Abstract
Background
Continuous evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) outpaces monovalent vaccine cross-protection to new viral variants. Consequently, bivalent coronavirus disease 2019 (COVID-19) vaccines including Omicron antigens were developed. The contrasting immunogenicity of the bivalent vaccines and the impact of prior antigenic exposure on new immune imprinting remains to be clarified.
Methods
In the large prospective ENFORCE cohort, we quantified spike-specific antibodies to 5 Omicron variants (BA.1 to BA.5) before and after BA.1 or BA.4/5 bivalent booster vaccination to compare Omicron variant-specific antibody inductions. We evaluated the impact of previous infection and characterized the dominant antibody responses.
Results
Prior to the bivalent fourth vaccine, all participants (N = 1697) had high levels of Omicron-specific antibodies. Antibody levels were significantly higher in individuals with a previous polymerase chain reaction positive (PCR+) infection, particularly for BA.2-specific antibodies (geometric mean ratio [GMR] 6.79, 95% confidence interval [CI] 6.05–7.62). Antibody levels were further significantly boosted in all individuals by receiving either of the bivalent vaccines, but greater fold inductions to all Omicron variants were observed in individuals with no prior infection. The BA.1 bivalent vaccine generated a dominant response toward BA.1 (adjusted GMR 1.31, 95% CI 1.09–1.57) and BA.3 (1.32, 1.09–1.59) antigens in individuals with no prior infection, whereas the BA.4/5 bivalent vaccine generated a dominant response toward BA.2 (0.87, 0.76–0.98), BA.4 (0.85, 0.75–0.97), and BA.5 (0.87, 0.76–0.99) antigens in individuals with a prior infection.
Conclusions
Vaccination and previous infection leave a clear serological imprint that is focused on the variant-specific antigen. Importantly, both bivalent vaccines induce high levels of Omicron variant-specific antibodies, suggesting broad cross-protection of Omicron variants.
Keywords: COVID-19, bivalent vaccines, immunogenicity, antibodies, booster vaccination
Comparison of antibody responses following bivalent BA.1 and BA.4/5 vaccination and impact of previous infection. Prior antigenic exposure leaves a clear serological imprint. Both bivalent vaccines induce high levels of Omicron variant-specific antibodies, suggesting broad cross-protection of Omicron variants.
Graphical Abstract
Graphical Abstract.
Coronavirus disease 2019 (COVID-19) vaccination has been highly successful in limiting severe disease from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections. As population immunity has increased, the emergence of viral variants capable of evading antibodies has become a major focus of continued viral monitoring. The latest major shift in viral populations occurred after the introduction of the Omicron variant (B.1.1.529) in November 2021 [1, 2]. In Denmark, high vaccination coverage (2 primary doses and a booster dose in 78% of the adult population) led to dismissal of all societal restrictions in January/February of 2022. This resulted in the rapid spread of primarily the Omicron BA.2 variant [3], with studies from Danish blood donor cohorts estimating that 66% of the adult population had been infected with Omicron by April 2022 [4]. Similar scenarios of high Omicron infection rates despite high vaccination coverage occurred in many countries. Consequently, a large part of the population has hybrid immunity stemming from both original wild-type-strain vaccination and Omicron-strain infection.
The viral Omicron lineage has continued to evolve with BA.1 and BA.3 as the initial variants, and descendant with subsequent emergence of first BA.2 and then BA.4 and BA.5 (spike protein sequence diversity for Omicron BA.1 to BA.5 in Supplementary Figure 1). Lately, further diversification in the Omicron lineage has resulted in the spread of the BA.2-derived BA.2.75, the BA.2-recombinant XBB, and the BA.5-derived BQ.1 [5]. Exposure to sequential viral spike antigens (from vaccination or infection) has been shown to impact the quality of antibodies and the viral variant coverage [6, 7].
As the Omicron variants quickly dominated the global viral landscape, vaccine manufactures (Pfizer-BioNTech and Moderna) accordingly adapted the mRNA-based vaccine platform to engineer bivalent vaccines containing both the original- and Omicron-antigen [8]. Two versions of the bivalent vaccine were developed, containing Omicron BA.1 or BA.4/5 antigen, and numerous countries debated which bivalent vaccine was optimal. In Denmark, a fourth dose vaccination campaign was initiated during the autumn of 2022 with first the BA.1 vaccine (BNT162b2 BA.1 and mRNA-1273.214), which was replaced by the BA.4/5 vaccine (BNT162b2 BA.4/5) as soon as it became available, due to the anticipation of greater immune protection. The safety and vaccine efficacy of these bivalent vaccines has been evaluated in population monitoring and phase 2/3 evaluations [8–10].
Recently, data have suggested that exposure to heterologous SARS-CoV-2 spike antigens can drive both de novo antibody formation as well as significant affinity maturation of existing B-cell clones [11, 12]. However, it is also clear that previous exposure to spike variants leaves an imprint on serum antibodies [12]. As viral evolution is continuing under human immune selective pressure, it becomes an important question to understand the impact of antigenic imprinting to design vaccination strategies for optimal population immunity.
In the prospective ENFORCE cohort, we therefore set out to quantify and compare Omicron variant-specific antibody responses in 1697 individuals receiving a BA.1 or BA.4/5 bivalent vaccine and to determine how previous SARS-CoV-2 infection impacts these responses. Furthermore, we evaluated signs of specific variant-directed antibody coverage to gain a better understanding of population immunity with a focus on achieving broad cross-protection of viral Omicron variants.
METHODS
ENFORCE (Danish National Cohort Study of Effectiveness and Safety of SARS-CoV-2 Vaccines) is an open-label, non-randomized, parallel group, phase IV study. The study enrolled Danish citizens (≥18 years) prior to their first SARS-CoV-2 vaccination (clinicaltrials.gov identifier: NCT04760132). Participants were included from February to August 2021 at 7 study sites, covering all 5 Danish regions. The safety and effectiveness of the SARS-CoV-2 vaccines, and the durability of the vaccine response is currently being evaluated for a 2-year period with frequent blood sampling. Details of the ENFORCE cohort has recently been published [13]. The ENFORCE study was approved by the Danish Medicines Agency (no. 2020-006003-42) and the National Committee on Health Research Ethics (no. 1-10-72-337-20). All participants provided written informed consent.
Study Design and Sample Collection
In the Danish vaccination program, people above 50 years of age, healthcare personnel, and individuals at increased risk of severe COVID-19 disease were invited to receive a fourth SARS-CoV-2 vaccine dose in the autumn of 2022. Participants from the ENFORCE cohort that met the following criteria were included in the study: (1) received a fourth bivalent SARS-CoV-2 vaccine dose from September to November 2022 and (2) had serum samples collected and antibody data quantified before (−14 to 0 days) and after (28 ± 8 days) the fourth bivalent vaccine dose. Study participants were separated into 2 groups: (1) participants with a previous polymerase chain reaction (PCR)-verified SARS-CoV-2 infection (previous PCR+) and (2) participants with no previous PCR-verified infection and not nucleocapsid seroconverted (no infection). Previous infections were classified into viral variants by PCR test date (Supplementary Table 1). Study participants with no previous PCR-verified infection, but who met our definition for nucleocapsid seroconversion were excluded from the analysis to avoid misclassification. Seroconversion was defined as a nucleocapsid Immunoglobulin G (IgG) level >3000 AU/mL and a > 2-fold increase in the level of nucleocapsid IgG compared to the level prior to the first vaccination.
Information on age, sex, medical history, vaccine priority group (as defined by the Danish COVID-19 vaccination program), vaccination dates, and vaccine type were collected at the time of the fourth vaccine dose and confirmed by the Danish National Patient Registry and the Danish Vaccination Registry. Data on any previous positive SARS-CoV-2 PCR tests were extracted from the Danish National Microbiology database (Statens Serum Institut, Copenhagen, Denmark).
Severe Acute Respiratory Syndrome Coronavirus 2 Spike IgG Antibody Profiling
Serum IgG antibodies to multiple SARS-CoV-2 spike variants were detected and quantified using the Meso Scale Diagnostics (MSD) V-PLEX SARS-CoV-2 panel 25 and 29 (IgG) kits (cat. no. K15583U-4 and K15624U-4, Meso Scale Diagnostics, Rockville, Maryland). The pre-coated SARS-CoV-2 spike antigen spots of interest were wild-type (Wuhan-Hu-1), Omicron BA.1, BA.2, BA.3, BA.4, and BA.5. The assay was performed as previously published [14, 15]. Briefly, unspecific antibody binding was blocked by MSD Blocker A. Serum samples were diluted 1:100 000 in MSD Diluent 100 and incubated in the plate wells along with MSD Reference Standard 1 to establish a calibration curve. Bound IgG was detected by MSD SULFO-TAG anti-human IgG antibody. GOLD Read Buffer B was added, and the plates were read by a MESO QuickPlex SQ 120 reader. Raw data were processed by MSD Discovery Workbench Software (version 4.0). Spike IgG concentrations were calculated for each variant by fitting the measured signals to the calibration curve and correcting for dilution. Quantifications were reported in arbitrary units per mL (AU/mL).
Data and Statistical Analysis
Participant characteristics were summarized as frequency and percent for the categorical variables and median and interquartile range (IQR) for the continuous variables.
To assess the immune status before the fourth vaccine dose and evaluate the impact of previous infection, serum antibody levels to wild-type spike and 5 Omicron spike variants (BA.1 to BA.5) were compared between those with a previous PCR+ infection and those with no prior infection using unpaired t tests of the log10 transformed antibody level. The estimates were back transformed to be presented as geometric means and geometric mean ratios (GMR) together with their 95% confidence intervals (CI). Similarly, antibody levels before and after vaccination were presented as geometric means and GMRs with 95% CI. To compare the change in antibody levels before and after the fourth dose, paired t tests stratified by prior infection status were used.
Univariable and multivariable linear regression was used to evaluate if the change in antibody levels on the log10 scale differed based on the target of the bivalent vaccines (BA.1 vs BA.4/5). Estimates of the mean differences were back transformed to be presented as the GMR with 95% CI. Multivariable models were adjusted for factors selected a priori and included sex (males, females), age group (<55, 55–64, 65–74, ≥75 years) and Charlson comorbidity index (CCI) score (0, 1–2, ≥3) [16]. The CCI score was calculated based on hospital admission records and discharge disease diagnoses (ICD-10 codes) in the 5 years prior to receiving the fourth dose as recorded in the Danish National Patient Registry [17]. Models were constructed separately for each of the specific spike antibody measurements (wild-type and Omicron BA.1 to BA.5) and by prior infection status.
To further characterize the hierarchy of serum polyclonal responses, antibody levels measured for each of the specific Omicron variants BA.1 to BA.5 were ranked. The Omicron variant with the highest antibody response was defined as dominant and was selected before and after vaccination for each participant. The proportion of participants with dominant responses to each of the 5 variants was then calculated. Similarly, the relative serum antibody induction before/after the fourth vaccine dose was ranked, and the proportion of participants with dominant antibody inductions to each of the variants was calculated.
RESULTS
Participant Selection
A total of 6972 individuals were enrolled into the ENFORCE study cohort between February and August 2021. During the first year, 688 participants withdrew from the study. Of the 6284 still under follow-up, 2352 received a fourth bivalent vaccine dose and provided serum samples for analysis both before and after vaccination. We subsequently excluded 18 (0.8%) who were PCR positive between their before and after fourth dose vaccination visit and 637 (27.1%) who had no previous PCR-verified infection but who had seroconverted for antibodies to nucleocapsid.
Thus, a total of 1697 participants were included in the analysis, whereof 672 (39.6%) received a bivalent BA.1 vaccine (303 (45.1%) mRNA-1273.214 from Moderna and 369 (54.9%) BNT162b2 BA.1 from Pfizer/BioNTech). The remaining 1025 (60.4%) received a bivalent BA.4/5 vaccine (only the BA.4/5 vaccine from Pfizer/BioNTech has been used in Denmark). The majority of the cohort, 63% (n = 1070), had a previous PCR-verified SARS-CoV-2 infection prior to their fourth dose. Based on PCR test date, 834 (77.9%) were estimated to have had a BA.2 infection (classification of the remaining previously infected participants in Supplementary Table 1). The time from positive PCR test to blood sampling prior to the fourth bivalent vaccine dose was a median of 227 days (IQR: 197–252). The participants demographics are shown in Table 1 grouped by bivalent vaccine and previous infection status. Importantly, the time in days from both first dose (median 539 days, IQR: 516–560) and from third dose (313, IQR: 301–327) were well balanced across the 4 groups.
Table 1.
Participant demographics
| Total (N = 1697) |
Bivalent BA.1 Previous PCR+ (N = 392) | Bivalent BA.1 No Previous Infection (N = 280) |
Bivalent BA.4/5 Previous PCR+ (N = 678) | Bivalent BA.4/5 No Previous Infection (N = 347) |
|
|---|---|---|---|---|---|
| Median (IQR) | |||||
| Age at fourth vaccine dose (y) | 69 (61, 76) | 70 (63, 78) | 73 (66, 81) | 64 (58, 72) | 70 (62, 76) |
| No. of persons (%) | |||||
| Age group | |||||
| < 55 | 132 (7.8) | 15 (3.8) | 8 (2.9) | 90 (13.3) | 19 (5.5) |
| 55–64 | 541 (31.9) | 108 (27.6) | 52 (18.6) | 272 (40.1) | 109 (31.4) |
| 65–74 | 503 (29.6) | 126 (32.1) | 89 (31.8) | 177 (26.1) | 111 (32.0) |
| ≥75 | 521 (30.7) | 143 (36.5) | 131 (46.8) | 139 (20.5) | 108 (31.1) |
| Sex | |||||
| Male | 732 (43.1) | 180 (45.9) | 130 (46.4) | 278 (41.0) | 144 (41.5) |
| Female | 965 (56.9) | 212 (54.1) | 150 (53.6) | 400 (59.0) | 203 (58.5) |
| Vaccine priority group | |||||
| 1. Individuals at increased risk | 263 (15.5) | 51 (13.0) | 52 (18.6) | 108 (15.9) | 52 (15.0) |
| 2. Healthcare workers | 92 (5.4) | 8 (2.0) | 6 (2.1) | 67 (9.9) | 11 (3.2) |
| 3. General population | 1342 (79.1) | 333 (84.9) | 222 (79.3) | 503 (74.2) | 284 (81.8) |
| CCI score at fourth vaccine dose | |||||
| 0 | 1255 (74.0) | 279 (71.2) | 190 (67.9) | 532 (78.5) | 254 (73.2) |
| 1–2 | 395 (23.3) | 101 (25.8) | 80 (28.6) | 134 (19.8) | 80 (23.1) |
| ≥3 | 47 (2.8) | 12 (3.1) | 10 (3.6) | 12 (1.8) | 13 (3.7) |
| Previous vaccines received | |||||
| Pfizer/BioNTech (BNT162b2) | 870 (51.3) | 183 (46.7) | 159 (56.8) | 358 (52.8) | 170 (49.0) |
| Moderna (mRNA-1273) | 766 (45.1) | 204 (52.0) | 118 (42.1) | 275 (40.6) | 169 (48.7) |
| AstraZeneca(ChAdOx1)/mRNA | 61 (3.6) | 5 (1.3) | 3 (1.1) | 45 (6.6) | 8 (2.3) |
| Fourth dose bivalent vaccine type | |||||
| Pfizer/BioNTech (BA.1 or BA.4/5) | 1394 (82.1) | 196 (50.0) | 173 (61.8) | 678 (100) | 347 (100) |
| Moderna (BA.1) | 303 (17.9) | 196 (50.0) | 107 (38.2) | 0 | 0 |
| Median (IQR) | |||||
| Days from first dose | 539 (516, 560) | 536 (515, 555) | 543 (524, 558) | 538 (513, 567) | 540 (520, 561) |
| Days from third (booster) dose | 313 (301, 327) | 309 (297, 321) | 313 (301, 324) | 315 (302, 332) | 314 (303, 328) |
| Days from last positive PCR test | 227 (197, 252) | 220 (188, 237) | NA | 232 (202, 257) | NA |
Participant demographics at the fourth bivalent vaccine dose grouped by bivalent vaccine and prior infection status.
Abbreviations: CCI, Charlson comorbidity index; IQR, interquartile range; PCR, polymerase chain reaction.
Antibody Levels Following Three Vaccine Doses and Impact of Previous Infection
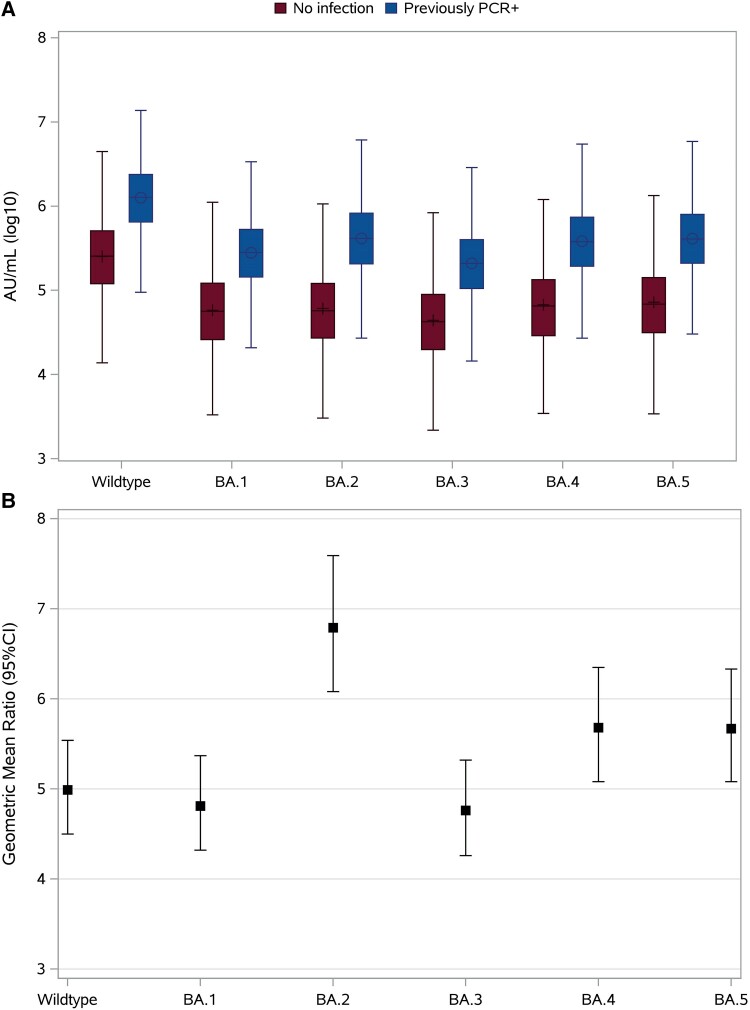
To assess the immune status before the fourth vaccine dose and evaluate the impact of previous infection, we plotted the serum levels of antibodies to both wild-type spike and the 5 Omicron spike variants (Figure 1A). All participants, irrespective of prior infection status, had high levels of both wild-type- and Omicron-specific antibodies prior to the fourth vaccine dose that exceeded the previously defined approximate cutoff (1 × 104 AU/mL) of COVID-19 vaccine hypo-responsiveness [17]. Additionally, the impact of a previous PCR+ infection was evident. The median antibody levels to wild-type spike were 1.25 × 106 AU/mL (95% CI 1.18–1.33) in previously infected individuals compared to 0.25 × 106 AU/mL (95% CI 0.23–0.27) in those with no infection (GMR 4.99, 95% CI 4.49–5.56, P < .0001). Overall, serum antibody levels for all variants were significantly higher in those with a previous PCR+ infection compared to those with no prior infection. The highest ratio was observed for BA.2-specific antibodies (GMR 6.79, 95% CI 6.05–7.62) (Figure 1B).
Figure 1.
Antibody levels in serum to wild-type spike and 5 Omicron spike variants BA.1 to BA.5 prior to the fourth vaccine dose. A, Panel shows the antibody levels in participants triple-vaccinated with no prior infection (Dark red box [+], n = 627) and participants triple-vaccinated with a previous PCR+ infection (Dark blue box [O], n = 1070). Data show the geometric mean with 95% confidence intervals. B, Panel displays the geometric mean ratio with 95% confidence intervals between previous PCR+ infected and non-infected. Abbreviations: CI, confidence interval; PCR, polymerase chain reaction.
Immune Responses of the Bivalent Fourth Dose
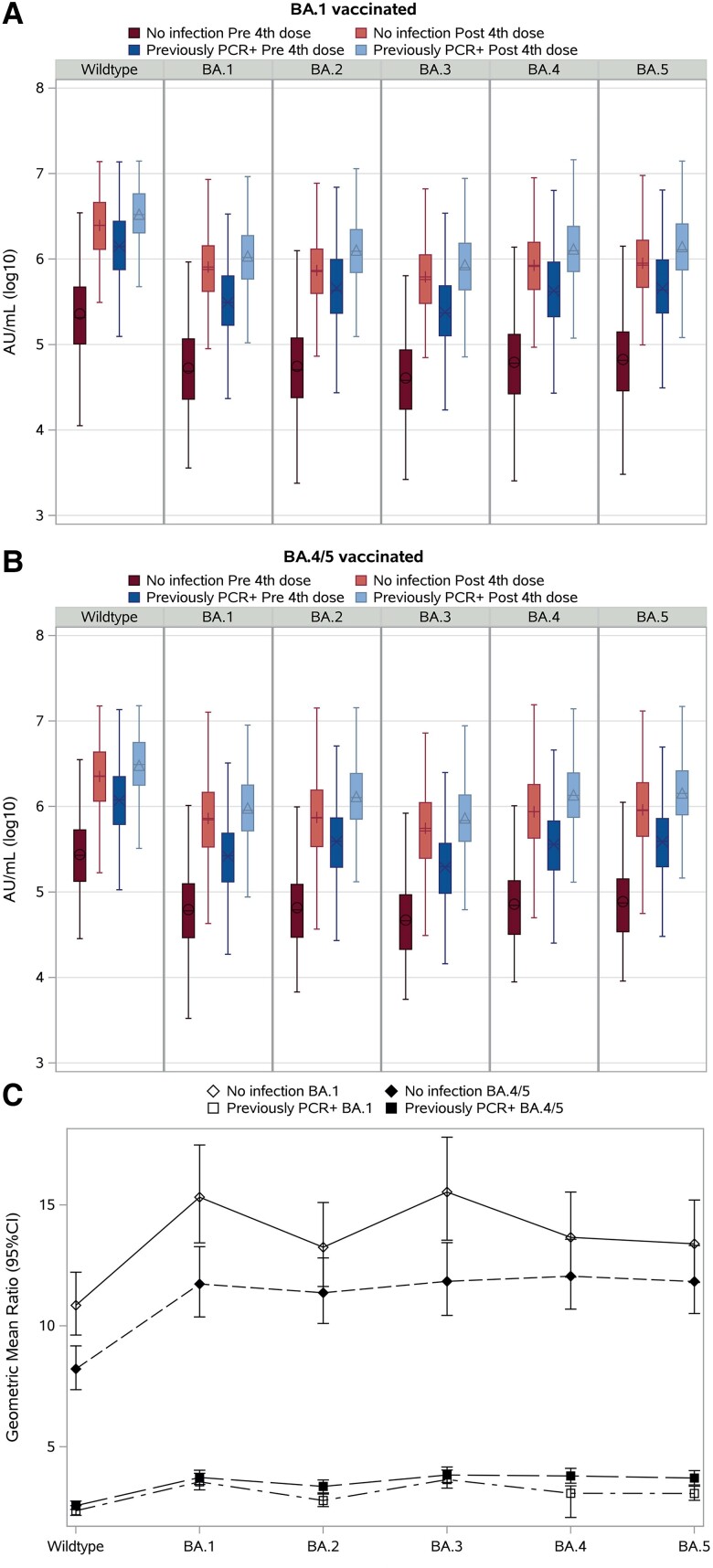
All participants benefited from vaccination with significant increases in antibody levels to all variants, irrespective of bivalent vaccine or prior infection status (Figure 2A and 2B). For participants with a previous PCR+ infection, we observed increases to the wild-type antigen by 2.3-fold (95% CI 2.1–2.6) and 2.6-fold (95% CI 2.4–2.8) and increases to the BA.5 antigen by 3.1-fold (95% CI 2.8–3.4) and 3.8-fold (95% CI 3.5–4.2) following bivalent BA.1 and BA.4/5 vaccination, respectively. In contrast, the vaccine response in individuals with no prior infection was of greater amplitude. Antibodies were induced to the wild-type antigen by 10.8-fold (95% CI 9.6–12.2) and 8.2-fold (95% CI 7.4–9.1) following bivalent BA.1 and BA.4/5 vaccination, respectively. Additionally, greater fold inductions following BA.1 versus BA.4/5 vaccination were found across all Omicron antigens in individuals with no prior infection. Most prominently, for the BA.1 and BA.3 antigens, respectively, there was a 15.3-fold (95% CI 13.4–17.5) and 15.5-fold (95% CI 13.5–18.8) difference in antibody induction after bivalent BA.1 vaccination and 11.7-fold (95% CI 10.4–13.3) and 11.8-fold (95% CI 10.4–13.4) after bivalent BA.4/5 vaccination (Figure 2C).
Figure 2.
Antibody levels in serum to wild-type spike and 5 Omicron spike variants BA.1 to BA.5 prior to and after the bivalent fourth vaccine dose. A, Panel shows participants vaccinated with the BA.1 bivalent vaccine with no prior infection (n = 280) before (dark red box [O]) and after (light red box [+]) bivalent vaccination, and participants vaccinated with the BA.1 bivalent vaccine with a previous PCR+ infection (n = 392) before (dark blue box [X]) and after (light blue box [Δ]) bivalent vaccination. B, Panel shows participants vaccinated with the BA.4/5 bivalent vaccine with no prior infection (n = 347) before (dark red box [O]) and after (light red box [+]) bivalent vaccination, and participants vaccinated with the BA.4/5 bivalent vaccine with a previous PCR+ infection (n = 678) before (dark blue box [X]) and after (light blue box [Δ]) bivalent vaccination. Data show the geometric mean with 95% confidence intervals. C, Panel shows the geometric mean ratio (before/after bivalent fourth vaccine dose) with 95% confidence intervals for each of the 4 groups to the 6 antigens. Abbreviations: CI, confidence interval; PCR, polymerase chain reaction.
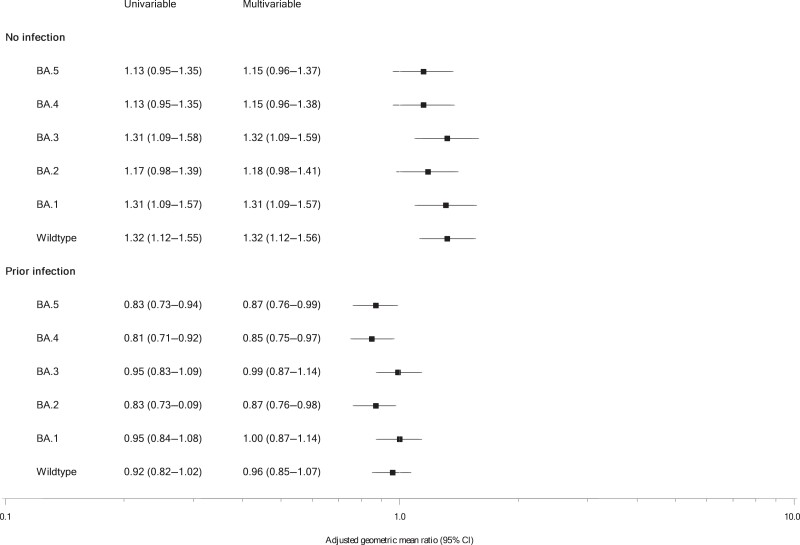
Linear regression modeling was used to further explore the immune response to the bivalent vaccines. In both the univariable and multivariable model, we found evidence for significantly different antibody responses based on prior infection status in those receiving a BA.1 vaccine compared to those receiving a BA.4/5 vaccine (Figure 3). In individuals with a previous PCR+ infection, we found a significant 15% lower response to the BA.4 antigen (adjusted GMR 0.85, 95% CI 0.75–0.97, P = .01) in those vaccinated with the BA.1 vaccine compared to those vaccinated with the BA.4/5 vaccine. Significantly lower responses to BA.2 (0.87, 0.76–0.98, P = .02) and BA.5 (0.87, 0.76–0.99, P = .03) antigens were also found. In contrast, in individuals with no prior infection, those vaccinated with the bivalent BA.1 vaccine had a significant 31% greater response to the BA.1 antigen (adjusted GMR 1.31, 95% CI 1.09–1.57, P = .004) compared to those receiving the bivalent BA.4/5 vaccine. Similar differences were observed for wild-type (1.32, 1.12–1.56, P = .0009) and BA.3 (1.32, 1.09–1.59, P = .005) antigens.
Figure 3.
Forest plot of the univariable and multivariable linear regression displaying antibody responses to the bivalent fourth vaccine dose (BA.1 vs BA.4/5) by prior infection status. Data show the adjusted geometric mean ratio with 95% confidence intervals.
Dominant Omicron Variant-specific Immune Responses
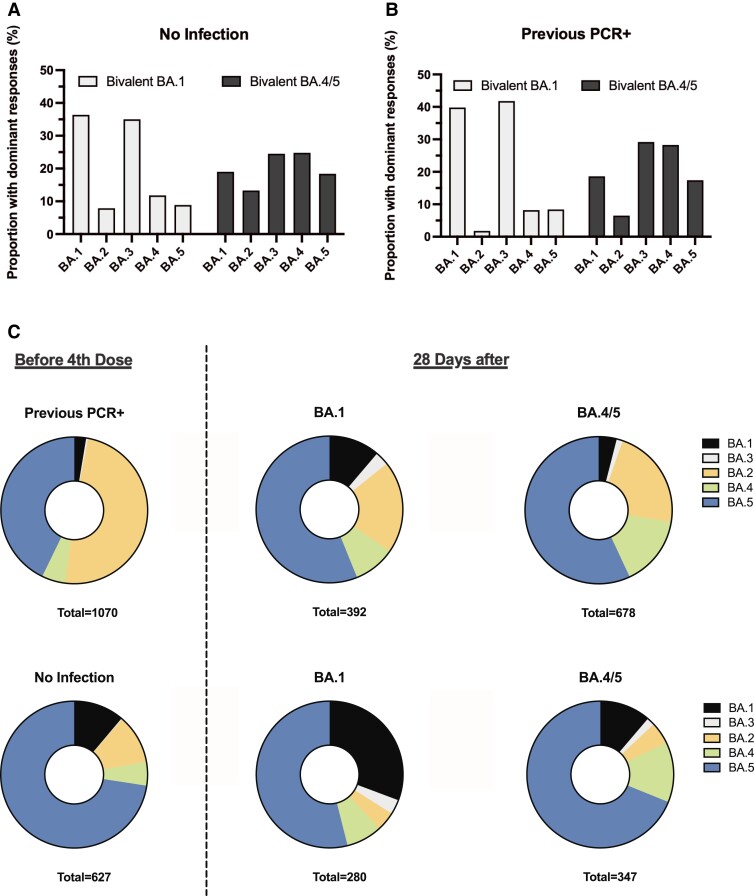
To provide a measure of the dominant antibody responses, we ranked the ratio before/after fourth dose in each participant to the 5 Omicron antigens. For participants with no infection vaccinated with the BA.1 vaccine, there was a marked dominant response to BA.1 (36.4%) and BA.3 antigens (35%). In contrast, in individuals with no infection vaccinated with the BA.4/5 vaccine, a broad response was captured by dominant responses to BA.4 (24.8%) and BA.3 (24.5%) antigens (Figure 4A). For participants with a previous PCR+ infection, a similar marked dominant response to BA.1 (39.8%) and BA.3 (41.8%) antigens was observed in those receiving the bivalent BA.1 vaccine (Figure 4B).
Figure 4.
Proportion of participants with dominant responses to the 5 Omicron antigens BA.1 to BA.5. A, Panel shows the dominant induced responses in participants with no prior infection following bivalent BA.1 and BA.4/5 vaccination. B, Panel shows the dominant induced responses in participants with a previous PCR+ infection following bivalent BA.1 and bivalent BA.4/5 vaccination. C, Panel shows pie charts with the proportion of participants with dominant serological responses before and after the bivalent fourth vaccine dose. Abbreviation: PCR, polymerase chain reaction.
Lastly, to enumerate the quantitative variant-specific responses after 4 mRNA vaccine doses, with and without the impact of previous infection, we ranked the serological response to the 5 Omicron variants of a given individual. In those with a previous PCR+ infection, 48.9% had a dominant response targeting the BA.2 antigen, whereas only 2.6% had a dominant response to the BA.1 antigen, clearly demonstrating the impact of previous infection. Following bivalent vaccination, the proportion with dominant responses to BA.1 remained at 3.9% in those receiving the BA.4/5 vaccine but increased to 11.2% in those receiving the BA.1 vaccine (Figure 4C top). In participants with no prior infection, 11.1% had a dominant response to BA.1 before the fourth dose. This proportion was significantly expanded so that 30.7% had a dominant BA.1 focused response after bivalent BA.1 vaccination (Figure 4C bottom).
DISCUSSION
In this study, we evaluated Omicron variant-specific antibody responses in 1697 individuals receiving either a BA.1 or BA.4/5 bivalent booster after having received 3 prior monovalent vaccine doses. Prior to the fourth bivalent booster dose, we identified increased levels of antibodies in individuals with a previous SARS-CoV-2 infection. Antibody levels to all Omicron variants were further significantly boosted by receiving either of the bivalent vaccines. However, the magnitude of the increase was greater for individuals with no prior infection. The BA.1 vaccine was found to dominate serological imprinting toward the BA.1 and BA.3 antigens, whereas the BA.4/5 vaccine enabled broad Omicron antigen imprinting.
In this cohort, we found high antibody levels more than 300 days after receiving a third vaccination with a monovalent mRNA vaccine, supporting the observation of slower antibody decline after the third dose (booster) than after the primary vaccination series (2 doses) [18, 19]. Furthermore, we observed significantly higher antibody levels in individuals with a previous PCR+ infection. In Denmark, a large spread of Omicron infections, predominantly fueled by the BA.2 variant, occurred in the period between the roll-out of the third and the fourth vaccine [3]. Consistent with this, the largest fold difference between those with and without previous infection was observed for the BA.2-directed antibody response, clearly indicating a strong serological imprint toward the dominant infecting variant, which is in line with previous observations [20]. Antibody responses following bivalent vaccination were also impacted by prior infection, as we observed higher inductions of BA.2, BA.4, and BA.5-specific antibodies in individuals with a previous PCR+ infection that received a BA.4/5 bivalent vaccine. The BA.2, BA.4, and BA.5 variants are antigenically more related than pre-Omicron variants and Omicron variants BA.1 and BA.3 [21]. The observation of a link between these variants is in line with other studies that have described a protective impact from previous BA.2 infection against BA.5 infection in a vaccinated population, suggesting enhancement of antigenic breadth and hence cross-neutralization [3, 7]. Nevertheless, both the BA.1 and BA.4/5 vaccine induced high levels of Omicron variant-specific antibodies, irrespective of prior infection status, supporting the use of both bivalent vaccines. This is consistent with the report of highly protective vaccine efficacy for both the bivalent BA.1 (74%) and BA.4/5 (80%) booster as a fourth dose compared to three vaccine doses/no fourth booster [22].
Interestingly, we set out to explore dominant responses to the Omicron-specific antigens following bivalent BA.1 and BA.4/5 vaccination. Irrespective of prior infection status, we observed the strongest directed response to the BA.1 antigen in recipients of the bivalent BA.1 vaccine (37.2% no infection vs 39.9% previous PCR+). These similar numbers suggest that despite immune memory of previous Omicron BA.2 infection, antibodies can be induced to preferentially target the new vaccine antigen. Whether these antibodies originate from de novo induced B cells or from further affinity maturation of existing antibodies cannot be deciphered in this data set. However, a recent study from mice found that in heterologous omicron boosted animals 25%–50% of the antibodies came from newly induced B cells [11]. In addition, a study in humans described strong antigenic imprinting by pre-existing memory B cells, but immunization with an antigen that was antigenically distant from the original strain induced new B cells [23]. Thus, despite repeated SARS-CoV-2 exposures to evolving antigens the antibody responses continuously evolve or induce a new functional direction.
Given the continuous viral evolution and rapid emergence of new viral variants of SARS-CoV-2, cross-reactive immune responses and broad antigenic coverage is highly desirable. Lately, newly emerged Omicron sub-lineages, including BA.2.75, XXB, and BQ.1, have once more called into question whether the antibody responses obtained by the bivalent vaccines demonstrate sufficient protection against these variants. As new viral variants emerge, it is therefore essential to continue monitoring the cross-neutralization potential of the bivalent vaccines.
As a limitation in this study, we investigated the 5 initial circulating Omicron variants (BA.1 to BA.5), and hence we have no data to conclude on any serological targeting of newer variants. Another limitation is the unavailability of viral sequencing data to confirm the variant that caused the SARS-CoV-2 infection. Alternatively, infections were classified into viral variants by the predominantly circulating variant at the time of the positive PCR test. Furthermore, despite adjusting for the demographic variables in our regression model, the roll-out of the bivalent vaccines contained some skewing as individuals at increased risk of severe COVID-19 disease were offered the bivalent fourth dose while the vaccine supply was dominated by the BA.1 bivalent vaccine.
In conclusion, prior antigenic exposure by vaccination or previous infection leaves a clear serological imprint that is focused on the variant-specific antigen. Bivalent BA.1 and BA.4/5 vaccination induces some immune imprinting as presented by Omicron variant-directed antibody inductions to the vaccine target. This study supports the roll-out of both bivalent vaccines as they appear to provide broad cross-protection of viral Omicron variants, essential for optimal population immunity.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Eva A M Baerends, Department of Infectious Diseases, Aarhus University Hospital, Aarhus, Denmark; Department of Clinical Medicine, Aarhus University, Aarhus, Denmark.
Joanne Reekie, Center of Excellence for Health, Immunity and Infections, Rigshospitalet, University of Copenhagen, Copenhagen, Denmark.
Signe R Andreasen, Department of Infectious Diseases, Aarhus University Hospital, Aarhus, Denmark; Department of Clinical Medicine, Aarhus University, Aarhus, Denmark.
Nina B Stærke, Department of Infectious Diseases, Aarhus University Hospital, Aarhus, Denmark; Department of Clinical Medicine, Aarhus University, Aarhus, Denmark.
Dorthe Raben, Center of Excellence for Health, Immunity and Infections, Rigshospitalet, University of Copenhagen, Copenhagen, Denmark.
Henrik Nielsen, Department of Infectious Diseases, Aalborg University Hospital, Aalborg, Denmark; Department of Clinical Medicine, Aalborg University, Aalborg, Denmark.
Kristine T Petersen, Department of Infectious Diseases, Aalborg University Hospital, Aalborg, Denmark.
Isik S Johansen, Department of Infectious Diseases, Odense University Hospital, Odense, Denmark; Department of Clinical Research, University of Southern Denmark, Odense, Denmark.
Susan O Lindvig, Department of Infectious Diseases, Odense University Hospital, Odense, Denmark; Department of Clinical Research, University of Southern Denmark, Odense, Denmark.
Lone W Madsen, Department of Infectious Diseases, Odense University Hospital, Odense, Denmark; Department of Clinical Research, University of Southern Denmark, Odense, Denmark.
Lothar Wiese, Department of Medicine, Zealand University Hospital, Roskilde, Denmark.
Mette B Iversen, Department of Medicine, Zealand University Hospital, Roskilde, Denmark.
Thomas Benfield, Department of Infectious Diseases, Copenhagen University Hospital—Amager and Hvidovre, Hvidovre, Denmark; Departments of Clinical Medicine and Public Health, University of Copenhagen, Copenhagen, Denmark.
Kasper K Iversen, Departments of Clinical Medicine and Public Health, University of Copenhagen, Copenhagen, Denmark; Department of Cardiology and Emergency Medicine, Herlev Hospital, Herlev, Denmark.
Fredrikke D Larsen, Department of Infectious Diseases, Aarhus University Hospital, Aarhus, Denmark; Department of Clinical Medicine, Aarhus University, Aarhus, Denmark.
Sidsel D Andersen, Department of Infectious Diseases, Aarhus University Hospital, Aarhus, Denmark; Department of Clinical Medicine, Aarhus University, Aarhus, Denmark.
Anna K Juhl, Department of Infectious Diseases, Aarhus University Hospital, Aarhus, Denmark; Department of Clinical Medicine, Aarhus University, Aarhus, Denmark.
Lisa L Dietz, Department of Infectious Diseases, Aarhus University Hospital, Aarhus, Denmark; Department of Clinical Medicine, Aarhus University, Aarhus, Denmark.
Astrid K Hvidt, Department of Infectious Diseases, Aarhus University Hospital, Aarhus, Denmark; Department of Clinical Medicine, Aarhus University, Aarhus, Denmark.
Sisse R Ostrowski, Department of Clinical Immunology, Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department of Clinical Medicine, University of Copenhagen, Copenhagen, Denmark.
Tyra G Krause, Department of Infectious Disease Epidemiology and Prevention, Statens Serum Institut, Copenhagen, Denmark.
Lars Østergaard, Department of Infectious Diseases, Aarhus University Hospital, Aarhus, Denmark; Department of Clinical Medicine, Aarhus University, Aarhus, Denmark.
Ole S Søgaard, Department of Infectious Diseases, Aarhus University Hospital, Aarhus, Denmark; Department of Clinical Medicine, Aarhus University, Aarhus, Denmark.
Jens Lundgren, Center of Excellence for Health, Immunity and Infections, Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Departments of Clinical Medicine and Public Health, University of Copenhagen, Copenhagen, Denmark; Department of Infectious Diseases, Copenhagen University Hospital—Rigshospitalet, Copenhagen, Denmark.
Martin Tolstrup, Department of Infectious Diseases, Aarhus University Hospital, Aarhus, Denmark; Department of Clinical Medicine, Aarhus University, Aarhus, Denmark.
Notes
Author contributions. J. L., L. Ø., O. S. S., N. B. S., J. R., D. R., T. G. K., and M. T. conceptualized the work. H. N., K. T. P., I. S. J., S. O. L., L. W. M., L. W., M. B. I., T. B., K. K. I., F. D. L., and S. R. O. did the clinical visits and collected the samples. E. B., S. R. A., S. D. A., A. K. J., L. L. D., and A. K. H. performed the laboratory analyses. E. B., J. R., and M. T. performed the data analysis and visualization. M. T. supervised and led the study. E. B., J. R., J. L., and M. T. drafted the manuscript and have accessed and verified the underlying data. All authors read and approved the final draft. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Acknowledgments. The ENFORCE study group members all contributed substantially to the study. A full list of members of the ENFORCE study group is provided as Supplementary Material and may be found at https://chip.dk/Research/Studies/ENFORCE/Study-Group.
Ethical approval. The study involved human participants and the protocol was approved by the Danish Medicines Agency (no. 2020-006003-42) and the National Committee on Health Research Ethics (no. 1-10-72-337-20). All participants provided written informed consent.
Data availability. Data from the ENFORCE cohort may be made available to researchers upon approval of an application to the ENFORCE scientific steering committee and further approval by relevant authorities. Applications for data must be sent to enforce.rigshospitalet@regionh.dk. If approval is granted, data will be provided as deidentified data. The ENFORCE protocol is available at https://www.enforce.dk/ and more detailed information about data access may be found at https://chip.dk/Research/Studies/ENFORCE/Study-Governance.
Financial support . The ENFORCE study is supported by a grant from the Danish Ministry of Health (SUM).
References
- 1. Dejnirattisai W, Huo J, Zhou D, et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 2022; 185:467–484.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu L, Iketani S, Guo Y, et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 2022; 602:676–81. [DOI] [PubMed] [Google Scholar]
- 3. Hansen CH, Friis NU, Bager P, et al. Risk of reinfection, vaccine protection, and severity of infection with the BA.5 Omicron subvariant: a nation-wide population-based study in Denmark. Lancet Infect Dis 2022; 23:167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Erikstrup C, Laksafoss AD, Gladov J, et al. Seroprevalence and infection fatality rate of the SARS-CoV-2 Omicron variant in Denmark: a nationwide serosurveillance study. Lancet Reg Health Eur 2022; 21:100479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Q, Iketani S, Li Z, et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 2023. 186:279–286.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hoffmann M, Behrens GMN, Arora P, et al. Effect of hybrid immunity and bivalent booster vaccination on omicron sublineage neutralisation. Lancet Infect Dis 2023; 23:25–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Muik A, Lui BG, Bacher M, et al. Exposure to BA.4/5 S protein drives neutralization of Omicron BA.1, BA.2, BA.2.12.1, and BA.4/5 in vaccine-experienced humans and mice. Sci Immunol 2022; 7:eade9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chalkias S, Harper C, Vrbicky K, et al. A bivalent Omicron-containing booster vaccine against COVID-19. N Engl J Med 2022; 387:1279–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hause AM, Marquez P, Zhang B, et al. Safety monitoring of bivalent COVID-19 mRNA vaccine booster doses among persons aged >/=12 years—United States, August 31–October 23, 2022. MMWR Morb Mortal Wkly Rep 2022; 71:1401–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chalkias S, Eder F, Essink B, et al. Safety, immunogenicity and antibody persistence of a bivalent Beta-containing booster vaccine against COVID-19: a phase 2/3 trial. Nat Med 2022; 28:2388–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schiepers A, van ’t Wout MFL, Greaney AJ, et al. Molecular fate-mapping of serum antibody responses to repeat immunization. Nature 2023; 615:482–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reynolds CJ, Gibbons JM, Pade C, et al. Heterologous infection and vaccination shapes immunity against SARS-CoV-2 variants. Science 2022; 375:183–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Staerke NB, Reekie J, Johansen IS, et al. Cohort profile: the Danish national cohort study of effectiveness and safety of SARS-CoV-2 vaccines (ENFORCE). BMJ Open 2022; 12:e069065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nielsen SS, Nielsen SSF, Vibholm LK, et al. SARS-CoV-2 elicits robust adaptive immune responses regardless of disease severity. EBioMedicine 2021; 68:103410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hvidt AK, Baerends EAM, Søgaard OS, et al. Comparison of vaccine-induced antibody neutralization against SARS-CoV-2 variants of concern following primary and booster doses of COVID-19 vaccines. Front Med (Lausanne) 2022; 9:994160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Quan H, Li B, Couris CM, et al. Updating and validating the Charlson comorbidity index and score for risk adjustment in hospital discharge abstracts using data from 6 countries. Am J Epidemiol 2011; 173:676–82. [DOI] [PubMed] [Google Scholar]
- 17. Sogaard OS, Reekie J, Johansen IS, et al. Characteristics associated with serological COVID-19 vaccine response and durability in an older population with significant comorbidity: the Danish Nationwide ENFORCE study. Clin Microbiol Infect 2022; 28:1126–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gilboa M, Regev-Yochay G, Mandelboim M, et al. Durability of immune response after COVID-19 booster vaccination and association with COVID-19 Omicron infection. JAMA Netw Open 2022; 5:e2231778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lau CS, Oh MLH, Phua SK, Liang YL, Aw TC. 210-day Kinetics of total, IgG, and neutralizing spike antibodies across a course of 3 doses of BNT162b2 mRNA vaccine. Vaccines (Basel) 2022; 10:1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reynolds CJ, Pade C, Gibbons JM, et al. Immune boosting by B.1.1.529 (Omicron) depends on previous SARS-CoV-2 exposure. Science 2022; 377:eabq1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rossler A, Netzl A, Knabl L, et al. BA.2 and BA.5 omicron differ immunologically from both BA.1 Omicron and pre-Omicron variants. Nat Commun 2022; 13:7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Andersson NW, Thiesson EM, Baum U, et al. Comparative effectiveness of the bivalent BA.4-5 and BA.1 mRNA-booster vaccines in the Nordic countries. medRxiv [Preprint]. January 19, 2023. Available from: 10.1101/2023.01.19.23284764. [DOI]
- 23. Alsoussi WB, Malladi SK, Zhou JQ, et al. , SARS-CoV-2 Omicron boosting induces de novo B cell response in humans. Nature 2023; 617:592–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.