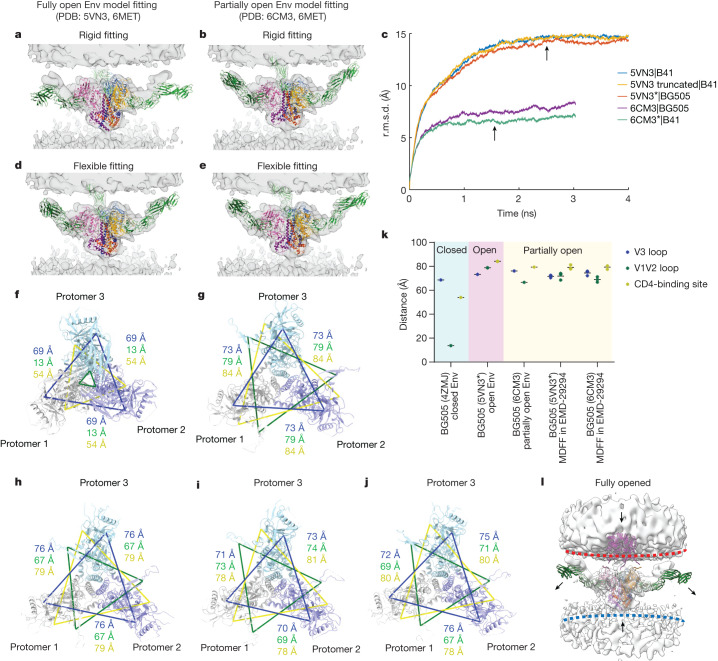
Fig. 4. MDFF analysis suggests that the Env trimer bound to three CD4 molecules is in a partially open conformation.
a,b, Rigid-body fitting of the SOSIP models (fully open Env (PDB: 5VN3; a)) and partially open Env (PDB: 6CM3; b)) containing full-length CD4 molecules (PDB: 6MET) into the cryo-ET density map of Env bound to three CD4 molecules. c, Time evolution of the backbone r.m.s.d. from five independent MDFF simulations into the same density, relative to their starting structure. MDFF simulations ran for around 4 ns on three models of open Env with CD4 (5VN3, truncated 5VN3 (SOSIP.651) and BG505 homology model 5VN3*) and for 3 ns on two models of partially open Env with CD4 (6CM3 and B41 homology model 6CM3*). The MDFF structures measured in i and j were selected after the MDFF simulation reached convergence (arrows; at 2.5 ns and 1.5 ns, respectively). d,e, The results of MDFF of the models in a (d) and b (e) into the same cryo-ET density map. f–k, Analysis of Env openness on the models of BG505 SOSIP: closed (f, 4ZMJ), open (g, 5VN3*), partially open (h, 6CM3), and MDFF-fitted models (5VN3* fitted into Electron Microscopy Data Bank (EMDB) EMD-29294 (i) and 6CM3 fitted into EMD-29294 (j)). CD4 was omitted for clarity. Protomers are coloured in grey, purple and teal. The interprotomer distance was measured between α-carbons of three residues from each protomer: His330 (blue; located at base of the V3 loop), Pro124 (green; located at base of the V1V2 loop) and Asp368 (yellow; located at CD4-binding site). The distance and their average were plotted in k. l, Hypothetical conformational changes of the Env–CD4 complex required to engage the co-receptor. To overcome the steric constraints in CD4 molecules, release of CD4 or membrane bending may facilitate the movement of Env towards the coreceptors embedded in the target membranes.