Abstract
Although water is essential for life, as per the United Nations, around 2 billion people in this world lack access to safely managed drinking water services at home. Herein we report the development of a two-dimensional (2D) fluorinated graphene oxide (FGO) and polyethylenimine (PEI) based three-dimensional (3D) porous nanoplatform for the effective removal of polyfluoroalkyl substances (PFAS), pharmaceutical toxins, and waterborne pathogens from contaminated water. Experimental data show that the FGO-PEI based nanoplatform has an estimated adsorption capacity (qm) of ∼219 mg g–1 for perfluorononanoic acid (PFNA) and can be used for 99% removal of several short- and long-chain PFAS. A comparative PFNA capturing study using different types of nanoplatforms indicates that the qm value is in the order FGO-PEI > FGO > GO-PEI, which indicates that fluorophilic, electrostatic, and hydrophobic interactions play important roles for the removal of PFAS. Reported data show that the FGO-PEI based nanoplatform has a capability for 100% removal of moxifloxacin antibiotics with an estimated qm of ∼299 mg g–1. Furthermore, because the pore size of the nanoplatform is much smaller than the size of pathogens, it has a capability for 100% removal of Salmonella and Escherichia coli from water. Moreover, reported data show around 96% removal of PFAS, pharmaceutical toxins, and pathogens simultaneously from spiked river, lake, and tap water samples using the nanoplatform.
1. Introduction
Drinking water is essential for human beings on this earth.1−4 As per the United Nations International Children’s Emergency Fund (UNICEF), one in four persons in this world does not have access to basic drinking water services.1 Based on a recent report by the Environmental Protection Agency (EPA), 45% of US tap water is contaminated with forever chemicals like per- and polyfluoroalkyl substances (PFAS).2 As per the EPA, the concentrations of per- and polyfluoroalkyl substances are higher than the EPA limit (70 ng L–1) in ground and surface water systems in 3186 locations in 50 states in the USA.2−6 Since 2016, as per US EPA instructions, the industry has been using short-chain homologues of the long-chain PFAS.2−6 In 2022, reported data by the US EPA indicated that the concentrations of short-chain perfluorobutanesulfonic acid (PFBS), perfluorobutanoic acid (PFBA), and tetrafluoro-heptafluoropropoxy propanoic acid (GenX) are increasing in environmental samples, which is alarming.2−6 Several studies indicate that short- as well as long-chain PFAS can play a major role in hepatic, hematological, and renal cancer.7−12 To tackle the PFAS contamination in water challenges, scientists are designing novel adsorbents with high affinity for per- and polyfluoroalkyl substances for PFAS removal from water.9−27 Because C–F bonds in per- and polyfluoroalkyl substances can participate in electrostatic, hydrophobic, and fluorophilic interactions, several efforts have been devoted to design new adsorbents to remove PFAS using dipole–dipole, ionic–dipolar, and fluorophilic interactions.12−22 However, several reported data show high removal capacity for long-chain per- and polyfluoroalkyl substances, and the removal efficiency decreased significantly for short-chain PFAS.12−22 Herein, we have designed a 2D fluorinated graphene oxide (FGO) and polyethylenimine (PEI) based three-dimensional (3D) porous nanoplatform for the effective removal of short-chain per- and polyfluoroalkyl substances like perfluorobutanoic acid (PFBA) and perfluorobutanesulfonic acid (PFBS), as well as long-chain per- and polyfluoroalkyl substances like perfluorohexanesulfonate (PFHxS) and perfluorononanoic acid (PFNA). We have also used only FGO and polyethylenimine (PEI) attached graphene oxide (PEI-GO) to understand the role of hydrophobic, electrostatic, and fluorophilic interactions in the short- and long-chain PFAS removal process.
According to the UN and WHO, every 2 min, a child dies from a water-borne illness in this world.1,29−40 On the other hand, about 7.2 million Americans get sick every year from diseases spread through waterborne pathogens.1,32−52 Because of the low infectious dose, disease transmission is facilitated heavily via the outbreaks of diarrhea, cholera, and typhoid.1,29−37 Recent reports indicate that direct costs of all illnesses caused by waterborne microorganisms are around $3.3 billion/year caused by 17 pathogens.1,40−52 Several antibiotics such as fluoroquinolone, tetracycline, fluoroquinolone, etc., are present in aquatic environments and drinking water.29−36 Recently, several reports indicated that antibiotic residues in water can produce antibiotic-resistant bacteria, which are a serious threat for our society.29−36 To tackle the challenges, scientists are designing a novel adsorbent-based technology that has the potential to improve access to safe drinking water by removing chemical and biological toxins.29−52 Because currently available antibiotics are polar molecules, several adsorbents have been designed to capture via hydrophobic as well as electrostatic interactions.35,36,43−52 However, because of the low removal efficacy, they are not very effective.35,36,43−52 To address the chemical and biological toxin-free drinking water problem, we have used the same FGO-PEI based three-dimensional (3D) porous nanoplatform for the effective removal of moxifloxacin antibiotics via fluorophilic, electrostatic, and hydrophobic interactions. To understand the role of fluorophilic and electrostatic interactions for removing different antibiotics, we have used tetracycline and moxifloxacin antibiotics, where moxifloxacin can only participate in fluorophilic interaction with the adsorbent. In addition, the same nanoplatform has been used for the removal of Salmonella and Escherichia coli waterborne pathogens from contaminated water samples. Because the pore size of the nanoplatform is much smaller than the size of pathogens, 100% of the Salmonella and Escherichia coli are expected to be captured during filtration.
2. Methods
2.1. Synthesis of Graphene Oxide from Graphite
We developed water-soluble graphene oxide from graphite using the improved Hummer’s method, as we and others have reported before.34−43 For the development of graphene oxide (GO), we oxidized graphite using strong oxidizing agents such as sodium nitrate, concentrated sulfuric acid, and potassium permanganate.34−39 Experimental details are reported in the Supporting Information.
2.2. Synthesis of Fluorinated Graphene
We developed fluorinated graphene oxide from GO using HF.24−28,33 For this purpose, GO and HF were autoclaved and maintained at 180 °C for 30 h. Experimental details are reported in the Supporting Information.
2.3. Synthesis of the 2D Fluorinated Graphene Oxide and PEI Based Three-Dimensional (3D) Porous Nanoplatform
We developed the 2D fluorinated graphene oxide and polyethylenimine (PEI) based 3D porous nanoplatform (FGO-PEI) using PEI as a cross-linking agent between two fluorinated graphene oxides. For this purpose, we used the EDC–NHS coupling procedure, as we have reported before.34−39 Experimental details are reported in the Supporting Information.
2.4. Determining Perfluorobutanoic Acid (PFBA) and Perfluorobutanesulfonic Acid (PFBS) Removal Amount Using LC–MS
For the determination of the removal amount of perfluorobutanoic acid (PFBA) and perfluorobutanesulfonic acid (PFBS) using the two-dimensional (2D) fluorinated graphene oxide and PEI based three-dimensional (3D) porous nanoplatform, we used LC–MS (Agilent technologies).11−21 Details are reported in the Supporting Information.
2.5. Surface Enhanced Raman Spectroscopy (SERS) Analysis for the Determination of the Concentration of Tetracycline and Moxifloxacin Antibiotics from Environmental Samples
For the measurement of SERS, we used the confocal Raman system with laser excitation of 670 nm gold nanoparticles as the plasmonic surface.27−30 Experimental details have been reported before.27−30 Details are reported in the Supporting Information.
2.6. Determining the Number of Bacteria Captured during Filtration Using the Colony Counting Technique
The number of live bacteria was counted in the water samples with a colony counter after 24 h at 37 °C incubation, as we have reported before.34−49,47 Details are reported in the Supporting Information.
3. Results and Discussion
3.1. Design and Characterization of the 2D Fluorinated Graphene Oxide and Polyethylenimine (FGO-PEI) Based 3D Porous Nanoplatform
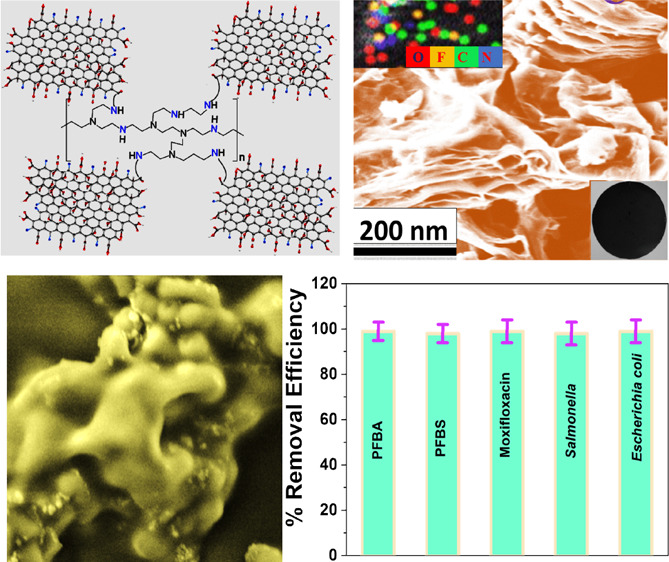
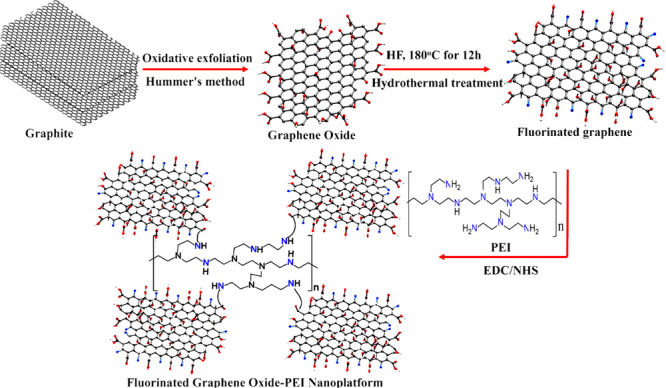
For the development of the 2D fluorinated graphene oxide and polyethylenimine based 3D porous nanoplatform, we used a three-step synthesis process as reported in Figure 1. For this purpose, in the first step, we developed water-soluble graphene oxide from graphite using the improved Hummer’s method, as we and others have reported before.34−43 At the end, the yellow mixture was washed with 5% HCl followed by ethanol to remove the metal impurities. The yellow suspension was centrifuged at very high speed several times until the pH was near neutral. The final solution was dried by lyophilization. After lyophilization, graphene oxides were characterized by tunneling electron microscopic (TEM), as shown in Figure 2A, which indicates a 2D structure. The energy dispersive spectrometry (EDS) mapping data reported as an inserted figure in Figure 2A clearly shows the presence of C and O for 2D GO.34−39 Similarly, the energy dispersive X-ray analysis data reported in Table 1 also show the presence of C and O in 2D GO. The X-ray diffraction (XRD) data for FGO, as reported in Figure 2E, show the presence of (002) and (100) reflection peaks.24−26,33 The reported broad diffraction peak observed from FGO is associated with the disorder induced by the fluorination process.34−39
Figure 1.
Scheme shows the synthetic path used for the development of the 2D fluorinated graphene oxide and polyethylenimine (PEI) based three-dimensional (3D) porous nanoplatform.
Figure 2.
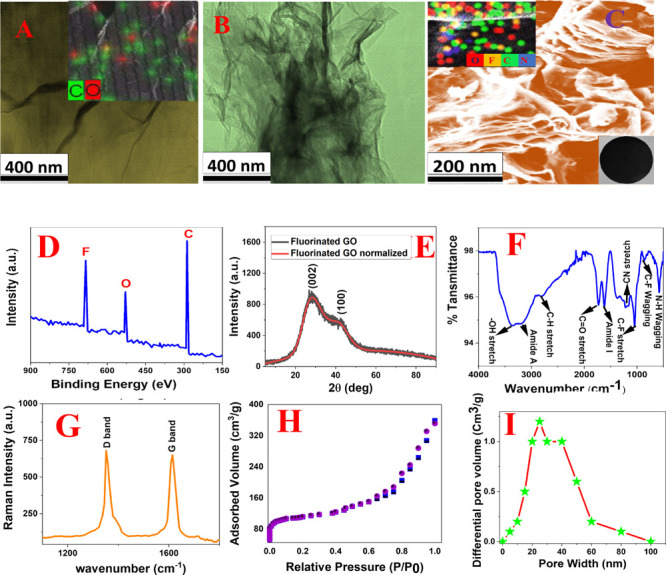
(A) TEM image shows the morphology of 2D graphene oxide. Inserted energy dispersive spectrometry (EDS) mapping shows the presence of C and O in 2D graphene oxide. (B) TEM image shows the morphology of 2D fluorinated graphene oxide. (C) SEM image shows the morphology of the 2D fluorinated graphene oxide and PEI based 3D nanoplatform. Inserted energy dispersive spectrometry (EDS) mapping shows the presence of C, O, N, and F in the 3D nanoplatform. Another inserted image shows the nanoplatform developed using the FGO-PEI based 3D nanoplatform. (D) The XPS spectrum from FGO confirms peaks at 288.5, 533.2, and 688.3 eV, which are due to C, O, and F, respectively. (E) XRD data from FGO show the presence of (002) and (100) reflection peaks. (F) FTIR spectra from the FGO-PEI based 3D nanoplatform shows the presence of −C–F, −C=O, −O–H, amide-I, and amide-A peaks. (G) Raman spectra from the FGO-PEI based nanoplatform shows the presence of D and G peaks. (H) N2 adsorption/desorption isotherm of the FGO-PEI based 3D nanoplatform. (I) Pore size distributions from the FGO-PEI based 3D nanoplatform show that pore size varies from 5 to 100 nm, with the highest pore density being around 30 nm.
Table 1. Elemental Composition (Weight %) for the 2D GO, 2D FGO, and FGO-PEI Based Nanoplatform.
| sample | wt % C | wt % O | wt % F | wt % N |
|---|---|---|---|---|
| 2D GO | 70 | 30 | 0 | 0 |
| 3D FGO | 70 | 12 | 18 | 0 |
| 3D platform | 67 | 5 | 18 | 10 |
In the second step, we developed fluorinated graphene oxide from GO using HF.24−28,33 Experimental details are reported in the Supporting Information. The resultant solution was naturally cooled to room temperature. Then the fluorinated graphene semisolid compound was obtained and filtered followed by washing with water to remove excess acid. The pure fluorinated graphene was finally freeze-dried through lyophilization for future use. After lyophilization, graphene oxides were characterized by TEM, as shown in Figure 2B, which indicates the 2D structure. The EDS mapping data (inserted picture in Figure 2B) and energy dispersive X-ray analysis (EDX) analysis data (Table 1) clearly show the presence of C, O, and F for 2D fluorinated graphene oxide. The X-ray diffraction (XRD) data for FGO (Figure 2E) show the presence of (002) and (100) reflection peaks.24−26,33 The reported broad diffraction peak observed from FGO is associated with the disorder induced by the fluorination process. The XPS spectrum from FGO (Figure 2D) confirms peaks at 288.5, 533.2, and 688.3 eV that are due to C, O, and F, respectively.
In the third step, we developed the 2D fluorinated graphene oxide and polyethylenimine (PEI) based 3D porous nanoplatform (FGO-PEI). For this purpose, PEI was used as a cross-linking agent between two fluorinated graphene oxides via the formation of amide bond as shown in Figure 1. For this purpose, we used the EDC–NHS coupling procedure, as we have reported before.34−39 Experimental details are reported in the Supporting Information.
For this purpose, fluorinated graphene was dispersed in DMF, and then the mixture was sonicated for 1 h. Then under a nitrogen atmosphere, 250 mL of EDC (1 mg/mL) and 150 mL of NHS (1 mg/mL) were added to the DMF suspended fluorinated graphene solution. After that, 400 mg of PEI was added to the resulting mixture and stirred for several hours at room temperature. Finally, the semisolid fluorinated graphene-PEI nanocomposite was used for the development of the nanoplatform, whose diameter is ∼5 cm and surface area is ∼78 cm2, as shown in the inserted figure in Figure 2C. After that, the nanoplatform was characterized by SEM, as shown in Figure 2C, which indicates the 3D porous structure. EDS mapping data reported as the inserted figure in Figure 2C clearly show the presence of C, O, F, and N in the 3D porous platform. Similarly, EDX analysis data reported in Table 1 also show the presence of C, O, N, and F in the 3D porous platform. Figure 2F shows the Fourier-transform infrared spectroscopy (FTIR) data from the 3D porous platform. FTIR spectra show the presence of amide A and O–H stretching vibrational band between 3000 and 3500 cm–1 range. The FTIR peak at 1120 cm–1 from the 3D porous platform van is due to the C–F bond stretching. The observed IR band at 1732 cm–1 is due to the C=O stretching of the carboxylic functional groups. Similarly, the observed IR band at 1618 cm–1 is due to the amide-I band, which indicates amide bond formation. Figure 2G shows the Raman spectra from the FGO-PEI based nanoplatform that clearly show the G-band peak at 1580 cm–1 due to the E2g mode of graphitic carbon and D-band peak at 1350 cm–1 due to the disordered graphite carbon atoms.34−39
Next, to determine the specific surface area for FGO-PEI based 3D porous nanoplatform, we used a Tristar II 3020 surface area analyzer that has the capability for surface area and porosity measurements. Experimental details have been reported by us and others previously.34−39 For this purpose, we used the Barrett–Joyner–Halenda (BJH) method for the determination of pore size distribution for the FGO-PEI based 3D porous nanoplatform.34−39Figure 2H shows the N2 adsorption/desorption isotherm of the FGO-PEI based 3D nanoplatform. From the measurement, we determined the specific surface area and pore volume for the FGO-PEI based nanoplatform. The specific surface area for the 3D porous nanoplatform is 360 m2 g–1, and the pore volume for the 3D porous nanoplatform is 0.590 cm3 g–1. Figure 2H shows the pore size distributions from the FGO-PEI based 3D nanoplatform. Using the BJH method, we determined that the pore size for the FGO-PEI 3D nanoplatform varies from 2 to 100 nm, with the highest pore density around 30 nm. Because the pore size and distribution are highly dependent on the method of preparation and experimental conditions, they may vary with different batches of experiments. For the measurement of the water flux for the FGO-PEI based 3D nanoplatform, we used the electronic balance method, as we have reported before.34−39 By determining the amount of permeated water through the FGO-PEI based 3D nanoplatform, we estimated that the water flux is around 281 L m–1 h–1 bar–1.
3.2. Capturing and Separating PFBA, PFBS, PFHxS, and PFNA from Environmental Samples
As we have discussed before, PFAS contamination is a serious concern for society due to its association with adverse health effects such as cancer, immunity decline, and liver damage.2−8 As a result, there is an urgent need for advanced systems that can separate PFAS from water.9−22 Next, to determine the capturing and separating efficiency of PFBA, PFBS, PFHxS, and PFNA from environmental samples using the FGO-PEI based 3D nanoplatform, we performed the following experiments. Initially, we infected the drinking water samples with PFBA, PFBS, PFHxS, and PFNA selectively and simultaneously. For the selective PFAS separation experiment, we spiked water samples with 1000 ng/L of PFBA, PFBS, PFHxS, or PFNA. For the mixture of PFBA, PFBS, PFHxS, and PFNA experiment, we spiked water samples with 250 ng/L concentration for each of them. After that, we performed filtration using the FGO-PEI nanoplatform. At the end, for the determination of the removal amount for PFBA and PFBS using the nanoplatform, we used LC–MS (Agilent technologies). Experimental details are reported in the Supporting Information. Before using the nanoplatform for the removal of PFAS, we performed a stability experiment for the nanoplatform at different pH values. For this purpose, we exposed the nanoplatform in water for a few weeks and determined whether any F is released from nanoplatform. We used LC–MS to determine the amount of F released. We did not observe any release of F from the nanoplatform even after being exposed to water for 21 days at pH 7. Similarly, we also performed the same experiment with the variation of pH between 6 and 10. We did not observe any release of F from the nanoplatform even after being exposed to water for 21 days and different pH values.
The removal efficiency (RE) for PFBA, PFBS, PFHxS, and PFNA toxic chemicals was determined using eq 1:9−24
| 1 |
where Cinitial is the concentration of PFBA, PFBS, PFHxS, or PFNA toxic chemicals before separation and Ct is the concentration of PFBA, PFBS, PFHxS, or PFNA toxic chemicals at time t during the separation process. The amount of PFBA, PFBS, PFHxS, and PFNA toxic chemicals removed by GO, FGO, PEI-GO, or PEI-FGO adsorbent was determined using eq 2:9−24
| 2 |
where qt is the amount of PFBA, PFBS, PFHxS, and PFNA toxic chemicals absorbed separately per gram of GO, FGO, PEI-GO, or PEI-FGO at time t and Cinitial is the concentration of PFBA, PFBS, PFHxS, or PFNA toxic chemicals before separation.
On the other hand, Ct is the concentration of toxic chemicals at time t during separation process and CA is the concentration of the GO, FGO, PEI-GO, or PEI-FGO adsorbent. For the determination of the rate of removal via adsorption of PFBA, PFBS, PFHxS, and PFNA toxic chemicals separately, we used Ho and McKay’s pseudo-second-order adsorption model as reported in eq 3:9−24
| 3 |
where qt is the amount of PFBA, PFBS, PFHxS, and PFNA toxic chemicals absorbed separately per gram of GO, FGO, PEI-GO, or PEI-FGO at time t. On the other hand, qe is the quantity of PFBA, PFBS, PFHxS, and PFNA toxic chemicals adsorbed separately at equilibrium, and kobs is the rate constant for the separation process via adsorption using the GO, FGO, PEI-GO, or PEI-FGO adsorber. For determining the maximum adsorption capacity for each PFBA, PFBS, PFHxS, and PFNA toxic chemical using the PEI-FGO adsorbent, we used the Langmuir adsorption model as reported in eq 4:9−24
| 4 |
where qe is the quantity of PFBA, PFBS, PFHxS, and PFNA toxic chemicals adsorbed separately at equilibrium; qm is the maximum capacity of each PFBA, PFBS, PFHxS, and PFNA toxic chemical adsorption by the PEI-FGO adsorbent at equilibrium; Ce is the concentration of each PFBA, PFBS, PFHxS, and PFNA toxic chemical; and b is the Langmuir constant for the adsorption process. For this experiment, we used 10 μg to 1000 μg/L toxic chemicals.
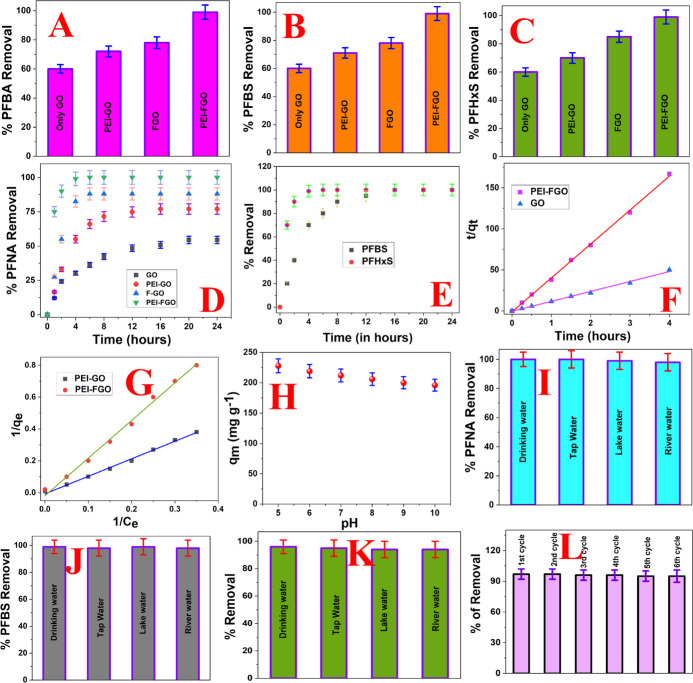
As shown in Figure 3A,B, reported experimental data indicate that the removal efficiency for short-chain per- and polyfluoroalkyl substances like PFBA or PFBS is ∼99% when the FGO-PEI nanoplatform has been used. Similarly, as shown in Figure 3C,D, reported experimental data indicate that the removal efficiency for long-chain per- and polyfluoroalkyl substances like PFHxS or PFNA is ∼99% when the FGI-PEI nanoplatform has been used. To compare the removal efficiency with the GO based nanoplatform, FGO based nanoplatform, and GO-PEI based nanoplatform, we have performed the PFBA, PFBS, PFHxS, and PFNA separation experiment individually from drinking water. As reported in Figure 3A, we have observed ∼45% separation of PFBA when only the GO based nanoplatform has been used. Furthermore, we have observed ∼70% separation of PFBA when the GO-PEI based nanoplatform has been used. On the other hand, we have observed ∼80% separation of PFBA when the FGO based nanoplatform has been used. Moreover, we have observed ∼99% separation of PFBA when the FGO-PEI based nanoplatform has been used. Similarly, as reported in Figure 3B–D, we have observed the highest separation efficiency for PFBS, PFHxS, and PFNA toxic chemicals when the FGO-PEI based nanoplatform has been used. On the other hand, the lowest separation efficiency for PFBS, PFHxS, and PFNA toxic chemicals are observed when only the GO based nanoplatform has been used. Furthermore, in all cases, the separation efficiency is higher when the FGO based nanoplatform has been used in comparison to the separation efficiency observed when the GO-PEI based nanoplatform has been used.
Figure 3.
(A) Perfluorobutanoic acid (PFBA) removal efficiency from drinking water using the 2D-GO based nanoplatform, PEI-attached 2D-GO (GO-PEI) based nanoplatform, and PEI-attached 2D F-GO (FGO-PEI) based nanoplatform. For this experiment, we used 1000 ng/L of PFBA infected drinking water. (B) Perfluorobutanesulfonic acid (PFBS) removal efficiency from drinking water using the 2D-GO based nanoplatform, GO-PEI based nanoplatform, and FGO-PEI based nanoplatform. For this experiment, we used 1000 ng/L of PFBS infected drinking water. (C) Perfluorohexanesulfonate (PFHxS) removal efficiency from drinking water using the 2D-GO based nanoplatform, GO-PEI based nanoplatform, and FGO-PEI based nanoplatform. For this experiment, we used 1000 ng/L of PFHxS infected drinking water. (D) Variation of perfluorononanoic acid (PFNA) removal efficiency with time for the GO based nanoplatform, PEI-attached 2D-GO (GO-PEI) based nanoplatform, and PEI-attached 2D F-GO (FGO-PEI) based nanoplatform. (E) Plots show the time-dependent removal efficiency for PFBS and PFHxS using the FGO-PEI based nanoplatform. For this experiment, we used 1000 ng/L of PFBS or PFHxS infected water samples. (F) Plot shows the variation of (t/qt) with time for PFNA using the PEI-FGO and GO adsorber separately, where qt is the quantity of PFNA removed per gram of the FGO-PEI or GO nanoplatform. (G) Plot shows the variation of 1/qe with 1/Ce for PFNA using the PEI-FGO and PEI-GO adsorber separately, where qe is the quantity of PFNA adsorbed at equilibrium and Ce is the concentration of PFNA. (H) Plot shows how the PFNA removal efficiency varies with pH when the FGO-PEI based nanoplatform was used. (I) PFNA removal efficiency from tap water, Mississippi river water, lake water, and drinking water using the FGO-PEI based nanoplatform. For this experiment, we used 1000 ng/L of PFBA infected water samples. (J) PFBS removal efficiency from tap water, Mississippi river water, lake water, and drinking water using the FGO-PEI based nanoplatform. For this experiment, we used 1000 ng/L of PFBS infected water samples. (K) Removal efficiency of PFBS, PFBA, PFHxS, and PFNA simultaneously from tap water, Mississippi river water, lake water, and drinking water using the FGO-PEI based nanoplatform. For this experiment, we used 250 ng/L of PFBS, 250 ng/L ng/L of PFBA, 250 ng/L of PFHxS, and 250 ng/L of PFNA infected water samples. (L) Plot shows how the removal efficiency for PFBS, PFBA, PFHxS, and PFNA together varies with the number of cycles of filtration when we used the FGO-PEI based nanoplatform. For this experiment, we used 250 ng/L of PFBS, 250 ng/L of PFBA, 250 ng/L of PFHxS, and 250 ng/L of PFNA infected water samples.
Figure 3D shows the kinetics of the PFNA removal process using GO, FGO, PEI-GO, and PEI-FGO adsorber individually. Time-dependent data show that the removal efficiency reached ∼75% within an hour when PEI-FGO has been used as adsorber and the removal efficiency is ∼100% within 4 h. On the other hand, the removal efficiency reached ∼89% after 8 h and then remained the same when FGO has been used as adsorber. Similarly, the removal efficiency reached ∼75% after 12 h and then remained the same when PEI-GO has been used as adsorber. Moreover, the removal efficiency reached ∼45% after 16 h and then remained the same when GO has been used as adsorber. All the above-reported data clearly indicate that the separation efficiency as well as the kinetics of separation is highest for the FGO-PEI based nanoplatform. The observed highest separation efficiency for the FGO-PI based nanoplatform can be due to the several facts as discussed below. Because graphene oxide is well-known to carry a negative surface charge due to the presence of oxygen-containing (−OH, −CO2H, epoxy) functional groups, it undergoes a repulsive electrostatic interaction with anionic per- and polyfluoroalkyl substances, and as a result, the removal efficiency is low when only graphene oxides are used, as reported in Figure 3A–D.
On the other hand, because of the presence of unique C–F bonds in FGO, it can interact with PFBA, PFBS, PFHx, or PFNA using dipole–dipole, ionic–dipolar, and fluorophilic interactions, and the toxin removal capability is highly increased. Because we have used FGO-PEI based nanoplatform, the electrostatic interactions between the carboxylate or sulfonate headgroup in per- and polyfluoroalkyl substances and the cationic amines of the PEI also can play important roles in enhancing the removing efficiency. The amine group from PEI also can participate in hydrophobic interactions with per- and polyfluoroalkyl substances, which will help to remove the toxin. Experimental data indicate ∼70% separation of PFBA, PFBS, PFHx, or PFNA when the GO-PEI based nanoplatform has been used, and on the other hand, we have observed ∼99% separation of per- and polyfluoroalkyl substances when the FGO-PEI based nanoplatform has been used. Our observed experimental data clearly show that fluorophilic interaction, which is known as C–F···F–C interaction, plays a very important role for the ∼99% separation of PFBA, PFBS, PFHx, or PFNA when the FGO-PEI based nanoplatform has been used. It is interesting to note from Figure 3A–D that the separation efficiency by the FGO based nanoplatform is higher (∼90%) for long-chain per- and polyfluoroalkyl substances like PFHxS or PFNA than for short-chain per- and polyfluoroalkyl substances (∼80%) like PFBA or PFBS. Similarly, Figure 3E shows the time-dependent removal efficiency for PFBS and PFHxS using the FGO-PEI based nanoplatform, which indicates that the rate of removal is much higher for long-chain per- and polyfluoroalkyl substances than short-chain per- and polyfluoroalkyl substances. The above data clearly indicate that the fluorophilic interactions and hydrophobic interactions for long-chain per- and poly fluoroalkyl substances like PFHxS or PFNA are much higher than those of short-chain per- and polyfluoroalkyl substances like PFBA or PFBS, which are due to the higher number of fluorine atoms.
Next, to determine the absorption kinetics, we have used Ho and McKay’s pseudo-second-order adsorption model,9−24 as we have discussed before. Using eqs 2 and 3 and Figure 3F, we have estimated the rate constant for the PFNA adsorption (kobs) process for FGO-PEI nanoplatform, which is ∼4.38 g mg–1 h–1. Similarly, using Figure 3G and eq 4, we have estimated that the maximum capacity of PFNA adsorption by the PEI-FGO adsorbent at equilibrium (qm) is ∼208.4 mg g–1. The experimentally observed maximum capacity by PEI-FGO is comparable with the ionic fluorogels reported by Kumarasamy et al. (qm = 217 mg g–1)12 and magnetic fluorinated polymer sorbents reported by Tan et al. (qm = 200 mg g–1).16 The experimentally observed value is much higher than the literature reported value for fluorine-rich calixarene based porous polymers19 and amine functionalized COF.18 On the other hand, the experimentally observed estimated maximum capacity of PFNA adsorption by the PEI-GO adsorbent at equilibrium is ∼123.8 mg g–1, which is comparable with the reported data by Lei et al.21 Similarly, the experimentally observed estimated maximum capacity of PFNA adsorption by the FGO adsorbent at equilibrium is ∼167.3 mg g–1, which is comparable to that of fluorine-rich calix [4] arene-based porous polymers (qm= 188.7 mg g–1).15 Reported data show that the qm value follows the order FGO-PEI > FGO > GO-PEI, which indicates that fluorophilic interactions are very important for the separation of PFAS. Reported data also indicate that electrostatic and hydrophobic interactions play important roles for the removal of PFAS.
Next, to find out whether the FGO-PEI based nanoplatform can be used for the separation PFBA or PFBS from the environmental samples selectively or simultaneously, we used tap water, lake water, Mississippi River water, and Mississippi reservoir water samples spiked with 1000 ng/L of PFBA, PFBS, PFHxS, or PFNA individually. Similarly, we also performed experiments using tap water, lake water, Mississippi River water, and Mississippi reservoir water samples spiked with 250 ng/L of PFBA, 250 ng/L of PFBS, 250 ng/L of PFHxS, and 250 ng/L PFNA simultaneously. Because the pH for tap water (pH = 7.5), lake water (pH = 6.7), Mississippi River water (pH = 7.9), and Mississippi reservoir water (pH = 8.1) samples varies between 6 and 9, we have determined how the maximum capacity of PFNA adsorption by FGO-PEI varies with the pH of water. As reported in Figure 3G, the qm values vary from 227 to 196 mg g–1 as the pH varies from 5 to 10. Because pKa values for PFBA, PFBS, PFHxS, and PFNA are less than 3,9−24 they will be mostly in anionic form in the water solution when the pH of the solution is between 5 and 11. Because, in acidic pH, the amine group is more protonated, the electrostatic interaction will be high between PFNA and FGO-PEI, and the maximum capacity of PFNA adsorption will be higher. In our case, the pH for tap water, lake water, Mississippi River water, and Mississippi reservoir water varies between 6.7 and 8.1 where qm values vary from 214 to 206 mg g–1. Because the qm value variation amount is small, we have neglected the pH variation effect for the removal efficiency for environmental samples we have studied. As shown in Figure 3J,K, reported experimental data indicate that the removal efficiency for PFBS is more than 95% for spiked environmental samples using the PEI-FGO based nanoplatform. As shown in Figure 3K, the simultaneous removal efficiency of PFBS, PFBA, PFHxS, and PFNA from tap water, Mississippi river water, lake water, and drinking water is more than 95% using the FGO-PEI based nanoplatform. All the reported data indicate that although environmental water from different sources contains different organic compounds and heavy metals and their pH values are different, our FGO-PEI based nanoplatform can be used for PFBS, PFBA, PFHxS, and PFNA separation from environmental samples selectively or simultaneously.
As reported in Figure 3L, the removal efficiency performance for the FGO-PEI based nanoplatform remains almost the same until five cycles of filtration of PFBS, PFBA, PFHxS, and PFNA in river water samples. After six cycles of filtration, we have noted that the removal efficiency decreases slightly, and it can be due to the fouling issue of the FGO-PEI based nanoplatform that occurs because of the accumulation of microorganisms on the surface from environmental water samples.
3.3. Capturing and Separating Tetracycline and Moxifloxacin Antibiotics from Environmental Samples
As we have discussed before, antibiotic contamination is a serious concern for society due to its association with adverse health effects such as the formation of antibiotic-resistant genes.30−40 It can cause several health problems such as immunity decline, genetic defects, and cancer.30−40 As a result, there is an urgent need to design a novel system that has the capability for the detection and removal of antibiotics from environmental samples.40−47 Next, to find out the capturing and separating efficiency of tetracycline and moxifloxacin antibiotics from environmental samples using the FGO-PEI based nanoplatform, we performed the following experiments. Initially, we infected the drinking water samples with tetracycline and moxifloxacin antibiotics selectively and simultaneously. For the selective antibiotic separation experiment, we spiked water samples with 1000 ng/L of tetracycline and moxifloxacin antibiotics; for the mixture of tetracycline and moxifloxacin antibiotics, we spiked water samples with 250 ng/L concentration for each of them. After that, we performed filtration using the GO-PEI, FGO, and FGO-PEI based nanoplatform. At the end, we used surface enhanced Raman spectroscopy (SERS) for determining the removal efficiency for tetracycline and moxifloxacin antibiotics selectively or simultaneously. For the measurement of SERS, we used the confocal Raman system with laser excitation of 670 nm and gold nanoparticles as the plasmonic surface. Experimental details are reported in the Supporting Information.36−39
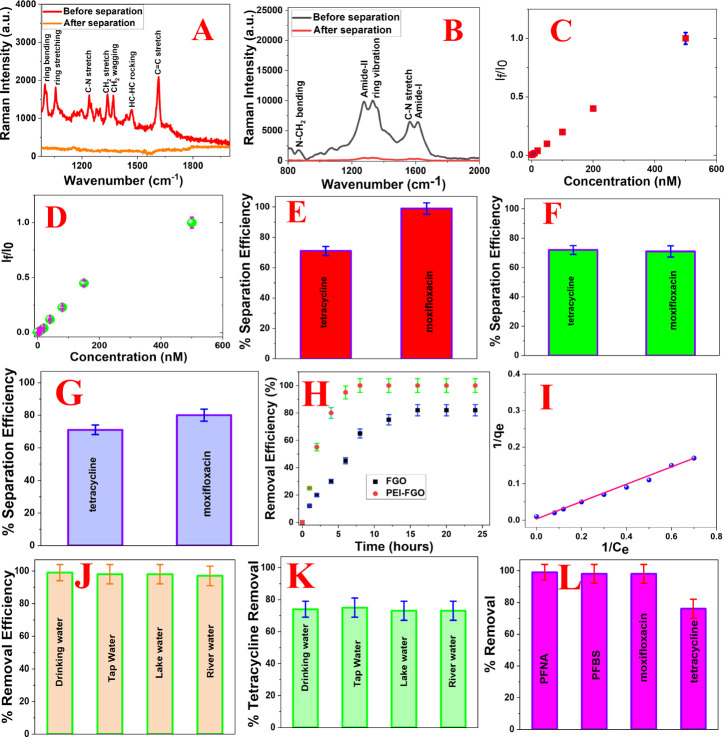
Figure 4A shows the SERS spectra from moxifloxacin antibiotic in water samples before separation. The observed Raman band at 1343 cm–1 is mainly due to the CH2 stretch vibration of the quinolone ring.40−42 Similarly, the observed Raman band at 1373 cm–1 is due to the CH2 wagging of the octahydro pyrrole pyridine group. vibration of the quinolone ring (νring) of Moxi.40−42 On the other hand, the observed Raman band at 1620 cm–1 is due to the C=C stretching.40−42 Similarly, we have also observed a Raman band at 1210 cm–1 due to the C–N stretching. Figure 4B shows the SERS spectra from tetracycline antibiotic in water samples before separation. In the SERS spectra, we have observed amide-I and amide-II Raman bands. We have also observed Raman bands due to the C=N stretching, ring vibration, and N–CH3 bending. To understand whether SERS can be used to quantify the concentration of moxifloxacin antibiotic, we performed a concentration (ng/L)-dependent Raman study. Figure 4C shows how the SERS intensity at 1620 cm–1 for C=C stretch vibration from moxifloxacin antibiotics varies with the concentration (ng/L), where I0 is the SERS intensity at 1620 cm–1 for C=C stretch vibration when the concentration is 1000 ng/L and If is the SERS intensity at 1620 cm–1 for C=C stretch vibration when the concentration varies from 1000 ng/L to 5 pg/L. The linear relation shows clearly that SERS can be used to quantify the concentration of moxifloxacin antibiotic when the concentration is between 1000 ng/L and 5 pg/L. Similarly, to understand whether SERS can be used to quantify the concentration of tetracycline antibiotic, we performed a concentration (ng/L)-dependent Raman study. Figure 4D shows how the SERS intensity at 1230 cm–1 for the amide-III band from tetracycline antibiotics varies with the concentration (ng/L), where I0 is the SERS intensity at 1230 cm–1 for the amide-III band when the concentration is 100 μg/L and If is the SERS intensity at 1230 cm–1 for the amide-III band when the concentration varies from 1000 ng/L to 5 pg/L. The linear relation shows clearly that SERS can be used to quantify the concentration of tetracycline antibiotic when the concentration is between 1000 ng/L and 5 pg/L.
Figure 4.
(A) Surface enhanced Raman spectra (SERS) from water samples of moxifloxacin antibiotics (1000 ng/L) before filtration. SERS from water samples after filtration using the FGO-PEI based nanoplatform. (B) SERS from water samples of tetracycline antibiotics (1000 ng/L) before filtration. SERS from water samples after filtration using the FGO-PEI based nanoplatform. (C) Plot shows how the SERS intensity at 1620 cm–1 for the C=C stretch from moxifloxacin antibiotics varies with the concentration (ng/L). I0 is the SERS intensity at 1620 cm–1 when the concentration is 1000 ng/L. If is the SERS intensity at 1620 cm–1 when the concentration varies from 1000 ng/L to 5 pg/L. (D) Plot shows how the SERS intensity at 1230 cm–1 for amide-III band from tetracycline antibiotics varies with the concentration (ng/L). I0is the SERS intensity at 1230 cm–1 when the concentration is 1000 ng/L. If is the SERS intensity at 1230 cm–1 when the concentration varies from 1000 ng/L to 5 pg/L. (E) Tetracycline and moxifloxacin antibiotic removal efficiency from drinking water using theFGO-PEI based nanoplatform. For this experiment, we used 1000 ng/L of antibiotic infected drinking water. (F) Tetracycline and moxifloxacin antibiotic removal efficiency from drinking water using the GO-PEI based nanoplatform. For this experiment, we used 1000 ng/L of each antibiotic separately. (G) Tetracycline and moxifloxacin antibiotic removal efficiency from drinking water using the FGO based nanoplatform. For this experiment, we used 1000 ng/L of antibiotic infected drinking water. (H) Variation of moxifloxacin antibiotic removal efficiency with time for the FGO and FGO-PEI based nanoplatform. (I) Plot shows the variation of 1/qe with 1/Ce for moxifloxacin antibiotics using the PEI-FGO adsorber, where qe is the quantity of moxifloxacin antibiotics absorbed at equilibrium and Ce is the concentration of moxifloxacin antibiotics. (J) Moxifloxacin antibiotic removal efficiency from tap water, Mississippi river water, lake water, and drinking water using the FGO-PEI based nanoplatform. For this experiment, we used 1000 ng/L of moxifloxacin antibiotic infected water samples. (K) Tetracycline antibiotic removal efficiency from tap water, Mississippi river water, lake water, and drinking water using the FGO-PEI based nanoplatform. For this experiment, we used 1000 ng/L of tetracycline antibiotic infected water samples. (L) Removal efficiency of PFNA, PFBS, tetracycline, and moxifloxacin antibiotics simultaneously from tap water, Mississippi river water, lake water, and drinking water using the FGO-PEI based nanoplatform. For this experiment, we used 250 ng/L of perfluorobutanesulfonic acid (PFBS), 250 ng/L of PFNA, 250 ng/L of tetracycline, and 250 ng/L of moxifloxacin antibiotic infected water samples.
As shown in Figure 4E, reported experimental data indicate that the removal efficiency for moxifloxacin antibiotic is ∼99% when FGO-PEI based nanoplatform has been used. On the other hand, as reported in Figure 4F, we have observed ∼71% separation of moxifloxacin antibiotic when the GO-PEI based nanoplatform has been used. Similarly, as reported in Figure 4G, we have observed ∼80% separation of moxifloxacin antibiotic when the FGO based nanoplatform has been used. Because we have used the FGO-PEI based nanoplatform, the electrostatic interactions between the carboxylate group in moxifloxacin antibiotic and the cationic amines of the PEI also can play important roles in enhancing the removing efficiency. The amine group from PEI also can participate in hydrophobic interactions with moxifloxacin antibiotic, which will help to remove the toxin. The observed highest separation efficiency for the FGO-PEI based nanoplatform can be due to the presence of unique C–F bonds in FGO, which can adsorb moxifloxacin antibiotic using dipole–dipole, ionic–dipolar, and fluorophilic interactions resulting in highly increased toxin removal capability. Because of the absence of a C–F bond in the GO-PEI based nanoplatform, the fluorophilic interaction between moxifloxacin antibiotic and nanoplatform is missing, which decreases the moxifloxacin antibiotic removal capability. Similarly, because of the absence of an amine group in the FGO based nanoplatform, the electrostatic interaction between the carboxylate group in moxifloxacin antibiotic and the amine group in the nanoplatform is missing, which decreases the moxifloxacin antibiotic removal capability. As shown in Figure 4E, reported experimental data indicate that the removal efficiency for tetracycline antibiotic is ∼73% when the FGO-PEI based nanoplatform has been used. Similarly, as reported in Figure 4F, we have observed ∼74% separation of tetracycline antibiotic when the GO-PEI based nanoplatform has been used. On the other hand, as reported in Figure 4G, we have observed ∼71% separation of tetracycline antibiotic when the FGO based nanoplatform has been used. It is interesting to note that the observed separation efficiencies for tetracycline antibiotic are very similar for the FGO-PEI based nanoplatform, FGO based nanoplatform, and GO-PEI based nanoplatform, which clearly indicate that fluorophilic interactions play a very important role for the ∼99% separation of moxifloxacin antibiotic using FGO-PEI.
As reported in Figure 4H, the time-dependent moxifloxacin antibiotic separation data show that the removal efficiency reached ∼100% within an hour when PEI-FGO has been used as adsorber. On the other hand, the removal efficiency reached ∼80% after 18 h and then remained the same when FGO has been used as adsorber. Similarly, the removal efficiency reached ∼71% after 20 h and then remained the same when PEI-GO has been used as adsorber. All the above-reported data clearly indicate that the separation efficiency and the kinetics of separation are highest for the FGO-PEI based nanoplatform in the case of moxifloxacin antibiotic, which clearly indicate that fluorophilic interactions play a very important role. Using eq 4 and reported experimental data in Figure 4I, we have estimated that the maximum capacity of moxifloxacin antibiotic adsorption by PEI-FGO adsorbent at equilibrium (qm) is ∼298.6 mg g–1. The experimentally observed maximum capacity by PEI-FGO is comparable to that of the MOF-808-SIPA system reported by Zhao et al. (qm = 287 mg g–1)44 and carbon aerogel sorbents reported by Zong et al. (qm = 197 mg g–1).45 On the other hand, the estimated maximum capacity of adsorption by the PEI-GO adsorbent at equilibrium is ∼166.7 mg g–1. Similarly, the estimated maximum capacity of moxifloxacin antibiotic adsorption by the FGO adsorbent at equilibrium is ∼217.6 mg g–1. Reported data show qm values in the order of FGO-PEI > FGO > GO-PEI, which indicate that fluorophilic interactions are very important for the separation of moxifloxacin antibiotics. Reported data also indicate that electrostatic and hydrophobic interactions play important roles for the removal of moxifloxacin antibiotics.
Next, to find out whether the FGO-PEI based nanoplatform can be used for the separation of tetracycline and moxifloxacin antibiotics from the environmental samples selectively or simultaneously, we have used tap water, lake water, Mississippi River water, and Mississippi reservoir water samples spiked with 1000 ng/L of tetracycline and moxifloxacin antibiotics. Because the pH for tap water (pH = 7.5), lake water (pH = 6.7), Mississippi River water (pH = 7.9), and Mississippi reservoir water (pH = 8.1) samples varies between 6 and 9, we have determined how the maximum capacity of moxifloxacin antibiotic adsorption by FGO-PEI varies with the pH of water. Because qm values vary from ∼293.5 to ∼306.5 mg g–1 with the change of pH from 6.5 to 9, we have neglected the pH variation effect for the removal efficiency for environmental samples we have studied. We also performed experiments using tap water, lake water, Mississippi River water, and Mississippi reservoir water samples spiked with a mixture of 250 ng/L of perfluorobutanesulfonic acid (PFBS), 250 ng/L of perfluorobutanoic acid (PFNA), 125 ng/L of tetracycline, and 125 ng/L of moxifloxacin antibiotics. As shown in Figure 4J, reported experimental data indicate that the removal efficiency for moxifloxacin antibiotics is more than 97% for spiked environmental samples. The reported data indicate that although environmental water from different sources contains different organic compounds and heavy metals as well as have different pH values, our FGO-PEI based nanoplatform can be used for moxifloxacin antibiotic separation from environmental samples selectively. As shown in Figure 4K, reported experimental data also indicate that the removal efficiency for tetracycline antibiotics is around 70% for the spiked environmental sample, which is very similar to the removal efficiency in the drinking water sample. Figure 4L shows the removal efficiency of PFBS, PFBA, tetracycline, and moxifloxacin antibiotic simultaneously from tap water, Mississippi river water, lake water, and drinking water using the FGO-PEI based nanoplatform. Reported data show that the removal efficiency is around 98% for PFBS, PFBA, and moxifloxacin antibiotics. On the other hand, the removal efficiency is around 70% for tetracycline antibiotics from the environmental sample. All the above data clearly indicate that fluorophilic interactions play a very important role for the separation of moxifloxacin antibiotic.
3.4. Capturing and Separating Salmonella and Escherichia coli Waterborne Pathogens from Environmental Samples
As we have discussed before, even in the 21st century, people in many countries in this world are still lacking water that is free from pathogens.30−45 Waterborne pathogens such as Salmonella and Escherichia coli bacteria are the leading causes of waterborne diseases.30−45 As a result, there is an urgent need to design a novel system that has the capability for the removal of waterborne pathogens from environmental samples.30−45 Next, to find out the capturing and separating efficiency of waterborne pathogens such as Salmonella and Escherichia coli bacteria from environmental samples using the FGO-PEI based nanoplatform, we performed the following experiments. Initially, different waterborne pathogens like Salmonella and Escherichia coli were cultured according to the ATCC protocol, as we have reported before.34−39,47 Experimental details are reported in the Supporting Information. Once different waterborne pathogens like Salmonella and Escherichia coli grew to 105 CFU/ml, we used them for water contamination separation experiments. Initially, we infected the drinking water sample with Salmonella and Escherichia coli bacteria selectively and simultaneously. For the selective pathogen’s separation experiment, we spiked water samples with 103 CFU/mL of Salmonella or scherichia coli bacteria. For the mixture of Salmonella and Escherichia coli bacteria experiment, we spiked water samples with 500 CFU/mL concentration for each of them. After that, we performed filtration using the FGO-PEI based nanoplatform. At the end, we used colony counting for determining the removal efficiency for Salmonella and Escherichia coli bacteria selectively or simultaneously, as we have reported before.34−39,47 Experimental details are reported in the Supporting Information.
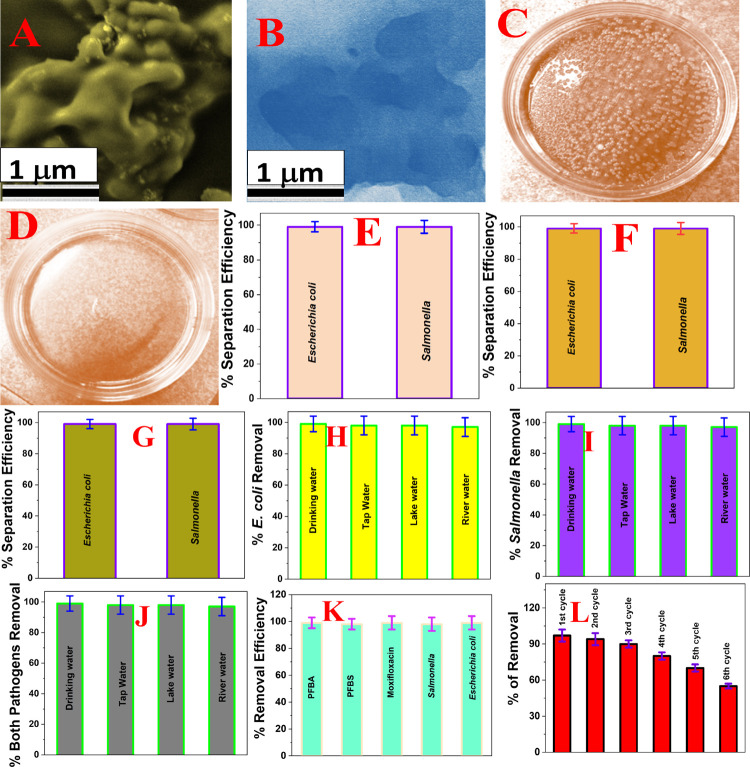
Figure 5E shows the Salmonella and Escherichia coli waterborne pathogen removal efficiency from drinking water using the FGO-PEI based nanoplatform. For this experiment, we used 103 CFU/mL of each bacterium. The SEM image in Figure 5A shows that Salmonella waterborne pathogens are captured by the FGO-PEI based nanoplatform during filtration. Similarly, the TEM image in Figure 5B shows that Escherichia coli waterborne pathogens are captured by the FGO-PEI based nanoplatform during filtration. Figure 5C shows the colony counting data indicating that Escherichia coli is present in water samples before filtration. On the other hand, colony counting data reported in Figure 5D show that no Escherichia coli is present in water samples after filtration, which indicates that 100% of the bacteria is captured during filtration. The observed 100% separation efficiency is due to the pore size of the nanoplatform being between 5 and 100 nm, which is much smaller than the size of Salmonella and Escherichia coli waterborne pathogens (>1000 nm); therefore, 100% of pathogens were captured by the nanoplatform because they cannot pass through the pore of the nanoplatform. Figure 5F shows the Salmonella and Escherichia coli waterborne pathogen removal efficiency from drinking water using the GO-PEI based nanoplatform. In this case, because the pore size of the membrane is between 5 and 120 nm, which is much smaller than the size of Salmonella and Escherichia coli waterborne pathogens (>1000 nm), 100% of pathogens were captured by the nanoplatform because they cannot pass through the pore of the membrane. Similarly, Figure 5G shows the Salmonella and Escherichia coli waterborne pathogen removal efficiency from drinking water using the FGO based nanoplatform. As reported in Figure 5G, 100% of pathogens were captured by the FGO nanoplatform. All the reported data clearly indicate that the removal efficiency is 100% for all nanoplatforms, with or without the presence of fluorine.
Figure 5.
(A) SEM image shows that Salmonella waterborne pathogens are captured by the FGO-PEI based nanoplatform during filtration. (B) TEM image shows that Escherichia coli waterborne pathogens are captured by the FGO-PEI based nanoplatform during filtration. (C) Colony counting data show that Escherichia coli is present in water samples before filtration. (D) Colony counting data show that no Escherichia coli is present in water samples after filtration, which indicates that 100% bacteria is captured during filtration. (E) Salmonella and Escherichia coli waterborne pathogen removal efficiency from drinking water using the FGO-PEI based nanoplatform. For this experiment, we used 103 CFU/mL of each bacterium. (F) Salmonella and Escherichia coli waterborne pathogen removal efficiency from drinking water using the GO-PEI based nanoplatform. For this experiment, we used 103 CFU/mL of each bacterium. G) Salmonella and Escherichia coli waterborne pathogen removal efficiency from drinking water using the FGO based nanoplatform. For this experiment, we used 103 CFU/mL of each bacterium. (H) Escherichia coli waterborne pathogen removal efficiency from tap water, Mississippi river water, lake water, and drinking water using the FGO-PEI based nanoplatform. For this experiment, we used 103 CFU/mL of bacteria infected water samples. (I) Salmonella waterborne pathogen removal efficiency from tap water, Mississippi river water, lake water, and drinking water using the FGO-PEI based nanoplatform. For this experiment, we used 103 CFU/mL of bacteria infected water samples. (J) Simultaneously Salmonella and Escherichia coli waterborne pathogen removal efficiency from tap water, Mississippi river water, lake water, and drinking water using the FGO-PEI based nanoplatform. For this experiment, we used 500 CFU/mL of bacteria infected water samples. (K). Removal efficiency of PFBS, PFBA, tetracycline, and moxifloxacin antibiotics and Salmonella and Escherichia coli waterborne pathogens simultaneously from tap water, Mississippi river water, lake water, and drinking water using the FGO-PEI based nanoplatform. For this experiment, we used water samples infected with 333 ng/L of PFBS, 333 ng/L of PFBA, 333 ng/L of moxifloxacin antibiotics, and 500 CFU/mL of each bacterium. (L) Plot shows how the removal efficiency PFBS, PFBA, tetracycline, and moxifloxacin antibiotics varies with the number of cycles of filtration when we have used the FGO-PEI based nanoplatform in the presence of Salmonella and Escherichia coli waterborne pathogens. For this experiment, we used water samples infected with 333 ng/L of PFBS, 333 ng/L of PFBA, 333 ng/L of moxifloxacin antibiotics, and 500 CFU/mL of each bacterium.
Next, to find out whether the FGO-PEI based nanoplatform can be used for the separation of Salmonella and Escherichia coli waterborne pathogens from the environmental samples selectively or simultaneously, we used tap water, lake water, Mississippi River water, and Mississippi reservoir water samples spiked with 103 CFU/mL of Salmonella or Escherichia coli. Similarly, we also performed experiments using tap water, lake water, Mississippi River water, and Mississippi reservoir water samples spiked with a mixture of 250 ng/L of PFBS, 250 ng/L of PFBA, and 250 ng/L of moxifloxacin antibiotics and 500 CFU/mL Salmonella and 500 CFU/mL Escherichia coli waterborne pathogens. Because the pH values for tap water (pH = 7.5), lake water (pH = 6.7), Mississippi River water (pH = 7.9), and Mississippi reservoir water (pH = 8.1) samples vary between 6 and 9, we have determined how the bacteria varies with the pH of water. Because the removal efficiency remains 100%, with the change of pH from 6.5 to 9, we have neglected the pH variation effect for the removal efficiency for environmental samples we have studied. As shown in Figure 5H, reported experimental data indicate that the removal efficiency for Salmonella waterborne pathogens is more than 99% for spiked environmental samples. The reported data indicate that although environmental water from different sources contains different organic compounds and heavy metals and their pH values are different, our FGO-PEI based nanoplatform can be used for Salmonella waterborne pathogen separation from environmental samples selectively. As shown in Figure 5I, reported experimental data indicate that the removal efficiency for Escherichia coli waterborne pathogens is more than 99% for spiked environmental samples. Similarly, as shown in Figure 5J, reported experimental data indicate that the removal efficiency for Escherichia coli and Salmonella waterborne pathogens together is more than 99% for spiked environmental samples. Figure 5K shows that the removal efficiency is more than 96% for PFBS, PFBA, and moxifloxacin antibiotics and Salmonella and Escherichia coli waterborne pathogens simultaneously from tap water, Mississippi river water, lake water, and drinking water using the FGO-PEI based nanoplatform. Figure 5L shows how the removal efficiency of PFBS, PFBA, tetracycline, and moxifloxacin antibiotics varies with the number of cycles of filtration in the presence of Salmonella and Escherichia coli waterborne pathogens. As shown in Figure 5L, the removal efficiency decreases significantly with the number of cycles, and it is due to the fouling issue of the FGO-PEI based nanoplatform that occurs because of the accumulation of Escherichia coli and Salmonella waterborne pathogens on the surface of environmental water samples. After around 10 cycles, all the pores are covered by Escherichia coli and Salmonella waterborne pathogens; as a result, we are not able to use the nanoplatform for filtration anymore.
4. Conclusions
In conclusion, our finding reveals that the FGO-PEI based porous nanoplatform has an estimated adsorption capacity of 219 mg g–1 for PFAS and can be used for effective removal of forever toxic chemicals like PFBA, PFBS, PFHxS, and PFNA. Similarly, the FGO-PEI based porous nanoplatform exhibits an estimated adsorption capacity of 298.6 mg g–1 for moxifloxacin antibiotic and can be used for effective removal of pharmaceutical toxins. The comparative PFNA capturing study using different types of nanoplatforms indicate estimated qm ∼167 mg g–1 for the FGO platform and qm ∼124 mg g–1 for the PEI-GO platform, indicating that fluorophilic, electrostatic, and hydrophobic interactions play an important role for capturing PFAS at high capacity. Similarly, a comparative antibiotic capturing study using moxifloxacin and tetracycline indicates that the nanoplatform can be used for 100% separation of moxifloxacin antibiotics with an estimated qm of ∼299 mg g–1. Experimental data demonstrate that because the pore size of the 3D nanoplatform is much smaller than Salmonella and Escherichia coli waterborne pathogens, it can be used for 100% separation of waterborne pathogens from contaminated water samples. Reported data using spiked tap water, Mississippi River water, and Mississippi reservoir water samples demonstrated that the removal efficiency is more than 96% for perfluoro PFBS, PFBA, and moxifloxacin antibiotics and Salmonella and Escherichia coli waterborne pathogens simultaneously using the FGO-PEI based nanoplatform. Although reported data clearly reveal the great potential of the PEI-attached 2D F-GO based nanoplatform for water-based environmental toxin removal applications, we are still at an early stage of development for the use in real-life applications. In the future, we plan to study how to use the nanoplatform for the complete removal of a wide range of PFAS, antibiotics, and pathogens at environmentally relevant concentrations from real water matrices.
Acknowledgments
The work described in this document was conducted in the Department of Chemistry at Jackson State University. The authors would like to acknowledge the support provided by the US Army Engineer Research and Development Center (ERDC) and the Military Engineering Research and Development Area under contract W912HZ-21C0040. Permission to publish was granted by the ERDC Geotechnical and Structures Laboratory. The research described and the resulting data presented herein were funded under CA BO340 497014 Project, “Graphene Applications for Military Engineering,” under Contract W912HZ-21C0040, managed by the US Army Engineer Research and Development Center.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c06360.
Detailed design and characterization of the mixed dimensional heterostructure and other experimental details (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- 2.1 billion people lack safe drinking water at home, more than twice as many lack safe sanitation https://www.unicef.cn/en/press-releases/21-billion-people-lack-safe-drinking-water-home-more-twice-many-lack-safe-sanitation2017.
- United States Environmental Protection Agency . Drinking Water Health Advisories for PFAS: Fact Sheet for Public Water Systems (PFOA, PFOS, GenX Chemicals and PFBS). https://www.epa.gov/system/files/documents/2022-06/drinking-water-ha-pfas-factsheet-water-system.pdf2022.
- Smalling K. L.; Romanok K. M.; Bradley P. M.; Morriss M. C.; Gray J. L.; Kanagy L. K.; Gordon S. E.; Williams B. M.; Breitmeyer S. E.; Jones D. K.; DeCicco L. A.; Eagles-Smith C. A.; Wagner T. Per- and Polyfluoroalkyl Substances (PFAS) in U.S. Tapwater: Comparison of Public-Supply and Underserved Private-Well Exposures and Associated Health Implications. Environ. Int. 2023, 178, 108033 10.1016/j.envint.2023.108033. [DOI] [PubMed] [Google Scholar]
- Turner S. W. D.; Rice J. S.; Nelson K. D.; Vernon C. R.; McManamay R.; Dickson K.; Marston L. Comparison of Potential Drinking Water Source Contamination across One Hundred U.S. Cities. Nat. Commun. 2021, 12, 7254. 10.1038/s41467-021-27509-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evich M. G.; Davis M. J. B.; McCord J. P.; Acrey B.; Awkerman J. A.; Knappe D. R. U.; Lindstrom A. B.; Speth T. F.; Tebes-Stevens C.; Strynar M. J.; Wang Z.; Weber E. J.; Henderson W. M.; Washington J. W. Per- and polyfluoroalkyl substances in the environment. Science 2022, 375, eabg9065 10.1126/science.abg9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartell S. M.; Vieira V. M. Critical review on PFOA, kidney cancer, and testicular cancer. J. Air Waste Manag. Assoc. 2021, 71, 663–679. 10.1080/10962247.2021.1909668. [DOI] [PubMed] [Google Scholar]
- Culp T. E.; Khara B.; Brickey K. P.; Geitner M.; Zimudzi T. J.; Wilbur J. D.; Jons S. D.; Roy A.; Paul M.; Ganapathy subramanian B.; Zydney A. L.; Kumar M.; Gomez E. D. Nanoscale control of internal inhomogeneity enhances water transport in desalination nanoplatforms. Science 2021, 371, 72–75. 10.1126/science.abb8518. [DOI] [PubMed] [Google Scholar]
- Hou D.; Li T.; Chen X.; He S.; Dai J.; Mofid S. A.; Hou D.; Iddya A.; Jassby D.; Yang R.; Hu L.; Ren Z. J. Hydrophobic nanostructured wood nanoplatform for thermally efficient distillation. Sci. Adv. 2019, 5, eaaw3203 10.1126/sciadv.aaw3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evich M. G.; Davis M. J. B.; McCord J. P.; Acrey B.; Awkerman J. A.; Knappe D. R. U.; Lindstrom A. B.; Speth T. F.; Tebes-Stevens C.; Strynar M. J.; Wang Z.; Weber E. J.; Henderson W. M.; Washington J. W. Per- and Polyfluoroalkyl Substances in the environment. Science 2022, 375 (6580), 1. 10.1126/science.abg9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.; Wei Z.; Gu C.; Yao Y.; Xue J.; Zhu H.; Kannan K.; Sun H.; Zhang T. First Evidence of Prenatal Exposure to Emerging Per- and Polyfluoroalkyl Substances Associated with E-Waste Dismantling: Chemical Structure-Based Placental Transfer and Health Risks. Environ. Sci. Technol. 2022, 56 (23), 17108–1711. 10.1021/acs.est.2c05925. [DOI] [PubMed] [Google Scholar]
- Ateia M.; Alsbaiee A.; Karanfil T.; Dichtel W. Efficient PFAS Removal by Amine-Functionalized Sorbents: Critical Review of the Current Literature. Environ. Sci. Technol. Lett. 2019, 6 (12), 688–695. 10.1021/acs.estlett.9b00659. [DOI] [Google Scholar]
- Kumarasamy E.; Manning I. M.; Collins L. B.; Coronell O.; Leibfarth F. A. Ionic Fluorogels for Remediation of Per- and Polyfluorinated Alkyl Substances from Water. ACS Cent. Sci. 2020, 6, 487–492. 10.1021/acscentsci.9b01224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román Santiago A.; Yin S.; Elbert J.; Lee J.; Shukla D.; Su X. Imparting Selective Fluorophilic Interactions in Redox Copolymers for the Electrochemically Mediated Capture of Short-Chain Perfluoroalkyl Substances. J. Am. Chem. Soc. 2023, 145 (17), 9508–9519. 10.1021/jacs.2c10963. [DOI] [PubMed] [Google Scholar]
- Tan X.; Zhong J.; Fu C.; Dang H.; Han Y.; Král P.; Guo J.; Yuan Z.; Peng H.; Zhang C.; Whittaker A. K. Amphiphilic Perfluoropolyether Copolymers for the Effective Removal of Polyfluoroalkyl Substances from Aqueous Environments. Macromolecules 2021, 54, 3447–3457. 10.1021/acs.macromol.1c00096. [DOI] [Google Scholar]
- Shetty D.; Jahović I.; Skorjanc T.; Erkal T. S.; Ali L.; Raya J.; Asfari Z.; Olson M. A.; Kirmizialtin S.; Yazaydin A. O.; Trabolsi A. Rapid and Efficient Removal of Perfluorooctanoic Acid from Water with Fluorine-Rich Calixarene-Based Porous Polymers. ACS Appl. Mater. Interfaces 2020, 12, 43160–43166. 10.1021/acsami.0c13400. [DOI] [PubMed] [Google Scholar]
- Tan X.; Dewapriya P.; Prasad P.; Chang Y.; Huang X.; Wang Y.; Gong X.; Hopkins T. E.; Fu C.; Thomas K. V.; Peng H.; Whittaker A. K.; Zhang C. Efficient Removal of Perfluorinated Chemicals from Contaminated Water Sources Using Magnetic Fluorinated Polymer Sorbents. Angew. Chem., Int. Ed. 2022, 61, e202213071 10.1002/anie.202213071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning I. M.; Chew N. G. P.; Macdonald H. P.; Miller K. E.; Strynar M. J.; Coronell O.; Leibfarth F. A. Hydrolytically Stable Ionic Fluorogels for High-Performance Remediation of Per- and Polyfluoroalkyl Substances (PFAS) from Natural Water. Angew. Chem., Int. Ed. 2022, 61, e202208150 10.1002/anie.202208150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W.; Xiao L.; Ling Y.; Ching C.; Matsumoto M.; Bisbey R. P.; Helbling D. E.; Dichtel W. R. Removal of GenX and Perfluorinated Alkyl Substances from Water by Amine-Functionalized Covalent Organic Frameworks. J. Am. Chem. Soc. 2018, 140, 12677–12681. 10.1021/jacs.8b06958. [DOI] [PubMed] [Google Scholar]
- Abbasian Chaleshtari Z.; Foudazi R. A Review on Per- and Polyfluoroalkyl Substances (PFAS) Remediation: Separation Mechanisms and Molecular Interactions. ACS EST Water 2022, 2 (12), 2258. 10.1021/acsestwater.2c00271. [DOI] [Google Scholar]
- Liu L.; Che N.; Wang S.; Liu Y.; Li C. Copper Nanoparticle Loading and F Doping of Graphene Aerogel Enhance Its Adsorption of Aqueous Perfluorooctanoic Acid. ACS Omega 2021, 6 (10), 7073–7085. 10.1021/acsomega.1c00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X.; Lian Q.; Zhang X.; Wang T.; Gee M.; Holmes W.; Jin S.; Ponnusamy S. K.; Gang D. S.; Zappi M. E. Removal of perfluorooctanoic acid via polyethyleneimine modified graphene oxide: Effects of water matrices and understanding mechanisms. Chemosphere 2022, 308, 136379 10.1016/j.chemosphere.2022.136379. [DOI] [PubMed] [Google Scholar]
- Xiao L.; Ling Y.; Alsbaiee A.; Li C.; Helbling D. E.; Dichtel W. R. β-Cyclodextrin Polymer Network Sequesters Perfluorooctanoic Acid at Environmentally Relevant Concentrations. J. Am. Chem. Soc. 2017, 139, 7689–7692. 10.1021/jacs.7b02381. [DOI] [PubMed] [Google Scholar]
- Ateia M.; Attia M. F.; Maroli A.; Tharayil N.; Alexis F.; Whitehead D. C.; Karanfil T. Rapid Removal of Poly- and Perfluorinated Alkyl Substances by Poly(Ethylenimine)-Functionalized Cellulose Microcrystals at Environmentally Relevant Conditions. Environ. Sci. Technol. Lett. 2018, 5, 764–769. 10.1021/acs.estlett.8b00556. [DOI] [Google Scholar]
- Chen X.; Fan K.; Liu Y.; Li Y.; Liu X.; Feng W.; Wang X. Recent Advances in Fluorinated Graphene from Synthesis to Applications: Critical Review on Functional Chemistry and Structure Engineering. Adv. Mater. 2021, 34, 2101665. 10.1002/adma.202101665. [DOI] [PubMed] [Google Scholar]
- Nair R. R.; Ren W. C.; Jalil R.; Riaz I.; Kravets V. G.; Britnell L.; Blake P.; Schedin F.; Mayorov A. S.; Yuan S. J.; et al. Fluorographene: A Two-Dimensional Counterpart of Teflon. Small 2010, 6, 2877–2884. 10.1002/smll.201001555. [DOI] [PubMed] [Google Scholar]
- Carpenter J.; Kim H.; Suarez J.; van der Zande A.; Miljkovic N. The Surface Energy of Hydrogenated and Fluorinated Graphene. ACS Appl. Mater. Interfaces. 2023, 15, 2429–2436. 10.1021/acsami.2c18329. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Jiang Y.; Sun J.; Wang Y.; Qian L.; Kim S. H.; Chen L. Inverse Relationship between Thickness and Wear of Fluorinated Graphene: “Thinner Is Better.. Nano Lett. 2022, 22 (14), 6018–6025. 10.1021/acs.nanolett.2c01043. [DOI] [PubMed] [Google Scholar]
- Itoh Y.; Chen S.; Hirahara R.; Konda T.; Aoki T.; Ueda T.; Shimada I.; Cannon J. J.; Shao C.; Shiomi J.; Tabata K. V.; Nijo H.; Sato K.; Aida T. Ultrafast water permeation through nanochannels with a densely fluorous interior surface. Science 2022, 376, 738–743. 10.1126/science.abd0966. [DOI] [PubMed] [Google Scholar]
- Kent A. G.; Vill A. C.; Shi Q.; Satlin M. J.; Brito I. L. Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial Hi-C. Nat. Commun. 2020, 11 (1), 4379. 10.1038/s41467-020-18164-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokdyk J. P.; Firnstahl A. D.; Walsh J. F.; Spencer S. K.; de Lambert J. R.; Anderson A. C.; Rezania L.-I. W.; Kieke B. A. Jr.; Borchardt M. A. Viral, bacterial, and protozoan pathogens and fecal markers in wells supplying groundwater to public water systems in Minnesota. USAWater Res. 2020, 178, 115814 10.1016/j.watres.2020.115814. [DOI] [PubMed] [Google Scholar]
- Wang P.; Wu D.; You X.; Li W.; Xie B. Distribution of antibiotics, metals and antibiotic resistance genes during landfilling process in major municipal solid waste landfills. Environ. Pollut. 2019, 255, 113222 10.1016/j.envpol.2019.113222. [DOI] [PubMed] [Google Scholar]
- Yu X.; Zhao W.; Lyu S.; Cai Z.; Yu G.; Wang H.; Barceló D.; Sui Q. Estimating the Mass of Pharmaceuticals Harbored in Municipal Solid Waste Landfills by Analyzing Refuse Samples at Various Ages and Depths. Environ. Sci. Technol. 2023, 57 (15), 6063–6071. 10.1021/acs.est.2c09455. [DOI] [PubMed] [Google Scholar]
- Yang X.; Jia X.; Ji X. Acid Induced Fluorinated Graphene Oxide. RSC Adv. 2015, 5, 9337–9340. 10.1039/C4RA13884H. [DOI] [Google Scholar]
- Gao Y.; Pramanik A.; Patibandla S.; Gates K.; Hill G.; Ignatius A.; Ray P. C. Development of Human Host Defense Antimicrobial Peptide-Conjugated Biochar Nanocomposites for Combating Broad-Spectrum Superbugs. ACS Appl. Bio Mater. 2020, 3, 7696–7705. 10.1021/acsabm.0c00880. [DOI] [PubMed] [Google Scholar]
- Pramanik A.; Gates K.; Gao Y.; Zhang Q.; Han F. X.; Begum S.; Rightsell C.; Sardar D.; Ray P. C. Composites Composed of Polydopamine Nanoparticles, Graphene Oxide, and ε-Poly- l -lysine for Removal of Waterborne Contaminants and Eradication of Superbugs. ACS Appl. Nano Mater. 2019, 2, 3339–3347. 10.1021/acsanm.9b00161. [DOI] [Google Scholar]
- Jones S.; Pramanik A.; Kanchanapally R.; Viraka Nellore B. P.; Begum S.; Sweet C.; Ray P. C. Multifunctional Three-Dimensional Chitosan/Gold Nanoparticle/Graphene Oxide Architecture for Separation, Label-Free SERS Identification of Pharmaceutical Contaminants, and Effective Killing of Superbugs. ACS Sustainable Chem. Eng. 2017, 5, 7175–7187. 10.1021/acssuschemeng.7b01351. [DOI] [Google Scholar]
- Sinha S. S.; Jones S.; Pramanik A.; Ray P. C. Nanoarchitecture Based SERS for Biomolecular Fingerprinting and Label-Free Disease Markers Diagnosis. Acc. Chem. Res. 2016, 49, 2725–2735. 10.1021/acs.accounts.6b00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramanik A.; Gao Y.; Gates K.; Begum S.; Ray P. C. Giant Chemical and Excellent Synergistic Raman Enhancement from a 3D MoS2–xOx–Gold Nanoparticle Hybrid. ACS Omega 2019, 4, 11112–11118. 10.1021/acsomega.9b00866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramanik A.; Mayer J.; Patibandla S.; Gates K.; Gao Ye; Davis D.; Seshadri R.; Ray P. C. Mixed-Dimensional Heterostructure Material-Based SERS for Trace Level Identification of Breast Cancer-Derived Exosomes. ACS-Omega 2020, 5 (27), 16602–16611. 10.1021/acsomega.0c01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. H.; Wei H.; Santiago P. J.; Thrift W. J.; Regan R.; Jiang S. Sensing Antibiotics in Wastewater Using Surface-Enhanced Raman Scattering. Environ. Sci. Technol. 2023, 57 (12), 4880–4891. 10.1021/acs.est.3c00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X.; Song Y.; Xu X.; Peng D.; Wang J.; Qie X.; Lin K.; Yu M.; Ge M.; Wang Y.; Zhang D.; Yang Q.; Wang M.; Huang W. E. Development of a Fast Raman-Assisted Antibiotic Susceptibility Test (FRAST) for the Antibiotic Resistance Analysis of Clinical Urine and Blood Samples. Anal. Chem. 2021, 93, 5098–5106. 10.1021/acs.analchem.0c04709. [DOI] [PubMed] [Google Scholar]
- Muhammad M.; Yan B.; Yao G.; Chao K.; Zhu C.; Huang Q. Surface-enhanced Raman spectroscopy for trace detection of tetracycline and Dicyandiamide in milk using transparent substrate of Ag nanoparticle arrays. ACS Appl. Nano Mater. 2020, 3, 7066–7075. 10.1021/acsanm.0c01389. [DOI] [Google Scholar]
- Mukherjee D.; Behera J.; Mondal S.; Pla S. C.; Volkmer D.; Das M. C. Critical Perspectives on Metal–Organic Frameworks and Their Composites for the Adsorptive Removal of Antibiotics from Wastewater Matrices. Cryst. Growth Des. 2023, 23 (11), 7612. 10.1021/acs.cgd.3c00892. [DOI] [Google Scholar]
- Zhao X.; Gao X.; Ding R.; Huang H.; Gao X.; Liu B. Post-synthesis introduction of dual functional groups in metal-organic framework for enhanced adsorption of moxifloxacin antibiotic. J. Colloid Interface Sci. 2023, 639, 59–67. 10.1016/j.jcis.2023.02.035. [DOI] [PubMed] [Google Scholar]
- Zong Y.; Ma S.; Gao J.; Xu M.; Xue J.; Wang M. Synthesis of Porphyrin Zr-MOFs for the Adsorption and Photodegradation of Antibiotics under Visible Light. ACS Omega 2021, 6 (27), 17228–17238. 10.1021/acsomega.1c00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domes C.; Domes R.; Popp J.; Pletz M. W.; Frosch T. Ultrasensitive Detection of Antiseptic Antibiotics in Aqueous Media and Human Urine Using Deep UV Resonance Raman Spectroscopy. Anal. Chem. 2017, 89, 9997–10003. 10.1021/acs.analchem.7b02422. [DOI] [PubMed] [Google Scholar]
- Pramanik A.; Dhar J. A.; Banerjee R.; Davis M.; Gates K.; Nie J.; Davis D.; Han F. X.; Ray P. C. WO3 Nanowire-Attached Reduced Graphene Oxide-Based 1D–2D Heterostructures for Near-Infrared Light-Driven Synergistic Photocatalytic and Photothermal Inactivation of Multidrug-Resistant Superbugs. ACS Appl. Bio Mater. 2023, 6 (2), 919–931. 10.1021/acsabm.3c00057. [DOI] [PubMed] [Google Scholar]
- Oberoi A. S.; Jia Y.; Zhang H.; Khanal S. K.; Lu H. Insights into the Fate and Removal of Antibiotics in Engineered Biological Treatment Systems: A Critical Review. Environ. Sci. Technol. 2019, 53, 7234–7264. 10.1021/acs.est.9b01131. [DOI] [PubMed] [Google Scholar]
- Wang S.; Yang S.; Quispe E.; Yang H.; Sanfiorenzo C.; Rogers S. W.; Wang K.; Yang Y.; Hoffmann M. R. Removal of antibiotic resistant bacteria and genes by UV-assisted electrochemical oxidation on degenerative TiO2 nanotube arrays. ACS ES&T Engg. 2021, 1 (3), 612–622. 10.1021/acsestengg.1c00011. [DOI] [Google Scholar]
- Waak M. B.; LaPara T. M.; Halle C.; Hozalski R. M. Nontuberculous mycobacteria in two drinking water distribution systems and the role of residual disinfection. Environ. Sci. Technol. 2019, 53 (15), 8563–8573. 10.1021/acs.est.9b01945. [DOI] [PubMed] [Google Scholar]
- Liu L.; Lopez E.; Dueñas Osorio L.; Stadler L.; Xie Y.; Alvarez P. J. J.; Li Q. The importance of system configuration for distributed direct potable water reuse. Nat. Sustainability 2020, 3, 548–555. 10.1038/s41893-020-0518-5. [DOI] [Google Scholar]
- Huang Y.; Wang X.; Xiang W.; Wang T.; Otis C.; Sarge L.; Lei Y.; Li B. Forward-looking roadmaps for long-term continuous water quality monitoring: bottlenecks, innovations, and prospects in a critical review. Environ. Sci. Technol. 2022, 56, 5334–5354. 10.1021/acs.est.1c07857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.