Abstract
The tumor suppressor TP53 is frequently inactivated in a mutation-independent manner in cancers and is reactivated by inhibiting its negative regulators. We here cotarget MDM2 and the nuclear exporter XPO1 to maximize transcriptional activity of p53. MDM2/XPO1 inhibition accumulated nuclear p53 and elicited a 25- to 60-fold increase of its transcriptional targets. TP53 regulates MYC, and MDM2/XPO1 inhibition disrupted the c-MYC–regulated transcriptome, resulting in the synergistic induction of apoptosis in acute myeloid leukemia (AML). Unexpectedly, venetoclax-resistant AMLs express high levels of c-MYC and are vulnerable to MDM2/XPO1 inhibition in vivo. However, AML cells persisting after MDM2/XPO1 inhibition exhibit a quiescence- and stress response–associated phenotype. Venetoclax overcomes that resistance, as shown by single-cell mass cytometry. The triple inhibition of MDM2, XPO1, and BCL2 was highly effective against venetoclax-resistant AML in vivo. Our results propose a novel, highly translatable therapeutic approach leveraging p53 reactivation to overcome nongenetic, stress-adapted venetoclax resistance.
Maximal p53 reactivation by combined MDM2 and XPO1 inhibition in AML abrogates MYC-mediated resistance to venetoclax in AML.
INTRODUCTION
Inactivation of tumor suppressors and activation of oncogenes are fundamental driving forces in cancer development, progression, and resistance to therapies. p53 is a critical tumor suppressor protein that governs transcriptional programs that regulate numerous cellular functions, such as cell cycle progression, senescence, and cell death (1). Inactivation of p53 occurs most frequently through missense and/or truncating mutations of the TP53 gene or by up-regulation of negative regulators, such as Mouse double minute 2 (MDM2) and MDM4. The proto-oncogene MYC controls a myriad of cellular processes across numerous cancers and leukemias by regulating cell cycle progression, mitochondrial and ribosomal biogenesis, and metabolic abnormalities (2). It is tightly controlled by p53 in a negative feedback loop to maintain cellular homeostasis under physiological conditions (3, 4). However, this interaction is often dysregulated in cancer and leukemia cells (5) by MYC amplification or overexpression, resulting in the inactivation of p53, as reported recently (6, 7). Conversely, DNA damage–inducing chemotherapeutic drugs or ionizing radiation activates TP53 and suppress MYC expression either through direct binding of p53 to the MYC promoter or by up-regulation of tumor-suppressive microRNAs such as miR-145 (8, 9).
Acute myeloid leukemias (AMLs) are a heterogeneous group of hematologic malignancies characterized by the inability of hematopoietic stem/progenitor cells to undergo cell death and differentiation, resulting in the accumulation of immature myeloid blasts in the bone marrow (BM) and peripheral blood (PB) (10). We previously reported the ability of BCL2/BCL-XL inhibition by ABT-737 (11) and of BCL2 inhibition by ABT-199 [venetoclax (Ven)] to overcome blocks in apoptosis in AML (12, 13). Ven in combination with hypomethylating agents (HMAs) has revolutionized the treatment of patients with AML, with response rates of 70 to 90% (14). Acquired resistance to Ven-based therapies is common, although resulting in a median overall survival (OS) of only 2.4 months after Ven/HMA failure (15). While TP53 mutations are relatively infrequent in de novo AML (~10%) compared to other cancers, p53 is frequently inactivated by MDM2 overexpression, which prompted us to preclinically and clinically investigate the effects of MDM2 inhibition on the restoration of p53 function (16, 17). Clinical trials of MDM2 inhibitors demonstrated overall responses in approximately 20% of patients with AML (18, 19), warranting alternative approaches to improve their efficacy. The normal function of p53 is critically important for the efficacy of Ven (20), and we recently reported that combinatorial inhibition of MDM2 and BCL2 in TP53 wild-type AML was highly effective in Ven-resistant (Ven-R) AML through the p53-mediated reduction of myeloid cell leukemia 1 (MCL1) (21, 22); however, additional resistance mechanisms to this combination treatment remain uncovered.
Exportin-1 (XPO1; also known as CRM1) is an important transporter of more than 220 cargo proteins, including tumor suppressors such as p53, RB transcriptional corepressor 1 (RB1), Forkhead box O3 (FOXO3a), protein phosphatase 2A, and BRCA1 DNA repair associated, from the nucleus to the cytoplasm (23). XPO1 also exports a wide range of mRNAs (over 3000) with an eIF4E-sensitive element in the 3′ untranslated region and a 7-methylguanosine cap at the 5′ end by direct binding of a pentatricopeptide repeat protein (24). These mRNAs encode various oncogenes such as MYC, CCND1, and MCL1. On the basis of its biological role as a regulator of oncogenes and tumor suppressor genes, XPO1 has been investigated as an attractive therapeutic target in various malignancies. Selinexor (Sel; KPT-330), a second-generation selective XPO1 inhibitor, has been Food and Drug Administration (FDA)–approved for treatment of multiple myeloma and diffuse large B cell lymphomas (25, 26). Sel has also been assessed in several early-phase clinical trials for AML, where it exhibited anti-leukemia activities (27, 28). We previously reported that up-regulation of XPO1 protein is an independent negative determinant of AML patient survival and that coinhibition of XPO1 and MDM2 accumulates nuclear p53 and induces synergistic apoptosis in AML cell lines in a p53-dependent manner (29). Detailed molecular mechanisms of action, biomarkers of sensitivity, and in vivo efficacy of this combination have yet to be investigated.
Here, we report the molecular sequelae of combined MDM2/XPO1 inhibition including notable nuclear accumulation of p53 resulting in greatly increased transcription of target genes. We next focused on the regulation of MYC by p53, as we found unexpected overexpression of c-MYC in AML cells refractory to BCL2 inhibition. This led us to hypothesize that the triple combination of MDM2, XPO1, and BCL2 inhibition would overcome different mechanisms of resistance to BCL2 inhibition, a major clinical problem. The triple combination demonstrated a 300% extension of survival in vivo in a Ven-resistant patient-derived xenograft (PDX) model of AML.
RESULTS
Dual inhibition of MDM2 and XPO1 results in nuclear p53 accumulation and synergistic anti-leukemia efficacy in AML with limited impact on normal hematopoietic cells
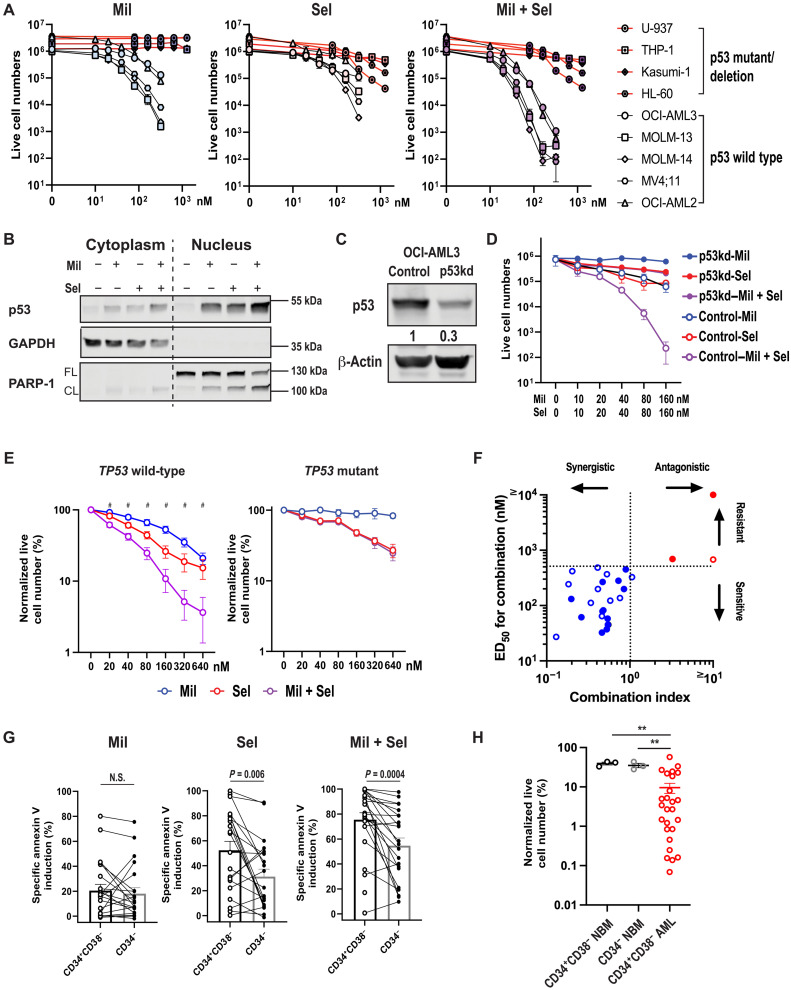
MDM2 inhibition with milademetan (Mil) reduced cell numbers and induced apoptosis in TP53 wild-type (OCI-AML3, MOLM-13, MOLM-14, MV4;11, and OCI-AML2) but not in TP53-mutant AML cells (U-937, THP-1, Kasumi-1, and HL-60 cells; Fig. 1A and fig. S1A), as expected. The XPO1 inhibitor Sel decreased cell numbers and induced apoptosis in both TP53 wild-type and mutant AML cells, with relatively higher half-maximal inhibitory concentration(IC50) and half-maximal concentration for apoptosis induction (ED50) values in cells with TP53 mutations (Fig. 1A, fig. S1A, and table S1). Mil + Sel induced synergistic cytoreduction and apoptosis (three- to four-log10 cytoreduction) in TP53 wild-type, but not in TP53-mutant or TP53-deleted, cells (Fig. 1A, fig. S1A, and table S1). The nuclear and cytoplasmic fractions from OCI-AML3 and MOLM-14 cells, which are inherently resistant to Ven, showed increased nuclear accumulation of p53 when treated with Mil + Sel, as compared to Mil or Sel alone, leading to enhanced poly (ADP-ribose) polymerase (PARP) cleavage (Fig. 1B and fig. S1B). Knockdown of TP53 in OCI-AML3 cells (Fig. 1C and fig. S1C) and MOLM-13 cells with CRISPR-engineered TP53-knockout or TP53-mutations (fig. S1D) almost completely abrogated the observed synergistic effects by Mil + Sel (Fig. 1D and fig. S1, C and E), confirming the dependency of efficacy of the dual MDM2 and XPO1 inhibition on wild-type TP53.
Fig. 1. Dual MDM2/XPO1 inhibition accumulates nuclear p53 and induces synergistic cytoreduction in TP53 wild-type AML stem/progenitor cells.
(A) TP53 wild-type and TP53-mutant AML cell lines were treated with Mil, Sel, or Mil + Sel. (B) Immunoblots of cytoplasmic and nuclear fractionations of OCI-AML3 cells treated with Mil, Sel, or Mil + Sel for 12 hours. (C) Immunoblots of p53 in OCI-AML3 cells transfected with control short hairpin RNA (shRNA; control) and shp53 (p53kd). (D) OCI-AML3 control/p53kd cells were treated with Mil, Sel, or Mil + Sel. (E) TP53 wild-type (N = 27, left) and TP53-mutant (N = 3, right) primary AML samples were treated with Mil, Sel, or Mil + Sel. Data represent means ± SEM normalized live cell numbers of primary AML samples treated. #, significant differences in normalized live cell numbers after Mil + Sel versus Mil and Mil + Sel versus Sel. (F) A two-dimensional plot for combination indices and concentrations for ED50 values. TP53 wild-type and TP53-mutant primary AML samples are shown in blue and red, respectively. Open circles represent primary AML samples resistant to Ven-based regimens. (G) Percentages of specific apoptosis in CD34+CD38− immature versus CD34− mature AML cells treated with Mil, Sel, or Mil + Sel. Paired t tests were used for statistical comparisons. (H) Live cell numbers in CD34+CD38− and CD34− normal BM (NBM) and CD34+CD38− AML cells treated with Mil + Sel. The percentages of the live cell numbers were normalized to those of untreated cells. Data represent means ± SEM of the normalized live cell numbers of samples treated. A Mil and Sel concentration of 160 nM was used. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; N.S., not significant. **P < 0.01.
Thirty primary AML samples, 23 of which (77%) were from patients categorized as adverse risk according to European LeukemiaNet criteria (30), were treated with Mil, Sel, or Mil + Sel (patient characteristics shown in fig. S2 and tables S2 and S3). These included 5 (17%) and 12 (40%) baseline (i.e., before Ven-based treatments) samples from patients who achieved complete remission (CR) by Ven-based therapies and those who had AML resistant to such treatments (i.e., after Ven-based treatments), respectively. Mil + Sel induced significantly higher levels of cytoreduction (two- to three-log10) compared to Mil or Sel alone and synergistic apoptosis in TP53 wild-type but not in TP53-mutant samples (Fig. 1, E and F). Among the recurrent gene mutations detected in these samples, TP53 mutations were identified as the sole determinant of resistance to Mil + Sel by multivariate analysis (table S4). Mil + Sel induced comparable apoptosis both in primary samples derived from patients with AML sensitive and resistant to Ven-based therapies, with comparable ED50 values for Mil + Sel between Ven-sensitive and Ven-R AML samples (fig. S1F).
Sel has reduced the frequency of leukemia stem/progenitor cells (LSPCs) in AML (31). Sel and Mil + Sel exerted increased apoptosis induction in immature AML CD34+CD38− cells as compared to more mature CD34− cells, but no difference was observed in cells treated with Mil (Fig. 1G). Notably, Mil + Sel induced significantly greater cytoreduction in CD34+CD38− AML cells compared to CD34+CD38− or CD34− normal BM cells (Fig. 1H). Together, dual inhibition of MDM2 and XPO1 induced synergistic apoptosis in AML cells carrying wild-type TP53 and in AML cells resistant to Ven, independent of recurrent mutations other than those of TP53. Dual MDM2/XPO1 inhibition was effective, especially against LSPCs, with modest to no discernable effects on normal hematopoietic stem cells (HSCs).
Dual MDM2 and XPO1 inhibition enhances activation of the p53 transcriptional program and disrupts the MYC transcriptional program
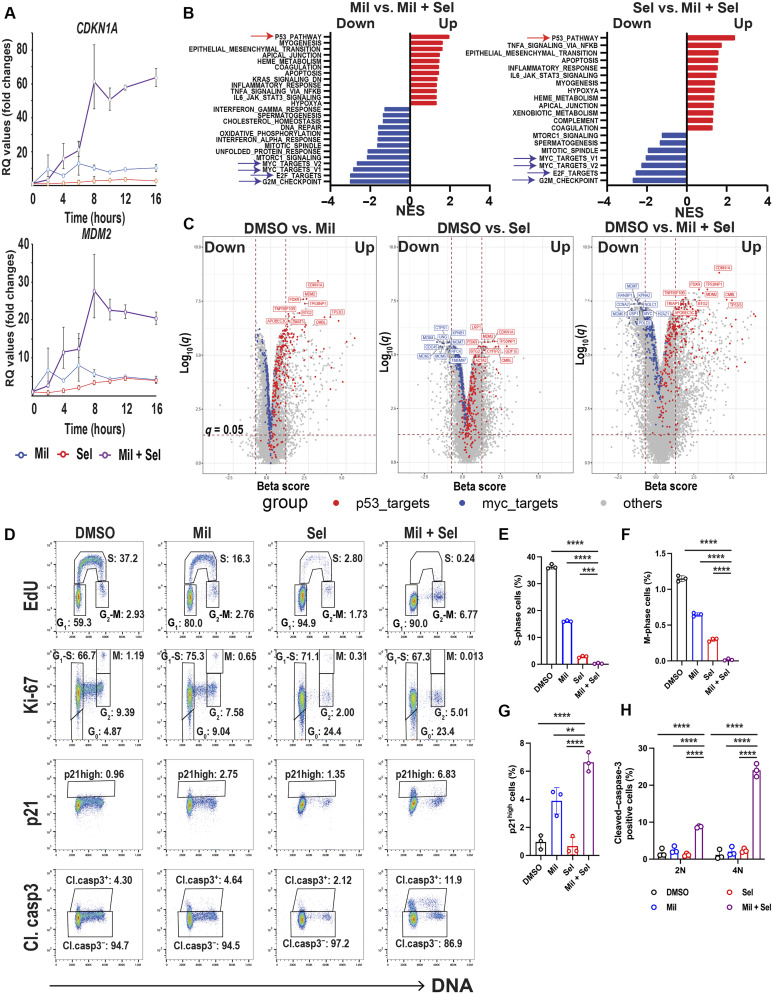
Expression levels of p53 target genes were highly induced by Mil + Sel compared to Mil or Sel alone in inherently Ven-resistant OCI-AML3 cells, peaking at 8 to 12 hours, including a 60-fold increase in CDKN1A and a 25-fold increase in MDM2 levels (Fig. 2A and fig. S3A). To explore the impact of dual MDM2/XPO1 inhibition on the transcriptome of TP53 wild-type AML cells, we performed RNA sequencing (RNA-seq) in OCI-AML3 cells treated with dimethyl sulfoxide (DMSO), Mil, Sel, or Mil + Sel. As expected, pathway analyses revealed p53 pathway to be most highly up-regulated following treatment with Mil or Mil + Sel compared to DMSO (Fig 2B and fig. S3, B to D). Although more than 200 cargo proteins and 3000 target mRNAs are XPO1 clients, RNA-seq identified the p53 pathway as the top Sel–up-regulated pathway in AML cells (fig. S3C). p53 pathway was also identified as the most up-regulated pathway when Mil + Sel was compared to Mil or Sel alone (Fig. 2B). Regarding other significantly up- or down-regulated pathways, apoptosis pathway was detected in Sel versus Mil + Sel as an up-regulated pathway but not in DMSO versus Sel, suggesting that the anti-AML effect by Sel alone is mostly cytostatic (Fig. 2B and fig. S3C). Fatty acid metabolism pathway was down-regulated only in DMSO versus Mil + Sel and not in any other comparison (fig. S3D), implicating that synergistic effects by the dual MDM2/XPO1 inhibition are partially mediated by inhibiting fatty acid metabolism. On the other hand, pathway analyses identified E2F targets, G2M checkpoint genes, and MYC targets as individually top down-regulated and shared by Mil + Sel versus control, Mil, or Sel (Fig 2B and fig. S3, B to D). We confirmed statistically significant down-regulation of MYC and its targets MCM2 and MCM4, which act as components of helicase complexes at the replication fork in S phase, and PLK1 by quantitative polymerase chain reaction (PCR; fig. S3E). These findings demonstrate an unexpected convergence of commonly up- and down-regulated pathways resulting from MDM2 or XPO1 inhibition alone and the dual MDM2/XPO1 inhibition, as the p53 pathway has not been reported as the most activated pathway in the literature upon XPO1 inhibition (23).
Fig. 2. Dual MDM2/XPO1 inhibition enhances p53 target transcription and dysregulates MYC transcriptional program, leading to cell cycle arrest.
(A) Relative quantitation (RQ) values by quantitative PCR for CDKN1A and MDM2 in OCI-AML3 cells treated with Mil, Sel, or Mil + Sel for the indicated time points. (B) Pathway analyses comparing Mil versus Mil + Sel and Sel versus Mil + Sel in RNA-seq in OCI-AML3 cells treated with Mil, Sel, or Mil + Sel for 12 hours. The top up-regulated and down-regulated pathways are indicated by red and blue arrows, respectively. NES, normalized enrichment score. (C) Volcano plots [beta score (magnitude) and q values (significance, −log10 scale)] from differential gene expression profiles in RNA-seq from OCI-AML3 cells DMSO versus Mil (left), Sel (middle), and versus Mil + Sel (right). The top 10 up-regulated TP53 targets and down-regulated MYC targets are indicated with red and blue colors, respectively. The remaining genes are indicated in gray. (D) Cell cycle analyses using multiparameter flow cytometry with Ki-67, p21, and cleaved caspase-3 in OCI-AML3 cells treated with Mil, Sel, or Mil + Sel for 24 hours. Caspase-3–negative cells were gated for EdU, Ki-67, and p21 panels. (E and F) Change in the percentages of cells in S (E) and M (F) phases in OCI-AML3 cells in indicated treatments. (G) The percentages of “p21 high” OCI-AML3 cells [rectangles shown in the third row of (D)] in indicated treatments. (H) Change of cell percentages of cleaved caspase-3–positive OCI-AML3 cells [rectangles shown in the fourth row of (D)] in indicated treatments for G1 (2N) and G2-M (4N) phases. A Mil and Sel concentration of 160 nM was used. **P < 0.01; ***P < 0.001; ****P < 0.0001.
Volcano plots clarified the differences in the magnitude and significance of individual p53 and MYC targets among the three treatment conditions, using an unbiased filtering approach from differential gene expression profiles (Fig 2C). As expected, significant up-regulation of TP53 targets, including CDKN1A, MDM2, FDXR, and TP53INP1 was observed following Mil compared to DMSO. These p53 targets were also shown to be up-regulated in our first clinical trial with MDM2 inhibitor RG7112 (18), attesting to the potential clinical relevance of this finding. Similarly, TP53 targets were up-regulated, and MYC targets were down-regulated by Sel treatment. Notably, the magnitude and significance of p53 target up-regulation were prominently enhanced, and in contrast, MYC targets were further down-regulated by Mil + Sel, compared to single agents (Fig 2C). Similar findings were observed in RNA-seq in CRISPR-engineered TP53 wild-type and p.R248Q mutant MOLM-13 cells after DMSO, Mil, Sel, or Mil + Sel treatment (fig. S4 and S5). XPO1 inhibition has been reported to impair cell growth through down-regulation of homeobox (HOX) genes in nucleophosmin 1 (NPM1) mutant (NPM1c) AML cells (32). We observed the down-regulation of HOX genes and of MEIS1 in NPM1c OCI-AML3 cells treated with Sel (fig. S6A). Mil + Sel–induced growth inhibition in NPM1c OCI-AML3 cells through p53 activation occurred within 24 hours, which was much faster than that induced by HOX gene silencing (7 to 9 days) (32), indicating a broader impact of p53 pathway activation than that of HOX silencing. The data demonstrate that the enhanced transcriptional activation of p53 by dual MDM2/XPO1 inhibition is accompanied by a similarly enhanced down-regulation of the MYC transcriptional program.
On the basis of these findings, we next assessed the impact of dual MDM2/XPO1 inhibition on the cell cycle in OCI-AML3 cells, as it significantly down-regulated cell cycle–regulating genes controlling S and G2-M phases. We developed a flow cytometry panel to simultaneously measure Ki-67, DNA, 5-ethynyl-2′-deoxyuridine (EdU), p21, and active (cleaved) caspase-3 levels. As expected, Mil delayed G1 to S transition, and Sel induced profound G1 cell cycle arrest with a significant increase in cells in G0 determined by low levels of Ki-67 with the 2N DNA content (Fig. 2, D to F, and fig. S6, B and C). In contrast, Mil + Sel treatment resulted in almost complete cell cycle arrest with significantly reduced S-phase cells, with G1 and G2-M blocks (Fig. 2, D to F, and fig. S4B). Mil + Sel increased p21 levels in both G1 and G2-M phases, with 2N and 4N DNA content at 24 hours, suggesting p53 reactivation–induced G0-G1 and G2-M arrest (Fig. 2, D and G), being consistent with previous findings (33, 34). Furthermore, elevated levels of cleaved caspase-3 were more profound at G2-M (4N) than at G1 (2N) (Fig. 2, D and H), indicating that apoptosis induction occurred first in G2-M, which was followed by late cell death in G1, as the G2-M population almost disappeared at a later time point (48 hours; fig. S6, D and E). Collectively, these data suggest that dual inhibition of MDM2 and XPO1 cooperatively induces cell cycle arrest and apoptosis across cell cycle phases.
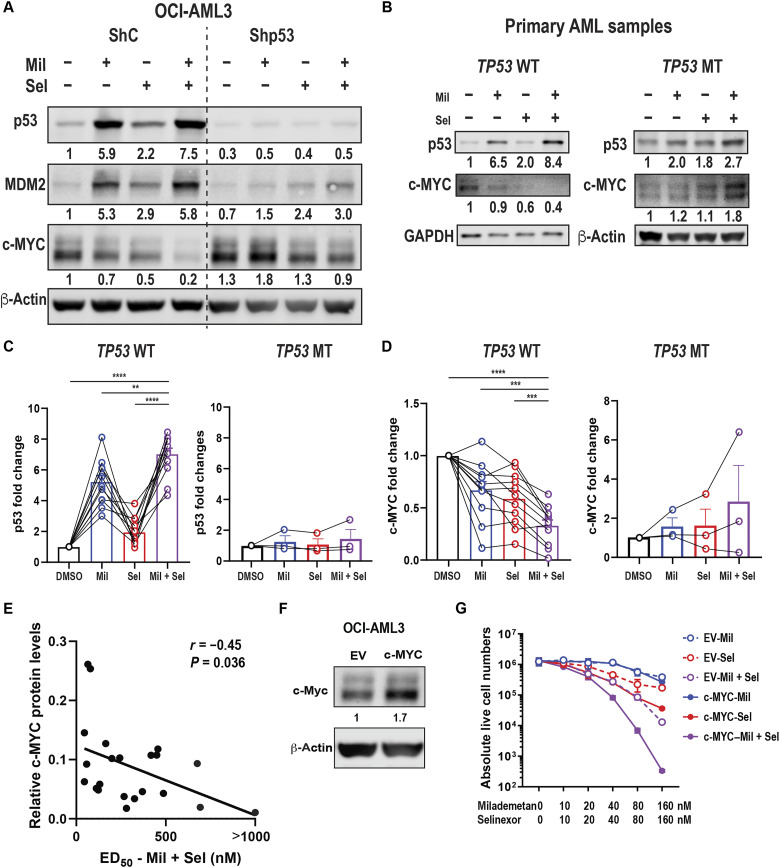
Dual inhibition of MDM2 and XPO1 reduces c-MYC in a p53-dependent manner; baseline c-MYC protein is biomarker for the efficacy of MDM2/XPO1 inhibition
We previously reported that XPO1 inhibition reduced c-MYC protein levels by blocking translation initiation of c-MYC (35). As expected, Mil + Sel profoundly up-regulated p53 targets and greatly down-regulated MYC mRNA and protein levels compared to Mil or Sel alone in OCI-AML3 ShC cells, but not in OCI-AML3 shp53 cells (Fig. 3A and fig. S7, A and B). Similarly, c-MYC protein levels were markedly reduced by Mil + Sel in TP53 wild-type MOLM-13 cells compared to those in TP53-knockout or TP53-mutant cells (fig. S7C). Enhanced reduction of c-MYC protein levels by Mil + Sel compared to controls was also observed in primary AML samples with wild-type but not with mutant TP53 (Fig. 3, B to D). Furthermore, baseline c-MYC protein levels in primary AML samples showed a statistically significant negative correlation with ED50 concentrations (r = −0.45, P = 0.036) for Mil + Sel (Fig. 3E), suggesting that c-MYC levels are a potential biomarker for Mil + Sel in AML. Mil + Sel exerted superior cytoreduction and apoptosis induction in c-MYC–overexpressing OCI-AML3 cells compared to empty vector controls, associated with a profound decrease in c-MYC protein levels (Fig. 3, F and G, and fig. S7, D and E). Together, pretreatment c-MYC protein levels may serve as a biomarker for the efficacy of dual MDM2 and XPO1 inhibition in AMLs.
Fig. 3. Dual MDM2/XPO1 inhibition–mediated c-MYC reduction is p53 dependent and baseline c-MYC protein levels correlate with sensitivity to Mil + Sel.
(A) Immunoblot of p53, MDM2, and c-MYC in OCI-AML3 cells transfected with shRNA for scramble control (ShC) and p53 knockdown (Shp53) treated with Mil, Sel, or Mil + Sel for 12 hours. (B) Immunoblot of p53, MDM2, and c-MYC of TP53 wild-type (WT) and mutant (MT) primary AML samples treated with Mil, Sel, or Mil + Sel. (C) Protein levels of p53 in 10 TP53 WT (left) and 3 TP53 MT (right) primary AML samples. (D) Protein levels of c-MYC in 10 and 3 WT (left) and MT (right) TP53 primary AML samples. Data in (C) and (D) represent means ± SEM values. (E) The correlational plot of baseline c-MYC protein levels and ED50 values for Mil + Sel in primary AML samples. (F) Immunoblot of c-MYC in OCI-AML3 cells transfected with empty vector (EV) control or MYC-overexpressing plasmids (c-MYC). (G) Live cell numbers in OCI-AML3 EV and c-MYC cells treated with Mil, Sel, or Mil + Sel for 72 hours. A Mil and Sel concentration of 160 nM was used. **P < 0.01; ***P < 0.001; ****P < 0.0001.
Anti-leukemia efficacy of MDM2 and XPO1 inhibition in Ven-R AML xenograft is associated with suppression of c-MYC, leading to restored sensitivity to Ven
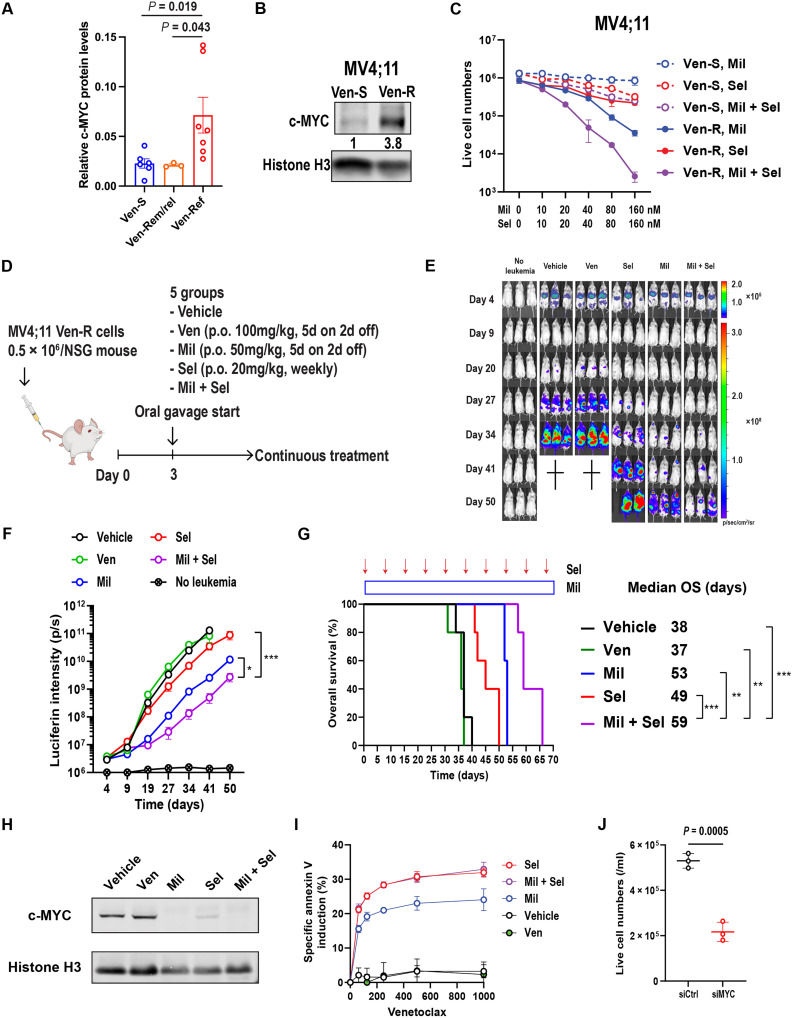
To further investigate the relevance of c-MYC protein in Ven-R AML, we compared protein levels of c-MYC in primary AML samples from patients who achieved CR (Ven-sensitive and Ven-S, collected before Ven-based therapies), those who initially achieved CR and later relapsed (Ven-Rem/rel, collected after Ven-based therapies), and those with disease primarily refractory to Ven-based therapy (Ven-Ref, collected after Ven-based therapies; table S3). We found significantly higher c-MYC protein levels in Ven-Ref primary AML samples than in Ven-S or Ven-Rem/rel samples (Fig. 4A and fig. S8A). c-MYC protein levels were significantly higher in MV4;11 Ven-R (21) than in Ven-S cells (Fig. 4B). Ven-R MV4;11 cells were also resistant to Ven or Ven + 5′-azacitidine (fig. S8, B and C), serving as a clinically relevant model of Ven/HMA-resistant AML. Furthermore, in line with c-MYC–overexpression in OCI-AML3 cells (Fig. 3F), MV4;11 Ven-R cells were more sensitive to Mil + Sel than parental cells (Fig 4C and fig. S8D), showing 1 to 2 log10 greater cytoreduction.
Fig. 4. Dual MDM2/XPO1 inhibition induces synergistic anti-leukemia effects in Ven-R AML cells.
(A) c-MYC protein levels in primary AML samples from patients sensitive to Ven-containing regimens (Ven-S; N = 6), those who had remission and then relapse (Ven-Rem/rel; N = 3), and those who had primarily refractory disease (Ven-Ref; N = 7). Data represent means ± SEM. (B) Immunoblot of c-MYC in MV4;11 Ven-S and resistant (Ven-R) cells. (C) Live cell numbers of MV4;11 Ven-S and Ven-R cells treated with Mil, Sel, or Mil + Sel for 72 hours. (D) Schematic diagram of in vivo xenograft model. (E) Images of leukemia burden measured by luciferin intensities in three representative mice from each treatment group. (F) Luciferin intensities of each treatment group (N = 8, 9, 8, 9, and 5 for vehicle, Ven, Mil, Sel, and Mil + Sel, respectively). (G) Survival curves of each treatment group and treatment durations of Mil (blue rectangle) and Sel (red arrows). (H) Immunoblots for c-MYC in MV4;11 Ven-R cells obtained from moribund mice in each in vivo treatment group. (I) Ven (48 hours)–specific annexin V induction in MV4;11 Ven-R cells after each in vivo treatment. (J) Live cell numbers of MV4;11 Ven-R cells treated with 200 nM of Ven (48 hours) with prior transfection with control siRNA (siCtrl) or MYC siRNA (siMYC) sequences. Histone H3 serves as the loading control in (B) and (H). **P < 0.01; ****P < 0.0001.
In a systemic model of MV4;11 Ven-R cells labeled with luciferase in NOD.Cg-Prkdcscid Prkdcscid/SzJ (NSG) mice (Fig. 4D), the leukemia burden was reduced, and OS was prolonged in the Mil + Sel group compared to vehicle, Ven, Mil, or Sel groups (Fig. 4, E to G). Although Sel or Mil + Sel treatments reduced white blood cell (WBC) counts, Mil + Sel did not induce changes in body weight, hemoglobin levels, or platelet counts relative to those in the control group without AML cells or the individual treatment groups (fig. S8, E and F). The data indicate that the Mil + Sel combination exerts superior anti-leukemia efficacy compared to the individual agents in Ven-R AML cells in vitro and in vivo. However, the Mil + Sel group eventually had aggressive AML and succumbed to the disease, suggesting that a modified therapeutic strategy is necessary to eradicate the residual persistent AML cells. c-MYC protein levels were essentially undetectable in Ven-resistant MV4;11 (MV4;11 VR) cells harvested from mice treated with Mil, Sel, or Mil + Sel after they became moribund (Fig. 4H), suggesting that c-MYC suppression by MDM2/XPO1 inhibition was still effective but resistance developed, nevertheless. These cells demonstrated partially restored sensitivity to Ven (Fig. 4I). Consistently, MYC knockdown by small interfering RNA (siRNA) increased the sensitivity to Ven in MV4;11 VR cells (Fig. 4J and fig. S8G). These findings suggest that c-MYC overexpression contributes to Ven resistance and that sustained reduction of c-MYC in residual disease could render AML cells susceptible to Ven again.
Triple inhibition of MDM2, XPO1, and BCL2 further enhances the anti-leukemia efficacy of MDM2 and XPO1 inhibition in vitro and in vivo
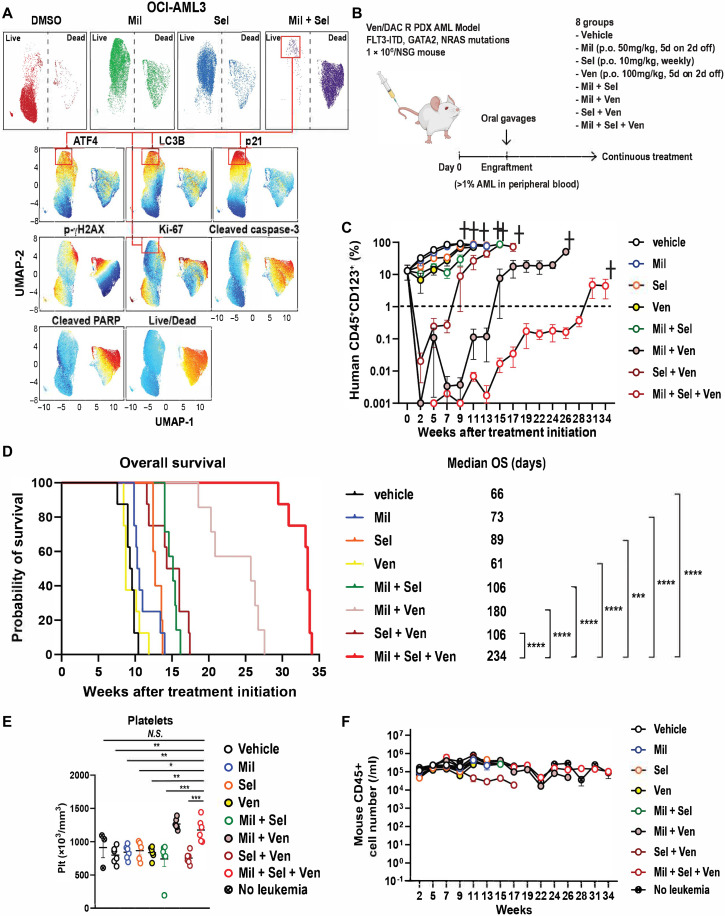
To further define the biological characteristics of residual resistant AML cells after dual inhibition of MDM2 and XPO1, we used high-parametric flow cytometry to determine cellular levels of active (cleaved) caspase-3, cleaved PARP, activating transcription factor 4 (ATF4), H2A.X variant histone (γH2AX), p21, microtubule associated protein 1 light chain 3 beta (LC3B), Ki-67, and an amine-reactive live/dead discriminating dye, which enabled us to evaluate apoptosis, autophagy, and integrated stress response (ISR) (36). Mil or Sel alone induced only moderate cell death, with elevated levels of p21, LC3B, and ATF4 and decreased Ki-67 levels in the surviving cell population. Mil + Sel induced profound cell death (Fig. 5A and fig. S9A), leaving alive a small but resistant fraction of AML cells with the highest levels of p21, along with high ATF4 and LC3B and very low Ki-67 (Fig. 5A, connected rectangles), suggesting a quiescent AML cell population protected by three distinct cell-protective stress response pathways (i.e., quiescence, ISR, and autophagy). This observation corresponded to the finding of CDKN1A as the most up-regulated p53 target gene in OCI-AML3 cells after Mil + Sel treatment (Fig. 2C). We reported previously that cell cycle arrest in MDM2 inhibitor–resistant cells could be overcome by BCL2 inhibition (21).
Fig. 5. MDM2/XPO1 inhibition leaves alive p21high/Ki-67low AML cells and triple inhibition of MDM2, XPO1 and BCL2 exhibits superior anti-leukemia effects in Ven-R AML cells.
(A) Uniform manifold approximation and projection (UMAP) data of target protein levels determined in OCI-AML3 cells with indicated treatments (160 nM of Mil and/or Sel, 48 hours). Top: Two major populations of live (left) and dead (right) cells under each treatment condition. Bottom: Protein levels on a color scale with residual AML cells after Mil + Sel highlighted in connected red rectangles. (B) Schematic of the in vivo PDX AML model. (C) Percentages of live human AML cell numbers in PB specimens from mice treated with the indicated drugs. (D) OS of mice with indicated treatments (N = 8, 8, 8, 5, 7, 7, 8, and 8 for vehicle, Ven, Mil, Sel, Mil + Sel, Mil + Ven, Sel + Ven. and Mil + Sel + Ven groups, respectively). (E) Murine platelet counts in PB specimens in each treatment group at week of 7. (F) Murine CD45+ cell numbers in PB specimens collected from indicated treatment groups over the entire treatment courses (N = 6). Data represent means ± SEM values in (C), (E), and (F). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Next, we simultaneously inhibited MDM2, XPO1, and BCL2, which led to an even more profound cytoreduction of OCI-AML3 cells and primary AML samples with wild-type TP53 and with clinical resistance to Ven-based therapies, compared to all other single agents and doublet combinations (fig. S9, B and C). Primary AML samples with mutant TP53 did not respond to the triple inhibition of MDM2, XPO1, and BCL2, confirming dependency of the efficacy of the triple combination on wild-type TP53 (fig. S9D). To investigate the efficacy of this triple combination therapy in vivo, we injected PDX AML cells, established from an AML patient resistant to Ven and decitabine (DAC) (37) into NSG mice, established engraftment, and treated mice with seven different treatment regimens, the triple combination of Mil, Sel [reduced doses from 20 to 10 mg/kg a week, which is a lower dose than previous studies (31, 38), based on suppression of WBC counts in the previous MV4;11 VR xenograft model], and Ven; a two-drug combination (Mil + Sel, Mil + Ven, or Sel + Ven); or the individual drugs as monotherapy (Fig. 5B). The group treated with the triple combination showed the deepest cytoreduction in circulating blasts (>4-log10; Fig. 5C and fig. S9E), and this cytoreduction was sustained with lower residual levels of circulating blasts compared to Mil + Ven group or all other treatment groups. The triple combination group showed significantly longer OS than any other group (Fig. 5D), reflecting the profound cytoreduction. The median OS ranged from 66 days in the control group to 234 days in the triple combination group. The triple combination did not affect mouse body weights throughout the course of the treatment (fig. S9F). The triple combination did not affect hemoglobin levels (fig. S9G). Triple combination– and Mil + Ven–treated groups showed higher murine CD45+ cell and platelet counts than any other treatment group, probably reflecting the recovery of normal hematopoiesis from myelosuppression secondary to AML infiltration (Fig. 5E and fig. S9H). The triple combination group demonstrated comparable levels of normal murine CD45+ cells compared to the control group lacking AML throughout the course of treatment (Fig. 5F), indicating hematologic tolerability of the triple combination in mice. These results suggest that the concomitant inhibition of MDM2, XPO1, and BCL2 is a feasible and highly effective treatment strategy that has the potential to overcome resistance to Ven.
Triple inhibition of MDM2, XPO1, and BCL2 overcomes Ven resistance by eradicating AML cell populations in quiescence with increased MCL1 and stress responses
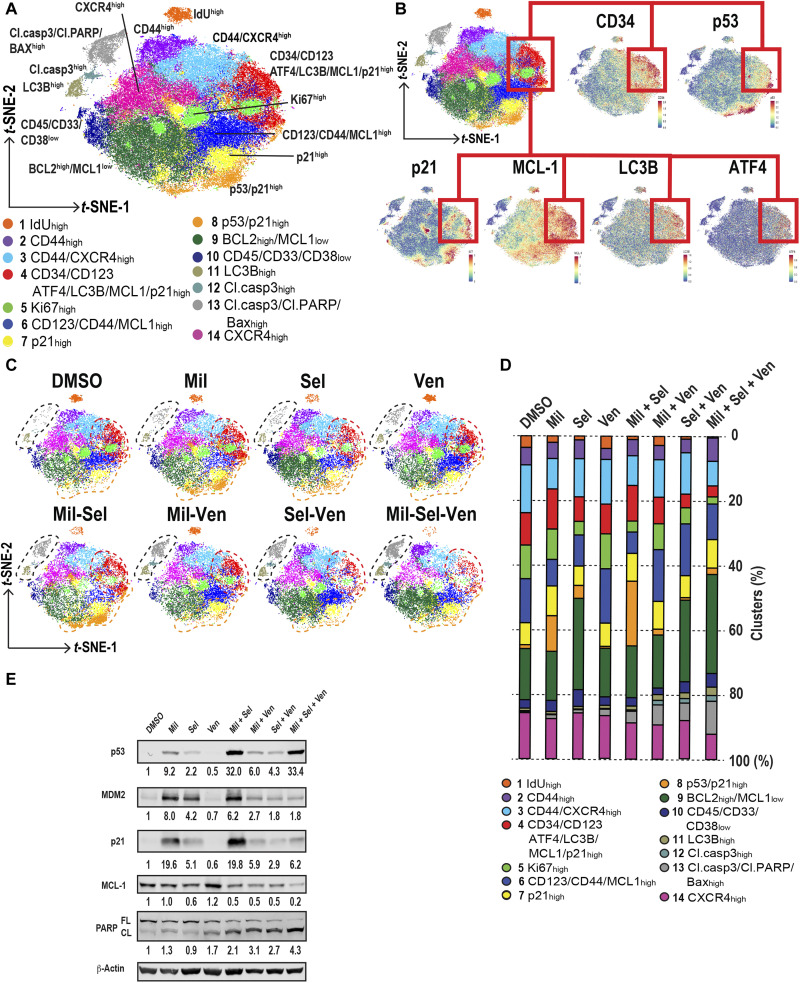
To gain insight into the mechanisms of action of the triple combination of Mil, Sel, and Ven against Ven/HMA-resistant AML at the single-cell level, we treated the PDX AML cells ex vivo with Mil, Sel, and/or Ven for 24 hours to assess the early impact on cellular responses, when the cell death was still modest, and used single-cell mass cytometry [cytometry by time of flight (CyTOF); fig. S10A]. As expected, the triple combination demonstrated the lowest number of live cells determined by 195-platinum (195Pt) positivity (fig. S10B). To annotate p53 reactivation or up-regulated stress response–associated cells, unsupervised clustering of 195Pt-negative live cells was performed incorporating all surface and intracellular markers (see Supplementary Materials and Method). Unsupervised clustering distinguished 14 cell clusters (Fig. 6A), and annotations were performed on the basis of the characteristic of protein levels (Fig 6B and fig. S10C). Specifically, AML cells in S phase were determined solely by increase of iododeoxyuridine (IdU; cluster 1); those proliferating were determined by increased Ki-67 (cluster 5), and those undergoing apoptosis were determined by increased cleaved caspase-3 and BAX (clusters 11, 12, and 13). p53 reactivation–associated clusters were defined by increased p53 and/or p21 (clusters 4, 7, and 8). Notably, cluster 4 also demonstrated enrichment of LSPC markers CD34 and CD123 and the highest MCL1 levels. The unsupervised clustering also revealed shifts in the distribution of those clusters by each treatment (Fig. 6, C and D). p53 reactivation by Mil or Mil + Sel decreased the number of IdU- and Ki-67–positive proliferating cells, consistent with the observed cell cycle perturbation by Mil + Sel in the previous cell cycle analyses (Fig. 2, D to F). However, p53 reactivation generated clusters with high p21 (clusters 4, 7, and 8). On the other hand, while Ven had no impact on clusters 1 and 5, Mil + Ven effectively decreased the number of p53 reactivation–associated clusters 7 and 8. Nevertheless, cluster 4 persisted after Mil + Ven, exhibiting high p21 along with increased MCL1, BCL-XL, ATF4, and LC3B (figs. S10C and S11). The triple combination decreased cluster 4, inducing apoptosis shown as increase in clusters 11, 12, and 13 (Fig. 6, C and D). These data suggest that the triple combination is sufficient to induce apoptosis in persistent AML cells harboring increased p21, BCL-XL, and MCL1 levels along with up-regulated stress responses. The high p21 levels induced by Mil + Sel in OCI-AML3 cells were decreased by adding Ven, along with a profound decrease in MCL1 and increase in cleaved PARP levels in the triple combination (Fig. 6E and fig. S10, D and E).
Fig. 6. Triple inhibition of MDM2, XPO1, and BCL2 induces apoptosis in p53 reactivation–responsive persistent AML cells with increased MCL1 along with stress responses.
(A) t-distributed stochastic neighbor embedding (t-SNE) plots generated by unsupervised clustering and dimension reduction using RphenoGraph. Clusters are annotated by numbers with distinct colors. (B) The t-SNE plot shown in (A) along with marker expression plots by t-SNE for CD34, p53, p21, MCL1, LC3B, and ATF4. Cluster 4 is connected by red lines and rectangles. (C) t-SNE plots of samples treated with each indicated treatment. Clusters 11, 12, and 13; cluster 4; and cluster 8 are circled by black, red, and orange dashed lines, respectively. (D) Proportions of each cluster in each sample with the indicated treatment in 195Pt-negative live cells. The colors correspond with the ones in the t-SNE plot shown in (A). (E) Immunoblots of indicated proteins collected from OCI-AML3 cells with indicated treatments (100 nM of Mil, Sel, and/or Ven for 24 hours).
To detect any TP53 mutations that emerged and investigate phenotypic changes in AML blasts after the triple combination, we performed single-cell DNA + protein sequencing (scDNA + protein seq) analysis on BM samples collected from mice before and after the triple combination. We did not detect any TP53 mutations in pre- and posttreatment samples. The AML cells after triple combination demonstrated increased immature phenotypic markers including CD117, CD123, CD45RA, and CD44 (clusters 5 to 8), consisting of clusters with high levels of monocytic markers CD11b, CD11c, and CD64 (clusters 5 and 6; figs. S12 and S13). The residual AML cells after the triple combination exhibited relatively low MCL1 levels compared to those with Ven or double combinations (fig. S14). Further proteomic characterizations will be required to better understand the detailed molecular changes in these residual AML cells. Collectively, the triple inhibition of MDM2, XPO1, and BCL2 counteracted the MDM2/XPO1 inhibition–induced increase in p21 and MCL1 with activated stress responses to overcome Ven resistance, thereby inducing profound cell kill and notable increase in survival of mice bearing Ven-R AML.
DISCUSSION
Inhibition of key p53-inactivating molecules, such as MDM2 and XPO1, or of the main anti-apoptotic protein BCL2 has met with limited clinical success as monotherapy (13, 18, 27). We have previously provided rationale for the combined inhibition of MDM2 and BCL2 in AML, and a clinical trial, combining idasanutlin and Ven as MDM2 and BCL2 inhibitors, respectively, showed promising results (21, 22). We also reported that combined MDM2 and XPO1 inhibition increased the nuclear levels of p53, and importantly that wild-type TP53 was critical for the efficacy of XPO1 inhibition in AML. Knockdown of or mutations of TP53 completely abrogated apoptosis in AML (29). Furthermore, we demonstrated that the combined inhibition of MDM2 and XPO1 synergistically and massively enhances the transcriptional activity of p53, which, in turn, resulted in significant reduction of c-MYC expression and its transcriptional signaling, which was attributable to the negative regulation of c-MYC by p53 (8, 9).
c-MYC has been reported to transform normal hematopoietic cells into myeloid leukemia (39, 40) and is essential for hematopoietic and leukemia stem cell (LSC) maintenance (41, 42). Single-cell RNA-seq identified MYC as one of the most up-regulated genes in HSC-like immature AML subpopulations (43). Several studies have shown the clinical impact of c-MYC expression in AML (44, 45), and c-MYC–targeted therapies have been investigated for various cancers, lymphomas, and leukemias (5, 46). Therapeutic targeting of bromodomain extraterminal proteins (BET) down-regulated c-MYC but demonstrated only modest activity in patients with AML (47, 48). Resistance mechanisms to BET inhibitors include the sustained MYC expression in LSCs despite the loss of Brd4 from the Myc enhancer (49) or the rapid restoration of MYC after BRD4 inhibition (50), suggesting that therapeutic combinations that leverage alternative pathways are needed to consistently suppress MYC expression and improve anti-leukemia efficacy. Dual targeting of p53 and c-MYC by MDM2 and BRD4 inhibitors exhibited promising efficacy in myeloid malignancies (51, 52), suggesting that p53 activation adds therapeutic benefit to p53-independent MYC inhibition in cancers. These studies collectively support our concept of maximal reactivation of p53 by dual targeting of MDM2 and XPO1, where the enhanced p53 activity potently inhibits c-MYC. Notably, significant up-regulation of MYC was observed in LSCs at the single-cell level (43), and we reported that MYC targets were significantly up-regulated in LSCs and minimal residual disease cells in adverse risk patients with AML (53), which underscores the importance of MYC in immature, therapy-resistant AML cells.
We identified elevated c-MYC protein levels in AML cells refractory to Ven-based therapies, the mechanisms of which are still not completely understood. We speculate that RAS pathway–activating gene mutations (e.g., NRAS, KRAS, HRAS, and PTPN11) may account for elevated c-MYC levels in Ven-R AML cells, as reported for RAS activation that transcriptionally induces MYC and/or stabilizes c-MYC proteins by posttranslational modifications (54, 55). Resistance factors to Ven/HMA include activation of signaling, including RAS mutations (56). Although the sample size was limited in the present study, five of seven AML samples resistant to Ven-based therapies had RAS pathway mutations (KRAS, NRAS, or PTPN11), whereas only one had a PTPN11 mutation in the Ven-S AML group (table S3). Consistently, the combination of MDM2 and XPO1 inhibitors was highly active in primary AML samples with RAS pathway mutations (fig. S2). Notably, we found that the combination of BCL2 and mitogen-activated protein kinase (MAPK) inhibition (cobimetinib) induced synergistic cell death in preclinical models, with a significant reduction of c-MYC and the perturbation of the MYC pathway in BCL2 + MAPK inhibition-sensitive AML cells (57), supporting the fact that the potential importance of targeting MYC by p53 activation may benefit RAS-mutated AMLs. Significant down-regulation of the fatty acid metabolism pathway by Mil + Sel may also explain mechanisms of action of MDM2/XPO1 inhibition in Ven-R AML (58).
Previously, we reported that p53 reactivation by MDM2 inhibition induced prosurvival G1 cell cycle arrest, which was effectively eliminated by adding Ven by MDM2 inhibition–mediated suppression of RAS pathway, leading to the reduction of MCL1 (21). In accordance with these findings, CDKN1A was one of the most up-regulated genes following p53 reactivation by combined MDM2 and XPO1 inhibition. Multiparametric flow cytometry of multiple stress response regulators identified high p21 and low Ki-67 levels along with increased stress response molecules ATF4 and LC3B in residual AML cells, suggesting that quiescent AML cells account, at least in part, for resistance to p53 reactivation–induced cell death. p21 was by far the highest up-regulated gene in the original MDM2 inhibitor trial (18). MV4;11 VR cells with acquired resistance to MDM2/XPO1 inhibition in vivo showed a sustained decrease in c-MYC, which, in turn, was associated with restored sensitivity to Ven, a finding supported by increased sensitivity to Ven by MYC knockdown.
The triple combination of MDM2, XPO1, and BCL2 inhibition exerted superior anti-leukemia activity in comparison to all single-agent and dual-agent combination treatments in Ven/HMA-resistant primary AML cells and markedly prolonged survival in the PDX AML model by 300 to 400% compared to single agents. Our previously reported data on the combination of BCL2 and MDM2 inhibition were reproduced in this study and clinically translated into nearly 50% response rates in older, unfit relapsed/refractory patients with AML (22). These findings are particularly relevant because resistance to Ven-based therapies has become one of the major clinical challenges, associated with extremely short survival after relapse from Ven/HMA (15). Using CyTOF, we identified a cluster of AML cells with concurrently high levels of p21, ATF4, LC3B, BCL-XL, and MCL1. We previously reported that the combinatorial inhibition of MDM2 and BCL2 significantly decreased MCL1 levels, thereby inducing synergistic anti-leukemia efficacy (21). Mil + Ven reduced the cluster with high MCL1 levels; however, it left behind a cell population with up-regulated p21, ATF4, LC3B, BCL-XL, and MCL1, which was reduced by the triple combination. Therapy-tolerant persister cells after BH3 mimetics treatment established increased ISR with ATF4 up-regulation (59), being another potential resistance mechanism to MDM2 + BCL2 inhibition.
Another potential concern of MDM2 + BCL2 inhibition is the emergence or selection of mutant TP53 clones, as we have shown in a recent clinical trial (22). Our scDNA + protein seq analysis did not detect any TP53 mutations before or after treatment, likely because of the absence of TP53-mutant clones in the PDX AML cells, as we have shown that TP53 mutations detected after MDM2 + BCL2 inhibitor therapy were preexisting and could be identified by duplex sequencing (22). A cellular hierarchy–based drug response framework has shown that Ven is one of the top effective agents in quiescent LSPCs compared to primed/cycling LSPCs or more mature AML fractions, and XPO1 and nuclear export–associated genes are highly expressed in cycling LSPCs (60). In the present study, Mil + Sel induced cell-kinetic quiescence with high p21 and low Ki-67, and the addition of Ven induced a pronounced cytoreduction in Ven-R AML cells in vitro and in vivo, potentially targeting both quiescent and cycling LSPCs. Unsupervised clustering also detected a distinct cluster with high levels of CD44 and CXCR4, having been associated with LSC survival or Ven resistance (61, 62), which requires further investigation as another relevant mechanism related to BM niche–mediated therapy resistance. We have successfully addressed drug resistance in fms related receptor tyrosine kinase 3 (FLT3) mutant AMLs by targeting CXCR4, CD44, VLA4, and SDF1 (63). Whether triple inhibition addresses Ven resistance in more differentiated monocytic AML and/or by altered metabolic features remains to be investigated. ScDNA + protein seq revealed that the residual AML cells after the triple combination of Mil + Sel + Ven in vivo predominantly consisted of immature cell clusters with increased CD123, CD117, and CD45RA levels (clusters 5 and 6). These clusters also demonstrated elevated levels of monocytic markers CD11b, CD11c, and CD64. Those clusters also showed elevated CD69, a metabolic gatekeeper in immune cells (64), regulating glucose uptake and glycolysis, and CD71 (transferrin receptor 1), a marker for iron uptake, suggesting a metabolic adaption that confers resistance to the triple combination. These findings suggest that these adapted AML cells demonstrate multiphenotypic characters of immature and monocytic blasts after the triple inhibition of MDM2, XPO1, and BCL2. Further studies are needed to fully characterize those subpopulations. In addition, we conducted flow cytometry experiments on residual CD45+ cells in PDX AML cells obtained from mice that became moribund during each in vivo treatment. Notably, those cells already adapted to each treatment after continuous in vivo treatments. In contrast to PDX AML samples after each in vitro treatment shown in Fig. 6, we did not see increases in LC3B, ATF4, p21, or MCL1 in those treated with and adapted to the triple combination (Mil + Sel + Ven). Rather, MCL1 levels were higher in the Ven-treated sample than in any other sample, and those in the triple combination–treated sample displayed relatively lower MCL1 levels, indicating that MCL1 may not be relevant to resistance mechanisms to the triple combination.
Last, the proposed combinations are highly translatable, as MDM2 inhibitors such as Mil (RAIN-32), HDM201, and APG-115 are in clinical trials (19), and Sel and Ven have already been FDA-approved. ED50 concentrations of Sel in this study ranged up to 240 nM, which is substantially lower than the Cmax (1 μM) observed in patient plasma (27), suggesting that doses of Sel can be reduced in Mil + Sel combination. Moreover, the reduced dose of Sel (20 to 10 mg/kg per week) was well tolerated in mice without any suppression of normal hematopoiesis, as documented by normal blood counts, with superior cytoreduction and survival of mice treated with the triple combination of Mil + Sel + Ven in vivo. The triple combination did not induce severe toxicities but rather improved murine (i.e., normal) CD45+ cell numbers and platelet counts compared to vehicle or single-agent treatments. We are also conducting early clinical studies with the central nervous system–sparing XPO1 inhibitor eltanexor, which may be more tolerable than Sel. Further investigation and validation of the triple MDM2, XPO1, and BCL2 inhibitor combination in a clinical trial are warranted.
MATERIALS AND METHODS
Experimental design
The objective of the study was to determine whether the dual MDM2/XPO1 inhibition results in the accumulation of nuclear p53 protein and induces synergistic anti-leukemia efficacy in AML cells including in primary AML samples, as a novel treatment approach to overcome Ven resistance. To that end, we used AML cell lines including Ven-resistant ones (OCI-AML3 and MV4;11 VR) and primary AML samples sensitive/resistant to Ven-based therapies along with or without TP53 mutations. We did not use statistical methods to predetermine sample sizes for primary AML samples but specified them on the basis of the incidence of mutant p53 AML samples, which is around 10% of AML samples in the institution. Samples obtained from patients with acute promyelocytic leukemia were excluded.
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board at the University of Texas MD Anderson Cancer Center (LAB02-395). Informed consent was obtained from all the patients. Primary AML samples from patients with AML and normal BM samples from healthy donors were obtained through the Leukemia Sample Bank Core at the MD Anderson Cancer Center. All but one primary AML samples (N = 33) were subjected to the next-generation sequencing-based 81 AML gene panel and cytogenetic analysis, which are routinely applied in the clinic of the Department of Leukemia at MD Anderson Cancer Center. Primary AML samples with more than 60% blast percentages were included. Samples with 30% or more spontaneous apoptosis when starting experiments were excluded.
Animal experiments
The animal study protocols were approved by the Institutional Animal Care and Use Committee at the University of Texas MD Anderson Cancer Center. All animals were obtained from the Department of Experimental Radiation Oncology or in-house breeding colonies at the MD Anderson Cancer Center.
Luciferase-labeled MV4;11 Ven-R cells (0.5 × 106 cells per mouse) were injected into 6-week-old male NSG mice. After confirming engraftment using in vivo bioluminescence imaging by IVIS 200 imaging system (Perkin Elmer, Waltham, MA, USA) on day 4 after inoculation and randomization based on body weights and luminescence intensities, the mice were administered vehicle (0.5% methylcellulose for Mil and 0.6% PVP K29/32 with 0.6% Pluronic F-68 for Sel), Mil (50 mg/kg, 5 consecutive days per week), Sel (20 mg/kg, once a week), Mil + Sel, or Ven (100 mg/kg, 5 days a week) by oral gavage until they became moribund. Leukemia burden was measured by luminescence at indicated time points.
PDX AML cells were obtained from an AML patient resistant to Ven and DAC therapy and were established in our laboratory (37). AML cells had FLT3-ITD (two ITDs with ratios of 0.235 and 0.119), GATA2 (p.R362*), and NRAS (p.G12D, <2% of variant allele frequency) mutations after Ven/DAC. One million PDX AML cells were injected into NSG mice (all 6-week-old female). At week 9 after inoculation, engraftment was confirmed by the presence of more than 1% of human CD45 (Pacific Blue, BioLegend, #304029) and CD123 (APC, BioLegend, #306012) double-positive cells among the sum of mouse CD45- and human CD45-positive populations measured by flow cytometry in PB specimens. Randomization was performed on the basis of the percentages of circulating leukemia cells and body weights. Oral gavage treatments included vehicle, Mil (50 mg/kg, 5 days a week), Sel (10 mg/kg, once a week), Ven (100 mg/kg, 5 days a week), Mil + Sel, Mil + Ven, Sel + Ven, and Mil + Sel + Ven. The treatment was administered until all mice in each group became moribund. Leukemia burden in PB was measured by flow cytometry (human CD45 and CD123 double-positive cells) biweekly. Hemoglobin levels and platelet counts in PB were measured using a hemocytometer (Horiba, Kyoto, Japan).
Cell lines and cell culture
The human AML cell lines MOLM-13 was purchased from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). OCI-AML3 cells were provided by M. Minden (Ontario Cancer Institute, Toronto, ON, Canada). MV4;11, OCI-AML2, MOLM-14, HL-60, Kasumi-1, U-937, and THP-1 cells were purchased from the American Type Culture Collection (Manassas, VA, USA). All cell lines were subjected to cell line authentications performed by the Cytogenetics and Cell Authentication Core at the University of Texas MD Anderson Cancer Center every 6 months. OCI-AML3 cells transfected with scrambled and TP53 short hairpin RNA (shRNA) were generated previously (65). CRISPR-engineered MOLM-13 cells with TP53 knockout and mutations were generated and described previously (66). MV4;11 VR cells were obtained through long-term exposure to Ven as described previously (21) and were subsequently transduced with lentivirus delivering enhanced green fluorescent protein (GFP)/luciferase on the pCD550A-1 plasmid as previously described (21). The top 5% GFP-positive cells were sorted using a FACSAria II cell sorter (BD Biosciences, Franklin Lakes, NJ, USA). All AML cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) along with penicillin (100 mg/ml) and streptomycin (100 U/ml) at 37°C in 5% CO2 in a humidified atmosphere. All primary AML samples from patients and normal BM mononuclear cells from healthy donors were freshly obtained and isolated by density-gradient centrifugation using Lymphocyte Separation Medium (Corning, Manassas, VA, USA). Isolated cells were subjected to red blood cell lysis using BD Pharm Lyse lysing solution (BD Biosciences) and maintained in RPMI 1640 medium supplemented with 10% FBS, thrombopoietin (20 ng/ml), interleukin-3 (20 ng/ml), FLT3-ligand (100 ng/ml), and stem cell factor (100 ng/ml) for indicated treatment durations. All cytokines were purchased from PeproTech (Rocky Hill, NJ, USA). AML samples that demonstrated spontaneous apoptosis of less than 30% determined by annexin V/4′,6-diamidino-2-phenylindole (DAPI) positivity were subjected to further analyses. BM mesenchymal stromal cells (MSCs) were isolated from BM aspirates of healthy donors and expanded as described previously (67).
Reagents
Mil (RAIN-32, formerly known as DS-3032b) and Sel (KPT-330) were obtained from Daiichi Sankyo (Tokyo, Japan) and Karyopharm Therapeutics (Newton, MA, USA), respectively, under the three-party material transfer agreement. Ven (ABT-199) was purchased from Chemgood (Henrico, VA, USA). Methyl Cellulose and polyethylene glycol, molecular weight 400 were purchased from Merck Millipore (Burlington, MA, USA). Phosal 50 PG was purchased from Lipoid LLC (Newark, NJ, USA). PVP K29/32 and Pluronic F-68 were provided from Karyopharm Therapeutics.
c-MYC overexpression in OCI-AML3 cells
To establish c-MYC–overexpressing OCI-AML3 cells, c-MYC protein expression cassettes were delivered into OCI-AML3 cells by lentiviral transduction. Briefly, a DNA sequence encoding human MYC was inserted into the multiple cloning site of the commercially available lentiviral transfer vector pCD550A-1 (Systems Biosciences, Palo Alto, CA, USA). After verification of the construction by Sanger sequencing, the lentivirus was prepared and used to transduce OCI-AML3 cells as described previously (21). In parallel, control OCI-AML3 cells were transduced with the lentivirus containing the empty vector. Infected cells were selected using puromycin at up to 1 μg/ml. Increased expression of c-MYC was verified by immunoblot analysis.
Apoptosis analysis
Annexin V and DAPI binding assays were performed to assess apoptosis as described previously (16). Cells were plated and treated with DMSO, Mil, Sel, and/or Ven for 72 hours unless otherwise specified. Counting beads (AccuCount, Spherotech, Lake Forest, IL, USA, #ACFP-70-10) were used to calculate absolute cell numbers. Live cell numbers were calculated from annexin V– and DAPI-negative cells using counting beads. For primary AML samples, simultaneous staining of anti-human CD45 (PE, BD Biosciences, #560975), CD34 (PE-Cy7, BioLegend, #343516), and CD38 (APC Fire 750, BioLegend, #356626) antibodies were also performed. FlowJo software (version 10, Ashland, OR, USA) was used for the analyses. CompuSyn software (version 1.0, ComboSyn) was used to calculate IC50 and ED50 values and combination indices.
RNA sequencing
OCI-AML3, TP53 wild-type, and p.R248Q-MOLM-13 cells were treated with DMSO, Mil, Sel, or Mil + Sel for 12 hours. Cells were then harvested, and total RNA was extracted from each sample using QIAGEN RNeasy extraction kit (Venlo, Netherlands; N = 3 per treatment) according to the manufacturer’s protocol. RNA-seq was performed as reported previously described (68).
Differential gene expression profiling and pathway analyses
Gene expression was quantified using the Kallisto/sleuth pipeline. Read quantification was performed using Kallisto (v. 0.44.0), a pseudo-alignment–based method to quantify RNA abundance at the transcript level in transcripts per million (TPM) counts (69). Kallisto quantification was used with the number of bootstraps set to 100 using Ensembl cDNA transcripts (Human assembly hg19, release GRC38.14) for indexing. Hierarchical clustering and principal components analysis of the samples were performed. Sleuth v0.30.0 was used to measure the abundance in TPM with covariates for treatment conditions and downstream differential gene expression (70) to leverage the bootstrap estimates of Kallisto and to output model-based, gene-level normalized TPM matrix. The abundance of the genes was calculated as the sum of the TPMs mapped to a given gene. When using the sleuth preparation, gene isoforms were aggregated with a target map file derived from the Refseq hg19 transcriptome. For each gene, both the likelihood ratio test and Wald test were performed on the condition parameter to obtain their respective false discovery rate (FDR)–corrected P values. Significantly altered genes were those passing the two tests at a FDR cutoff less than 0.1. The plot_transcript_heatmap function in Sleuth package was used to visualize the cluster analysis. EnhancedVolcano R package (https://github.com/kevinblighe/EnhancedVolcano) was used to generate the volcano plots. This package is a visual tool for displaying differentially expressed genes among overall gene expression levels. Beta scores (log2) of −1 and 1 (0.5-fold and 2-fold, respectively) and a q value of 0.05 were used as threshold lines for the magnitude of change in gene expression and significance, respectively. HALLMARK gene sets were used for pathway enrichment and functional classification of differentially expressed genes.
Immunoblot analysis and subcellular fractionation
Lysis at a density of 1 × 106 cells/50 μl for AML cell lines and that of 2 × 106/50 μl for primary AML samples were performed in protein lysis buffer [0.25 M tris-HCl, 2% SDS, 4% β-mercaptoethanol, 10% glycerol, and 0.02% bromophenol blue, supplemented with 2-mercaptoethanol and 100× Protease/Phosphatase Inhibitor Cocktail (Cell Signaling Technology, Danvers, MA, USA, #5872)]. Protein lysates were incubated at 95°C for 5 min for denaturing. Immunoblot analysis was performed as reported previously (71). Briefly, an equal amount of protein lysate was loaded onto a 10 to 12% SDS-PAGE gel (Bio-Rad Laboratories, Hercules, CA, USA) and quantitated using an Odyssey imaging system (LI-COR Biosciences, Lincoln, NE, USA). Antibodies used for Western blotting in this study include against p53 (DO-1; Santa Cruz Biotechnology, Dallas, TX, USA, #sc-126), MDM2 (D-12; Santa Cruz Biotechnology, #sc-5304), PARP-1 (F-2; Santa Cruz Biotechnology, #sc-8007), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 14C10; Cell Signaling Technology, #2118), β-actin (13E5; Cell Signaling Technology, #4970), c-MYC (D84C12; Cell Signaling Technology, #5605), histone H3 (Cell Signaling Technology, #9715), MCL1 (D35A5; Cell Signaling Technology, #5453), α-tubulin (11H10; Cell Signaling Technology, #2125), p21 (12D1; Cell Signaling Technology, #2947).
Nuclear and cytoplasmic proteins were extracted using a subcellular fractionation kit (ProteoExtract; EMD Millipore, Billerica, MA, USA) according to the manufacturer’s protocol. Protein lysates were subjected to Western blot using anti-p53, anti-PARP, and GAPDH antibodies.
Cell cycle analysis with concomitant staining of multiple intracellular proteins
OCI-AML3 Cells treated with DMSO, Mil, Sel, or Mil + Sel were harvested for cell cycle analysis using Click-iT Plus EdU incorporation kit (Thermo Fisher Scientific, #C10634) according to the manufacturer’s protocol. Briefly, after EdU incorporation, harvested cells were first labeled with fixable live and dead stain Ghost BV540 (Tonbo Biosciences, San Diego, CA, USA) at room temperature (RT) for 15 min. Cells were then fixed using the fixative reagent and washed with 1% bovine serum albumin (BSA) in PBS (1% BSA), followed by permeabilization using the saponin-based buffer. After Click-iT reactions, cells were stained with other intracellular antibodies for subsequent flow cytometry analysis. Antibodies used were those against Ki-67 (PE-Cy7; BD Biosciences, #561283), cleaved caspase-3 [fluorescein isothiocyanate (FITC); BD Biosciences, #559341], and p21 (AF700, R&D Systems, Minneapolis, MN, USA, #IC1047N). FxCycle Violet stain (Thermo Fisher Scientific, #F10347) was used to stain DNA contents.
Multiparameter flow cytometry for multiple stress responses and uniform manifold approximation and projection analysis
A previously described protocol for multiple modes of cell death (72) was modified and a multiparameter flow cytometry assay for multiple stress responses and cell death modes was established (73). Harvested OCI-AML3 cells treated with DMSO, Mil, Sel, or Mil + Sel were labeled with LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (ThermoFisher Scientific, L34957) at RT for 20 min. Washed cells were then fixed in freshly prepared 1.6% paraformaldehyde during vortexing and incubated for 10 min at RT. Washed cells were then permeabilized in 90% ice-cold methanol (100 μl of cell suspension +900 μl of methanol) during vortexing and incubated for 30 min at −20°C. Cells were washed using cell staining buffer (1% BSA in PBS) and subsequently stained with an unconjugated ATF4 antibody (Proteintech, Rosemont, IL, USA, 10835-1-AP) at 4°C for 30 min. Cells were washed twice with cell staining buffer and stained with a secondary antibody (goat–anti-rabbit AF647, Thermo Fisher Scientific) at 4°C for 30 min. Washed cells were incubated with other fluorophore-conjugated antibodies against LC3B (PE, clone D11; Cell Signaling Technology, #8899), γ-H2A.X (PE-Cy7, clone 2F3; BioLegend, #613420), Ki-67 (BV421; BD Biosciences, #562899), cleaved caspase-3 (FITC; BD Biosciences, #559341), and cleaved PARP (AF594; BD Biosciences, #564130) at 4°C for 30 min for subsequent flow cytometry analysis.
For cluster analysis, we used t-distributed stochastic neighbor embedding (t-SNE) (74) and uniform manifold approximation and projection (75) for dimension reduction, implemented using the Cytofkit package in R (version 3.6.3). The analyses were performed using all the markers except for the parameters used in manual gating to identify and export downstream subpopulations.
Single-cell mass cytometry (CyTOF)
Frozen PDX AML cells engrafted and propagated in NSG mice were thawed and incubated in MEMα medium supplemented with 20% FBS, 5% heparin, MgSO4, and DNase I (10 U/ml) for 15 min. Washed cells were cocultured with MSCs and treated with DMSO, Mil, Sel, Ven, doublet combinations, or triple combination for 24 hours. A concentration of 100 nM was used as the drug concentration. After drug exposure, cells were harvested and stained with metal-tagged antibodies against cell surface proteins listed below, barcoded using Cell-IDTM 20-Plex Pd Barcoding Kit (Standard BioTools, South San Francisco, CA, USA), pooled, stained with metal-tagged antibodies against intracellular proteins, and subjected to CyTOF analysis as described previously (76, 77). Briefly, viable single cells determined by 191/193Ir and cisplatin were gated with FlowJo software and exported as flow cytometry standard data for subsequent analyses using Cytofkit software (78) after debarcoding using Debarcoder software (Standard BioTools). Cell populations in each sample were identified and embedded by RPhenograph and visualized using t-SNE dimension reduction. We used the following antibodies: BCL-XL (141 Pr, clone 54H6; CST, 2764BF), cleaved caspase-3 (142 Nd, clone D3E9;Standard BioTools, 3142004A), BCL2 (144 Nd, clone 100; BioLegend, 658702), CD123 (145 Nd, clone 7G3; BD Biosciences, 554527), CD34 (148 Nd, clone 581; BD Biosciences, 555820); LC3B (150 Nd, clone D11; CST 3868 custom), p21 (153 Eu, 12D1; CST 2947BF), CD45 (154 Sm, clone HI30; Standard BioTools 3154001B), CD33 (158 Gd, clone WM53; Standard BioTools, 3158001B), ATF4 (160 Gd, polyclonal; ProteinTech 10835-1-AP), p53 (165 Ho, clone 184721; R&D, MAB1355), CD44 (166 Er, clone BJ18; Standard BioTools, 3166001B), CD38 (168 Er, HIT2; BioLegend, 303502), CXCR4 (172 Yb, clone 12G5; BioLegend, 306502), BAX (173 Yb, clone 2D2; BioLegend, 633602), Ki-67 (175 Lu, clone Ki-67; BioLegend, 350502), and MCL1 (176 Yb, clone D2W9E; CST, 94296BF).
Quantitative real-time PCR
The mRNA expression levels in AML cell lines were quantified using the TaqMan gene expression assays listed below, including CDKN1A (Hs00355782_m1), MDM2 (Hs01066930_m1), PMAIP1 (Hs00560402_m1), CMBL (Hs00540853_m1), BAX (Hs00180269_m1), BBC3 (Hs00248075_m1), FAS (Hs00236330_m1), MYC (Hs00153408_m1), MCM2 (Hs01091564_m1), MCM4 (Hs00907398_m1), PLK1 (Hs00983227_m1), 18S ribosomal RNA (rRNA; Hs03928985_g1), and B2M (Hs00187842_m1; Thermo Fisher Scientific). 18S rRNA or B2M was used as internal controls.
Transfection of siRNAs
MV4;11 VR cells were transfected with a pool of either control siRNAs or MYC siRNAs (Horizon Discovery, Cambridge, UK) by electroporation using a Nucleofector 2b Device with Cell Line Nucleofector Kit L (Lonza, Basel, Switzerland) according to the manufacturer’s recommended protocol. Transfected cells were incubated for 24 hours and subjected to Ven treatments or Western blotting.
Single-cell DNA + protein sequencing
PDX AML cells obtained from BM in moribund mice receiving vehicle and the triple combination of Mil + Sel + Ven were collected and subjected to scDNA + protein seq. The cells were labeled with oligo-tagged 45 surface antibodies (TotalSeq-D Human Heme Oncology Cocktail, V1.0; BioLegend, #399906). Each antibody in the panel is conjugated to an oligonucleotide that contains a capture sequence, clone-specific barcode sequence, and a PCR handle compatible with Mission Bio primers (South San Francisco, CA, USA) for cell barcoding and library preparation. An average of 4455 labeled cells per sample were loaded onto a Mission Bio Tapestri instrument on which emulsions were created that isolate single cells in individual subnanoliter droplets. The emulsions were then incubated to lyse the cells and release nuclear DNA. PCR reagents and biotinylated oligo primers were introduced into each droplet. Multiplex PCR was performed to amplify the DNA regions targeted by the primers with 279 amplicons covering 37 genes and the captured antibodies in the protein panel. During this PCR step, each cell’s amplified DNA and protein product were barcoded with a sequence unique to that single cell. After PCR, emulsions were broken, and protein libraries were separated from DNA libraries by biotinylated oligo selection. Sequencing was performed on the Illumina NovaSeq 6000 platform using 150-nt paired end sequencing for each cell and for the protein panel. Data analyses were performed using Mission Bio’s Tapestri Insight software and our platform developed in house.
Quantification and statistical analysis
Statistical analyses were performed using the two-tailed Student’s t test, one-way analysis of variance (ANOVA), or Mann-Whitney U test with the Prism statistical software program (version 9.0; GraphPad Software, Boston, MA, USA). Multivariate analyses were performed using STATA SE software (version 13.0; StataCorp, College Station, TX, USA). The Kaplan-Meier method was used to generate survival curves, and the log-rank test was used for the comparison of the two groups. A P value less than 0.05 was considered statistically significant. In the figure legends, *, **, ***, and **** indicate P values less than 0.05, 0.01, 0.001, and 0.0001, respectively, unless otherwise specified. All experiments were performed with three replicates, and the error bars in the figure legends represent means ± SD values unless otherwise specified.
Acknowledgments
We thank N. Hail, Jr., for helping edit the manuscript. We also thank S. Colla and P. Lockyer at The University of Texas MD Anderson Cancer Center for the help in measuring the complete blood counts of mouse blood samples, G. Al-Atrash at The University of Texas MD Anderson Cancer Center for help with obtaining normal BM samples, and A. Lesegretain at Daiichi Sankyo Inc. for coordinating collaborative work with Daiichi Sankyo Inc. We acknowledge the Research Medical Library at The University of Texas MD Anderson Cancer Center for the help with editing.
Funding: This work was supported by The Paul and Mary Haas Chair in Genetics (M.A.), Leukemia Specialized Programs of Research Excellence (CA100632; M.A.), Leukemia Specialized Programs of Research Excellence Developmental Research Project (Y.N., J.I., and M.A.), Cancer Prevention Research Institute of Texas (RP130397; M.A.), NIH Cancer Center Support Grant (P30CA016672) and the University of Texas MD Anderson Cancer Center MDS/AML Moon Shot (M.A.), Shared Instrumentation Award from the Cancer Prevention Research Institution of Texas (CPRIT; RP121010), National Cancer Institute R21 (CA267401) (Y.N., M.M., J.I., and M.A.), Research Fellowship, The Uehara Memorial Foundation (Y.N.), Oversea Research Fellowship, Japan Society for the Promotion of Science (Y.N.), and Research Funding: Daiichi-Sankyo Inc. (Y.N., J.I., and M.A.). E.A. is a TRIUMPH Fellow in the CPRIT Training Program (RP210028).
Author contributions: Conceptualization: Y.N., J.I., K.K., and M.A. Methodology: Y.N., E.A., and M.M. Investigation: Y.N., E.A., R.H.M., L.B.O., D.A.S., S.K., M.M., and T.P. Visualization: Y.N., E.A., and M.M. Funding acquisition: Y.N., J.I., and M.A. Project administration: Y.N., E.A., VRR, P.Y.M., W.T., B.Z.C., S.B., B.L.E., M.K., and T.S. Supervision: J.I., K.K., and M.A. Writing (original draft): Y.N. Writing (review and editing): J.I., K.K., N.G.D., and M.A.
Competing interests: T.S. is an employee of Daiichi Sankyo Inc. N.G.D. has been involved in consultancy in Daiichi Sankyo Inc. The authors declare that they have no other competing interests.
Data and materials availability: All of the materials used in the study except reagents obtained under the material transfer agreement between the University of Texas MD Anderson Cancer Center and pharmaceutical companies will be provided upon appropriate requests (contact: mandreef@mdanderson.org). RNA-seq datasets are available in a publicly available Dryad platform (https://doi.org/10.5061/dryad.bzkh189g0). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S14
Tables S1 to S4
REFERENCES AND NOTES
- 1.A. Vazquez, E. E. Bond, A. J. Levine, G. L. Bond, The genetics of the p53 pathway, apoptosis and cancer therapy. Nat. Rev. Drug Discov. 7, 979–987 (2008). [DOI] [PubMed] [Google Scholar]
- 2.V. Llombart, M. R. Mansour, Therapeutic targeting of “undruggable” MYC. EBioMedicine 75, 103756 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.J. S. Ho, W. Ma, D. Y. Mao, S. Benchimol, p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol. Cell. Biol. 25, 7423–7431 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.T. J. Phesse, K. B. Myant, A. M. Cole, R. A. Ridgway, H. Pearson, V. Muncan, G. R. van den Brink, K. H. Vousden, R. Sears, L. T. Vassilev, A. R. Clarke, O. J. Sansom, Endogenous c-Myc is essential for p53-induced apoptosis in response to DNA damage in vivo. Cell Death Differ. 21, 956–966 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.C. Lourenco, D. Resetca, C. Redel, P. Lin, A. S. MacDonald, R. Ciaccio, T. M. G. Kenney, Y. Wei, D. W. Andrews, M. Sunnerhagen, C. H. Arrowsmith, B. Raught, L. Z. Penn, MYC protein interactors in gene transcription and cancer. Nat. Rev. Cancer 21, 579–591 (2021). [DOI] [PubMed] [Google Scholar]
- 6.C. L. Xu, B. Sang, G. Z. Liu, J. M. Li, X. D. Zhang, L. X. Liu, R. F. Thorne, M. Wu, SENEBLOC, a long non-coding RNA suppresses senescence via p53-dependent and independent mechanisms. Nucleic Acids Res. 48, 3089–3102 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Y. C. Feng, X. Y. Liu, L. Teng, Q. Ji, Y. Wu, J. M. Li, W. Gao, Y. Y. Zhang, T. La, H. Tabatabaee, X. G. Yan, M. F. B. Jamaluddin, D. Zhang, S. T. Guo, R. J. Scott, T. Liu, R. F. Thorne, X. D. Zhang, L. Jin, c-Myc inactivation of p53 through the pan-cancer lncRNA MILIP drives cancer pathogenesis. Nat. Commun. 11, 4980 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M. Sachdeva, S. Zhu, F. Wu, H. Wu, V. Walia, S. Kumar, R. Elble, K. Watabe, Y.-Y. Mo, p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc. Natl. Acad. Sci. U.S.A. 106, 3207–3212 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. R. Porter, B. E. Fisher, L. Baranello, J. C. Liu, D. M. Kambach, Z. Nie, W. S. Koh, J. Luo, J. M. Stommel, D. Levens, E. Batchelor, Global inhibition with specific activation: How p53 and MYC redistribute the transcriptome in the DNA double-strand break response. Mol. Cell 67, 1013–1025.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.E. Papaemmanuil, M. Gerstung, L. Bullinger, V. I. Gaidzik, P. Paschka, N. D. Roberts, N. E. Potter, M. Heuser, F. Thol, N. Bolli, G. Gundem, P. Van Loo, I. Martincorena, P. Ganly, L. Mudie, S. McLaren, S. O’Meara, K. Raine, D. R. Jones, J. W. Teague, A. P. Butler, M. F. Greaves, A. Ganser, K. Döhner, R. F. Schlenk, H. Döhner, P. J. Campbell, Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 374, 2209–2221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.M. Konopleva, R. Contractor, T. Tsao, I. Samudio, P. P. Ruvolo, S. Kitada, X. Deng, D. Zhai, Y.-X. Shi, T. Sneed, M. Verhaegen, M. Soengas, V. R. Ruvolo, T. McQueen, W. D. Schober, J. C. Watt, T. Jiffar, X. Ling, F. C. Marini, D. Harris, M. Dietrich, Z. Estrov, J. McCubrey, W. S. May, J. C. Reed, M. Andreeff, Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell 10, 375–388 (2006). [DOI] [PubMed] [Google Scholar]
- 12.R. Pan, L. J. Hogdal, J. M. Benito, D. Bucci, L. Han, G. Borthakur, J. Cortes, D. J. DeAngelo, L. Debose, H. Mu, H. Döhner, V. I. Gaidzik, I. Galinsky, L. S. Golfman, T. Haferlach, K. G. Harutyunyan, J. Hu, J. D. Leverson, G. Marcucci, M. Müschen, R. Newman, E. Park, P. P. Ruvolo, V. Ruvolo, J. Ryan, S. Schindela, P. Zweidler-McKay, R. M. Stone, H. Kantarjian, M. Andreeff, M. Konopleva, A. G. Letai, Selective BCL-2 inhibition by ABT-199 causes on-target cell death in acute myeloid leukemia. Cancer Discov. 4, 362–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M. Konopleva, D. A. Pollyea, J. Potluri, B. Chyla, L. Hogdal, T. Busman, E. McKeegan, A. H. Salem, M. Zhu, J. L. Ricker, W. Blum, C. D. DiNardo, T. Kadia, M. Dunbar, R. Kirby, N. Falotico, J. Leverson, R. Humerickhouse, M. Mabry, R. Stone, H. Kantarjian, A. Letai, Efficacy and biological correlates of response in a Phase II study of venetoclax monotherapy in patients with acute myelogenous leukemia. Cancer Discov. 6, 1106–1117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.C. D. DiNardo, B. A. Jonas, V. Pullarkat, M. J. Thirman, J. S. Garcia, A. H. Wei, M. Konopleva, H. Dohner, A. Letai, P. Fenaux, E. Koller, V. Havelange, B. Leber, J. Esteve, J. Wang, V. Pejsa, R. Hajek, K. Porkka, A. Illes, D. Lavie, R. M. Lemoli, K. Yamamoto, S. S. Yoon, J.-H. Jang, S. P. Yeh, M. Turgut, W. J. Hong, Y. Zhou, J. Potluri, K. W. Pratz, Azacitidine and venetoclax in previously untreated acute myeloid leukemia. N. Engl. J. Med. 383, 617–629 (2020). [DOI] [PubMed] [Google Scholar]
- 15.A. Maiti, C. R. Rausch, J. E. Cortes, N. Pemmaraju, N. G. Daver, F. Ravandi, G. Garcia-Manero, G. Borthakur, K. Naqvi, M. Ohanian, N. J. Short, Y. Alvarado, T. M. Kadia, K. Takahashi, M. Yilmaz, N. Jain, S. Kornblau, G. Montalban Bravo, K. Sasaki, M. Andreeff, P. Bose, A. Ferrajoli, G. C. Issa, E. J. Jabbour, L. Masarova, P. A. Thompson, S. Wang, S. Konoplev, S. A. Pierce, J. Ning, W. Qiao, J. S. Welch, H. M. Kantarjian, C. D. DiNardo, M. Y. Konopleva, Outcomes of relapsed or refractory acute myeloid leukemia after frontline hypomethylating agent and venetoclax regimens. Haematologica 106, 894–898 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.K. Kojima, M. Konopleva, I. J. Samudio, M. Shikami, M. Cabreira-Hansen, T. McQueen, V. Ruvolo, T. Tsao, Z. Zeng, L. T. Vassilev, M. Andreeff, MDM2 antagonists induce p53-dependent apoptosis in AML: Implications for leukemia therapy. Blood 106, 3150–3159 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.J. Ishizawa, K. Nakamaru, T. Seki, K. Tazaki, K. Kojima, D. Chachad, R. Zhao, L. Heese, W. Ma, M. C. J. Ma, C. DiNardo, S. Pierce, K. P. Patel, A. Tse, R. E. Davis, A. Rao, M. Andreeff, Predictive gene signatures determine tumor sensitivity to MDM2 inhibition. Cancer Res. 78, 2721–2731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.M. Andreeff, K. R. Kelly, K. Yee, S. Assouline, R. Strair, L. Popplewell, D. Bowen, G. Martinelli, M. W. Drummond, P. Vyas, M. Kirschbaum, S. P. Iyer, V. Ruvolo, G. M. González, X. Huang, G. Chen, B. Graves, S. Blotner, P. Bridge, L. Jukofsky, S. Middleton, M. Reckner, R. Rueger, J. Zhi, G. Nichols, K. Kojima, Results of the Phase I trial of RG7112, a small-molecule MDM2 antagonist in leukemia. Clin. Cancer Res. 22, 868–876 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.M. Konopleva, G. Martinelli, N. Daver, C. Papayannidis, A. Wei, B. Higgins, M. Ott, J. Mascarenhas, M. Andreeff, MDM2 inhibition: An important step forward in cancer therapy. Leukemia 34, 2858–2874 (2020). [DOI] [PubMed] [Google Scholar]
- 20.T. Nechiporuk, S. E. Kurtz, O. Nikolova, T. Liu, C. L. Jones, A. D’Alessandro, R. Culp-Hill, A. d’Almeida, S. K. Joshi, M. Rosenberg, C. E. Tognon, A. V. Danilov, B. J. Druker, B. H. Chang, S. K. McWeeney, J. W. Tyner, The TP53 apoptotic network is a primary mediator of resistance to BCL2 inhibition in AML cells. Cancer Discov. 9, 910–925 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R. Pan, V. Ruvolo, H. Mu, J. D. Leverson, G. Nichols, J. C. Reed, M. Konopleva, M. Andreeff, Synthetic lethality of combined Bcl-2 inhibition and p53 activation in AML: Mechanisms and superior antileukemic efficacy. Cancer Cell 32, 748–760.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.N. G. Daver, M. Dail, J. S. Garcia, B. A. Jonas, K. W. L. Yee, K. R. Kelly, N. Vey, S. Assouline, G. J. Roboz, S. Paolini, D. A. Pollyea, A. Tafuri, J. M. Brandwein, A. Pigneux, B. L. Powell, P. Fenaux, R. L. Olin, G. Visani, G. Martinelli, M. Onishi, J. Wang, W. Huang, C. Green, M. G. Ott, W.-J. Hong, M. Y. Konopleva, M. Andreeff, Venetoclax and idasanutlin in relapsed/refractory AML: A nonrandomized, open-label phase 1b trial. Blood 141, 1265–1276 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.A. S. Azmi, M. H. Uddin, R. M. Mohammad, The nuclear export protein XPO1 - from biology to targeted therapy. Nat. Rev. Clin. Oncol. 18, 152–169 (2021). [DOI] [PubMed] [Google Scholar]
- 24.L. Volpon, B. Culjkovic-Kraljacic, H. S. Sohn, A. Blanchet-Cohen, M. J. Osborne, K. L. B. Borden, A biochemical framework for eIF4E-dependent mRNA export and nuclear recycling of the export machinery. RNA 23, 927–937 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.A. Chari, D. T. Vogl, M. Gavriatopoulou, A. K. Nooka, A. J. Yee, C. A. Huff, P. Moreau, D. Dingli, C. Cole, S. Lonial, M. Dimopoulos, A. K. Stewart, J. Richter, R. Vij, S. Tuchman, M. S. Raab, K. C. Weisel, M. Delforge, R. F. Cornell, D. Kaminetzky, J. E. Hoffman, L. J. Costa, T. L. Parker, M. Levy, M. Schreder, N. Meuleman, L. Frenzel, M. Mohty, S. Choquet, G. Schiller, R. L. Comenzo, M. Engelhardt, T. Illmer, P. Vlummens, C. Doyen, T. Facon, L. Karlin, A. Perrot, K. Podar, M. G. Kauffman, S. Shacham, L. Li, S. Tang, C. Picklesimer, J.-R. Saint-Martin, M. Crochiere, H. Chang, S. Parekh, Y. Landesman, J. Shah, P. G. Richardson, S. Jagannath, Oral selinexor-dexamethasone for triple-class refractory multiple myeloma. N. Engl. J. Med. 381, 727–738 (2019). [DOI] [PubMed] [Google Scholar]
- 26.N. Kalakonda, M. Maerevoet, F. Cavallo, G. Follows, A. Goy, J. S. P. Vermaat, O. Casasnovas, N. Hamad, J. M. Zijlstra, S. Bakhshi, R. Bouabdallah, S. Choquet, R. Gurion, B. Hill, U. Jaeger, J. M. Sancho, M. Schuster, C. Thieblemont, F. De la Cruz, M. Egyed, S. Mishra, F. Offner, T. P. Vassilakopoulos, K. Warzocha, D. McCarthy, X. Ma, K. Corona, J.-R. Saint-Martin, H. Chang, Y. Landesman, A. Joshi, H. Wang, J. Shah, S. Shacham, M. Kauffman, E. Van Den Neste, M. A. Canales, Selinexor in patients with relapsed or refractory diffuse large B-cell lymphoma (SADAL): A single-arm, multinational, multicentre, open-label, phase 2 trial. Lancet Haematol. 7, e511–e522 (2020). [DOI] [PubMed] [Google Scholar]
- 27.R. Garzon, M. Savona, R. Baz, M. Andreeff, N. Gabrail, M. Gutierrez, L. Savoie, P. M. Mau-Sorensen, N. Wagner-Johnston, K. Yee, T. J. Unger, J.-R. Saint-Martin, R. Carlson, T. Rashal, T. Kashyap, B. Klebanov, S. Shacham, M. Kauffman, R. Stone, A phase 1 clinical trial of single-agent selinexor in acute myeloid leukemia. Blood 129, 3165–3174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.W. Fiedler, J. Chromik, S. Amberg, M. Kebenko, F. Thol, V. Schlipfenbacher, A. Christine Wilke, F. Modemann, M. Janning, H. Serve, A. Ganser, C. Bokemeyer, S. Theile, U. Deppermann, A. L. Kranich, M. Heuser, A Phase II study of selinexor plus cytarabine and idarubicin in patients with relapsed/refractory acute myeloid leukaemia. Br. J. Haematol. 190, e169–e173 (2020). [DOI] [PubMed] [Google Scholar]
- 29.K. Kojima, S. M. Kornblau, V. Ruvolo, A. Dilip, S. Duvvuri, R. E. Davis, M. Zhang, Z. Wang, K. R. Coombes, N. Zhang, Y. H. Qiu, J. K. Burks, H. Kantarjian, S. Shacham, M. Kauffman, M. Andreeff, Prognostic impact and targeting of CRM1 in acute myeloid leukemia. Blood 121, 4166–4174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.H. Döhner, E. Estey, D. Grimwade, S. Amadori, F. R. Appelbaum, T. Büchner, H. Dombret, B. L. Ebert, P. Fenaux, R. A. Larson, R. L. Levine, F. Lo-Coco, T. Naoe, D. Niederwieser, G. J. Ossenkoppele, M. Sanz, J. Sierra, M. S. Tallman, H.-F. Tien, A. H. Wei, B. Lowenberg, C. D. Bloomfield, Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129, 424–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.J. Etchin, J. Montero, A. Berezovskaya, B. T. Le, A. Kentsis, A. L. Christie, A. S. Conway, W. C. Chen, C. Reed, M. R. Mansour, C. E. Ng, S. Adamia, S. J. Rodig, I. A. Galinsky, R. M. Stone, B. Klebanov, Y. Landesman, M. Kauffman, S. Shacham, A. L. Kung, J. C. Wang, A. Letai, A. T. Look, Activity of a selective inhibitor of nuclear export, selinexor (KPT-330), against AML-initiating cells engrafted into immunosuppressed NSG mice. Leukemia 30, 190–199 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.L. Brunetti, M. C. Gundry, D. Sorcini, A. G. Guzman, Y.-H. Huang, R. Ramabadran, I. Gionfriddo, F. Mezzasoma, F. Milano, B. Nabet, D. L. Buckley, S. M. Kornblau, C. Y. Lin, P. Sportoletti, M. P. Martelli, B. Falini, M. A. Goodell, Mutant NPM1 maintains the leukemic state through HOX expression. Cancer Cell 34, 499–512.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.W. R. Taylor, G. R. Stark, Regulation of the G2/M transition by p53. Oncogene 20, 1803–1815 (2001). [DOI] [PubMed] [Google Scholar]
- 34.K. Kojima, M. Konopleva, I. J. Samudio, W. D. Schober, W. G. Bornmann, M. Andreeff, Concomitant inhibition of MDM2 and Bcl-2 protein function synergistically induce mitochondrial apoptosis in AML. Cell Cycle 5, 2778–2786 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Y. Tabe, K. Kojima, S. Yamamoto, K. Sekihara, H. Matsushita, R. E. Davis, Z. Wang, W. Ma, J. Ishizawa, S. Kazuno, M. Kauffman, S. Shacham, T. Fujimura, T. Ueno, T. Miida, M. Andreeff, Ribosomal biogenesis and translational flux inhibition by the Selective Inhibitor of Nuclear Export (SINE) XPO1 antagonist KPT-185. PLOS ONE 10, e0137210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.M. Muftuoglu, P. Y. Mak, V. Ruvolo, Y. Nishida, P. P. Ruvolo, B. Z. Carter, M. Andreeff, High dimensional interrogation of stress response patterns and cell death modes in AML. Blood 136, 15 (2020). [Google Scholar]
- 37.B. Z. Carter, P. Y. Mak, W. Tao, M. Warmoes, P. L. Lorenzi, D. Mak, V. Ruvolo, L. Tan, J. Cidado, L. Drew, M. Andreeff, Targeting MCL-1 dysregulates cell metabolism and leukemia-stroma interactions and resensitizes acute myeloid leukemia to BCL-2 inhibition. Haematologica 107, 58–76 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.M. A. Fischer, S. Y. Friedlander, M. P. Arrate, H. Chang, A. E. Gorska, L. D. Fuller, H. E. Ramsey, T. Kashyap, C. Argueta, S. Debler, M. Byrne, M. T. Villaume, A. C. Shaver, W. Senapedis, Y. Landesman, E. Baloglu, S. Shacham, M. R. Savona, Venetoclax response is enhanced by selective inhibitor of nuclear export compounds in hematologic malignancies. Blood Adv. 4, 586–598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.H. Luo, Q. Li, J. O’Neal, F. Kreisel, M. M. Le Beau, M. H. Tomasson, c-Myc rapidly induces acute myeloid leukemia in mice without evidence of lymphoma-associated antiapoptotic mutations. Blood 106, 2452–2461 (2005). [DOI] [PubMed] [Google Scholar]
- 40.E. Bulaeva, D. Pellacani, N. Nakamichi, C. A. Hammond, P. A. Beer, A. Lorzadeh, M. Moksa, A. Carles, M. Bilenky, S. Lefort, J. Shu, B. T. Wilhelm, A. P. Weng, M. Hirst, C. J. Eaves, MYC-induced human acute myeloid leukemia requires a continuing IL-3/GM-CSF costimulus. Blood 136, 2764–2773 (2020). [DOI] [PubMed] [Google Scholar]
- 41.C. Bahr, L. von Paleske, V. V. Uslu, S. Remeseiro, N. Takayama, S. W. Ng, A. Murison, K. Langenfeld, M. Petretich, R. Scognamiglio, P. Zeisberger, A. S. Benk, I. Amit, P. W. Zandstra, M. Lupien, J. E. Dick, A. Trumpp, F. Spitz, A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature 553, 515–520 (2018). [DOI] [PubMed] [Google Scholar]
- 42.L. Zhang, J. Li, H. Xu, X. Shao, L. Fu, Y. Hou, C. Hao, W. Li, K. Joshi, W. Wei, Y. Xu, F. Zhang, S. Dai, P. Breslin, J. Zhang, J. Zhang, Myc-Miz1 signaling promotes self-renewal of leukemia stem cells by repressing Cebpα and Cebpδ. Blood 135, 1133–1145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.P. van Galen, V. Hovestadt, M. H. Wadsworth Ii, T. K. Hughes, G. K. Griffin, S. Battaglia, J. A. Verga, J. Stephansky, T. J. Pastika, J. Lombardi Story, G. S. Pinkus, O. Pozdnyakova, I. Galinsky, R. M. Stone, T. A. Graubert, A. K. Shalek, J. C. Aster, A. A. Lane, B. E. Bernstein, Single-cell RNA-Seq reveals AML hierarchies relevant to disease progression and immunity. Cell 176, 1265–1281.e24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.M. Ohanian, U. Rozovski, R. Kanagal-Shamanna, L. V. Abruzzo, S. Loghavi, T. Kadia, A. Futreal, K. Bhalla, Z. Zuo, Y. O. Huh, S. M. Post, P. Ruvolo, G. Garcia-Manero, M. Andreeff, S. Kornblau, G. Borthakur, P. Hu, L. J. Medeiros, K. Takahashi, M. J. Hornbaker, J. H. Zhang, G. M. Nogueras-Gonzalez, X. L. Huang, S. Verstovsek, Z. Estrov, S. Pierce, F. Ravandi, H. M. Kantarjian, C. E. Bueso-Ramos, J. E. Cortes, MYC protein expression is an important prognostic factor in acute myeloid leukemia. Leuk. Lymphoma 60, 37–48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.L. Gao, A. Saeed, S. Golem, D. Zhang, J. Woodroof, J. McGuirk, S. Ganguly, S. Abhyankar, T. L. Lin, W. Cui, High-level MYC expression associates with poor survival in patients with acute myeloid leukemia and collaborates with overexpressed p53 in leukemic transformation in patients with myelodysplastic syndrome. Int. J. Lab. Hematol. 43, 99–109 (2021). [DOI] [PubMed] [Google Scholar]
- 46.S. E. Ahmadi, S. Rahimi, B. Zarandi, R. Chegeni, M. Safa, MYC: A multipurpose oncogene with prognostic and therapeutic implications in blood malignancies. J. Hematol. Oncol. 14, 121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.C. Berthon, E. Raffoux, X. Thomas, N. Vey, C. Gomez-Roca, K. Yee, D. C. Taussig, K. Rezai, C. Roumier, P. Herait, C. Kahatt, B. Quesnel, M. Michallet, C. Recher, F. Lokiec, C. Preudhomme, H. Dombret, Bromodomain inhibitor OTX015 in patients with acute leukaemia: A dose-escalation, phase 1 study. Lancet Haematol. 3, e186–e195 (2016). [DOI] [PubMed] [Google Scholar]
- 48.G. Borthakur, O. Odenike, I. Aldoss, D. A. Rizzieri, T. Prebet, C. Chen, R. Popovic, D. A. Modi, R. H. Joshi, J. E. Wolff, B. A. Jonas, A phase 1 study of the pan-bromodomain and extraterminal inhibitor mivebresib (ABBV-075) alone or in combination with venetoclax in patients with relapsed/refractory acute myeloid leukemia. Cancer 127, 2943–2953 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.C. Y. Fong, O. Gilan, E. Y. Lam, A. F. Rubin, S. Ftouni, D. Tyler, K. Stanley, D. Sinha, P. Yeh, J. Morison, G. Giotopoulos, D. Lugo, P. Jeffrey, S. C. Lee, C. Carpenter, R. Gregory, R. G. Ramsay, S. W. Lane, O. Abdel-Wahab, T. Kouzarides, R. W. Johnstone, S.-J. Dawson, B. J. Huntly, R. K. Prinjha, A. T. Papenfuss, M. A. Dawson, BET inhibitor resistance emerges from leukaemia stem cells. Nature 525, 538–542 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.P. Rathert, M. Roth, T. Neumann, F. Muerdter, J.-S. Roe, M. Muhar, S. Deswal, S. Cerny-Reiterer, B. Peter, J. Jude, T. Hoffmann, Ł. M. Boryń, E. Axelsson, N. Schweifer, U. Tontsch-Grunt, L. E. Dow, D. Gianni, M. Pearson, P. Valent, A. Stark, N. Kraut, C. R. Vakoc, J. Zuber, Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature 525, 543–547 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.S. A. Abraham, L. E. Hopcroft, E. Carrick, M. E. Drotar, K. Dunn, A. J. Williamson, K. Korfi, P. Baquero, L. E. Park, M. T. Scott, F. Pellicano, A. Pierce, M. Copland, C. Nourse, S. M. Grimmond, D. Vetrie, A. D. Whetton, T. L. Holyoake, Dual targeting of p53 and c-MYC selectively eliminates leukaemic stem cells. Nature 534, 341–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.A.-L. Latif, A. Newcombe, S. Li, K. Gilroy, N. A. Robertson, X. Lei, H. J. S. Stewart, J. Cole, M. T. Terradas, L. Rishi, L. McGarry, C. McKeeve, C. Reid, W. Clark, J. Campos, K. Kirschner, A. Davis, J. Lopez, J.-I. Sakamaki, J. P. Morton, K. M. Ryan, S. W. G. Tait, S. A. Abraham, T. Holyoake, B. Higgins, X. Huang, K. Blyth, M. Copland, T. J. T. Chevassut, K. Keeshan, P. D. Adams, BRD4-mediated repression of p53 is a target for combination therapy in AML. Nat. Commun. 12, 241 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.G. C. Issa, C. B. Benton, V. Mohanty, Y. F. Shen, Z. Alaniz, F. Wang, P. A. Futreal, W. Wang, J. L. Jorgensen, N. Navin, M. Y. Konopleva, K. Chen, M. Andreeff, Identification of gene expression signatures in leukemia stem cells and minimal residual disease following treatment of adverse risk acute myeloid leukemia. Blood 134, 2717 (2019). [Google Scholar]
- 54.E. Kerkhoff, R. Houben, S. Löffler, J. Troppmair, J.-E. Lee, U. R. Rapp, Regulation of c-myc expression by Ras/Raf signalling. Oncogene 16, 211–216 (1998). [DOI] [PubMed] [Google Scholar]
- 55.W.-B. Tsai, I. Aiba, Y. Long, H.-K. Lin, L. Feun, N. Savaraj, M. T. Kuo, Activation of Ras/PI3K/ERK pathway induces c-Myc stabilization to upregulate argininosuccinate synthetase, leading to arginine deiminase resistance in melanoma cells. Cancer Res. 72, 2622–2633 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.C. D. DiNardo, I. S. Tiong, A. Quaglieri, S. MacRaild, S. Loghavi, F. C. Brown, R. Thijssen, G. Pomilio, A. Ivey, J. M. Salmon, C. Glytsou, S. A. Fleming, Q. Zhang, H. Ma, K. P. Patel, S. M. Kornblau, Z. Xu, C. C. Chua, X. Chen, P. Blombery, C. Flensburg, N. Cummings, I. Aifantis, H. Kantarjian, D. C. S. Huang, A. W. Roberts, I. J. Majewski, M. Konopleva, A. H. Wei, Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood 135, 791–803 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.L. Han, Q. Zhang, M. Dail, C. Shi, A. Cavazos, V. R. Ruvolo, Y. Zhao, E. Kim, M. Rahmani, D. H. Mak, S. S. Jin, J. Chen, D. C. Phillips, P. B. Koller, R. Jacamo, J. K. Burks, C. DiNardo, N. Daver, E. Jabbour, J. Wang, H. M. Kantarjian, M. Andreeff, S. Grant, J. D. Leverson, D. Sampath, M. Konopleva, Concomitant targeting of BCL2 with venetoclax and MAPK signaling with cobimetinib in acute myeloid leukemia models. Haematologica 105, 697–707 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.B. M. Stevens, C. L. Jones, D. A. Pollyea, R. Culp-Hill, A. D’Alessandro, A. Winters, A. Krug, D. Abbott, M. Goosman, S. Pei, H. Ye, A. E. Gillen, M. W. Becker, M. R. Savona, C. Smith, C. T. Jordan, Fatty acid metabolism underlies venetoclax resistance in acute myeloid leukemia stem cells. Nat. Cancer 1, 1176–1187 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.H. Kalkavan, M. J. Chen, J. C. Crawford, G. Quarato, P. Fitzgerald, S. W. G. Tait, C. R. Goding, D. R. Green, Sublethal cytochrome c release generates drug-tolerant persister cells. Cell 185, 3356–3374.e22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.A. G. X. Zeng, S. Bansal, L. Jin, A. Mitchell, W. C. Chen, H. A. Abbas, M. Chan-Seng-Yue, V. Voisin, P. van Galen, A. Tierens, M. Cheok, C. Preudhomme, H. Dombret, N. Daver, P. A. Futreal, M. D. Minden, J. A. Kennedy, J. C. Y. Wang, J. E. Dick, A cellular hierarchy framework for understanding heterogeneity and predicting drug response in acute myeloid leukemia. Nat. Med. 28, 1212–1223 (2022). [DOI] [PubMed] [Google Scholar]
- 61.L. Jin, K. J. Hope, Q. Zhai, F. Smadja-Joffe, J. E. Dick, Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat. Med. 12, 1167–1174 (2006). [DOI] [PubMed] [Google Scholar]
- 62.X. Yu, L. Munoz-Sagredo, K. Streule, P. Muschong, E. Bayer, R. J. Walter, J. C. Gutjahr, R. Greil, M. L. Concha, C. Müller-Tidow, T. N. Hartmann, V. Orian-Rousseau, CD44 loss of function sensitizes AML cells to the BCL-2 inhibitor venetoclax by decreasing CXCL12-driven survival cues. Blood 138, 1067–1080 (2021). [DOI] [PubMed] [Google Scholar]
- 63.S. Piya, H. Mu, S. Bhattacharya, P. L. Lorenzi, R. E. Davis, T. McQueen, V. Ruvolo, N. Baran, Z. Wang, Y. Qian, C. M. Crews, M. Konopleva, J. Ishizawa, M. J. You, H. Kantarjian, M. Andreeff, G. Borthakur, BETP degradation simultaneously targets acute myelogenous leukemia stem cells and the microenvironment. J. Clin. Invest. 129, 1878–1894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D. Cibrián, F. Sánchez-Madrid, CD69: From activation marker to metabolic gatekeeper. Eur. J. Immunol. 47, 946–953 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.J. Moffat, D. A. Grueneberg, X. Yang, S. Y. Kim, A. M. Kloepfer, G. Hinkle, B. Piqani, T. M. Eisenhaure, B. Luo, J. K. Grenier, A. E. Carpenter, S. Y. Foo, S. A. Stewart, B. R. Stockwell, N. Hacohen, W. C. Hahn, E. S. Lander, D. M. Sabatini, D. E. Root, A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 (2006). [DOI] [PubMed] [Google Scholar]
- 66.S. Boettcher, P. G. Miller, R. Sharma, M. McConkey, M. Leventhal, A. V. Krivtsov, A. O. Giacomelli, W. Wong, J. Kim, S. Chao, K. J. Kurppa, X. Yang, K. Milenkowic, F. Piccioni, D. E. Root, F. G. Rucker, Y. Flamand, D. Neuberg, R. C. Lindsley, P. A. Janne, W. C. Hahn, T. Jacks, H. Döhner, S. A. Armstrong, B. L. Ebert, A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science 365, 599–604 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.M. Studeny, F. C. Marini, J. L. Dembinski, C. Zompetta, M. Cabreira-Hansen, B. N. Bekele, R. E. Champlin, M. Andreeff, Mesenchymal stem cells: Potential precursors for tumor stroma and targeted-delivery vehicles for anticancer agents. J. Natl. Cancer Inst. 96, 1593–1603 (2004). [DOI] [PubMed] [Google Scholar]
- 68.J. Ishizawa, S. F. Zarabi, R. E. Davis, O. Halgas, T. Nii, Y. Jitkova, R. Zhao, J. St-Germain, L. E. Heese, G. Egan, V. R. Ruvolo, S. H. Barghout, Y. Nishida, R. Hurren, W. Ma, M. Gronda, T. Link, K. Wong, M. Mabanglo, K. Kojima, G. Borthakur, N. MacLean, M. C. J. Ma, A. B. Leber, M. D. Minden, W. Houry, H. Kantarjian, M. Stogniew, B. Raught, E. F. Pai, A. D. Schimmer, M. Andreeff, Mitochondrial ClpP-mediated proteolysis induces selective cancer cell lethality. Cancer Cell 35, 721–737.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.N. L. Bray, H. Pimentel, P. Melsted, L. Pachter, Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 70.H. Pimentel, N. L. Bray, S. Puente, P. Melsted, L. Pachter, Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690 (2017). [DOI] [PubMed] [Google Scholar]
- 71.J. Ishizawa, K. Kojima, D. Chachad, P. Ruvolo, V. Ruvolo, R. O. Jacamo, G. Borthakur, H. Mu, Z. Zeng, Y. Tabe, J. E. Allen, Z. Wang, W. Ma, H. C. Lee, R. Orlowski, D. D. Sarbassov, P. L. Lorenzi, X. Huang, S. S. Neelapu, T. McDonnell, R. N. Miranda, M. Wang, H. Kantarjian, M. Konopleva, R. E. Davis, M. Andreeff, ATF4 induction through an atypical integrated stress response to ONC201 triggers p53-independent apoptosis in hematological malignancies. Sci. Signal. 9, ra17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.D. Bergamaschi, A. Vossenkamper, W. Y. J. Lee, P. Wang, E. Bochukova, G. Warnes, Simultaneous polychromatic flow cytometric detection of multiple forms of regulated cell death. Apoptosis 24, 453–464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.M. Muftuoglu, Z. Alaniz, D. Mak, A. J. Lin, J. K. Burks, B. Z. Carter, M. Andreeff, High-dimensional single-cell proteomic analysis reveals therapeutic vulnerabilities in relapsed/refractory AML by concomitant analysis of cell surface antigens, signaling pathways and anti-apoptotic proteins. Blood 136, 26 (2020). [Google Scholar]
- 74.E.-A. D. Amir, K. L. Davis, M. D. Tadmor, E. F. Simonds, J. H. Levine, S. C. Bendall, D. K. Shenfeld, S. Krishnaswamy, G. P. Nolan, D. Pe’er, viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.E. Becht, L. McInnes, J. Healy, C.-A. Dutertre, I. W. H. Kwok, L. G. Ng, F. Ginhoux, E. W. Newell, Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol., (2019). [DOI] [PubMed] [Google Scholar]
- 76.B. Z. Carter, P. Y. Mak, H. Mu, H. Zhou, D. H. Mak, W. Schober, J. D. Leverson, B. Zhang, R. Bhatia, X. Huang, J. Cortes, H. Kantarjian, M. Konopleva, M. Andreeff, Combined targeting of BCL-2 and BCR-ABL tyrosine kinase eradicates chronic myeloid leukemia stem cells. Sci. Transl. Med. 8, 355ra117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.L. Han, P. Qiu, Z. Zeng, J. L. Jorgensen, D. H. Mak, J. K. Burks, W. Schober, T. J. McQueen, J. Cortes, S. D. Tanner, G. J. Roboz, H. M. Kantarjian, S. M. Kornblau, M. L. Guzman, M. Andreeff, M. Konopleva, Single-cell mass cytometry reveals intracellular survival/proliferative signaling in FLT3-ITD-mutated AML stem/progenitor cells. Cytometry A 87, 346–356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.H. Chen, M. C. Lau, M. T. Wong, E. W. Newell, M. Poidinger, J. Chen, Cytofkit: A bioconductor package for an integrated mass cytometry data analysis pipeline. PLOS Comput. Biol. 12, e1005112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S14
Tables S1 to S4