Abstract
The G protein-coupled receptor ADGRE5 (CD97) binds to various metabolites that play crucial regulatory roles in metabolism. However, its function in the antiviral innate immune response remains to be determined. In this study, we report that CD97 inhibits virus-induced type-I interferon (IFN-I) release and enhances RNA virus replication in cells and mice. CD97 was identified as a new negative regulator of the innate immune receptor RIG-I, and RIG-1 degradation led to the suppression of the IFN-I signaling pathway. Furthermore, overexpression of CD97 promoted the ubiquitination of RIG-I, resulting in its degradation, but did not impact its mRNA expression. Mechanistically, CD97 upregulates RNF125 expression to induce RNF125-mediated RIG-I degradation via K48-linked ubiquitination at Lys181 after RNA virus infection. Most importantly, CD97-deficient mice are more resistant than wild-type mice to RNA virus infection. We also found that sanguinarine-mediated inhibition of CD97 effectively blocks VSV and SARS-CoV-2 replication. These findings elucidate a previously unknown mechanism through which CD97 negatively regulates RIG-I in the antiviral innate immune response and provide a molecular basis for the development of new therapeutic strategies and the design of targeted antiviral agents.
Keywords: CD97, RNA virus, IFN-I, Ubiquitination, RIG-I
Subject terms: Infection, RIG-I-like receptors, Viral infection
Introduction
Zoonotic infectious diseases caused by recently discovered viruses have become important public health problems that seriously threaten the health and life of human beings. Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), which is transmitted among a variety of animals and is spreading throughout the world [1]. Particularly among elderly COVID-19 patients, severe respiratory infections, including pneumonia and acute respiratory distress syndrome (ARDS), substantially increases the risk of death [2]. However, the pathogenic mechanism underlying rapid viral infection is not yet fully understood, and few targeted therapies are available for these diseases. Therefore, identifying key molecules that reduce viral replication offers novel insights for the development of new broad-spectrum drugs, which is an important and effective strategy to combat sudden infectious diseases.
Sufficient evidence demonstrates that IFN-I and interferon-stimulated genes (ISGs), which are upregulated via an innate immune signaling pathway, are key molecules in clearing invasive viruses [3, 4]. The regulatory mechanisms underlying the production of IFN-I and host targeting of viruses for degradation are important but remain largely unknown. IFN-I signaling pathway activation depends on the identification of pathogen-associated molecular patterns (PAMPs) by host pattern recognition receptors (PRRs) [5, 6]. There are several types of PRRs, including RIG-I-like receptors (RLRs) and melanoma differentiation-associated gene 5 (MDA5) [7]. RIG-I contains a tandem caspase-recruiting domain (CARD), which is responsible for initiating downstream signaling, and a central DExD/H box RNA helicase domain, which is responsible for recognizing viral RNA [8, 9]. RIG-I undergoes a conformational change and is recruited to the mitochondrial antiviral signaling (MAVS) protein through the CARD domain when pathogenic RNA binds to a helicase domain. MAVS activates downstream adaptors, including TANK binding kinase 1 (TBK1)/inhibition of IκB kinase ε (IKKε), interferon regulatory factor 3 (IRF3), and nuclear factor κB (NF-κB), which stimulate the production of proinflammatory cytokines and upregulate the expression of IFN-I to defend against viral infection [10]. Precise regulation of RIG-I signaling is crucial for effective viral clearance without pathogenic immunopathology.
Multiple viruses have evolved strategies to suppress the host innate immune system by targeting the degradation of PRRs or adaptors to promote viral proliferation. RIG-I can be downregulated by foot-and-mouth disease virus (FMDV) or influenza A virus (IAV), leading to a decrease in IFN-I production [11, 12]. Several types of posttranslational modifications, such as phosphorylation [13, 14], SUMOylation [15], and polyubiquitination [16], modulate RIG-I activation and impact RIG-I function during viral infection. Notably, an increasing number of studies have focused on the regulation of RIG-I functions by the ubiquitination of posttranslational modifications, which modify the IFN-I signaling response. Multiple E3 ubiquitin ligases, such as MEX3C [17], REUL [18, 19], TRIM25 [20], and TRIM4 [21], modulate the K63-linked polyubiquitination of RIG-I to positively regulate the IFN-I signaling pathway. Conversely, c-Cbl [22], CHIP [23], RNF122 [24], and RNF125 [25] negatively regulate RIG-I by catalyzing the K48-linked polyubiquitination of RIG-I-mediated signaling and promoting its proteasomal degradation. However, some host proteins, such as G3BP1 and Riok3, play dual roles and can enhance immune responses while facilitating viral replication by degrading RIG-I via E3 ligase-mediated ubiquitination after viral infection [26, 27]. However, the regulatory mechanisms involved in RIG-I degradation-mediated antiviral innate immune responses remain to be investigated. The discovery of host genes that influence viral replication will provide new insights for the design of targeted antiviral drugs.
CD97 is a member of the adhesion G protein-coupled receptor (AGPCR) family, a group of receptors characterized by seven-transmembrane (TM7) domains. AGPCRs are widely expressed in various species and play diverse roles in cell adhesion and signaling [28]. The AGPCR family includes subfamilies such as ADGRE, ADGRB, and ADGRG, with CD97 belonging to the ADGRE subfamily. CD97 is composed of extensive extracellular domains, encompassing numerous protein-folding regions and para-end GPCR autoproteolysis-inducing (GAIN) domains. It also consists of a TM7 region and an intracellular domain. The GAIN domain facilitates automatic catalytic cleavage at the GPCR proteolysis site (GPS), leading to the formation of an extracellular N-terminal fragment (NTF) and an intracellular C-terminal fragment (CTF). NTF and CTF cooperate to participate in receptor mediation cell function [29]. The extracellular domains of CD97 can interact with various ligands, including CD55 [30], chondroitin sulfate [31], CD90 [32], and integrin [33], to initiate intracellular signaling pathways. CD97 is widely expressed in both immune and nonimmune cells [34]. It plays a crucial role in promoting spleen dendritic cell homeostasis and stabilizing the immunological synapse between dendritic cells and T cells [35, 36]. However, the full extent of its role in antiviral innate immunity and defense against viruses remains unexplored. In the current study, we discovered that CD97 suppresses IFN-I production by promoting RNF125-mediated RIG-I degradation after RNA virus infection to promote viral replication in vitro and in mouse models in vivo. Our research revealed that CD97 negatively regulates IFN-I synthesis during the innate immune response to viral infection, a previously unknown function.
Materials and Methods
Mice
B6;129P2-Adgre5tm1Dgen/JC57BL/6J mice [37] were purchased from the Jackson Laboratory (JAX stock code 005788). Genotyping was performed using an optimized JAX protocol (28954). The following primers were used: moIMR0008, 5′-GACGAGTTCTTCTGAGGGGATCGATC-3′; moIMR0789, 5′-ATGGTCAGCTTGAATTAGCCAGTTG-3′; and moIMR0790, 5′-AGCAGCGGGGCTCCATCAGGTTCTG′.
Viral infection
Six- to eight-week-old mice were inoculated with VSV via intraperitoneal injection for in vivo infection as previously described [38]. Mouse weight was monitored for 14 days after intraperitoneal injection of VSV (2 × 107 PFU per mouse), and survival was monitored for 7 days after intraperitoneal injection of VSV (1 × 108 PFU per mouse). Mice were intranasally inoculated with SARS-CoV-2-susceptible mouse strains (500 TCID50 per mouse) via intraperitoneal injection. qPCR was performed to measure the expression of IFN-β, IFN-α4, VSV-G, and SARS-CoV-2, as determined by copy number in organs. Samples from virus-infected mice were used for H&E staining at the indicated time points. The levels of the IFN-β protein in VSV-infected mice were measured using an IFN-β ELISA detection kit.
Cell culture and preparation of macrophages
For peritoneal-derived macrophage acquisition, peritoneal lavage was collected from the mice after their death, peritoneal macrophages were isolated, and the volume of the samples was adjusted to 10 mL with medium. A total of 2 × 106 cells were cultured in 6-well culture plates in RPMI 1640 medium containing 10% fetal bovine serum and antibiotics (100 mg/mL streptomycin and 100 U/mL penicillin). As previously mentioned, nonadherent cells were eliminated by thorough washing with RPMI 1640 medium [39]. For the acquisition of bone marrow-derived macrophages, the femur and tibia excised from mice were separated, and the bone marrow was flushed out of the bones with cold RPMI 1640 medium. The RPMI 1640 medium containing the bone marrow was filtered through a 70-μm sieve and centrifuged twice. The supernatant was discarded, and the activated macrophages were cultured in RPMI 1640 medium containing 25 ng/mL GM-CSF. Seven days after induction, the plate was gently scraped with a cell scraper, and the cells were used in subsequent experiments.
SiRNA silencing and qRT‒PCR
SiRNA (final concentration: 30 pM) was transfected with reagent provided by the manufacturer into Caco-2 cells. Every siRNA sequence was prepared by Beijing Qingke Biotechnology Co., Ltd. The siRNA sequences are shown in Table S1. As explained by Hou et al. [40], RT‒qPCR (qPCR) was used to measure the levels of mRNA. The 2-△△CT method was used to calculate the relative fold change in mRNA expression. All primers were designed by Primer Premier 6.0 software. The primer sequences are shown in Table S1.
Immunoprecipitation and immunoblotting
The indicated plasmids were transfected into HEK-293T or HeLa cells for 36 h. PBS-washed cells were then resuspended in commercial lysis buffer on ice for 20 min. Immunoprecipitation was performed using anti-Flag or anti-HA magnetic beads at 4 °C for 2 h. For IP of endogenous proteins, commercial antibody-bound magnetic beads were used. Then, three lysis buffer washes of the beads were performed. Using SDS loading buffer, the immunoprecipitates were eluted and then denatured for 10 min at 95 °C. Proteins that had been extracted were electrophoresed in a 10% SDS gel and then transferred to polyvinylidene difluoride (PVDF) membranes. Tris-buffered saline-Tween (TBST) with 5% nonfat dry milk was used to block the blots. Primary antibodies against the targeted proteins were used. As previously described, the membranes were washed, and then, the manufacturer′s instructions were followed to visualize the membranes using enhanced chemiluminescence reagents. Information on the antibodies used in this study is listed in Table S2.
Dual-luciferase reporter assay
HEK-293T (2 × 105) cells were seeded in 24-well plates and transfected with pRL-TK (a Renilla luciferase plasmid; 10 ng), firefly luciferase (100 ng), and various concentrations of the appropriate control or protein-expressing plasmid(s). The cells were collected 24 h after transfection, and the manufacturers’ recommendations were followed to measure the firefly and Renilla luciferase activities using a dual-luciferase reporter assay system kit and a SpectraMax M5 microplate reader. The relative fluorescence intensity of firefly luciferase was normalized against that of Renilla luciferase.
Generation of stable CD97-expressing or knockout cell lines
We used a four-plasmid lentiviral packaging system consisting of the eukaryotic expression vectors pLVX-IRES-puro/lentiCRISP v2, pLP1, pLP2, and pLP/VSV-G to stably express or knock out CD97 in HeLa cells. The vector pLVX-IRES-puro carries a Flag tag on the C-terminus, and primers were created to clone CD97 (GenBank no. NM_078481.4) into this plasmid. Gene editing with CRISPR‒Cas9 was used to produce CD97-KO cells, which were purchased from CRISPR Direct (http://crispr.dbcls.jp). Stable cell lines and lentiviruses were generated using the methods described in earlier studies [39].
Immunofluorescence assays
As previously described [41], transfected cells were cultured on coverslips before being fixed with 4% paraformaldehyde and after being washed twice with ice-cold PBS. Following fixation, the cells were permeabilized with PBS containing 0.2% Triton X-100 and washed three times with PBS. The cell nuclei were then counterstained with DAPI, and the cells were visualized using a Leica SP8 confocal microscope.
Viral infection in vitro
To measure gene and signaling molecule expression, cells were infected with VSV and SARS-CoV-2. As previously described [42], one hour after viral infection in serum-free medium, the culture supernatant was collected, and the cells were transferred back to the medium with 2% fetal bovine serum (FBS). At the indicated time, total cells and/or culture medium were harvested for viral titration, qPCR, immunoblotting, or immunofluorescence assays.
Ethics statement
All animal experimentation was performed strictly following the State Council-approved People’s Republic of China Regulations on the Administration of Experimental Animal Affairs (1 November 1988). The mice were handled according to the Shandong Normal University Institutional Animal Care and Use Committee (IACUC) policies on animal use. The work with live SARS-CoV-2 virus was performed inside biosafety cabinets in the ABSL-3 biological safety laboratory at the Changchun Veterinary Research Institute.
Quantification and statistical analysis
Statistical analysis was performed using GraphPad Prism software (version 8.0; GraphPad, San Diego, CA, USA). Statistical significance was determined via two-way analysis of variance (ANOVA), and one-way ANOVA was performed to compare multiple samples. The results from at least three independent experiments are presented as the mean and standard deviation (SD), and a P value of 0.05 or lower was regarded as statistically significant.
Results
CD97 promotes virus replication in vitro
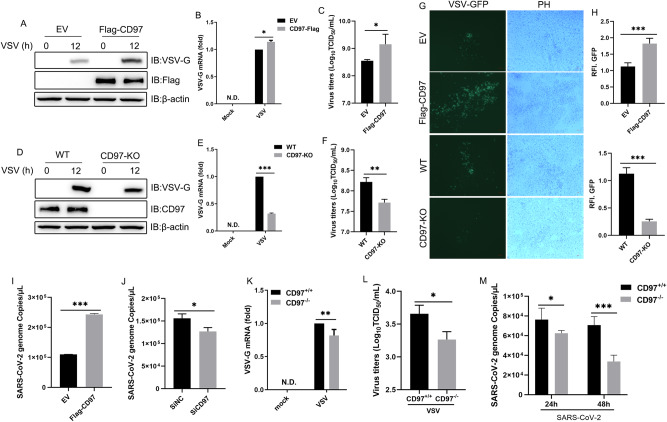
CD97 functions as a G protein-coupled receptor and is extensively expressed in both immune and nonimmune cells [34]. However, it is unclear whether CD97 plays a role in the host innate immune response against viral infection. To investigate the biological function of CD97, we first constructed CD97-overexpressing HeLa cells using a lentivirus packaging system. Immunoblotting analysis demonstrated the stable expression of Flag-CD97 in the CD97-overexpressing cell line, which was validated using a commercial Flag antibody (Fig. S1A). Although the expected CD97 band with a size of 70–100 kDa could be detected, we also observed a significantly larger and unexpected molecular weight band of Flag-CD97, exceeding 150 kDa (Fig. S1A). To validate the accuracy of the CD97 recombinant vector, we constructed a vector with the same sequence, incorporating a GFP tag. Confocal microscopy revealed that compared to the control, the GFP-CD97 vector predominantly localized to the cytomembrane rather than diffusing throughout the entire cell (Fig. S1B), which is consistent with a previously reported study [43]. In addition, many CD97-positive HeLa cells were found in CD97-overexpressing cell lines by flow cytometry using a CD97 monoclonal antibody (Fig. S1D). Furthermore, we employed flag magnetic beads to precipitate Flag from HeLa cells and analyzed the protein through mass spectrometry, which confirmed the overexpression of CD97 corresponding to the band (data not shown). Therefore, we speculated that exogenous CD97 expression might exist in the form of dimers or multimers, resembling the characteristics observed in reported G protein-coupled receptors [44]. CD97-overexpressing HeLa cells were utilized to evaluate the levels of VSV. Enhanced protein and mRNA levels of VSV-G were observed in Flag-CD97 cells compared to empty vector cells (Fig. 1A, B). Then, viral titration measured at the 50% tissue culture infectious dose (TCID50) per milliliter of VSV-infected cells was markedly increased in Flag-CD97 cells (Fig. 1C). To validate the critical role of CD97 in viral replication, we generated HeLa cells with CD97 knockout using the CRISPR/Cas9 system. The knockout of CD97 was confirmed through both immunoblot analysis and flow cytometry (Fig. S1C and E). In contrast to the outcome observed with overexpression, CD97 knockout markedly decreased VSV-G protein and mRNA expression levels and viral titration (Fig. 1D–F). Furthermore, CD97-promoted VSV replication was proven with VSV-GFP fused with green fluorescent protein expression by using a fluorescence microscope (Fig. 1G, H). In line with the findings that CD97 promotes VSV replication, the number of SARS-CoV-2 genome copies was increased by CD97 in Caco-2 cells (Fig. 1I, J). Subsequently, we evaluated the replication of VSV and SARS-CoV-2 (mouse-susceptible C57MA14 strain) in peritoneal-derived macrophages (PMs) isolated from CD97-knockout (CD97−/−) mice and their wild-type counterparts (CD97+/+ mice). The CD97-knockout mouse model had been previously generated and studied [37], and we confirmed the homozygous status of these mice through PCR analysis (Fig. S1F). Uniformly, a lower viral replication rate was observed in VSV- and SARS-CoV-2-infected CD97−/− PMs than in CD97+/+ PMs (Fig. 1K‒M), indicating that CD97 potentially mediates RNA virus replication. Collectively, these results demonstrated that CD97 facilitated RNA virus replication in vitro.
Fig. 1.
CD97 promotes RNA virus replication in vitro. A and B Immunoblot and qPCR analysis of VSV-G in VSV (MOI = 1)-infected HeLa cells after CD97 lentivirus packaging overexpression and D and E CD97 knockout. C and F Analysis of virus titers based on the TCID50 after VSV (MOI = 1) infection for 24 h in CD97-overexpressing and CD97-knockout cells. G Phase-contrast (PH) and fluorescence microscopy analysis of the cells infected with VSV-GFP (MOI = 1) for 24 h. Scale bars: 100 µm. H Quantitative analysis of the relative fluorescence intensity (RFI) of GFP shown in (G) using ImageJ software. I and J q-PCR analysis of the number of SARS-CoV-2 genome copies in SARS-CoV-2 (MOI = 0.1)-infected CD97-overexpressing and CD97-knockdown Caco-2 cells infected for 48 h. K and L q-PCR and TCID50 assay of the indicated gene levels and virus titers, respectively, of CD97+/+ and CD97−/− PMs after VSV infection. M qPCR analysis of the number of SARS-CoV-2 genome copies in PMs. The mean and SD from three independent experiments are shown. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001
CD97 negatively regulates virus-triggered IFN-I signaling
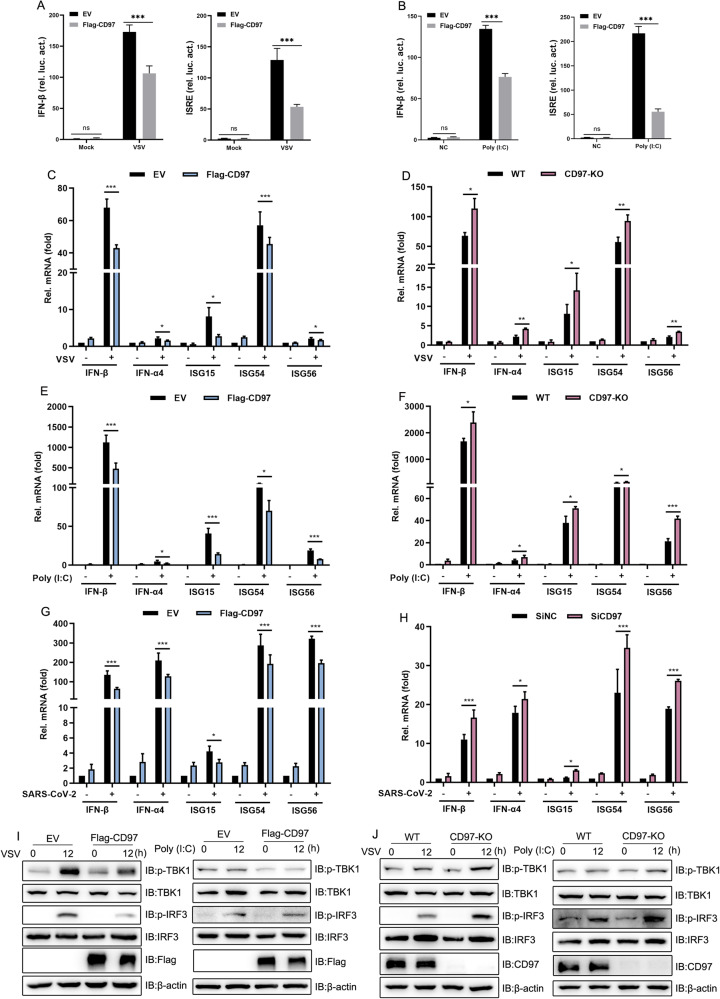
To further understand the antiviral function of CD97, we investigated the impact of CD97 on the virus-induced IFN-I response. Above all, IFN-β and ISRE (interferon-stimulated response element) luciferase reporters were employed to evaluate the activation of IFN-I signaling through VSV infection and RNA virus analog poly(I:C) stimulation. As shown in Figs. 2A, B, IFN-β and ISRE luciferase activity was significantly inhibited by CD97. Furthermore, CD97-overexpressing HeLa cells were found to produce significantly lower levels of IFN-β, IFN-a4, ISG15, ISG56, and ISG54 transcripts than control HeLa cells after VSV infection and poly(I:C) stimulation (Fig. 2C and E). In contrast, IFN and ISG transcript levels were notably increased in CD97-deficient HeLa cells (Fig. 2D and F). Subsequently, similar trends in IFN and ISG transcript levels were found in SARS-CoV-2-infected Caco-2 cells (Fig. 2G, H), in which CD97 was overexpressed or knocked down, respectively.
Fig. 2.
CD97 inhibits IFN-I production induced by viral infection. A and B Luciferase reporter assays were conducted with HEK-293T cells transfected for 24 h with IFN-β, ISRE, pRL-TK, CD97, or control plasmids and then infected with VSV or stimulated with poly(I:C) (10 μg/mL) for 12 h. C and D qPCR analysis of the indicated gene levels in CD97-overexpressing or CD97- knockout HeLa cells infected with VSV (MOI = 1) for 12 h. E and F qPCR analysis of the indicated genes in CD97-overexpressing or CD97-knockout HeLa cells stimulated with poly(I:C) for 12 h. G and H CD97-overexpressing and CD97-knockdown Caco-2 cells infected with SARS-CoV-2 (MOI = 0.1) for 48 h. The mean and SD from three independent experiments are shown. *P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001. I and J Immunoblot analysis of the indicated protein in CD97-overexpressing and CD97-knockout HeLa cells infected with VSV and stimulated with poly(I:C) (10 μg/mL) for 12 h
Furthermore, repressed IFN-I signaling, as indicated by the low phosphorylation rate of both TBK1 (Ser172) and IRF3 (Ser386), was observed in CD97-overexpressing cells after VSV infection or poly(I:C) transfection (Fig. 2I), and the opposite results were found when CD97 expression was knocked out (Fig. 2J). Altogether, these data suggested that CD97 negatively regulated RNA virus-triggered antiviral IFN-I signaling responses.
CD97-deficient primary macrophages promote IFN-I signaling pathway activation
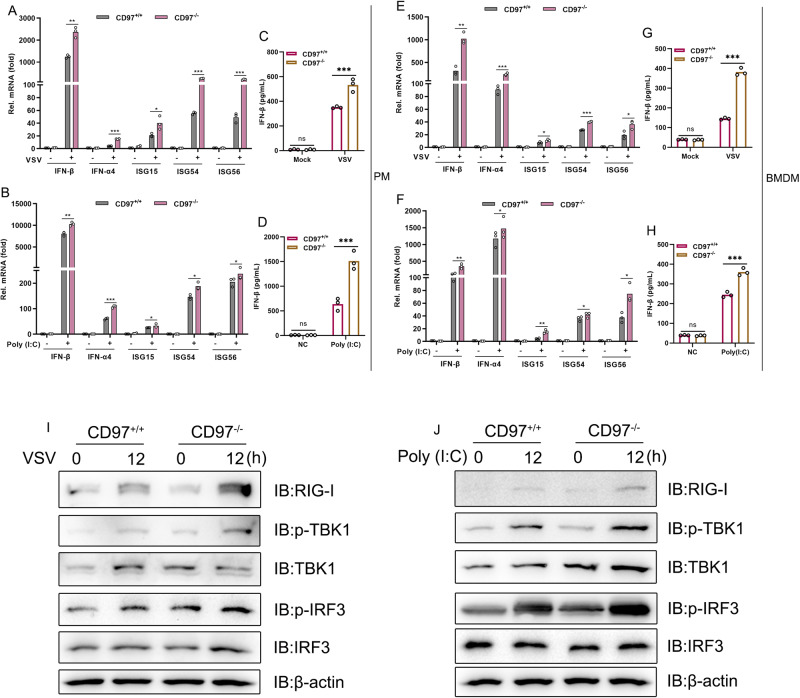
Next, the functional importance of CD97 in the IFN-I signaling pathway in primary macrophages was characterized. PMs and bone marrow-derived macrophages (BMDMs) were successfully isolated from CD97+/+ and CD97−/− littermate mice and challenged with viruses and virus analogs. A much higher number of IFN-β, IFN-α4, ISG15, ISG54, and ISG56 transcripts were found in the CD97−/− PMs than in the CD97+/+ PMs after VSV infection (Fig. 3A) and after poly(I:C) stimulation (Fig. 3B), and similar rates of IFN-β production were found (Fig. 3C, D). Similar results were observed with BMDMs (Fig. 3E–H). Subsequently, CD97−/− PMs exhibited significantly increased phosphorylation of TBK1 (Ser172) and IRF3 (Ser386) compared with CD97+/+ PMs (Fig. 3I, J). Altogether, these data demonstrated that CD97 negatively regulated RNA virus-induced IFN-I signaling in primary macrophages.
Fig. 3.
CD97 inhibits IFN-I signaling pathway activation induced by virus or virus analogs in primary cells. A and B qPCR analysis of indicated genes in CD97+/+ and CD97−/− PMs A infected with VSV (MOI = 1) for 12 h; B stimulated with poly(I:C) (10 μg/mL) for 12 h. C and D ELISA results of IFN-β levels in CD97+/+ and CD97−/− PMs under the same conditions as in (A) and (B). E and F qPCR analysis of the indicated genes in CD97+/+ and CD97−/− BMDMs under the same conditions as in (A) and (B). G and H ELISA results of IFN-β levels in CD97+/+ and CD97−/− BMDMs under the same conditions as in (E) and (F). The mean and SD from three independent experiments are shown. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. I and J Immunoblot analysis of the indicated proteins in CD97+/+ and CD97−/− PMs infected with VSV (MOI = 1) for 12 h and stimulated with poly(I:C) (10 μg/mL) for 12 h
CD97 interacts with and degrades RIG-I to suppress IFN-I production during RNA virus infection
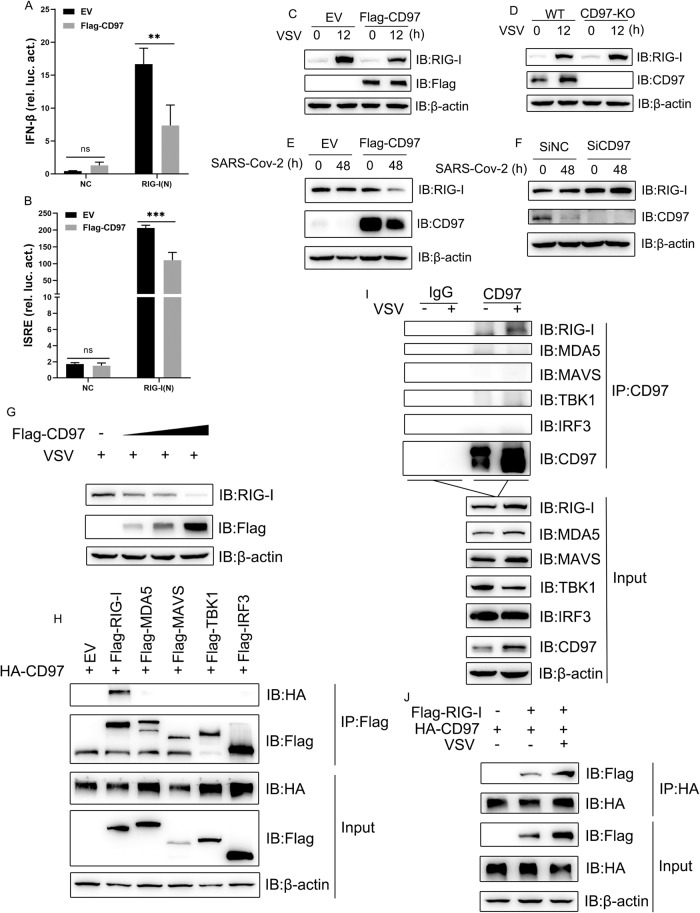
CD97 negatively regulates the production of IFN-I during RNA virus infection. We speculated that CD97 may regulate an innate immune component. To test this hypothesis, we first used an IFN-β and ISRE luciferase reporter to determine whether RIG-I is involved in CD97-mediated regulation of IFN-I. A luciferase reporter assay showed that CD97 blocked the IFN and ISRE promoters that are activated by RIG-I (Fig. 4A, B), indicating that CD97 may act on RIG-I. As expected, CD97 overexpression led to a much lower level of endogenous RIG-I expression after VSV or SARS-CoV-2 infection, while knockdown/knockout of CD97 had the opposite effect (Fig. 4C–F, and Fig. 3I, J). Additionally, CD97 overexpression decreased the endogenous RIG-I protein level in a dose-dependent manner (Fig. 4G). Moreover, at both exogenous and endogenous levels, CD97 interacted with RIG-I but not MDA5, TBK1, MAVS, or IRF3 (Fig. 4H, I). The interaction between CD97 and RIG-I was enhanced after VSV infection (Fig. 4J). Altogether, these results indicated that CD97 bound to RIG-I and inhibited its expression after RNA virus infection.
Fig. 4.
CD97 interacts with RIG-I and promotes its degradation. A and B Luciferase reporter assays were performed using HEK-293T cells transfected with IFN-β, ISRE, pRL-TK, CD97, or control plasmids plus the indicated plasmids for 36 h. The mean and SD from three independent experiments are shown. **P ≤ 0.01, ***P ≤ 0.001, and ns, not significant. C and D Immunoblot analysis of the indicated protein in CD97-overexpressing and CD97-knockout HeLa cells infected with VSV (MOI = 1) for 12 h. E and F Immunoblot analysis of the indicated proteins in SARS-CoV-2-infected CD97-overexpressing and CD97-knockdown Caco-2 cells. G Immunoblot analysis of the indicated proteins after transfection with different concentrations of Flag-CD97 (0.6, 1.2, or 1.8 μg/mL) or empty plasmid for 24 h in HeLa cells infected with VSV (MOI = 1) for 12 h. H Cotransfection of HA-CD97 with the indicated plasmids or control plasmid in HEK-293T cells and coimmunoprecipitation analysis of the interaction. I Immunoprecipitation and immunoblot analysis of the endogenous interaction between CD97 and the indicated proteins in HeLa cells infected with VSV for 12 h. J Coimmunoprecipitation analysis of the interaction between RIG-I and CD97 in cotransfected HEK-293T cells with or without VSV infection
CD97 degrades RIG-I via the ubiquitin‒proteasome pathway
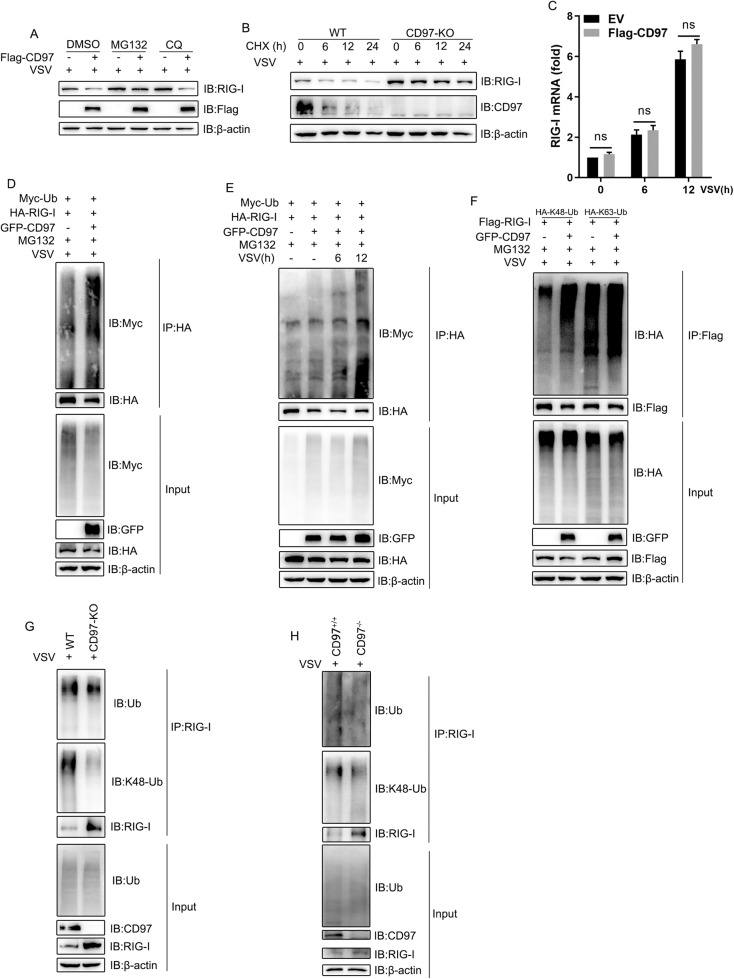
Considering that protein degradation includes mainly the ubiquitin‒proteasome and autophagy‒lysosome pathways, we evaluated the degradation mechanism using the corresponding inhibitors MG132 and chloroquine (CQ). The results showed that MG132 but not CQ was able to prevent CD97 from degrading RIG-I (Fig. 5A). Moreover, CD97 knockout clearly relieved RIG-I degradation induced by the protein synthesis inhibitor cycloheximide (CHX) (Fig. 5B), and CD97 negligibly affected RIG-I mRNA transcription (Fig. 5C). These results demonstrated that CD97 degraded RIG-I via the proteasome pathway. As ubiquitination is essential in this degradation pathway, with CD97 overexpression, we evaluated the polyubiquitination of RIG-I. The results corroborated that CD97 induced higher levels of polyubiquitination of RIG-I and gradually accelerated ubiquitination of RIG-I after VSV infection, as measured at different times after infection (Fig. 5D, E). We also found that CD97 increased the K48- but not the K63-linked ubiquitination rate (Fig. 5F). In contrast, CD97-deficient HeLa cells and PMs showed depleted endogenous RIG-1 ubiquitination and K48-specific ubiquitination (Fig. 5G, H), suggesting that CD97-induced K48-linked polyubiquitination of RIG-I to augment its degradation. Collectively, these results suggested that CD97 potentiated the ubiquitination of RIG-I, leading to its degradation, which reduces IFN-I signaling pathway activity against RNA virus infection.
Fig. 5.
CD97 promotes RIG-I degradation by the ubiquitin proteasome pathway. A HEK-293T cells transfected with Flag-CD97 or empty vector for 36 h, treated with DMSO, MG132 (10 μM), or CQ (20 μM) and infected with VSV for 12 h. B Immunoblot analysis of the indicated proteins in CD97-knockout HeLa cells subjected to VSV infection, followed by CHX (100 μg/mL) treatment for the indicated duration. C qPCR analysis of RIG-I in CD97-overexpressing HeLa cells infected with VSV for the indicated durations. The mean and SD from three independent experiments are shown; ns, not significant. D and E The ubiquitination of RIG-I was analyzed by cotransfecting HEK-293T cells with the indicated plasmids, treating them with MG132 and infecting them with VSV (MOI = 1) for 12 h or the indicated durations. F Different lysine-binding sites (K48 and K63) in ubiquitin were analyzed by cotransfecting with the indicated plasmids. G The protein lysates of WT and CD97-KO HeLa cells were analyzed to measure the ubiquitination level by precipitating endogenous RIG-I using the indicated antibodies. H Ubiquitination of RIG-I in CD97+/+ and CD97−/− PMs infected with VSV
CD97 promotes RNF125-mediated ubiquitination and RIG-I degradation
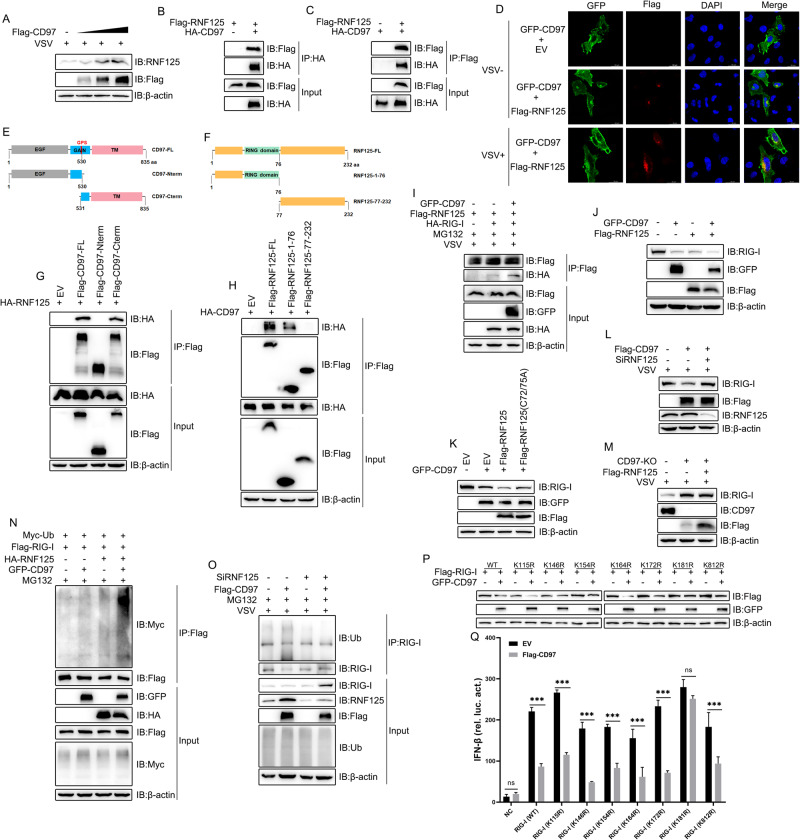
Protein degradation via the proteasome pathway through the action of E3 ubiquitin ligases is an important cellular behavior [45]. To investigate the potential functions of CD97-controlled E3 ligases, we analyzed transcriptomic high-throughput sequencing data obtained from virus-infected CD97-overexpressing cells and found that the mRNA expression of RNF125 was significantly upregulated (data not shown). The upregulated mRNA of RNF125 was identified by qPCR upon CD97 overexpression during mock and VSV infection (Fig. S2A). Immunoblotting showed that the RNF125 level gradually increased when different doses of CD97 were administered (Fig. 6A and Fig. S2B), indicating that CD97 upregulated the expression of RNF125 after viral infection. RNF125 has been demonstrated to promote the degradation of RIG-I by catalyzing its K48-linked polyubiquitination [25]. Therefore, we propose that RNF125 may be an E3 ligase targeting RIG-I that interacts with and is controlled by CD97. Next, a co-IP assay was performed to show that CD97 is associated with both exogenous and endogenous RNF125 (Fig. 6B, C, and Fig. S2C), and an immunofluorescence assay revealed that CD97 colocalized with RNF125 (Fig. 6D). The levels of interaction and colocalization were elevated after VSV infection (Fig. 6D and Fig. S2C). RNF125 carries an N-terminal RING-finger domain, C2HC zinc finger domain, and a C-terminal domain [25], while CD97 harbors an N-terminal domain including variable EGF modules and a seven-transmembrane C-terminal domain [46]. We constructed multiple CD97- and RNF125-truncation mutants (Fig. 6E, F), and co-IP analyses showed that CD97-FL, a C-terminal mutant but not an N-terminal mutant, interacted with RNF125 (Fig. 6G). Moreover, CD97 coprecipitated with RNF125-FL, in which the N-terminal 1-76 amino acids but not the C-terminal 77-232 amino acids (Fig. 6H). We found that CD97 strengthened the interaction between RIG-I and RNF125 after viral infection (Fig. 6I). Altogether, these data indicated that CD97 positively regulated the interaction between RNF125 and RIG-I.
Fig. 6.
CD97 promotes RNF125-mediated ubiquitination and degradation of RIG-I. A Immunoblot analysis of the indicated protein at different concentrations (0.6, 1.2, or 1.8 μg/mL) of Flag-CD97 or an empty plasmid in HeLa cells after VSV infection for 12 h. B and C Coimmunoprecipitation analysis of the interaction between RNF125 and CD97 in cotransfected HEK-293T cells. D Immunofluorescence analysis showing colocalization of RNF125 and CD97 in cells with or without VSV infection. Scale bars: 30 µm. E CD97 mutant model. F RNF125 mutant model. G and H Coimmunoprecipitation analysis of the interaction between RNF125 and CD97 by transfection with the indicated mutant plasmids for 36 h in HEK-293T cells. I Coimmunoprecipitation analysis showing that the interaction between RNF125 and RIG-I was affected by CD97 in HEK-293T cells. J Immunoblot analysis of RIG-I as indicated after cotransfection of GFP-CD97 and Flag-RNF125 plasmids into HEK-293T cells and culture for 36 h. K Flag-RNF125 or Flag-RNF125 (C72/75A) was transfected with GFP-CD97 or vector in HEK-293T cells, and RIG-I levels were measured by immunoblotting. L CD97-overexpressing HeLa cells transfected with RNF125 siRNA after VSV infection were used to measure RIG-I levels by immunoblotting. M CD97-knockout HeLa cells transfected with the Flag-RNF125 plasmid were infected with VSV, and the RIG-I level was measured. N The ubiquitination of RIG-I was analyzed by cotransfecting the indicated plasmids into HEK-293T cells for use in a coimmunoprecipitation analysis. O The endogenous ubiquitination of RIG-I was analyzed in HeLa cells after transfection with Flag-CD97 or RNF125 siRNA and VSV infection. P and Q Wild-type RIG-I or RIG-I mutated at lysine sites (K–R) were transfected with or without CD97 to identify action sites by immunoblot or luciferase assays; specifically, the inhibitory effect of RIG-I was measured. The mean and SD from three independent experiments are shown. ***P ≤ 0.001, ns, not significant
Next, we found that RNF125 antagonizes CD97, which led to the degradation of RIG-I (Fig. 6J). In addition, an RNF125 ligase inactive mutant (C72/75A) failed to antagonize CD97 (Fig. 6K), and RNF125 knockdown promoted the CD97-mediated degradation of RIG-I (Fig. 6L). In contrast, overexpression of RNF125 blunted the decrease in RIG-I levels in CD97-knockout HeLa cells (Fig. 6M). Furthermore, cotransfection of CD97 and RNF125 resulted in a significantly higher ubiquitination rate than transfection with either CD97 or RNF125 alone (Fig. 6N), and RNF125 knockdown inhibited exogenous CD97-induced RIG-I ubiquitination (Fig. 6O). These results suggested that CD97 enhanced the RNF125-mediated ubiquitination of RIG-I. We further analyzed the most conserved lysine residues in RIG-I among various species and constructed plasmids in which these lysine residues were individually replaced with an arginine residue (K/R). Luciferase analyses showed that all RIG-I-mutant expression plasmids increased the promotor activity of IFN-β, and CD97 abolished the activity of all RIG-I mutants except K181R (Fig. 6P). Moreover, CD97 decreased the expression levels of all exogenous RIG-I mutants except K181R (Fig. 6Q), suggesting that K181 is needed for CD97-induced RIG-I ubiquitination. Altogether, these data suggested that Lys181 was essential for CD97- and RNF125-mediated ubiquitination and that CD97 enhanced RNF125-mediated Lys48-linked degradation after the ubiquitination of RIG-I.
CD97 deficiency promotes IFN-I production and attenuates viral replication after viral infection in vivo
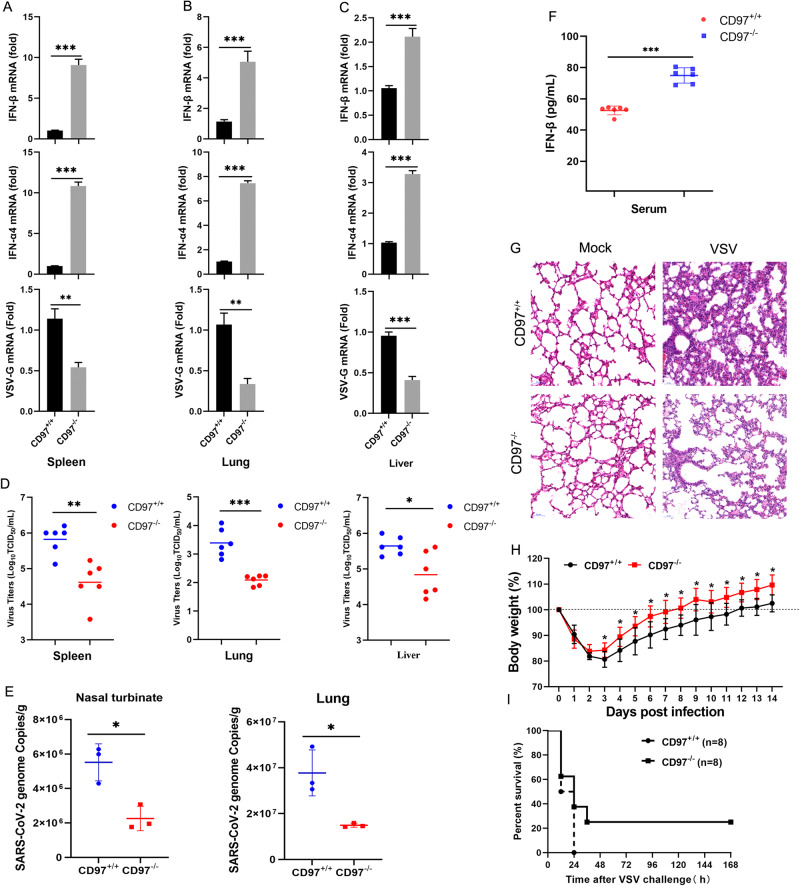
To evaluate the physiological relevance of this work, we studied the importance of CD97 in antiviral host defense in vivo. CD97+/+ and CD97−/− mice were intraperitoneally injected with VSV for 36 h, and then, we measured the change in the VSV-G mRNA level and titer in spleens, lungs, and livers. The results showed that compared to those in the CD97+/+ mice, the fold change in VSV-G mRNA expression and VSV titer was significantly lower in the CD97−/− mice (Fig. 7A–D). Most importantly, we found that CD97 knockout led to a decreased number of genome copies in the nasal turbinates and lungs of mice subjected to nasal inhalation of SARS-CoV-2 (mouse-susceptible C57MA14 strain) (Fig. 7E). Moreover, we found higher IFN-β and IFN-α4 mRNA expression levels in the organs of the CD97−/− mice than in CD97+/+ mice (Fig. 7A–C), and higher IFN-β protein levels was observed in the serum of CD97−/− mice (Fig. 7F). Next, pathological injury manifested by thickened alveolar walls following VSV infection was found in CD97+/+ mice and its subsequent amelioration was found in CD97−/− mice (Fig. 7G). Furthermore, mouse weight was monitored for 14 days after mice were intraperitoneally injected with VSV 2 × 107 PFU per mouse, and the number of surviving mice within 7 days of an intraperitoneal injection of VSV 1 × 108 PFU per mouse was determined. All of the mice showed a weight increase on Day 1, and this increase was the greatest on Day 3. As expected, CD97−/− mice showed faster weight gain from Day 3 to Day 14 (Fig. 7H). High concentrations of VSV injection were lethal for all control mice within 24 h of infection, but two CD97−/− mice survived, one survived to 36 h, and the other survived to the 7th day (Fig. 7I). Altogether, these findings showed that CD97-deficient mice showed a robust IFN response to RNA virus infection, limited viral replication, slowed infection, and increased survival.
Fig. 7.
CD97 deficiency attenuates viral replication in vivo after virus infection. A–C CD97+/+ and CD97−/− mice were infected for 36 h by intraperitoneal injection of VSV (2 × 107 PFU per mouse), and then qPCR analysis of the indicated genes in the spleen, liver, and lungs was performed. D TCID50 analysis of viral titer. E qPCR analysis of the number of SARS-CoV-2 copies in the nasal turbinates and lungs of CD97+/+ and CD97−/− mice 4 days after infection via nasal inhalation. F ELISA analysis of IFN-β levels in mouse serum after intraperitoneal injection of VSV. G Hematoxylin and eosin-stained sections of the lungs from CD97+/+ and CD97−/− mice infected with VSV. Scale bars: 100 µm. H and I Body weight and survival (Kaplan‒Meier curves) were monitored (n = 8 per group) in CD97+/+ and CD97−/− mice injected intraperitoneally with VSV (2 × 107 PFU per mouse and 1 × 108 PFU per mouse). The mean and SD from three independent experiments are shown. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001
Sanguinarine decreases the CD97 level to attenuate viral replication
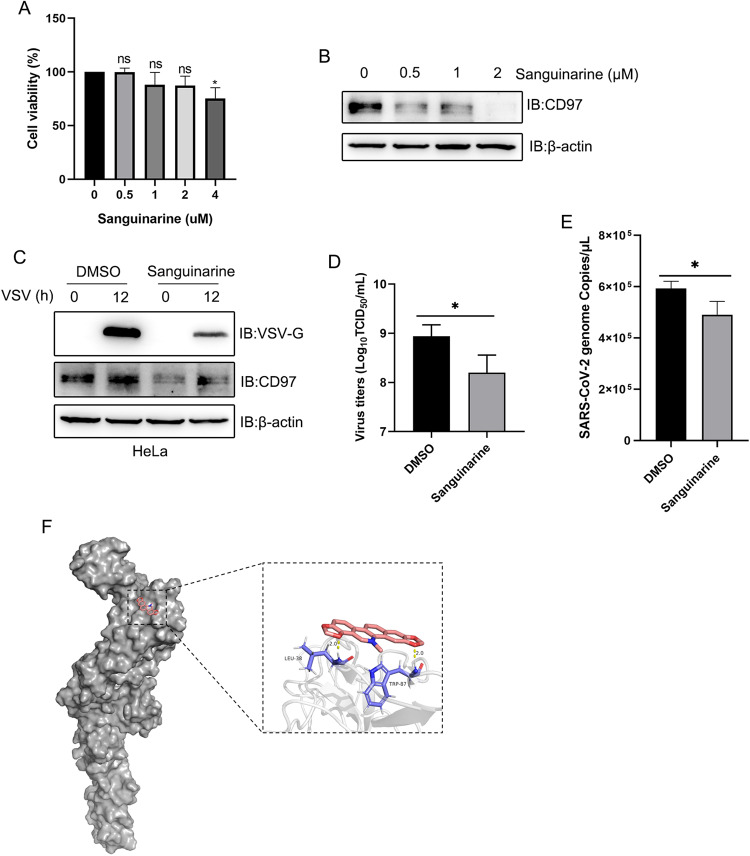
Small-molecule drugs targeting host genes to treat disease are increasingly critical in the fight against viral infection. We wondered whether drugs that target CD97 play an antiviral role. However, to date, drugs that directly target CD97 have not been developed. Fortunately, Chunhui et al. identified sanguinarine, a small-molecule drug that targets CD97 [47]. Therefore, we assessed the antiviral and CD97-inhibitory effects of sanguinarine. First, we discovered that sanguinarine (at 0.5, 1, and 2 μM) significantly inhibited the expression of CD97 but did not affect cell viability (Fig. 8A, B). Furthermore, we found that sanguinarine (0.5 μM) markedly reduced the expression levels of VSV-G and the virus titer (Fig. 8C, D) and decreased the number of SARS-CoV-2 genome copies (Fig. 8E), indicating that sanguinarine attenuated virus propagation. To gain insights into the potential interaction between CD97 and sanguinarine, a molecular docking study was performed using AutoDock Vina software and visualized by PyMOL software. We found that CD97 docked to sanguinarine with a binding energy of −7.05 kcal/mol, and in the binding site, Leu 38 and Trp 87 of CD97 formed hydrogen bonds with sanguinarine (Fig. 8F). Altogether, these data demonstrated that sanguinarine can inhibit CD97 and exert an antiviral effect.
Fig. 8.
Sanguinarine attenuates viral replication. A CCK-8 analysis of cell viability challenged with sanguinarine at the indicated dose. B Immunoblot analysis of the indicated proteins after challenge with sanguinarine. C Immunoblot analysis of the indicated proteins after sanguinarine treatment and VSV infection. D The viral titer of VSV was detected by TCID50. E q-PCR analysis of the number of SARS-CoV-2 genome copies after challenge with DMSO and sanguinarine. F In silico docking of ligands with CD97. The CD97 (7DO4) protein is in gray to highlight the binding cavity, sanguinarine is represented by the pink line, and the yellow dotted line represents the coordination interactions between sanguinarine and protein atoms
Discussion
CD97, also known as ADGRE5, belongs to the EGF-TM7 family of adhesion GPCRs and is highly conserved in eukaryotes. It has also been reported to be an oncogene in hepatocellular carcinoma [43], thyroid cancer [48], gastric cancer [49, 50], and prostate cancer [51] mediated through a variety of regulatory mechanisms. A recent study demonstrated that CD97 regulates the mechanosensing of red blood cells to promote splenic dendritic cell homeostasis [35]. However, the function of CD97 in viral replication is unclear. Here, we demonstrated that CD97 increases the RNA virus replication rates in cells and mice.
Increasing evidence has demonstrated that G protein-coupled receptors play positive or negative regulatory antiviral roles. For example, metabotropic glutamate receptor subtype 2 (mGluR2) increases the SARS-CoV-2 infection rate in nasal turbinates and lungs by increasing virus internalization [44]; G protein-coupled receptor 54 (GPR54) mediates immune evasion and increases the viral replication rate mediated through a negative feedback loop involving TBK1 signaling [52]; and the metabolite-sensing G protein-coupled receptor TGR5 broadly protects host cells from VSV, HSV-1, and NDV infection [53]. Thus, GPCRs function through complex regulatory mechanisms; they contribute by internalizing viruses or by regulating intracellular molecules. CD97 is widely expressed in both immune and nonimmune cells [34]. It plays a vital role in maintaining the homeostasis of spleen dendritic cells and reinforcing the stability of the immunological synapse formed between dendritic cells and T cells [35, 36]. However, how CD97 regulates the antiviral innate immune signaling response remains unclear. In this study, we determined that CD97 inhibits IFN-I production induced by VSV and SARS-CoV-2, thereby promoting viral replication. The phosphorylation activation of IRF3 and TBK1 during RNA virus infection is necessary for the generation of IFN-I [54]. We proved that CD97 decreases the phosphorylation of TBK1 and IRF3 in both VSV infection and poly (I:C) stimulation in HeLa cells. Analogously, compared to CD97+/+ macrophages, CD97−/− PMs or BMDMs exhibited significantly increased IFN-I mRNA and protein levels as well as increased phosphorylation of TBK1 and IRF3. This is the first report to show that CD97 suppresses the IFN-I signaling pathway response to facilitate viral replication.
In this study, the CD97 target negatively regulates RIG-I. RIG-I, a receptor protein that detects viral RNA in the cytoplasm, is essential for the recognition of cytoplasmic dsRNA and the activation of the IFN-I signaling pathway [55]. RIG-I-deficient cells and mice are more vulnerable to VSV, IAV, hepatitis C virus (HCV), and Japanese encephalitis virus (JEV) infection, indicating an essential role for RIG-I in the IFN-I signaling pathway [56, 57]. However, RIG-I-mediated antiviral innate immune signaling is governed by many host proteins. For instance, NLRP12 interacts with the ubiquitin ligase TRIM25 to prevent TRIM25-mediated K63-linked ubiquitination of RIG-I [58]; Siglec-G induces recruitment of SHP2 and the E3 ubiquitin ligase c-Cbl to RIG-I, leading to RIG-I degradation via K48-linked ubiquitination [22]; and the p97 complex is able to directly bind both nonubiquitinated RIG-I and the E3 ligase RNF125, promoting the K48-linked ubiquitination of RIG-I [59]. In this study, our data showed that CD97 downregulated the expression of RIG-I to repress the IFN-I signaling pathway after RNA virus infection. In investigating the mechanism by which CD97 acts on RIG-I, we discovered that CD97 degrades RIG-I via the RNF125-mediated ubiquitin‒proteasome pathway. RNF125, an E3 ubiquitin ligase, has been reported to recognize RIG-I and thus mediate proteasome pathway degradation [25]. Sufficient evidence has demonstrated that host genes, including CypA [60] and Siglec-G [22], interact with E3 ubiquitin ligases to target the adaptor regulation of IFN-I, which is an important strategy to maintain metabolic homeostasis. In this study, our data showed that CD97 antagonizes the action of RNF125 to inhibit RIG-I function. Specifically, CD97 enhances the RNF125-mediated ubiquitination of RIG-I. Moreover, we constructed CD97 mutants containing the C-terminus and N-terminus to precipitate with RNF125, which indicated that the C-terminus but not the N-terminus interacted with RNF125. CD97 functions as a mediator in signaling pathways related to tumor aggressiveness and the formation of immunological synapses, both involving its internalization [36, 43]. Therefore, in the context of viral infections, CD97 may be internalized and expressed within the cytoplasm. During this process, it interacts with RNF125, facilitating the association between RNF125 and RIG-I, leading to the ubiquitination and subsequent degradation of RIG-I.
While the study clarified that CD97 induces the degradation of RIG-I, it remains uncertain whether CD97 consistently induces RIG-I degradation or if this process occurs exclusively when CD97 is activated. To investigate the underlying mechanism, we introduced the ligand recombinant human CD55 protein to activate CD97 [61] and subsequently assessed the expression of RIG-I. The results indicated that the expression of RIG-I exhibited a very modest decline (data not shown), indicating that CD97 degrades RIG-I in a viral-dependent manner. Further investigation is needed to understand how the interaction between viruses and CD97 results in the degradation of RIG-I and whether other ligands of CD97 play a functional role. In addition, we conducted an investigation into whether CD97 is activated by viral infection. The results showed that VSV infection for 6 and 12 h resulted in an upregulation of CD97 expression (Fig. S3A). Similarly, the protein expression level of CD97 was elevated upon SARS-CoV-2 infection for 24 h (Fig. S3B). These findings suggest that CD97 becomes activated during viral infections, leading to the inhibition of RIG-I expression. However, unexpectedly, the expression of CD97 was inhibited at 24 h post-VSV infection and 48 h post-SARS-CoV-2 infection (Fig. S3A, B). Although different viruses, different cell lines, and different doses all affect the interaction between the virus and the host, it is noteworthy that a consistent pattern emerges in both scenarios, wherein CD97 is upregulated during the early phase and downregulated during the late phase of infection. We hypothesized that upregulated CD97 expression during the early stages of infection may facilitate virus replication by inhibiting the IFN-I signaling pathway. Conversely, as the host mounts a defense against the virus, it increases IFN production, leading to the degradation of the protein that promotes viral replication, including CD97, as a countermeasure to curb viral replication.
The COVID-19 epidemic has substantially impacted societal systems and individuals worldwide. The extensive usage of SARS-CoV-2 vaccines and drugs has resulted in spike protein mutation, significantly increasing SARS-CoV-2 transmission and making prevention and control more challenging [62]. Traditional small-molecule drugs used to treat SARS-CoV-2 primarily target the RdRp and 3CL proteases, but they show an effectiveness rate of less than 50% and lead to drug resistance susceptibility [63]. The discovery of small-molecule drugs or techniques targeting host protein targets has played a more important role in viral infection than virus-targeted antiviral medications. Host targets with broad antiviral activity should be the basis for the development of antiviral small-molecule medicines. For example, IFITM has been shown to significantly inhibit the replication of SARS-CoV-2 and IAV [64]. Additionally, medicines should possess a minimal number of side effects and high targeting efficiency. To date, the mechanism underlying CD97 action in antiviral innate immunity in vivo has been unclear. We discovered that CD97-deficient mice exhibit normal growth and development and show increased resistance to VSV and SARS-CoV-2 infection. In addition, sanguinarine was shown to inhibit the expression of CD97 and decrease VSV and SARS-CoV-2 propagation, which may provide a reference for the development of molecules targeted for use as broad-spectrum antiviral drugs.
In conclusion, we described the mechanism by which CD97 potentiates the degradation of RIG-I to counteract innate immunity and facilitate viral replication (Fig. 9). Our findings demonstrated that CD97 associates with RNF125 and promotes RIG-I degradation via K48-linked ubiquitination after RNA virus infection, which ultimately prevents the antiviral signaling response. More intriguingly, we found that an important compound, sanguinarine, functions as an inhibitor of CD97 to inhibit viral replication. The findings of this study demonstrate for the first time that CD97 is essential for viral infection and for suppressing host innate immunity.
Fig. 9.
Schematic showing that CD97 inhibits the antiviral IFN-I response by negatively regulating RIG-I. CD97 interacts with RNF125 and enhances RNF125-mediated ubiquitinated degradation of RIG-I to inhibit the RNA virus-induced antiviral innate immune signaling pathway. Sanguinarine, an inhibitor of CD97, suppresses viral replication
Supplementary information
Acknowledgements
The study was partially supported by grants from the National Natural Science Fund of China (32072834, 31972665), Special fund support for Taishan Scholar Project (H. H, tspd20181207), Shandong Provincial Natural Science Foundation, China (ZR2021MC050), and Jinan Innovation Team (202228060).
Author contributions
HBH, YWG, and HMW conceptualized the study; HBH, HSC, and PLH designed the experiments; XFW performed SARS-CoV-2 infection in vitro and in vivo experiments; HSC, ABX, and HW performed VSV infection in vivo and pathological experiments; WQH directed animal experiments; HSC, WJQ, RKY, XW, and DCH performed most of the immunoblotting, immunofluorescence, and qPCR experiments; XYL and GMZ designed and constructed the recombinant plasmids of CD97; HSC, PLH, and XFW collected and analyzed the data; WYS and TCW participated in discussions; HSC wrote the original draft; HBH and PLH edited and revised the original draft. All the authors reviewed and approved the final manuscript.
Data availability
The authors assert that all data supporting the conclusions of this study can be located in the paper and its supplementary information files or can be obtained from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Huasong Chang, Peili Hou, Xuefeng Wang.
Contributor Information
Hongmei Wang, Email: hongmeiwang@sdnu.edu.cn.
Yuwei Gao, Email: yuwei0901@outlook.com.
Hongbin He, Email: hongbinhe@sdnu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-023-01103-z.
References
- 1.Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–20. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levin AT, Hanage WP, Owusu-Boaitey N, Cochran KB, Walsh SP, Meyerowitz-Katz G. Assessing the age specificity of infection fatality rates for COVID-19: systematic review, meta-analysis, and public policy implications. Eur J Epidemiol. 2020;35:1123–38. doi: 10.1007/s10654-020-00698-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–5. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Hou P, Ma W, Wang X, Wang H, Yu Z, et al. SARS-CoV-2 ORF10 suppresses the antiviral innate immune response by degrading MAVS through mitophagy. Cell Mol Immunol. 2022;19:67–78. doi: 10.1038/s41423-021-00807-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–20. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 6.Rehwinkel J, Reis e Sousa C. RIGorous detection: exposing virus through RNA sensing. Science. 2010;327:284–286. doi: 10.1126/science.1185068. [DOI] [PubMed] [Google Scholar]
- 7.Loo Y-M, Gale M. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–92. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang F, Ramanathan A, Miller MT, Tang G-Q, Gale M, Patel SS, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–7. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–22. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou F, Sun L, Zheng H, Skaug B, Jiang Q-X, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–61. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weerawardhana A, Uddin MB, Choi J-H, Pathinayake P, Shin SH, Chathuranga K, et al. Foot-and-mouth disease virus non-structural protein 2B downregulates the RLR signaling pathway via degradation of RIG-I and MDA5. Front Immunol. 2022;13:1020262. doi: 10.3389/fimmu.2022.1020262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gack MU, Albrecht RA, Urano T, Inn K-S, Huang IC, Carnero E, et al. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–49. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallach D, Kovalenko A. Phosphorylation and dephosphorylation of the RIG-I-like receptors: a safety latch on a fateful pathway. Immunity. 2013;38:402–3. doi: 10.1016/j.immuni.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 14.Wies E, Wang MK, Maharaj NP, Chen K, Zhou S, Finberg RW, et al. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38:437–49. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu M-M, Liao C-Y, Yang Q, Xie X-Q, Shu H-B. Innate immunity to RNA virus is regulated by temporal and reversible sumoylation of RIG-I and MDA5. J Exp Med. 2017;214:973–89. doi: 10.1084/jem.20161015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J Biol Chem. 2009;284:807–17. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- 17.Kuniyoshi K, Takeuchi O, Pandey S, Satoh T, Iwasaki H, Akira S, et al. Pivotal role of RNA-binding E3 ubiquitin ligase MEX3C in RIG-I-mediated antiviral innate immunity. Proc Natl Acad Sci USA. 2014;111:5646–51. doi: 10.1073/pnas.1401674111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao D, Yang Y-K, Wang R-P, Zhou X, Diao F-C, Li M-D, et al. REUL is a novel E3 ubiquitin ligase and stimulator of retinoic-acid-inducible gene-I. PLoS ONE. 2009;4:e5760.. doi: 10.1371/journal.pone.0005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oshiumi H, Miyashita M, Inoue N, Okabe M, Matsumoto M, Seya T. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe. 2010;8:496–509. doi: 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 20.Gack MU, Shin YC, Joo C-H, Urano T, Liang C, Sun L, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–20. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 21.Yan J, Li Q, Mao A-P, Hu M-M, Shu H-B. TRIM4 modulates type I interferon induction and cellular antiviral response by targeting RIG-I for K63-linked ubiquitination. J Mol Cell Biol. 2014;6:154–63. doi: 10.1093/jmcb/mju005. [DOI] [PubMed] [Google Scholar]
- 22.Chen W, Han C, Xie B, Hu X, Yu Q, Shi L, et al. Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell. 2013;152:467–78. doi: 10.1016/j.cell.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Zhao K, Zhang Q, Li X, Zhao D, Liu Y, Shen Q, et al. Cytoplasmic STAT4 promotes antiviral type I IFN production by blocking CHIP-mediated degradation of RIG-I. J Immunol. 2016;196:1209–17. doi: 10.4049/jimmunol.1501224. [DOI] [PubMed] [Google Scholar]
- 24.Wang W, Jiang M, Liu S, Zhang S, Liu W, Ma Y, et al. RNF122 suppresses antiviral type I interferon production by targeting RIG-I CARDs to mediate RIG-I degradation. Proc Natl Acad Sci USA. 2016;113:9581–6. doi: 10.1073/pnas.1604277113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arimoto K-I, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci USA. 2007;104:7500–5. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, Ru Y, Ren J, Bai J, Wei J, Fu S, et al. G3BP1 inhibits RNA virus replication by positively regulating RIG-I-mediated cellular antiviral response. Cell Death Dis. 2019;10:946.. doi: 10.1038/s41419-019-2178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen Y, Tang K, Chen D, Hong M, Sun F, Wang S, et al. Riok3 inhibits the antiviral immune response by facilitating TRIM40-mediated RIG-I and MDA5 degradation. Cell Rep. 2021;35:109272.. doi: 10.1016/j.celrep.2021.109272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yona S, Lin H-H, Siu WO, Gordon S, Stacey M. Adhesion-GPCRs: emerging roles for novel receptors. Trends Biochem Sci. 2008;33:491–500. doi: 10.1016/j.tibs.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Hsiao C-C, Wang W-C, Kuo W-L, Chen H-Y, Chen T-C, Hamann J, et al. CD97 inhibits cell migration in human fibrosarcoma cells by modulating TIMP-2/MT1- MMP/MMP-2 activity-role of GPS autoproteolysis and functional cooperation between the N- and C-terminal fragments. FEBS J. 2014;281:4878–91. doi: 10.1111/febs.13027. [DOI] [PubMed] [Google Scholar]
- 30.Hamann J, Stortelers C, Kiss-Toth E, Vogel B, Eichler W, van Lier RA. Characterization of the CD55 (DAF)-binding site on the seven-span transmembrane receptor CD97. Eur J Immunol. 1998;28:1701–7. doi: 10.1002/(SICI)1521-4141(199805)28:05<1701::AID-IMMU1701>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 31.Kwakkenbos MJ, Pouwels W, Matmati M, Stacey M, Lin H-H, Gordon S, et al. Expression of the largest CD97 and EMR2 isoforms on leukocytes facilitates a specific interaction with chondroitin sulfate on B cells. J Leukoc Biol. 2005;77:112–9. doi: 10.1189/jlb.0704402. [DOI] [PubMed] [Google Scholar]
- 32.Wandel E, Saalbach A, Sittig D, Gebhardt C, Aust G. Thy-1 (CD90) is an interacting partner for CD97 on activated endothelial cells. J Immunol. 2012;188:1442–50. doi: 10.4049/jimmunol.1003944. [DOI] [PubMed] [Google Scholar]
- 33.Wang T, Ward Y, Tian L, Lake R, Guedez L, Stetler-Stevenson WG, et al. CD97, an adhesion receptor on inflammatory cells, stimulates angiogenesis through binding integrin counterreceptors on endothelial cells. Blood. 2005;105:2836–44.. doi: 10.1182/blood-2004-07-2878. [DOI] [PubMed] [Google Scholar]
- 34.Jaspars LH, Vos W, Aust G, Van Lier RA, Hamann J. Tissue distribution of the human CD97 EGF-TM7 receptor. Tissue Antigens. 2001;57:325–31. doi: 10.1034/j.1399-0039.2001.057004325.x. [DOI] [PubMed] [Google Scholar]
- 35.Liu D, Duan L, Rodda LB, Lu E, Xu Y, An J, et al. CD97 promotes spleen dendritic cell homeostasis through the mechanosensing of red blood cells. Science. 2022;375:eabi5965.. doi: 10.1126/science.abi5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cerny O, Godlee C, Tocci R, Cross NE, Shi H, Williamson JC, et al. CD97 stabilises the immunological synapse between dendritic cells and T cells and is targeted for degradation by the Salmonella effector SteD. PLoS Pathog. 2021;17:e1009771.. doi: 10.1371/journal.ppat.1009771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veninga H, Becker S, Hoek RM, Wobus M, Wandel E, van der Kaa J, et al. Analysis of CD97 expression and manipulation: antibody treatment but not gene targeting curtails granulocyte migration. J Immunol. 2008;181:6574–83. doi: 10.4049/jimmunol.181.9.6574. [DOI] [PubMed] [Google Scholar]
- 38.Hou J, Han L, Zhao Z, Liu H, Zhang L, Ma C, et al. USP18 positively regulates innate antiviral immunity by promoting K63-linked polyubiquitination of MAVS. Nat Commun. 2021;12:2970.. doi: 10.1038/s41467-021-23219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S, Hou P, Pan W, He W, He D C, Wang H, et al. DDIT3 targets Innate Immunity via the DDIT3-OTUD1-MAVS pathway to promote bovine viral diarrhea virus replication. J Virol. 2021;95. 10.1128/JVI.02351-20. [DOI] [PMC free article] [PubMed]
- 40.Hou P, Zhao M, He W, He H, Wang H. Cellular microRNA bta-miR-2361 inhibits bovine herpesvirus 1 replication by directly targeting EGR1 gene. Vet Microbiol. 2019;233:174–83. doi: 10.1016/j.vetmic.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Hou P, Wang X, Wang H, Wang T, Yu Z, Xu C, et al. The ORF7a protein of SARS-CoV-2 initiates autophagy and limits autophagosome-lysosome fusion via degradation of SNAP29 to promote virus replication. Autophagy. 2022. 10.1080/15548627.2022.2084686. [DOI] [PMC free article] [PubMed]
- 42.Wang X, Zhao Y, Yan F, Wang T, Sun W, Feng N, et al. Viral and host transcriptomes in SARS-CoV-2-infected human lung cells. J Virol. 2021;95:e0060021.. doi: 10.1128/JVI.00600-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin Y, Xu X, Tang J, Zhang W, Zhangyuan G, Ji J, et al. CD97 promotes tumor aggressiveness through the traditional G protein-coupled receptor-mediated signaling in hepatocellular carcinoma. Hepatology. 2018;68:1865–78. doi: 10.1002/hep.30068. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Yang G, Wang X, Wen Z, Shuai L, Luo J, et al. SARS-CoV-2 uses metabotropic glutamate receptor subtype 2 as an internalization factor to infect cells. Cell Discov. 2021;7:119.. doi: 10.1038/s41421-021-00357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng N, Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem. 2017;86:129–57. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- 46.Yang L-Y, Liu X-F, Yang Y, Yang L-L, Liu K-W, Tang Y-B, et al. Biochemical features of the adhesion G protein-coupled receptor CD97 related to its auto-proteolysis and HeLa cell attachment activities. Acta Pharmacol Sin. 2017;38:56–68. doi: 10.1038/aps.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma C, Lv Q, Teng S, Yu Y, Niu K, Yi C. Identifying key genes in rheumatoid arthritis by weighted gene co-expression network analysis. Int J Rheum Dis. 2017;20:971–9. doi: 10.1111/1756-185X.13063. [DOI] [PubMed] [Google Scholar]
- 48.Ward Y, Lake R, Martin PL, Killian K, Salerno P, Wang T, et al. CD97 amplifies LPA receptor signaling and promotes thyroid cancer progression in a mouse model. Oncogene. 2013;32:2726–38. doi: 10.1038/onc.2012.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li C, Liu D-R, Li G-G, Wang H-H, Li X-W, Zhang W, et al. CD97 promotes gastric cancer cell proliferation and invasion through exosome-mediated MAPK signaling pathway. World J Gastroenterol. 2015;21:6215–28. doi: 10.3748/wjg.v21.i20.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu D, Li C, Trojanowicz B, Li X, Shi D, Zhan C, et al. CD97 promotion of gastric carcinoma lymphatic metastasis is exosome dependent. Gastric Cancer. 2016;19:754–66. doi: 10.1007/s10120-015-0523-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward Y, Lake R, Yin JJ, Heger CD, Raffeld M, Goldsmith PK, et al. LPA receptor heterodimerizes with CD97 to amplify LPA-initiated RHO-dependent signaling and invasion in prostate cancer cells. Cancer Res. 2011;71:7301–11. doi: 10.1158/0008-5472.CAN-11-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang H, Xiong Q, Wang N, Chen R, Ren H, Siwko S, et al. Kisspeptin/GPR54 signaling restricts antiviral innate immune response through regulating calcineurin phosphatase activity. Sci Adv. 2018;4:eaas9784.. doi: 10.1126/sciadv.aas9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong Q, Huang H, Wang N, Chen R, Chen N, Han H, et al. Metabolite-sensing G protein coupled receptor TGR5 protects host from viral infection through amplifying type I interferon responses. Front Immunol. 2018;9:2289.. doi: 10.3389/fimmu.2018.02289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630.. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 55.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 56.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–5. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 57.Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 58.Chen S-T, Chen L, Lin D S-C, Chen S-Y, Tsao Y-P, Guo H, et al. NLRP12 regulates anti-viral RIG-I activation via interaction with TRIM25. Cell Host Microbe. 2019;25. 10.1016/j.chom.2019.02.013. [DOI] [PMC free article] [PubMed]
- 59.Hao Q, Jiao S, Shi Z, Li C, Meng X, Zhang Z, et al. A non-canonical role of the p97 complex in RIG-I antiviral signaling. EMBO J. 2015;34:2903–20. doi: 10.15252/embj.201591888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu W, Li J, Zheng W, Shang Y, Zhao Z, Wang S, et al. Cyclophilin A-regulated ubiquitination is critical for RIG-I-mediated antiviral immune responses. ELife. 2017;6. 10.7554/eLife.24425. [DOI] [PMC free article] [PubMed]
- 61.Park GB, Kim D. MicroRNA-503-5p inhibits the CD97-mediated JAK2/STAT3 pathway in metastatic or paclitaxel-resistant ovarian cancer cells. Neoplasia. 2019;21:206–15. doi: 10.1016/j.neo.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Planas D, Bruel T, Staropoli I, Guivel-Benhassine F, Porrot F, Maes P, et al. Resistance of Omicron subvariants BA.2.75.2, BA.4.6, and BQ.1.1 to neutralizing antibodies. Nat Commun. 2023;14:824.. doi: 10.1038/s41467-023-36561-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Foo CS, Abdelnabi R, Vangeel L, De Jonghe S, Jochmans D, Weynand B, et al. Ivermectin does not protect against SARS-CoV-2 infection in the Syrian hamster model. Microorganisms. 2022;10. 10.3390/microorganisms10030633. [DOI] [PMC free article] [PubMed]
- 64.Shi G, Kenney AD, Kudryashova E, Zani A, Zhang L, Lai KK, et al. Opposing activities of IFITM proteins in SARS-CoV-2 infection. EMBO J. 2021;40:e106501.. doi: 10.15252/embj.2020106501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors assert that all data supporting the conclusions of this study can be located in the paper and its supplementary information files or can be obtained from the corresponding author upon reasonable request.