Abstract
The two lysis genes cph1 and cpl1 of the Streptococcus pneumoniae bacteriophage Cp-1 coding for holin and lysozyme, respectively, have been cloned and expressed in Escherichia coli. Synthesis of the Cph1 holin resulted in bacterial cell death but not lysis. The cph1 gene was able to complement a lambda Sam mutation in the nonsuppressing E. coli HB101 strain to produce phage progeny, suggesting that the holins encoded by both phage genes have analogous functions and that the pneumococcal holin induces a nonspecific lesion in the cytoplasmic membrane. Concomitant expression of both holin and lysin of Cp-1 in E. coli resulted in cell lysis, apparently due to the ability of the Cpl1 lysozyme to hydrolyze the peptidoglycan layer of this bacterium. The functional analysis of the cph1 and cpl1 genes cloned in a pneumococcal mutant with a complete deletion of the lytA gene, which codes for the S. pneumoniae main autolysin, provided the first direct evidence that, in this gram-positive-bacterium system, the Cpl1 endolysin is released to its murein substrate through the activity of the Cph1 holin. Demonstration of holin function was achieved by proving the release of pneumolysin to the periplasmic fraction, which strongly suggested that the holin produces a lesion in the pneumococcal membrane.
Two main mechanisms to liberate phage progeny from bacterium-infected cells have been postulated. Some small phages have developed a mechanism of liberation based on the synthesis of a single protein that has no lytic activity; e.g., the icosahedral Escherichia coli phage φX174 possesses a gene encoding a protein that induces cell lysis and progeny liberation through activation of the host lytic system (2). However, in most phages, a late gene codes for a cell wall lytic enzyme that acts in a coordinated way with a protein named holin, which is thought to produce holes in the cytoplasmic membrane through which the lytic enzyme reaches the cell wall peptidoglycan. This two-protein mechanism of lysis has been well-documented in gram-negative bacteria, mainly in lambda and lambdoid phages (29). The gene coding for the holin is normally located immediately upstream of the lysin gene, and in the case of lambda phage, the operon controlling the lysis process is formed by three genes: S, which encodes the holin, R, which encodes the lysin, and Rz, which encodes a protein of unknown function that is not essential for lysis.
The mechanism of phage lysis in gram-positive bacteria has been much less studied. Recent data suggest that the system involving the activity of two phage genes to induce host lysis might also be widespread among gram-positive bacteria (29). A dual translational start motif, similar to that of the S lambda gene, has been demonstrated for the holin gene in the Bacillus subtilis phage φ29 (27), and it has been proposed that conformational changes in the two proteins encoded by the holin gene may serve as regulatory mechanisms (26). A common feature of most holins described so far is a lack of sequence similarity, although they share distinctive features, namely, high hydrophobicity, two or three predicted transmembrane domains separated by β-turns, and highly charged C-terminal ends (29). The study of phage lysis genes has been complex, since the proteins encoded by these genes are designed to kill bacteria and are usually expressed at low levels; consequently, the cloning of such genes on multicopy plasmids requires us to pay particular attention to negative regulation.
On the other hand, the lytic enzymes encoded by Streptococcus pneumoniae and its virulent and temperate phages, either N-acetylmuramoyl-l-alanine amidases (amidases, hereinafter) or lysozymes, have been well-characterized in our laboratory. These lytic enzymes show a modular organization where the N-terminal domain determines enzyme specificity and the C-terminal domain, which has several repeated motifs, is responsible for substrate binding, mainly to choline, a structural component of the cell wall teichoic acids (12). Nevertheless, very few data are known about the two-step lysis system in pneumococci.
The two-step lysis system of the temperate pneumococcal phage EJ-1, which consists of a holin and an amidase, has been recently studied in detail with E. coli (3). Remarkably, the concomitant expression of the genes coding for these two proteins led to lysis of E. coli and Pseudomonas putida through hydrolysis of the peptidoglycans of these gram-negative bacteria. We have now studied the functional activities of the holin and the lysozyme encoded by the pneumococcal phage Cp-1 in a heterologous system as well as in S. pneumoniae.
MATERIALS AND METHODS
Bacterial strains, plasmids, phage, and growth conditions.
The S. pneumoniae strains used were wild-type R6, a derivative of R36A (Rockefeller University), as a lawn for the phage; R6st, a streptomycin-resistant strain used for infection in liquid medium; and M31, a ΔlytA mutant (21). The E. coli strains used were HB101 [F− hsdS20(rB− mB−) recA13 ara-14 proA2 leuB lacY1 galK2 rpsL20 xyl-5 mtl-1 supE44], C600 (F− thr-1 leuB6 thi-1 lacY1 supE44 rfbD1 fhuA21), and LE392 [F− hsdR574 (rK− mK+) supE44 supF58 lacY1 galK2 galT22 metB1 trpR55] (20). Plasmid pNM185 is a broad-host-range gene expression vector (15), pLSE1 is a shuttle vector between gram-positive and -negative bacteria (18), and pGL80 is a pBR325 derivative constitutively expressing the lytA gene, which encodes the pneumococcal LytA autolysin (6). Bacteriophage Cp-1 has been described elsewhere (19). E. coli strains were grown in Luria-Bertani (LB) medium at 37 or 30°C with shaking. Where appropriate, 50 μg of kanamycin per ml or 100 μg of ampicillin per ml was added to the culture medium. S. pneumoniae was grown without shaking in C medium supplemented with 0.8 mg of yeast extract per ml and 0.8 μg of tetracycline per ml or 1 μg of lincomycin per ml (10). Growth was monitored with a Coleman nephelometer.
Preparation of phage DNA.
Bacteriophage Cp-1 was propagated in strain R6st and purified by two equilibrium bandings in a cesium chloride gradient as previously described (4), except that pneumococcal cells were grown and infected in medium 3 (11). Deproteinized phage DNA was prepared by phenol extraction after treatment with proteinase K, as described elsewhere (19).
Plasmid isolation and transformation procedures.
The preparation of pneumococcal DNA and the transformation procedure for S. pneumoniae have been described elsewhere (28). Transformation of E. coli cells was carried out by the RbCl method (20).
Construction of plasmids.
PCR using Cp-1 DNA as the template was performed to generate DNA fragments containing the cph1 gene alone or the cassette cph1-cpl1. In these fragments the genes retained their own ribosome-binding sites. We used appropriate oligonucleotides to create SacII and SacI restriction sites at the 5′ and 3′ ends of the PCR fragments for cloning into pNM185. An analogous strategy, but one that generated at the 5′ and 3′ ends of the PCR fragments EcoRI and HindIII restriction sites, respectively, was followed to clone the same fragments into pLSE1.
Complementation of phage lambda Sam7 lethal function.
E. coli HB101 and HB101 transformants containing plasmid pAMR11 or pAMR12 and the suppressing strain E. coli LE392 were grown at 30°C in LB medium supplemented with 0.2% maltose, 10 mM MgSO4, and, when required, 50 μg of kanamycin per ml. When the cultures reached an optical density at 600 nm of about 0.8, samples (200 μl) were removed and infected at 37°C for 30 min with different dilutions of a preparation of bacteriophage lambda cI857 Sam7 (Lambda DNA Packaging System; Promega) in the presence or absence of 0.2 mM 3-methyl-benzoate (3-MB). The samples were mixed with 5 ml of soft agar containing 10 mM MgSO4 and, when required, 0.2 mM 3-MB and 50 μg of kanamycin per ml and poured onto LB plates containing, when needed, 50 μg of kanamycin per ml. The plates were incubated at 30°C (37°C for the control strain E. coli LE392), and the number of plaques was determined. Addition of 0.2 mM 3-MB to E. coli HB101 containing pAMR11 induces expression of cph1 at levels that are sublethal for the cells unless the R lysin of lambda cI857 Sam7 is also present. Thus, complementation of the defective lambda S gene leads to phage plaques (8).
Detection of pneumolysin.
Samples of M31(pLSE1) or M31(pAMR11) were taken to prepare protoplasts according to a published procedure (25). For the assay of pneumolysin, 0.5 ml (packed volume) of fresh sheep blood cells was suspended in 10 ml of 0.15 M NaCl; 0.5-ml portions of this suspension were incubated with the above-described periplasmic fractions (0.5 ml), and amounts of released hemoglobin were determined spectrophotometrically at 541 nm in the supernatant fluids after centrifugation.
Molecular biology techniques.
Standard molecular biology techniques for DNA isolation, restriction analysis, labeling, cloning, Southern blotting, and hybridization were as described previously (20).
Nucleotides and enzymes.
Oligonucleotides and deoxynucleoside triphosphates were from Pharmacia. T4 DNA ligase was from Boehringer Mannheim Biochemicals. T4 polynucleotide kinase and Bal 31 were from Amersham Corp. Proteinase K was from Sigma. Membranes for Southern and Northern blots were purchased from Schleicher & Schuell. Radioactive compounds were from Amersham Corp.
RESULTS
Analysis of ORFs involved in lytic functions in Cp-1 phage.
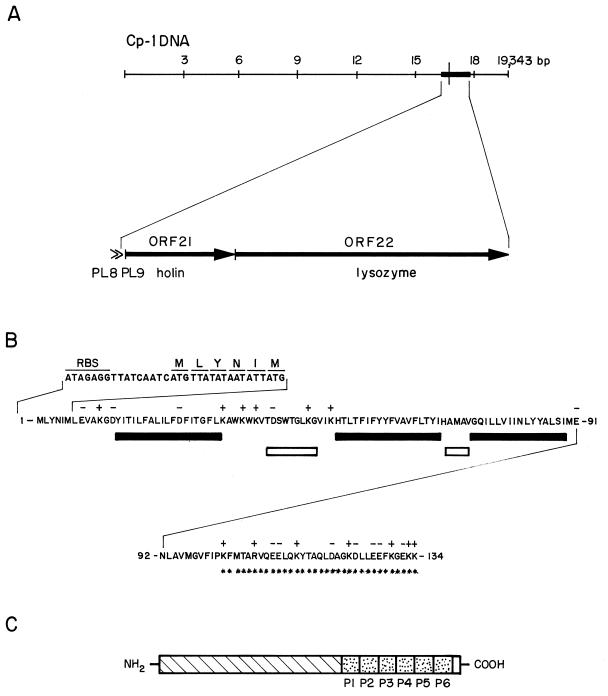
The cpl1 gene coding for the lytic enzyme of the phage Cp-1 has been previously cloned and expressed in E. coli. The Cpl1 protein was purified and characterized as a lysozyme that requires the presence of choline in the cell wall substrate for activity (5). The elucidation of the complete DNA sequence of the Cp-1 genome has allowed us to recognize the cpl1 gene as open reading frame 22 (ORF22) (14). Upstream of the cpl1 gene, we have found that ORF21 (cph1) codes for a putative protein of 134 amino acids (15.4 kDa) that exhibits most of the characteristics associated with holin sequences previously described (Fig. 1A). It has been observed that the stop codon of cph1 overlaps with the start codon of cpl1, a trait shared by most of the two-component phage lytic systems. Interestingly, the cph1 holin gene lacks the dual start motif for translation previously found in lambdoid, Salmonella P22, and B. subtilis φ29 phages (29). The standard motif consists of two ATG codons separated by one or two codons, at least one of which is lysine. Instead, the cph1 gene contains two methionine triplets at positions 1 and 6; the lysine triplet is not found between both ATG codons, and only one ribosome-binding site is clearly identified upstream of the start codon (Fig. 1B). Cph1 exhibits a hydrophylic C-terminal end with several charged amino acids, whereas the N-terminal part has a net negative charge. Analysis of the predicted structure also reveals three potential hydrophobic transmembrane regions, according to the PredictProtein program from the EMBL server. Two amino acid sequences are predicted to form β-turns flanking the second putative transmembrane region, although the first β-turn, amino acids 39 to 47, has been determined with a higher degree of reliability than the second one, amino acids 69 to 72. The same three transmembrane domains were predicted for Cph1 when the TMbase program of the Baylor College of Medicine-Human Genome Center server was employed. In the global mapping of Cp-1 promoters we demonstrated the existence of two tandem promoters, separated by 78 nucleotides and located 5′ from the cph1 gene (14). The location of the lysis genes in the Cp-1 genome is depicted in Fig. 1, where the structural and functional regions of the corresponding proteins are also shown.
FIG. 1.
Lysis genes of Cp-1. (A) Localizations of ORF21 and ORF22, which encode holin and lysozyme, respectively. PL8 and PL9, represented by carets, are the late tandem promoters preceding the holin gene and are named according to the nomenclature given to the Cp-1 genome (14). (B) Amino acid sequence, translational start region, and predicted secondary structure of the Cph1 holin. RBS, ribosome-binding site. Filled and open bars represent putative transmembrane domains and β-turns, respectively. Positive (+) and negative (−) charges of amino acids are indicated above the sequence. Asterisks indicate the charged C-terminal domain. (C) Modular organization of Cpl1 lysozyme. The hatched region represents the N-terminal domain, and the six dotted boxes, P1 to P6, indicate choline-binding domains.
Cloning of cph1 and cpl1 in E. coli.
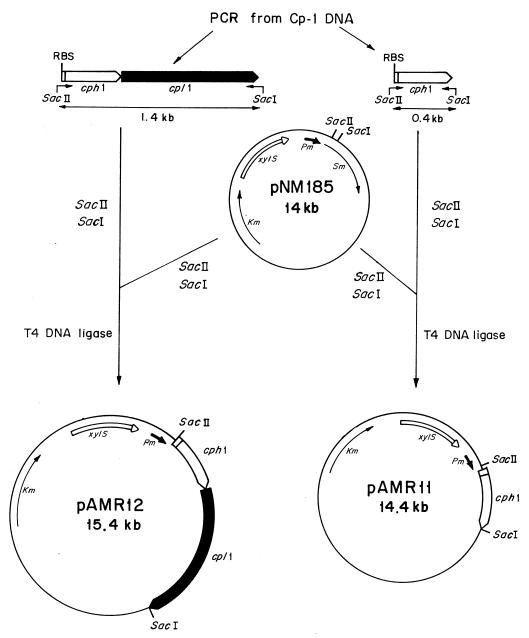
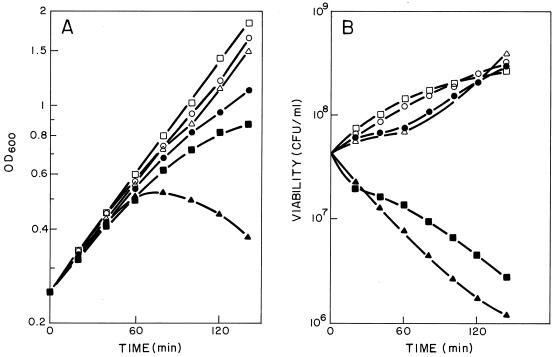
To test whether the product of the cph1 gene is a protein that induces damage in the membrane of the host cell to facilitate the activity of the lytic enzyme(s) on the cell wall, we decided to clone this gene in a plasmid vector (pNM185) capable of replicating in a wide variety of bacteria although not in S. pneumoniae. The cph1 gene or a cassette containing the two lytic genes of Cp-1 (cph1 and cpl1) were expressed under the control of a positively regulated promoter (Pm) of the meta pathway operon of the TOL plasmid (15) (Fig. 2). Transcription of the genes from Pm is specifically induced by the product of the xylS regulator gene only when effector molecules like 3-MB are present. When the HB101(pAMR11) clone, which contains the holin gene, was induced by 2 mM 3-MB, a one-log-unit drop in the number of viable cells, accompanied by a threefold increase in cell mass, was observed (Fig. 3). In contrast, the expression of cpl1 alone in this heterologous system did not affect either growth or cell viability (23). However, when we analyzed the physiological effect induced in HB101(pAMR12), which contains the genes coding for holin and the Cpl1 lysozyme, a decrease in cell turbidity and a 1.5-log-unit drop in cell viability were observed after 2 h of induction. These results strongly suggested that the expression of the Cph1 holin in E. coli might produce a lesion in the cytoplasmic membrane that dramatically affects cell viability and allows the Cpl1 lysozyme to reach the cell wall. This assumption is also supported by the experiment shown in Fig. 4, where E. coli cells expressing concomitantly the holin Cph1 and the amidase LytA (from pAMR11 and pGL80, respectively) lysed 2 h after induction. These observations also suggested that the major pneumococcal autolysin, LytA, as well as the Cp-1 lysozyme Cpl1, can recognize and degrade some links of the cell wall of E. coli.
FIG. 2.
Schematic representation of the construction of pAMR11 and pAMR12. Plasmids are drawn with the relevant elements and restriction sites indicated. Thin and thick lines represent vector-derived sequences and Cp-1-derived sequences, respectively. Arrows represent the direction of transcription of the genes. Pm indicates an inducible promoter of the TOL plasmid, and xylS encodes the cognate regulator of Pm. Oligonucleotides used for isolation of the genes by PCR are marked as bent arrows. Sm and Km indicate the genes conferring streptomycin and kanamycin resistance, respectively, and RBS indicates the ribosome-binding sites.
FIG. 3.
Effects of expression of the cph1 and cpl1 genes on the growth of the E. coli. (A) Cultures of HB101(pNM185) (○ and •), HB101(pAMR11) (□ and ▪), and HB101(pAMR12) (▵ and ▴) that were uninduced (open symbols) or induced at time zero with 2 mM 3-MB (filled symbols). Cultures were incubated at 30°C. OD600, optical density at 600 nm. (B) Viabilities of the same cultures obtained after plating two appropriate dilutions on LB medium.
FIG. 4.
Effects of cph1 and lytA genes on the growth of E. coli. Shown are optical densities at 600 nm (OD600) of cultures of HB101(pAMR11, pGL80) that were uninduced (○) or induced at time zero with 2 mM 3-MB (•). Cultures were incubated at 30°C.
The cph1 product complements an S-negative lysis-defective lambda phage mutant.
The above-described results suggested that the cytoplasmic membrane is the target of Cph1, as has already been documented for the S protein (holin) coded for by the lambda phage. Sam7 lambda phage carry an amber mutation in the S gene and, consequently, cannot induce lysis of the infected host unless suppressing E. coli cells are used, since the holin has been demonstrated to be essential for the induction of the lytic activity of the product of R lysin gene (16). To further document the physiological role of Cph1, we carried out complementation tests using nonsuppressing HB101 cells harboring different recombinant plasmids and infected with lambda phage Sam7. As expected, HB101 control cells that did not contain any plasmid were not lysed but they produced plaques when expressing the cph1 gene (Table 1). Interestingly, lambda Sam7 was unable to form lysis plaques on HB101 cells expressing only the cpl1 gene.
TABLE 1.
Complementation of the function of the lambda Sam7 gene by the cph1 gene product
| E. coli strain | Gene(s) of interest | Phage lambda Sam7a
|
|
|---|---|---|---|
| With 3-MBb | Without 3-MB | ||
| LE392 | supF58 | + | + |
| HB101 | − | − | |
| HB101(pCIP100) | cpl1 | − | − |
| HB101(pAMR11) | cph1 | + | − |
| HB101(pAMR12) | cph1 and cpl1 | + | − |
The number of PFU per 100 μl of lambda phage was determined as described in Materials and Methods. + and − indicate that the ratio of plaques obtained for a particular strain and for the reference strain E. coli LE392 was >0.5:1 and <10−6:1, respectively.
3-MB was added to a final concentration of 0.2 mM.
Cloning and expression of the cph1 and cpl1 genes in S. pneumoniae.
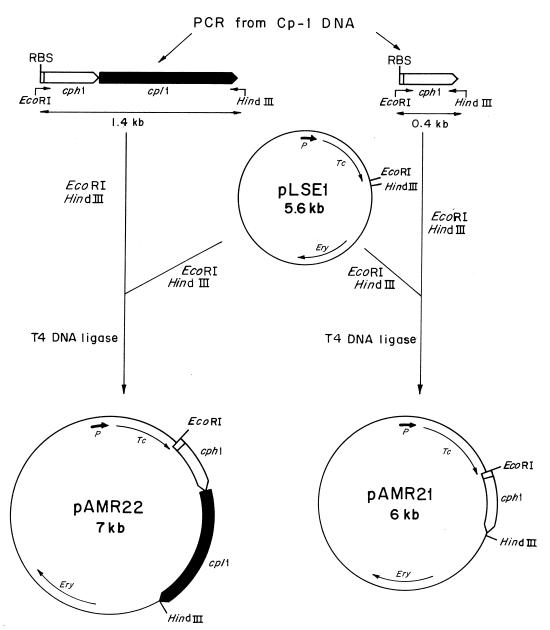
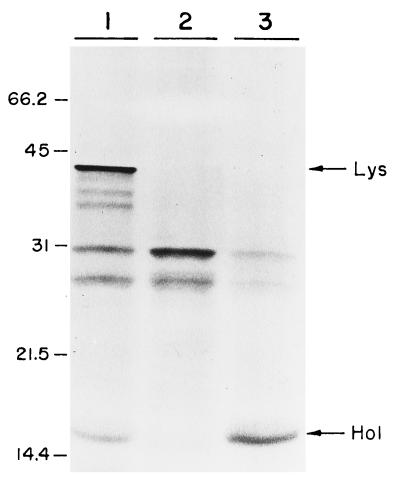
There are few examples in the literature illustrating a two-gene lysis system that facilitates the liberation of phage progeny into a medium, and to the best of our knowledge, it has not been directly proved for S. pneumoniae that the phage endolysin is released to its murein substrate through a lesion produced in the membrane by a holin. Vector plasmids containing regulated promoters are not available in the pneumococcal system, which is a limitation in achieving an appropriate and controlled expression of deleterious genes like those involved in lytic functions. We have failed to prepare pneumococcal transformants with plasmids harboring cph1 alone or with cpl1 under the control of its own promoter, probably because of the lethal effects of these lytic enzymes when their genes are cloned in homologous systems. To overcome this problem, we used pLSE1 in which promoterless cph1 and cpl1 genes were cloned under the control of the tetp promoter that is located 1.6 kb upstream of the cloned gene(s). The strategy followed to construct the recombinant plasmids is summarized in Fig. 5. We first constructed the recombinant plasmids pAMR21 and pAMR22 in E. coli C600, and then we transformed the M31 pneumococcal strain. This mutant provides a suitable background, since the single lytic activity to be analyzed corresponds to that expressed by the cloned lytic genes. Furthermore, these plasmids containing the isolated cph1 gene or the tandem cph1 and cpl1 genes were also used for electrophoretic identification of the Cp-1 lysis proteins in an in vitro transcription-translation assay. As judged from the electrophoretic mobilities, pAMR21 carried a gene that encoded a 15.5-kDa protein, which is in good agreement with the proposed molecular mass of the Cp-1 holin. On the other hand, pAMR22 carried genes that coded for two proteins of 15.5 and 41 kDa, which correspond to the predicted molecular masses of the holin and lysozyme of Cp-1, respectively (14) (Fig. 6).
FIG. 5.
Schematic representation of the construction of pAMR21 and pAMR22. Plasmids are drawn with the relevant elements and restriction sites indicated. Thin lines represent vector-derived sequences, and thick lines represent Cp-1-derived sequences. Arrows indicate the direction of transcription of the genes. P represents the promoter of the tetracycline resistance gene. Oligonucleotides used for isolation of the genes by PCR are indicated as bent arrows. Tc and Ery indicate the genes conferring tetracycline and erythromycin resistance, respectively, and RBS indicates the ribosome-binding sites.
FIG. 6.
In vitro transcription-translation of plasmids pLSE1, pAMR21, and pAMR22. Samples containing pAMR22 (lane 1), pLSE1 (lane 2), and pAMR21 (lane 3) were loaded onto a sodium dodecyl sulfate–15% polyacrylamide gel. Arrows indicate the positions of the holin (Hol) and the lysozyme (Lys). Standard size markers, in kilodaltons, are shown on the left.
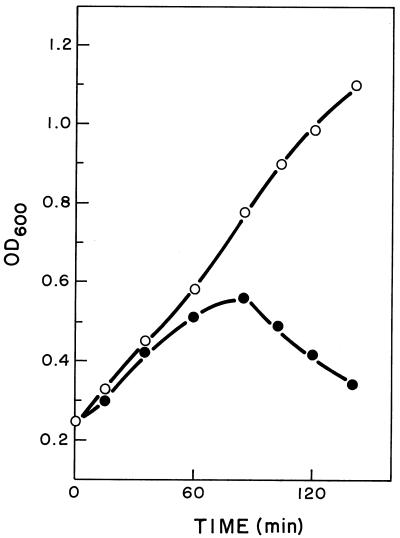
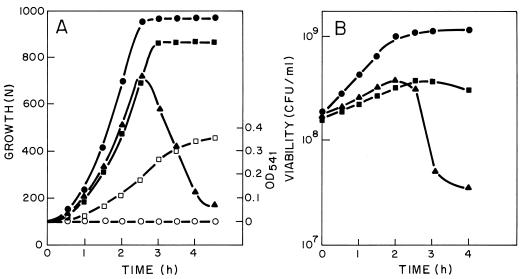
The physiological response of the pneumococcus to the expression of the lytic genes of Cp-1 was analyzed by incubating at 37°C the M31 strains harboring pAMR21 or pAMR22. For M31(pAMR21) a decrease in the growth rate relative to that of the control culture that harbors pLSE1 was observed (Fig. 7A). Nevertheless, it is noticeable that there is no apparent reduction in cell viability under these experimental conditions (Fig. 7B).
FIG. 7.
Effects of cph1 and cpl1 genes on the growth of S. pneumoniae. (A) Cultures of M31(pLSE1) (•), M31(pAMR21) (▪), and M31(pAMR22) (▴) were incubated at 37°C. Pneumolysin release to the periplasmic fraction was measured, as explained in Materials and Methods, for M31(pLSE1) (○) and M31(pAMR21) (□) cultures. N, nephelometric units; OD541, optical density at 541 nm. (B) Viabilities of the same cultures obtained after plating two appropriate dilutions on C medium.
We had previously reported that cells of the M31(pAPE10) strain that contained the cpl1 gene cloned under the control of the tetp promoter grew as normal “diplo” cells that autolysed only at the end of the stationary phase of growth, whereas cells of the M31 strain formed short chains and did not lyse (17). In contrast, M31(pAMR22) cells that express the genes cph1 and cpl1 lysed before the culture reached the stationary phase of growth (Fig. 7A). When pneumococcal cells harboring pAMR21 or pAMR22 were grown in the presence of an antibiotic, either tetracycline or lincomycine, added immediately after the overnight cultures were diluted out (which represents the time zero of these assays), we did not observe any noticeable change from the results reported in Fig. 7. These observations confirmed the constitutive expression of the tet promoter (18).
To further illustrate that the Cph1 holin produced a lesion in the cytoplasmic membrane, allowing the liberation of some intracellular proteins to the periplasmic space, we determined the periplasmic amounts of pneumolysin, a pneumococcal protein of cytoplasmic localization (9), in M31(pAMR21). The results clearly demonstrated that a significant amount of pneumolysin was released from M31(pAMR21) cells but, as expected, this protein was not present in the supernatant of a control culture of M31(pLSE1) (Fig. 7A).
DISCUSSION
We had already demonstrated that the pneumococcal phage Cp-1 coded for a choline-dependent lysozyme (5), and we have now found that ORF21, located immediately upstream of the lysozyme gene, codes for a protein with traits typical of a holin (29). These genes are transcribed from two tandem promoters found upstream of the cph1 gene and are expressed from late phage transcripts that, according to the results of previous Northern blot analyses, use a putative transcription terminator located downstream of cpl1 (14). The observation that the product of the cph1 gene can functionally complement a lambda S mutant (Table 1) provides further support for the hypothesis that the cytoplasmic membrane is the target of the putative Cph1 protein and suggests that this protein is the holin of Cp-1 since the S and Cph1 proteins played similar physiological roles. These results also demonstrate that the holin-dependent induction of a lesion in the inner membrane was nonspecific, as has already been suggested for other systems (26).
The structural characteristics of Cph1 and the genetic organizations of the two lysis genes of Cp-1 also correspond to typical features of holins (29). Nevertheless, the dual start motif characteristic of some holin genes, which has been demonstrated to control posttransductionally the activities of the proteins encoded by these genes (29), has not been found in the holin of Cp1. Alternative mechanisms of regulation of holins have been postulated for P1 phage, where a protein, named antiholin, functions as an inhibitor of the phage holin (24), and for c2 phage, where two proteins showing structural characteristics of holins have been found (13). In the latter case, a gene like that encoding holin has been mapped upstream of the lytic enzyme gene of c2 but the gene encoding the second holin appears to be localized far from the two lysis genes. However, an analysis of the complete nucleotide sequence of Cp-1 (14) strongly suggested that only two genes are involved in lysis functions in this phage, and the transcriptional data discussed above revealed a plausible coordinated mechanism of transcriptional regulation that involves stepwise activities of the holin and the lysozyme, i.e., the holin forms a hole in the membrane that facilitates the access of the lysozyme to the cell wall.
Expression of the cph1 gene in E. coli resulted in a significant inhibition of growth after 60 min of induction. Growth inhibition correlates with the loss in cell viability, probably due to the lesion produced in the inner membrane by the Cph1 protein, as was previously demonstrated for the Ejh holin of the pneumococcal phage EJ-1 (3). Interestingly, the coordinated expression of the cph1 and cpl1 genes in E. coli resulted in a partial cell lysis but the expression of cpl1 alone did not lyse the host cell even after a long period of incubation (5). These observations indicate that the pneumococcal lytic enzymes can be liberated to the periplasmic space in this heterologous system only in the presence of the Cph1 holin, which may produce a lesion in the cytoplasmic membrane of E. coli. These observations also suggest that the Cpl1 lysozyme might hydrolyze the peptidoglycan of E. coli, as has already been demonstrated for the LytA and Ejl amidases (3). The lysozymes coded for by gene 14 of B. subtilis phage φ29 and Lactococcus lactis phage φLC3, two phages of gram-positive bacteria, have also been shown to degrade the murein of E. coli (8). The findings reported here indicate that pneumococcal cell wall lytic enzymes, other than amidases, can also degrade the murein of E. coli. This ability has been ascribed to the lower level of structural complexity of murein from gram-negative bacteria, since solubilized pneumococcal murein lacking choline is also a substrate of the host LytA amidase (7). Considering these results together, we can postulate that the lytic enzymes of S. pneumoniae and its bacteriophages exhibit a host range of activities wider than previously thought, although the murein-hydrolase activities of these enzymes are strongly dependent, in a homologous system, on the presence of choline residues in the cell wall substrate. We have suggested that this dependence, directly linked to the evolutionary acquisition of a characteristic substrate-binding domain, may represent a remarkable advantage for enzymes that interact with polymeric substrates in improving their catalytic efficiencies (22).
In the absence of genetic tools in S. pneumoniae to develop a vector with regulated promoters to control the expression of the cloned lysis genes, these genes were successfully expressed in a homologous system when they were cloned under the control of the tetp of the vector plasmid pLSE1. Using these experimental conditions we did not observe a reduction in cell viability when an autolysin-deficient M31 strain was transformed with pAMR21, although the lysis of the culture was triggered at the mid-log phase of growth only when the cph1-cpl1 cassette was introduced in M31. We had previously reported that cells of M31(pAPE10), which has the cpl1 gene cloned under the control of the tetp promoter, grew as normal diplo cells that autolysed only at the end of the stationary phase of growth but that cells of M31 formed short chains and did not lyse (17). Therefore, these results indicate that an alteration in the cytoplasmic membrane is required to facilitate the lytic activity of the lysozyme on the cell wall substrate. It has been suggested that the secondary structures of the holins described so far lead to the formation of host-independent membrane lesions, which should explain the growth inhibition observed when the cph1 gene was expressed in the homologous or heterologous systems analyzed here. Simultaneous expression of cph1 and cpl1 genes should result in cell lysis due to the liberation of lysozyme to the periplasmic space, which takes advantage of the alteration produced in the cytoplasmic membrane as the result of a homo-oligomeric structure generated when the holin is inserted in the host membrane (30). The current model for holin function is that killing is apparently independent of endolysin activity, that is, the holin kills the cells by forming the hole, irrespective of endolysin release (29). This situation does not appear to be the case for S. pneumoniae under the experimental conditions used in our work. The survival of M31(pAMR21) cells might be ascribed to a low expression of holin. Alternatively, we cannot rule out that some peculiar characteristics of the pneumococcal system may allow the microorganism to repair the cytoplasmic membrane by means of the alterations produced by the activity of the holin.
The biochemical and physiological characteristics of holins still remain to be elucidated. Holins usually do not show similarity in their primary structures, although they exhibit characteristic secondary structures. However, significant amino acid sequence similarities have been observed between the Ejh holin of the pneumococcal phage EJ-1 and those of two temperate phages (φLC3 and Tuc2009) of Lactococcus lactis (1, 3). This is not the case for Cph1 holin, which does not exhibit amino acid similarity with the proteins reported in the data banks. Furthermore, this holin is larger than the Ejh holin since it exhibits three potential transmembrane regions rather than the two found in the latter. All these observations indicate that, in remarkable contrast to the genes encoding the pneumococcal murein hydrolases (either lysozymes or amidases), the ORFs that encode holins show no similarity. As already suggested for lambdoid phage evolution (29), this finding further documents an independent evolution of the two genes implicated in the lysis system of the pneumococcal phages. The approach reported in this work provides an appropriate framework to analyze for S. pneumoniae the precise roles of the proteins encoded by the lysis genes of the pneumococcal phages.
ACKNOWLEDGMENTS
We thank E. García, J. L. García, and E. Díaz for helpful comments. We also thank E. Cano and M. Carrasco for their technical assistance and A. Hurtado, V. Muñoz, and M. Fontenla for the art work.
This work was supported by grant PB93-0015-C02-01 from the DGICYT. A.C.M. was the recipient of a fellowship from the DGICYT.
REFERENCES
- 1.Arendt E K, Dali C, Fitzgerald G F, Van de Guchte M. Molecular characterization of lactococcal bacteriophage Tuc2009 and identification and analysis of the genes encoding lysin, a putative holin, and two structural proteins. Appl Environ Microbiol. 1994;60:1875–1883. doi: 10.1128/aem.60.6.1875-1883.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bläsi U, Halfman G, Lubitz W. Induction of autolysis of Escherichia coli by φX174 gene E product. In: Nombela C, editor. Microbial cell wall synthesis and autolysis. Amsterdam, The Netherlands: Elsevier Science Publishers; 1984. pp. 213–218. [Google Scholar]
- 3.Díaz E, Munthali M, Lünsdorf H, Höltje J-V, Timmis K N. The two-step lysis system of pneumococcal bacteriophage EJ-1 is functional in Gram-negative bacteria: triggering of the major pneumococcal autolysin in Escherichia coli. Mol Microbiol. 1996;19:667–681. doi: 10.1046/j.1365-2958.1996.399929.x. [DOI] [PubMed] [Google Scholar]
- 4.García E, Gómez A, Ronda C, Escarmís C, López R. Pneumococcal bacteriophage Cp-1 contains a protein bound to the 5′ termini of its DNA. Virology. 1983;128:92–104. doi: 10.1016/0042-6822(83)90321-5. [DOI] [PubMed] [Google Scholar]
- 5.García J L, García E, Arrarás A, García P, Ronda C, López R. Cloning, purification, and biochemical characterization of the pneumococcal bacteriophage Cp-1 lysin. J Virol. 1987;61:2573–2580. doi: 10.1128/jvi.61.8.2573-2580.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.García P, García J L, García E, López R. Nucleotide sequence and expression of the pneumococcal autolysin gene from its own promoter in Escherichia coli. Gene. 1986;43:265–272. doi: 10.1016/0378-1119(86)90215-5. [DOI] [PubMed] [Google Scholar]
- 7.García-Bustos J F, Tomasz A. Teichoic acid-containing muropeptides from Streptococcus pneumoniae as substrates for the pneumococcal autolysin. J Bacteriol. 1987;169:447–453. doi: 10.1128/jb.169.2.447-453.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henrich B, Binishofer B, Bläsi U. Primary structure and functional analysis of the lysis genes of Lactobacillus gasseri bacteriophage φadh. J Bacteriol. 1995;177:723–732. doi: 10.1128/jb.177.3.723-732.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson M K. Cellular location of pneumolysin. FEMS Microbiol Lett. 1977;2:243–245. [Google Scholar]
- 10.Lacks S, Hotchkiss R D. A study of the genetic material determining an enzyme activity in pneumococcus. Biochim Biophys Acta. 1960;39:508–517. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 11.López R, Ronda C, García P, Escarmís C, García E. Restriction cleavage maps of the DNAs of Streptococcus pneumoniae bacteriophages containing protein covalently bound to their 5′ ends. Mol Gen Genet. 1984;197:67–74. doi: 10.1007/BF00327924. [DOI] [PubMed] [Google Scholar]
- 12.López R, García E, García P, García J L. The pneumococcal cell wall degrading enzymes: a modular design to create new lysins? Microb Drug Resist. 1997;3:199–211. doi: 10.1089/mdr.1997.3.199. [DOI] [PubMed] [Google Scholar]
- 13.Lubbers M W, Waterfield N R, Beresford T P J, Le Page R W F, Jarvis A W. Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Appl Environ Microbiol. 1995;61:4348–4356. doi: 10.1128/aem.61.12.4348-4356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martín A C, López R, García P. Analysis of the complete nucleotide sequence and functional organization of the genome of Streptococcus pneumoniae bacteriophage Cp-1. J Virol. 1996;70:3678–3687. doi: 10.1128/jvi.70.6.3678-3687.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mermod N, Ramos J L, Lehrbach P R, Timmis K N. Vector for regulated expression of cloned genes in a wide range of gram-negative bacteria. J Bacteriol. 1986;167:447–454. doi: 10.1128/jb.167.2.447-454.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reader R W, Siminovitch L. Lysis defective mutants of bacteriophage lambda: genetics and physiology of S cistron mutants. Virology. 1971;43:607–622. doi: 10.1016/0042-6822(71)90286-8. [DOI] [PubMed] [Google Scholar]
- 17.Romero A, López R, García P. Lytic action of cloned pneumococcal phage lysis genes in Streptococcus pneumoniae. FEMS Microbiol Lett. 1993;108:87–92. doi: 10.1016/0378-1097(93)90492-k. [DOI] [PubMed] [Google Scholar]
- 18.Ronda C, García J L, López R. Characterization of genetic transformation in Streptococcus oralis NCTC 11427. Expression of pneumococcal amidase in S. oralis using a new shuttle vector. Mol Gen Genet. 1988;215:53–57. doi: 10.1007/BF00331302. [DOI] [PubMed] [Google Scholar]
- 19.Ronda C, López R, García E. Isolation and characterization of a new bacteriophage, Cp-1, infecting Streptococcus pneumoniae. J Virol. 1981;48:721–730. doi: 10.1128/jvi.40.2.551-559.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 21.Sánchez-Puelles J M, Ronda C, García J L, García P, López R, García E. Searching for autolysin functions. Characterization of a pneumococcal mutant deleted in the lytA gene. Eur J Biochem. 1986;158:289–293. doi: 10.1111/j.1432-1033.1986.tb09749.x. [DOI] [PubMed] [Google Scholar]
- 22.Sánchez-Puelles J M, Sanz J M, García J L, García E. Cloning and expression of gene fragments encoding the choline-binding domain of pneumococcal murein hydrolases. Gene. 1990;89:69–75. doi: 10.1016/0378-1119(90)90207-8. [DOI] [PubMed] [Google Scholar]
- 23.Sanz J M, García J L. Structural studies of the lysozyme coded by the pneumococcal phage Cp-1. Conformational changes induced by choline. Eur J Biochem. 1990;187:409–416. doi: 10.1111/j.1432-1033.1990.tb15319.x. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt C, Velleman M, Arber W. Three functions of bacteriophage P1 involved in cell lysis. J Bacteriol. 1996;178:1099–1104. doi: 10.1128/jb.178.4.1099-1104.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seto H, Tomasz A. Protoplast formation and leakage of intramembrane cell components: induction by the competence activator substance of pneumococci. J Bacteriol. 1975;121:344–353. doi: 10.1128/jb.121.1.344-353.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steiner M, Bläsi U. Charged amino-terminal amino acids affect the lethal capacity of lambda lysis protein S107 and S105. Mol Microbiol. 1993;8:525–533. doi: 10.1111/j.1365-2958.1993.tb01597.x. [DOI] [PubMed] [Google Scholar]
- 27.Tedin K, Resch A, Steiner M, Bläsi U. Dual translational start motif evolutionarily conserved in the holin gene of Bacillus subtilis phage φ29. Virology. 1995;206:479–484. doi: 10.1016/s0042-6822(95)80063-8. [DOI] [PubMed] [Google Scholar]
- 28.Tomasz A. Requirement for protein synthesis during induction of competence. J Bacteriol. 1970;101:860–871. doi: 10.1128/jb.101.3.860-871.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zagotta M T, Wilson D B. Oligomerization of the bacteriophage lambda S protein in the inner membrane of E. coli. J Bacteriol. 1990;172:912–921. doi: 10.1128/jb.172.2.912-921.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]