Abstract
Glycerol monolaurate (GML) is a surfactant that has been found to inhibit the post-exponential phase activation of virulence factor production and the induction of β-lactamase in Staphylococcus aureus. It has been suggested that signal transduction is the most probable target for GML (S. J. Projan, S. Brown-Skrobot, P. M. Schlievert, F. Vandenesch, and R. P. Novick, J. Bacteriol. 176:4204–4209, 1994). We found that GML suppresses growth of vancomycin-resistant Enterococcus faecalis on plates with vancomycin and blocks the induction of vancomycin resistance, which involves a membrane-associated signal transduction mechanism, either at or before initiation of transcription. Given the surfactant nature of GML and the results of previous experiments, we suggest that GML blocks signal transduction. In contrast, GML has no effect on the induction of erythromycin-inducible macrolide resistance in S. aureus, which does not involve signal transduction.
In previous studies, we have observed that glycerol monolaurate (GML) at subinhibitory concentrations prevents the synthesis of staphylococcal exoproteins and does so at the level of transcription (6, 7). Since GML is a mild surfactant, it is likely to act at the cell membrane and therefore to inhibit either of the two membrane-related processes involved in exoprotein production, namely signal transduction and secretion. We have shown previously that it blocks the induction but not the secretion of β-lactamase or the constitutive synthesis of the enzyme, suggesting that signal transduction is the target (6). However, its effects on signal transduction are clearly not universal—GML does not block the agr signal transduction system, suggesting that it acts on some other, unknown regulatory pathway required for exoprotein synthesis; it has at most a minimal effect on the induction of competence in Bacillus subtilis, and it does not appear to affect chemotaxis in this species (unpublished data).
We have therefore begun a series of studies to test other membrane functions for sensitivity to GML, and we have chosen to start with the induction of vancomycin resistance in enterococci, since this involves one of the few well-characterized two-component signal transduction systems involving a transmembrane receptor in gram-positive cocci (1). GML, incidentally, has no significant effect on gram-negative bacteria, presumably because of its inability to penetrate the outer membrane. The vancomycin resistance system has significant clinical importance, because it has eliminated the only available treatment for multiple-drug-resistant Enterococcus faecalis and threatens to spread to multiple-drug-resistant Staphylococcus aureus strains for which vancomycin is the only available antibiotic. In E. faecalis, the most common form of vancomycin resistance is provided by expression of a vancomycin-inducible plasmid gene cluster. Induction is via a well-characterized signal transduction system, in which VanS is the membrane-associated histidine protein kinase, VanR is a transcriptional activator (response regulator), and the pathway determines the synthesis of peptidoglycan precursors whose assembly is not inhibited by vancomycin (1). We have observed that GML inhibits the induction of vancomycin resistance in E. faecalis at the level of transcription, whereas it does not inhibit the induction of erythromycin resistance in S. aureus, which does not involve signal transduction. These results are consistent with the possibility that GML acts at the level of signal transduction.
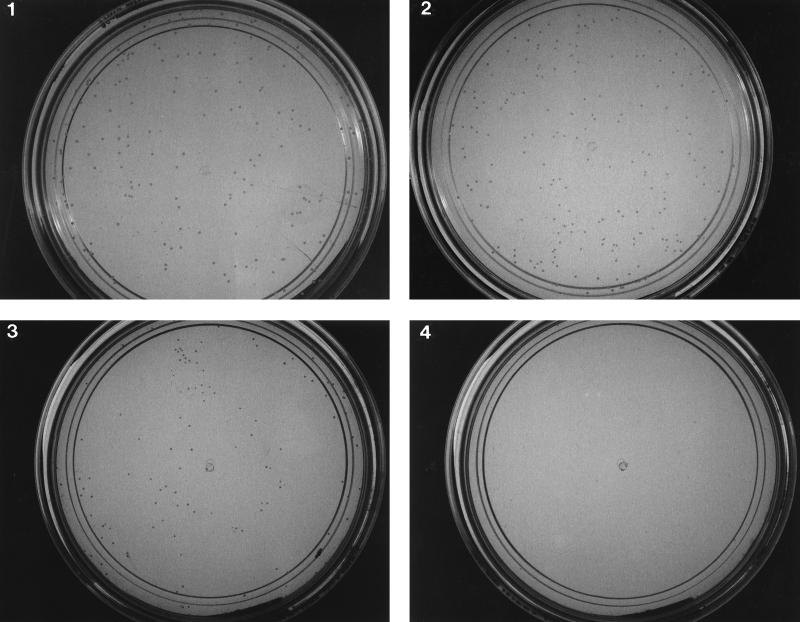
Effect of GML on vancomycin resistance on plates.
The E. faecalis strains used in this work, were kindly provided by P. Courvalin (1). BM4138(pAT90) is a derivative of E. faecalis BM4138 that carries plasmid pAT90. pAT90 contains the intergenic region between vanS and vanH, which includes the vanA promoter, transcriptionally fused to the chloramphenicol transacetylase (CAT) gene. JH2-2(pAT80) is a derivative of E. faecalis JH2-2 (4) that carries plasmid pAT80. pAT80 contains the entire VanA gene cluster (vanR, vanS, vanH, vanA, and vanX) responsible for resistance to vancomycin. In order to test the effect of GML on growth of vancomycin-resistant colonies, JH2-2(pAT80), which contains the entire VanA system, was plated on brain heart infusion (BHI) agarose supplemented with either vancomycin (50 μg/ml) or GML (7.5 μg/ml), both agents, or neither, and colony formation was scored after 18 h at 37°C. We used agarose in plates instead of agar, since previous unpublished observations showed that GML is not active in standard agar-based media, but remains active if agarose is substituted for agar. As shown in Fig. 1 JH2-2(pAT80) grew similarly on BHI agarose with vancomycin (481 colonies), GML (389 colonies), or without additives (404 colonies). On plates containing both GML and vancomycin, colony counts were reduced by a factor of 2 (217 colonies), and the colonies were so small as to be barely visible. These results indicate that GML inhibits the expression of vancomycin resistance and suggest that this inhibition is at the level of induction.
FIG. 1.
Effect of GML on vancomycin resistance in E. faecalis. Bacteria [JH2-2(pAT80)] were grown in BHI broth until they reached a turbidity of 40 Klett units, centrifuged, washed and resuspended in BHI broth, and diluted and plated on BHI agarose plates with or without GML and vancomycin. Plates were incubated for 34 h at 37°C and then photographed. Plates: 1, no GML, no vancomycin; 2, GML (7.5 μg/ml), no vancomycin; 3, no GML, vancomycin (50 μg/ml); 4, GML (7.5 μg/ml), vancomycin (50 μg/ml).
Effect of GML on induction of the vanA promoter.
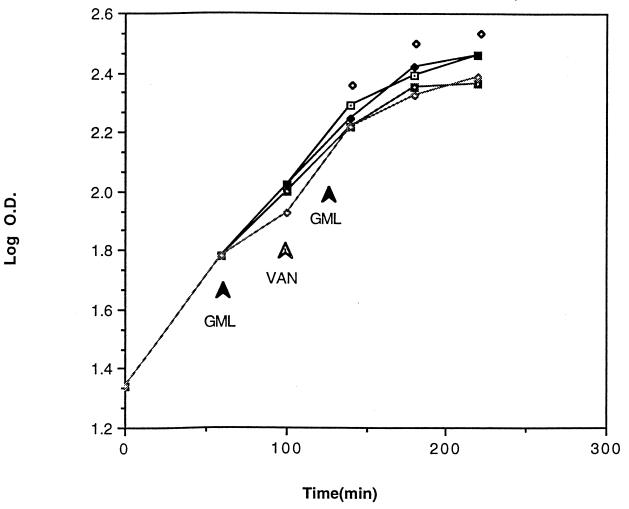
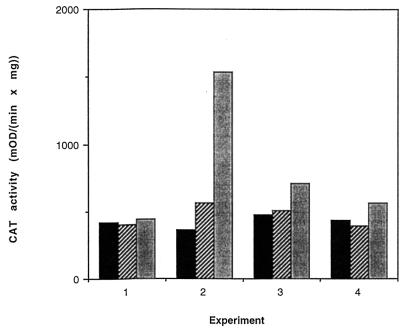
BM4138(pAT90) cells were taken from a fresh overnight plate, resuspended in BHI broth, and grown in a 96-well microplate with shaking in a programmed microplate reader. Cell growth was monitored by optical density; the growth curves obtained from microplates were comparable to growth curves obtained from standard flask cultures. GML and vancomycin were added according to the scheme presented in Fig. 2. As can be seen, at the concentrations that we used, neither GML nor vancomycin significantly affected the exponential growth rate of bacteria. At each time point, cells from one well of each experiment were removed, centrifuged, washed, treated with xylene to permeabilize the cell membrane, and used immediately for the CAT assay. The CAT assay was modified from a previously described procedure (8) and performed with a microplate reader (Molecular Devices), with a dual-kinetic (405/650-nm) program. CAT activities of samples are shown in Fig. 3 and are represented as Vmax per mg of bacterial dry weight. As previously shown (1), vancomycin at subinhibitory concentrations (3 μg/ml) induces transcription from the vanA promoter, measured as an increase in CAT activity. GML blocked the increase in CAT activity, indicating that it inhibits induction either at or before the initiation of transcription.
FIG. 2.
Effect of GML on growth of E. faecalis in microtiter plates. Bacteria [JH2-2(pAT80)] were grown in microplate wells (200 μl/well). GML and vancomycin (3 μg/ml) were added as indicated. Cell density (optical density [O.D.]) was monitored at 650 nm. Samples (whole contents of certain wells) were removed 40, 80, 120 min after addition of vancomycin and assayed for CAT activity (Fig. 3). Each cell density point represents the mean of two independent measurements. Experiment 1 had no GML and no vancomycin (⧫). Experiment 2 had no GML and vancomycin at 100 min (⊡). Experiment 3 had GML at 10 μg/ml at 60 min and 15 μg/ml at 120 min and vancomycin at 100 min (◘). Experiment 4 had GML at 20 μg/ml at 60 min and 10 μg/ml at 120 min and vancomycin at 100 min (◊).
FIG. 3.
Effect of GML on induction of vancomycin resistance. Samples, taken as indicated in the legend to Fig. 2, were treated with xylene (vortexed for 20 s) and directly assayed for CAT activity with a microplate reader. CAT activity was expressed as Vmax per mg of dry weight (milli-optical density unit [mOD]). Numbers 1 to 4 correspond to the experiment numbers explained in the legend to Fig. 2. Each CAT activity value represents the mean of two independent measurements. Bars represent times after vancomycin addition of 40 (▪), 80 (▨), and 120 ( ) min.
Effect of GML on MLS induction.
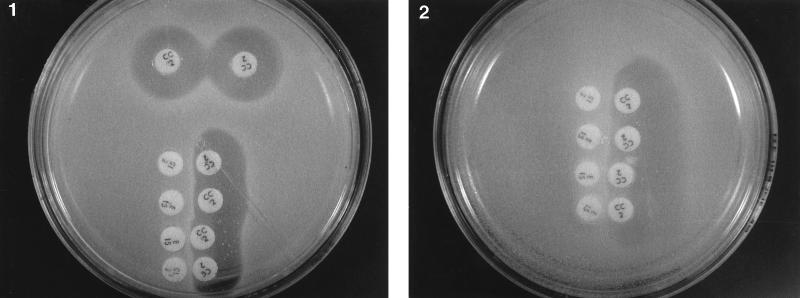
To investigate the possibility that GML may affect an induction pathway that does not involve signal transduction, we repeated the classical experiment of Weisblum and DeMohn (9) with erythromycin induction of resistance to macrolide-lincosamide-streptogramin B (MLS) antibiotics. In this experiment, erythromycin was shown to induce resistance to erythromycin and to other MLS antibiotics that are not themselves inducers (e.g., clindamycin). The resistance to MLS antibiotics is specified by the ermC gene, which encodes rRNA methylase. The induction of this resistance occurs by translational attenuation, changing the conformation of ermC mRNA from inactive to active as a result of complex formation between erythromycin, sensitive ribosomes, and mRNA (1a, 10). Such a mechanism of induction obviously does not include signal transduction. We used S. aureus RN2442, a derivative of RN450 containing plasmid pE194, which confers inducible resistance to erythromycin (2, 3). Growth of bacteria up to the erythromycin disc and on the side of the clindamycin disc adjacent to the erythromycin disc indicates induction of both resistances. As shown in Fig. 4, erythromycin induction of clindamycin and erythromycin resistances was unaffected by the presence of GML, even at a GML concentration that was growth inhibitory (20 μg/ml).
FIG. 4.
Effect of GML on erythromycin-inducible resistance to clindamycin. Bacteria (RN2442) were spread on plates (5 × 107 cells/plate) containing agar (plate 1) or agarose plus GML (20 μg/ml) (plate 2), and antibiotic discs (erythromycin, 15 μg/disc; clindamycin, 2 μg/disc) were placed 1 cm apart as shown. Plates were incubated at 37°C for 16 h and photographed.
Conclusions.
We have shown that GML inhibits the activation by vancomycin of the vanA promoter. This was demonstrated directly by means of a vanA-cat transcriptional fusion and by inhibition of colony formation. The most likely interpretation of these results is that GML blocks signal transduction in the VanS-VanR pathway, as it appears to do in the β-lactamase induction system. Induction of vancomycin resistance by vancomycin occurs via the well-characterized VanS-VanR signal transduction pathway. However, vancomycin is not the ligand for VanS, and it has been suggested that some cell wall intermediate that accumulates in the presence of vancomycin is the actual inducer (5). We have not ruled out the possibility that GML acts by interfering with accumulation of this inducer rather than by blocking VanS directly.
Currently the exact mechanism of GML action is unknown. We propose that the lipid structure of GML makes the cell membrane the most likely target. Membrane-bound GML could interfere with signal transduction by directly binding to transmembrane receptors, by changing the membrane structure (perhaps affecting conformation of the receptor), or by some other means, such as interfering with the normal function of the receptor. Direct action on target genes seems to be unlikely, which is consistent with our finding that GML has no effect on MLS induction, which does not involve signal transduction.
REFERENCES
- 1.Arthur M, Molinas C, Courvalin P. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faeciumBM4147. J Bacteriol. 1992;174:2582–2591. doi: 10.1128/jb.174.8.2582-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Dubnau D, Hahn J, Grandi G, Gryczan T J. Translational attenuation of ermC: a deletion analysis. Mol Gen Genet. 1982;186:204–216. doi: 10.1007/BF00331851. [DOI] [PubMed] [Google Scholar]
- 2.Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J Bacteriol. 1982;150:804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iordanescu S. Three distinct plasmids originating in the same Staphylococcus aureusstrain. Arch Roum Pathol Exp Microbiol. 1976;35:111–118. [PubMed] [Google Scholar]
- 4.Jacob A E, Hobbs S J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974;117:360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai M H, Kirsch D R. Induction signals for vancomycin resistance encoded by the vanA gene cluster in Enterococcus faecium. Antimicrob Agents Chemother. 1996;40:1645–1648. doi: 10.1128/aac.40.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Projan S J, Brown-Skrobot S, Schlievert P M, Vandenesch F, Novick R P. Glycerol monolaurate inhibits the production of β-lactamase, toxic shock syndrome toxin-1, and other staphylococcal exoproteins by interfering with signal transduction. J Bacteriol. 1994;176:4204–4209. doi: 10.1128/jb.176.14.4204-4209.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlievert P M, Deringer J R, Kim M H, Projan S J, Novick R P. Effect of glycerol monolaurate on bacterial growth and toxin function. Antimicrob Agents Chemother. 1992;36:626–631. doi: 10.1128/aac.36.3.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw W V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- 9.Weisblum B, Demohn V. Erythromycin-inducible resistance in Staphylococcus aureus: survey of antibiotic classes involved. J Bacteriol. 1969;98:447–452. doi: 10.1128/jb.98.2.447-452.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weisblum B, Horinouchi S. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci USA. 1980;77:7079–7083. doi: 10.1073/pnas.77.12.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]