Abstract
Neolamarckia cadamba (Roxb.) Bosser is a medicinally important, fast-growing, timber-yielding tree species. In the present study, the virome of N. cadamba was explored using the publicly available N. cadamba transcriptome datasets and a putative novel virus, tentatively named as cadamba cryptic virus 1 (CdbCV1), was identified. CdbCV1 contained two genome segments, each coding for a single protein. CdbCV1 RNA1 (1564 nt) encoded for an RNA dependent RNA polymerase (RdRp) protein while CdbCV1 RNA2 (1492 nt) encoded for a coat protein (CP). Phylogenetic and sequence similarity analyses revealed the relatedness of CdbCV1 to pepper cryptic virus 1 and pittosporum cryptic virus 1. Based on the species demarcation criteria, genome organization and phylogeny, CdbCV1 can be regarded a new member of the genus Deltapartitivirus.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13337-023-00845-8.
Keywords: Neolamarckia cadamba, Deltapartitivirus, Novel, Public domain
Neolamarckia cadamba (Roxb.) Bosser (commonly known as Kadam or Cadamba; family: Rubiaceae), native to Southeast Asia, is an underexploited miracle medicinal tree species found globally in tropical and subtropical climates. The tree is fast-growing with a straight, cylindrical trunk and broad canopy. N. cadamba has been used in traditional medicine for centuries. It has antimicrobial, anti-inflammatory, and antioxidant properties, and have shown therapeutic potential in the treatment of various diseases/disorders including diabetes, anaemia, stomatitis, leprosy, cancer and other infectious diseases [5]. The timber of N. cadamba, highly valued for its strength and durability, is used for construction purposes, furniture, and boat making. Besides, the tree is used in paper production and its leaves are used as fodder for livestock. N. cadamba is one of the best landscape trees for urban greening, and suitable for woody forage produce [5]. However, N. cadamba is susceptible to several insect pests such as defoliators, mealy bugs, scale insects, and diseases including die-back, damping-off and root rots [3].
Among the biotic constraints, viral pathogens cause huge losses worldwide owing to the economically important diseases they incite in a wide range of plant species [11]. In the Next Generation Sequencing (NGS) era, the enormous transcriptomic and genomic data generated globally and available publicly provides an opportunity for the exploration of species-specific viromes through various computational tools. Alongside, novel viral sequences that may be present in the NGS data will also be unveiled, a process known as ‘Data Driven Virus Discovery’ (DDVD) [9]. Our previous DDVD studies have identified several putative novel plant viral sequences associated with targeted plant species [12, 13]. In the present study, we explored the N. cadamba transcriptome datasets for novel viral sequences and identified a putative novel delatapartitivirus.
Four transcriptome libraries (SRR1107780, SRR1107781, SRR1107782, SRR7130587) of N. cadamba available in National Centre for Biotechnology Information’s (NCBI) Sequence Read Archive (SRA) database were searched for putative novel plant viral reads using RNA-dependent RNA polymerase (RdRp) search tool of Serratus explorer [6] as described in Sidharthan et al. [13]. For genome recovery of identified novel virus, raw reads of virus-positive library were retrieved, trimmed using Trimmomatic v. 0.36 [1] (quality score: 25), de novo assembled using rnaSPAdes v 3.15.4 [2] and mapped against related viral sequence using blastx option of ncbi blast + v 2.10.1 [4] in Galaxy Australia server (https://usegalaxy.org.au/). Recovered genome segments were annotated based on the predicted motifs in the encoded proteins and BLAST analysis as described in Sidharthan et al. [13]. Phylogenetic analysis was carried out after MUSCLE alignment of protein sequences encoded by the recovered viral genome segments along with other related viral sequences using neighbourhood-joining method (NJ) and Poisson model with 1000 bootstrap replicates in MEGA 7 v 7.0.26 [8]. Sequence identity matrices were created using Sequence Demarcation Tool v 1.2 [10]. Trimmed reads were mapped onto recovered genome segments and mean coverage values were obtained as described in Sidharthan et al. [13].
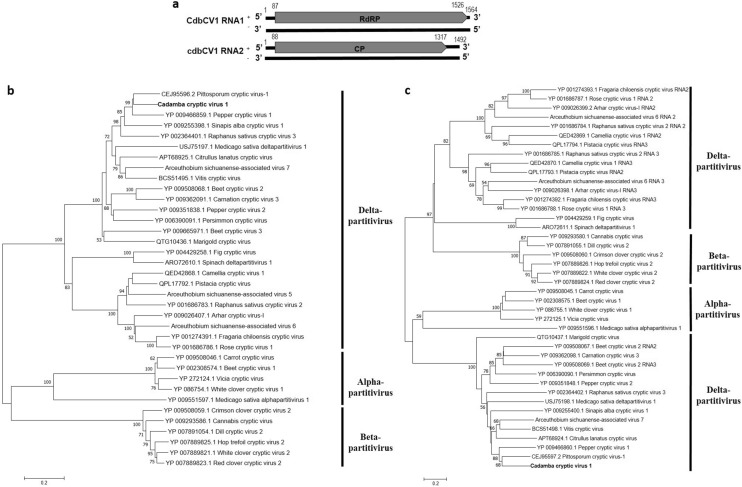
Reads of a putative novel deltapartitivirus was identified in the library SRR7130587 derived from the Bioproject PRJNA464271 [7]. The identified putative novel virus was tentatively named as cadamba cryptic virus 1 (CdbCV1). After assembly and BLAST analysis against related viral sequences, two genome segments of CdbCV1 were recovered. CdbCV1 RNA1 (1564 nt) contains a single open reading frame (ORF) of 1440 nt encoding for a 54.3 kDa protein (479 aa) with viral RNA dependent RNA polymerase (RdRp) motif (PF00680). CdbCV1 RNA2 (1492 nt) contains a single ORF of 1230 nt encoding for a 47.1 kDa protein (409 aa) with no predicted motif (Fig. 1a). BLAST analysis showed that CdbCV1 proteins 1 and 2 shared a maximum of 74.74% and 51.08% sequence identities (at 100% query coverage), respectively with RdRp (AVL84362.1) and coat protein (CP) (AVV48359.1) sequences of pepper cryptic virus 1 (PCV-1). Phylogenetic analysis based on RdRp and CP sequences grouped CdbCV1 with PCV-1 and pittosporum cryptic virus 1 (PiCV1) (Fig. 1b, c). Sequence identity analysis revealed that CdbCV1 RdRp and CP sequences shared a maximum of 76.0% and 53.9% identities, respectively with the corresponding sequences of PiCV1 (Fig. S1, S2). Reads of CdbCV1 could not be detected in other three N. cadamba transcriptome libraries in MEGABLAST searches.
Fig. 1.
Genome organization of cadamba cryptic virus 1 (CdbCV1) identified in the current study (a). Phylogenetic relationship of CdbCV1 with other alpha-, beta- and deltapartitiviruses based on RdRp (b) and CP amino acid sequences (c). Bootstrap values more than 50% are only indicated. CdbCV1 is shown in bold
Deltapartitiviruses are plant-infecting double stranded (ds) RNA genome containing viruses with two or three genome segments [15]. Members of deltapartitiviruses are regarded as new species if they share ≤ 90% and ≤ 80% sequence similarities, respectively for RdRp and CP sequences with other known members [15]. Based on this species demarcation criteria, CdbCV1 can be regarded as a novel deltapartitivirus. A consensus statement report by Simmonds et al. [14] stated that viruses that are known only by sequences from metagenomics data should be incorporated into the official International Committee on Taxonomy of Viruses (ICTV) taxonomy scheme. Thus, the identified virus can be regarded as a bona fide one. As the putative novel virus identified in the current study is solely based on bioinformatic analysis of transcriptome data, host assignment of the virus should be regarded cautiously until further validation. The current study paves way for further studies on understanding biological properties and distribution of the identified virus.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank the GCR, the Director, ICFRE-IFB, Hyderabad, the Head, Division of Plant Pathology and the Director, ICAR-CAFRI, Jhansi for their support. We thank Galaxy Australia, a service provided by the Australian Biocommons and its partners. The authors are also thankful to the original submitters of datasets that are explored in the study.
Data availability
The viral genome sequences described in the study are available in GenBank with accession numbers BK064372 to BK064373, and are provided in the ESM_1.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
This work does not contain any animal or human participants.
Informed consent
This work does not contain any animal or human participants.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
V. Kavi Sidharthan, Email: kavi.icfre@gmail.com.
Mushineni Ashajyothi, Email: aashjyo18@gmail.com.
References
- 1.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushmanova E, Antipov D, Lapidus A, Prjibelski AD. rnaSPAdes: a de novo transcriptome assembler and its application to RNA-Seq data. GigaScience. 2019;8(9):giz100. doi: 10.1093/gigascience/giz100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung AYC, Ajik M, Nilus R, Hastie A, Ong RC, Chey VK. New records of insects associated with Laran (Neolamarckia cadamba) in Sabah. Sepilok Bull. 2009;10:45–64. [Google Scholar]
- 4.Cock PJ, Chilton JM, Grüning B, Johnson JE, Soranzo N. NCBI BLAST + integrated into galaxy. Gigascience. 2015;4:13742–s14015. doi: 10.1186/s13742-015-0080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devgan M, Bhatia L, Kumar H. Anthocephalus cadamba: a comprehensive review. Res J Pharm Technol. 2012;5(12):1478–1483. [Google Scholar]
- 6.Edgar RC, Taylor J, Lin V, Altman T, Barbera P, Meleshko D, Lohr D, Novakovsky G, Buchfink B, Al-Shayeb B, Banfield JF. Petabase-scale sequence alignment catalyses viral discovery. Nature. 2022;602(7895):142–147. doi: 10.1038/s41586-021-04332-2. [DOI] [PubMed] [Google Scholar]
- 7.He JJ, Ye R, Chen T, Xi QY, Luo JY, Zhang HJ, Wu JH, Sun JJ, Zhang YL. Exploration of miRNAs in Neolamarckia cadamba and their potential cross-kingdom functions. ExRNA. 2020;2(1):1–9. doi: 10.1186/s41544-019-0043-8. [DOI] [Google Scholar]
- 8.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lauber C, Seitz S. Opportunities and challenges of data-driven virus discovery. Biomolecules. 2022;12(8):1073. doi: 10.3390/biom12081073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muhire BM, Varsani A, Martin DP. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE. 2014;9(9):e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubio L, Galipienso L, Ferriol I. Detection of plant viruses and disease management: relevance of genetic diversity and evolution. Front Plant Sci. 2020;11:1092. doi: 10.3389/fpls.2020.01092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sidharthan VK, Chaturvedi KK, Baranwal VK. Diverse RNA viruses in a parasitic flowering plant (spruce dwarf mistletoe) revealed through RNA-seq data mining. J Gen Plant Pathol. 2022;88(2):138–144. doi: 10.1007/s10327-021-01049-y. [DOI] [Google Scholar]
- 13.Sidharthan VK, Vanamala G, Rajeswari V, Baranwal VK. Identification of a putative novel cholivirus in the transcriptome of Gymnema sylvestre R. Br. Arch Microbiol. 2023;205(5):186. doi: 10.1007/s00203-023-03517-9. [DOI] [PubMed] [Google Scholar]
- 14.Simmonds P, Adams MJ, Benkő M, Breitbart M, Brister JR, Carstens EB, Davison AJ, Delwart E, Gorbalenya AE, Harrach B, Hull R. Consensus statement: virus taxonomy in the age of metagenomics. Nat Rev Microbiol. 2017;15:161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- 15.Vainio EJ, Chiba S, Ghabrial SA, Maiss E, Roossinck M, Sabanadzovic S, Suzuki N, Xie J, Nibert M, ICTV Report Consortium ICTV virus taxonomy profile: Partitiviridae. J Gen Virol. 2018;99:17–18. doi: 10.1099/jgv.0.000985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The viral genome sequences described in the study are available in GenBank with accession numbers BK064372 to BK064373, and are provided in the ESM_1.