Abstract
The role of exogenous metabolites as putative signal molecules mediating and/or regulating the carbon starvation adaptation program in Vibrio sp. strain S14 was investigated. Addition of the stationary-phase supernatant extract (SSE) of Vibrio sp. strain S14 to logarithmic-phase cells resulted in a significant number of carbon starvation-induced proteins being up-regulated. Halogenated furanones, putative antagonists of acylated homoserine lactones (AHLs), inhibited the synthesis of proteins specifically induced upon carbon starvation. The effect of the furanone was the opposite of that caused by SSE with respect to the up- and down-regulation of protein expression, indicating that both the furanone and the putative signalling molecules were acting on the same regulatory pathway. Culturability was rapidly lost when Vibrio sp. strain S14 was starved in the presence of the furanone at a low concentration. The furanone also had a negative effect on the ability of carbon-starved cells to mount resistance against UV irradiation and hydrogen peroxide exposure. The SSE of Vibrio sp. strain S14 had the ability to provide cross-protection against the loss in viability caused by the furanone. We have further demonstrated that the SSE taken from low- as well as high-cell-density cultures of Vibrio sp. strain S14 induced luminescence in Vibrio harveyi. Taken together, the results in this report provide evidence that Vibrio sp. strain S14 produces extracellular signalling metabolites during carbon and energy starvation and that these molecules play an important role in the expression of proteins crucial to the development of starvation- and stress-resistant phenotypes.
The adaptation to nutrient depletion by gram-negative bacteria such as Escherichia coli and Vibrio spp. involves a highly organized series of intracellular events that enable them to adapt to starvation conditions, as indicated by physiological experiments and analyses of the synthesis rates of individual proteins (21). In response to starvation, the marine Vibrio sp. strain S14 becomes more resistant to a variety of environmental challenges, such as visible and UV light, heavy metal exposure, and temperature shifts, a phenomenon referred to as cross-protection against secondary stresses (27). Starved cells markedly increase their nutrient-scavenging capacity and instantaneously respond to nutrient addition by increased RNA and protein synthesis in order to efficiently initiate the outgrowth response (8).
High-resolution two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) analysis of Vibrio sp. strain S14 has revealed that the carbon starvation response initially involves the induction of about 160 proteins and the repression of about 140 proteins (31). This significant reorganization suggests that several regulatory mechanisms are involved in orchestrating the response into relevant phenotypic expressions. Recent findings have shown that in Vibrio sp. strain S14, the immediate down-regulation of macromolecular synthesis upon depletion of exogenous carbon is accomplished via a RelA-ppGpp-mediated stringent response (7). Mutational analyses indicate that the enzymatic activities encoded by homologs to RelA and SpoT, the mediators of the stringent response, affect the synthesis rate of a majority of the proteins within the carbon starvation stimulon. Importantly, an altered activity of the SpoT homolog significantly impairs some starvation survival-related characteristics (29, 31).
Although many studies have focused on understanding the regulation of gene expression and the phenotypic consequences, there is limited understanding of how bacteria sense environmental changes and thereby produce signals for the genetic machinery to respond appropriately. Currently, there is a wide interest in the role of signal molecules in gene regulation. Small molecules have been shown to be involved in mediating gene expression in a number of microorganisms. Butyrolactones have been recognized as regulators of sporulation and antibiotic production in actinomycetes (3, 17). In Myxococcus xanthus, multicellular fruiting body early development requires the generation of and response to an extracellular A signal, a specific set of amino acids which function either individually or in a combination as a cell density signal during starvation. These compounds allow the cell to sense that a minimal cell density has been reached, thereby allowing development to proceed (19). Additionally, the E signal comprising one or more branched-chain fatty acids needs to be transmitted between cells to complete M. xanthus development (6). It has been suggested that in E. coli, the high-energy compound acetyl phosphate, produced intracellularly, plays a regulatory role during stasis (26). In E. coli, transient accumulation of 1,5-anhydroglucitol in the medium during the stationary phase was observed, and this compound has been proposed to be a metabolic signal of nutritional conditions (33). Stationary-phase supernatant extracts (SSE) of E. coli have also been demonstrated to inhibit SdiA-mediated expression of ftsQAZ, an operon whose genes encode functions required for cell division. SdiA is a member of the LuxR family of transcriptional activators (11). Recently, N-acyl homoserine lactones (AHLs) have been shown to play a widespread role in intercellular communication among gram-negative bacteria by controlling a number of cell density-dependent factors (9, 10, 14, 34, 35). Homoserine lactone (HSL) has also been proposed to be a key signal for inducing the transcription of rpoS in E. coli (18).
We have initiated a research project to study the role of putative extracellular signal molecules in the regulation of the carbon starvation response of Vibrio sp. strain S14. We assessed the expression of starvation-specific protein responders following exposure of the cells to compounds present in the nonpolar extract of cell-free stationary-phase supernatants. In addition, furanones, which have recently been found to specifically inhibit several AHL and AHL-like molecule-regulated phenotypes in several bacterial species (12, 13, 22, 24), were added to starvation cultures in order to study their effect on the starvation-specific adaptation program. Furanones are produced as secondary metabolites by the red alga Delisea pulchra (5). In this study, we used two furanones to investigate interference with the postulated signalling pathway in Vibrio sp. strain S14.
MATERIALS AND METHODS
Strains, growth conditions, and starvation conditions.
The marine Vibrio sp. strain S14 (CCUG 15956) has been described previously (1, 23). Detailed biochemical tests and 16S rRNA sequence analysis have shown that Vibrio sp. strain S14 displays a high degree of similarity to Photobacterium (Vibrio) angustum (36). The strain was cultured in a marine minimal medium prepared (MMMx, where the superscript x indicates the glucose concentration in micrograms per milliliter) as previously described (30). Culture flasks were inoculated with fresh overnight colonies, and the culture was kept in exponential growth at 28°C by recurrent dilutions until the maximal growth rate was established (usually five to seven generations). Carbon starvation conditions were achieved by pelleting cells grown in MMM400 and resuspending them in MMM0 unless otherwise stated and were statically incubated at 24°C in darkness. Growth was monitored as a function of optical density (OD). Viable counts were measured as CFU on half-strength LB20 agar (30) by the drop plate method (15).
Preparation and extraction of supernatants.
Two liters of a Vibrio sp. strain S14 MMM1000 culture was used to prepare each crude extract. For the SSE, cells were grown 5 h into stationary phase (OD at 610 nm [OD610] = 0.751) before they were removed by centrifugation and filtration (Millipore GS filter; pore size, 0.2 μm). Similarly, a log-phase supernatant extract was prepared by removing the cells from a logarithmically growing culture (OD610 = 0.343). The supernatant (2 liters) was sequentially extracted with 3 300-ml volumes of dichloromethane. The extracts were combined and taken to dryness in vacuo. The extract was analyzed for the presence of acylated HSLs by gas chromatography (GC)-mass spectrometry (MS) and 1H and 13C nuclear magnetic resonance spectroscopy (NMR).
Chemical analyses of supernatant extracts.
Samples for GC-MS were dissolved in ethyl acetate containing naphthalene (Aldrich Chemical Co., Milwaukee, Wis.) as an external standard at a concentration of 10 μg ml−1. GC-MS was performed with a Hewlett-Packard model 5980 series II gas chromatograph and a polyimide-coated fused-silica capillary column (BPX5; 12-m length, 0.22-mm inside diameter, 0.15-μm 5% phenyl [equivalent] polysilphenylene-siloxane stationary phase; SGE Pty Ltd., Melbourne, Australia). All injections were performed in the splitless mode with an inlet pressure of 100 kPa. The injection port was at 280°C, and the interface was at 300°C. The gas chromatograph was held at 50°C for 1.5 min and ramped at 20°C min−1 to 300°C. Helium was used as the carrier gas. The MS was performed on a Hewlett-Packard model 5971 mass selective detector in positive electron ionization mode. Ions (m/z 71 and 143) characteristic of the acylated HSLs N-butanoyl-l-homoserine lactone (BHL), N-hexanoyl-l-homoserine lactone (HHL), N-octanoyl-l-homoserine lactone (OHL), and N-decanoyl-l-homoserine lactone (DHL) and the ion characteristic of the external standard (m/z 128) were monitored in selected ion monitoring mode.
The samples for NMR were dissolved in deuterated chloroform (CDCl3), and 1H and 13C NMR spectra were recorded on a Bruker AM300 spectrometer. Spectral data were compared with authentic 1H and 13C NMR spectra of the known acylated HSLs BHL, HHL, OHL, DHL, N-(3-hydroxy)-butanoyl-l-homoserine lactone (HBHL), and N-(3-oxo)-hexanoyl-l-homoserine lactone and published data for N-(3R-hydroxy-7-cis-tetradecanoyl)-l-homoserine lactone (32).
Extraction and purification of D. pulchra secondary metabolites.
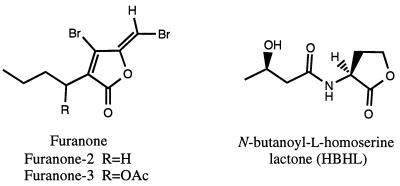
Crude extract from D. pulchra was prepared by dichloromethane extraction of freeze-dried plants. Furanone-2 ([5Z]-3-butyl-4-bromo-5-[bromomethylidene]-2[5H]-furanone]) and furanone-3 ([1′R,5Z]-3-[1′-acetoxybutyl]-4-bromo-5-[bromomethylidene]-2[5H]-furanone]) were isolated and purified by vacuum liquid chromatography and high-performance liquid chromatography as previously described (5). The structural similarities of these compounds to the HBHL of Vibrio harveyi are presented in Fig. 1.
FIG. 1.
Chemical structures of two furanone molecules (furanone-2 and furanone-3) extracted from the marine macroalga D. pulchra and the bioluminescence autoinducer from V. harveyi (HBHL).
2D-PAGE.
Samples were labeled and processed as previously described (31). To study the effect of SSE on the synthesis of proteins during logarithmic growth, a stock solution of SSE, freshly diluted in ethanol, was added to a culture (OD420 = 0.35) at a final concentration of 50 μg ml−1 2.5 h before [35S]methionine labeling was initiated. The final concentration of ethanol in the experiment was 0.3% (vol/vol). Ethanol controls were tested and found not to alter the expression of proteins. To study the effect of furanone-2 during carbon starvation, a stock solution of furanone-2 was added to the culture to reach a final concentration of either 5, 10, or 35 μg ml−1 just prior to centrifugation of the growing cells. The cell pellet was resuspended in MMM0 with the appropriate concentration of furanone-2. One hour after the onset of starvation, samples were labeled with [35S]methionine for 10 min. Five minutes after the addition of furanone-2, log-phase culture samples were labeled for 10 min. Isoelectric focusing, second-dimension resolution, and spot quantification were performed as previously described (31). The relative rate of protein synthesis (parts per million) was calculated by normalization of the individual spot density values to the sum of the spot density values of all detected spots in the gel.
Criteria used to distinguish different groups of proteins.
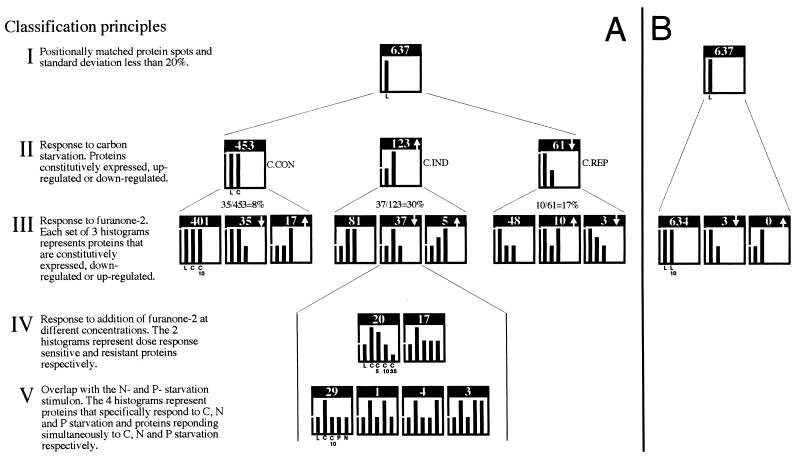
To distinguish proteins with unaltered synthesis rates from those that were induced or repressed in the presence of either SSE or furanone-2, we used a classification criterion based on the relative difference in the relative protein synthesis rates. If the standard deviation of the mean density values of the protein spots was greater than 20% based on duplicate and triplicate samples, the proteins were excluded from the comparison. Of the 1,187 proteins initially matched to all 2D-PAGE gels, this procedure excluded 550 proteins. The synthesis rates of the remaining 637 matched proteins were evenly distributed over the whole range of spot density values (parts per million), and therefore the analysis is likely to give an unbiased comparison.
Stress assays.
Stress resistance assays were performed after Vibrio sp. strain S14 cells were starved for 24 h for a carbon and energy source in the presence of 5 μg of furanone-2 ml−1. The following stresses were included in this study: (i) UV stress, achieved by irradiation at 254 nm for 0, 3, 5, 8, and 12 s, and (ii) H2O2 stress, achieved by incubating the 24-h-starved culture with 250 μM, 500 μM, or 1 mM H2O2 (final concentration) for 0, 30, or 60 min in 1.5-ml Eppendorf tubes. Stock solutions of H2O2 were freshly prepared from the commercially available 30% (vol/vol) stock (May & Baker Australia Pty Ltd.). The samples were treated with catalase (500 U ml−1; Sigma Chemical Co.) for 5 min before plating to facilitate removal of excess H2O2. The stress resistance was assessed by comparing the CFU of Vibrio sp. strain S14 at appropriate times to time zero of the stress assay.
Competition experiment.
Exponentially growing Vibrio sp. strain S14 cells were subjected to carbon starvation conditions as described above. The competition assay was performed when the cells had been simultaneously exposed to furanone-2 (5 μg ml−1) and SSE (50 μg ml−1). Viable counts were measured as CFU on half-strength LB20 agar (30) by the drop plate method (15). The results were expressed as percent CFU/milliliter by comparing the CFU of Vibrio sp. strain S14 at different times into the starvation period to CFU at time zero of starvation.
Bioluminescence assay.
V. harveyi grown in LB20 broth overnight with shaking at 24°C was diluted 1:50 in fresh LB20 broth. Twenty-microliter aliquots of the diluted culture of V. harveyi were added to 200-μl samples in 96-well opaque microtiter trays. The samples consisted of 100 μl of cell-free Vibrio sp. strain S14 cultures and 100 μl of fresh LB20 broth. V. harveyi supernatant was used as a positive control, and fresh medium, corresponding to the supernatant that was used, served as the negative control. The medium control did not induce bioluminescence at the time when the test samples and the positive control induced bioluminescence. Luminescence was monitored as counts per second, using a Microbeta Plus liquid scintillation counter (Wallac Inc.) in the luminescence mode. The results are reported as a relative value by dividing the counts per second of the sample by the counts per second of the corresponding negative control at the time when the difference in the luminescence between the samples and the negative control was maximal.
RESULTS
SSE preferentially up-regulates carbon starvation-induced proteins.
Several cases for the role of extracellular molecules in mediating adaptive responses have been proposed (11, 25). To investigate if components in the SSE induce the carbon starvation response, we determined the synthesis rate of individual proteins 2.5 h after the addition of SSE to a logarithmically growing culture (Fig. 2 and 6). This was achieved by pulse-labeling the translational products with [35S]methionine and subsequently analyzing the relative amount of [35S]methionine incorporated into the individual proteins by means of 2D-PAGE and quantitative image analysis. Protein expression was assessed during logarithmic growth in the presence of SSE at 50 μg ml−1. For comparative purposes, samples were also prepared from 1-h carbon-, 1-h nitrogen-, and 1-h phosphorus-starved cells (no SSE added).
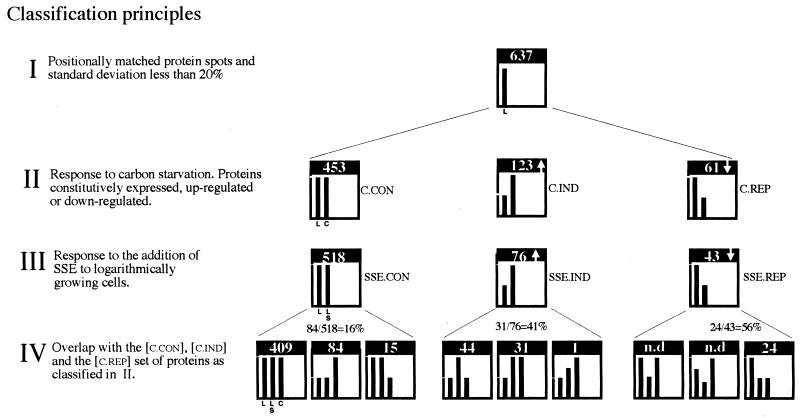
FIG. 2.
Classification and enumeration of proteins based on change in the relative synthesis rate in response to carbon starvation (II) and to the addition of SSE during logarithmic growth (III) and overlap with carbon starvation-induced proteins (IV). The difference in the heights of the bars in each graph represents the relative difference in the relative synthesis rate (±20%) for the number of proteins that are indicated in each graph. The twofold difference in the heights of the bars indicates the minimum change considered significant. Bars are labeled in the first histogram of each row, and the labels are applicable to the other histograms in the same row. Conditions compared: logarithmic growth (L), addition of SSE at 50 μg ml−1 during logarithmic growth (LS), and 1 h of carbon starvation (C).
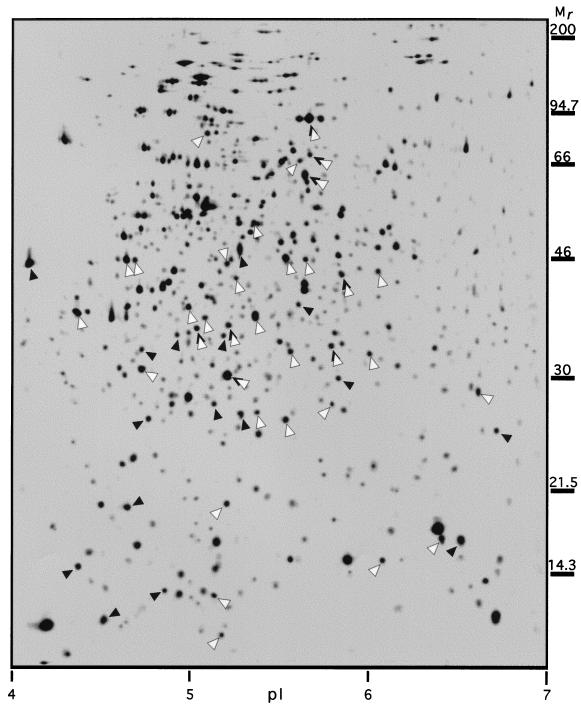
FIG. 6.
Standard 2D-PAGE image showing the positions of some carbon starvation-induced proteins as synthesized 1 h after the onset of carbon starvation in Vibrio sp. strain S14. Proteins found to be reduced in the relative synthesis rate when furanone-2 was added at 10 μg ml−1 immediately prior to the onset of carbon starvation are marked with closed arrowheads; proteins increased in the relative synthesis rate when SSE was added at 50 μg ml−1 to logarithmically growing cells are marked with open arrowheads. The threshold value used to determine whether the synthesis rate of a protein was considered to be altered by any of the treatments was set to a factor of 1.8.
From approximately 1,800 protein spots detected in each gel, 637 were chosen for quantitative comparison (Fig. 2). This initial selection was based on whether the spots could be positionally matched to all gels and if the difference in values (parts per minute) between duplicate or triplicate samples exceeded 20% of the mean value (Fig. 2, I). The 637 proteins were divided into different classes on the basis of the magnitude of their response to carbon starvation (Fig. 2, II). The criterion used to determine the change in the synthesis rate of a protein was based on whether the mean value differed by at least a factor of 1.8.
In response to carbon starvation, of the 637 proteins, 123 were induced ([C.IND]), 61 were repressed ([C.REP]), and 453 were constitutively expressed ([C.CON]) (Fig. 2, II). When SSE was added at 50 μg ml−1 to the logarithmically growing culture, of the 637 proteins compared, 76 were found to be up-regulated ([SSE.IND]), 43 were down-regulated ([SSE.REP]), and 518 were unaltered ([SSE.CON]) (Fig. 2, III). Of the 76 [SSE.IND] proteins, 31 were found to belong to the set of carbon starvation-induced proteins and 44 were found to belong to the set of proteins identified as constitutively expressed upon carbon starvation. Given that the number of [C.IND] proteins was 123 and that the number of [C.CON] proteins was 453 (Fig. 2, II), these figures correspond to a 25% overlap between [SSE.IND] and [C.IND] proteins, whereas the overlap between [SSE.IND] and [C.CON] proteins was 10%. Of the 43 [SSE.REP] proteins, 24 were found to belong to the [C.REP] proteins, corresponding to a 56% overlap (Fig. 2, IV).
Vibrio sp. strain S14 cells starved in the presence of furanone-2 lose viability.
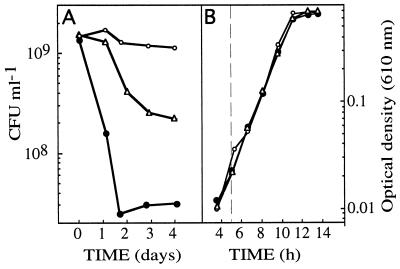
The viability of carbon-starved Vibrio sp. strain S14 cells, in the presence of furanone-2 at different concentrations, was determined to investigate if the furanone interferes with the starvation-induced phenotypes. This hypothesis relies on the specific interference of AHL-regulated systems by furanones in several bacteria and the possibility that HSL, AHL, or structurally similar molecules are signals of adaptive responses. It was found that when carbon-starved Vibrio sp. strain S14 was exposed to low concentrations of furanone-2 (5 μg ml−1), the viability was reduced by a factor of approximately 30 over a period of 4 days, as judged by the CFU values. This is considerably faster than the control, which exhibited 1% loss in CFU during the same period (Fig. 3A).
FIG. 3.
Effect on culturability of carbon-starved Vibrio sp. strain S14, assessed by CFU (A) and growth rate of Vibrio sp. strain S14 assessed by OD (B) after the addition of furanone-2 (closed circles) and furanone-3 (triangles) at 5 μg ml−1. Open circles denote control cultures without the addition of furanones. Experiments were repeated at least three times, each time in triplicate. The results presented are average values of the triplicate data from a representative experiment.
To investigate whether this effect could reflect general alterations in metabolic functions, growth rate was monitored in the presence of the same concentrations of furanone-2 as in the starvation viability test. No effect on growth rate was observed when furanone-2 was included in the growth media at 5 μg ml−1 (Fig. 3B) or 10 μg ml−1 (data not shown), while a small reduction in growth rate was observed at 15 μg ml−1 (data not shown). Once the growth rate had been stabilized after the addition of furanone-2 at 15 μg ml−1, it was constant throughout the logarithmic growth phase.
In another set of experiments, the effect on viability during starvation and the growth rate of Vibrio sp. strain S14 of a second furanone (furanone-3), structurally similar to furanone-2 (Fig. 1) and also produced by D. pulchra, was tested. The addition of 5 μg of furanone-3 ml−1 had a negative effect on viability but significantly less than that caused by furanone-2, reflecting the specificity displayed by these components (Fig. 3A). The growth rate, however, was unaltered in the presence of furanone-3 (Fig. 3B).
Vibrio sp. strain S14 cells starved in the presence of furanone-2 lose the ability to mount resistance against UV and H2O2 stresses.
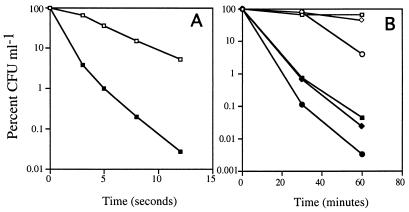
To determine if carbon-starved Vibrio sp. strain S14 is affected in its ability to mount cross-protection against environmental stresses in the presence of furanone-2, UV irradiation and hydrogen peroxide stress assays were performed. It has been reported that Vibrio sp. strain S14 acquires maximum cross-protection against UV irradiation after 40 h of starvation (27). However, the stress assays were performed after 24 h, as cells exposed to furanone-2 lose viability with time. To account for the loss in viability, the results are reported as percent CFU/milliliter by comparing the CFU at different times to CFU at time zero of the stress assay.
While 36.8% of 24-h carbon-starved cells were able to mount resistance against UV irradiation for 5 s in the absence of furanone-2 as judged by CFU, only 1% of the cells were culturable in the presence of furanone-2 at 5 μg ml−1 at the same time (Fig. 4A). The difference in cross-protection became more pronounced with increased time of UV irradiation. After 12 s, 5.2% of the cells from the control sample were culturable, compared with 0.027% of the cells exposed to furanone-2 (188-fold higher).
FIG. 4.
Effect of furanone-2 on the ability of Vibrio sp. strain S14 to acquire cross-protection against UV irradiation and hydrogen peroxide stresses during carbon starvation. Cells were starved in the presence or absence of furanone-2 (5 μg ml−1) for 24 h. The stress assays were performed at 24 h of carbon starvation. (A) Effect of furanone-2 on UV irradiation cross-protection acquired by Vibrio sp. strain S14 during carbon starvation. Open squares, control, i.e., cells starved in the absence of furanone-2; closed squares, cells starved for 24 h in the presence of furanone-2. (B) Effect of furanone-2 on hydrogen peroxide cross-protection acquired by Vibrio sp. strain S14 during carbon starvation. Open symbols, control samples, i.e., cells starved in the absence of furanone-2; closed symbols, samples exposed to furanone-2. Squares, 250 μM H2O2; diamonds, 500 μM H2O2; circles, 1 mM H2O2. Experiments were repeated three times, each time in triplicate. The results presented are average values of the triplicate data from a representative experiment.
The ability of carbon-starved Vibrio sp. strain S14 cells to develop cross-protection against oxidative stress invoked by hydrogen peroxide in the presence of furanone-2 at 5 μg ml−1 was also tested. After 24 h of carbon starvation with or without furanone-2, cells were exposed to 250 μM, 500 μM, or 1 mM H2O2 for 60 min. In the presence of 250 μM H2O2, 66% of cells not exposed to furanone-2 were culturable, as opposed to 0.044% of the cells exposed to furanone-2 after 60 min (Fig. 4B). Similarly, while 42.3% of cells from the control culture exposed to 500 μM H2O2 were culturable, only 0.023% of cells starved in the presence of furanone-2 were viable (Fig. 4B).
Furanone-2 specifically inhibits the induction of carbon starvation-induced proteins.
To investigate whether the reduction in viability of starved cells exposed to furanone-2 could be attributed to the failure to regulate the expression of proteins within the carbon starvation stimulon, we compared the synthesis rates of a large number of proteins in cells exposed to different concentrations of furanone-2. Protein expression was assessed during logarithmic growth in the presence of furanone-2 at 10 μg ml−1 and after 1 h of carbon and energy source starvation in the presence of furanone-2 at various concentrations.
The 637 proteins previously compared (Fig. 5) were divided into different classes, first on the basis of the magnitude of their response to carbon starvation (Fig. 5A, II) and second on the basis of the magnitude of their response to the exposure for furanone-2 (Fig. 5A and B, III; Fig. 6). Of the 123 [C.IND] proteins, 37 were found to be significantly less, or not, induced when furanone-2 was added at the onset of starvation at a concentration of 10 μg ml−1 (Fig. 5A, III). In the set of 123 [C.IND] proteins, only 5 were induced and 81 were unaltered during starvation in the presence of furanone-2 (Fig. 5A, III). Of the 61 [C.REP] proteins, 10 were increased, 3 were decreased, and 48 remained unaltered in their relative synthesis rates during starvation in the presence of furanone-2 (Fig. 5A, III). Of the 453 [C.CON] proteins, only 35 were decreased, 17 were increased, and 401 remained unaltered in their relative synthesis rates during starvation in the presence of furanone-2 (Fig. 5A, III). On addition of furanone-2 to logarithmically growing cells, only 3 of the 637 matched proteins were found to be significantly less expressed than in the untreated culture (Fig. 5B). No proteins were found to be increased in the rate of synthesis (Fig. 5B).
FIG. 5.
(A) Classification and enumeration of proteins based on change in the relative synthesis rate in response to carbon starvation (II), the addition of furanone-2 to carbon-starved cells (III and IV), and starvation for nitrogen or phosphorus (V). (B) Classification and enumeration of proteins based on change in the relative synthesis rate in response to the addition of furanone-2 to logarithmically growing Vibrio sp. strain S14 (III). Bars are labeled in the first histogram of each row and are applicable to the other histograms of the same row. The difference in the heights of the bars in each graph represents the relative difference in the relative synthesis rate (±20%) for the number of proteins that are indicated in each histogram. Conditions compared: logarithmic growth (L), 1-h carbon starvation (C), 1-h carbon starvation in the presence of furanone-2 at 5, (C5), 10 (C10), and 35 (C35) μg ml−1, 1-h nitrogen (N) and 1-h phosphorus (P) starvation, and synthesis 5 min after the addition of furanone-2 at 10 μg ml−1 to logarithmically growing cells (L10).
When the set of 31 carbon starvation-induced proteins identified as induced by SSE (Fig. 2, IV) was compared with that of the 37 carbon starvation-induced proteins identified as down-regulated by the furanone-2 (Fig. 5A, III), it was found that 12 proteins were common. Additionally, we compared the synthesis rates of individual proteins in cells exposed to different concentrations of furanone-2. Twenty of the 37 proteins exhibited a dose-response relationship, whereas 17 were less affected by either lower or higher concentrations of furanone-2 (Fig. 5A, IV). Furthermore, 29 of the 37 proteins that were identified as furanone-2-sensitive carbon starvation proteins were induced by neither phosphorus starvation nor nitrogen starvation (Fig. 5A, V). Interestingly, 11 of the 12 proteins common to SSE induction and furanone-2 repression were found to be uniquely induced by carbon starvation, i.e., not induced by nitrogen or phosphorus starvation.
Vibrio sp. strain S14 cells starved in the presence of furanone-2 and SSE do not lose viability.
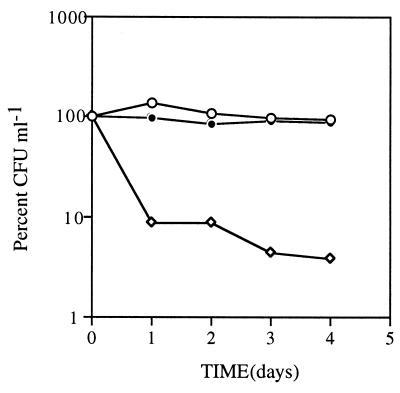
The viability of Vibrio sp. strain S14 cells starved in the presence of furanone-2 (5 μg ml−1) and SSE (50 μg ml−1) was determined to investigate if the components in the SSE were able to provide protection against the loss in viability caused by furanone-2. The hypothesis relies on the results from the 2D-PAGE analyses, which showed that there was a significant overlap of proteins that were induced by SSE and repressed by furanone-2, suggesting that both components were acting on the same regulatory pathway. When Vibrio sp. strain S14 was starved in the presence of furanone-2 (5 μg ml−1) and SSE (50 μg ml−1), there was a 13% drop in viability across the 4-day period (Fig. 7). On the contrary, the control, cells exposed to furanone-2 (5 μg ml−1) alone during the same period, exhibited a 94% reduction in viability, suggesting that the SSE had the ability to provide cross-protection against furanone-2 (Fig. 7).
FIG. 7.
Effect of the addition of SSE (50 μg ml−1) to Vibrio sp. strain S14 cells starved in the presence of furanone-2 (5 μg ml−1). The SSE and the furanone-2 were added to logarithmic-phase cells at time zero of carbon starvation. Open circles, control cultures starved in the absence of both furanone-2 and SSE; diamonds, cells exposed to furanone-2 (5 μg ml−1); closed circles, cells exposed to both furanone-2 and SSE. Experiments were repeated at least three times, each time in triplicate. The results are presented as average values of the triplicate data from a representative experiment.
Vibrio sp. strain S14 stationary-phase supernatants induce bioluminescence in V. harveyi.
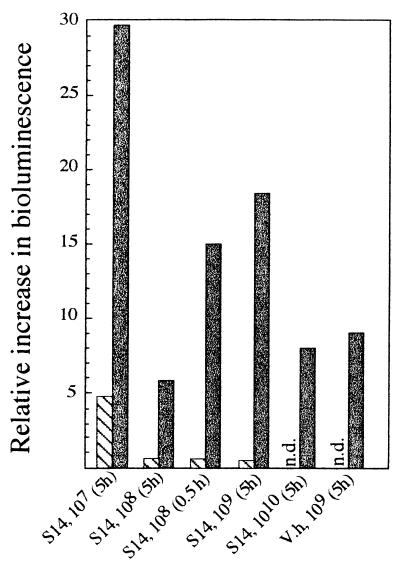
To investigate whether Vibrio sp. strain S14 excretes components that can mediate the expression of AHL-regulated systems, Vibrio sp. strain S14 cell-free supernatants were tested for the ability to induce bioluminescence in V. harveyi. The supernatant from 5-h carbon-starved Vibrio sp. strain S14 cells induced luminescence in V. harveyi as well as or better than the corresponding positive control (V. harveyi stationary-phase supernatants) (Fig. 8). This result was consistent regardless of whether the carbon starvation situation was achieved by allowing the culture to enter stationary phase by the means of glucose depletion or by washing a log-phase culture and resuspending the cells in MMM0 (data not shown). In these experiments, log-phase supernatants were not active or significantly less active than their stationary-phase counterparts (Fig. 8).
FIG. 8.
Fold increase in bioluminescence from V. harveyi after the addition of supernatant from 5- and 0.5-h carbon-starved (solid grey bars) and logarithmically growing (hatched bars) Vibrio sp. strain S14 (S14) and 5-h carbon-starved V. harveyi cells (V.h). Supernatants from Vibrio sp. strain S14 cells starved for 5 h were prepared from cultures allowed to run out of glucose at 3.5 × 107, 2.4 × 108, 2.5 × 109, and 6.5 × 1010 CFU ml−1, whereas the supernatant from 0.5-h starved cells was collected at 1.6 × 108 CFU ml−1. Supernatants from starved V. harveyi were collected from cultures allowed to run out of glucose at 4.9 × 109 CFU ml−1. Supernatants from logarithmically growing cells that were not tested are indicated by n.d. (not determined). Experiments were repeated three times, each time in triplicate. The results reported are average values of the triplicate data from a representative experiment.
There were no indications that samples from cell densities higher than 3.5 × 107 CFU ml−1 induced luminescence to a greater extent. The supernatant taken from a Vibrio sp. strain S14 stationary-phase culture, allowed to run out of glucose at a cell density of 3.5 × 107 CFU ml−1, was as active as the supernatants taken at cell densities of 108, 109, and 1010 CFU ml−1 (Fig. 8). No major difference was detected with regard to the time of starvation (0.5 or 5 h) before cells were removed to prepare the supernatant (Fig. 8).
Chemical analyses.
GC-MS was used to determine the presence of AHLs which could be assayed without derivatization. These AHLs (BHL, HHL, OHL, and DHL) were not detected by GC-MS at the limit of detection for quantitative analysis of BHL of 2 ng (i.e., 2 μl at 1 μg ml−1).
AHLs were not detected in the crude extract of the SSE of Vibrio sp. strain S14 by 1H or 13C NMR spectroscopy. The homoserine lactone ring has characteristic signals in the 1H and 13C NMR spectra for the nethine proton (δ 4.55) at C-2 (49.2 ppm) and the methylene protons (δ 4.28, 4.47) at C-4 (66.1 ppm) (4, 32). These signals were not detected in the extracts tested.
DISCUSSION
Many gram-negative bacteria, when subjected to starvation, become more resistant to various environmental challenges (21). The marine Vibrio sp. strain S14 can survive in a carbon and energy source-depleted environment for up to 6.5 years (28). This ability to keep the cellular machinery intact without access to an exogenous energy source is considered to reflect an extensive reorganization of the cellular makeup, initiated after the onset of carbon starvation. To address the possibility that the regulation of this process is partially mediated by signal molecules produced by Vibrio sp. strain S14, we determined the effect of SSE on protein expression. Several observations have recently been made providing evidence for the role of extracellular molecules or signals in adaptive responses similar to those of starvation and outgrowth (2, 11, 20, 25).
As evident from the global analysis, the addition of SSE during logarithmic growth preferentially influenced the synthesis rate of proteins classified as either induced or repressed upon carbon starvation (Fig. 2, III and IV; Fig. 6). Clearly, SSE addition during logarithmic growth altered the synthesis rates of the protein responders in analogy with the changes that were observed in response to carbon starvation (Fig. 2, II). The results presented in Fig. 2 indicate that not all [C.IND] or [C.REP] proteins were affected by SSE. It is probable that the efficacy of the compounds in the SSE to regulate some carbon starvation-regulated proteins may have been overridden by other more prevailing signals abundant in the cells, possibly as a result of the supply of glucose at the time of SSE addition. However, it is not expected that one regulator or signal mediates the expression of all carbon starvation responders. Therefore, the experimental design may underestimate the relative specificity of the SSE to regulate proteins normally regulated in response to carbon starvation.
In support of the observation that a signal molecule(s) with a role in the regulation of the carbon starvation response is produced and excreted upon starvation of Vibrio sp. strain S14, we found that the addition of SSE (50 μg ml−1) inhibited the growth rate in logarithmically growing cells. Conversely, addition of crude extract (50 μg ml−1) prepared from log-phase supernatants to log-phase cells caused a significantly less pronounced inhibition of growth, and the cells subsequently resumed normal growth (data not shown).
Furanones, which are acylated brominated lactones, are produced as secondary metabolites by the red macroalga D. pulchra. They specifically inhibit colonization in marine bacteria and have also been shown to specifically interfere with AHL and AHL-like molecule-regulated phenotypes. It has been proposed that the furanones compete with AHL (or AHL-like) signals for binding to the regulatory receptor site. The concentrations of the furanones used to inhibit the AHL and AHL-like molecule-regulated phenotypes is highly species dependent and varies from 5 to 100 μg ml−1. These furanone concentrations do not inhibit the growth rate of the organism while affecting the specific phenotype (12, 13, 22, 24).
The data obtained in our studies demonstrate that the presence of furanone-2 at a concentration that did not affect the growth rate (5 μg ml−1) (Fig. 3) leads to impaired viability during carbon starvation. Additionally, cells incubated with furanone-2 for 24 h during carbon starvation were unable to develop cross-protection against UV irradiation and H2O2 exposure (Fig. 4). These observations suggest that the reduction in viability is a result of the interference by furanone-2 with a regulatory system that is essential for successful adaptation to starvation. Cells starved in the presence of furanone-3, on the other hand, were considerably less impaired in viability (Fig. 3). This gives further evidence that the loss in viability caused by furanone-2 cannot be attributed to a general toxicity effect, especially considering that the two molecules are structurally similar (Fig. 1).
The possibility that furanone-2 can interfere with a regulatory system critical in the response to starvation was further supported by several results obtained from the global analysis of protein expression upon carbon starvation. Thirty percent of the proteins that were identified as carbon starvation induced were shown not to be induced, or to be significantly less induced, when furanone-2 was included in the starvation regimen. In contrast, the fraction of proteins that were repressed among those identified as constitutively expressed upon starvation amounted to 8% (Fig. 5A, II and III). Furthermore, 17% of the proteins identified as carbon starvation repressed were overexpressed when furanone-2 was included in the starvation regimen, whereas the fraction of overexpressed proteins among those identified as constitutively expressed upon starvation was as low as 4% (Fig. 5A, II and III). This finding suggests that the postulated regulatory system inhibited by furanone-2 plays a role in the regulation of a subpopulation of the set of proteins induced and repressed by carbon starvation.
The experimental approach that we used does not directly discriminate between whether the effects seen on the rate of synthesis of proteins that do not belong to the carbon starvation stimulon are indirectly caused by the effects on the expression of proteins within the carbon starvation stimulon or if furanone-2 directly interferes with separate regulatory systems. If the latter model is correct, it is anticipated that the rate of synthesis of the proteins that do not belong to the carbon starvation stimulon should also be altered as a result of the addition of furanone-2 during logarithmic growth. As only three proteins are affected by furanone-2 during exponential phase (Fig. 5B), the role of the system interfered with by furanone-2 is suggested to be restricted to the regulation of phenotypes expressed upon carbon starvation. The majority of the carbon starvation-induced proteins down-regulated by furanone-2 were not induced by either nitrogen or phosphorus starvation (Fig. 5A, V). Therefore, the effect caused by furanone-2 is unlikely to be due to a mechanism generally activated upon growth arrest. As we have previously shown that neither nitrogen nor phosphorus starvation promotes the development of starvation- and stress-resistant cells (16, 27), this finding is in accordance with the reduced viability of starved cells exposed to furanone-2.
Of the 37 proteins down-regulated by furanone-2, 12 were found to be common to the 31 carbon starvation-induced proteins up-regulated by SSE. This finding clearly suggests that the effects seen upon the addition of furanone-2 as well as of SSE are associated with the action of a specific regulatory mechanism involved in the regulation of carbon starvation-induced proteins. This hypothesis, that both furanone-2 and SSE act on the same regulatory pathway, is further strengthened by the results showing that the presence of SSE at 50 μg ml−1 is able to override the negative effect on viability caused by furanone-2 at 5 μg ml−1. Interestingly, 11 (92%) of the 12 proteins that were up-regulated by SSE and down-regulated by furanone-2 were found to belong to a core set of the carbon starvation stimulon which includes proteins that are not induced by nitrogen or phosphorus starvation. Given that neither nitrogen nor phosphorus starvation promotes increased starvation and stress resistance, this finding may account for the reduced viability observed when Vibrio sp. strain S14 was starved in the presence of furanone-2.
The induction of bioluminescence in V. harveyi by the addition of cell-free cultures shows that Vibrio sp. strain S14 selectively, upon carbon starvation, produces a molecule(s) that induces the AHL-regulated phenotype in V. harveyi. Moreover, this putative signal is produced and secreted in amounts sufficiently high to trigger the bioluminescence, even if the cell density does not exceed 3.5 × 107 cells ml−1 (Fig. 8). These findings indicate that the production of the signal in Vibrio sp. strain S14 is not explicitly dependent on a positive feedback mechanism, as would be the case in quorum-sensing signalling systems, but rather suggest that the synthesis is controlled by some other starvation-specific signals. Huisman and Kolter (18) proposed that ppGpp may serve as one such signal. In view of the experimental procedure used to prepare SSE, and the fact that stationary-phase but not log-phase supernatant induced bioluminescence in V. harveyi, it is further implied that the regulatory signal is a nonpolar molecule(s) that is excreted upon carbon starvation. In addition, stationary/starvation-phase cell-free cultures from Vibrio sp. strain S14 have the ability to induce swarming in an swrI (homologous to luxI) mutant strain of Serratia liquefaciens (data not shown). This finding further suggests that Vibrio sp. strain S14 produces and secretes a molecule capable of interfering with AHL-regulated systems. To test the hypothesis that HSL might be the intracellular signal, as has been suggested for E. coli (18), we exposed logarithmically growing cultures to up to 10 mM commercially available HSL (Sigma). However, the nonacylated homoserine lactone did not cause any reduction in the growth rate (data not shown).
The identity of the proposed signal molecule(s) produced by Vibrio sp. strain S14 during starvation requires further studies. Data obtained from 1H NMR, 13C NMR, and GC-MS analyses suggest that no AHLs are excreted by Vibrio sp. strain S14. However, other low-molecular-weight compounds (e.g., cyclic dipeptides) have been found to interfere with AHL-regulated phenotypes (4). Hence, it is probable that Vibrio sp. strain S14 produces a molecule(s) that is not AHL but has the capability to interfere with the AHL-regulated systems. We are presently using bioassay-guided fractionations and chemical analyses in an attempt to isolate the putative signal molecules in Vibrio sp. strain S14.
The role of extracellular molecules in adaptive responses in nondifferentiating bacteria has been largely unexplored. Although numerous microorganisms have been shown to produce extracellular molecules as quorum sensors, there have been very few reports of studies in which small molecules have been important irrespective of cell density. This study provides evidence that one or several extracellular metabolites synthesized upon carbon and energy starvation may play a pivotal role in the regulation of proteins crucial to the development of phenotypes that confer starvation and stress resistance in Vibrio sp. strain S14.
ACKNOWLEDGMENTS
This study was supported by grants from the Australian Research Council, the Swedish National Environmental Protection Agency (EU Biotechnology Programme), and the Centre for Marine Biofouling and Bio-Innovation (The University of New South Wales, Sydney, Australia).
REFERENCES
- 1.Albertson N H, Nyström T, Kjelleberg S. Exoprotease activity of two marine bacteria during starvation. Appl Environ Microbiol. 1990;56:218–223. doi: 10.1128/aem.56.1.218-223.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batchelor S E, Cooper M, Chhabra S R, Glover L A, Stewart G S A B, Williams P, Prosser J I. Cell density-regulated recovery of starved biofilm populations of ammonia-oxidizing bacteria. J Bacteriol. 1997;63:2281–2286. doi: 10.1128/aem.63.6.2281-2286.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibb M. The regulation of antibiotic production in Streptomyces coelicolor A3(2) Microbiology. 1996;142:1335–1344. doi: 10.1099/13500872-142-6-1335. [DOI] [PubMed] [Google Scholar]
- 4.de Nys, R. 1997. Unpublished data.
- 5.de Nys R, Wright A D, Konig G M, Sticher O. New halogenated furanones from the marine alga Delisea pulchra (cf. fimbriata) Tetrahedron. 1993;49:11213–11220. [Google Scholar]
- 6.Downard J, Toal D. Branched-chain fatty acids: the case for a novel form of cell-cell signalling during Myxococcus xanthus development. Mol Microbiol. 1995;16:171–175. doi: 10.1111/j.1365-2958.1995.tb02290.x. [DOI] [PubMed] [Google Scholar]
- 7.Flärdh K, Axberg T, Albertson N H, Kjelleberg S. Stringent control during carbon starvation of marine Vibrio sp. strain S14: molecular cloning, nucleotide sequence, and deletion of the relA gene. J Bacteriol. 1994;176:5949–5957. doi: 10.1128/jb.176.19.5949-5957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flärdh K, Kjelleberg S. Glucose upshift of long-term carbon-starved marine Vibrio strain sp. S14 causes severe amino acid starvation and induction of the stringent response. J Bacteriol. 1994;176:5897–5903. doi: 10.1128/jb.176.19.5897-5903.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuqua C, Winans S, Greenberg E. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 10.Fuqua W, Winans S, Greenberg P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Lara J, Shang L H, Rothfield L I. An extracellular factor regulates expression of sdiA, a transcriptional activator of cell division genes in Escherichia coli. J Bacteriol. 1996;178:2742–2748. doi: 10.1128/jb.178.10.2742-2748.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Givskov M, de Nys R, Manefield M, Gram L, Maximillen R, Eberl L, Molin S, Steinberg P D, Kjelleberg S. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J Bacteriol. 1996;178:6618–6622. doi: 10.1128/jb.178.22.6618-6622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gram L, de Nys R, Maximillen R, Givskov M, Steinberg P D, Kjelleberg S. Inhibitory effects of secondary metabolites from the red alga Delisea pulchra on swarming motility of Proteus mirabilis. Appl Environ Microbiol. 1996;62:4284–4287. doi: 10.1128/aem.62.11.4284-4287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray K M. Intercellular communication and group behavior in bacteria. Trends Microbiol. 1997;5:184–188. doi: 10.1016/S0966-842X(97)01002-0. [DOI] [PubMed] [Google Scholar]
- 15.Hoben H J, Somsegaran P. Comparison of the pour, spread, and drop plate methods for enumeration of Rhizobium spp. in inoculants made from presterilized peat. Appl Environ Microbiol. 1982;44:1246–1247. doi: 10.1128/aem.44.5.1246-1247.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmquist L, Kjelleberg S. Changes in viability, respiratory activity and morphology of the marine Vibrio sp. strain S14 during starvation for individual nutrient and subsequent recovery. FEMS Microbiol Ecol. 1993;12:215–224. [Google Scholar]
- 17.Horinuchi S, Beppu T. Autoregulatory factors and communication in Actinomycetes. Annu Rev Microbiol. 1992;46:377–398. doi: 10.1146/annurev.mi.46.100192.002113. [DOI] [PubMed] [Google Scholar]
- 18.Huisman G W, Kolter R. Sensing starvation: a homoserine-lactone-dependent signaling pathway in Escherichia coli. Science. 1994;265:537–539. doi: 10.1126/science.7545940. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan H, Plamann L. A Myxococcus xanthus cell density-sensing system required for multicellular development. FEMS Microbiol Lett. 1996;139:89–95. doi: 10.1111/j.1574-6968.1996.tb08185.x. [DOI] [PubMed] [Google Scholar]
- 20.Kaprelyants A S, Kell D. Do bacteria need to communicate with each other for growth? Trends Microbiol. 1996;4:237–242. doi: 10.1016/0966-842X(96)10035-4. [DOI] [PubMed] [Google Scholar]
- 21.Kjelleberg S. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. [Google Scholar]
- 22.Manefield, M., L. Gram, L. Harris, and S. Kjelleberg. 1997. Unpublished data.
- 23.Mårdén P, Nyström T, Kjelleberg S. Uptake of leucine by a marine Gram-negative heterotrophic bacterium during exposure to starvation conditions. FEMS Microbiol Ecol. 1987;45:233–241. [Google Scholar]
- 24.Maximillen, R., R. de Nys, C. Holmström, L. Gram, M. Givskov, K. Crass, S. Kjelleberg, and P. D. Steinberg. 1997. Unpublished data.
- 25.Nikolaev Y A. Involvement of exometabolites in stress adaptation of Escherichia coli. Microbiology. 1997;66:28–31. [Google Scholar]
- 26.Nyström T. The glucose starvation stimulon of Escherichia coli: induced and repressed synthesis of enzymes of central metabolic pathways and role of acetyl phosphate in gene expression and starvation survival. Mol Microbiol. 1994;12:833–843. doi: 10.1111/j.1365-2958.1994.tb01069.x. [DOI] [PubMed] [Google Scholar]
- 27.Nyström T, Olsson R M, Kjelleberg S. Survival, stress resistance, and alterations in protein expression in the marine Vibrio sp. strain S14 during starvation for different individual nutrients. Appl Environ Microbiol. 1992;58:55–65. doi: 10.1128/aem.58.1.55-65.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Östling J. Ph.D thesis. Göteborg, Sweden: Göteborg University; 1996. [Google Scholar]
- 29.Östling J, Flärdh K, Kjelleberg S. Isolation of a carbon starvation regulatory mutant in a marine vibrio. J Bacteriol. 1995;177:6978–6982. doi: 10.1128/jb.177.23.6978-6982.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Östling J, Goodman A, Kjelleberg S. Behaviour of IncP-1 plasmids and a miniMu transposon in a marine Vibrio sp.: isolation of starvation inducible lac operon fusions. FEMS Microbiol Ecol. 1991;86:83–94. [Google Scholar]
- 31.Östling J, Holmquist L, Kjelleberg S. Global analysis of the carbon starvation response of a marine vibrio disrupted in genes homologous to relA and spoT. J Bacteriol. 1996;178:4901–4908. doi: 10.1128/jb.178.16.4901-4908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schripsema J, de Rudder K E E, van Vliet T B, Lankhorst P P, de Vroom E, Kijne J W, van Brussel A A N. Bacteriocin small of Rhizobium leguminosarum belongs to the class of N-acyl-l-homoserine lactone molecules, known as autoinducers and as quorum sensing co-transcription factors. J Bacteriol. 1996;178:366–371. doi: 10.1128/jb.178.2.366-371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shiga Y, Mizuno H, Akanuma H. Conditional synthesis and utilization of 1,5-anhydroglucitol in Escherichia coli. J Bacteriol. 1993;175:7138–7141. doi: 10.1128/jb.175.22.7138-7141.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swift S, Bainton N, Winson M. Gram-negative bacterial communication by N-acyl homoserine lactones: a universal language? Trends Microbiol. 1994;2:193–198. doi: 10.1016/0966-842x(94)90110-q. [DOI] [PubMed] [Google Scholar]
- 35.Swift S, Throup J P, Williams P, Salmond G P C, Stewart G S A B. Quorum sensing: a population-density component in the determination of bacterial phenotype. Trends Biochem Sci. 1996;21:214–219. [PubMed] [Google Scholar]
- 36.Zabkar, S. (University of New South Wales, Sydney, Australia). 1997. Personal communication.