Abstract
The robustness of NMR coherence transfer in proximity of a paramagnetic center depends on the relaxation properties of the nuclei involved. In the case of Iron-Sulfur Proteins, different pulse schemes or different parameter sets often provide complementary results. Tailored versions of HCACO and CACO experiments significantly increase the number of observed Cα/C’ connectivities in highly paramagnetic systems, by recovering many resonances that were lost due to paramagnetic relaxation. Optimized 13C direct detected experiments can significantly extend the available assignments, improving the overall knowledge of these systems. The different relaxation properties of Cα and C’ nuclei are exploited in CACO vs COCA experiments and the complementarity of the two experiments is used to obtain structural information. The two [Fe2S2]+ clusters containing NEET protein CISD3 and the one [Fe4S4]2+ cluster containing HiPIP protein PioC have been taken as model systems. We show that tailored experiments contribute to decrease the blind sphere around the cluster, to extend resonance assignment of cluster bound cysteine residues and to retrieve details on the topology of the iron-bound ligand residues.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10858-023-00425-4.
Keywords: Paramagnetic NMR, 13C NMR, Iron-Sulfur proteins, Optimized 13C experiments, Transverse relaxation, NEET proteins
Introduction
The direct detection of 13C NMR is routinely used in biomolecular NMR to complement 1H detected experiments and to obtain information that cannot be obtained via 1H detection (Bermel et al. 2006a; Felli and Pierattelli 2022). Usually, the gain in sensitivity is given by the polarization transfer from 1H to 13C spins, which largely overcomes the signal loss due to the 1H transverse relaxation during the INEPT transfer and, in many applications, provides better results than 13C-start experiments (Shimba et al. 2004; Bermel et al. 2009; Richter et al. 2010; Pontoriero et al. 2020; Vögeli et al. 2005; Pritchard and Hansen 2019). However, in paramagnetic metalloproteins, nuclear relaxation rates depend on the square of the gyromagnetic ratio (γ2) of the nucleus investigated, on the r−6 dependence from metal-to-nucleus distance and, for the first coordination sphere, on the amount of unpaired electron spin density delocalized from the metal ion(s) to the nuclear spins (Miao et al. 2022; Pell et al. 2019; Trindade et al. 2022; Bertini et al. 2017). Therefore, the choice of the most efficient experiment is not trivial: complementary results are often obtained when the same experiment is recorded with different pulse schemes (Arnesano et al. 2003; Gelis et al. 2003; Invernici et al. 2020; Ciofi-Baffoni et al. 2014) and “non-systematic” assignment strategies, tailored according to the relaxation properties of specific spin systems, need to be defined (Trindade et al. 2021a).
In protein NMR, backbone Cα-C’ connectivities are very useful for sequence-specific and site-specific assignments, because of the correlation between chemical shift values and amino acid type (Mao et al. 2014). Many experiments have been designed to identify homonuclear Cα-C’ connectivities (Bertini et al. 2005a; Lee et al. 2005; Machonkin et al. 2002; Kostic et al. 2002; Bax 2011), which can also be used to address macromolecular dynamics and interactomics (Stenstrom et al. 2022; Mori et al. 2010; Ferrage et al. 2006). In paramagnetic systems, relaxation losses easily make a 13C-start experiment comparable and eventually more sensitive than a corresponding 1H-start experiment (Pontoriero et al. 2020; Balayssac et al. 2006). The intrinsic asymmetry of CACO experiments can also be exploited: CACO is based on a single coherence transfer step from the excited nucleus to the observed nucleus, therefore transferring magnetization from Cα to C’ or vice versa, in the COCA experiment, would call into the scene different relaxation rates. As a consequence, complementary information are obtained from Cα to C’ and C’ to Cα transfer pathways (Machonkin et al. 2002; Bertini et al. 2005b), contributing to characterize the first coordination sphere of a metal center. In the absence of magnetic anisotropies of the metal ions, when no structural information can be obtained from chemical shifts (Pintacuda et al. 2007; Zhu et al. 2023; Wu et al. 2022; Herath et al. 2021; Muntener et al. 2022; Parker et al. 2020), the proximity of the metal center can be monitored only by mapping the paramagnetic relaxation of nuclear spins nearby and each individual spin system has a different behaviour, depending on its topology vis-à-vis the metal center (Trindade et al. 2021a).
Within this frame, we considered the behaviour of two different paramagnetic metalloproteins: CISD3 and PioC, both containing iron-sulfur clusters. At variance with other metalloproteins, in which the electron relaxation is driven by the coordination number of the metal ion (Spronk et al. 2018; Ravera et al. 2021; Beniamino et al. 2022; Trindade et al. 2021b), the electronic correlation times in iron-sulfur proteins are determined by the magnetic coupling among the iron ions. Therefore, different FeS clusters affect their protein environments at different extent (Blondin and Girerd 1990; Banci et al. 2018, 1990; close to the molecular mechanism understanding 2018; Bennett et al. 2022; Azam et al. 2020; Gervason et al. 2019; Stegmaier et al. 2019; Mulliez et al. 1999; Trindade et al. 2023). CISD3 (CDGSH Iron Sulfur Domain-3) is a 93 aminoacids protein containing two [Fe2S2]+ clusters (Karmi et al. 2018; Tamir et al. 2015). Each cluster is coordinated by a 3-Cys, 1-His biding motif, embedded into a highly conserved domain, called CDGSH domain. The protein is a monomer and the two CDGSH domains are similar but not equivalent. Both clusters are in the reduced [Fe2S2]+ form, formally containing one Fe3+ and one Fe2+ ion (Golinelli-Cohen et al. 2016; Gee et al. 2021; Camponeschi et al. 2022). The crystallographic structure of the protein is available only for the H75C/H113C mutant (Lipper et al. 2018). We used CISD3 because an extended assignment of the protein has been recently obtained from a combination of standard and tailored 1H and 13C detected experiments (Silva et al. 2023). The system is particularly challenging because [Fe2S2]+ clusters are efficient relaxing agents (Camponeschi et al. 2021; Cai et al. 2017; Pochapsky et al. 2001; Valer et al. 2022) and paramagnetism affects about 60% of residues in CISD3 (Grifagni et al. 2023), preventing the identification of the NMR signals of the cluster-bound residues and the resolution of the NMR solution structure. PioC, a High Potential Iron Protein (HiPIP) from Rhodopseudomonas palustris TIE-1 (Bird et al. 2014), is a very small protein (54 amino acids) that contains a single [Fe4S4]2+ cluster. The electron relaxation times of iron ions in [Fe4S4]2+ clusters are shorter than the [Fe2S2]+ case, for this reason the detection of Cα/C’ connectivities of iron-bound cysteine residues can be attempted and a high-quality NMR structure has been obtained (Trindade et al. 2021c) (PDB ID: 6XYV).
Together, these two iron-sulfur proteins will assess the performances of CACO experiments in systems characterized by different relaxation properties. On the one hand, we will show how the optimization and the combination of 1H and 13C start experiments contribute to improve the resonance assignment in highly paramagnetic systems. On the other hand, when signal losses due to paramagnetism are less severe and a complete resonance assignment can be obtained, the structural dependence of paramagnetic relaxation enhancements can be exploited in many intriguing ways.
Results and discussion
Tailored HCACO experiments contribute to resonance assignment: the case of CISD3
CISD3 is a small protein that contains two [Fe2S2]+ clusters. Triple resonance NMR experiments recorded using routine parameter sets, permit the identification of only ca 40% of the protein residues, consistent with a detectability threshold of about 9 Å from each iron ion of the cluster (Camponeschi et al. 2021). Our interest here is to discuss the complementarity of various CACO experiments (Bertini et al. 2005a; Bermel et al. 2003, 2005a). In particular, the comparison of 1H vs 13C start CACO experiments promise to reveal unique information on the metal to nucleus distances of the spin systems that are affected by the paramagnetic center.
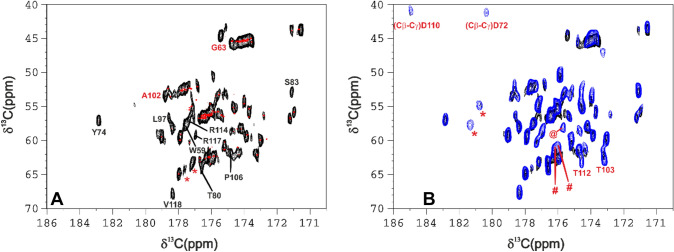
First, we compared two HCACO experiments recorded with the same pulse sequence (Fig. 1) but using two different parameter sets. One experiment was recorded with a conventional parameter set, optimized for a small/medium sized protein, the other was recorded by optimizing all delays for the detection of fast relaxing signals and for minimizing signal losses. While 43 signals are observed in the “routine” version of the HCACO and then sequence specifically assigned, thirteen additional signals appeared in the “paramagnetic” spectrum. Out of them, nine signals have been sequence specifically assigned (black-labelled peaks in Fig. 2A) using a combination of standard and tailored triple resonance NMR experiments (Grifagni et al. 2023). These residues are at Hα-Fe distances in the range 7–8.7 Å (except for Thr 80 and Ser 83, both in the linker region between the two CDGSH domains, where some conformational rearrangements in solution might take place), indicating that the blind sphere of the optimized HCACO experiment has decreased. The four peaks identified but not yet assigned in the tailored HCACO are also expected to be within the same Hα-Fe distance range, this significantly restricts the number of possible candidates for their assignment. Moreover, all nuclear spins within this distance range from the paramagnetic center should experience paramagnetic relaxation but no paramagnetic shift (Banci et al. 2018; Camponeschi et al. 2019), therefore chemical shift values can also be used for signal assignment. In the tailored CACO spectrum of Fig. 2A, the signal at 175.4/44.5 ppm can be safely attributed to a glycine residue. There are three unassigned glycine residues in CISD3, all located in proximity to an iron ion. Only Gly 63 has an Hα more than 6 Å apart from the closest iron ion. This is the only distance compatible with the observed HCACO peak, and therefore we assign this signal to Gly 63. Likewise, the peak at 178.7/53.5 ppm is assigned because these chemical shifts are characteristic of alanine residues and the only alanine residue not yet assigned is Ala 102, which has the Hα at 6.1 Å from the iron ion. We are then left with two signals at relatively downfield Cα values (labelled with red asterisks in Fig. 2A), whose Cα/C’ shifts might be consistent with valine, phenilalanine or leucine. The only candidates for the assignment are Val 61, Phe 76 and Leu 82, which have Hα-Fe distances of 5.7 Å, 7.0 Å and 7.9 Å, respectively. Among them, we might speculate that Val 61 is probably too close to the iron ion to be observable and, therefore, Phe 76 and Leu 82 are the most likely candidates for the two unassigned peaks of Fig. 2A.
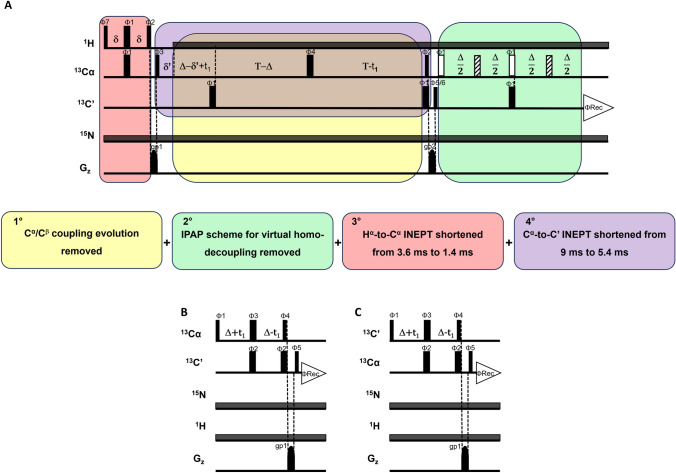
Fig. 1.
A Standard HCACO experiment. White and dashed 180° pulses indicate IPAP scheme for virtual homo-decoupling of Cα/C coherence. The colored panels highlight parts of the pulse scheme that have been modified during the optimization process. The delays utilized in the routine HCACO experiment are the following δ = 1.8 ms, δ’ = 1.1 ms, Δ = 4.5 ms, Τ = 14.2 ms. The first modification, highlighted in yellow, eliminates the Cα/Cβ coupling evolution period. T is set equal to Δ = 4.5 ms, the two 180° Cα and C’ pulses are applied together. The next modification, highlighted in green, removes the IPAP scheme for virtual homo-decoupling. The entire block is removed and acquisition begins immediately after the last 90 °C’ pulse. In the subsequent step, highlighted in red, the Hα-to-Cα INEPT delay δ was shortened from 1.8 ms to 0.7 ms. The last modification, highlighted in purple, shortens the Cα-to-C’ INEPT delay ∆ from 4.5 ms to 2.7 ms. The phase cycle, maintained throughout the modifcations, is: Φ1 = x; Φ2 = y; Φ3 = x, − x; Φ4 = x, x, x, x, x, x, x, x, y, y, y, y, y, y, y, y; Φ5 = x, x, x, x, − x, − x, − x, − x; Φ6 = − y, − y, − y, − y, y, y, y, y; Φ7 = x, x, − x, − x; ΦRec = x, − x, − x, x, − x, x, x, − x, − x, x, x, − x, x, − x, − x, x. B The CACO-AP and C COCA-AP pulse schemes implemented with a Cα-to-C' INEPT delay equal to Δ = 2.7 ms. The phase cycle for the two experiments is the following: Φ1 = x; Φ2 = y; Φ3 = x, − x; Φ4 = x, x, x, x, y, y, y,y; Φ5 = x, x, − x, − x; ΦRec = x, − x, − x, x, − x, x, x, − x. Gradient strenght was gp1 = 30%, gp2 = 50%. PFG gradients used in all the pulse sequences had a sine bell shape and duration of 1 ms. The period set for the dissipation of circulating currents was 200 μs
Fig. 2.
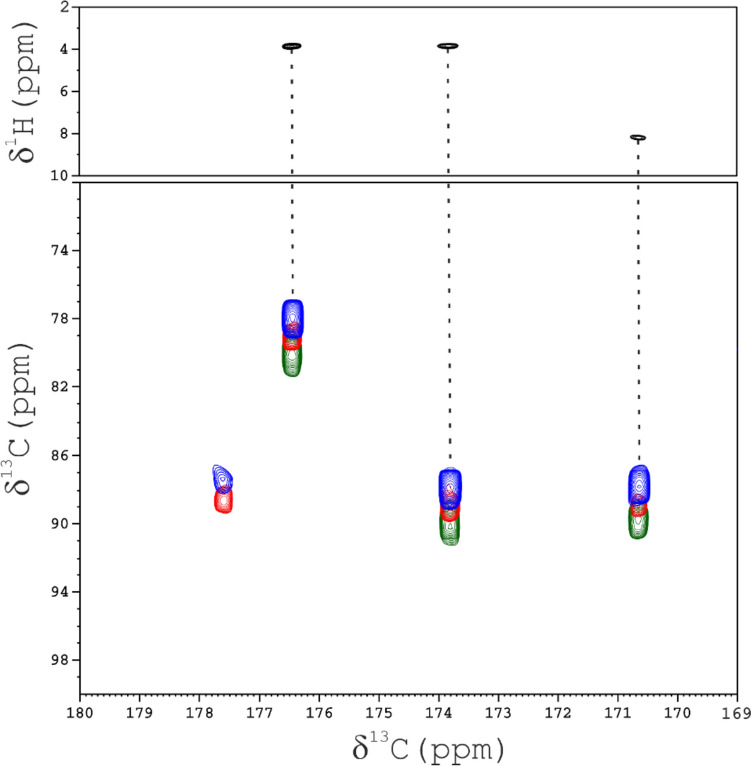
175 MHz, 298 K NMR spectra of reduced CISD3. A HCACO spectra obtained using the sequence reported in Fig. 1A. The spectrum shown in red (standard dataset) has been obtained with the following parameters: acquisition (t2max) and recycle delays 92 ms and 1 s; δ = 1.8 ms, δ’ = 1.1 ms, ∆ = 4.5 ms, T = 14.2 ms. 32 scans each FID were recorded over a 1024 × 660 data point matrix. Total experimental time was 6.5 h. The spectrum shown in black (paramagnetic tailored dataset) has been obtained with acquisition (t2max) and recycle delays 46 ms and 400 ms; δ = 1.4 ms, δ’ = 1.1 ms, T = ∆ = 2.7 ms. 1024 scans each FID were recorded over a 512 × 50 data point matrix. Total experimental time was 6.5 h. Assignment reported in black shows residues that could be sequence specifically assigned with a set of paramagnetic tailored experiments60, labels in red indicate peaks that were identified, and some of them assigned, in this experiment. B HCACO (black) vs CACO-AP (blue). The spectrum shown in black is the same reported in panel A, the spectrum shown in blue has been obtained with the pulse sequence reported in Fig. 1B, with acquisition (t2max) and recycle delays 24.5 ms and 300 ms. The Cα-to-C’ INEPT period was set to ∆ = 2.7 ms. 1024 scans each FID were recorded over a 512 × 70 data point matrix. Total experimental time was 6.5 h. Labels show peaks that were observed only in the CACO-AP and discussed throughout the text
1H start vs 13C start experiments
To compare the features of 13C- vs 1H-excitation, we performed a CACO-AP experiment, in which both the initial Hα-to-Cα INEPT and the final Cα-to-C’ IPAP are removed, and signal detected as a C’ antiphase doublet, phased in dispersion mode (Bertini et al. 2005a). The pulse sequence for this experiment is reported in Fig. 1B. As expected, for signals that are not affected by the paramagnetic center, the sensitivity of the CACO-AP experiment is lower than the HCACO. Nevertheless, Fig. 2B shows nine additional signals in the CACO-AP, compared to the 1Hα-start HCACO. These resonances have not been observed in other 1H start experiments (with a few exceptions discussed here below), because their scalar coupled 1H nuclei are too close to the metal to be observed, with estimated metal-to-proton distances shorter than 7.0 Å. Protonless experiments, in which fast relaxing protons are never involved into the coherence transfer pathway (Balayssac et al. 2006), provide the best coherence pathways to circumvent signal losses due to paramagnetic relaxation enhancement. As outlined previously in the case of tailored HCACO, signals identification within a given experiment is associated to a well-defined distance range between electron and nuclear spins. When a structure is available, the predicted distance range and the chemical shifts information allow us to propose some assignments. Among signals exclusively observed in the CACO-AP experiment (Fig. 2B), two of them fall in the peculiar CACO region of threonine residues. There are three unassigned threonine residues in CISD3: Thr 99, Thr 103 and Thr 112, all belonging to the C-term CDGSH domain and close to the C-term cluster (cluster 2, hereafter). The signal at 173.1/62.9 ppm is completely missing in the tailored HCACO, however in the CON experiment (data not shown), there is a non-assigned correlation between a C’ at 173.1 ppm and a, still unassigned, N backbone atoms at 116.6 ppm. This pattern suggests that this CACO peak belongs to a threonine residue with an Hα very close to the metal center but with the C’ nucleus relatively far from the iron ion, enough to allow the detection of its CON peak. The nucleus-to-iron distances of all unassigned amino acids, which are reported in Table S1, are well consistent with the assignment to Thr 103, connected with Gln 104, whose HN group has not been assigned. The other threonine peak, observed in the CACO-AP at 174.5/63.3 ppm, is barely detectable also in the HCACO experiment but it does not have any possible connectivity in the CON spectrum, indicating a different orientation of this spin system versus the cluster, compared with Thr 103. We expect for this residue larger Fe-Hα and shorter Fe–C’ distances than Thr 103. According to Table S1, this is the case of Thr 112. By exclusion, Thr 99 would remain unobserved, because Thr 99 has shorter metal-to-carbon distances than the other two threonine residues. The two signals at 184.9/41.1 and 180.2/41.3 ppm can be attributed to Cβ-Cγ connectivities of aspartate residues. According to the crystallographic structure of the H75C/H113C double mutant, both carboxylate groups of Asp 72 and Asp 110, which are the only two aspartate residues of the protein, are at about 6 Å from the iron and, therefore, they might be observable in a CACO-AP experiment. Their residue specific assignment is much more tentative: both carboxylate groups are expected to form H-bonds with Ser 74 and Thr 112, respectively. At variance with Asp 72, the side chain of Asp 110 is also very close to the guanidinium moiety of Arg 105, which might explain the unusual downfield shift of aspartate Cγ signals and suggest the assignment proposed in Fig. 2B. There are five additional signals observed in the CACO-AP experiment that, following previous considerations, belong to residues with Cα/C’ distances in the range 5.0/6.5 Å from the metal center, i.e. the distance range experienced by the herewith assigned threonine backbone Cα/C’ and aspartate side chains Cβ/Cγ signals. Possible candidates, according to Table S1, are: Pro 46, Arg 64, Asp 72, Ser 74, Phe 76, Pro 84, Lys 101, Asp 110, which are in the proper distance range and are still unassigned. Among possible candidates we do not consider the iron-bound cysteine and histidine residues, because their Cα should experience considerable hyperfine shift (Querci et al. 2023). The two signals at 176.2/61.5 ppm and 175.9/60.9 ppm, labelled with #, are in the Pro-Phe region and possible candidates are Pro 46, Pro 84 and Phe 76. On the other hand, the signal at 175.7/57.9 ppm, labelled with @, is within the Arg-Lys region and, therefore, candidates are Arg 64 and Lys 101. There is no possible assignment based on chemical shifts predictions for the two peaks at 181.2/57.7 ppm and 180.7/55.0 ppm, labelled with *. Unusual C’ shift values may arise from the network of H-bonds around the two clusters: it has been shown that H-bonds are important to modulate the reduction potential of individual iron ions (Langen et al. 1992) and contribute to the delocalization of unpaired spin density from the iron ions to the backbone nitrogen atoms involved in H-bonds (Lin et al. 2009; Xia et al. 1999; Trindade et al. 2020). Unpaired electron delocalization might be partly transferred also to the C’ atom of the preceding residue. Ser 74, Asp 110 and also the two previously mentioned Arg 64 and Lys 101 might experience this effect. Even if these peaks are unassigned, they can be utilized to map intermolecular interactions. Indeed, they clearly arise from the immediate proximity to the cluster and are probably the NMR observables closest to the cluster that can be acquired.
Optimization of the HCACO experiment: complete resonance assignment of iron bound residues in HiPIP protein PioC
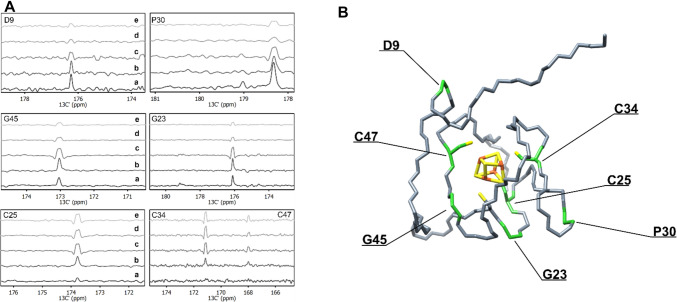
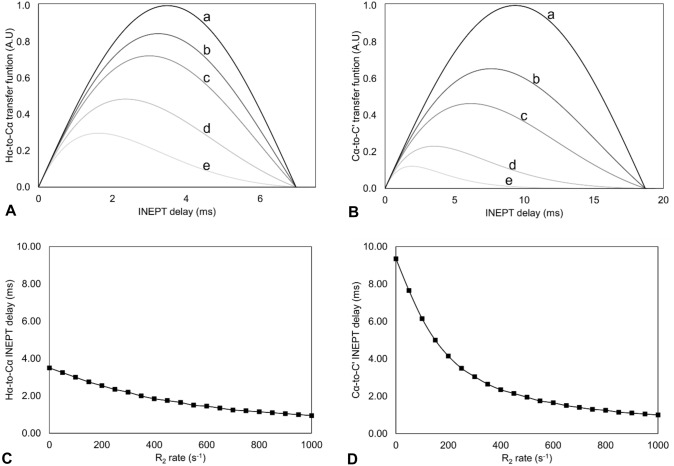
We considered as a test system for the optimization of the HCACO experiment, the HiPIP protein PioC, containing a [Fe4S4]2+ cluster. At variance with CISD3, PioC is a very stable protein, its NMR assignment is almost complete and signals from residues that bind the metal center can be observed. PioC is therefore a paradigmatic case for the quantitative assessment of HCACO performances. A series of HCACO experiments, recorded with the same acquisition parameters (same number of scans, t1 increments, acquisition and recycle delays) was recorded at 1.2 GHz, and the peak intensities are reported for some selected peaks in Fig. 3A. The lower panels show the Cα/C’ connectivities of Fe-bound cysteines, that are the residues most affected by paramagnetic relaxation; the middle panels report peak intensities of residues that are close to the paramagnetic center but do not belong to the first coordination sphere; the upper panels show peaks unaffected by paramagnetic relaxation. Figure 3B shows the location of these residues within the protein frame. In the routine experiment (traces a), Cys residues are missing or only barely visible, the two Gly residues are weak and signals far from the paramagnetic center are recorded with good sensitivity. The stepwise implementation of the pulse sequence is schematically shown in Fig. 1A and it allows one to analyse the contribution of each parameter to the overall effects. The removal, in the F1 dimension, of the constant time needed to refocus the homonuclear Cα/Cβ coupling (traces b), significantly contributes to revive signals affected by paramagnetic relaxation at the expenses of the sensitivity of signals not affected by paramagnetism. Similar effects are observed when the final IPAP scheme is replaced and signals detected as C’ antiphase dispersion doublets (traces c). These two modifications are essential for the detectability of cysteine signals, but they also improve the intensities of the residues affected by paramagnetic relaxation at lower extent, like the two glycine residues Gly 23 and Gly 45 (Fig. 3A middle panels): whenever a significant paramagnetic contribution to relaxation occurs, these two implementations do contribute to increase the number of observable peaks. Further improvements can be obtained by shortening the delay of the Hα-to-Cα INEPT (d) and of the Cα-to-C’ INEPT (e). The sensitivity gain in these cases is strictly dependent on the behavior of the relaxation weighted coherence transfer functions, which are reported in Fig. 4. In Fig. 3A (lower panels), we observe that, when Hα-to-Cα and Cα-to-C’ INEPT delays are decreased from their “standard” values of 3.6 ms and 9.0 ms to, respectively, 1.4 ms and 5.4 ms, signals of Fe bound Cys 25, Cys 34 and Cys 47 increase in intensity, although at different extent. On the other hand, signals of Gly 45 and Gly 23 decrease the intensity. The impact of relaxation rates on the coherence transfer affects both the efficiency of the transfer and the choice of the optimal delay. As shown in Fig. 4C and D, optimal transfer delays decrease at increasing R2 values and, as a consequence, at decreasing metal-to-nucleus distances. In the case of the experiments reported in Fig. 3A, delays were calibrated according to very fast nuclear relaxation, with the aim to identify signals close to the detection limit. Indeed, we observe an increase in signal intensity for Cα/C’ of Cys residues at the expenses, in this case, of the intensity of those signals that are affected by paramagnetism at a lower extent.
Fig. 3.
A Stacked rows extracted from five different HCACO experiments for seven residues of the HIPIP protein PioC. Each panel show a selected row of the HCACO experiment recorded with modified versions of the experiments, as presented in Fig. 1A and re-summarized here: a: standard pulse sequence; b: no Cα/Cβ coupling; c: no Cα/Cβ coupling & no IPAP; d: no Cα/Cβ coupling & no IPAP & shorter Hα/Cα INEPT period; e: no Cα/Cβ coupling & no IPAP & shorter Hα/Cα INEPT & shorter Ca/C' INEPT evolution. B Lowest energy NMR conformer of PioC (PDB ID: 6XYV). Residues shown in panels A are labeled and marked in green
Fig. 4.
Calculated relaxation (R2)-weighted transfer functions for the A Hα-to-Cα and B Cα-to-C’ INEPT steps of HCACO pulse sequence, simulating Hα and Cα nuclear spins with increasing transverse relaxation rates. a: neglected R2 relaxation (R2 = 0 s−1) b: R2 = 50 s−1, c: R2 = 100 s−1, d: R2 = 250 s−1 and e: R2 = 500 s−1. The maximum of the transfer function decreases with increasing transverse relaxation rates. Best calculated INEPT delay for C Hα-to-Cα and D Cα-to-C’ coherence transfer at different R2 rates
Essentially, Hα-to-Cα and Cα-to-C’ polarization transfers act here as relaxation-based filters that select signals according to transverse relaxation rates. Figure 5 shows optimized HCACO-AP and CACO-AP spectra recorded on PioC, in the spectral region where Cα/C’ connectivities of cluster bound cysteines have been assigned. Four signals, from the four iron-bound cysteine residues are observed in the CACO-AP experiment. The optimized HCACO-AP experiment, recorded with an Hα-to-Cα INEPT transfer period of 1.4 ms, gives only three signals; indicating that the HαyCαz coherence of Cys 22 relaxes faster than the other three cysteine residues and its signal is lost prior to 13Cα t1 evolution. However, a further decrease of the transfer delay from 1.4 ms to 1 ms, succeeded to revive Cys 22 Cα/C’ connectivity also in an optimized HCACO-AP experiment (blue contour plot, in Fig. 5).
Fig. 5.
CACO spectra of PioC acquired at 175 MHz at 298 K. The Cα carrier of the F1 dimension is placed at 86 ppm, in order to excite selectively the hyperfine shifted Cα Cys signals. Green contours: HCACO-AP recorded with 1.4 ms as Hα-to-Cα INEPT transfer; blue contours: HCACO-AP recorded with 1 ms as Hα-to-Cα INEPT transfer; red contours: CACO-AP experiment. For the sake of clarity, red and blue spectra have been arbitrarily shifted in the F1 dimension. The top panel shows the H(CA)CO-AP version of the experiment, recorded with acquisition, recycle and Hα-to-Cα INEPT transfer delay of 46 ms, 250 ms and 1.4 ms, where the Hα/C’ connectivities are obtained
The HCACO can be easily modified to evolve Hα instead of Cα during t1, i.e. an H(CA)CO experiments (Serber et al. 2000; Bermel et al. 2006b; Duma et al. 2003). This was particularly useful in the case of PioC, where the “missing piece” of the complete resonance assignment involve Hα of Cys residues. Three of the four cysteines’ Hα protons could be unambiguously assigned with this experiment. In particular, the Hα at 8.18 ppm was previously observed in a 13C-HSQC experiment and, because of the severe overlap of three Cα cysteines’ shifts, it was previously assigned to Hα of Cys 22. In the H(CA)CO-AP experiment (Fig. 5, upper panel), the Hα/C’ correlation allows us to correctly assign the Hα at 8.18 ppm to Cys 47, leaving the Hα of Cys 22 as the only unassigned resonance of the first coordination sphere of the cluster.
Mapping the geometry of cluster bound residues: CACO-AP vs COCA-AP
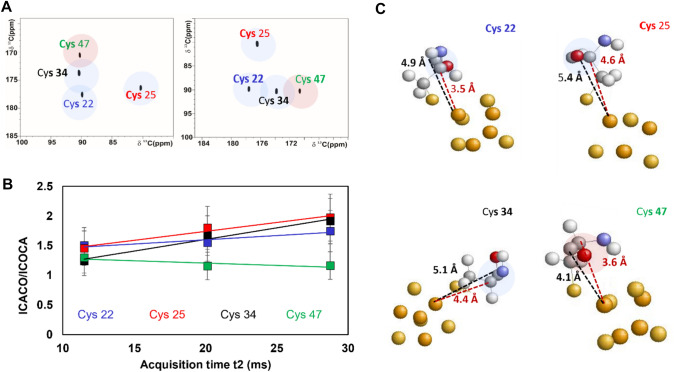
The relaxation properties of Cβ, Cα and C’ of iron bound cysteines depend on the orientation of backbone and side chain atoms with respect to the iron ions; different topologies of metal bound residues originate distinctive spectroscopic features. A promising tool to study this effect is the comparison of COCA-AP and CACO-AP experiments, shown in Fig. 6.
Fig. 6.
A COCA-AP (left-half) and CACO-AP (right-half) spectra. B ICACO/ICOCA peak intensity ratios related to increasing acquisition time in the direct dimension of the spectrum (t2). Peak intensities have been calculated by performing the experiments using a 46 ms acquisition time in the direct dimension and then considering for FT a smaller number of effective data points. C Molecular view of PioC’s cluster binding residues, atoms are coloured as follow: hydrogen (white), carbon (gray), oxygen (red), nitrogen (blue), iron (orange) and sulfur (yellow); different topologies of Cα-C’ vector are presented. C’-Fe and Cα-Fe distance are marked in black and red dashed lines, respectively
The two pulse sequences are identical and only the Cα/C’ frequencies are swapped (Fig. 1B, C). The experiments are based on a single coherence transfer step from the excited nucleus to the observed nucleus; performing the transfer from Cα to C’ or viceversa would call into the scene different relaxation rates. Therefore, the relative intensity of COCA and CACO peaks depends on the different topologies of metal binding side chains. We recorded, on the [Fe4S4]2+ protein PioC, COCA-AP and CACO-AP under the same experimental conditions using recycle, coherence transfer and t1 acquisition delays of 500 ms, 5.4 ms and 2.7 ms respectively. Considering only the relaxation dependent part, peak intensities are modulated by:
| 1 |
| 2 |
Two parameters contribute to the asymmetry of peak intensities: (i) The different values of indirect and direct acquisition periods: in a COCA-AP experiment, the observable signals relax with R2C’ during constant time 2∆ and R2Cα during t2 while the opposite occurs in the CACO-AP experiment; (ii) The choice of the recycle delay: the amount of initial magnetization depends on the progressive saturation of the excited nucleus, this will affect CACO and COCA spectra at different extents.
In order to discuss how the signal intensity in the two experiments is modulated by the geometry of the ligands, we maintained a 5.4 ms 2∆ constant time for the indirect dimension and measured the experimental intensity ratios, ICACO/ICOCA, at different acquisition times t2, as reported in Fig. 6B. When t2 is only 10 ms, the four cysteines have a similar behaviour, with CACO experiment being about 40% more sensitive than COCA. The increase of the evolution period t2 affects the ICACO/ICOCA ratio at different extents: for Cys 22, Cys 25 and Cys 34, the ratio increases at longer t2 values while an opposite trend is observed for Cys 47. According to Eqs. (1, 2) and keeping constant the recycle delay, the slopes experimentally obtained depend on the difference between R2Cα and R2C’: Cys 22, Cys 25 and Cys 34 have a positive value of ∆R2Cα−C’, while for Cys 47 the ∆R2Cα−C’ is negligible or negative. There is a correlation between this finding and the metal-to-carbon distances observed in the NMR structure of PioC (Trindade et al. 2021c): the orientation of Cys 47 is such that C’ and Cα are at similar distances from the closest iron ion and in particular the C’ atom is only 4.1 Å from the iron ion. For Cys 22, 25 and 34 the topologies are different, with significantly larger Fe–C’ distances, ≥ 4.9 Å (Fig. 6C). We may speculate that, in the case of Cys 47, both Cα and C’ relaxation rates are dominated by the paramagnetic contributions (Querci et al. 2023), and additional contributions to relaxation may result in negative values of ∆R2Cα−C’. In the other cases, C’ is too far from the Fe ion electron-nucleus dipolar relaxation significantly affect only the Cα nuclei. The consequence is that the C’yCαz coherence, acquired during t2 in a CACO experiment, relaxes much slower than the CαyC’z. CACO is therefore the best experiment to observe the Cα/C’ connectivities of Cys 22, Cys 25 and Cys 34 whereas for Cys 47 the two experiments have the same sensitivity suggesting that, as a general strategy in paramagnetic systems, COCA should be used as a complementary experiment in order to observe Cα/C’ peaks that might escape detection in CACO.
Conclusions
A complete, multinuclear NMR assignment provides unique site specific information which are essential to study conformational changes and intermolecular interactions. In metalloproteins, the development of experimental approaches to recover the NMR information in the proximity of a paramagnetic center represents a relevant topic, rejuvenated also in the last years by the extensive use of paramagnetic probes to obtain structural information from paramagnetic NMR (Su and Chen 2019; Dasgupta et al. 2021; Parigi et al. 2023; Ravera et al. 2022; Clore and Iwahara 2009; Clore 2015; Ott et al. 2021; Abelein et al. 2022; Zambelli et al. 2023).
We have analyzed here the performances of HCACO, CACO and COCA, three well known experiments, part of a very successful battery of 13C detected experiments (Felli and Pierattelli 2022). We have discussed the relevant modifications needed to adapt these experiments to study site-specific, fast relaxing systems. We also provided examples on how these experiments contribute to improve the available assignments and, therefore, to map the proximity of a metal center. Different coherence transfer pathways call into the scene specific relaxation mechanisms, this should be taken into account for the selection of the experiments to be performed. Apparently redundant experiments, such as CACO and COCA, do contain complementary information which can eventually be converted into structural information.
In the case of PioC, we have corrected the assignment of cluster bound cysteine residues and shown that, even when extended assignment and NMR structure are available, these experiments provide additional information and structural constraints. In the case of CISD3, the optimization of HCACO to paramagnetic system provides a 30% increase of the observed Cα/C’ signals, a further similar improvement is obtained when the 1H start experiment is replaced by the 13C start CACO experiment. HCACO and CACO-AP are complementary and reduce the blind sphere around the [Fe2S2]+ cluster from ca 9 to 6 Å.
These experiments are important in the frame of non-systematic sequence-specific assignment strategies: within tailored NMR experiments, the identification of a signal is related to the distance of these spins from the paramagnetic center. This is applicable in all paramagnetic metalloproteins; indeed the only relevant variable is the range of metal to proton distance where conventional NMR experiments do not work properly and complementary experiments must be designed. The information obtained are important to study the interaction of paramagnetic metalloproteins with small ligand molecules (Zuo et al. 2023) and protein partners, particularly for CISD3 and its homologues (Ferecatu et al. 2014; Karmi et al. 2022).
Materials and methods
Cisd3 and PioC protein expression and purification
PioC and CISD3 (Ala 37–Leu 127) genes were expressed and purified, as previously reported (Grifagni et al. 2023; Trindade et al. 2021c, 2020). Uniformly 15N, 13C labelled PioC and CISD3 were expressed in the M9 minimal media with the addition of 1.2 g/L of ammonium sulfate (15NH4, 99%) and 3.0 g/L of [U–13C6] D-glucose.
All the purification steps were performed in an anaerobic glovebox, as previously done for other FeS proteins (Maione et al. 2020).
NMR experiments
All NMR spectra were performed on a Bruker Avance Neo spectrometer operating at a field of 16.4 Tesla and on a Bruker Ascend Neo spectrometer operating at 28.2 T. Both spectrometers were equipped with a TXO probe head optimised for 13C carbon direct detection. Experiments were collected at 298 K.
13C-HCACO and CACO-AP acquired on CISD3 protein
The standard HCACO (Duma et al. 2003; Bermel et al. 2005b) used a recycle delay of 1 s, acquisition time t2 of 92 ms and a Hα-to-Cα INEPT transfer period of 3.6 ms. A 1024 × 660 data point matrix was acquired, with 32 transients for each increment and a spectral width of 31 ppm and 44 ppm for direct (F2) and indirect (F1) dimensions. For paramagnetic optimized HCACO, the experiment was recorded shortening the Hα– to –Cα INEPT transfer period to 1.4 ms. The acquisition time of the direct dimension, t2, was shortened to 46 ms and a recycle delay of 400 ms was used together with 1024 scans. A data matrix of 512 × 50 point was collected, with a spectral width of 31 ppm and 44 ppm for F2 and F1 dimensions. The CACO-AP (Bertini et al. 2005a) was recorded with 300 ms of recycle delay and with a acquisition time of 24.5 ms. The Cα-to-C’ INEPT transfer delay was shortened to 5.4 ms instead of 9 ms. The collected data matrix was 512 × 70 with 1024 scans for each increment, with a spectral width of 59 ppm and 60 ppm for F2 and F1 dimensions. Cα carrier was set to 46 ppm for the standard HCACO experiment and to 53.2 ppm for the paramagnetic optimized HCACO and CACO-AP. All the experiments were recorded using waltz65 and garp4 decoupling scheme for 1H and 15N decoupling, respectively. Smoothed square shape for all gradients was used. Q5 and Q3-shaped pulses, with a duration of 300 μs and 231 μs respectively (Emsley and Bodenhausen 1992), were used for 13C band-selective π/2 and π flip angle pulses. Except for HCACO experiments, acquired with the IPAP scheme for virtual decoupling of CαzC’y component of the magnetization, CACO-AP was processed as pseudo-absorbed signals in the direct dimension, as reported elsewhere (Bertini et al. 2005a). Before Fourier transformation, a square sine weighting function, shifted by 90°, was applied in both dimensions to all the spectra. The evaluation of signal intensity was performed considering the volume of each cross-peak in the spectra (Mori et al. 2008), obtained through an integral operation using Computed-Automated NMR Assignment (CARA) software (Keller 2004).
13C-HCACO, CACO-AP and COCA-AP acquired on PioC protein
Optimized HCACO-AP was acquired with recycle delay of 250 ms, acquisition time of 46 ms and an Hα-to-Cα INEPT transfer delay of 1.4 ms. The total data matrix for the experiment was 512 × 128 point, with 1024 transients for each increment and a spectral width of 31 and 80 ppm for F2 and F1 dimensions. A further optimized HCACO-AP was acquired with a data matrix of 512 × 100, with 7168 scans for each increment, and a Hα-to-Cα INEPT transfer delay of 1 ms. The recycle delay and acquisition time for the experiment were 250 ms and 46 ms, respectively. The spectral width for F2 and F1 dimensions were 21 ppm and 64 ppm, respectively. CACO-AP and COCA-AP experiments were recorded both with recycle delay, acquisition time and Cα-to-C’ INEPT transfer delay of 500 ms, 47 ms and 5.4 ms, respectively. A data matrix of 2048 × 120 points, with 128 transients for each increment, was collected and a spectral width of 123 ppm and 64 ppm for direct and indirect dimensions has been used. The H(CA)CO-AP experiment was acquired with recycle delay, acquisition time and Hα-to-Cα INEPT transfer delay of 250 ms, 46 ms and 1.4 ms. A data matrix of 512 × 220 point was collected, with 512 transients for each increment. The spectral width for direct and indirect dimension were 31 ppm, 64 ppm and 11 ppm, respectively. Shaped pulses, 1H and 15N decoupling, processing parameters were as described above. The optimization of HCACO experiment was monitored at 28.2 T, using a series of experiments, recorded with the same acquisition parameters. Experiments were recorded with 128 ns, 1 s recycle delay, 9.6 ms and 50 ms as t1 and t2 acquisition dimensions. Each experiment was 5 h. Throughout this series of experiments, 15N CPD was achieved using a p5m4sp180 decoupling scheme. An 8 ms, smoothed adiabatic chirp pulse with 8.4 kHz sweep width was used.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The support of the CERM/CIRMMP center of Instruct-ERIC is gratefully acknowledged. This work was supported in part by the project “Potentiating the Italian Capacity for Structural Biology Services in Instruct-ERIC" (ITACA.SB, Project no. IR0000009) within the call MUR 3264/2021 PNRR M4/C2/L3.1.1, funded by the European Union—Next Generation EU. LQ is a PhD student under the Tuscany Health Ecosystem-ECS_00000017 (CUP B83C22003920001), spoke 7, funded by the European Union—Next Generation EU. This work was funded by national funds through FCT–Fundação para a Ciência e a Tecnologia, I.P. (FCT), Project MOSTMICRO-ITQB with refs UIDB/04612/2020 and UIDP/04612/2020, and LS4FUTURE Associated Laboratory (LA/P/0087/2020).
Author contributions
MP, FC, LQ and ROL wrote the main manuscript text. LQ and JMS prepared figures. DG and IBT prepared the samples. LQ, DG, IBT and JMS collected and analyzed the data. All authors reviewed the manuscript.
Funding
Open access funding provided by Università degli Studi di Firenze within the CRUI-CARE Agreement.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abelein A, Ciofi-Baffoni S, Mörman C, Kumar R, Giachetti A, Piccioli M, Biverstål H. Molecular structure of Cu(II)-bound Amyloid-β monomer implicated in inhibition of peptide self-assembly in Alzheimer's disease. JACS Au. 2022;2:2571–2584. doi: 10.1021/jacsau.2c00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnesano F, Banci L, Bertini I, Felli IC, Luchinat C, Thompsett AR. A strategy for the NMR characterization of type II copper(II) proteins: the case of the copper trafficking protein CopC from Pseudomonas syringae. J Am Chem Soc. 2003;125:7200–7208. doi: 10.1021/ja034112c. [DOI] [PubMed] [Google Scholar]
- Azam T, Przybyla-Toscano J, Vignols F, Couturier J, Rouhier N, Johnson MK. [4Fe-4S] cluster trafficking mediated by Arabidopsis mitochondrial ISCA and NFU proteins. J Biol Chem. 2020;295:18367–18378. doi: 10.1074/jbc.RA120.015726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balayssac S, Bertini I, Luchinat C, Parigi G, Piccioli M. 13C direct detected NMR increases the detectability of residual dipolar couplings. J Am Chem Soc. 2006;128:15042–15043. doi: 10.1021/ja0645436. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Luchinat C. The 1H NMR parameters of magnetically coupled dimers - The Fe2S2 proteins as an example. Struct Bonding. 1990;72:113–135. doi: 10.1007/BFb0058197. [DOI] [Google Scholar]
- Banci L, Camponeschi F, Ciofi-Baffoni S, Piccioli M. The NMR contribution to protein–protein networking in Fe–S protein maturation. J Biol Inorg Chem. 2018;23:665–685. doi: 10.1007/s00775-018-1552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax A. Triple resonance three-dimensional protein NMR: before it became a black box. J Magn Reson. 2011;213:442–445. doi: 10.1016/j.jmr.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniamino Y, Cenni V, Piccioli M, Ciurli S, Zambelli B. The Ni(II)-Binding Activity of the Intrinsically Disordered Region of Human NDRG1, a Protein Involved in Cancer Development. Biomolecules. 2022 doi: 10.3390/biom12091272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett SP, Crack JC, Puglisi R, Pastore A, Le Brun NE. Native mass spectrometric studies of IscSU reveal a concerted, sulfur-initiated mechanism of iron-sulfur cluster assembly. Chem Sci. 2022;14:78–95. doi: 10.1039/D2SC04169C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermel W, Bertini I, Felli IC, Kümmerle R, Pierattelli R. 13C direct detection experiments on the paramagnetic oxidized monomeric copper, zinc superoxide dismutase. J Am Chem Soc. 2003;125:16423–16429. doi: 10.1021/ja037676p. [DOI] [PubMed] [Google Scholar]
- Bermel W, Bertini I, Duma L, Emsley L, Felli IC, Pierattelli R, Vasos PR. Complete assignment of heteronuclear protein resonances by protonless NMR spectroscopy. Angew Chem Int Ed. 2005;44:3089–3092. doi: 10.1002/anie.200461794. [DOI] [PubMed] [Google Scholar]
- Bermel W, Bertini I, Duma L, Felli IC, Emsley L, Pierattelli R, Vasos PR. Complete assignment of heteronuclear protein resonances by protonless NMR spectroscopy. Angewandte Chemie. 2005;44:3089–3092. doi: 10.1002/anie.200461794. [DOI] [PubMed] [Google Scholar]
- Bermel W, Bertini I, Felli I, Piccioli M, Pierattelli R. 13C-detected protonless NMR spectroscopy of proteins in solution. Prog Nucl Magn Reson Spectrosc. 2006;48:25–45. doi: 10.1016/j.pnmrs.2005.09.002. [DOI] [Google Scholar]
- Bermel W, Bertini I, Felli IC, Lee YM, Luchinat C, Pierattelli R. Protonless NMR experiments for sequence-specific assignment of backbone nuclei in unfolded proteins. J Am Chem Soc. 2006;128:3918–3919. doi: 10.1021/ja0582206. [DOI] [PubMed] [Google Scholar]
- Bermel W, Bertini I, Felli IC, Pierattelli R. Speeding up 13C direct detection biomolecular NMR experiments. J Am Chem Soc. 2009;131:15339–15345. doi: 10.1021/ja9058525. [DOI] [PubMed] [Google Scholar]
- Bertini I, Jimenez B, Piccioli M. 13C direct detected experiments: optimization for paramagnetic signals. J Magn Reson. 2005;174:125–132. doi: 10.1016/j.jmr.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Bertini I, Jimenez B, Piccioli M, Poggi L. Asymmetry in 13C–13C COSY spectra provides information on ligand geometry in paramagnetic proteins. J Am Chem Soc. 2005;127:12216–12217. doi: 10.1021/ja051058m. [DOI] [PubMed] [Google Scholar]
- Bertini I, Luchinat C, Parigi G, Ravera E. NMR of paramagnetic molecules. Amsterdam: Elsevier; 2017. pp. 1–508. [Google Scholar]
- Bird LJ, Saraiva IH, Park S, Calcada EO, Salgueiro CA, Nitschke W, Louro RO, Newman DK. Nonredundant roles for cytochrome c2 and two high-potential iron-sulfur proteins in the photoferrotroph Rhodopseudomonas palustris TIE-1. J Bacteriol. 2014;196:850–858. doi: 10.1128/JB.00843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondin G, Girerd JJ. Interplay of electron exchange and electron-transfer in metal polynuclear complexes in proteins or chemical-models. Chem Rev. 1990;90:1359–1376. doi: 10.1021/cr00106a001. [DOI] [Google Scholar]
- Cai K, Tonelli M, Frederick RO, Markley JL. Human mitochondrial ferredoxin 1 (FDX1) and ferredoxin 2 (FDX2) both bind cysteine desulfurase and donate electrons for iron-sulfur cluster biosynthesis. Biochemistry. 2017;56:487–499. doi: 10.1021/acs.biochem.6b00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camponeschi F, Muzzioli R, Ciofi-Baffoni S, Piccioli M, Banci L. Paramagnetic (1)H NMR spectroscopy to investigate the catalytic mechanism of radical S-adenosylmethionine enzymes. J Mol Biol. 2019;431:4514–4522. doi: 10.1016/j.jmb.2019.08.018. [DOI] [PubMed] [Google Scholar]
- Camponeschi F, Gallo A, Piccioli M, Banci L. The long-standing relationship between paramagnetic NMR and Iron-Sulfur proteins: the mitoNEET example. An old method for new stories or the other way around? Magn Reson Discuss. 2021;2:203–211. doi: 10.5194/mr-2-203-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camponeschi F, Piccioli M, Banci L. The intriguing mitoNEET: functional and spectroscopic properties of a unique [2Fe-2S] cluster coordination geometry. Molecules. 2022;27:8218. doi: 10.3390/molecules27238218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofi-Baffoni S, Gallo A, Muzzioli R, Piccioli M. The IR-(1)(5)N-HSQC-AP experiment: a new tool for NMR spectroscopy of paramagnetic molecules. J Biomol NMR. 2014;58:123–128. doi: 10.1007/s10858-013-9810-2. [DOI] [PubMed] [Google Scholar]
- Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM. Practical aspects of paramagnetic relaxation enhancement in biological macromolecules. Methods Enzymol. 2015;564:485–497. doi: 10.1016/bs.mie.2015.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta R, Gupta K, de Groot HJM, Ubbink M. The resting oxidized state of small laccase analyzed with paramagnetic NMR spectroscopy. ChemPhysChem. 2021;22:733–740. doi: 10.1002/cphc.202100063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duma L, Hediger S, Lesage A, Emsley L. Spin-state selection in solid-state NMR. J Magn Reson. 2003;164:187–195. doi: 10.1016/S1090-7807(03)00187-3. [DOI] [PubMed] [Google Scholar]
- Emsley L, Bodenhausen G. Optimization of shaped selective pulses for Nmr using a quaternion description of their overall propagators. J Magn Reson. 1992;97:135–148. [Google Scholar]
- Felli IC, Pierattelli R. (13)C direct detected NMR for challenging systems. Chem Rev. 2022;122:9468–9496. doi: 10.1021/acs.chemrev.1c00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferecatu I, Goncalves S, Golinelli-Cohen MP, Clemancey M, Martelli A, Riquier S, Guittet E, Latour JM, Puccio H, Drapier JC, Lescop E, Bouton C. The diabetes drug target MitoNEET governs a novel trafficking pathway to rebuild an Fe-S cluster into cytosolic aconitase/iron regulatory protein 1. J Biol Chem. 2014;289:28070–28086. doi: 10.1074/jbc.M114.548438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrage F, Pelupessy P, Cowburn D, Bodenhausen G. Protein backone dynamic through 13C'-13Calpha cross-relaxation in NMR spectroscopy. J Am Chem Soc. 2006;128:11072–11078. doi: 10.1021/ja0600577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee LB, Pelmenschikov V, Mons C, Mishra N, Wang H, Yoda Y, Tamasaku K, Golinelli-Cohen MP, Cramer SP. NRVS and DFT of MitoNEET: understanding the special vibrational structure of a [2Fe-2S] cluster with (Cys)(3)(His)(1) ligation. Biochemistry. 2021;60:2419–2424. doi: 10.1021/acs.biochem.1c00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelis I, Katsaros N, Luchinat C, Piccioli M, Poggi L. A simple protocol to study blue copper proteins by NMR. Eur J Biochem. 2003;270:600–609. doi: 10.1046/j.1432-1033.2003.03400.x. [DOI] [PubMed] [Google Scholar]
- Gervason S, Larkem D, Mansour AB, Botzanowski T, Muller CS, Pecqueur L, Le Pavec G, Delaunay-Moisan A, Brun O, Agramunt J, Grandas A, Fontecave M, Schunemann V, Cianferani S, Sizun C, Toledano MB, D'Autreaux B. Physiologically relevant reconstitution of iron-sulfur cluster biosynthesis uncovers persulfide-processing functions of ferredoxin-2 and frataxin. Nat Commun. 2019;10:3566. doi: 10.1038/s41467-019-11470-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golinelli-Cohen MP, Lescop E, Mons C, Goncalves S, Clemancey M, Santolini J, Guittet E, Blondin G, Latour JM, Bouton C. Redox control of the human iron-sulfur repair protein MitoNEET activity via its iron-sulfur cluster. J Biol Chem. 2016;291:7583–7593. doi: 10.1074/jbc.M115.711218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifagni D, Silva JM, Cantini F, Piccioli M, Banci L. Relaxation-based NMR assignment: spotlights on ligand binding sites in human CISD3. J Inorg Biochem. 2023;239:112089. doi: 10.1016/j.jinorgbio.2022.112089. [DOI] [PubMed] [Google Scholar]
- Herath ID, Breen C, Hewitt SH, Berki TR, Kassir AF, Dodson C, Judd M, Jabar S, Cox N, Otting G, Butler SJ. A chiral lanthanide tag for stable and rigid attachment to single cysteine residues in proteins for NMV. Chemistry. 2021;27:13009–13023. doi: 10.1002/chem.202101143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Invernici M, Trindade IB, Cantini F, Louro RO, Piccioli M. Measuring transverse relaxation in highly paramagnetic systems. J Biomol NMR. 2020;74:431–442. doi: 10.1007/s10858-020-00334-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmi O, Marjault HB, Pesce L, Carloni P, Onuchic JN, Jennings PA, Mittler R, Nechushtai R. The unique fold and lability of the [2Fe-2S] clusters of NEET proteins mediate their key functions in health and disease. J Biol Inorg Chem. 2018;23:599–612. doi: 10.1007/s00775-018-1538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmi O, Marjault HB, Bai F, Roy S, Sohn YS, Darash Yahana M, Morcos F, Ioannidis K, Nahmias Y, Jennings PA, Mittler R, Onuchic JN, Nechushtai R. A VDAC1-mediated NEET protein chain transfers [2Fe-2S] clusters between the mitochondria and the cytosol and impacts mitochondrial dynamics. Proc Natl Acad Sci USA. 2022;119:e2121491119. doi: 10.1073/pnas.2121491119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R. The computer aided resonance assignment tutorial. Goldau: CANTINA Verlag; 2004. [Google Scholar]
- Kostic M, Pochapsky SS, Pochapsky TC. Rapid recycle (13)C', (15)N and (13)C, (13)C' heteronuclear and homonuclear multiple quantum coherence detection for resonance assignments in paramagnetic proteins: example of Ni(2+)-containing acireductone dioxygenase. J Am Chem Soc. 2002;124:9054–9055. doi: 10.1021/ja0268480. [DOI] [PubMed] [Google Scholar]
- Langen R, Jensen GM, Jacob U, Stephen PJ, Warshel A. Protein control of iron-sulfur cluster redox potentials. J Biol Chem. 1992;267:25625–25627. doi: 10.1016/S0021-9258(18)35647-3. [DOI] [PubMed] [Google Scholar]
- Lee D, Vögeli B, Pervushin K. Detection of C', Ca correlations in proteins using a new time- and sensitivity-optimal experiment. J Biomol NMR. 2005;31:273–278. doi: 10.1007/s10858-005-2361-4. [DOI] [PubMed] [Google Scholar]
- Lin IJ, Xia B, King DS, Machonkin TE, Westler WM, Markley JL. Hyperfine-shifted (13)C and (15)N NMR signals from Clostridium pasteurianum rubredoxin: extensive assignments and quantum chemical verification. J Am Chem Soc. 2009;131:15555–15563. doi: 10.1021/ja905928x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipper CH, Karmi O, Sohn YS, Darash-Yahana M, Lammert H, Song L, Liu A, Mittler R, Nechushtai R, Onuchic JN, Jennings PA. Structure of the human monomeric NEET protein MiNT and its role in regulating iron and reactive oxygen species in cancer cells. Proc Natl Acad Sci U S A. 2018;115:272–277. doi: 10.1073/pnas.1715842115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machonkin TE, Westler WM, Markley JL. 13C–13C 2D NMR: a novel strategy for the study of paramagnetic proteins with slow electronic relaxation times. J Am Chem Soc. 2002;124:3204–3205. doi: 10.1021/ja017733j. [DOI] [PubMed] [Google Scholar]
- Maione V, Grifagni D, Torricella F, Cantini F, Banci L. CIAO3 protein forms a stable ternary complex with two key players of the human cytosolic iron-sulfur cluster assembly machinery. J Biol Inorg Chem. 2020;25:501–508. doi: 10.1007/s00775-020-01778-z. [DOI] [PubMed] [Google Scholar]
- Mao B, Tejero R, Baker D, Montelione GT. Protein NMR structures refined with Rosetta have higher accuracy relative to corresponding X-ray crystal structures. J Am Chem Soc. 2014;136:1893–1906. doi: 10.1021/ja409845w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Q, Nitsche C, Orton H, Overhand M, Otting G, Ubbink M. Paramagnetic chemical probes for studying biological macromolecules. Chem Rev. 2022;122:9571–9642. doi: 10.1021/acs.chemrev.1c00708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Jimenez B, Piccioli M, Battistoni A, Sette M. The solution structure of the monomeric copper, zinc superoxide dismutase from Salmonella enterica: structural insights to understand the evolution toward the dimeric structure. Biochemistry. 2008;47:12954–12963. doi: 10.1021/bi801252e. [DOI] [PubMed] [Google Scholar]
- Mori M, Kateb F, Bodenhausen G, Piccioli M, Abergel D. Towards structural dynamics: protein motions viewed by chemical shift modulations and direct detection of C'N multiple-quantum relaxation. J Am Chem Soc. 2010;132:3594–3600. doi: 10.1021/ja9103556. [DOI] [PubMed] [Google Scholar]
- Mulliez E, Ollagnier-de Choudens S, Meier C, Cremonini M, Luchinat C, Trautwein AX, Fontecave M. Iron-sulfur interconversions in the anaerobic ribonucleotide reductase from Escherichia coli. J Biol Inorg Chem. 1999;4:614–620. doi: 10.1007/s007750050385. [DOI] [PubMed] [Google Scholar]
- Muntener T, Joss D, Haussinger D, Hiller S. Pseudocontact shifts in biomolecular NMR spectroscopy. Chem Rev. 2022;122:9422–9467. doi: 10.1021/acs.chemrev.1c00796. [DOI] [PubMed] [Google Scholar]
- Ott JC, Suturina EA, Kuprov I, Nehrkorn J, Schnegg A, Enders M, Gade LH. Observability of paramagnetic NMR signals at over 10 000 ppm chemical shifts. Angew Chem Int Ed Engl. 2021;60:22856–22864. doi: 10.1002/anie.202107944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parigi G, Ravera E, Piccioli M, Luchinat C. Paramagnetic NMR restraints for the characterization of protein structural rearrangements. Curr Opin Struct Biol. 2023;80:102595. doi: 10.1016/j.sbi.2023.102595. [DOI] [PubMed] [Google Scholar]
- Parker D, Suturina EA, Kuprov I, Chilton NF. How the ligand field in lanthanide coordination complexes determines magnetic susceptibility anisotropy, paramagnetic NMR Shift, and relaxation behavior. Acc Chem Res. 2020;53:1520–1534. doi: 10.1021/acs.accounts.0c00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pell AJ, Pintacuda G, Grey CP. Paramagnetic NMR in solution and the solid state. Prog Nucl Magn Reson Spectrosc. 2019;111:1–271. doi: 10.1016/j.pnmrs.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Perard J, de Ollagnier Choudens S. Iron-sulfur clusters biogenesis by the SUF machinery: close to the molecular mechanism understanding. J Biol Inorg Chem. 2018;23:581–596. doi: 10.1007/s00775-017-1527-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintacuda G, John M, Su XC, Otting G. NMR structure determination of protein-ligand complexes by lanthanide labeling. Acc Chem Res. 2007;40:206–212. doi: 10.1021/ar050087z. [DOI] [PubMed] [Google Scholar]
- Pochapsky TC, Kostic M, Jain N, Pejchal R. Redox-dependent conformational selection in a Cys4Fe2S2 ferredoxin. Biochemistry. 2001;40:5602–5614. doi: 10.1021/bi0028845. [DOI] [PubMed] [Google Scholar]
- Pontoriero L, Schiavina M, Murrali MG, Pierattelli R, Felli IC. Monitoring the interaction of alpha-synuclein with calcium ions through exclusively heteronuclear nuclear magnetic resonance experiments. Angew Chem Int Ed Engl. 2020;59:18537–18545. doi: 10.1002/anie.202008079. [DOI] [PubMed] [Google Scholar]
- Pritchard RB, Hansen DF. Characterising side chains in large proteins by protonless (13)C-detected NMR spectroscopy. Nat Commun. 2019;10:1747. doi: 10.1038/s41467-019-09743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querci L, Trindade IB, Invernici M, Silva JM, Cantini F, Louro RO, Piccioli M. NMR of paramagnetic proteins: 13C derived paramagnetic relaxation enhancements are an additional source of structural information in solution. Magnetochemistry. 2023;9:66. doi: 10.3390/magnetochemistry9030066. [DOI] [Google Scholar]
- Ravera E, Gigli L, Czarniecki B, Lang L, Kummerle R, Parigi G, Piccioli M, Neese F, Luchinat C. A quantum chemistry view on two archetypical paramagnetic pentacoordinate nickel(II) complexes offers a fresh look on their NMR spectra. Inorg Chem. 2021;60:2068–2075. doi: 10.1021/acs.inorgchem.0c03635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravera E, Gigli L, Fiorucci L, Luchinat C, Parigi G. The evolution of paramagnetic NMR as a tool in structural biology. Phys Chem Chem Phys. 2022;24:17397–17416. doi: 10.1039/D2CP01838A. [DOI] [PubMed] [Google Scholar]
- Richter C, Kovacs H, Buck J, Wacker A, Furtig B, Bermel W, Schwalbe H. 13C-direct detected NMR experiments for the sequential J-based resonance assignment of RNA oligonucleotides. J Biomol NMR. 2010;47:259–269. doi: 10.1007/s10858-010-9429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serber Z, Richter C, Moskau D, Bohlen JM, Gerfin T, Marek D, Haberli M, Baselgia L, Laukien F, Stern AS, Hoch JC, Dotsch V. New carbon-detected protein NMR experiments using CryoProbes. J Am Chem Soc. 2000;122:3554–3555. doi: 10.1021/ja991371m. [DOI] [Google Scholar]
- Shimba N, Kovacs H, Stern AS, Nomura AM, Shimada I, Hoch JC, Craik CS, Dotsch V. Optimization of 13C direct detection NMR methods. J Biomol NMR. 2004;30:175–179. doi: 10.1023/B:JNMR.0000048855.35771.11. [DOI] [PubMed] [Google Scholar]
- Silva JM, Grifagni D, Cantini F, Piccioli M. (1)H, (13)C and (15)N assignment of the human mitochondrial paramagnetic iron-sulfur protein CISD3. Biomol Nmr Assign. 2023;17:17–22. doi: 10.1007/s12104-022-10113-3. [DOI] [PubMed] [Google Scholar]
- Spronk C, Zerko S, Gorka M, Kozminski W, Bardiaux B, Zambelli B, Musiani F, Piccioli M, Basak P, Blum FC, Johnson RC, Hu H, Merrell DS, Maroney M, Ciurli S. Structure and dynamics of Helicobacter pylori nickel-chaperone HypA: an integrated approach using NMR spectroscopy, functional assays and computational tools. J Biol Inorg Chem. 2018;23:1309–1330. doi: 10.1007/s00775-018-1616-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmaier K, Blinn CM, Bechtel DF, Greth C, Auerbach H, Muller CS, Jakob V, Reijerse EJ, Netz DJA, Schunemann V, Pierik AJ. Apd1 and Aim32 are prototypes of bishistidinyl-coordinated non-rieske [2Fe-2S] proteins. J Am Chem Soc. 2019;141:5753–5765. doi: 10.1021/jacs.8b13274. [DOI] [PubMed] [Google Scholar]
- Stenstrom O, Champion C, Lehner M, Bouvignies G, Riniker S, Ferrage F. How does it really move? Recent progress in the investigation of protein nanosecond dynamics by NMR and simulation. Curr Opin Struct Biol. 2022;77:102459. doi: 10.1016/j.sbi.2022.102459. [DOI] [PubMed] [Google Scholar]
- Su XC, Chen JL. Site-specific tagging of proteins with paramagnetic ions for determination of protein structures in solution and in cells. Acc Chem Res. 2019;52:1675–1686. doi: 10.1021/acs.accounts.9b00132. [DOI] [PubMed] [Google Scholar]
- Tamir S, Paddock ML, Darash-Yahana-Baram M, Holt SH, Sohn YS, Agranat L, Michaeli D, Stofleth JT, Lipper CH, Morcos F, Cabantchik IZ, Onuchic JN, Jennings PA, Mittler R, Nechushtai R. Structure-function analysis of NEET proteins uncovers their role as key regulators of iron and ROS homeostasis in health and disease. Biochim Biophys Acta. 2015;1853:1294–1315. doi: 10.1016/j.bbamcr.2014.10.014. [DOI] [PubMed] [Google Scholar]
- Trindade IB, Invernici M, Cantini F, Louro RO, Piccioli M. (1)H, (13)C and (15)N assignment of the paramagnetic high potential iron-sulfur protein (HiPIP) PioC from Rhodopseudomonas palustris TIE-1. Biomol Nmr Assign. 2020;14:211–215. doi: 10.1007/s12104-020-09947-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade IB, Invernici M, Cantini F, Louro RO, Piccioli M. Sequence-specific assignments in NMR spectra of paramagnetic systems: a non-systematic approach. Inorg Chim Acta. 2021;514:119984. doi: 10.1016/j.ica.2020.119984. [DOI] [Google Scholar]
- Trindade IB, Hernandez G, Lebegue E, Barriere F, Cordeiro T, Piccioli M, Louro RO. Conjuring up a ghost: structural and functional characterization of FhuF, a ferric siderophore reductase from E. coli. J Biol Inorg Chem. 2021;26:313–326. doi: 10.1007/s00775-021-01854-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade IB, Invernici M, Cantini F, Louro RO, Piccioli M. PRE-driven protein NMR structures: an alternative approach in highly paramagnetic systems. FEBS J. 2021;288:3010–3023. doi: 10.1111/febs.15615. [DOI] [PubMed] [Google Scholar]
- Trindade IB, Coelho A, Cantini F, Piccioli M, Louro RO. NMR of paramagnetic metalloproteins in solution: Ubi venire, quo vadis? J Inorg Biochem. 2022;234:111871. doi: 10.1016/j.jinorgbio.2022.111871. [DOI] [PubMed] [Google Scholar]
- Trindade IB, Firmino MO, Noordam SJ, Alves AS, Fonseca BM, Piccioli M, Louro RO. Protein interactions in Rhodopseudomonas palustris TIE-1 reveal the molecular basis for resilient photoferrotrophic iron oxidation. Molecules. 2023 doi: 10.3390/molecules28124733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valer L, Rossetto D, Parkkila T, Sebastianelli L, Guella G, Hendricks AL, Cowan JA, Sang L, Mansy SS. Histidine ligated iron-sulfur peptides. ChemBioChem. 2022;23:e202200202. doi: 10.1002/cbic.202200202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögeli B, Kovacs H, Pervushin K. Simultaneous 1H- or 2H-, 15N- and multiple-band-selective 13C-decoupling during acquisition in 13C-detected experiments with proteins and oligonucleotides. J Biomol NMR. 2005;31:1–9. doi: 10.1007/s10858-004-5857-4. [DOI] [PubMed] [Google Scholar]
- Wu FJ, Rieder PS, Abiko LA, Rossler P, Gossert AD, Haussinger D, Grzesiek S. Nanobody GPS by PCS: an efficient new NMR analysis method for g protein coupled receptors and other large proteins. J Am Chem Soc. 2022;144:21728–21740. doi: 10.1021/jacs.2c09692. [DOI] [PubMed] [Google Scholar]
- Xia B, Pikus JD, Xia W, McClay K, Steffan RJ, Chae YK, Westler WM, Markley JL, Fox BG. Detection and classification of hyperfine-shifted 1H, 2H, and 15N resonances of the Rieske ferredoxin component of toluene 4-monooxygenase. Biochemistry. 1999;38:727–739. doi: 10.1021/bi981851a. [DOI] [PubMed] [Google Scholar]
- Zambelli B, Basak P, Hu H, Piccioli M, Musiani F, Broll V, Imbert L, Boisbouvier J, Maroney MJ, Ciurli S. The structure of the high-affinity nickel-binding site in the Ni, Zn-HypA•UreE2 complex. Metallomics. 2023 doi: 10.1093/mtomcs/mfad003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Yang DT, Gronenborn AM. Ligand-capped Cobalt(II) multiplies the value of the double-histidine motif for PCS NMR studies. J Am Chem Soc. 2023;145:4564–4569. doi: 10.1021/jacs.2c12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo K, Kranjc A, Capelli R, Rossetti G, Nechushtai R, Carloni P. Metadynamics simulations of ligands binding to protein surfaces: a novel tool for rational drug design. Phys Chem Chem Phys. 2023;25:13819–13824. doi: 10.1039/D3CP01388J. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.