Abstract
Rapid advancements in high-throughput biological techniques have facilitated the generation of high-dimensional omics datasets, which have provided a solid foundation for precision medicine and prognosis prediction. Nonetheless, the problem of missing heritability persists. To solve this problem, it is essential to explain the genetic structure of disease incidence risk and prognosis by incorporating interactions. The development of the Bayesian theory has provided new approaches for developing models for interaction identification and estimation. Several Bayesian models have been developed to improve the accuracy of model and identify the main effect, gene-environment (G×E) and gene-gene (G×G) interactions. Studies based on single-nucleotide polymorphisms (SNPs) are significant for the exploration of rare and common variants. Models based on the effect heredity principle and group-based models are relatively flexible and do not require strict constraints when dealing with the hierarchical structure between the main effect and interactions (M-I). These models have a good interpretability of biological mechanisms. Machine learning-based Bayesian approaches are highly competitive in improving prediction accuracy. These models provide insights into the mechanisms underlying the occurrence and progression of complex diseases, identify more reliable biomarkers, and develop higher predictive accuracy. In this paper, we provide a comprehensive review of these Bayesian approaches.
Keywords: Bayesian, gene-environment interactions, gene-gene interactions, effect heredity, machine learning, review
Complex diseases, such as cancer, are critical non-communicable chronic diseases that seriously endanger the health and quality of life of people. They consistently rank among the leading causes of death, contributing significantly to the global burden of disease (1). In 2020, the World Health Organization reported an estimated 19.29 million new cancer cases worldwide and 9.89 million cancer-related deaths (2). In clinical practice, surgical treatment, radiotherapy, and chemotherapy are the most common treatments used for malignant tumors. In recent years, targeted therapies, such as epidermal growth factor receptor (EGFR) inhibitors and immunotherapies (including PD-1 and PD-L1 inhibitors), have opened new frontiers in precision therapy, providing better treatment outcomes for specific patients (3,4). However, the survival of patients with cancer has not improved significantly. This can be attributed to several critical potential targets that may not have been detected or fully understood, immune escape and drug resistance (5).
Accordingly, there is an urgent need to identify the important factors associated with the progression and prognosis of cancer to guide the prevention and treatment of highly heterogeneous cancer types and facilitate prognostic assessment. However, models based on traditional clinical factors, such as treatment, stage, age, and sex often perform poorly (6). Fortunately, the rapid development of genome sequencing technology has led to the generation of high-dimensional omics data encompassing various types, including single-nucleotide polymorphisms (SNPs), methylation, microRNA, mRNA, lncRNA, and proteomics data (7). Notably, previous research has shown that biomarkers screened from different omics data using appropriate biostatistical methods can reliably predict the occurrence and prognosis of various diseases (8-10).
In recent years, the development of diagnostic and prognostic interaction models for complex diseases has become a popular research topic (11). Studies have shown that diagnostic and prognostic models that only include the main effects cannot significantly improve model performance (12). The interactions are critical for the diagnosis and prognosis of complex diseases beyond the role of the main effects of genetic or environmental factors (13). In incidence risk studies, models that integrate gene-environment (G×E) and gene-gene (G×G) interactions may provide additional genetic evidence for a high incidence of cancer in specific exposure groups (14,15). These interactions should not be ignored in prognostic studies. Integrating G×G and G×E interactions into prognostic models is important to improve model performance. For example, Marley et al. showed that the interactions between citrus intake and genes may influence melanoma risk (16). Other studies found that two-way and three-way interactions between DNA methylation and smoking significantly affected the prognosis of lung cancer, which is expected to be a methylation-specific drug target (14,17,18).
Models developed in the Bayesian framework that integrate interactions can utilize the qualities of Bayesian theory to provide new ideas for studying interactions (19-21). Firstly, this class of models can adaptively incorporate prior information (22,23). Secondly, Bayesian methods can flexibly incorporate the principle of effect heredity between the main terms and interactions, and the developed models can be used to obtain good interpretability of biological mechanisms (22). Thirdly, Bayesian approaches can be combined with machine learning (ML) methods, such as neural networks and factorization machines (FMs), to develop novel approaches with higher predictive accuracy (24,25). However, Bayesian approaches that incorporate interactions involve many challenges, such as the high dimensionality of the data and its resulting computational cost, the incorporation of dependence on the main effect and interaction (M-I), and the complexity of the posterior inference (26).
In the past years, the analysis of interactions under the Bayesian framework has been developed to allow for two-order and higher-order models (27,28), linear and nonlinear (29), binary and count (30), with continuous traits and censored survival outcomes (22,31). Although several Bayesian methods have been proposed for the estimation and identification of interactions, relevant comprehensive reviews are lacking. In this study, we review Bayesian approaches that involve interactions. The purpose of this review is to provide a survey of approaches that can be used within the Bayesian framework to detect G×G and G×E interactions related to the occurrence or prognosis of complex diseases.
Bayesian Approaches in Interaction Studies
We summarize four classes of approaches (SNP-based, effect heredity-based, group-based, and ML-based) within the Bayesian framework for detecting interactions associated with the occurrence and development of complex diseases. Although we focused on the Bayesian approaches used in human genetics, we also incorporated Bayesian approaches used in animal and plant genetics (32), which are closely related to human genetics. In addition, some ML-based Bayesian methods are not particularly developed for medical research; however, they are generally similar in terms of data dimensions and research strategies to the methods used in human genetics data. Therefore, they are also included in this study.
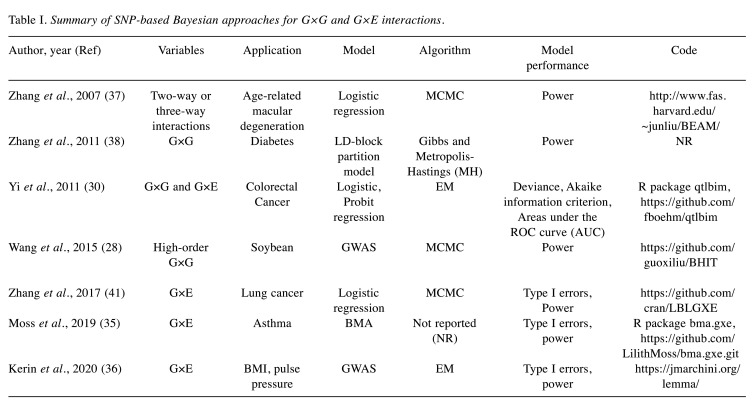
SNP-based approaches. SNP-based studies are characterized by certain unique challenges. First, SNP-based studies usually incorporate genotype SNPs with strong linkage disequilibrium (LD); hence, strong correlations may not only exist between the main terms and interactions but also among the main terms (33). Secondly, SNP data typically include a higher number of genotypes with lower frequencies, which generate predictors of near-zero variation (34). Over the past few years, a series of SNP-based Bayesian interaction analysis approaches have been developed to explore the key roles of rare haplogroups and G×G and G×E interactions on the occurrence of complex diseases. Their relevant characteristics in this study are summarized in Table I.
Table I. Summary of SNP-based Bayesian approaches for G×G and G×E interactions.
Moss et al. extended the Bayes model averaging (BMA) approach and proposed a BMA 2DF approach (35). The method used the multivariate Wald test with 2DF to test the G×E interaction and the main effects of genes. Kerin et al. proposed a Bayesian whole genome regression model to jointly model the main genetic effects and G×E interactions in a large-scale dataset (36). It built a linear environment mixed model analysis for the assumption that the main effects and the G×E interaction effects follow two separate mixtures of Gaussian priors and estimated the environmental scores (ES). The ES can be used to both estimate the proportion of phenotypic variance attributable to G×E effects and to test for G×E effects at genetic variants across the genome (36). In addition, stratification by minor allele frequency and LD can be analyzed to better explore the genetic structure of the disease. Zhang et al. proposed a Bayesian epistasis association mapping (BEAM) algorithm to identify the critical main terms, low-order and high-order interactions in genome-wide case-control studies (37). The BEAM algorithm has two components: a Bayesian epistasis inference tool that is implemented with Markov Chain Monte Carlo (MCMC) and a test B statistic that can be adapted to the correlation structure of candidate markers to assess statistical significance. In addition, they extended the BEAM algorithm in 2011 and developed the block-based BEAM2 algorithm, dividing SNPs into LD-blocks and selecting the main effects and interactions within the blocks associated with the disease (38). Wang et al. developed a Bayesian method for genome-wide association studies (GWAS) that allows for a Bayesian model on both continuous and discrete data. This method is suitable for detecting higher-order interactions associated with phenotypes (28). The above methods are mostly applicable to genome-wide association studies in large-scale cohorts.
Several Bayesian regression models have been proposed to detect variants and interactions. Biswas and the team developed a series of Logistic Bayesian LASSO (LBL) approaches, including LBL-G×E-I, LBL-G×E-D, LBL-G×E, LBLc-G×E, and LBLc-G×E-G×S to detect phenotype-related interactions in case-control studies (39-43). The three methods, LBL-G×E-I, LBL-G×E-D, and LBL-G×E, were differentiated based on whether the assumption of G-E independence was made. LBL-G×E-I assumed G-E independence in the control population (39). LBL-G×E-D allowed for G-E dependence by modeling haplotype frequencies as a function of covariates using a multinomial logistic regression model (41). LBL-G×E allowed for uncertainty in the assumption of G-E independence by allowing Markov chains to move between LBL-G×E-I and LBL-G×E-D via the reversible jump MCMC (41). LBLc-G×E and LBLc-G×E-G×S were developed to accommodate complex sampling designs (43). LBLc-G×E incorporated stratified variables as covariates with their main effects in the model, whereas LBLc-G×E-G×S incorporated the interaction between stratified variables and haplotypes in the model (43).
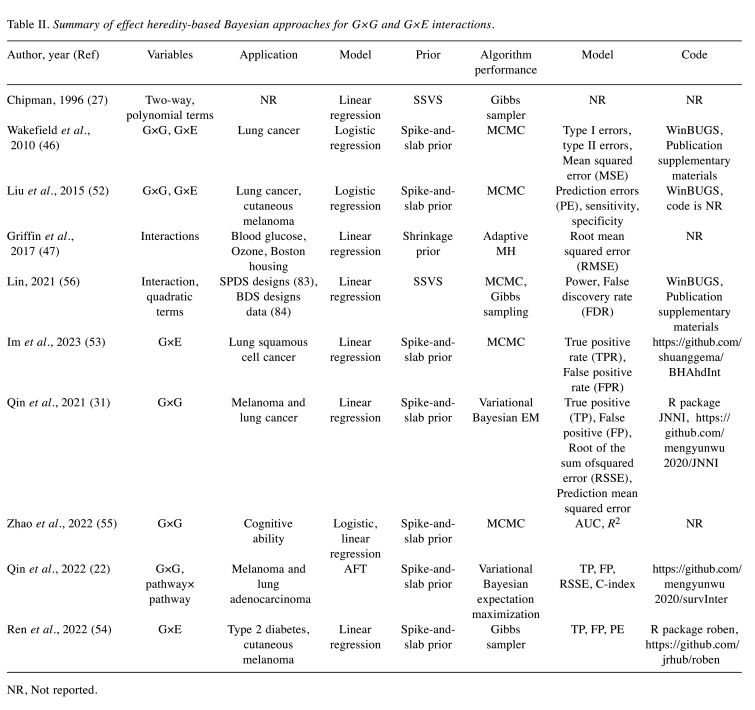
Effect heredity-based approaches. The effect heredity is widely used in studying interaction identification. It incorporates the main terms and interaction dependencies into the analysis (27). Strong heredity indicates that the interaction can only be active if all its main effects are active, whereas weak heredity indicates that the interaction can be active if one of the two main effects is active (27). Models that follow the hereditary principle are generally easier to interpret (44). These models allow us to reliably estimate the main and interaction effects, which increase the power for detecting real signals (30). In contrast, models that violate the hereditary principle are unreliable and perform poorly (31,45). To address the problem of variable selection in interaction studies, two priors dominate the Bayesian literature: spike-and-slab (also referred to as stochastic search variable selection, SSVS) and shrinkage priors (46-49). The spike-and-slab prior is a mixture of two discrete distributions that can be assigned to the model parameters using an indicator variable with different distributions (spike or slab prior distributions) to fit different effects (46). The shrinkage prior allows the regression coefficients to be constrained by imposing a continuous prior distribution (47). The relevant characteristics of these studies are summarized in Table II.
Table II. Summary of effect heredity-based Bayesian approaches for G×G and G×E interactions.
NR, Not reported.
Spike-and-slab prior. Chipman developed priors for interaction identification based on the spike-and-slab prior and two alternative priors, and followed the effect heredity principle (27,50,51). This approach has been extended and applied to case-control studies. Wakefield et al. established a special case of the Bayesian mixture model of G×E (46). Their study included a small number of statistically significant environmental variables. Therefore, it was assumed that the main effect of environmental variables was activated, and only the dependence between the main effect of the genes and the interaction effects needed to be considered (46). Liu et al. proposed a Bayesian hierarchical mixture model that incorporates the effect heredity principle to enhance the study of genetic and environmental main effects as well as G×E and G×G interactions (52).
Researchers have conducted numerous innovative studies to explore the main interaction effects associated with continuous response variables. Im et al. developed a Bayesian approach for a finite mixture regression based on environmental factors, imaging features, and imaging × environment interactions, following the effect heredity principle (53). In addition, another study expanded the method to the least absolute deviation regression to construct a robust Bayesian variable selection approach that can effectively account for heavy-tailed errors and outliers in the response variable while following the heredity of M-I effects (54). Zhao et al. proposed a Bayesian interaction selection model, which worked not only for continuous response but also for qualitative response variables. Furthermore, the model could accurately identify the primary effect features and interactions under hereditary conditions (55). Prior distribution can accommodate correlations between brain topological information and modalities to improve biological plausibility. Qin et al. developed a structured Bayesian approach using a linear model that considered both low- and high-level interactions (31). They integrated the “main effects/interactions-network” information by constructing an adjacency matrix.
They further developed a two-level Bayesian interaction analysis method based on a log-normal accelerated failure time model (AFT) (22). This method allowed the analysis of both low-level gene-gene interactions and high-level pathway-pathway interactions in the model. To improve the interpretability of the results, the effect heredity principle was followed not only between M-I but also between low-level gene-gene interactions and high-level pathway-pathway interactions. Thus, high-level pathway-pathway interactions could only be selected if at least, one low-level gene-gene interaction was selected.
The effect heredity-based approach utilizing spike-and-slab prior had been extended in other fields. For example, a Bayesian model that allowed variable selection and followed the principle of effect heredity was proposed by Lin et al. This model can be used for any split-plot and blocked screening design (56). In addition to the main effects, two-way interaction effects and quadratic effects were included in the model. Particularly, Lin et al. developed strong effect heredity models and two types of weak effect heredity models: the strict weak and relaxed weak effect heredity models.
Shrinkage prior. Griffin et al. developed a simple method to constrain the hierarchical relationship between M-I using strong and weak effect heredity (47). They incorporated the local shrinkage parameters of the two main effects into the local shrinkage parameters of the interaction. In the case of strong heredity, the local shrinkage parameters of the interaction term tended to be large when the local shrinkage parameters of the two main effects were large. In the case of weak heredity, the local shrinkage parameter of the interaction term tends to be large when one of the local shrinkage parameters of the two main effects is large. This study also incorporated these hierarchical priors into a generalized additive model. Yi et al. developed Bayesian models based on logistic and Probit regression, using different shrinkage parameters of Student-t prior distributions for the main and interaction effects, in order to follow the relationship between the main and interaction effects, and implemented a fast and stable expectation-maximization (EM) algorithm for fitting models (30).
Group-based approaches. An alternative analysis strategy commonly used to identify interactions is a group-based approach. Group-based Bayesian approaches employ two types of analysis strategies. The first is that once a group is selected, all variables in that group are included in the final model. Gu et al. proposed a Bayesian two-step Lasso approach to select the main terms and interactions for censored survival outcomes (57). In the first step, the Bayesian group Lasso was used to select the important marker groups, and in the second step, Bayesian adaptive Lasso was used to identify individual biomarkers. Groups were selected when both the main terms and interactions were statistically significant. The Bayesian group Lasso not only serves as an initial screening step to reduce the parameter space but also has a similar role to the strong effect heredity principle for variable selection, as proposed by Chipman (27,57).
The other analysis strategy is that whether the group is activated or not is solely determined by a few variables within the group. In other words, for an active group, not all variables within that group are active. For example, Chen et al. developed a Bayesian sparse group selection approach that used two nested binary indicator variables to indicate whether the group and variables were active or not (58). When none of the variables within the group were active, the group was not activated. However, when a variable within the group was active, the group was activated. Although the analysis strategy was not specifically developed for detecting interactions, it can incorporate the interaction and its two main effects within each group. Similarly, other group-based Bayesian variable selection approaches can be used as alternatives to the Bayesian group Lasso and Bayesian sparse group selection approach (59). Several studies employing systematic reviews have been conducted for group-based Bayesian variable selection approaches (55).
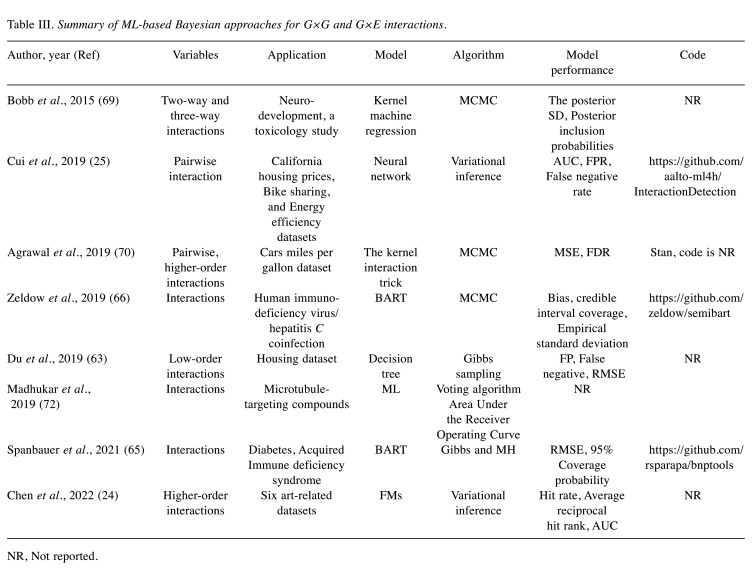
ML-based approaches. Neural networks, decision tree models, regression tree models, linear kernel functions, nonlinear kernel functions, and FMs are commonly used ML techniques (60,61). Generally, models based on ML techniques have a higher prediction accuracy. Novel methods constructed by combining the Bayesian framework with these methods are highly competitive in detecting low- and high-order linear or non-linear interactions. The relevant characteristics of these studies are summarized in Table III. A neural network is a powerful predictive method in ML, and has the potential to identify interactions (62). Cui et al. developed a Bayesian neural network based on neural networks and Hessian to estimate global interactions by aggregating local interactions from trained Bayesian neural networks, thereby improving the estimation accuracy of the interaction effects (25).
Table III. Summary of ML-based Bayesian approaches for G×G and G×E interactions.
NR, Not reported.
Du et al. developed a Bayesian decision tree method to detect low-order interactions (63). It solved the general problem of detecting spurious interactions by introducing Dirichlet process forests. The Bayesian additive regression tree (BART) is a nonparametric ensemble ML model that can reliably estimate regression relationships and has good prediction accuracy when non-linear relationships and interactions are present (29,64). Spanbauer et al. developed a mixed BART model to achieve the goal of precision medicine by extending the BART with a generalized random effect matrix (65). Zeldow et al. developed a new Bayesian semi-parametric model based on BART (semi-BART) to obtain an interpretable coefficient (66).
In previous statistical genomics studies, kernel machine regression (KMR) was mainly used to test the overall effect of a genetic pathway or to study the role of a gene in the presence of possible interactions (67,68). Bobb et al. developed a hierarchical variable selection method in the Bayesian KMR that can efficiently handle highly correlated predictors (69). Agrawal et al. developed a kernel interaction trick that computes the exact posterior of the main effects and the interactions between selected main effects with reduced runtime by orders of magnitude over MCMC applications (70). Furthermore, the interaction could handle not only pairwise interactions but also higher-order multi-way interactions.
FMs are generic-supervised learning methods widely used in ML to efficiently model feature interactions (71). Chen et al. proposed a Bayesian feature interaction selection method based on FMs. This method effectively reduced the number of interactions and extended the model to detect higher-order interactions in higher-order decomposers (24). Significantly, the method incorporates the principle of effect heredity into a second-order interaction study.
Madhukar et al. developed an efficient and accurate platform, BANDIT, based on a Bayesian ML approach to identify drug interaction targets, accelerate drug discovery, and guide clinical application (72). The researchers tested BANDIT and showed that it achieved a high accuracy in identifying shared target interactions and discovering novel potential targets for cancer therapies.
Discussion
As can be observed, several Bayesian approaches are available to detect G×E or/and G×G interactions using omics data. By summarizing the existing literature, the current study is particularly useful for researchers performing interaction studies. SNP data is a special kind of omics data with a strong LD between SNPs, and usually includes a higher number of genotypes with lower frequencies (73,74). Therefore, we first summarized the Bayesian interaction approaches that specifically handle SNPs. Although statistical approaches, such as LBL, are well developed and demonstrate good performance, two core problems remain that are not solved in the existing Bayesian approaches. First, many exposures change over time (e.g., environmental tobacco smoke and pharmaceutical drugs) (75) and the nature of exposure could be related to the G×E interaction. Thus, the G×E interaction may be dynamically changing, and to the best of our knowledge, present Bayesian approaches do not incorporate them. Second, millions of SNPs exist in humans. When only second-order interactions are considered and higher-order interactions are exempted, it can pose a huge computational challenge owing to the existence of hundreds of millions of G×G SNPs (76).
We further summarize other Bayesian approaches for detecting interactions, including effect heredity-based and group-based approaches. Previous studies improved the spike-and-slab and shrinkage priors, and the strong and weak effect heredity of the main terms and interactions were incorporated into the effect heredity-based approaches. However, certain challenges persist. Reasonable values of the hyperparameters in spike-and-slab priors are crucial for identifying and estimating variables. The “semi-automatic” procedure for hyperparameter selection has been proposed; however, in the case of high-dimensional and highly correlated predictors, it needs to be discussed extensively (19,50,77). When using the shrinkage prior with correlated predictors, the multimodality of the posterior may lead to sampling difficulties, particularly slow convergence of the MCMC (78). Furthermore, some existing studies on effect heredity require further studies because they have only incorporated the dependence between the main effects and two-way interactions and did not involve the dependence between terms and three-way interactions. Some group-based approaches perform similarly to strong effect heredity-based approaches (57,79). While the details of these approaches are different, in many cases, there are close conceptual links between them. However, group-based approaches may involve a more complex structure. To summarize, the group-based and effect heredity-based approaches have improved the interpretability of results and have accurately detected interactions of biological significance that affect complex diseases. However, they may perform poorly in terms of prediction accuracy compared with the ML-based approaches.
Finally, we summarized the Bayesian approaches for interaction detection based on ML. ML-based approaches mainly emphasize improving the predictive accuracy of the models (80); hence, the interpretability of the results is poor. Only the Bayesian feature interaction selection method based on FMs incorporates the heredity of the main effects and two-way interactions (24). In addition, the prediction accuracy of ML methods is influenced by the tuning parameters, which may require professional skills and is time consuming (81). Moreover, the application of methods such as neural networks in the detection of complex disease interactions is limited because they are generally processed as black-boxes (82). In the future, new Bayesian ML-based approaches in which interactions are learned using white-box ML models could be considered.
Conclusion
In conclusion, novel biostatistical methods are expected to eliminate the above limitations for future Bayesian approaches to G×G and G×E interactions. Methods based on a Bayesian framework may provide more accurate results, drive ongoing advances in interaction detection, and explain missed heritability during the development of complex diseases. More importantly, they can provide a novel approach for theoretical studies in areas such as precision medicine. Unfortunately, Bayesian approaches are relatively rare to detect the interactions of censored survival outcomes. This is because the complex survival time distribution and hazard function make it difficult to infer the posterior distribution via the conjugate distribution. Unlike the classical estimation methods, Bayesian approaches based on the Cox proportional hazards model are unable to eliminate the baseline hazard, and the posterior sampling and convergence are more challenging. Therefore, future exploration is required.
Conflicts of Interest
The Authors declare no conflicts of interest.
Authors’ Contributions
Na Sun: Collection and writing. Yu Wang: Collection. Jiadong Chu and Qiang Han: Drawing of tables. Yueping Shen: Designed and critically revised the work. All Authors have participated actively in this study and agree to the content of the manuscript and being listed as authors on the paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China under Grant Number 81973143.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Cancer today. Who. Available at: https://gco.iarc.fr/today/home. [Last accessed on November 1, 2023]
- 3.Ayati A, Moghimi S, Salarinejad S, Safavi M, Pouramiri B, Foroumadi A. A review on progression of epidermal growth factor receptor (EGFR) inhibitors as an efficient approach in cancer targeted therapy. Bioorg Chem. 2020;99:103811. doi: 10.1016/j.bioorg.2020.103811. [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest. 2015;125(9):3384–3391. doi: 10.1172/JCI80011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christensen CL, Mikse OR, Cherniack AD, Beauchamp EM, Pugh TJ, Wilkerson MD, Fecci PE, Butaney M, Reibel JB, Soucheray M, Cohoon TJ, Janne PA, Meyerson M, Hayes DN, Shapiro GI, Shimamura T, Sholl LM, Rodig SJ, Freeman GJ, Hammerman PS, Dranoff G, Wong KK. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013;3(12):1355–1363. doi: 10.1158/2159-8290.CD-13-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Guo X, Wang G, Ma W, Liu R, Han X, Li L, Baklaushev VP, Bryukhovetskiy AS, Wang W, Wang X, Zhang C. Real-world study of the incidence, risk factors, and prognostic factors associated with bone metastases in women with uterine cervical cancer using surveillance, epidemiology, and end results (SEER) data analysis. Med Sci Monit. 2018;24:6387–6397. doi: 10.12659/MSM.912071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30(9):418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Sun S, Guo W, Wang Z, Wang X, Zhang G, Zhang H, Li R, Gao Y, Qiu B, Tan F, Gao Y, Xue Q, Gao S, He J. Development and validation of an immune-related prognostic signature in lung adenocarcinoma. Cancer Med. 2020;9(16):5960–5975. doi: 10.1002/cam4.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia L, Zhu Y, Zhang C, Deng S, Deng Y, Yang Z, Mei J, Liu L. Decreased expression of EFCC1 and its prognostic value in lung adenocarcinoma. Ann Transl Med. 2019;7(22):672. doi: 10.21037/atm.2019.10.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhuang Z, Chen L, Mao Y, Zheng Q, Li H, Huang Y, Hu Z, Jin Y. Diagnostic, progressive and prognostic performance of m(6)A methylation RNA regulators in lung adenocarcinoma. Int J Biol Sci. 2020;16(11):1785–1797. doi: 10.7150/ijbs.39046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Lin Y, Shu Y, He J, Gao W. Interaction between N(6)-methyladenosine (m(6)A) modification and noncoding RNAs in cancer. Mol Cancer. 2020;19(1):94. doi: 10.1186/s12943-020-01207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aschard H, Chen J, Cornelis MC, Chibnik LB, Karlson EW, Kraft P. Inclusion of gene-gene and gene-environment interactions unlikely to dramatically improve risk prediction for complex diseases. Am J Hum Genet. 2012;90(6):962–972. doi: 10.1016/j.ajhg.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McAllister K, Mechanic LE, Amos C, Aschard H, Blair IA, Chatterjee N, Conti D, Gauderman WJ, Hsu L, Hutter CM, Jankowska MM, Kerr J, Kraft P, Montgomery SB, Mukherjee B, Papanicolaou GJ, Patel CJ, Ritchie MD, Ritz BR, Thomas DC, Wei P, Witte JS. Current challenges and new opportunities for gene-environment interaction studies of complex diseases. Am J Epidemiol. 2017;186(7):753–761. doi: 10.1093/aje/kwx227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang R, Shen S, Wei Y, Zhu Y, Li Y, Chen J, Guan J, Pan Z, Wang Y, Zhu M, Xie J, Xiao X, Zhu D, Li Y, Albanes D, Landi MT, Caporaso NE, Lam S, Tardon A, Chen C, Bojesen SE, Johansson M, Risch A, Bickeböller H, Wichmann HE, Rennert G, Arnold S, Brennan P, McKay JD, Field JK, Shete SS, Le Marchand L, Liu G, Andrew AS, Kiemeney LA, Zienolddiny-Narui S, Behndig A, Johansson M, Cox A, Lazarus P, Schabath MB, Aldrich MC, Dai J, Ma H, Zhao Y, Hu Z, Hung RJ, Amos CI, Shen H, Chen F, Christiani DC. A large-scale genome-wide gene-gene interaction study of lung cancer susceptibility in Europeans with a trans-ethnic validation in Asians. J Thorac Oncol. 2022;17(8):974–990. doi: 10.1016/j.jtho.2022.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R, Lai L, Dong X, He J, You D, Chen C, Lin L, Zhu Y, Huang H, Shen S, Wei L, Chen X, Guo Y, Liu L, Su L, Shafer A, Moran S, Fleischer T, Bjaanaes MM, Karlsson A, Planck M, Staaf J, Helland Å, Esteller M, Wei Y, Chen F, Christiani DC. SIPA1L3 methylation modifies the benefit of smoking cessation on lung adenocarcinoma survival: an epigenomic-smoking interaction analysis. Mol Oncol. 2019;13(5):1235–1248. doi: 10.1002/1878-0261.12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marley AR, Li M, Champion VL, Song Y, Han J, Li X. Citrus-Gene interaction and melanoma risk in the UK Biobank. Int J Cancer. 2022;150(6):976–983. doi: 10.1002/ijc.33862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji X, Lin L, Shen S, Dong X, Chen C, Li Y, Zhu Y, Huang H, Chen J, Chen X, Wei L, He J, Duan W, Su L, Jiang Y, Fan J, Guan J, You D, Shafer A, Bjaanaes MM, Karlsson A, Planck M, Staaf J, Helland Å, Esteller M, Wei Y, Zhang R, Chen F, Christiani DC. Epigenetic-smoking interaction reveals histologically heterogeneous effects of TRIM27 DNA methylation on overall survival among early-stage NSCLC patients. Mol Oncol. 2020;14(11):2759–2774. doi: 10.1002/1878-0261.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Song Y, Li Y, Wei Y, Shen S, Zhao Y, You D, Su L, Bjaanaes MM, Karlsson A, Planck M, Staaf J, Helland Å, Esteller M, Shen H, Christiani DC, Zhang R, Chen F. A trans-omics assessment of gene-gene interaction in early-stage NSCLC. Mol Oncol. 2023;17(1):173–187. doi: 10.1002/1878-0261.13345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chipman H. New York, NY, Springer New York. 2006. Prior distributions for Bayesian analysis of screening experiments. In: Screening: Methods for experimentation in industry, drug discovery, and genetics. Dean A, Lewis S (eds.) pp. pp. 236–267. [Google Scholar]
- 20.Wolc A, Dekkers JCM. Application of Bayesian genomic prediction methods to genome-wide association analyses. Genet Sel Evol. 2022;54(1):31. doi: 10.1186/s12711-022-00724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu J, Sun NA, Hu W, Chen X, Yi N, Shen Y. The application of Bayesian methods in cancer prognosis and prediction. Cancer Genomics Proteomics. 2022;19(1):1–11. doi: 10.21873/cgp.20298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin X, Ma S, Wu M. Two-level Bayesian interaction analysis for survival data incorporating pathway information. Biometrics. 2023;79(3):1761–1774. doi: 10.1111/biom.13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beis G, Papasotiriou I. Is network meta-analysis a revolutionary statistical tool for improving the reliability of clinical trial results? A brief overview and emerging issues arising. In Vivo. 2023;37(3):972–984. doi: 10.21873/invivo.13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YF, Wang Y, Ren PJ, Wang M, de Rijke M. Bayesian feature interaction selection for factorization machines. Artif Intell. 2022;302:103589. doi: 10.1016/j.artint.2021.103589. [DOI] [Google Scholar]
- 25.Cui T, Marttinen P, Kaski S. Learning global pairwise interactions with Bayesian neural networks. 2019;arXiv:190108361. doi: 10.48550/arXiv.1901.08361. [DOI] [Google Scholar]
- 26.Kim G, Lai CQ, Arnett DK, Parnell LD, Ordovas JM, Kim Y, Kim J. Detection of gene-environment interactions in a family-based population using SCAD. Stat Med. 2017;36(22):3547–3559. doi: 10.1002/sim.7382. [DOI] [PubMed] [Google Scholar]
- 27.Chipman H. Bayesian variable selection with related predictors. Can J Stat. 1996;24(1):17–36. doi: 10.2307/3315687. [DOI] [Google Scholar]
- 28.Wang J, Joshi T, Valliyodan B, Shi H, Liang Y, Nguyen HT, Zhang J, Xu D. A Bayesian model for detection of high-order interactions among genetic variants in genome-wide association studies. BMC Genomics. 2015;16:1011. doi: 10.1186/s12864-015-2217-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chipman HA, George EI, McCulloch RE. BART: Bayesian additive regression trees. Ann Appl Stat. 2010;4(1):266–298. doi: 10.1214/09-aoas285. [DOI] [Google Scholar]
- 30.Yi N, Kaklamani VG, Pasche B. Bayesian analysis of genetic interactions in case-control studies, with application to adiponectin genes and colorectal cancer risk. Ann Hum Genet. 2011;75(1):90–104. doi: 10.1111/j.1469-1809.2010.00605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin X, Ma S, Wu M. Gene-gene interaction analysis incorporating network information via a structured Bayesian approach. Stat Med. 2021;40(29):6619–6633. doi: 10.1002/sim.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10(6):392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, Lander ES. Linkage disequilibrium in the human genome. Nature. 2001;411(6834):199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 34.Surakka I, Horikoshi M, Mägi R, Sarin AP, Mahajan A, Lagou V, Marullo L, Ferreira T, Miraglio B, Timonen S, Kettunen J, Pirinen M, Karjalainen J, Thorleifsson G, Hägg S, Hottenga JJ, Isaacs A, Ladenvall C, Beekman M, Esko T, Ried JS, Nelson CP, Willenborg C, Gustafsson S, Westra HJ, Blades M, de Craen AJ, de Geus EJ, Deelen J, Grallert H, Hamsten A, Havulinna AS, Hengstenberg C, Houwing-Duistermaat JJ, Hyppönen E, Karssen LC, Lehtimäki T, Lyssenko V, Magnusson PK, Mihailov E, Müller-Nurasyid M, Mpindi JP, Pedersen NL, Penninx BW, Perola M, Pers TH, Peters A, Rung J, Smit JH, Steinthorsdottir V, Tobin MD, Tsernikova N, van Leeuwen EM, Viikari JS, Willems SM, Willemsen G, Schunkert H, Erdmann J, Samani NJ, Kaprio J, Lind L, Gieger C, Metspalu A, Slagboom PE, Groop L, van Duijn CM, Eriksson JG, Jula A, Salomaa V, Boomsma DI, Power C, Raitakari OT, Ingelsson E, Järvelin MR, Thorsteinsdottir U, Franke L, Ikonen E, Kallioniemi O, Pietiäinen V, Lindgren CM, Stefansson K, Palotie A, McCarthy MI, Morris AP, Prokopenko I, Ripatti S, ENGAGE Consortium The impact of low-frequency and rare variants on lipid levels. Nat Genet. 2015;47(6):589–597. doi: 10.1038/ng.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moss LC, Gauderman WJ, Lewinger JP, Conti DV. Using Bayes model averaging to leverage both gene main effects and G × E interactions to identify genomic regions in genome-wide association studies. Genet Epidemiol. 2019;43(2):150–165. doi: 10.1002/gepi.22171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kerin M, Marchini J. Inferring gene-by-environment interactions with a Bayesian whole-genome regression model. Am J Hum Genet. 2020;107(4):698–713. doi: 10.1016/j.ajhg.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Liu JS. Bayesian inference of epistatic interactions in case-control studies. Nat Genet. 2007;39(9):1167–1173. doi: 10.1038/ng2110. [DOI] [PubMed] [Google Scholar]
- 38.Zhang BY, Zhang J, Liu JS. Block-based Bayesian epistasis association mapping with application to WTCCC type 1 diabetes data. Ann Appl Stat. 2011;5(3):2052–2077. doi: 10.1214/11-AOAS469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biswas S, Xia S, Lin S. Detecting rare haplotype-environment interaction with logistic Bayesian LASSO. Genet Epidemiol. 2014;38(1):31–41. doi: 10.1002/gepi.21773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Biswas S. An improved version of logistic Bayesian LASSO for detecting rare haplotype-environment interactions with application to lung cancer. Cancer Inform. 2015;14(Suppl 2):11–16. doi: 10.4137/CIN.S17290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Lin S, Biswas S. Detecting rare and common haplotype-environment interaction under uncertainty of gene-environment independence assumption. Biometrics. 2017;73(1):344–355. doi: 10.1111/biom.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papachristou C, Biswas S. Comparison of haplotype-based tests for detecting gene-environment interactions with rare variants. Brief Bioinform. 2020;21(3):851–862. doi: 10.1093/bib/bbz031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Hofmann JN, Purdue MP, Lin S, Biswas S. Logistic Bayesian LASSO for genetic association analysis of data from complex sampling designs. J Hum Genet. 2017;62(9):819–829. doi: 10.1038/jhg.2017.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J, Lim J, Kim Y, Jang W. Bayesian variable selection with strong heredity constraints. Journal of the Korean Statistical Society. 2018;47(3):314–329. doi: 10.1016/j.jkss.2018.03.003. [DOI] [Google Scholar]
- 45.Bien J, Taylor J, Tibshirani R. A LASSO for hierarchical interactions. Ann Stat. 2013;41(3):1111–1141. doi: 10.1214/13-AOS1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakefield J, De Vocht F, Hung RJ. Bayesian mixture modeling of gene-environment and gene-gene interactions. Genet Epidemiol. 2010;34(1):16–25. doi: 10.1002/gepi.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Griffin J, Brown P. Hierarchical shrinkage priors for regression models. Bayesian Anal. 2017;12(1):135–159. doi: 10.1214/15-ba990. [DOI] [Google Scholar]
- 48.Ishwaran H, Rao JS. Spike and slab variable selection: Frequentist and Bayesian strategies. Ann Stat. 2005;33(2):730–773. doi: 10.1214/009053604000001147. [DOI] [Google Scholar]
- 49.Williams J, Xu S, Ferreira MAR. BGWAS: Bayesian variable selection in linear mixed models with nonlocal priors for genome-wide association studies. BMC Bioinformatics. 2023;24(1):194. doi: 10.1186/s12859-023-05316-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.George EI, McCulloch RE. Variable selection via Gibbs sampling. J Am Stat Assoc. 1993;88(423):881–889. doi: 10.1080/01621459.1993.10476353. [DOI] [Google Scholar]
- 51.Raftery AE, Madigan D, Hoeting JA. Bayesian model averaging for linear regression models. J Am Stat Assoc. 1997;92(437):179–191. doi: 10.1080/01621459.1997.10473615. [DOI] [Google Scholar]
- 52.Liu C, Ma J, Amos CI. Bayesian variable selection for hierarchical gene-environment and gene-gene interactions. Hum Genet. 2015;134(1):23–36. doi: 10.1007/s00439-014-1478-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Im Y, Huang Y, Tan A, Ma S. Bayesian finite mixture of regression analysis for cancer based on histopathological imaging-environment interactions. Biostatistics. 2023;24(2):425–442. doi: 10.1093/biostatistics/kxab038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ren J, Zhou F, Li XX, Ma SG, Jiang Y, Wu C. Robust Bayesian variable selection for gene–environment interactions. Biometrics. 2023;79(2):684–694. doi: 10.1111/biom.13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao Y, Wu B, Kang J. Bayesian interaction selection model for multimodal neuroimaging data analysis. Biometrics. 2023;79(2):655–668. doi: 10.1111/biom.13648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin CY. Stochastic search variable selection for split-plot and blocked screening designs. J Qual Technol. 2021;53(1):72–87. doi: 10.1080/00224065.2019.1651621. [DOI] [Google Scholar]
- 57.Gu X, Yin G, Lee JJ. Bayesian two-step Lasso strategy for biomarker selection in personalized medicine development for time-to-event endpoints. Contemp Clin Trials. 2013;36(2):642–650. doi: 10.1016/j.cct.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen RB, Chu CH, Yuan SS, Wu YN. Bayesian sparse group selection. J Comput Graph Stat. 2016;25(3):665–683. doi: 10.1080/10618600.2015.1041636. [DOI] [Google Scholar]
- 59.Xu XF, Ghosh M. Bayesian variable selection and estimation for Group Lasso. Bayesian Anal. 2015;10(4):909–936. doi: 10.1214/14-ba929. [DOI] [Google Scholar]
- 60.He X, Chua T-S. Neural factorization machines for sparse predictive analytics. 2017;arXiv:1708.05027. doi: 10.48550/arXiv.1708.05027. [DOI] [Google Scholar]
- 61.Dreiseitl S, Ohno-Machado L. Logistic regression and artificial neural network classification models: a methodology review. J Biomed Inform. 2002;35(5-6):352–359. doi: 10.1016/S1532-0464(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 62.Tsang M, Cheng D, Liu Y. Detecting statistical interactions from neural network weights. 2017;arXiv:170504977. doi: 10.48550/arXiv.1705.04977. [DOI] [Google Scholar]
- 63.Du J, Linero AR. Interaction detection with Bayesian decision tree ensembles. 2019;arXiv:1809.08524v1. doi: 10.48550/arXiv.1809.08524. [DOI] [Google Scholar]
- 64.Logan BR, Maiers MJ, Sparapani RA, Laud PW, Spellman SR, McCulloch RE, Shaw BE. Optimal donor selection for hematopoietic cell transplantation using Bayesian machine learning. JCO Clin Cancer Inform. 2021;5:494–507. doi: 10.1200/CCI.20.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spanbauer C, Sparapani R. Nonparametric machine learning for precision medicine with longitudinal clinical trials and Bayesian additive regression trees with mixed models. Stat Med. 2021;40(11):2665–2691. doi: 10.1002/sim.8924. [DOI] [PubMed] [Google Scholar]
- 66.Zeldow B, Lo Re V 3rd, Roy J. A semiparametric modeling approach using Bayesian additive regression trees with an application to evaluate heterogeneous treatment effects. Ann Appl Stat. 2019;13(3):1989–2010. doi: 10.1214/19-AOAS1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu D, Lin X, Ghosh D. Semiparametric regression of multidimensional genetic pathway data: least-squares kernel machines and linear mixed models. Biometrics. 2007;63(4):1079–1088. doi: 10.1111/j.1541-0420.2007.00799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maity A, Lin X. Powerful tests for detecting a gene effect in the presence of possible gene-gene interactions using garrote kernel machines. Biometrics. 2011;67(4):1271–1284. doi: 10.1111/j.1541-0420.2011.01598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bobb JF, Valeri L, Claus Henn B, Christiani DC, Wright RO, Mazumdar M, Godleski JJ, Coull BA. Bayesian kernel machine regression for estimating the health effects of multi-pollutant mixtures. Biostatistics. 2015;16(3):493–508. doi: 10.1093/biostatistics/kxu058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Agrawal R, Trippe B, Huggins J, Broderick T. The Kernel interaction trick: Fast Bayesian discovery of pairwise interactions in high dimensions. 2022;arXiv:1905.06501. doi: 10.48550/arXiv.1905.06501. [DOI] [Google Scholar]
- 71.Rendle S. 2010 IEEE International conference on data mining. Factorization machines. ; pp. pp. 995–1000. [DOI] [Google Scholar]
- 72.Madhukar NS, Khade PK, Huang L, Gayvert K, Galletti G, Stogniew M, Allen JE, Giannakakou P, Elemento O. A Bayesian machine learning approach for drug target identification using diverse data types. Nat Commun. 2019;10(1):5221. doi: 10.1038/s41467-019-12928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Slatkin M. Linkage disequilibrium—understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 2008;9(6):477–485. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Los Campos G, Grueneberg A, Funkhouser S, Pérez-Rodríguez P, Samaddar A. Fine mapping and accurate prediction of complex traits using Bayesian Variable Selection models applied to biobank-size data. Eur J Hum Genet. 2023;31(3):313–320. doi: 10.1038/s41431-022-01135-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aschard H, Lutz S, Maus B, Duell EJ, Fingerlin TE, Chatterjee N, Kraft P, Van Steen K. Challenges and opportunities in genome-wide environmental interaction (GWEI) studies. Hum Genet. 2012;131(10):1591–1613. doi: 10.1007/s00439-012-1192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li P, Guo MZ, Wang CY, Liu XY, Zou Q. An overview of SNP interactions in genome-wide association studies. Brief Funct Genomics. 2015;14(2):143–155. doi: 10.1093/bfgp/elu036. [DOI] [PubMed] [Google Scholar]
- 77.Perrakis K, Ntzoufras I. Stochastic Search Variable Selection (SSVS) Wiley StatsRef: Statistics Reference. 2015;Online:1–6. doi: 10.1002/9781118445112.stat07829. [DOI] [Google Scholar]
- 78.Piironen J, Vehtari A. Sparsity information and regularization in the horseshoe and other shrinkage priors. Electron J Stat. 2017;11(2):5018–5051. doi: 10.1214/17-ejs1337si. [DOI] [Google Scholar]
- 79.Lai WT, Chen RB. A review of Bayesian group selection approaches for linear regression models. Wiley Interdiscip Rev Comput Stat. 2021;13(4):e1513. doi: 10.1002/wics.1513. [DOI] [Google Scholar]
- 80.Sato M, Morimoto K, Kajihara S, Tateishi R, Shiina S, Koike K, Yatomi Y. Machine-learning approach for the development of a novel predictive model for the diagnosis of hepatocellular carcinoma. Sci Rep. 2019;9(1):7704. doi: 10.1038/s41598-019-44022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Idri A, Hosni M, Abnane I, Carrillo de Gea JM, Fernández Alemán JL. Impact of parameter tuning on machine learning based breast cancer classification. In: Rocha Á, Adeli H, Reis L, Costanzo S (eds). New Knowledge in Information Systems and Technologies. WorldCIST’19 2019. Advances in Intelligent Systems and Computing, vol 932. Cham, Switzerland, Springer. doi: 10.1007/978-3-030-16187-3_12. [DOI] [Google Scholar]
- 82.Liang Y, Li SG, Yan CG, Li MZ, Jiang CJ. Explaining the black-box model: A survey of local interpretation methods for deep neural networks. Neurocomputing. 2021;419:168–182. doi: 10.1016/j.neucom.2020.08.011. [DOI] [Google Scholar]
- 83.Lin CY, Yang P. Response surface methodology using split-plot definitive screening designs. J Qual Technol. 2015;47(4):351–362. doi: 10.1080/00224065.2015.11918139. [DOI] [Google Scholar]
- 84.Jones B, Nachtsheim CJ. Blocking schemes for definitive screening designs. Technometrics. 2016;58(1):74–83. doi: 10.1080/00401706.2015.1013777. [DOI] [Google Scholar]