at least 1. Analysis was performed by TopGO (version 2.30.1) (82), relying on Fisher’s test and Weight01 method to take into account ontology hierarchy; node size was set at 15. Cutoffs of 0.01 on the P value and 1.75 on the enrichment threshold were applied to select significantly enriched GO terms. The top 8 categories according to P value for up-regulated and down-regulated genes were visualized by barplots.
Overlap analysis with gene-disorder knowledge bases
MIM (gene-disorder association for genetic diseases), Orphanet (gene-disorder association for rare diseases), and Decipher knowledge bases were retrieved from Mart (Biomart version 100; www.ensembl.org/biomart/martview/). The SFARI gene list was downloaded from the SFARI website (https://gene.sfari.org/database/human-gene/, downloaded in April 2020). The evidence-based knowledge base for autism AutismKB was retrieved from [(77), accessed in April 2020]. Overlap analysis was performed between each considered knowledge base and the results of stage-wise DEA (7Dup versus WBS comparison), selecting DEGs with FDR < 0.1 and FC > 2 and testing them either as a single set or split in up and down-regulated genes. Overlap analysis was performed using GeneOverlap R package (version 1.14.0; https://github.com/shenlab-sinai/GeneOverlap) and considering the tested genes for each DEA as universe. Results were visualized by dotplot, reporting the number of overlapping genes for odds ratio (OR) > 1 and the dot for P values < 0.1. Dot size and color represent the P value and the OR, respectively.
Single-cell transcriptome analysis
Organoids collected from different genotypes and time points were dissociated by incubation with a solution of Papain (30 U/ml; Worthington, LS03126) and deoxyribonuclease I (3 U/μl; Zymo Research) for 30 to 45 min depending on organoid size. Dissociated suspensions were passed once through 0.4-mm Flowmi cell strainers, resuspended in PBS, and counted using TC20 automatic cell counter (Bio-Rad). Resulting single-cell suspension was mixed with reverse transcription–PCR master mix at a density of 1000 cells/μl and loaded together with Chromium Single-Cell 3′ gel beads and partitioning oil into a Chromium Single Cell 3′ Chip. The gel beads were coated with unique primers bearing 10x cell barcodes, unique molecular identifiers and poly(dT) sequences. The chip was then loaded onto a Chromium instrument (10x Genomics) for single-cell gel beads in emulsion (GEM) generation and barcoding. Amplified cDNAs were fragmented, and adapter and sample indices were incorporated into finished libraries, following the manufacturer’s instructions. The final libraries were quantified by Qubit system (Thermo Fisher Scientific) and calibrated with an in-house control sequencing library. The size profiles of the preamplified cDNA and sequencing libraries were examined by Agilent Bioanalyzer 2100 using a High Sensitivity DNA chip (Agilent). Two indexed libraries were equimolarly pooled and sequenced on Illumina NovaSeq 6000 platform using the v2 Kit (Illumina, San Diego, CA) with a customized paired-end, dual indexing format according to the recommendation by 10x Genomics. Using proper cluster density, a coverage of around 250 M reads per sample (2000 to 5000 cells) was obtained corresponding to at least 50,000 reads per cell.
Twenty-one biological samples (day 50; four WBS, four CTL, and four 7Dup) and (day 100; three WBS, three CTL, and three 7Dup) were examined by single-cell analysis with a coverage around 250 M reads per sample (2000 to 5000 cells) and at least 50,000 reads per cell as target. A total of 97,794 cells were retrieved after quality check with a minimum of 700 genes per cell and mitochondrial RNA more than 5% per cell to avoid low-quality cells. Libraries from single-cell sequencing were aligned relying on the CellRanger v2.1 pipeline and using hg38 as reference.
Before downstream analyses, data deriving from the 21 samples were integrated by Conos (83); the resulting clusters, common among all samples, were considered as shared populations and used as such for the scGen variational autoencoder algorithm (84). On the integrated dataset, uniform manifold approximation and projection (UMAP) dimensionality reduction as implemented in Scanpy (85) was applied. Clusters were identified by applying the Leiden algorithm, a community detection algorithm that has been optimized to identify communities that are more coherent with the biological phenotype and more reliably identify cell populations. The resolution parameter value was optimized by surveying the stability of the resulting clusters. This resulted in the identification of final clusters.
Cluster annotation in cell populations was obtained by a combination of the following approaches: (i) scanpy’s rank_genes_groups to identify the most characterizing genes per clusters; (ii) SCINA semisupervised annotations algorithm (86), using as source labels coming from three publicly available datasets (22, 31, 87); and (iii) overlap of cluster marker genes (log2FC > 0.5, q <0.05 compared to all other clusters) with cell type marker genes identified in a published single-cell dataset (31). Significance of overlap between marker sets was determined using Fisher’s exact test; (iv) projection of a single-cell fetal cortex data in the UMAP by ingest algorithm. The obtained information was lastly manually curated using a self-hosted cellxgene [chanzuckerberg/cellxgene: An interactive explorer for single-cell transcriptomics data (github.com), cellxgeneVIP (88), and cellxgeneGateway (https://github.com/Novartis/cellxgene-gateway)]. Dotplots showing a panel of cell population specific genes across clusters was produced using the Scanpy DotPlot function.
Pseudo-time analysis was performed on clusters identified by the Leiden algorithm with a resolution of 9. The resulting clusters were stratified by conditions/stage and then aggregated by their median to generate a new structure composed of “supercells.” Supercells with low contribution of reads for any genotype were discarded. The Palantir algorithm (89) was applied on control supercells (CTL) and then propagated.
For the neuron only cluster we applied the CellRank algorithm (32) using as starting point the lower pseudo-time calculated in the whole population. We have recalculated the distance neighbor, the dimensionality reduction, and the subsequent calculation only on the neuronal cells and calculated the transition matrix for every genotype separated considering pseudo-time (89), RNA velocity (33), and similarity. We identify the terminal state using the CFLARE (Clustering and Filtering Left and Right Eigenvectors) to identify the eigenvalue and eigenvector pairs that are more probable and infer the terminal states. For the comparison with sh datasets and 7q11.23 microduplication syndrome (7Dup), we downscaled the number of cells for 7Dup to be comparable in numbers, process not needed for the other conditions given the already comparable numbers. For panel 3F, we used Milo (90) and plot the differences between genotypes in the mature neuron clusters with P < 0.05.
Overlap with fetal marker genes
To refine the annotation of the subregions relative to the neural maturity clusters, we performed an overlap of the marker genes with the ones coming from a fetal dataset (91), choosing GW22 sample. Fetal dataset was downloaded from UCSC cell data browser, and the signatures were derived both from the reanalyzed data and the article.
The top 50 markers were then retrieved for each fetal cluster, as defined by the original annotation (91); those not having top 50 significant marker genes (KS_PFC; KS_Outlier) or not of interest for the comparison with brain organoid were not considered in the overlap. Similarly, the top 50 marker genes were retrieved for the subregions of interest in the brain organoid neural maturity cluster, considering only the more differentiated clusters, identified by the external position in the force directed graph and the CellRank algorithm (32); subclusters 1 and 6, not having 50 significant marker genes, were excluded from the overlap. Overlap analysis was performed with the multintersect function of the overlapper R package (https://github.com/plger/overlapper/) comparing the organoid and fetal top marker lists and considering as the universe the genes used in brain organoid single-cell analysis. The results are reported by a dotplot showing the number of overlapping genes; the dot is shown for overlaps having a P < 0.01; dot size shows the P value and dot color the enrichment.
Animals
All animal experiments were done in accordance with the Italian Laws (D.L.vo 116/92 and following additions), which enforces EU 86/609 Directive (Council Directive 86/609/EEC of 24 November 1986), and were approved by institutional ethics committee (organismo preposto al benessere degli animali, OPBA committee) and the Italian Ministry of Health (Authorization 1073/16-PR). Mice with hemizygous deletion (Gtf2i+/−) or duplication (Gtf2i+/Dup) of Gtf2i were generated as previously described (36) and were maintained in an outbred CD-1 background. Mice were housed in groups of five maximum in a normal 12-hour light:12-hour dark cycle, and food and water were provided ad libitum.
In utero electroporation
In utero electroporation was carried out as described (92). Timed pregnant mice were anesthetized using Avertin (1.25% solution, 0.02 ml/g of bodyweight) and the uterine horns exposed through a laparotomy. One microliter of endo-free purified (2 μg/μl) pCAG-EGFP plasmid (Addgene, catalog no. 89684) mixed with 0.05% Fast Green (Sigma-Aldrich, catalog no. F7252) in PBS was injected through the uterine wall into the lateral ventricle of the embryos. The DNA-injected embryos were held through the uterus, parallel to the embryonic anteroposterior axis. Then, five 40-V pulses of 50-ms duration at 1-s intervals were delivered across the embryonic head using 3-mm-diameter electrodes (Harvard Apparatus, catalog no. 45-0487) connected to a square wave electroporator (Harvard Apparatus, catalog no. EC1 45-0052). The uterine horns were placed back, and the laparotomy was closed with sutures. The embryonic development could continue until E17.5 or P7.5. Following the surgery, Carprofen (0.5 mg/ml, 5 mg/kg of bodyweight) was administered subcutaneously for the next two postoperative days.
LSD1 inhibitor administration
The LSD1 inhibitor (N-[4-[(1S,2R)-2-aminocyclopropyl]phenyl]-4-(4-methylpiperazin-1-yl)benzamide;dihydrochloride) was manufactured in the house by the drug discovery unit and was dissolved in 60% 5% glucose and 40% polyethylene glycol. Each mouse received four administrations of LSD1 inhibitor (10 mg/kg) or vehicle two times per week via oral gavage or one administration of LSD1 inhibitor or vehicle only for the bulk transcriptomic profiling experiment. Three hours following the last administration, the mouse performed the behavioral test.
Bulk transcriptomic analysis of mouse cortex
RNA-seq FASTQ data were quantified at the gene level using Salmon (version 1.3.0). Mouse Genecode M25 was used as reference for quantification and annotation. Five biological replicates were analyzed for each of the considered biological conditions (LSD1 inhibitor and vehicle, somatosensory, and prefrontal cortex, acute, and chronic treatment). One cortex for the prefrontal cortex chronic LSD1 inhibitor treatment was excluded from downstream analysis because identified as outlier in from quality control evaluations (sample-to-sample correlation). A total of 39 samples were included in DEA.
Protein-coding nonpredicted genes with expression levels higher than 2 cpm in at least four samples were tested for differential expression by edgeR. DEA comparing treated versus untreated mice was implemented separately for acute and chronic treatment, specifying the brain area as covariable in the statistical model. Significantly modulated genes were selected setting an FDR lower than 10%. Analyses were performed in R, version 4.0.3.
Behavioral experiments
Three-chamber sociability test was done as previously described (39). Mice with 2 to 4 months of age were acclimatized to the experimental room for at least 1 hour before the test. The test apparatus consisted of a three-compartment box of transparent polyvinyl chloride with each compartment of 20 cm by 40 cm by 22 cm (Ugo Basile). The test mouse habituated to the apparatus for 5 min. In the social preference session, the mouse could spend time either in the compartment with an empty metal grid enclosure (object) or in the compartment with a conspecific for 10 min. In the subsequent social novelty session, the mouse could spend time either with the familiar mouse from the previous session or with a novel mouse for 10 min. The apparatus was cleaned thoroughly with 70% ethanol, and the compartments were alternated between mice. The data were acquired and analyzed using Any-maze behavioral tracking software (Stoelting). In the washout experiments, treated mice were left undisturbed in their home cages for 2 weeks and subsequently performed again the behavioral test as described above.
Collection of biological materials
Blood samples were collected from the tail before drug administration and at the end of the behavioral experiments. In the washout experiments, an additional blood sample was collected at the end of the fourth administration.
Following the behavioral experiments, the mice were euthanized by cervical dislocation, the brain was rapidly removed, and hippocampus, prefrontal, and somatosensory cortices were dissected and snap-frozen in dry ice.
Immunofluorescence
Cortical organoids
COs were harvested at the indicated time point and fixed overnight at 4°C in 4% paraformaldehyde/PBS solution (Santa Cruz Biotechnology). After rinsing with PBS, organoids were embedded in 2% low melting agarose dissolved in PBS; upon agarose solidification, blocks were put in 70% ethanol and kept at 4°C before paraffin embedding, sectioning, and routine hematoxylin/eosin staining. Deparaffinization and rehydration were achieved by consecutive passages of 5 min each in the following solutions: 2× histolemon (Carlo Erba), 100% ethanol, 95% ethanol, 80% ethanol, and water. Sections were then incubated for 45 min at 95°C in 10 mM sodium citrate (Normapur)/0.05% Tween 20 (Sigma-Aldrich) buffer for simultaneous antigen retrieval and permeabilization and then left to cool for at least 2 hours at room temperature. To immunolabel the markers of interest, a blocking solution made of 5% donkey serum (ImmunoResearch) in PBS was applied for 30 min to the slides, while primary antibodies diluted in blocking solution were subsequently added, performing overnight incubation at 4°C. Secondary antibodies and 4′,6-diamidino-2-phenylindole (DAPI) were diluted in PBS and applied to the sections for 1 hour and 5 min, respectively. After each incubation step, washing steps (3× for 5 min) with PBS buffer were performed. After a final rinse in deionized water, slides were air-dried and mounted using Mowiol mounting medium. Image analysis was done in ImageJ, and the quantification was done by counting the number of positive cells relative to total DAPI-positive nuclei.
Animals
At the corresponding developmental stages, the whole head (E17.5) or extracted brains (P7.5) were collected, immersed in 4% paraformaldehyde/PBS at 4°C for 4 hours to overnight for fixation. Brains were washed twice with PBS, embedded into 3% low melting point agarose, then sectioned into 50 to 60 μm using a vibratome (Leica, catalog no. VT1000S), and mounted for direct visualization of EGFP or processed for immunofluorescence as it follows. Brain sections were washed briefly in PBS and blocked in PBS+ (PBS, 5% normal donkey serum, and 0.25% Triton X-100) for 2 hours at room temperature. Sections were incubated with primary antibodies diluted in PBS+ overnight at 4°C. Following, three 5-min washes in PBS, secondary antibody diluted in PBS+ was added for 1 hour at room temperature (1:500). Sections were then washed 3× for 5 min in PBS and incubated for 5 min in DAPI (Invitrogen, catalog no. D1306) with a dilution of 1:5000 in PBS. After another 5-min wash in PBS, sections were mounted onto glass slides, coverslipped, and imaged using a confocal microscope (Leica SP8).
For image analysis and quantification of proliferative and neuronal markers, we used custom made scripts in ImageJ. The choice of quantification and normalization method was based on manual inspection of image/immunostaining quality and/or marker expression. Specifically, for Phh3, Ki-67, and Bcl11b, we quantified the number of positive neurons and normalized it against the area covered by DAPI-positive neurons. On the other hand, for Tbr2 and Cux1, we quantified the average intensity of the positive cells and normalized it against the average intensity of the background.
Analysis of neuronal morphology
Morphology of entire neurons was visualized by the expression of pCAG-EGFP plasmid. Low–cell density dorsal telencephalic areas were used to determine morphological characteristics. Neurons transfected with plasmid DNA were selected by GFP expression, and images were acquired at ×60 magnification using a Leica SP8 confocal microscope. GFP-positive cells were traced with the NeuronJ plugin in ImageJ (FIJI package) for analysis of neuronal dendrite morphology following the developers’ instructions (93). After the neurites were traced, they were labeled as primary (emanating directly from the soma), secondary (branching from a primary), or tertiary (branching from a secondary), and a text file was generated containing measurements of the lengths of all the neurites. The Snapshot tool in NeuronJ was used to save the tracings as an image file, and these images were analyzed with the Sholl analysis plugin in ImageJ. The range of measurement was set using the straight-line tool traced from the center of the soma to the outermost neurite. Dendrite intersections were analyzed from a starting radius of 10 μm (to exclude the soma from analysis) with 5-μm steps to the outer radius. The resulting numbers of intersections per cell were used to calculate the mean and SEM for each radius interval.
Statistics and reproducibility
Data are presented as means ± SEM, unless otherwise indicated. Distribution of the raw data was tested for normality of distribution; statistical analyses were performed using the Student’s t test with Holm-Bonferroni correction for multiple comparisons and one-way analysis of variance (ANOVA) with Tukey correction for multiple comparisons or using a mixed linear regression approach. Statistical analyses were performed in Prism (GraphPad) and in R 4.2.0. Full statistical details for each figure panel are included in the legends. Sample() sizes were estimated empirically on the basis of previous studies. Behavioral experiments were performed by researchers who were blinded to the experimental conditions.
Acknowledgments
We are grateful to F. Pisati from IFOM tissue processing facility, E. Tam (University of Toronto) for assistance with mouse genotyping, A. Vitriolo for assistance with the statistics, IEO Genomic Unit team, IEO Imaging Facility team, and Cogentech’s Animal Facility.
Funding: We thank EPIGEN Flagship Project of the Italian National Research Council (CNR) (to G.T.); the European Research Council (ERC DISEASEAVATARS no.616441 to G.T. and ERC PoC 713652–LSDiASD to G.T.); EDC-MixRisk, European Union’s Horizon 2020 research and innovation programme (grant no 634880 to G.T.); ENDpoiNTs, European Union’s Horizon 2020 research and innovation programme (grant no 825759 to G.T.); Telethon (GGP14265 and GGP19226 to G.T. and GGP19295 to A.L.-T.); Fondazione Cariplo (2017-0886 to A.L.-T.); Fondazione Umberto Veronesi (to R.S.); Ministero della Salute, RC 2019–ERANET NEURON RRC-2019-2366750 -ALTRUISM (to M.M.); ERA-NET NEURON (ADNPinMED to P.F.-B.); AIRC grant no. IG-2018-21834 and EPIC-XS (project no. 823839, funded by the Horizon 2020 programme of the European Union to T.B.); and The Leverhulme Trust and The Horizon 2020 Innovative Training Network EpiSyStem (Marie Skłodowska-Curie Actions; to A.S.).
Author contributions: A.L.-T., R.S., and G.T. conceptualized and designed the experiments and wrote the manuscript. A.L.-T., S.T., and N.C. developed organoids differentiation protocol. A.L.-T., S.T., N.C., F.T., A.S., and R.S. developed organoid generation and maintenance. C.E.V. and C.C. developed data analysis pipeline, analyzed respectively single-cell and bulk transcriptomic data, and wrote the manuscript. R.S. analyzed behavioral experiments and data analysis. P.F.-B. performed in utero electroporation and immunostainings in mice. E.T. did the cell culture and library preparations. M.M. did the sample preparation for proteomics. W.T.G. provided fibroblasts. A.C. and T.B. provided the proteomic analysis of organoids. C.M. and M.V. designed and manufactured LSD1 inhibitor. L.O. provided Gtf2i mice and analyzed GTF2I expression in murine fetal samples. G.T. supervised the work.
Competing interests: G.T. is listed as an inventor on the granted patent no. US 11,369,577 and patent application no. EP15801762.4 owned by Istituto Europeo di Oncologia, IRCCS Casa Sollievo della Sofferenza, Università degli studi di Milano. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD036770. The bulk and single-cell transcriptomic data are available through Annotare (EMBL-EBI) with access codes as follows; bulk RNA mice, E-MTAB-13475; bulk RNA organoid, E-MTAB-13477; single-cell RNA organoid: E-MTAB-13478.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Tables S1 to S3
REFERENCES AND NOTES
- 1.B. R. Pober, Williams-Beuren syndrome. N. Engl. J. Med. 362, 239–252 (2010). [DOI] [PubMed] [Google Scholar]
- 2.S. J. Sanders, A. G. Ercan-Sencicek, V. Hus, R. Luo, M. T. Murtha, D. Moreno-De-Luca, S. H. Chu, M. P. Moreau, A. R. Gupta, S. A. Thomson, C. E. Mason, K. Bilguvar, P. B. S. Celestino-Soper, M. Choi, E. L. Crawford, L. Davis, N. R. D. Wright, R. M. Dhodapkar, M. DiCola, N. M. DiLullo, T. V. Fernandez, V. Fielding-Singh, D. O. Fishman, S. Frahm, R. Garagaloyan, G. S. Goh, S. Kammela, L. Klei, J. K. Lowe, S. C. Lund, A. D. McGrew, K. A. Meyer, W. J. Moffat, J. D. Murdoch, B. J. O’Roak, G. T. Ober, R. S. Pottenger, M. J. Raubeson, Y. Song, Q. Wang, B. L. Yaspan, T. W. Yu, I. R. Yurkiewicz, A. L. Beaudet, R. M. Cantor, M. Curland, D. E. Grice, M. Günel, R. P. Lifton, S. M. Mane, D. M. Martin, C. A. Shaw, M. Sheldon, J. A. Tischfield, C. A. Walsh, E. M. Morrow, D. H. Ledbetter, E. Fombonne, C. Lord, C. L. Martin, A. I. Brooks, J. S. Sutcliffe, E. H. Cook, D. Geschwind, K. Roeder, B. Devlin, M. W. State, Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 70, 863–885 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.N. Van der Aa, L. Rooms, G. Vandeweyer, J. van den Ende, E. Reyniers, M. Fichera, C. Romano, B. Delle Chiaie, G. Mortier, B. Menten, A. Destrée, I. Maystadt, K. Männik, A. Kurg, T. Reimand, D. McMullan, C. Oley, L. Brueton, E. M. H. F. Bongers, B. W. M. van Bon, R. Pfund, S. Jacquemont, A. Ferrarini, D. Martinet, C. Schrander-Stumpel, A. P. A. Stegmann, S. G. M. Frints, B. B. A. de Vries, B. Ceulemans, R. F. Kooy, Fourteen new cases contribute to the characterization of the 7q11.23 microduplication syndrome. Eur. J. Med. Genet. 52, 94–100 (2009). [DOI] [PubMed] [Google Scholar]
- 4.A. López-Tobón, S. Trattaro, G. Testa, The sociability spectrum: Evidence from reciprocal genetic copy number variations. Mol. Autism. 11, 50 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.R. L. Collins, J. T. Glessner, E. Porcu, M. Lepamets, R. Brandon, C. Lauricella, L. Han, T. Morley, L.-M. Niestroj, J. Ulirsch, S. Everett, D. P. Howrigan, P. M. Boone, J. Fu, K. J. Karczewski, G. Kellaris, C. Lowther, D. Lucente, K. Mohajeri, M. Nõukas, X. Nuttle, K. E. Samocha, M. Trinh, F. Ullah, U. Võsa; Epi25 Consortium; Estonian Biobank Research Team, M. E. Hurles, S. Aradhya, E. E. Davis, H. Finucane, J. F. Gusella, A. Janze, N. Katsanis, L. Matyakhina, B. M. Neale, D. Sanders, S. Warren, J. C. Hodge, D. Lal, D. M. Ruderfer, J. Meck, R. Mägi, T. Esko, A. Reymond, Z. Kutalik, H. Hakonarson, S. Sunyaev, H. Brand, M. E. Talkowski, A cross-disorder dosage sensitivity map of the human genome. Cell 185, 3041–3055.e25 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.B. J. Crespi, P. L. Hurd, Cognitive-behavioral phenotypes of Williams syndrome are associated with genetic variation in the GTF2I gene, in a healthy population. BMC Neurosci. 15, 127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.A. Antonell, M. Del Campo, L. F. Magano, L. Kaufmann, J. M. de la Iglesia, F. Gallastegui, R. Flores, U. Schweigmann, C. Fauth, D. Kotzot, L. A. Pérez-Jurado, Partial 7q11.23 deletions further implicate GTF2I and GTF2IRD1 as the main genes responsible for the Williams-Beuren syndrome neurocognitive profile. J. Med. Genet. 47, 312–320 (2010). [DOI] [PubMed] [Google Scholar]
- 8.N.-O. Chimge, A. V. Makeyev, F. H. Ruddle, D. Bayarsaihan, Identification of the TFII-I family target genes in the vertebrate genome. Proc. Natl. Acad. Sci. U.S.A. 105, 9006–9010 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.L. Poitras, M. Yu, C. Lesage-Pelletier, R. B. Macdonald, J.-P. Gagné, G. Hatch, I. Kelly, S. P. Hamilton, J. L. R. Rubenstein, G. G. Poirier, M. Ekker, An SNP in an ultraconserved regulatory element affects Dlx5/Dlx6 regulation in the forebrain. Development 137, 3089–3097 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.A. L. Roy, Pathophysiology of TFII-I: Old guard wearing new hats. Trends Mol. Med. 23, 501–511 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A. Adamo, S. Atashpaz, P.-L. Germain, M. Zanella, G. D’Agostino, V. Albertin, J. Chenoweth, L. Micale, C. Fusco, C. Unger, B. Augello, O. Palumbo, B. Hamilton, M. Carella, E. Donti, G. Pruneri, A. Selicorni, E. Biamino, P. Prontera, R. McKay, G. Merla, G. Testa, 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nat. Genet. 47, 132–141 (2015). [DOI] [PubMed] [Google Scholar]
- 12.J. Mariani, G. Coppola, P. Zhang, A. Abyzov, L. Provini, L. Tomasini, M. Amenduni, A. Szekely, D. Palejev, M. Wilson, M. Gerstein, E. L. Grigorenko, K. Chawarska, K. A. Pelphrey, J. R. Howe, F. M. Vaccarino, FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 162, 375–390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.B. Paulsen, S. Velasco, A. J. Kedaigle, M. Pigoni, G. Quadrato, A. J. Deo, X. Adiconis, A. Uzquiano, R. Sartore, S. M. Yang, S. K. Simmons, P. Symvoulidis, K. Kim, K. Tsafou, A. Podury, C. Abbate, A. Tucewicz, S. N. Smith, A. Albanese, L. Barrett, N. E. Sanjana, X. Shi, K. Chung, K. Lage, E. S. Boyden, A. Regev, J. Z. Levin, P. Arlotta, Autism genes converge on asynchronous development of shared neuron classes. Nature 602, 268–273 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D. Velmeshev, L. Schirmer, D. Jung, M. Haeussler, Y. Perez, S. Mayer, A. Bhaduri, N. Goyal, D. H. Rowitch, A. R. Kriegstein, Single-cell genomics identifies cell type-specific molecular changes in autism. Science 364, 685–689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.C. E. Villa, C. Cheroni, C. P. Dotter, A. López-Tóbon, B. Oliveira, R. Sacco, A. Ç. Yahya, J. Morandell, M. Gabriele, M. R. Tavakoli, J. Lyudchik, C. Sommer, M. Gabitto, J. G. Danzl, G. Testa, G. Novarino, CHD8 haploinsufficiency links autism to transient alterations in excitatory and inhibitory trajectories. Cell Rep. 39, 110615 (2022). [DOI] [PubMed] [Google Scholar]
- 16.C. Cheroni, S. Trattaro, N. Caporale, A. López-Tobón, E. Tenderini, S. Sebastiani, F. Troglio, M. Gabriele, R. B. Bressan, S. M. Pollard, W. T. Gibson, G. Testa, Benchmarking brain organoid recapitulation of fetal corticogenesis. Transl. Psychiatry 12, 520 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A. M. Paşca, S. A. Sloan, L. E. Clarke, Y. Tian, C. D. Makinson, N. Huber, C. H. Kim, J.-Y. Park, N. A. O’Rourke, K. D. Nguyen, S. J. Smith, J. R. Huguenard, D. H. Geschwind, B. A. Barres, S. P. Paşca, Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.S.-J. Yoon, L. S. Elahi, A. M. Pașca, R. M. Marton, A. Gordon, O. Revah, Y. Miura, E. M. Walczak, G. M. Holdgate, H. C. Fan, J. R. Huguenard, D. H. Geschwind, S. P. Pașca, Reliability of human cortical organoid generation. Nat. Methods 16, 75–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.P.-L. Germain, G. Testa, Taming human genetic variability: Transcriptomic meta-analysis guides the experimental design and interpretation of iPSC-based disease modeling. Stem Cell Rep. 8, 1784–1796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D. V. Hansen, J. H. Lui, P. R. L. Parker, A. R. Kriegstein, Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561 (2010). [DOI] [PubMed] [Google Scholar]
- 21.A. A. Pollen, T. J. Nowakowski, J. Chen, H. Retallack, C. Sandoval-Espinosa, C. R. Nicholas, J. Shuga, S. J. Liu, M. C. Oldham, A. Diaz, D. A. Lim, A. A. Leyrat, J. A. West, A. R. Kriegstein, Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.A. López-Tobón, C. E. Villa, C. Cheroni, S. Trattaro, N. Caporale, P. Conforti, R. Iennaco, M. Lachgar, M. T. Rigoli, B. Marcó de la Cruz, P. Lo Riso, E. Tenderini, F. Troglio, M. De Simone, I. Liste-Noya, G. Macino, M. Pagani, E. Cattaneo, G. Testa, Human cortical organoids expose a differential function of GSK3 on cortical neurogenesis. Stem Cell Rep. 13, 847–861 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.A. Gordon, S.-J. Yoon, S. S. Tran, C. D. Makinson, J. Y. Park, J. Andersen, A. M. Valencia, S. Horvath, X. Xiao, J. R. Huguenard, S. P. Pașca, D. H. Geschwind, Long-term maturation of human cortical organoids matches key early postnatal transitions. Nat. Neurosci. 24, 331–342 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.B. J. O’Roak, L. Vives, W. Fu, J. D. Egertson, I. B. Stanaway, I. G. Phelps, G. Carvill, A. Kumar, C. Lee, K. Ankenman, J. Munson, J. B. Hiatt, E. H. Turner, R. Levy, D. R. O’Day, N. Krumm, B. P. Coe, B. K. Martin, E. Borenstein, D. A. Nickerson, H. C. Mefford, D. Doherty, J. M. Akey, R. Bernier, E. E. Eichler, J. Shendure, Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–1622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.S. Tutukova, V. Tarabykin, L. R. Hernandez-Miranda, The role of neurod genes in brain development, function, and disease. Front. Mol. Neurosci. 14, 662774 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.M. Takeichi, The cadherin superfamily in neuronal connections and interactions. Nat. Rev. Neurosci. 8, 11–20 (2007). [DOI] [PubMed] [Google Scholar]
- 27.P. A. Thomson, N. R. Wray, A. M. Thomson, D. R. Dunbar, M. A. Grassie, A. Condie, M. T. Walker, D. J. Smith, D. J. Pulford, W. Muir, D. H. R. Blackwood, D. J. Porteous, Sex-specific association between bipolar affective disorder in women and GPR50, an X-linked orphan G protein-coupled receptor. Mol. Psychiatry 10, 470–478 (2005). [DOI] [PubMed] [Google Scholar]
- 28.J. Berg, S. A. Sorensen, J. T. Ting, J. A. Miller, T. Chartrand, A. Buchin, T. E. Bakken, A. Budzillo, N. Dee, S.-L. Ding, N. W. Gouwens, R. D. Hodge, B. Kalmbach, C. Lee, B. R. Lee, L. Alfiler, K. Baker, E. Barkan, A. Beller, K. Berry, D. Bertagnolli, K. Bickley, J. Bomben, T. Braun, K. Brouner, T. Casper, P. Chong, K. Crichton, R. Dalley, R. de Frates, T. Desta, S. D. Lee, F. D’Orazi, N. Dotson, T. Egdorf, R. Enstrom, C. Farrell, D. Feng, O. Fong, S. Furdan, A. A. Galakhova, C. Gamlin, A. Gary, A. Glandon, J. Goldy, M. Gorham, N. A. Goriounova, S. Gratiy, L. Graybuck, H. Gu, K. Hadley, N. Hansen, T. S. Heistek, A. M. Henry, D. B. Heyer, D. Hill, C. Hill, M. Hupp, T. Jarsky, S. Kebede, L. Keene, L. Kim, M.-H. Kim, M. Kroll, C. Latimer, B. P. Levi, K. E. Link, M. Mallory, R. Mann, D. Marshall, M. Maxwell, M. McGraw, D. McMillen, E. Melief, E. J. Mertens, L. Mezei, N. Mihut, S. Mok, G. Molnar, A. Mukora, L. Ng, K. Ngo, P. R. Nicovich, J. Nyhus, G. Olah, A. Oldre, V. Omstead, A. Ozsvar, D. Park, H. Peng, T. Pham, C. A. Pom, L. Potekhina, R. Rajanbabu, S. Ransford, D. Reid, C. Rimorin, A. Ruiz, E. S. Lein, Human neocortical expansion involves glutamatergic neuron diversification. Nature 598, 151–158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R. D. Hodge, T. E. Bakken, J. A. Miller, K. A. Smith, E. R. Barkan, L. T. Graybuck, J. L. Close, B. Long, N. Johansen, O. Penn, Z. Yao, J. Eggermont, T. Höllt, B. P. Levi, S. I. Shehata, B. Aevermann, A. Beller, D. Bertagnolli, K. Brouner, T. Casper, C. Cobbs, R. Dalley, N. Dee, S.-L. Ding, R. G. Ellenbogen, O. Fong, E. Garren, J. Goldy, R. P. Gwinn, D. Hirschstein, C. D. Keene, M. Keshk, A. L. Ko, K. Lathia, A. Mahfouz, Z. Maltzer, M. McGraw, T. N. Nguyen, J. Nyhus, J. G. Ojemann, A. Oldre, S. Parry, S. Reynolds, C. Rimorin, N. V. Shapovalova, S. Somasundaram, A. Szafer, E. R. Thomsen, M. Tieu, G. Quon, R. H. Scheuermann, R. Yuste, S. M. Sunkin, B. Lelieveldt, D. Feng, L. Ng, A. Bernard, M. Hawrylycz, J. W. Phillips, B. Tasic, H. Zeng, A. R. Jones, C. Koch, E. S. Lein, Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.M. E. Ko, C. M. Williams, K. I. Fread, S. M. Goggin, R. S. Rustagi, G. K. Fragiadakis, G. P. Nolan, E. R. Zunder, FLOW-MAP: A graph-based, force-directed layout algorithm for trajectory mapping in single-cell time course datasets. Nat. Protoc. 15, 398–420 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.T. J. Nowakowski, A. Bhaduri, A. A. Pollen, B. Alvarado, M. A. Mostajo-Radji, E. Di Lullo, M. Haeussler, C. Sandoval-Espinosa, S. J. Liu, D. Velmeshev, J. R. Ounadjela, J. Shuga, X. Wang, D. A. Lim, J. A. West, A. A. Leyrat, W. J. Kent, A. R. Kriegstein, Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.M. Lange, V. Bergen, M. Klein, M. Setty, B. Reuter, M. Bakhti, H. Lickert, M. Ansari, J. Schniering, H. B. Schiller, D. Pe’er, F. J. Theis, CellRank for directed single-cell fate mapping. Nat. Methods 19, 159–170 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.G. La Manno, R. Soldatov, A. Zeisel, E. Braun, H. Hochgerner, V. Petukhov, K. Lidschreiber, M. E. Kastriti, P. Lönnerberg, A. Furlan, J. Fan, L. E. Borm, Z. Liu, D. van Bruggen, J. Guo, X. He, R. Barker, E. Sundström, G. Castelo-Branco, P. Cramer, I. Adameyko, S. Linnarsson, P. V. Kharchenko, RNA velocity of single cells. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.C. Gagliardi, M. C. Bonaglia, A. Selicorni, R. Borgatti, R. Giorda, Unusual cognitive and behavioural profile in a Williams syndrome patient with atypical 7q11.23 deletion. J. Med. Genet. 40, 526–530 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.A. Karmiloff-Smith, H. Broadbent, E. K. Farran, E. Longhi, D. D’Souza, K. Metcalfe, M. Tassabehji, R. Wu, A. Senju, F. Happé, P. Turnpenny, F. Sansbury, Social cognition in Williams syndrome: Genotype/phenotype insights from partial deletion patients. Front. Psychol. 3, 168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.C. B. Mervis, J. Dida, E. Lam, N. A. Crawford-Zelli, E. J. Young, D. R. Henderson, T. Onay, C. A. Morris, J. Woodruff-Borden, J. Yeomans, L. R. Osborne, Duplication of GTF2I results in separation anxiety in mice and humans. Am. J. Hum. Genet. 90, 1064–1070 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.H. Colman, C. Giannini, L. Huang, J. Gonzalez, K. Hess, J. Bruner, G. Fuller, L. Langford, C. Pelloski, J. Aaron, P. Burger, K. Aldape, Assessment and prognostic significance of mitotic index using the mitosis marker phospho-histone H3 in low and intermediate-grade infiltrating astrocytomas. Am. J. Surg. Pathol. 30, 657–664 (2006). [DOI] [PubMed] [Google Scholar]
- 38.M. G. van Oijen, R. H. Medema, P. J. Slootweg, G. Rijksen, Positivity of the proliferation marker Ki-67 in noncycling cells. Am. J. Clin. Pathol. 110, 24–31 (1998). [DOI] [PubMed] [Google Scholar]
- 39.S. S. Moy, J. J. Nadler, A. Perez, R. P. Barbaro, J. M. Johns, T. R. Magnuson, J. Piven, J. N. Crawley, Sociability and preference for social novelty in five inbred strains: An approach to assess autistic-like behavior in mice. Genes Brain Behav. 3, 287–302 (2004). [DOI] [PubMed] [Google Scholar]
- 40.P. Licznerski, H.-A. Park, H. Rolyan, R. Chen, N. Mnatsakanyan, P. Miranda, M. Graham, J. Wu, N. Cruz-Reyes, N. Mehta, S. Sohail, J. Salcedo, E. Song, C. Effman, S. Effman, L. Brandao, G. N. Xu, A. Braker, V. K. Gribkoff, R. J. Levy, E. A. Jonas, ATP synthase c-subunit leak causes aberrant cellular metabolism in fragile X syndrome. Cell 182, 1170–1185.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A. Michalon, M. Sidorov, T. M. Ballard, L. Ozmen, W. Spooren, J. G. Wettstein, G. Jaeschke, M. F. Bear, L. Lindemann, Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron 74, 49–56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.P. Vianello, O. A. Botrugno, A. Cappa, R. Dal Zuffo, P. Dessanti, A. Mai, B. Marrocco, A. Mattevi, G. Meroni, S. Minucci, G. Stazi, F. Thaler, P. Trifiró, S. Valente, M. Villa, M. Varasi, C. Mercurio, Discovery of a novel inhibitor of histone lysine-specific demethylase 1A (KDM1A/LSD1) as orally active antitumor agent. J. Med. Chem. 59, 1501–1517 (2016). [DOI] [PubMed] [Google Scholar]
- 43.S. Faletti, D. Osti, E. Ceccacci, C. Richichi, B. Costanza, L. Nicosia, R. Noberini, G. Marotta, L. Furia, M. R. Faretta, S. Brambillasca, M. Quarto, L. Bertero, R. Boldorini, B. Pollo, S. Gandini, D. Cora, S. Minucci, C. Mercurio, M. Varasi, T. Bonaldi, G. Pelicci, LSD1-directed therapy affects glioblastoma tumorigenicity by deregulating the protective ATF4-dependent integrated stress response. Sci. Transl. Med. 13, eabf7036 (2021). [DOI] [PubMed] [Google Scholar]
- 44.R. Loomes, L. Hull, W. P. L. Mandy, What is the male-to-female ratio in autism spectrum disorder? A systematic review and meta-analysis. J. Am. Acad. Child Adolesc. Psychiatry 56, 466–474 (2017). [DOI] [PubMed] [Google Scholar]
- 45.L. M. Serfilippi, D. R. S. Pallman, B. Russell, Serum clinical chemistry and hematology reference values in outbred stocks of albino mice from three commonly used vendors and two inbred strains of albino mice. Contemp. Top. Lab. Anim. Sci. 42, 46–52 (2003). [PubMed] [Google Scholar]
- 46.N. N. Parikshak, R. Luo, A. Zhang, H. Won, J. K. Lowe, V. Chandran, S. Horvath, D. H. Geschwind, Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.A. J. Willsey, S. J. Sanders, M. Li, S. Dong, A. T. Tebbenkamp, R. A. Muhle, S. K. Reilly, L. Lin, S. Fertuzinhos, J. A. Miller, M. T. Murtha, C. Bichsel, W. Niu, J. Cotney, A. G. Ercan-Sencicek, J. Gockley, A. R. Gupta, W. Han, X. He, E. J. Hoffman, L. Klei, J. Lei, W. Liu, L. Liu, C. Lu, X. Xu, Y. Zhu, S. M. Mane, E. S. Lein, L. Wei, J. P. Noonan, K. Roeder, B. Devlin, N. Sestan, M. W. State, Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155, 997–1007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.T. Dixon-Salazar, J. L. Silhavy, S. E. Marsh, C. M. Louie, L. C. Scott, A. Gururaj, L. Al-Gazali, A. A. Al-Tawari, H. Kayserili, L. Sztriha, J. G. Gleeson, Mutations in the AHI1 gene, encoding jouberin, cause Joubert syndrome with cortical polymicrogyria. Am. J. Hum. Genet. 75, 979–987 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.F. Tran Mau-Them, L. Guibaud, L. Duplomb, B. Keren, K. Lindstrom, I. Marey, F. Mochel, M. J. van den Boogaard, R. Oegema, C. Nava, A. Masurel, T. Jouan, F. E. Jansen, M. Au, A. H. Chen, M. Cho, Y. Duffourd, E. Lozier, F. Konovalov, A. Sharkov, S. Korostelev, B. Urteaga, P. Dickson, M. Vera, J. A. Martínez-Agosto, A. Begemann, M. Zweier, T. Schmitt-Mechelke, A. Rauch, C. Philippe, K. van Gassen, S. Nelson, J. M. Graham, J. Friedman, L. Faivre, H. J. Lin, C. Thauvin-Robinet, A. Vitobello, De novo truncating variants in the intronless IRF2BPL are responsible for developmental epileptic encephalopathy. Genet. Med. 21, 1008–1014 (2019). [DOI] [PubMed] [Google Scholar]
- 50.J. T. Glessner, M. P. Reilly, C. E. Kim, N. Takahashi, A. Albano, C. Hou, J. P. Bradfield, H. Zhang, P. M. A. Sleiman, J. H. Flory, M. Imielinski, E. C. Frackelton, R. Chiavacci, K. A. Thomas, M. Garris, F. G. Otieno, M. Davidson, M. Weiser, A. Reichenberg, K. L. Davis, J. I. Friedman, T. P. Cappola, K. B. Margulies, D. J. Rader, S. F. A. Grant, J. D. Buxbaum, R. E. Gur, H. Hakonarson, Strong synaptic transmission impact by copy number variations in schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 107, 10584–10589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.A. R. Sanders, I. Rusu, J. Duan, J. E. Vander Molen, C. Hou, S. G. Schwab, D. B. Wildenauer, M. Martinez, P. V. Gejman, Haplotypic association spanning the 22q11.21 genes COMT and ARVCF with schizophrenia. Mol. Psychiatry 10, 353–365 (2005). [DOI] [PubMed] [Google Scholar]
- 52.J. A. Weiner, J. D. Jontes, Protocadherins, not prototypical: A complex tale of their interactions, expression, and functions. Front. Mol. Neurosci. 6, 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.E. Flaherty, T. Maniatis, The role of clustered protocadherins in neurodevelopment and neuropsychiatric diseases. Curr. Opin. Genet. Dev. 65, 144–150 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.A. Karmiloff-Smith, M. S. C. Thomas, M. H. Johnson, Introduction: Karmiloff-Smith from Piaget to neuroconstructivism, in Thinking Developmentally from Constructivism to Neuroconstructivism, A. Karmiloff-Smith, M. S. C. Thomas, M. H. Johnson, Eds. (Routledge, 2018), pp. 1–20. [Google Scholar]
- 55.B. Barak, Z. Zhang, Y. Liu, A. Nir, S. S. Trangle, M. Ennis, K. M. Levandowski, D. Wang, K. Quast, G. L. Boulting, Y. Li, D. Bayarsaihan, Z. He, G. Feng, Neuronal deletion of Gtf2i, associated with Williams syndrome, causes behavioral and myelin alterations rescuable by a remyelinating drug. Nat. Neurosci. 22, 700–708 (2019). [DOI] [PubMed] [Google Scholar]
- 56.T. Chailangkarn, C. A. Trujillo, B. C. Freitas, B. Hrvoj-Mihic, R. H. Herai, D. X. Yu, T. T. Brown, M. C. Marchetto, C. Bardy, L. McHenry, L. Stefanacci, A. Järvinen, Y. M. Searcy, M. DeWitt, W. Wong, P. Lai, M. C. Ard, K. L. Hanson, S. Romero, B. Jacobs, A. M. Dale, L. Dai, J. R. Korenberg, F. H. Gage, U. Bellugi, E. Halgren, K. Semendeferi, A. R. Muotri, A human neurodevelopmental model for Williams syndrome. Nature 536, 338–343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.A. T. N. Tebbenkamp, L. Varela, J. Choi, M. I. Paredes, A. M. Giani, J. E. Song, M. Sestan-Pesa, D. Franjic, A. M. M. Sousa, Z.-W. Liu, M. Li, C. Bichsel, M. Koch, K. Szigeti-Buck, F. Liu, Z. Li, Y. I. Kawasawa, C. D. Paspalas, Y. S. Mineur, P. Prontera, G. Merla, M. R. Picciotto, A. F. T. Arnsten, T. L. Horvath, N. Sestan, The 7q11.23 protein DNAJC30 interacts with ATP synthase and links mitochondria to brain development. Cell 175, 1088–1104.e23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.M. Zanella, A. Vitriolo, A. Andirko, P. T. Martins, S. Sturm, T. O’Rourke, M. Laugsch, N. Malerba, A. Skaros, S. Trattaro, P.-L. Germain, M. Mihailovic, G. Merla, A. Rada-Iglesias, C. Boeckx, G. Testa, Dosage analysis of the 7q11.23 Williams region identifies BAZ1B as a major human gene patterning the modern human face and underlying self-domestication. Sci. Adv. 5, eaaw7908 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.M. Mihailovich, P.-L. Germain, R. Shyti, D. Pozzi, R. Noberini, Y. Liu, D. Aprile, E. Tenderini, F. Troglio, S. Trattaro, S. Fabris, U. Ciptasari, M. T. Rigoli, N. Caporale, G. D Agostino, A. Vitriolo, D. Capocefalo, A. Skaros, A. Franchini, S. Ricciardi, I. Biunno, A. Neri, N. N. Kasri, T. Bonaldi, R. Aebersold, M. Matteoli, G. Testa, 7q11.23 CNV alters protein synthesis and REST-mediated neuronal intrinsic excitability. bioRxiv 2022.10.10.511483 [Preprint]. 10 October 2022. 10.1101/2022.10.10.511483. [DOI]
- 60.S. T. Schafer, A. C. M. Paquola, S. Stern, D. Gosselin, M. Ku, M. Pena, T. J. M. Kuret, M. Liyanage, A. A. Mansour, B. N. Jaeger, M. C. Marchetto, C. K. Glass, J. Mertens, F. H. Gage, Pathological priming causes developmental gene network heterochronicity in autistic subject-derived neurons. Nat. Neurosci. 22, 243–255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.A. L. Roy, Biochemistry and biology of the inducible multifunctional transcription factor TFII-I: 10 years later. Gene 492, 32–41 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.D. W. Kim, B. H. Cochran, Extracellular signal-regulated kinase binds to TFII-I and regulates its activation of the c-fos promoter. Mol. Cell. Biol. 20, 1140–1148 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.M. Ku, S. Y. Sokol, J. Wu, M. I. Tussie-Luna, A. L. Roy, A. Hata, Positive and negative regulation of the transforming growth factor beta/activin target gene goosecoid by the TFII-I family of transcription factors. Mol. Cell. Biol. 25, 7144–7157 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.A. Adamo, B. Sesé, S. Boue, J. Castaño, I. Paramonov, M. J. Barrero, J. C. Izpisua Belmonte, LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat. Cell Biol. 13, 652–659 (2011). [DOI] [PubMed] [Google Scholar]
- 65.L. A. Martin, E. Iceberg, G. Allaf, Consistent hypersocial behavior in mice carrying a deletion of Gtf2i but no evidence of hyposocial behavior with Gtf2i duplication: Implications for Williams-Beuren syndrome and autism spectrum disorder. Brain Behav. 8, e00895 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.A. Maiques-Diaz, T. C. Somervaille, LSD1: Biologic roles and therapeutic targeting. Epigenomics 8, 1103–1116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.M. Rapanelli, J. B. Williams, K. Ma, F. Yang, P. Zhong, R. Patel, M. Kumar, L. Qin, B. Rein, Z.-J. Wang, B. Kassim, B. Javidfar, L. Couto, S. Akbarian, Z. Yan, Targeting histone demethylase LSD1 for treatment of deficits in autism mouse models. Mol. Psychiatry 27, 3355–3366 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.F. Cavallo, F. Troglio, G. Fagà, D. Fancelli, R. Shyti, S. Trattaro, M. Zanella, G. D’Agostino, J. M. Hughes, M. R. Cera, M. Pasi, M. Gabriele, M. Lazzarin, M. Mihailovich, F. Kooy, A. Rosa, C. Mercurio, M. Varasi, G. Testa, High-throughput screening identifies histone deacetylase inhibitors that modulate GTF2I expression in 7q11.23 microduplication autism spectrum disorder patient-derived cortical neurons. Mol. Autism. 11, 88 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.M. Jabbi, Q. Chen, N. Turner, P. Kohn, M. White, J. S. Kippenhan, D. Dickinson, B. Kolachana, V. Mattay, D. R. Weinberger, K. F. Berman, Variation in the Williams syndrome GTF2I gene and anxiety proneness interactively affect prefrontal cortical response to aversive stimuli. Transl. Psychiatry 5, e622 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.H. H. Li, M. Roy, U. Kuscuoglu, C. M. Spencer, B. Halm, K. C. Harrison, J. H. Bayle, A. Splendore, F. Ding, L. A. Meltzer, E. Wright, R. Paylor, K. Deisseroth, U. Francke, Induced chromosome deletions cause hypersociability and other features of Williams-Beuren syndrome in mice. EMBO Mol. Med. 1, 50–65 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.T. L. Procyshyn, J. Spence, S. Read, N. V. Watson, B. J. Crespi, The Williams syndrome prosociality gene GTF2I mediates oxytocin reactivity and social anxiety in a healthy population. Biol. Lett. 13, 20170051 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.B. M. vonHoldt, E. Shuldiner, I. J. Koch, R. Y. Kartzinel, A. Hogan, L. Brubaker, S. Wanser, D. Stahler, C. D. L. Wynne, E. A. Ostrander, J. S. Sinsheimer, M. A. R. Udell, Structural variants in genes associated with human Williams-Beuren syndrome underlie stereotypical hypersociability in domestic dogs. Sci. Adv. 3, e1700398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.T. Bourgeron, From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat. Rev. Neurosci. 16, 551–563 (2015). [DOI] [PubMed] [Google Scholar]
- 74.C. Redies, N. Hertel, C. A. Hübner, Cadherins and neuropsychiatric disorders. Brain Res. 1470, 130–144 (2012). [DOI] [PubMed] [Google Scholar]
- 75.S. C. Taylor, S. L. Ferri, M. Grewal, Z. Smernoff, M. Bucan, J. A. Weiner, T. Abel, E. S. Brodkin, The role of synaptic cell adhesion molecules and associated scaffolding proteins in social affiliative behaviors. Biol. Psychiatry 88, 442–451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.P. Shannon, A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski, T. Ideker, Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.C. Yang, J. Li, Q. Wu, X. Yang, A. Y. Huang, J. Zhang, A. Y. Ye, Y. Dou, L. Yan, W. Zhou, L. Kong, M. Wang, C. Ai, D. Yang, L. Wei, AutismKB 2.0: A knowledgebase for the genetic evidence of autism spectrum disorder. Database 2018, bay106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.N. A. Kulak, G. Pichler, I. Paron, N. Nagaraj, M. Mann, Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324 (2014). [DOI] [PubMed] [Google Scholar]
- 79.S. Tyanova, T. Temu, J. Cox, The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016). [DOI] [PubMed] [Google Scholar]
- 80.S. Tyanova, T. Temu, P. Sinitcyn, A. Carlson, M. Y. Hein, T. Geiger, M. Mann, J. Cox, The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 81.J. Cox, M. Y. Hein, C. A. Luber, I. Paron, N. Nagaraj, M. Mann, Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.A. Alexa, J. Rahnenführer, T. Lengauer, Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607 (2006). [DOI] [PubMed] [Google Scholar]
- 83.N. Barkas, V. Petukhov, D. Nikolaeva, Y. Lozinsky, S. Demharter, K. Khodosevich, P. V. Kharchenko, Joint analysis of heterogeneous single-cell RNA-seq dataset collections. Nat. Methods 16, 695–698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.M. Lotfollahi, F. A. Wolf, F. J. Theis, scGen predicts single-cell perturbation responses. Nat. Methods 16, 715–721 (2019). [DOI] [PubMed] [Google Scholar]
- 85.F. A. Wolf, P. Angerer, F. J. Theis, SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.L. Zhang, Z. Choi, M. Wang, M. Guo, S. Modrusan, S. Kapur, H. Brugarolas, SCINA: Semi-supervised analysis of single cells in silico. Genes 10, 531 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.A. A. Pollen, A. Bhaduri, M. G. Andrews, T. J. Nowakowski, O. S. Meyerson, M. A. Mostajo-Radji, E. Di Lullo, B. Alvarado, M. Bedolli, M. L. Dougherty, I. T. Fiddes, Z. N. Kronenberg, J. Shuga, A. A. Leyrat, J. A. West, M. Bershteyn, C. B. Lowe, B. J. Pavlovic, S. R. Salama, D. Haussler, E. E. Eichler, A. R. Kriegstein, Establishing cerebral organoids as models of human-specific brain evolution. Cell 176, 743–756.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.K. Li, Z. Ouyang, D. Lin, M. Mingueneau, W. Chen, D. Sexton, B. Zhang, Cellxgene VIP unleashes full power of interactive visualization, plotting and analysis of scRNA-seq data in the scale of millions of cells. bioRxiv 2020.08.28.270652 [Preprint]. 14 April 2020. 10.1101/2020.08.28.270652. [DOI]
- 89.M. Setty, V. Kiseliovas, J. Levine, A. Gayoso, L. Mazutis, D. Pe’er, Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol. 37, 451–460 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.E. Dann, N. C. Henderson, S. A. Teichmann, M. D. Morgan, J. C. Marioni, Differential abundance testing on single-cell data using k-nearest neighbor graphs. Nat. Biotechnol. 40, 245–253 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.A. Bhaduri, M. G. Andrews, W. Mancia Leon, D. Jung, D. Shin, D. Allen, D. Jung, G. Schmunk, M. Haeussler, J. Salma, A. A. Pollen, T. J. Nowakowski, A. R. Kriegstein, Cell stress in cortical organoids impairs molecular subtype specification. Nature 578, 142–148 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.T. Saito, In vivo electroporation in the embryonic mouse central nervous system. Nat. Protoc. 1, 1552–1558 (2006). [DOI] [PubMed] [Google Scholar]
- 93.E. Meijering, M. Jacob, J. C. F. Sarria, P. Steiner, H. Hirling, M. Unser, Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 58A, 167–176 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Tables S1 to S3